Abstract
Colorectal cancer is the third most common type of cancer and the fourth leading cause of cancer-associated mortality worldwide. Serine/arginine-rich splicing factor 1 (SRSF1) is a well-characterized oncogenic factor that promotes tumorigenesis by controlling a number of alternative splicing events. However, there is limited network analysis, from a global aspect, to study the effect of SRSF1 on colorectal cancer. In the present study, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of available gene regulation data from The Cancer Genome Atlas database revealed the enriched functions and signaling pathways of SRSF1. Subsequently, Oncomine analysis was performed, which demonstrated that SRSF1 was upregulated in a number of types of colon cancer. From overlapping the analysis of 2,678 SRSF1-related genes and 3,625 colorectal cancer genes in GeneCards, 468 genes were identified as SRSF1-related colorectal cancer genes. The GO results revealed that these overlapped genes were primarily enriched in metabolic processes, response to DNA damage, regulation of the cell cycle and a number of additional biological processes. KEGG pathway analysis revealed that SRSF1-related colorectal cancer genes were associated with the cell cycle, deregulated signaling pathways associated with cancer progression and colorectal cancer signaling pathways. In addition, the Search Tool for the Retrieval of Interacting Genes/Proteins database and Cytoscape analysis demonstrated that 468 SRSF1-related colorectal cancer genes exhibit potential interaction networks in which these genes were enriched in DNA metabolic processes, cell cycle regulation and regulation of apoptosis. The results of the present study suggested that SRSF1 exhibited an increased degree of interaction with key molecules, including NUF2 NDC80 kinetochore complex component, kinesin family member 2C, structural maintenance of chromosomes 3, ATM serine/threonine kinase, BRCA1 DNA repair associated, protein kinase DNA-activated catalytic polypeptide, heat shock protein 90 alpha family class A member 1, ras homolog family member A, and phosphatase and tensin homolog. Collectively, the bioinformatics analysis of the present study indicated that SRSF1 may have key functions in the progression and development of colorectal cancer.
Keywords: serine/arginine-rich splicing factor 1, colorectal cancer, bioinformatics
Introduction
Serine/arginine splicing factor 1 (SRSF1, also known as SF2/ASF) is a member of the SR protein family of splicing regulators. SRSF1 regulates alternative splicing through recognizing and binding the exonic splicing enhancers (ESEs) (1). In addition, SRSF1 is involved in the regulation of mRNA stability (2), mRNA export (3), nonsense-mediated mRNA decay (4), translation (5–7) and microRNA processing (8). It has been demonstrated that SRSF1 is a well-characterized oncogenic factor that promotes tumorigenesis by controlling a number of alternative splicing (AS) events (9). A previous study has identified that SRSF1 promotes the skipping of RON proto-oncogene (RON) exon 11 to generate RONΔ11, which stimulates cell migration and invasion (10). The expression level of SRSF1 is upregulated in human breast cancer, and SRSF1 overexpression promotes the transformation of mammary cells by regulating a number of AS events (11). Additionally, the overexpression of SRSF1 has been identified in colon, thyroid, small intestine, kidney, lung, liver and pancreas tumors (12).
Colorectal cancer, as the third most common type of cancer and the fourth leading cause of cancer-associated mortality, affects >1,000,000 people and causes >500,000 mortalities every year worldwide (13). Histological types of colon cancer include adenocarcinoma, scirrhous tumors and neuroendocrine (14).
Treatments for patients with colon cancer include combinations of surgery, radiation therapy, chemotherapy and targeted therapy (13). Cancer of the colon may be cured with surgery when localized to the bowel; however, cancer that has spread or cancer recurrence following surgery are typically incurable (13). Thus, the development of effective biomarkers and specific therapeutic targets is required.
SRSF1 serves a key function in the tumorigenesis of lung and breast cancer (9,11,15,16), but there is limited information demonstrating the association between SRSF1 and colorectal cancer. SRSF1 may regulate alternative splicing of tumor-related Ras-related C3 botulinum toxin substrate 1b (Rac1b) by promoting the inclusion of alternative exon 3b in colorectal cells (17). Furthermore, alternative splicing of solute carrier family member 39 member 14 in colorectal cancer was identified to be regulated in the Wnt signaling pathway, and this was hypothesized to occur via the regulation of SRSF protein kinase 1 and SRSF1 (18). However, there is a limited amount of network analysis from a global aspect to study the effect of SRSF1 on colorectal cancer.
In the present study, the enriched functions and signaling pathways of SRSF1 were investigated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of available gene regulation data. Additionally, Oncomine analysis was performed, which revealed that SRSF1 was upregulated in a number of types of colon cancer. Furthermore, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database and Cytoscape analysis demonstrated that SRSF1-related colorectal cancer genes have potential interaction networks in which these genes were enriched in DNA metabolic processes, cell cycle regulation and regulation of apoptosis.
Materials and methods
The expression levels of SRSF1 in a number of types of colon cancer were obtained from the Oncomine dataset (https://www.oncomine.org/resource/login.html). The immunohistochemical staining results of SRSF1 in colorectal cancer and normal tissues were selected from the Human Protein Atlas (http://www.proteinatlas.org). cBioPortal (http://www.cbioportal.org/) was applied to identify the SRSF1 co-expressed genes. Briefly, the threshold selected was the absolute value of Pearson correlation coefficient >0.4. Furthermore, Venny 2.1 (BioinfoGP, CNB-CSIC, http://bioinfogp.cnb.csic.es/tools/venny/index.html) was utilized to overlap the identified gene set with a colon cancer-related gene list, screened using GeneCards. The resulting overlapped genes were defined as SRSF1-related genes in colon cancer. The GO and KEGG analysis were conducted with all genes on Database for Annotation, Visualization and Integrated Discovery platform (http://david.abcc.ncifcrf.gov/summary.jsp). Fisher exact P-values were calculated using SPSS 19.0 (IBM Corp., Armonk, NY, USA) for each enriched functional category and pathway, and enriched functional categories and pathways with P<0.05 were presented. The online tool STRING (http://string-db.org) was used to identify the potential interaction networks of protein products of these genes. The Cytoscape 2.8.3 software (Institute of Systems Biology, Seattle, WA, USA) was further utilized to construct interaction networks and sub-networks among these SRSF1-associated gene products in colon cancer. The gene regulation networks and protein interactions were visualized by using Cytoscape 2.8.3 software.
Results
Network of SRSF1-related genes
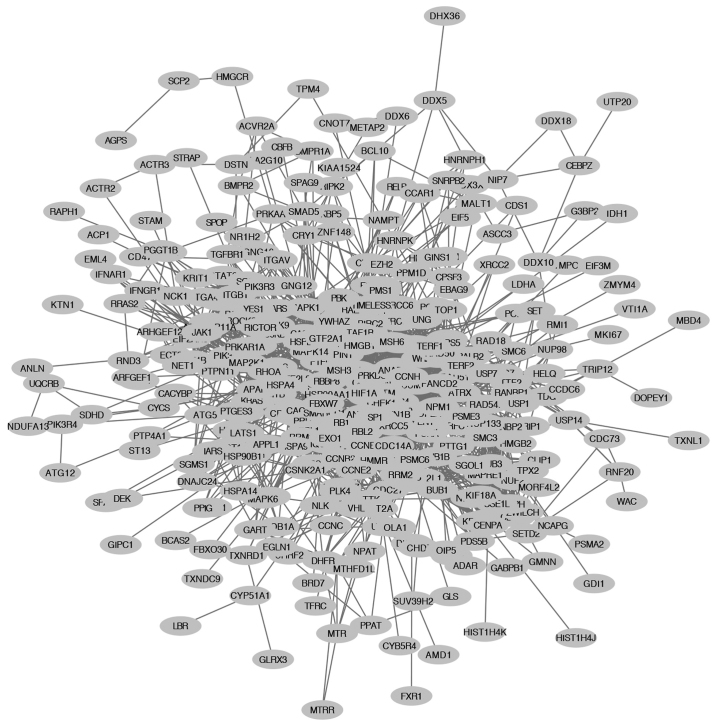
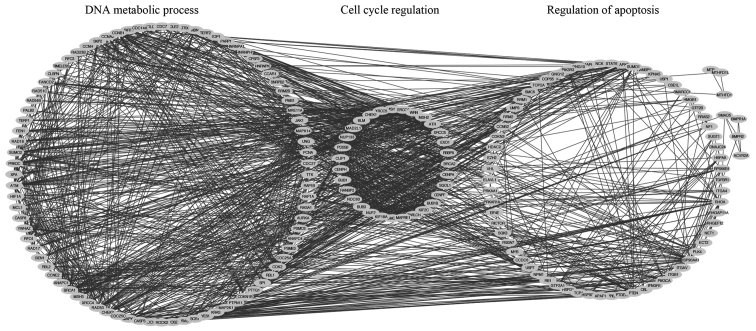
The co-expression network of SRSF1-regulated genes, from the colorectal cancer gene expression profiling database, was established using cBioPortal by setting the Pearson correlation efficient at >0.4. A total of 2,678 genes were identified to be SRSF1-related. Protein-protein interaction networks of 913/2,678 genes were constructed using the STRING database analysis (Fig. 1).
Figure 1.
Interaction network of SRSF1-related genes. The interactions of 2,678 SRSF1-related genes were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins database. The resulting complicated multicentric interaction network contained 913 genes. SRSF1, serine/arginine-rich splicing factor 1.
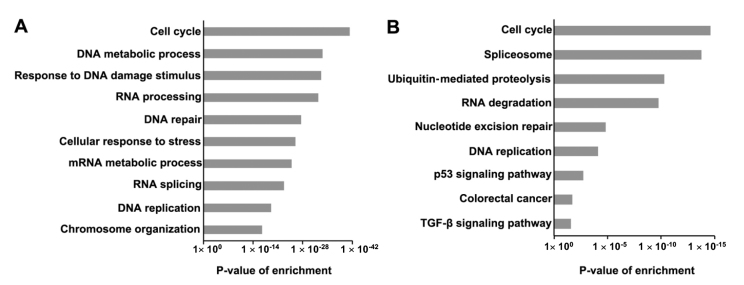
GO analysis for the SRSF1-related proteins revealed that SRSF1-regulated cancers genes were markedly enriched in a number of functions, including regulation of the cell cycle, DNA metabolic processes, RNA processing and DNA replication (Fig. 2A). The results indicated that SRSF1 is involved in cancer cell proliferation. In addition, KEGG pathway analysis revealed that the SRSF1-related genes were markedly enriched in cell cycle, spliceosome, ubiquitin-mediated proteolysis, RNA degradation, nucleotide excision repair, DNA replication and p53 signaling pathway (Fig. 2B). Notably, SRSF1-regulated genes participated in the progression of colorectal cancer, suggesting that SRSF1 may affect colorectal cancer development.
Figure 2.
Analysis of SRSF1-related genes. (A) Gene Ontology analysis (biological process) of 913 SRSF1-related genes is presented. Fisher exact P-values were calculated for each enriched functional category. Enriched functional categories with P<0.05 are presented. (B) Kyoto Encyclopedia of Genes and Genomes pathway of SRSF1-related genes. SRSF1, serine/arginine-rich splicing factor 1; TGF-β, transforming growth factor β.
SRSF1 serves a key function in colorectal cancer progression
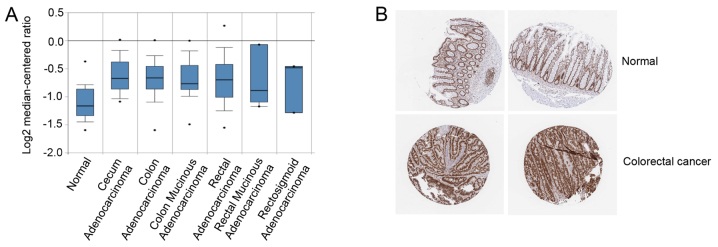
The association between SRSF1 and colorectal cancer was further analyzed. Using Oncomine analysis, the expression level of SRSF1 was determined in colon cancer. It was identified that SRSF1 was upregulated in a number of types of colon cancer, including cecum adenocarcinoma, colon adenocarcinoma and colon mucinous adenocarcinoma (Fig. 3A). In addition, the immunohistochemistry staining results of SRSF1, obtained from the Human Protein Atlas database (http://www.proteinatlas.org) (19), were analyzed, which revealed that SRSF1 was upregulated in colorectal cancer, compared with normal tissues (Fig. 3B) (http://www.proteinatlas.org/ENSG00000136450-SRSF1/cancer/tissue/colorectal+cancer). Taken together, SRSF1 may be involved in colorectal cancer progression.
Figure 3.
Expression level of SRSF1 in a number of types of colon cancer. (A) The mRNA expression level results of SRSF1 in a number of types of colon cancer were obtained from the Oncomine dataset. The median, upper and lower quartiles were plotted, and the whiskers indicate the data range. The points that are >1.5× the interquartile range are marked outliers. (B) Representative immunohistochemical staining of SRSF1 (HPA061301, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in two samples of colorectal cancer and normal tissues obtained from Human Protein Atlas. SRSF1, serine/arginine-rich splicing factor 1.
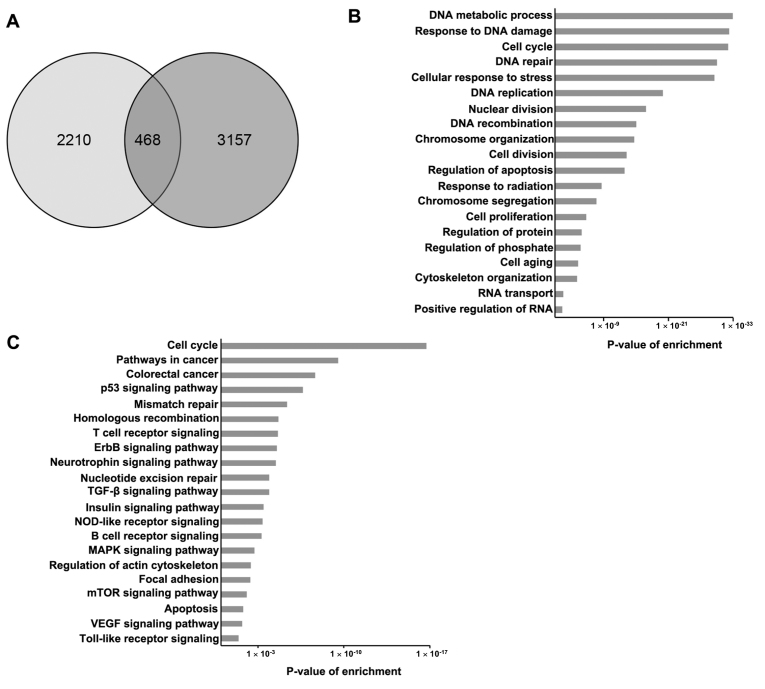
Overlapping analysis of 2,678 SRSF1-related genes and 3,625 genes defined as colorectal cancer genes in GeneCards identified 468 genes as SRSF1-related colorectal cancer genes (Fig. 4A). Subsequently, GO and KEGG pathway analysis was performed to classify the 468 overlapped genes. The GO results demonstrated that the overlapped genes were enriched in a number of biological processes, including metabolic processes, response to DNA damage and cell cycle regulation (Fig. 4B). Furthermore, KEGG pathway analysis revealed that SRSF1-related colorectal cancer genes were involved in the regulation of the cell cycle, deregulated signaling pathways associated with cancer progression and the colorectal cancer signaling pathway (Fig. 4C).
Figure 4.
Interaction network of SRSF1-related genes and colon cancer associated genes. (A) Overlapping analysis of 2,678 SRSF1-related genes (identified using The Cancer Genome Atlas) and 3,625 colon cancer genes (identified using GeneCards). (B) Gene Ontology analysis (biological process) of 468 SRSF1-related colon cancer genes. Fisher exact P-values were plotted for each enriched functional category. Enriched functional categories with P<0.05 were shown. (C) Kyoto Encyclopedia of Genes and Genomes pathway of SRSF1-related colon cancer genes. Fisher exact P-values were plotted for each enriched pathway. Enriched functional pathways with P<0.05 were shown. SRSF1, serine/arginine-rich splicing factor 1; ErbB, Epidermal growth factor receptor; TGF, transforming growth factor; NOD, nucleotide-binding oligomerization domain; MAPK, mitogen-activated protein kinase; mTOR, mechanistic target of rapamycin; VEGF, vascular endothelial growth factor.
Networks of SRSF1-related colorectal cancer genes
The STRING database and Cytoscape analysis was conducted on SRSF1-related colorectal cancer genes to reveal potential interaction networks. GO demonstrated that SRSF1-associated colorectal cancer genes were enriched in the functions of DNA metabolic processes, cell cycle regulation and regulation of apoptosis (Fig. 5).
Figure 5.
Interactions of SRSF1-related colon cancer genes were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins database and Cytoscape. SRSF1, serine/arginine-rich splicing factor 1.
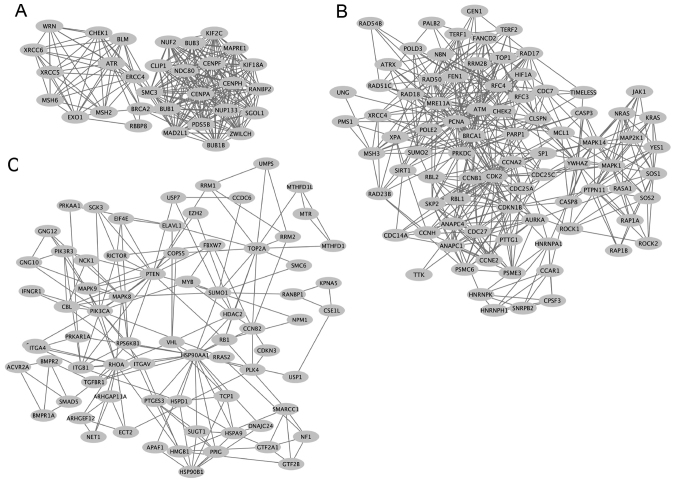
A sub-network of SRSF1-related colorectal cancer genes was identified to be enriched in cell cycle regulation. NUF2 NDC80 kinetochore complex component (NUF2), kinesin family member 2c (KIF2C), structural maintenance of chromosome 3 (SMC3), MAD2 mitotic arrest deficient-like 1 (MAD2L1) and ZWILCH kinetochore protein (ZWILCH) genes were identified to exhibit a high degree of interaction in colorectal cancer (Fig. 6A). In the sub-network of SRSF1-related colorectal cancer genes that were involved in DNA metabolic processes, ATM serine/threonine kinase (ATM), BRCA1 DNA repair associated (BRCA1), protein kinase DNA activated catalytic polypeptide (PRKDC), proliferating cancer nuclear antigen (PCNA), topoisomerase (DNA) 1 (TOP1) and MRE11 homolog double strain break repair nuclease (MRE11A) genes revealed a high connectivity value (Fig. 6B). Furthermore, heat shock protein 90 alpha family class A member 1 (HSP90AA1), Ras homolog family member A (RHOA), phosphatase and tensin homolog (PTEN) and ribosomal protein S6 kinase B1 (RPS6KB1) exhibited a high degree of interaction in another sub-network that regulated apoptosis (Fig. 6C).
Figure 6.
Sub-networks of SRSF1-related colon cancer genes. (A) Network of SRSF1-regulated genes involved in cell cycle regulation. (B) Network of SRSF1-regulated cancer-associated genes that serve roles in DNA metabolic processes. (C) Network of SRSF1-regulated genes involved in apoptosis regulation. SRSF1, serine/arginine-rich splicing factor 1.
Discussion
Serine/Arginine-rich (SR) proteins are a family of RNA-binding proteins with a serine/arginine rich domain. SR proteins regulate alternative splicing of a number of genes that are involved in cell cycle regulation, cell proliferation, apoptosis, epithelial-mesenchymal transition and drug resistance (10,12,20–22). As a member of SR proteins, SRSF1 (SF2/ASF) functions as a proto-oncogene that is generally upregulated in various types of cancer (12,23). In addition, SRSF1 may promote tumorigenesis by controlling the alternative splicing of a number of cancer-related genes, including RPS6KB1 the tumor suppressor bridging integrator 1, the tyrosine kinase receptor for macrophage-stimulating protein, cyclin D1, and vascular endothelial growth factor (24,25).
In the present study, bioinformatics analysis was performed systematically to determine the effect of SRSF1 on colorectal cancer. Gene expression data were obtained from The Cancer Genome Atlas database. Subsequently, potential SRSF1-regulated genes in colorectal cancer were obtained using cBioPortal online tools, followed by GO, KEGG and protein interactome analyses. Oncomine analysis was performed which revealed that SRSF1 is upregulated in a number of types of colon cancer. A total of 468 genes were identified as SRSF1-related colorectal cancer genes by the overlapping analysis of 2,678 SRSF1-related genes and 3,625 colorectal cancer genes (identified using GeneCards). The GO results suggested that these SRSF1-associated colorectal cancer genes were primarily enriched in metabolic processes, response to DNA damage and regulation of the cell cycle. In addition, KEGG pathway analysis revealed that SRSF1-related colorectal cancer genes were closely associated with cell cycle, deregulated signaling pathways associated with cancer progression and colorectal cancer signaling pathways. STRING database and Cytoscape analysis demonstrated that 468 SRSF1-related colorectal cancer genes exhibited potential interaction networks with functions of DNA metabolic process, cell cycle regulation and regulation of apoptosis. Notably, a panel of genes (NUF2, KIF2C, SMC3, MAD2L1 and ZWILCH) responsible for cell cycle regulation were identified to be highly associated with the expression level of SRSF1. NUF2 is a component of NDC80 kinetochore complex and is known to be critical for stable spindle microtubule-kinetochore attachment (26). The expression level of NUF2 has been identified to be upregulated and associated with poor prognosis in patients with colorectal cancer (27). Previous studies have revealed that NUF2 serves a function in a number of types of human cancer including lung, colorectal, gastric, prostate, urinary bladder, renal carcinoma and ovarian cancer (28–33). KIF2C (Kinesin family member 2C, also known as the mitotic centromere-associated kinesin, MACK) is the best characterized member of the kinesin-13 family, which is critical in the regulation of microtubule dynamics. KIF2C has been identified to be aberrantly regulated in cancer cells, which is associated with increased malignance, invasiveness, metastasis, drug resistance, chromosomal instability and remodeling of the microtubule cytoskeleton (34,35).
ATM, BRCA1, PRKDC, PCNA, TOP1 and MRE11A genes, involved in DNA metabolic processes, exhibit a high degree of interaction with SRSF1. ATM is an important cell cycle checkpoint kinase and a key regulator of the DNA double-strand-break response (36). ATM may regulate a number of downstream proteins, including p53, BRCA1, checkpoint kinase 2, checkpoint clamp loader component RAD17, cell cycle checkpoint control protein RAD9 and Nibrin. ATM and serine/threonine-protein kinase ATR, which are hypothesized to be regulators of cell cycle checkpoint signaling pathways (36).
Genes that regulate apoptosis, including HSP90AA1, RHOA, PTEN and RPS6KB1, were highly associated with the expression level of SRSF1. RHOA is a member of Rho GTPases, which are involved in major aspects of cancer development, including cell proliferation, apoptosis, cell polarity, adhesion, migration and invasion (37). Overexpression of RHOA is associated with invasion and poor prognosis in colorectal cancer (38).
The results of the present study, which was conducted using bioinformatics analysis, indicated that SRSF1 may have key functions in the progression and development of colorectal cancer by interaction networks. Additional studies may verify this potential regulatory network by performing molecular biology experiments.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 31400726) and the Natural Science Foundation of Liaoning Province of China (grant no. 2015020301).
References
- 1.Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 2.Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/S1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 5.Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–312. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al. SRSF1-regulated alternative splicing in breast cancer. Mol Cell. 2015;60:105–117. doi: 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 14.PDQ Cancer Information Summaries. Bethesda (MD): 2002. Colon Cancer Treatment (PDQ (R)): Health Professional Version. [Google Scholar]
- 15.de Miguel FJ, Sharma RD, Pajares MJ, Montuenga LM, Rubio A, Pio R. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 2014;74:1105–1115. doi: 10.1158/0008-5472.CAN-13-1481. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonçalves V, Henriques AF, Pereira JF, Costa Neves A, Moyer MP, Moita LF, Gama-Carvalho M, Matos P, Jordan P. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. Rna. 2014;20:474–482. doi: 10.1261/rna.041376.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsen K, Mansilla F, Schepeler T, Øster B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR, Laurberg S, et al. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol Cell Proteomics. 2011;10:M110.002998. doi: 10.1074/mcp.M110.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteom. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 20.He X, Ee PL, Coon JS, Beck WT. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res. 2004;10:4652–4660. doi: 10.1158/1078-0432.CCR-03-0439. [DOI] [PubMed] [Google Scholar]
- 21.He X, Arslan AD, Pool MD, Ho TT, Darcy KM, Coon JS, Beck WT. Knockdown of splicing factor SRp20 causes apoptosis in ovarian cancer cells and its expression is associated with malignancy of epithelial ovarian cancer. Oncogene. 2011;30:356–365. doi: 10.1038/onc.2010.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ezponda T, Pajares MJ, Agorreta J, Echeveste JI, López-Picazo JM, Torre W, Pio R, Montuenga LM. The oncoprotein SF2/ASF promotes non-small cell lung cancer survival by enhancing survivin expression. Clin Cancer Res. 2010;16:4113–4125. doi: 10.1158/1078-0432.CCR-10-0076. [DOI] [PubMed] [Google Scholar]
- 24.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 25.Silipo M, Gautrey H, Tyson-Capper A. Deregulation of splicing factors and breast cancer development. J Mol Cell Biol. 2015;7:388–401. doi: 10.1093/jmcb/mjv027. [DOI] [PubMed] [Google Scholar]
- 26.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi Y, Takano A, Miyagi Y, Tsuchiya E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y, Daigo Y. Cell division cycle-associated protein 1 overexpression is essential for the malignant potential of colorectal cancers. Int J Oncol. 2014;44:69–77. doi: 10.3892/ijo.2013.2177. [DOI] [PubMed] [Google Scholar]
- 28.Hayama S, Daigo Y, Kato T, Ishikawa N, Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S, Nakamura Y. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66:10339–10348. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- 29.Numnum TM, Makhija S, Lu B, Wang M, Rivera A, Stoff-Khalili M, Alvarez RD, Zhu ZB, Curiel DT. Improved anti-tumor therapy based upon infectivity-enhanced adenoviral delivery of RNA interference in ovarian carcinoma cell lines. Gynecol Oncol. 2008;108:34–41. doi: 10.1016/j.ygyno.2007.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko N, Miura K, Gu Z, Karasawa H, Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390:1235–1240. doi: 10.1016/j.bbrc.2009.10.127. [DOI] [PubMed] [Google Scholar]
- 31.Sethi G, Pathak HB, Zhang H, Zhou Y, Einarson MB, Vathipadiekal V, Gunewardena S, Birrer MJ, Godwin AK. An RNA interference lethality screen of the human druggable genome to identify molecular vulnerabilities in epithelial ovarian cancer. PLoS One. 2012;7:e47086. doi: 10.1371/journal.pone.0047086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiraishi T, Terada N, Zeng Y, Suyama T, Luo J, Trock B, Kulkarni P, Getzenberg RH. Cancer/Testis Antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu P, Shangguan J, Zhang L. Downregulation of NUF2 inhibits tumor growth and induces apoptosis by regulating lncRNA AF339813. Int J Clin Exp Pathol. 2015;8:2638–2648. [PMC free article] [PubMed] [Google Scholar]
- 34.Sanhaji M, Friel CT, Wordeman L, Louwen F, Yuan J. Mitotic centromere-associated kinesin (MCAK): A potential cancer drug target. Oncotarget. 2011;2:935–947. doi: 10.18632/oncotarget.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa K, Kamohara Y, Tanaka F, Haraguchi N, Mimori K, Inoue H, Mori M. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br J Cancer. 2008;98:1824–1829. doi: 10.1038/sj.bjc.6604379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Domenico EG, Romano E, Del Porto P, Ascenzioni F. Multifunctional role of ATM/Tel1 kinase in genome stability: from the DNA damage response to telomere maintenance. Biomed Res Int. 2014;2014:787404. doi: 10.1155/2014/787404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Wu Q, Zhang LH, Zhao YX, Wu X. The role of RhoA in vulvar squamous cell carcinoma: A carcinogenesis, progression, and target therapy marker. Tumour Biol. 2016;37:2879–2890. doi: 10.1007/s13277-015-4087-6. [DOI] [PubMed] [Google Scholar]
- 38.Jeong D, Park S, Kim H, Kim CJ, Ahn TS, Bae SB, Kim HJ, Kim TH, Im J, Lee MS, et al. RhoA is associated with invasion and poor prognosis in colorectal cancer. Int J Oncol. 2016;48:714–722. doi: 10.3892/ijo.2015.3281. [DOI] [PubMed] [Google Scholar]