Abstract
Angiotensin-converting enzyme 2 (ACE2) has protective effects on a wide range of morbidities associated with elevated angiotensin-II signaling. Most tissues, including pancreatic islets, express ACE2 mainly from the proximal promoter region. We previously found that hepatocyte nuclear factors 1α and 1β stimulate ACE2 expression from three highly conserved hepatocyte nuclear factor 1 binding motifs in the proximal promoter region. We hypothesized that other highly conserved motifs would also affect ACE2 expression. By systematic mutation of conserved elements, we identified five regions affecting ACE2 expression, of which two regions bound transcriptional activators. One of these is a functional FOXA binding motif. We further identified the main protein binding the FOXA motif in 832/13 insulinoma cells as well as in mouse pancreatic islets as FOXA2.
Keywords: ACE2, FOXA2, promoter, transcription factor, pancreas
Systematic mutation of conserved ACE2 promoter elements combined with electrophoretic mobility shift assay identified FOXA transcription factors (e.g., FOXA2 in pancreatic islets) as essential for basal ACE2 transcription.
Angiotensin-converting enzyme 2 (ACE2) that hydrolyzes angiotensin (Ang)–II to Ang- (1-7) has previously been investigated by many research groups as a natural inhibitor of Ang-II signaling. We have, for example, demonstrated that overexpression of ACE2 in the brain and pancreas counteracts neurogenic hypertension and hyperglycemia, respectively [1–3]. Other research groups have reported beneficial effects of ACE2 on pulmonary hypertension and fibrosis [4–6], heart failure [7, 8], diabetic kidney disease [9, 10], and diabetic retinopathy [11]. Elevation of ACE2 activity may therefore have therapeutic potential for treating conditions characterized by elevated Ang-II levels. As cellular ACE2 concentrations are determined by the balance between synthesis and degradation, ACE2 levels can be increased by stimulating synthesis or inhibiting degradation. We recently investigated whether ACE2 levels in pancreatic islets could be increased by blocking its ADAM17-mediated shedding from cells [12]. We found that ADAM17 levels are too low to markedly affect ACE2 levels. On the other hand, ACE2 levels were directly proportional to the ACE2 synthesis rates in insulinoma cells. Studies assessing the transcriptional regulation of ACE2 are therefore highly warranted.
Regulation of the ACE2 gene promoter is relatively poorly understood. The only independently verified induction of transcription by interaction between promoter elements and transcription factors binding to these elements are hepatocyte nuclear factors 1α and 1β (HNF1α and HNF1β), which bind to evolutionarily conserved motifs in the proximal ACE2 promoter region [13, 14]. In several but not all tissues, the expression of ACE2 is higher in female than in male animals [12, 15, 16]. The mechanism in mice was reported to be mediated by estrogen and an estrogen response element (ERE) [16]. Induction of ACE2 expression by cellular stress involving AMP-activated protein kinase activation has also been described in human hepatoma Huh7 cells [17]. The AMP-activated protein kinase activator AICAR led to increased ACE2 expression and binding of the histone deacetylase SIRT1 to a well-conserved DNA element far upstream (~14.5 kb) of the ACE2 coding region. We investigated the generality of the effects of estrogen and AICAR by assessing whether they affected ACE2 expression in an insulinoma cell line.
We previously characterized the distal and proximal ACE2 promoter regions. We found that the proximal promoter region is the most active promoter in tissues such as heart, pancreas, brain, and kidney, whereas the lung has expression primarily from the distal promoter region. We further determined that HNF1α and HNF1β stimulate ACE2 gene expression by binding to three highly conserved promoter motifs in the proximal promoter region. As there are additional highly conserved promoter motifs in the mammalian ACE2 promoter regions, we hypothesized that these conserved motifs are important cis-regulatory elements for ACE2 expression. We explored the hypotheses by mutating conserved elements and determining how these mutations affect promoter activity and transcription factor binding. We identified several new cis-regulatory sites affecting ACE2 expression and characterized one of these as a functional binding site for forkhead box transcription factors. We finally determined that FOXA2 is the major transcription factor from 832/13 insulinoma cells and mouse pancreatic islets binding to the site.
1. Materials and Methods
A. Cell Line and Animal Experiments
The rat insulinoma cell line 832/13 was a generous gift from Dr. Christopher B. Newgard, Duke University Medical Center (Durham, NC). Cells were maintained and transfected as previously described [13, 18]. For experiments involving estradiol, we used media with charcoal-stripped fetal bovine serum and no phenol red. Mouse islets were isolated as previously described [13]. Animal experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee at Louisiana State University Health Sciences Center.
B. Plasmids
The luciferase reporters hACE2 −454/−1 Luc and hACE2 −1699/−1 Luc containing the −454/−1 and −1699/−1 sequences of the human ACE2 promoter were previously described [13]. Similar plasmids containing the −3919/−1 and −5943/−1 sequences of the human ACE2 promoter inserted between XhoI and MluI sites in pGL3-Basic were made. The numbers indicate bases upstream of the start codon. Conserved sites were replaced with restriction enzyme sites using in vitro mutagenesis. For overexpression of EGFP-ERα and EGFP-ERβ fusion proteins, cells were transfected with plasmids pEGFP-C1-ERα [19] and pEGFP-C1-ERβ purchased from Addgene (catalog nos. 28230 and 28237, respectively). For overexpression of FOXA1, FOXA2, and FOXA3, we transfected cells with plasmids pGCDNsam-Foxa1-IRES-GFP, pGCDNsam-Foxa2-IRES-GFPs, and pGCDNsam-Foxa3-IRES-GFP purchased from Addgene (catalog nos. 33007, 33008 and 33009, respectively). A COUP-TFII expression plasmid was purchased from Origene (catalog no. sc108069). A FOXO1 expression plasmid, pcDNA3-FKHR, was a kind gift from Dr. Andrew Hollenbach (Louisiana State University Health Sciences Center, New Orleans, LS). A plasmid containing the open reading frame for human PPARγ was purchased from Origene (SC 124177). We generated a new expression plasmid for PPARγ by transferring the open reading frame as a NotI/NotI fragment to the pCMV6-XL5 plasmid from Origene. As an empty control plasmid, we used pCMV6-XL5.
C. Transfections and Luciferase Activities
Cells were cotransfected using Lipofectamine 2000 (Thermo Fisher Scientific) with 1 µg luciferase reporter plasmids, 0.25 µg Renilla luciferase control plasmid phRL-TK, ±1 µg expression plasmids for transcription factors as previously described [13]. Promoter strength was quantified as the firefly luciferase activity divided by the Renilla luciferase activity and expressed as relative light units. The empty luciferase reporter pGL3-Basic without inserts turned out to be responsive to FOXA transcription factors and AICAR due to cryptic binding sites. To separate effects on the ACE2 promoter from effects on the vector backbone, we corrected for the backbone effects by subtracting the activities of empty pGL3-Basic from activities with ACE2-luciferase reporters. Each transfection experiment was conducted four times.
D. Bioinformatics
The BKL TRANSFAC program from Biobase Gmbh was used to identify potential transcription factor binding sites.
E. Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was conducted with 32P-labeled, double-stranded DNA probes as previously reported [13] with extracts generated with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Base sequences for the DNA probes are listed in Table 1. Antibodies used were an antibody recognizing both FOXA1 and FOXA2 (sc-6553X, RRID: AB_2104865; Santa Cruz Biotechnology), anti-FOXA3 (sc-5361X, RRID: AB_647544; Santa Cruz Biotechnology), anti-FOXO1 (sc-11350X, RRID: AB_640607; Santa Cruz Biotechnology), anti-PPARγ (sc-7196X, RRID: AB_654710; Santa Cruz Biotechnology), and anti–COUP-TFII (PP-H7147-00, RRID: AB_2155627; R&D Systems).
Table 1.
Sequences of Oligonucleotides for EMSA
| Site | Forward Oligonucleotide | Reverse Oligonucleotide |
|---|---|---|
| R3 Wt | ggt ttt agt cta ggg aaa gtc att cag | ctg aat gac ttt ccc tag act aaa acc |
| R3 Mut | ggt ttt agt ctg aat tca gtc att cag | ctg aat gac tga att cag act aaa acc |
| R4 Wt | gcc caa ccc aag ttc aaa ggc tga taa | tta tca gcc ttt gaa ctt ggg ttg ggc |
| R4 Mut | gcc caa ccc aag tta gat ctc tga taa | tta tca gag atc taa ctt ggg ttg ggc |
| R6 Wt | ctc agc aga ttg ttt act gtg ttc ttc | gaa gaa cac agt aaa caa tct gct gag |
| R6 Mut | ctc agc aga gga tcc act gtg ttc ttc | gaa gaa cac agt gga tcc tct gct gag |
| R8 Wt | aca tat ctg tcc tct cca gga tga act | agt tca tcc tgg aga gga cag ata tgt |
| R8 Mut | aca tat ctg tcc tgc ggc cgc tga act | agt tca gcg gcc gca gga cag ata tgt |
| R12 Wt | tga ttt ggc cat aaa gtg aca gga gag | ctc tcc tgt cac ttt atg gcc aaa tca |
| R12 Mut | tga ttt ggc cac ccg ggg aca gga gag | ctc tcc tgt ccc cgg gtg gcc aaa tca |
| Mouse R6 | tcc agc agc ttg ttt act gtt ctc ttc | gaa gag aac agt aaa caa gct gct gga |
| Human −1363/−1337 | cct gga aga ctt gtt ttt ctg ggt gaa | ttc acc cag aaa aac aag tct tcc agg |
| Mouse −1390/−1364 | cct gga aga ctt gtt ttt ctg gat gga | tcc atc cag aaa aac aag tct tcc agg |
| Human ACE2 putative ERE | gtg tta agg tca aac ttc cct tta cca | tgg taa agg gaa gtt tga cct taa cac |
| Mouse ACE2 putative ERE | gca tca agg tca aac tct ctg tgt ttt | aaa aca cag aga gtt tga cct tga tgc |
| Vitellogenin-A2 ERE | cca aag tca ggt cac agt gac ctg atc | gat cag gtc act gtg acc tga ctt tgg |
Sequences are listed in the 5′ → 3′ direction.
F. Quantitative Reverse Transcription Polymerase Chain Reaction
Rat ACE2 messenger RNA (mRNA), rat β-actin mRNA, mouse ACE2 mRNA, mouse FOXA2 mRNA, and mouse 18S ribosomal RNA were quantified with SYBR green–based real-time quantitative reverse transcription polymerase chain reaction. Primer sequences are listed in Table 2. Abundance of transcripts from the mouse proximal and distal promoter regions was determined by Taqman-based quantitative reverse transcription polymerase chain reaction, as previously described [13].
Table 2.
Sequences of Oligonucleotides for Quantitative Reverse Transcription Polymerase Chain Reaction
| mRNA | Forward Primer | Reverse Primer |
|---|---|---|
| Rat ACE2 | atg ccg acc aaa gca tta aag t | atg atc gga ata ggt aca ttt cgt t |
| Rat β-actin | aga tga ccc aga tca tgt ttg aga | cca gag gca tac agg gac aac |
| Mouse ACE2 | gag gat aag cct aaa atc agc tct tg | tcg gaa cag gaa cat ttc gtt |
| Mouse FOXA2 | gga ccc caa gac ata ccg ac | atc ttg ttg ggg ctc tgc tg |
| Mouse 18S ribosomal RNA | cgg aca gga ttg aca gat tg | caa atc gct cca cca act aa |
Sequences are listed in the 5′ → 3′ direction.
G. Statistical Analysis
Data are presented as means ± SE. Data were analyzed by analysis of variance with post hoc contrasts compared with Bonferroni correction for multiple contrasts.
2. Results
A. Estradiol Has Minimal Effect on ACE2 Expression in 832/13 Cells
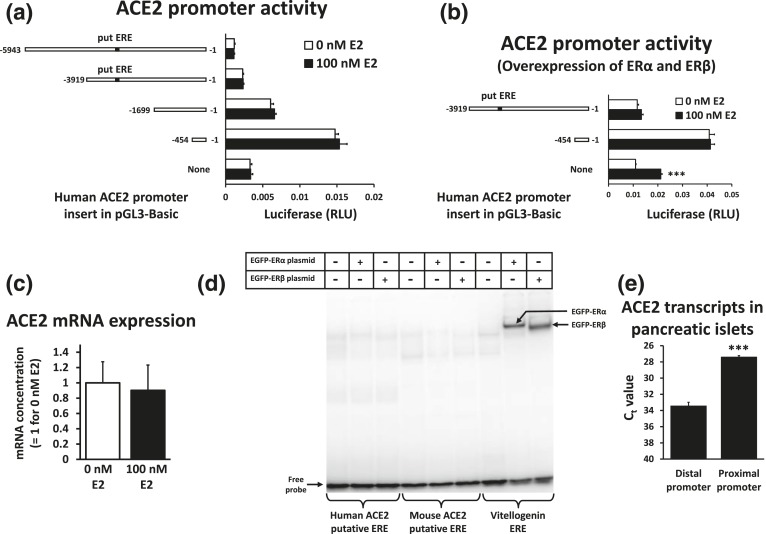
We used the rat insulinoma cell line 832/13 to study ACE2 promoter activity. The ACE2 promoter is active in the context of luciferase reporters in this cell line. Although lower than ACE2 expression in mouse islets, the endogenous ACE2 mRNA expression is clearly detectable in this cell line [13]. We previously defined the distal (−1509/−928) and proximal (−454/−1) human ACE2 promoter regions based on homology with other mammalian species [13], yet there are other conserved elements outside these regions that may affect transcription. An example is the DNA sequence AGGTCAAACTCTCTG at position −2986/−2972 of the mouse ACE2 promoter that has been identified as an ERE [16]. The homologous sequence for the human ACE2 promoter is AGGTCAAACTTCCCT (−2639/−2625). As ACE2 expression in pancreatic islets from female mice was higher than in islets from male mice [12], we tested whether estradiol can increase responsiveness of human ACE2 promoter sequences inserted into luciferase reporters in 832/13 cells. These cells are derived from male rats [20], and they can respond to estradiol, for example, in estradiol-mediated inhibition of expression of fatty acid synthase and the carbohydrate response element binding protein in lipogenic conditions [21, 22]. We observed no statistically significant estradiol response from any of the promoter constructs, including the ones containing the putative ERE [Fig. 1(a)]. To ensure that the lack of an estrogen response was not due to an insufficient level of estrogen receptors in our experimental conditions, we conducted a transfection experiment with luciferase reporters in which both ERα and ERβ were overexpressed as transcriptionally active fusion proteins with enhanced green fluorescent protein [19]. Green fluorescence was observed primarily in the cell nuclei (data not shown). Under these conditions, the background activity of the empty pGL3-Basic plasmid became estrogen responsive, likely due to cryptic binding sites, but not promoter activity driven by ACE2 promoter regions [Fig. 1(b)]. Endogenous ACE2 expression in 832/13 cells overexpressing ERα and ERβ was likewise unaffected by estradiol [Fig. 1(c)], at least under the culture conditions that we used. To test whether the putative EREs from the ACE2 promoters of mice and humans really have the ability to bind estrogen receptors, we conducted an EMSA of these regions compared with the well-established ERE from the vitellogenin A2 gene from Xenopus laevis [23] [Fig. 1(d)]. Although ERα and ERβ expressed as enhanced green fluorescent protein fusion proteins in 832/13 cells both bind to the vitellogenin A2 ERE, there is no visible binding to the putative ERE sequences. We conclude that these regions from the mouse and human ACE2 promoters are not bona fide EREs.
Figure 1.
The proximal ACE2 promoter is active in 832/13 cells and pancreatic islets. (a) Luciferase reporters containing human ACE2 promoter segments were transfected into 832/13 cells. A putative ERE (put ERE) is indicated. Transfected cells were treated with 0 or 100 nM estradiol (E2) for 44 hours. (b) Luciferase reporters containing human ACE2 promoter segments were transfected into 832/13 cells together with 0.5 μg EGFP-ERα and 0.5 μg EGFP-ERβ expression plasmid per well. A put ERE is indicated. Transfected cells were treated with 0 or 100 nM estradiol (E2) for 44 hours. ***P < 0.001 vs 0 nM E2. (c) 832/13 cells were transfected with EGFP-ERα and 0.5 μg EGFP-ERβ expression plasmids and subsequently treated with 0 or 100 nM E2 for 48 hours in four experiments. The content of ACE2 mRNA relative to β-actin mRNA was determined. (d) An EMSA was done with the probes for the put EREs from the mouse and human ACE2 promoters as well as the ERE from the vitellogenin A2 gene. Nuclear extracts were from 832/13 cells that were untransfected or transfected with either EGFP-ERα or EGFP-ERβ expression plasmids and grown in the presence of 100 nM E2. (e) In mouse pancreatic islets, the concentration of ACE2 mRNA with transcriptional initiation in the distal and proximal promoter regions was compared with specific Taqman-based quantitative reverse transcription polymerase chain reaction assays. Islets were isolated from four mice. The cycle threshold values (Ct) for the same threshold with RNA diluted to 20 ng/µL are indicated. ***P < 0.001 vs distal promoter region. RLU, relative light unit.
It is further remarkable that only the proximal ACE2 promoter region (−454/−1) shows strong promoter activity. Longer constructs decrease promoter activity [Fig. 1(a)], suggesting that regions upstream of the proximal promoter region contain elements inhibiting transcription. To test the relative strength of the promoter regions in mouse islets, we used previously developed Taqman-based quantitative reverse transcription polymerase chain reaction assays that directly compare the concentration of ACE2 mRNA derived from the distal and proximal promoter region [13]. We isolated RNA from mouse pancreatic islets and observed that the proximal promoter region is also the most active, as ACE2 transcripts from this region are approximately 66-fold more abundant than transcripts from the distal promoter region [Fig. 1(e)].
B. Identification of Cis-Regulatory Elements in the Proximal Promoter Region
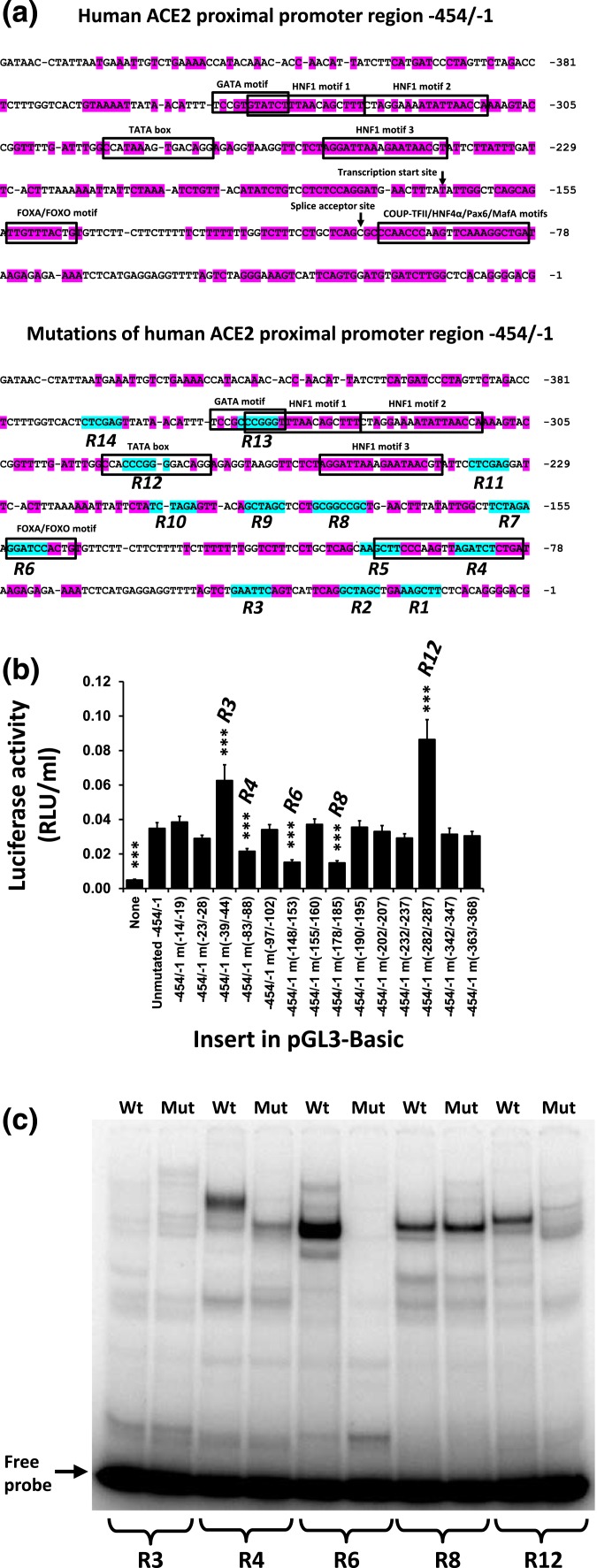
Because the proximal promoter is the most active in 832/13 cells and pancreatic islets (Fig. 1) [13], and since HNF1α and HNF1β exert their effects through conserved elements, we decided to search for other important transcription factor binding sites among highly conserved sequences of the proximal promoter region. In the context of −454/−1 human ACE2 promoter driving ACE2 expression, we mutated 14 conserved motifs as indicated in Fig. 2(a). We termed the wild-type regions containing these motifs R1 to R14. Three of these mutations (−83/−88 in R4, −148/−153 in R6, and −178/−185 in R8) led to significant downregulation of expression, whereas two mutations (−39/−44 in R3 and −282/−287 in R12) increased expression [Fig. 2(b)]. These well-conserved regions are therefore putative cis-regulatory elements binding activators and inhibitors, respectively, of ACE2 expression. To see, if the mutations altered binding of proteins to the promoter regions, we performed EMSA of double-stranded DNA corresponding to the unmutated and mutated regions [Table 1 and Fig. 2(c)]. Mutations of R3 and R8 gave subtle band changes that are difficult to interpret. Mutation of R12 resulted in the disappearance of a pronounced band, suggesting that it represents a transcriptional inhibitor of ACE2 expression. Mutations of regions R4 and R6 caused intense bands to disappear, suggesting that these bands represent transcriptional activators. We set out to characterize these elements further.
Figure 2.
Identification of cis-regulatory elements controlling ACE2 expression. (a) The conserved regions of the human proximal ACE2 promoter region and bases that were mutated in luciferase reporters by in vitro mutagenesis are indicated. Pink shading indicates bases conserved among seven placental mammalian species (human, mouse, rabbit, dog, panda, horse, and cow), and blue shading indicates the introduced mutations of 14 regions (R1 to R14). (b) 832/13 cells were transfected with luciferase reporters containing wild-type or mutated sequences of the human ACE2 proximal promoter region. Luciferase activities were determined 44 hours after transfection. R3, R4, R6, R8, and R12 denote the promoter regions, where mutations significantly affected luciferase activities. ***P < 0.001 vs the unmutated, wild-type −454/−1 promoter sequence. (c) An EMSA was conducted with nuclear extracts from 832/13 cells and with DNA probes that were wild-type and mutated sequences of five regions with putative cis-regulatory elements.
C. COUP-TFII Can Bind To But Is Not the Major Factor Binding To the –101/−79 Motif
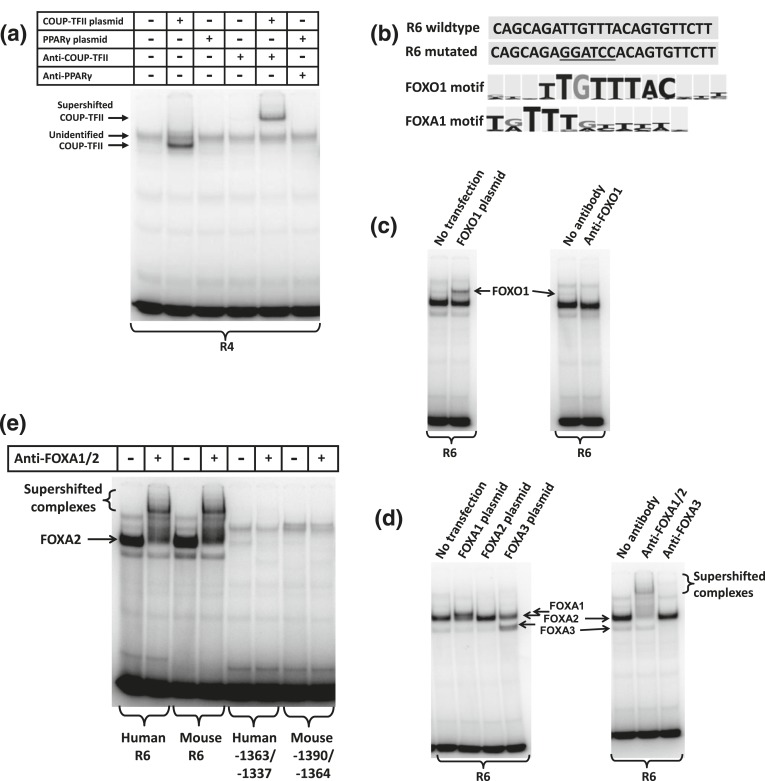
The −101/−79 motif is highly conserved among mammalian species. According to the BKL TRANSFAC program, several transcription factors have binding motifs with similarity to this conserved ACE2 promoter region, including COUP-TFII, HNF4α, PPARγ, and MafA. When COUP-TFII is overexpressed in 832/13 cells, a band appears in EMSA that can be supershifted with an antibody against COUP-TFII [Fig. 3(a)]. This demonstrates that COUP-TFII can bind to this DNA motif. However, the major band was not shifted with the COUP-TFII antibody, meaning that it contains a different transcription factor. Neither were we able to shift or alter band intensity with antibodies against PPARγ [Fig. 3(a)], E2A, NeuroD1, USF1, USF2, MafA, HNF4, Lef1, GATA4, or C/EBPγ (Supplemental Fig. 1 (273.3KB, pdf) ). The identity of this putative transcription factor remains elusive.
Figure 3.
Forkhead box transcription factors bind to the ACE2 proximal promoter region. (a) An EMSA was conducted with the R4 DNA probe. Nuclear extracts were from untransfected 832/13 cells or 832/13 cells transfected with COUP-TFII or PPARγ expression plasmids. Antibodies against COUP-TFII and PPARγ were included in the binding reactions as indicated. (b) The R6 region has similarity to FOXO1 and FOXA1 motifs as indicated by the BKL TRANSFAC program, whereas the mutation destroys the similarity. (c) An EMSA was done with the R6 probe and nuclear extracts from 832/13 cells that were untransfected or transfected with a FOXO1 expression plasmid (left panel). The right panel shows the effect on the band pattern when an antibody against FOXO1 is included. (d) An EMSA was done with the R6 probe and nuclear extracts from 832/13 cells that were untransfected or transfected with FOXA1, FOXA2, or FOXA3 expression plasmids (left panel). The right panel shows the effects on the band pattern when antibodies against the FOXA transcription factors are included in the binding reactions. (e) An EMSA was conducted with a nuclear extract from 832/13 cells. The probes were the human and mouse R6 regions as well as regions in the distal promoter region with a putative FOXA binding site. An antibody recognizing FOXA1 and FOXA2 was included in the binding reactions as indicated.
D. Forkhead Box Transcription Factors Bind To a FOXA Motif in the Proximal Promoter Region
The region R6 contains the region −153/−144, which has high similarity to FOXA and FOXO binding motifs [Fig. 3(b)]. FOXO1 is able to bind to this motif, as a band above the major band becomes intensified after FOXO1 overexpression, whereas inclusion of a FOXO1 antibody in the binding reaction diminishes the band intensity [Fig. 3(c)]. In this case, the antibody seemed to inhibit binding of FOXO1 to the R6 motif. Overexpression of FOXA1 and FOXA3 results in intense bands appearing above and below the major band [Fig. 3(d)]. The major band may further be intensified with FOXA2 overexpression [Fig. 3(d)]. An antibody recognizing both FOXA1 and FOXA2 is able to supershift the intense bands appearing with FOXA1 and FOXA2 overexpression, whereas the band appearing with FOXA3 overexpression is supershifted with a FOXA3-specific antibody [Fig. 3(d)]. The relative mobility FOXA3 > FOXA2 > FOXA1 is in accordance with the molecular weights where FOXA1 > FOXA2 > FOXA3, at least when the proteins are not posttranslationally modified. The EMSA clearly demonstrates that FOXA1, FOXA2, and FOXA3 are all capable of binding to the R6 region and that FOXA2 is the most pronounced protein in 832/13 nuclear extracts that bind to the motif. The homologous R6 region from the mouse genome also binds primarily FOXA2 from 832/13 nuclear extracts [Fig. 3(e)]. This contrasts with a conserved region (human −1363/−1337 and mouse −1390/−1364) in the distal ACE2 promoter region that, according to the BKL TRANSFAC program, also has similarity to FOXA binding sites but where FOXA2 only has low binding affinity [Fig. 3(e)].
E. The –153/−144 Motif Is a Functional FOXA Binding Site
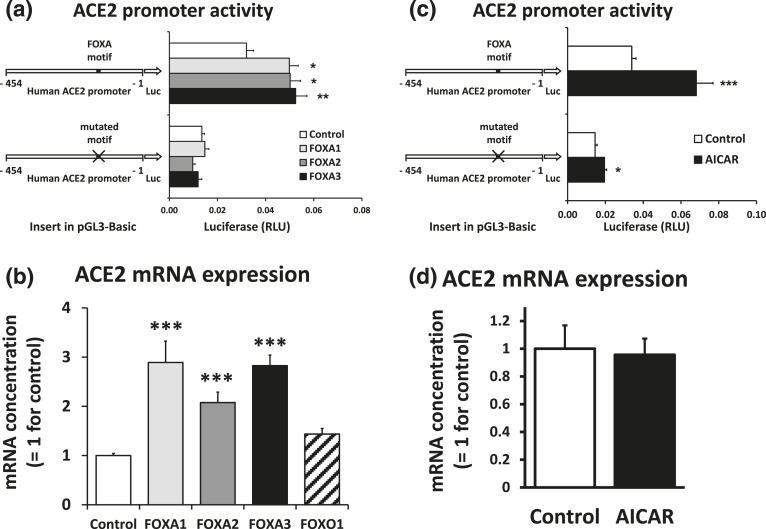
To test whether the –153/−144 FOXA motif is a functional FOXA binding site, we compared expression of luciferase reporters with either the wild-type or mutated –153/−144 motif when FOXA1, FOXA2, or FOXA3 was overexpressed. Overexpression of the FOXA transcription factors significantly induced promoter activity with the wild-type motif. The transcription factors had no effect when the FOXA motif was mutated [Fig. 4(a)]. The reduction in expression caused by the mutation in the control condition is consistent with the reduction observed with the R6 mutation in Fig. 2(b). The endogenous ACE2 mRNA expression in 832/13 cells was also significantly induced by overexpression of FOXA1, FOXA2, and FOXA3, whereas FOXO1 overexpression had less effect [Fig. 4(b)]. We conclude that the proximal ACE2 promoter region contains a functional FOXA binding site.
Figure 4.
FOXA transcription factors stimulate ACE2 expression. (a) 832/13 cells were transfected with luciferase reporters containing the wild-type human −454/−1 ACE2 promoter sequence or the promoter sequence in which the FOXA motif was mutated. Cells were cotransfected with expression plasmids for FOXA1, FOXA2, FOXA3, or a control plasmid. Luciferase activities were determined 44 hours after transfection and corrected for the background activities of empty pGL3-Basic. *P < 0.05, **P < 0.01 vs control. (b) 832/13 cells were transfected with expression plasmids for forkhead box transcription factors or a control plasmid. RNA was isolated 22 hours after transfection. The content of ACE2 mRNA relative to β-actin mRNA was determined. ***P < 0.001 vs control. (c) 832/13 cells were transfected with luciferase reporters containing the wild-type human −454/−1 ACE2 promoter sequence or the promoter sequence in which the FOXA motif was mutated. Transfected cells were treated with 0.5 mM AICAR or dimethyl sulfoxide (DMSO) vehicle as control for 44 hours. Luciferase activities were corrected for the background activities of empty pGL3-Basic. *P < 0.05, ***P < 0.001 vs control. (d) 832/13 cells were treated with 0.5 mM AICAR or DMSO vehicle as control for 48 hours. The content of ACE2 mRNA relative to β-actin mRNA was determined.
Cellular stress mediated by the AMP-activated protein kinase activator AICAR was reported to induce ACE2 expression in Huh7 hepatoma cells via activation of the NAD+-dependent deacetylase SIRT1 [17]. As FOXA2 is a substrate for SIRT1 deacetylation [24, 25], we investigated the role of AICAR on the activity of the human ACE2 proximal promoter region. In a luciferase reporter with the wild-type −454/−1 ACE2 promoter sequence, AICAR significantly induced luciferase expression [Fig. 4(c)]. While still being induced by AICAR, mutation of the FOXA site significantly diminished the induction (P = 0.03 for the interaction between plasmid and AICAR presence). On the other hand, AICAR had no effect on the endogenous expression of ACE2 mRNA in 832/13 cells [Fig. 4(d)]. AICAR-mediated ACE2 induction must therefore be cell type specific.
F. FOXA2 Is the Major Islet Transcription Factor Binding to the FOXA Motif
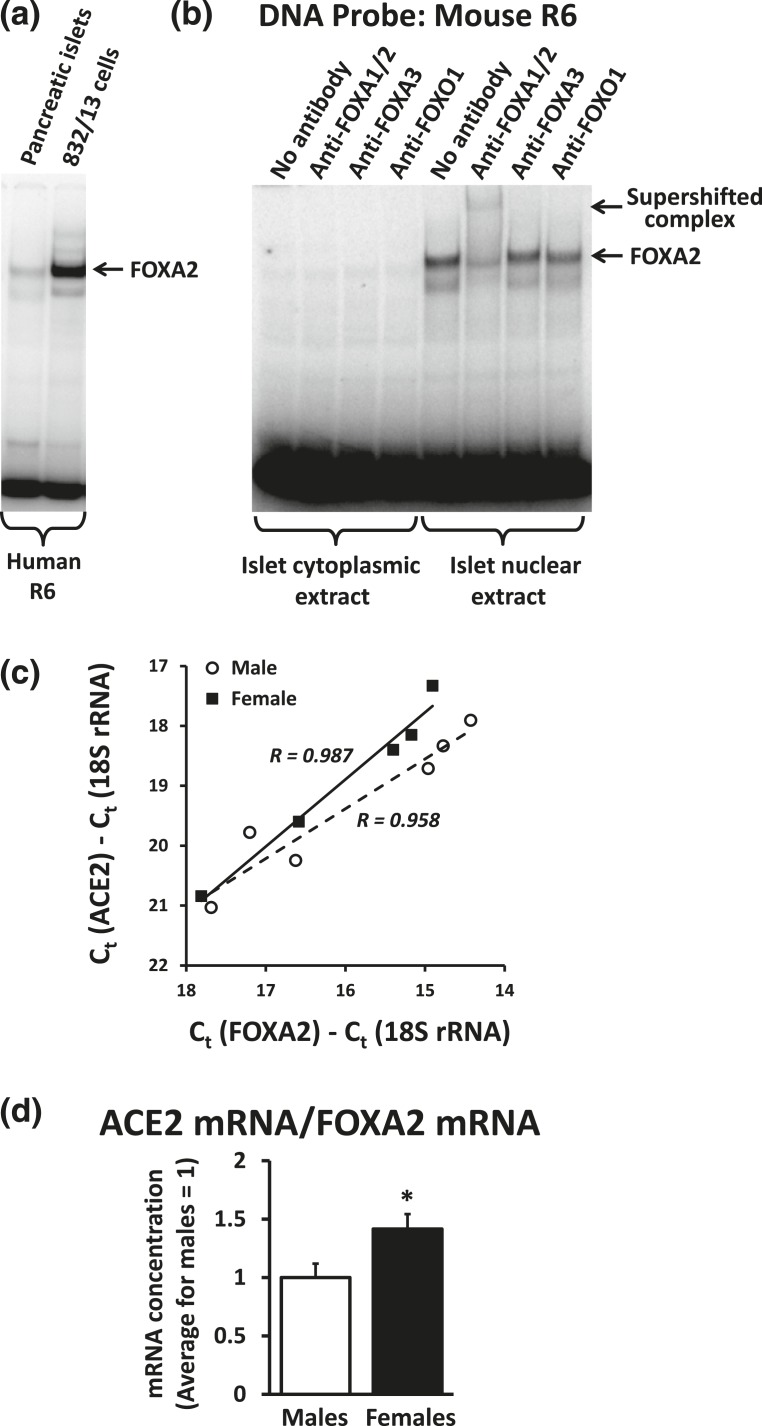
A nuclear extract from mouse pancreatic islets contains a protein binding to the human R6 region with the same mobility as FOXA2 from 832/13 cells [Fig. 5(a)]. The major band binding to the mouse R6 region can further be supershifted with the antibody against FOXA1 and FOXA2 [Fig. 5(b)]. We conclude that FOXA2 is the major protein in mouse islets binding to the FOXA motif in the proximal ACE2 promoter region.
Figure 5.
FOXA2 is the major protein in mouse islets binding the FOXA motif in the proximal ACE2 promoter. (a) An EMSA with the human R6 probe was conducted using nuclear extracts from mouse pancreatic islets and 832/13 cells. (b) An EMSA with the mouse R6 probe was conducted using cytoplasmic and nuclear extracts from mouse pancreatic islets. Antibodies against FOXA transcription factors were included in binding reactions as indicated. (c) The concentrations of FOXA2 mRNA and ACE2 mRNA relative to 18S ribosomal RNA were determined in freshly isolated pancreatic islets from six male and five female mice. We depict the ACE2 values against the FOXA2 values for each mouse. The solid and stippled lines are the regression lines for females and males, respectively. The concentrations of FOXA2 and ACE2 showed significant (P < 0.01) correlation for both males and females as indicated by the correlation coefficients R. (d) The content of ACE2 mRNA relative to FOXA2 mRNA was calculated. *P < 0.05 vs males.
FOXA transcription factors have been reported to be involved in sexual dimorphism in gene regulation in liver cancer [26]. We wished to test if a sex difference in FOXA2 expression in mouse islets is related to a sex difference in ACE2 expression. We determined the concentration of ACE2 mRNA and FOXA2 mRNA in pancreatic islets isolated from six males and five females of the C57Bl/6 background of ages 78 to 129 days. We did not observe statistically significant sex differences in ACE2 or FOXA2 expression normalized to 18S ribosomal RNA, possibly due to the high variance of the data. However, we noticed that there was a significant correlation (P < 0.01) between the FOXA2 and ACE2 mRNA expression for both males and females [Fig. 5(c)]. Calculation of the ACE2 mRNA expression per FOXA2 mRNA expression actually shows a 42% increase (P = 0.046) in females relative to males [Fig. 5(d)]. For a given concentration of FOXA2, there thus seems to be a higher expression of ACE2 mRNA in females than in males.
3. Discussion
ACE2 has shown beneficial effects on a wide variety of conditions where an overactive renin angiotensin system and Ang-II signaling play a role [1–11]. Our laboratory has, for example, demonstrated a glycemia-protective effect of pancreatic ACE2 [1, 2, 27] and a pivotal role in the regulation of autonomic function and blood pressure in hypertension [1, 3, 28, 29]. ACE2 is therefore an attractive therapeutic target as an endogenous regulator of renin angiotensin system signaling. Furthermore, its hydrolysis product, Ang- [1–7] tends to have effects opposite those of Ang-II. Compared with current pharmaceutical agents causing the renin angiotensin system blockade, ACE2 is expected to have less side effects such as dry cough associated with angiotensin-converting enzyme inhibitors. Another attractive property of ACE2 is that its Michaelis-Menten constant Km is orders of magnitude higher than physiological concentrations of Ang-II [18, 30]. The hydrolysis rate of Ang-II will therefore increase proportionally with the Ang-II concentration, suggesting that ACE2 has little effect at low Ang-II concentrations but exerts strong physiological effects at morbidities with elevated Ang-II concentrations. This may explain why overexpression of ACE2 lowers blood glucose levels and blood pressure in disease states but not in normoglycemic or normotensive animals [1, 28, 29].
We are interested in the regulation of ACE2 in pancreatic islets due to its glycemia-protective properties. Inhibiting ACE2 degradation is one possible way of elevating ACE2 levels. The protease ADAM17 can decrease ACE2 levels by shedding its extracellular domain, including the catalytic site. However, the levels of ADAM17 in mouse pancreatic islets are too low to markedly affect ACE2 content [12]. On the other hand, an increase in the ACE2 synthesis rate is directly proportional to cellular ACE2 levels [12]. A better understanding of the transcriptional regulation of ACE2 is therefore useful.
Little is known of the regulation of ACE2 from the regions in which transcription is initiated (i.e., the proximal and distal promoter regions). HNF1β was discovered to stimulate ACE2 transcription [14]. We extended this observation by determining that both HNF1α and HNF1β bind to three evolutionarily conserved HNF1 binding sites in the proximal promoter region [13]. In the current study, we have demonstrated that several of the additional evolutionarily conserved motifs in the ACE2 gene promoter are cis-regulatory elements. Highly conserved motifs at positions −153/−144 and –101/−79 of the human ACE2 promoter are required for basal transcription in 832/13 cells. Although the −101/−79 motif can bind COUP-TFII, it binds a potential transcriptional activator in 832/13 cells whose identity remains elusive. On the other hand, the –153/−144 motif is a functional binding site for FOXA transcription factors. The major factor binding the −153/−144 motif in both 832/13 insulinoma cells and mouse pancreatic islets is FOXA2. We further observed a correlation between the FOXA2 mRNA and ACE2 mRNA in mouse pancreatic islets. Although we cannot exclude the possibility of other factors affecting both ACE2 and FOXA2 expression, the correlation is consistent with the notion that FOXA2 stimulates ACE2 expression.
ACE2 expression is stimulated by AICAR in Huh-7 cells [17] and by estradiol in adipose tissue and 3T3-L1 adipocytes [16]. The effects were claimed to be mediated by conserved elements outside the proximal and distal promoter region, as we define these promoter regions. The putative ERE in the mouse ACE2 promoter as well as its human homolog did not bind estrogen receptors in vitro and are therefore unlikely to directly mediate estrogen responsiveness. We did not observe estradiol-mediated induction of ACE2 promoter activity or endogenous ACE2 mRNA expression in 832/13 cells, even when ERα and ERβ are overexpressed. The previously observed estrogen induction of ACE2 expression [16] must therefore be cell or tissue specific. We previously measured higher ACE2 enzymatic activity in females than in males [12]. This may be partly due to an elevated ACE2 transcription rate in females as indicated by higher ACE2 mRNA levels per FOXA2 mRNA concentration in females than in males. Other factors than estrogen mediating sex-specific effects on ACE2 expression could be androgens or a gene-dosage effect, as the Ace2 gene is located on the X chromosome. We did not observe stimulatory effects of AICAR on endogenous ACE2 mRNA levels in 832/13 cells showing that the effects must be cell or tissue specific. The AICAR effect in Huh7 cells was claimed to be mediated by a conserved element capable of binding SIRT1 that is located 14 kb upstream of the ACE2 promoter regions. It is in fact located closer to the Tmem27 gene than the Ace2 gene. Although it was described as an ACE2 promoter [17], it is more likely an enhancer, as no transcription is known to be initiated in this region. Interestingly, AICAR stimulated proximal ACE2 promoter activity in a luciferase reporter in a FOXA motif-dependent manner. As the reporter does not contain the upstream enhancer, AICAR/SIRT1 may exert effects on ACE2 expression through several genomic elements. Positive and negative effects may further cancel out, resulting in no net AICAR effect on endogenous 832/13 ACE2 expression.
Genome-wide mapping of transcription factor binding sites by ChIP-seq can be seen in the University of California, Santa Cruz genome browser for the ENCODE project [31]. In the February 2009 (GRCh37/hg19) assembly for the human genome, there are several transcription factor binding sites listed in the distal promoter region and the upstream enhancer region but none in the proximal promoter region. Yet, in mouse kidney, heart, brain, pancreas, and pancreatic islets, most ACE2 transcripts originate from the proximal promoter region (Fig. 1 and Pedersen et al. [13]). In the context of luciferase reporters, the proximal promoter regions are active promoters in both human HEK 293T cells and rat 832/13 cells, whereas the distal promoter regions did not by themselves promote transcription [13]. Our new results in Fig. 1 illustrate that elements upstream of the human proximal ACE2 promoter region can actually repress transcription in 832/13 cells. A limitation of current data listed for ACE2 in the ENCODE project is that the cells used for ChIP-seq experiments exhibit low ACE2 expression. We speculate that there is repression of the proximal promoter region. This is supported by a lack, according to data from the University of California, Santa Cruz genome browser, of a DNaseI hypersensitive region at the proximal promoter region, even though there is such a hypersensitive region at the distal promoter region. A potential mechanism is suggested by a recent study describing binding of the chromatin remodeler BRG1 and the forkhead box transcription factor FOXM1 to four regions upstream of the ACE2 proximal promoter region in mouse hearts after ventricular pressure overload but not to the proximal promoter region itself [32]. Furthermore, overexpression of both BRG1 and FOXM1 reduced ACE2 promoter activity in mouse cardiac endothelial cells [32].
The Cistrome data browser [33] allows visualization of FOXA2 binding to the ACE2 gene in mouse pancreas and mouse pancreatic β-cells. There are FOXA2 binding peaks overlapping the FOXA site of the mouse proximal ACE2 promoter region, in addition to several other binding peaks in the gene (e.g., located in introns). Whether FOXA2 regulation mainly occurs through the site in the proximal promoter region could be tested in future research using genome editing.
Most ACE2 transcripts in mouse kidney, heart, brain, whole pancreas, and pancreatic islets are generated from the proximal ACE2 promoter region [Fig. 1(d) and Pedersen et al. [13]]. The advantage of using the 832/13 cell line for ACE2 promoter studies is that the proximal ACE2 promoter is active in the context of luciferase reporters and that the cells actually express ACE2 in measurable quantities. The limitation is that expression in 832/13 cells is markedly lower than in highly expressing tissue such as pancreatic islets and kidney. Factors ensuring high expression in these tissues are therefore of great interest for future research.
We have now established that both HNF1α and FOXA2 stimulate ACE2 expression (Fig. 4) [13]. It was reported that high-fat diets and high concentrations of free fatty acids lead to depletion of HNF1α and FOXA2 in pancreatic β-cells and are associated with hyperglycemia and impaired glucose tolerance [34]. Although that might be thought to lead to a reduction of ACE2 in pancreatic islets in models of type 2 diabetes, we actually observed retention of islet ACE2 expression with the progression of diabetes. That is explained by the finding that β-cells are not the main cell type expressing ACE2 in islets [12]. Recent immunohistochemistry data suggest that ACE2 expression is at least partially colocalized with glucagon expression (i.e., in α-cells) [27]. FOXA2 is furthermore a well-known regulator of α-cell differentiation and glucagon synthesis. The role of FOXA2 in regulating islet ACE2 expression may therefore be more important in α-cells than in β-cells.
In summary, we have identified new cis-regulatory elements in the ACE2 proximal promoter region, including a functional forkhead box transcription factor binding site that is a binding motif for FOXA2 in pancreatic islets.
Acknowledgments
Acknowledgments
This work was supported by an Established Investigator Award from the American Heart Association to E.L. (12EIA8030004) and by a predoctoral fellowship from the American Heart Association Greater SouthEast Affiliate to H.C. (14PRE18830012).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ACE2
- angiotensin-converting enzyme 2
- Ang
- angiotensin
- EMSA
- electrophoretic mobility shift assay
- ERE
- estrogen response element
- HNF1α
- hepatocyte nuclear factor 1α
- HNF1β
- hepatocyte nuclear factor 1β
- mRNA
- messenger RNA.
References and Notes
- 1.Bindom SM, Hans CP, Xia H, Boulares AH, Lazartigues E. Angiotensin I–converting enzyme type 2 (ACE2) gene therapy improves glycemic control in diabetic mice. Diabetes. 2010;59:2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhabra KH, Xia H, Pedersen KB, Speth RC, Lazartigues E. Pancreatic angiotensin-converting enzyme 2 improves glycemia in angiotensin II-infused mice. Am J Physiol Endocrinol Metab. 2013;304:E874–E884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriramula S, Xia H, Xu P, Lazartigues E. Brain-targeted angiotensin-converting enzyme 2 overexpression attenuates neurogenic hypertension by inhibiting cyclooxygenase-mediated inflammation. Hypertension. 2014;65:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamazato Y, Ferreira AJ, Hong K-H, Sriramula S, Francis J, Yamazato M, Yuan L, Bradford CN, Shenoy V, Oh SP, Katovich MJ, Raizada MK. Prevention of pulmonary hypertension by angiotensin-converting enzyme 2 gene transfer. Hypertension. 2009;54:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shenoy V, Ferreira AJ, Qi Y, Fraga-Silva RA, Díez-Freire C, Dooies A, Jun JY, Sriramula S, Mariappan N, Pourang D, Venugopal CS, Francis J, Reudelhuber T, Santos RA, Patel JM, Raizada MK, Katovich MJ. The angiotensin-converting enzyme 2/angiogenesis-(1-7)/Mas axis confers cardiopulmonary protection against lung fibrosis and pulmonary hypertension. Am J Respir Crit Care Med. 2010;182:1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Molina-Molina M, Abdul-Hafez A, Uhal V, Xaubet A, Uhal BD. Angiotensin converting enzyme-2 is protective but downregulated in human and experimental lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008;295:L178–L185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oudit GY, Penninger JM. Recombinant human angiotensin-converting enzyme 2 as a new renin-angiotensin system peptidase for heart failure therapy. Curr Heart Fail Rep. 2011;8:176–183. [DOI] [PubMed] [Google Scholar]
- 8.Iwata M, Cowling RT, Yeo SJ, Greenberg B. Targeting the ACE2-Ang-(1-7) pathway in cardiac fibroblasts to treat cardiac remodeling and heart failure. J Mol Cell Cardiol. 2011;51:542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadarajah R, Milagres R, Dilauro M, Gutsol A, Xiao F, Zimpelmann J, Kennedy C, Wysocki J, Batlle D, Burns KD. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int. 2012;82:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu CX, Hu Q, Wang Y, Zhang W, Ma ZY, Feng JB, Wang R, Wang XP, Dong B, Gao F, Zhang MX, Zhang Y. Angiotensin-converting enzyme (ACE) 2 overexpression ameliorates glomerular injury in a rat model of diabetic nephropathy: a comparison with ACE inhibition. Mol Med. 2011;17:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez JM II, Hu P, Caballero S, Moldovan L, Verma A, Oudit GY, Li Q, Grant MB. Adeno-associated virus overexpression of angiotensin-converting enzyme-2 reverses diabetic retinopathy in type 1 diabetes in mice. Am J Pathol. 2016;186:1688–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen KB, Chodavarapu H, Porretta C, Robinson LK, Lazartigues E. Dynamics of ADAM17-mediated shedding of ACE2 applied to pancreatic islets of male db/db mice. Endocrinology. 2015;156:4411–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen KB, Chhabra KH, Nguyen VK, Xia H, Lazartigues E. The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim Biophys Acta. 2013;1829:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senkel S, Lucas B, Klein-Hitpass L, Ryffel GU. Identification of target genes of the transcription factor HNF1beta and HNF1alpha in a human embryonic kidney cell line. Biochim Biophys Acta. 2005;1731:179–190. [DOI] [PubMed] [Google Scholar]
- 15.Gupte M, Thatcher SE, Boustany-Kari CM, Shoemaker R, Yiannikouris F, Zhang X, Karounos M, Cassis LA. Angiotensin converting enzyme 2 contributes to sex differences in the development of obesity hypertension in C57BL/6 mice. Arterioscler Thromb Vasc Biol. 2012;32:1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Shoemaker R, Thatcher SE, Batifoulier-Yiannikouris F, English VL, Cassis LA. Administration of 17β-estradiol to ovariectomized obese female mice reverses obesity-hypertension through an ACE2-dependent mechanism. Am J Physiol Endocrinol Metab. 2015;308:E1066–E1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke NE, Belyaev ND, Lambert DW, Turner AJ. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin Sci (Lond). 2014;126:507–516. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen KB, Sriramula S, Chhabra KH, Xia H, Lazartigues E. Species-specific inhibitor sensitivity of angiotensin-converting enzyme 2 (ACE2) and its implication for ACE2 activity assays. Am J Physiol Regul Integr Comp Physiol. 2011;301:R1293–R1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol Endocrinol. 2000;14:518–534. [DOI] [PubMed] [Google Scholar]
- 20.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130:167–178. [DOI] [PubMed] [Google Scholar]
- 21.Tiano JP, Delghingaro-Augusto V, Le May C, Liu S, Kaw MK, Khuder SS, Latour MG, Bhatt SA, Korach KS, Najjar SM, Prentki M, Mauvais-Jarvis F. Estrogen receptor activation reduces lipid synthesis in pancreatic islets and prevents β cell failure in rodent models of type 2 diabetes. J Clin Invest. 2011;121:3331–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiano JP, Mauvais-Jarvis F. Molecular mechanisms of estrogen receptors’ suppression of lipogenesis in pancreatic β-cells. Endocrinology. 2012;153(7):2997–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott REM, Wu-Peng XS, Yen PM, Chin WW, Pfaff DW. Interactions of estrogen- and thyroid hormone receptors on a progesterone receptor estrogen response element (ERE) sequence: a comparison with the vitellogenin A2 consensus ERE. Mol Endocrinol. 1997;11:1581–1592. [DOI] [PubMed] [Google Scholar]
- 24.van Gent R, Di Sanza C, van den Broek NJF, Fleskens V, Veenstra A, Stout GJ, Brenkman AB. SIRT1 mediates FOXA2 breakdown by deacetylation in a nutrient-dependent manner. PLoS One. 2014;9:e98438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R-H, Xu X, Kim H-S, Xiao Z, Deng C-X. SIRT1 deacetylates FOXA2 and is critical for Pdx1 transcription and β-cell formation. Int J Biol Sci. 2013;9:934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chodavarapu H, Chhabra KH, Xia H, Shenoy V, Yue X, Lazartigues E. High-fat diet-induced glucose dysregulation is independent of changes in Islet ACE2 in mice. Am J Physiol Regul Integr Comp Physiol. 2016;311:R1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia H, Sriramula S, Chhabra KH, Lazartigues E. Brain angiotensin-converting enzyme type 2 shedding contributes to the development of neurogenic hypertension. Circ Res. 2013;113:1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Xia H, Cai Y, Halabi CM, Becker LK, Santos RAS, Speth RC, Sigmund CD, Lazartigues E. Brain-selective overexpression of human angiotensin-converting enzyme type 2 attenuates neurogenic hypertension. Circ Res. 2009;106:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, Godbout K, Parsons T, Baronas E, Hsieh F, Acton S, Patane M, Nichols A, Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J Biol Chem. 2002;277(17):14838–14843. [DOI] [PubMed] [Google Scholar]
- 31.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2012;41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Feng X, Zhou Q, Cheng W, Shang C, Han P, Lin C-H, Chen H-SV, Quertermous T, Chang C-P. Pathological Ace2-to-Ace enzyme switch in the stressed heart is transcriptionally controlled by the endothelial Brg1-FoxM1 complex. Proc Natl Acad Sci USA 2016;113:E5628–E5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, Zhu M, Wu J, Shi X, Taing L, Liu T, Brown M, Meyer CA, Liu XS. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2016;45(D1):D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtsubo K, Chen MZ, Olefsky JM, Marth JD. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat Med. 2011;17(9):1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]