Preface
Genome-scale bisulfite sequencing approaches have opened the door to ecological and evolutionary studies of DNA methylation in many organisms. These approaches can be powerful. However, they introduce new methodological and statistical considerations, some of which are particularly relevant to non-model systems. Here, we highlight how these considerations influence a study’s power to link methylation variation with a predictor variable of interest. Relative to current practice, we argue that sample sizes will need to increase to provide robust insights. We also provide recommendations for overcoming common challenges and an R Shiny app to aid in study design.
Keywords: DNA methylation, bisulfite sequencing, cell type heterogeneity, population structure, mixed effects models, ecological epigenetics
Introduction
DNA methylation – the covalent addition of methyl groups to cytosine bases – is a gene regulatory mechanism of well-established importance in development, disease, and the response to environmental conditions1–5. In addition, shifts in DNA methylation are thought to contribute to the speciation process and the evolution of trait differences between taxa6–8, in support of the idea that gene regulation plays a key role in evolutionary change. Because of its contribution to phenotypic diversity, interest in DNA methylation from the ecology and evolutionary biology communities is high4,5,9–16. This interest has been further encouraged by the development of sodium bisulfite sequencing, a cost-effective approach that allows researchers to measure genome-wide DNA methylation levels at base-pair resolution in essentially any organism17–19.
Approaches that rely on sodium bisulfite treatment of DNA followed by high-throughput sequencing produce what are collectively called “bisulfite sequencing (BS) data sets.” These data sets have properties (discussed in the following section) that differ in key ways from other common types of sequencing-based functional genomic data, such as RNA-seq data. Consequently, several statistical approaches have been developed that are specifically tailored to BS data sets20–23 (Box 1). However, the development, application, and evaluation of these tools has primarily focused on biomedical questions or model systems, with an emphasis on case-control studies and experimental manipulations in a restricted set of species24–27. In contrast, ecologists and evolutionary biologists often study non-model organisms, environmental gradients that do not follow a case-control design, and natural populations characterized by complex kin or population structure. They are also typically more limited in their ability to sample pure cell types, and may be interested in effects that are smaller than those reported in the context of major perturbations like cancer or pathogen infection28,29. Notably, all of these properties can affect statistical power for differential methylation analysis (the identification of site or region-specific associations between DNA methylation levels and a predictor variable of interest), one of the most common uses of BS data.
Box 1. Modeling approaches for bisulfite sequencing data.
Binomial regression
A binomial distribution intuitively describes bisulfite sequencing data generated for a given sample, i, at a given site: the number of methylated counts (m) represents the number of ‘successes’ in an experiment with t trials and p probability of success. Here, t translates to the total read depth and p to the (unobserved) true methylation level.
| (1) |
However, bisulfite sequencing data are overdispersed (i.e., show greater variance than expected) relative to binomial expectations. Thus, using a binomial regression to model bisulfite sequence data can result in an extremely high rate of false positives and is not recommended20,36.
Beta binomial regression
To account for overdispersion, beta binomial regressions have been proposed for bisulfite sequencing data20–22. Here, the parameter pi from the binomial setting (equation 1) is itself treated as a random variable that follows a two-parameter beta distribution.
| (2) |
The beta distribution is then re-parameterized as a beta binomial with parameters ti, πi (equal to αi/(αi, + βi)), and γ to capture overdispersion.
| (3) |
Here, πi is the analog of the binomial probability of success (pi) and can be interpreted as the underlying true methylation level (note that the binomial distribution is a special case of the beta binomial distribution when γ=0). πi is passed through a logit link function in order to transform probability values (which are bounded between 0 and 1) to a continuous space for linear modeling. Transformed values are modeled as a function of an intercept (β0), the predictor variable of interest (xi), and its effect size (βx).
Linear mixed effects models
While beta-binomial regressions have become a popular tool for modeling bisulfite sequencing data, these models are not appropriate for data sets that contain related individuals or population structure. Such data sets require approaches that can account for genetic covariance (i.e., nonindependence) among samples, such as linear mixed effects models.
| (4) |
Here, y is a vector of continuously distributed methylation levels (obtained by normalizing m/t) and g is a vector of random effects with a covariance structure determined by the genetic relatedness among individuals in the sample (described by K, a user-defined n × n pairwise relatedness matrix) and the heritability of the DNA methylation trait (h2, which can be decomposed into its genetic and environmental components).I is an n × n identity matrix.
Binomial mixed effects models
Linear mixed models are flexible and fast, but discard information about total read depth when counts are normalized. Binomial mixed effects models overcome this constraint by controlling for genetic covariance while modeling raw counts.
| (5) |
Where ε, g, σe2, and σg2 are described as in eq. (4). This model essentially combines the linear mixed model with the beta binomial regression. The variable p now reflects the vector of true methylation levels for all samples and is passed through a logit link function for linear modeling. The genetic covariance, as well as the overdispersion, is captured by the random effects component.
Summary of model properties
| Method | Models the count- based nature of the data |
Models genetic covariance |
Programs that implement the method |
|---|---|---|---|
| Binomial regression | Yes* | No | R and many others |
| Beta-binomial regression | Yes | No | DSS22, MOABS21, RadMeth20 |
| Linear mixed effects model | No | Yes | GEMMA52, EMMA53, EMMAX99, FaST-LMM55 |
| Binomial mixed effects model | Yes | Yes | MACAU36 |
Binomial regression is never recommended. Because bisulfite sequencing data are overdispersed relative to the assumptions of this model, binomial regression analyses tend to generate many false positives.
Our goal in this review is to outline methodological considerations for differential methylation analysis of BS data sets. We tailor our discussion specifically to concerns that commonly arise in ecological and evolutionary studies and that, except where noted, are generalizable across taxa. We first consider how high-throughput BS data are generated, and how this process leads to several idiosyncrasies that must be taken into account during analysis. Next, we identify four properties common to ecological and evolutionary data sets that can influence power: moderate effect sizes, kinship/population structure, taxonomic differences in DNA methylation patterns, and cell type heterogeneity. We analyze both simulated and published empirical data sets to demonstrate how these four features can affect the power and biological interpretation of differential methylation analysis. We also discuss the advantages and disadvantages of conducting differential methylation analyses on individual CpG sites versus larger genomic intervals. Finally, we provide recommendations for handling each issue, with the aim of facilitating robust, well-powered studies of DNA methylation’s role in ecological and evolutionary processes.
Properties of bisulfite sequencing data sets
High-throughput BS protocols, such as whole genome bisulfite sequencing (WGBS19) or reduced representation bisulfite sequencing (RRBS17), rely on the differential sensitivity of methylated versus unmethylated cytosines to the chemical sodium bisulfite (Figure 1). Specifically, treatment of DNA with sodium bisulfite converts unmethylated cytosines to uracil (replicated as thymine after PCR) but leaves methylated cytosines unchanged (in vertebrates, most DNA methylation occurs at cytosines in CG motifs, while, in other taxa, cytosines in CHG and CHH are also commonly methylated13,30,31). DNA methylation level estimates at a given site can thus be obtained via high-throughput sequencing of bisulfite converted DNA, by comparing the relative count of reads that contain a cytosine (C), which reflect an originally methylated DNA base, to the count of reads that contain a thymine (T), which reflect an originally unmethylated version of the same base. Current BS protocols require low amounts of DNA, avoid the use of species-specific arrays, and can be applied to organisms without a reference genome32, making them an increasingly popular choice for ecologists and evolutionary biologists33.
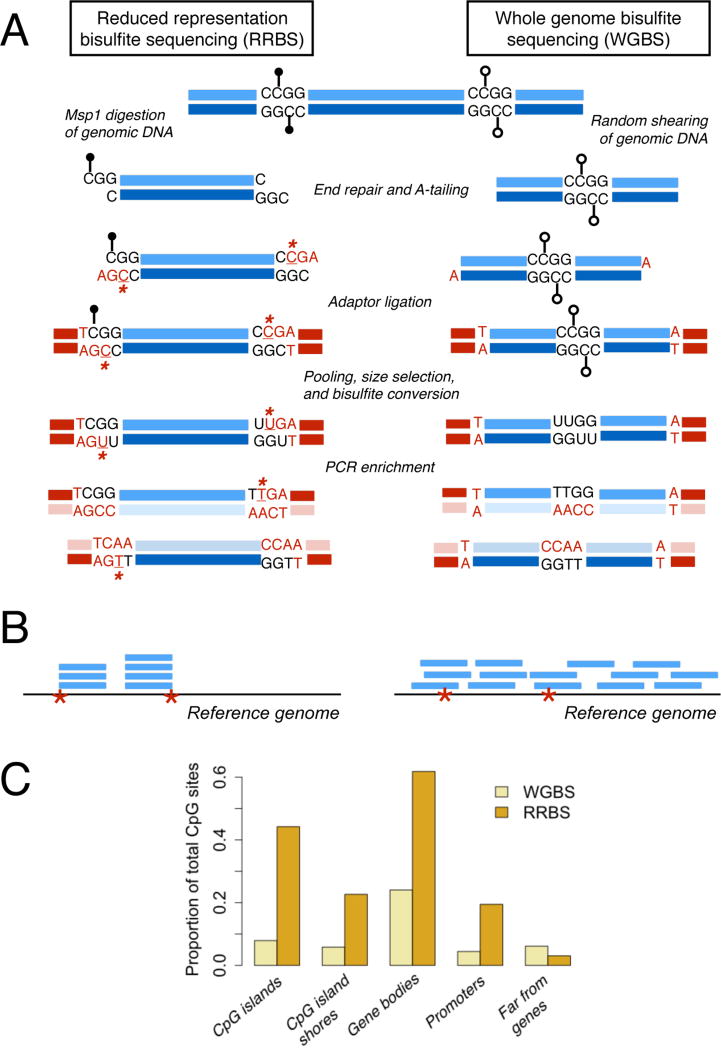
Figure 1. Overview of reduced representation bisulfite sequencing (RRBS; left) and whole genome bisulfite sequencing (WGBS; right).
(A) Steps to prepare an RRBS or WGBS library from genomic DNA. Black lollipops: methylated CpG sites; open lollipops: unmethylated CpG sites. Bases introduced during library preparation due to end repair or A-tailing are colored red; unmethylated cytosines that can be used to estimate conversion efficiency are underlined and marked with an asterisk. RRBS fragments start and end with the Msp1 digest sites (CCGG) flanking the initial piece of genomic DNA. (B) Read pileups after mapping RRBS and WGBS libraries to a reference (red asterisks=Msp1 digestion sites). Reads from RRBS libraries cover a small fraction of the genome. Further, because genomic DNA is fragmented with Msp1 and then size selected, all retained fragments should start and end with an Msp1 recognition site and be enriched for CpG sites. Sequencing reads that are shorter than the original fragment length will localize to the Msp1 recognition site associated with either the 5’ or 3’ end of the original fragment. (C) Bar charts compare the proportion of measured CpG sites that fall in gene bodies (between the TSS and the TES), promoters (2 kb upstream of the TSS), CpG islands, and regions far from genes (>100 kb from any annotated TSS or TES) in simulated RRBS and WGBS experiments given the same sequencing effort (20 million reads; read depths commonly used in WGBS studies typically exceed those of RRBS studies, however).
High-throughput BS data sets have a number of unique properties that influence both study design and data analysis. First, the raw data are binomially distributed count data, in which both the number of methylated reads (unconverted “C” bases) and the total read depth (number of methylated “C” bases plus unmethylated “T” bases) at each site contain useful information34,35 (note that in real data sets, these count data are usually over-dispersed due to biological variability20–22). For example, a site where 5 of 10 reads are methylated and a site where 50 of 100 reads are methylated both have estimated methylation levels of 50%. However, confidence in the methylation level estimate is higher for the second site, where total read depth is much greater. Information about relative confidence can be leveraged by modeling the raw count data rather than transforming counts to proportions or percentages, and several software packages now implement beta-binomial or binomial mixed effects models that do so20–22,36 (Box 1). These approaches provide a more powerful alternative to tests that assume continuously varying percentages or proportions (e.g., t-tests, Mann-Whitney U tests, linear models). They also control for count overdispersion, a known property of BS data that violates the assumptions of commonly used, but extremely false positive-prone20,36, binomial models.
Retaining read depth information during analysis relates to a second property of BS data: often, some samples have low read depth or missing data at a CpG site where other samples have high read depth (especially in RRBS data sets, where read coverage is affected by the sample-specific efficiency and specificity of the restriction enzyme digest: Figure 1, Supplementary Figure 1). Unlike RNA-seq data sets where read depth variation within a sample captures biological information (i.e., once normalized, lower read counts indicate lower expression levels), within-sample read depth variance in BS data sets is purely technical. Both read depth and effective sample size will thus vary across sites in the same data set, and will often do so systematically across different regions of the genome (particularly in RRBS data sets, due to variation in CpG density: Supplementary Figure 1).
Finally, the efficacy of the bisulfite conversion step can vary across samples or groups of samples prepared together, creating global batch effects. Though conversion efficiency is typically high (>98% of unmethylated cytosines converted to thymine37–39), small differences in conversion efficiency can have significant effects on genome-wide estimates of DNA methylation levels (see Fig S2 for an example from37). In particular, samples with low conversion efficiencies will tend to have upwardly biased estimates of DNA methylation levels relative to samples with higher conversion efficiencies, because fewer unmethylated Cs were converted to Ts. Thus, sample-specific bisulfite conversion rates should be directly estimated and taken into account (e.g., as a model covariate) in downstream analyses. However, we do not recommend estimating site-specific conversion rates, as these estimates are highly dependent on sequencing depth (because conversion occurs prior to sequencing, any observed relationship between sequencing depth and bisulfite conversion rate only reflects estimation error; Supplementary Figure 2). Estimates of sample-specific conversion rates can be obtained using CpG sites in the constitutively unmethylated chloroplast genome in plants13,19,40, an unmethylated DNA spike in (e.g., lambda phage DNA37–39), CHH and CHG sites (in species or cell types where CHH and CHG methylation is rare41,42), or the unmethylated cytosines added during RRBS library construction43 (Figure 1A). Empirical comparisons in a baboon RRBS data set36 suggests that spike-ins, CHH/CHG, and RRBS read end estimates roughly agree, but CHH/CHG estimates tend to be underestimated relative to the other methods and spike-ins seem to best capture a sample prep-related batch effect (Supplementary Figure 2).
Effect sizes in ecological and evolutionary studies
A primary determinant of power in differential methylation analysis is the distribution of true effect sizes. However, it is not obvious what the distributions of effect sizes for questions of ecological and evolutionary interest are likely to be. While effect size distributions and power analyses have been published for human disease case-control studies24–26, comparable information is not readily available for most other settings. Small or moderate epigenetic changes may still impact gene expression levels and consequently be of interest44,45; however, they will require larger sample sizes to detect.
To aid researchers in choosing appropriate sample sizes, we estimated effect sizes in BS data sets from plants, hymenopteran insects, and mammals that address a range of ecological and evolutionary questions, including: (i) developmental and demographic effects (eusocial insect caste differentiation41; age37); (ii) ecological effects (resource availability, including both large differences46 and more modest ones37); (iii) genetic effects (cis-acting methylation quantitative trait loci47); and (iv) species differences48,49 (Table 1). For comparison, we also include a data set contrasting cancer cells with normal tissue from the same donors28, which produces some of the largest effect sizes for differential methylation observed to date.
Table 1.
RRBS and WGBS data sets reanalyzed in this study
| Species | Predictor of interest |
Contrast | Method | Citation |
|---|---|---|---|---|
| Dog (Canis lupus familiaris) and wolf (Canis lupus) | Species differences | dog versus wolf | RRBS | 49 |
| Human (Homo sapiens) | Gestational famine during the Dutch Hunger Winter | famine-exposed versus same sex unexposed sibling | RRBS | 46 |
| Yellow baboon (Papio cynocephalus) | Age | continuous age values | RRBS | 37 |
| Yellow baboon | Resource base | wild-feeding versus human refuse-supplemented | RRBS | 37 |
| Clonal raider ant (Cerapachys biroi) | Caste | reproductive phase versus brood care phase | WGBS | 41 |
| Human | Cancer status | normal versus colorectal tumor samples (paired) | WGBS | 28 |
| Human, orangutan (Pongo abelii), gorilla (Gorilla gorilla), and chimpanzee (Pan troglodytes) | Species differences | human versus other great apes | WGBS | 48 |
| Mouseear cress (Arabidopsis thaliana) | Local genetic variation | nearby (putatively cis-acting) genotype | WGBS | 47 |
We first reanalyzed each data set using a uniform analysis pipeline (Supplementary Materials) and estimated two measures of effect size: (i) the mean difference in methylation levels between groups of samples, for binary comparisons (Figure 2A) and (ii) the proportion of variance explained by the variable of interest (Supplementary Figure 3). This analysis provides an empirical picture of how effect size distributions vary across differential methylation analyses. For example, local genetic variants tend to have large effects on DNA methylation levels, while environmental effects are consistently more modest (Figure 2A; Supplementary Figure 3). To understand how these differences impact power, we simulated BS data sets across a range of typical effect sizes and estimated the sample size required to identify differentially methylated sites in each case. All simulations presented in the main text assume that 10% of the sites in each dataset are true positives, but results from parallel analyses with varying proportions of true positives are shown in Supplementary Figure 4.
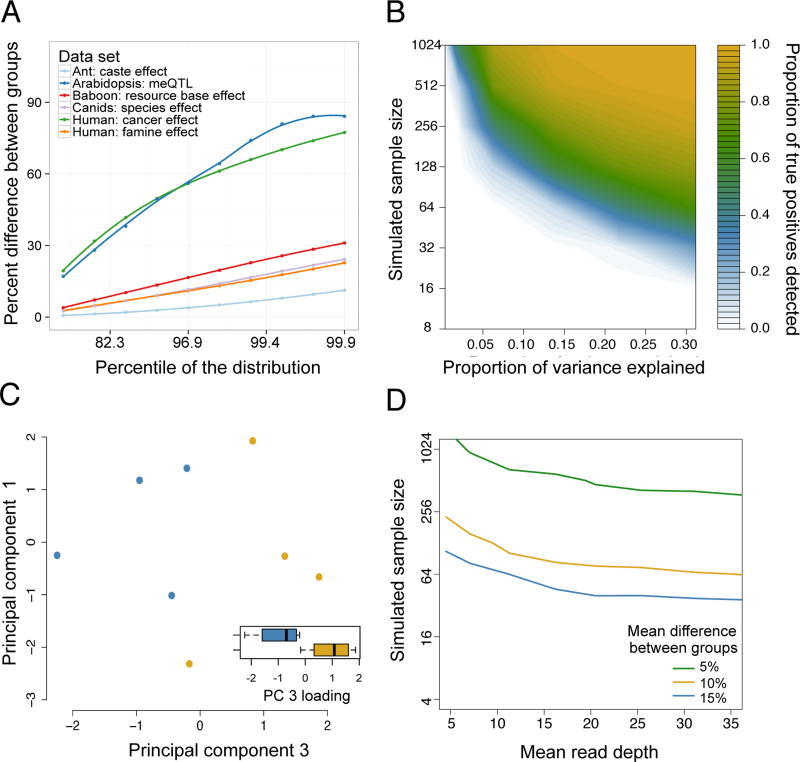
Figure 2. Estimates of effect sizes and their impact on the power of differential methylation analysis.
(A) The maximum percent difference in mean DNA methylation levels between two sample groups (y-axis), for selected percentiles of sites (x-axis, ranked from smallest to largest percent difference) in reanalyzed data sets (Table 1) with binary predictor variables. Mean differences are based on raw values, without correction for covariates. We show the largest percentiles here because those effects are most likely to be detected and of interest. (B) Power to detect differentially methylated sites at a 5% FDR in simulated RRBS datasets (sample size is plotted on a log scale). The magnitude of the effect of interest on DNA methylation levels (x-axis) is represented as the proportion of variance explained. (C) In a small ant data set (n=8), site-by-site analyses are underpowered to detect differential methylation between reproductive phase (blue dots) versus brood care phase (yellow dots) individuals, but PCA separates samples by caste (t-test for PC 3, which explains 21.7% of the overall variance: p = 0.022). In 1000 permutations, only 8.8% of permutations separate as cleanly on any of the first five PCs, suggesting that the original analysis was power-limited. Whiskers on boxplots represent the values for the third and first quartiles, plus or minus 1.5× the interquartile range, respectively. (D) The sample size and mean read depth combinations required to achieve 25% power (i.e., detect 25% true positives) in simulated RRBS datasets, for 3 different effect sizes. Increases in read depth do not affect power beyond ~20× coverage, and sample size or effect size increases always increase power more (Supplementary Figure 5).
Our simulations suggest that answering many ecological and evolutionary questions will require sample sizes that exceed those used in most current studies (Figure 2B; Supplementary Table 1). For example, to identify sites where the predictor variable explains 15% of the variance in DNA methylation levels (a mean difference between sample groups of 13–14% in our simulations) with 50% power requires an estimated 125 samples (250 samples for 80% power and 500 samples for 95% power). To accommodate the costs of larger sample sizes, we recommend choosing a reduced representation or capture-based approach rather than WGBS, and/or reducing per sample read depth. Indeed, consistent with results from a previous study25, we find almost no benefit to power after sequencing beyond a moderate read depth (~15–20x); in contrast, adding samples always increases power (Figure 2D; Supplementary Figure 5). In all cases, we strongly recommend against pooling DNA samples from multiple individuals into a single library, as this approach reduces power by collapsing the number of biological replicates.
Global analysis approaches that test for patterns in an entire data set, such as principal components analysis (PCA) or hierarchical clustering, may also be helpful in analyzing low powered data sets. These approaches are particularly useful when a predictor variable is associated with small changes in DNA methylation levels at any given locus, but such changes are common genome-wide. For example, in two published data sets (focused on the epigenetic effects of dominance rank in rhesus macaques and caste differences in clonal raider ants38,41), sample sizes were very small. The macaque study (n=3 high-ranking versus n=3 low-ranking animals) did not attempt site-by-site analysis, while the raider ant study (n=4 pools of reproductive phase ants versus n=4 pools of brood care phase ants) found no evidence for caste effects on DNA methylation using site-by-site paired t-tests. As shown in Figure 2B (see also Supplementary Figure 6), this result could have stemmed from low power. In support of this possibility, global analysis separates the sample groups of interest in both data sets. Specifically, the macaque study reported that hierarchical clustering distinguishes between high-ranking (n=3) and low-ranking (n=3) individuals, with increased separation when focusing on CpG sites near genes differentially expressed with rank38. Similarly, when we re-analyzed the clonal raider ant data set, we found that PCA separates reproductive and brood care individuals along principal component 3 (t-test for separation along PC 3: p=0.022; Figure 2C). Together, these results emphasize the potential utility of global analysis approaches in small studies.
Kinship and population structure
Ecological and evolutionary studies often focus on natural populations that contain related individuals or complex population structure. Accounting for these sources of variance is important because DNA methylation levels are often heritable50. In humans, where genetic effects on DNA methylation have been best studied, average estimated heritability levels are 18%-20% in whole blood50. As a result, more closely related individuals will tend to exhibit more similar DNA methylation patterns than unrelated individuals. Analyses that do not take genetic relationships into account can therefore produce spurious associations if the predictor of interest also covaries with kinship or ancestry. For example, samples are often collected along transects where climatic variables (e.g., temperature, altitude, rainfall) covary with genetic structure47,51. Genetic effects on DNA methylation could thus masquerade as climatic effects if genetic sources of variance are not also modeled.
Fortunately, this problem is structurally parallel to problems that have already been addressed in genotype-phenotype association studies, phylogenetic comparative analyses, and research on other functional genomic traits. The most straightforward solution is to use mixed effects models, which can incorporate a matrix of pairwise kinship or shared ancestry estimates to account for genetic similarity52–55 (Box 1). Specifically, this matrix is treated as the variance-covariance matrix for the heritable (genetic) component of a random effect variable (the environmental component is usually assumed to be independent across samples, so its variance-covariance is given by the identity matrix). The kinship matrix thus contributes to the predicted value of a heritable response variable, but does not affect the value of nonheritable response variables. Notably, while most approaches for controlling for relatedness implement linear mixed models that are only appropriate for continuous response variables52–54, recently developed binomial mixed models can be used to achieve the same task using count data36 (Box 1). These approaches avoid the need for transforming BS data from counts to proportions or ratios, thus preserving information about sequencing depth for each site-sample combination. Additionally, recent tools for calling SNP genotypes directly from BS reads (e.g., BisSNP56 and BS-SNPer57) can help with constructing kinship/relatedness matrices, although not without error (Box 2).
Box 2. Calling genotypes from bisulfite sequencing data.
Like other high-throughput sequencing assays100,101, bisulfite sequencing studies generate sequencing reads that contain information about genetic variation. Calling variants or genotypes from these data may be of interest for detecting genetic effects on DNA methylation levels (i.e., methylation quantitative trait loci, or meQTL), verifying sample identity, or controlling for genetic relatedness in downstream analyses. However, typical SNP-calling algorithms are not well suited to bisulfite sequencing data because the C to T conversion obscures true C/T polymorphisms. Several recently developed software packages attempt to overcome these challenges56,57. To assess the performance of one such program, BisSNP56, we analyzed a whole genome bisulfite sequencing data set for 29 Arabidopsis thaliana accessions47 where SNP calls were also available from whole genome sequencing through the 1001 Genomes Project and, for a subset of these individuals (n=25), genotype array data102.
Using BisSNP under default recommendations (Supplementary Materials), we identified 235,338 biallelic variable sites. This set was highly skewed to transitions (n=234,512 transitions, 99.65% of all called sites). Only 45% (n=106,925) of variants called using BisSNP represent putatively ‘true’ variants that were also identified in the 1001 Genomes resequencing data, but transversions were much more likely to be ‘true’ variants than transitions (90.3% compared to 45.3%). More stringent variant call filtering (variant quality ≥50 rather than ≥30) increased the proportion of likely true variants to 50.3%, but at the cost of retaining only 4.7% of the original sites. However, for previously identified variants in the BisSNP call set, BisSNP genotype calls and genotype array data agreed 87.5% of the time, with transversions agreeing more often than transitions (93.1% compared to 87.4%). Thus, BisSNP appears to provide relatively high-quality genotyping information for known variants.
However, our analyses do suggest that BisSNP genotypes provide a reliable way to verify sample identity and capture population structure. Using the set of biallelic SNPs that were identified by BisSNP, the 1001 Genomes Project, and the array data (n=3,553 SNPs overlapped between all 3 methods for n=25 accessions), a neighbor joining tree103 clearly clusters samples by accession. The single exception was a WGBS sample that may be mislabeled, as the BisSNP calls clustered separately from the resequencing and array genotype calls for this accession. Further, the pairwise genetic covariance matrix generated from BisSNP calls was highly consistent with the genetic covariance matrix generated from whole genome resequencing data (Mantel test r = 0.873, p < 10−6). Perhaps more importantly, the differences we did detect had marginal effects on differential DNA methylation analysis. Specifically, when we analyzed possible methylation quantitative trait loci (meQTL) in the Arabidopsis data set (Supplementary Materials), meQTL effect sizes were highly consistent between analyses using BisSNP calls to estimate population structure and analyses using whole genome resequencing data (Spearman’s rho=0.925, p<10−15).
Taxonomic differences in DNA methylation
Most research on DNA methylation to date has focused on humans and a handful of model systems. However, ecologists and evolutionary biologists study a wider range of species, and patterns of DNA methylation can vary dramatically among taxa13,30. Striking examples include the broad use of non-CpG (CHH and CHG) DNA methylation in plants relative to animals4,13,30, increased capacity for transgenerational epigenetic inheritance in plants15,58,59, and increased use of DNA methylation as a transposable element silencing mechanism in large eukaryotic genomes13,30. This variation means that patterns typical of one taxonomic group cannot necessarily be extrapolated to others (see3,4,12,13,30,31,60 for recent comparative studies and reviews of taxon-specific patterns). Here, we focus on how differences in the distribution of CpG DNA methylation levels across the genome can impact power and analysis strategies.
To provide some intuition about these differences, we synthesized data from published studies of flowering plants, hymenopteran insects, canids, humans, and non-human primates (Table 1). We estimated the mean and variance of methylation levels at each CpG site in each data set (Figure 3A–B; Supplementary Figure 7). Consistent with expectations, vertebrates show largely hypermethylated genomes, except in tumor samples where normal patterns are extensively perturbed28. In contrast, Arabidopsis genomes include many more hypomethylated sites, and the ant genome—typical of hymenopteran insects12,60—is almost completely unmethylated (Figure 3). Based on these observed values, we performed additional simulations (Supplementary Materials), with a particular focus on understanding how variance impacts power (because it is unlikely that a predictor variable of interest will significantly explain variation in DNA methylation levels at a locus where there is little variation to begin with). Importantly, the degree to which genomes are composed of relatively monomorphic (low variance) versus high variance sites systematically varies due to both taxon and sequencing strategy (Figure 3A–B, Supplementary Figure 7).
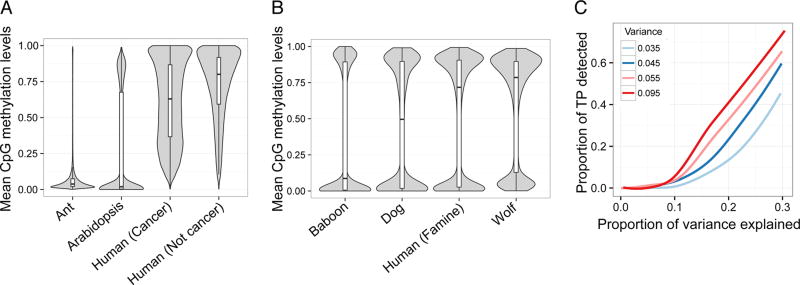
Figure 3. Properties of CpG methylation levels vary across data sets and influence power.
For each (A) WGBS and (B) RRBS data set, we plotted the distribution of mean DNA methylation levels at each CpG site with a median coverage >10× across all samples in the study. Whiskers on boxplots represent the values for the third and first quartiles, plus or minus 1.5× the interquartile range, respectively. (C) Power to detect differentially methylated sites (at a 5% FDR) in simulated RRBS datasets. The proportion of simulated true positives (TP) detected is plotted on the y-axis. Power increases as a function of the simulated effect size (represented as the proportion of variance explained; x-axis) and the variance in DNA methylation levels (colors). For all simulations, mean DNA methylation levels were held constant. The levels of variance in DNA methylation levels explored here (0.035, 0.045, 0.055, and 0.095) represent common values observed in real bisulfite sequencing data sets (Supplementary Figure 7).
Our simulations suggest that, all else being equal, power to detect differential methylation in BS data is limited by variance. Specifically, for any given sample size with a fixed mean DNA methylation level, power increases as a function of the underlying variance in DNA methylation levels (Figure 3C). These results suggest that analyses of low variance genomes (e.g., those of hymenopteran insects) may require larger sample sizes to detect a given effect than analyses of more variable systems, such as plants or mammals. An alternative approach is to filter out low variance sites prior to data analysis, which reduces the multiple testing burden. Notably, such filtering will also affect the relative representation of sites in genes, promoters, CpG islands, and other functional compartments of the genome because some of these compartments are consistently more variable than others (Supplementary Figure 7).
In the current literature, differences in the genome-wide distribution of DNA methylation levels across taxa have led to taxon-biased analysis approaches. For example, in hypomethylated insect genomes, several studies41,61,62 have used a binomial test to classify sites into ‘unmethylated’ or ‘methylated’ categories (i.e., all sites that do not pass a given significance threshold are considered ‘unmethylated’). Our simulations (Supplementary Materials) suggest that this approach not only loses information about quantitative variation, but is also sensitive to technical aspects of the data, such as sequencing depth. For example, using a binomial test approach, a site with an observed methylation level of 15% would be considered ‘unmethylated’ at a read depth of 20x, but ‘methylated’ at a read depth of 26× (Supplementary Figure 8). This problem likely accounts for the report of high rates of ‘sample-specific DNA methylation’ (where a site is methylated in one sample, but unmethylated in all other samples) in one recent study41. Indeed, our re-analysis of the same data shows that 77% of putative sample-specific sites can be more parsimoniously explained by greater read depth in the “outlier” sample (Supplementary Figure 8). Such problems can be readily avoided by not binarizing DNA methylation levels, which are intrinsically continuous traits, and by using count-based models that account for variation in sequencing depth20–22,36.
Discretizing DNA methylation levels into “genotype”-like data for population analyses, which has also been proposed49,63,64, can suffer from the same technical biases. Fortunately, most analyses that can be applied to discretized data can also be run on observed DNA methylation levels without artificial categorization, including differential methylation analysis (Box 1) and variance partitioning within and between populations65,66. Researchers interested in epigenetic inheritance67,68 have also analyzed DNA methylation levels as discrete states (e.g., ‘epialleles’35,64,69) in considering the evolutionary dynamics of epigenetic inheritance. However, because epialleles are thought to ‘mutate’ at a much faster rate than sequence variants and are not limited to discretized states, they are unlikely to behave like biallelic variants in classical population genetics70. There are ongoing efforts to modify classical population genetic models to take this hypervariability into account63,70, and we believe that the development of approaches that directly model the continuous nature of DNA methylation data would be particularly valuable in this regard.
Cell type heterogeneity
Epigenetic patterns vary substantially across cell types, contributing to differences in gene expression and biological function among different tissues. Because most sampled tissues also contain multiple cell types, putatively differentially methylated sites could, in some cases, be more parsimoniously explained by variation in cell type proportions rather than a direct effect of the variable of interest on DNA methylation71. Controlling for cell type heterogeneity is therefore a major concern in differential methylation analysis71, and a particular challenge for biologists working under field conditions or with non-model organisms where isolating purified cell types is not an option.
Three broad approaches can be used to confront this challenge. First, cellular composition can be phenotyped for use as a downstream statistical control using microscopy, flow cytometry, or, for animal blood samples, Giemsa or Wright-Giemsa stained blood smears. The ability to leverage these strategies will vary across species and collection conditions. However, some are already commonly applied in field studies (especially blood smears72–75), suggesting these approaches are feasible in at least some cases. The resulting estimates, or a composite measure (e.g., the first several principal components of variation in cell type proportions) can be incorporated as covariates in downstream analysis.
Second, if no measures of cell type heterogeneity are available for the samples of interest, another option is to use epigenomic profiles from sorted cells76,77 (‘reference epigenomes’) to predict the composition of mixed samples (a process known as ‘deconvolution’71,78). Even if obtained from different individuals or populations (and likely even if obtained from a closely related species), this approach can provide reasonable control for cell type heterogeneity79. Reference epigenome-based deconvolution is an active area of research, and several software packages exist to execute it79,80. Data from reference epigenomes can also be used to test if sites that are differentially methylated with respect to the predictor of interest are also differentially methylated by cell type, which would suggest the two are confounded8,37. However, if the between-sample compositional differences that would be required to produce the observed levels of differential methylation are not biologically plausible, tissue heterogeneity is unlikely to completely explain observed differentially methylated sites8.
Finally, if data from sorted cell populations are unavailable, researchers can apply methods that attempt to account for cell type heterogeneity without the need for reference information81–83. Both previously developed approaches, such as surrogate variable analysis (SVA: originally designed for differential gene expression analysis82), and approaches specifically developed with DNA methylation data in mind (e.g., FaST-LMM-EWASher81 and RefFreeEWAS83), have been suggested for this purpose. In applying them, researchers will need to evaluate whether the assumptions of these methods are met in their data. Because they were designed for batch correction, these approaches tend to assume that the largest sources of variance in DNA methylation levels (e.g., the top PCs) are due to cell type heterogeneity rather than differential methylation associated with the predictor of interest. Under this assumption, the only true positive associations that are detectable will tend to be both rare and of large effect. However, some predictors (e.g., environmental or disease perturbations) may truly have widespread, but modestly sized effects. For example, an analysis of resource base effects in baboon whole blood identified an association with DNA methylation levels at 1014 sites, after ruling out tissue heterogeneity confounds based on blood smear counts and comparisons against purified cell populations37. In comparison, FaST-LMM-EWASher detected a single differentially methylated site in the same data set. Recent comparisons of reference-based and reference-free methods suggest that reference-based approaches are consistently better powered79,80, and reference-free methods should only be used when sorted cell profiles are not available (SVA and RefFreeEWAS are recommended in these cases79,80). Such results suggest that researchers should consider investing in the generation of a small set of reference epigenomes, if possible for their system.
Site versus region-based analyses
DNA methylation levels are spatially correlated, such that CpG sites that are near each other (within a few hundred base pairs84,85) will tend to have more similar DNA methylation levels than those that are farther apart. In addition, regulatory regions such as promoters and CpG islands are characterized by a high density of CpG sites. Hence, spatially contiguous differentially methylated regions (DMRs) will generally be of greater functional interest than individual CpG sites. They also provide some reassurance that a signal of differential methylation is not a statistical or technical artifact.
Many strategies for DMR identification have been reported in the literature. Although they differ in modeling approach (Supplementary Table 2), all focus on identifying consecutive differentially methylated sites or regions with a specific number or density of differentially methylated sites. Such approaches have several advantages25,27. In addition to their potential functional relevance, region-based analyses can borrow strength across spatially contiguous sites, and some specifically incorporate coverage information to place higher weight on deeply sequenced sites21,23,86–88. In principle, taking a region-based approach could also reduce the multiple hypothesis testing burden (there should be fewer regions in a genome than individual sites). However, we note that in a false discovery rate framework89,90, DMR analyses will only be more powerful if they proportionally increase the number of true positives relative to null expectations.
Current region-based methods also have some limitations. First, they are less flexible than site-specific analyses, and generally do not enable users to control for other covariates or test the effects of continuous predictor variables (Supplementary Table 2). In addition, we are not aware of any region-based approach that controls for population structure, a known source of confounding. Second, some of the most commonly used DMR methods have been developed with long stretches of contiguously measured CpG sites in mind (e.g., WGBS data23,88,91,92). For RRBS or capture data, where stretches of interrogated sites are patchier, adjusting default parameters for window size or number of CpG sites in a putative DMR will therefore be necessary. For example, the default settings in BSmooth, one of the most popular DMR finding algorithms, perform DMR identification over windows that contain at least 70 CpG sites. In human WGBS data, 70 CpG sites can be captured in a window of 2.94 kb, on average. However, the patchiness of typical RRBS data sets means that a mean region size of 34.5 kb is necessary to capture stretches of 70 CpG sites, well beyond the range expected for spatial autocorrelation of DNA methylation levels. Finally, strategies for DMR identification have focused most immediately on CpG DMRs. Identifying CHG or CHH DMRs is indeed possible20,93–95, however, because the distribution, density, and variance of CHG and CHH sites differ from CpG sites, identifying non-CpG DMRs may also require careful adjustments to “off-the-shelf” settings.
One possible strategy to overcome the relative sparsity of RRBS data compared with WGBS data is to first identify differentially methylated sites and then aggregate them into DMRs (as in 18,37). A direct comparison between this approach and a “DMR-first” approach in simulated data indicates that they identify generally overlapping sets of DMRs, especially for longer stretches of sites (Supplementary Materials; Supplementary Figure 9). These results suggest a possible compromise between the modeling flexibility afforded by site-by-site analysis and prioritization of the most interesting candidate regions via DMR identification.
Conclusions and tools
Like most other genomic technologies, high-throughput BS approaches were first optimized in research contexts that afford a high degree of control (e.g., experimental case-control studies in model systems) and in systems that boast extensive genomic resources (e.g., humans). However, for ecologists and evolutionary biologists, these approaches often become most exciting when they can be extended to a much more diverse set of species and populations—even if these extensions come with complications. We believe that the biological insights to be gained from studies of DNA methylation in diverse taxa have substantial potential.
However, maximizing the yield from these studies will require careful consideration of taxon-specific characteristics, the use of analysis methods appropriate to a data set’s structure, and realistic assessments of power. In particular, our results reveal that, with sample sizes that are currently applied by many ecologists and evolutionary biologists, differential methylation analyses will tend to be moderately or lowly powered. Such studies may still have the potential to reveal interesting and important biology, but researchers should be aware that they are likely to detect only the largest effect sizes (as is also true for other types of genomic analysis96). Notably, how tightly small effects on differential methylation are linked to differences in downstream phenotypes, such as gene expression, remains somewhat unclear. While this relationship can be investigated for a few loci using experimental manipulation of DNA methylation levels in reporter assays97 or, more recently, CRISPR-dCas9 manipulation98, genome-wide tests are still missing from the literature.
Finally, to help quantify how sample size, effect size, population structure, and modeling approach affect BS data analysis, we have developed an R Shiny application to perform power analyses like those presented here. This app allows BS data to be simulated with user-specified properties, is coupled with a set of statistical analysis options to evaluate study power, and outputs the simulated count data for maximal flexibility. The app is freely available at www.tung-lab.org/protocols-and-software.html.
Supplementary Material
Acknowledgments
We thank Kasper Hansen and Irene Hernando-Herraez for providing processed file formats from their previously published work. We also thank Noah Snyder-Mackler, Luis Barreiro, and Xiang Zhou for helpful comments and suggestions, Mine Cetinkaya-Rundel for coding suggestions on the R Shiny app, Mackenzie Gavery for beta-testing it, the Baylor College of Medicine Human Genome Sequencing Center for access to the current version of the baboon genome assembly (Panu 2.0), and three anonymous reviewers for helpful comments on a previous version of this work. This work was supported by NIH R21-AG049936 and 1R01GM102562 to JT, NSF BCS-1455808 to JT and AJL. PAPD is supported by NIH K12GM000678 from the Training, Workforce Development & Diversity division of the National Institute of General Medical Sciences.
Footnotes
Data accessibility
Summaries of data availability and accession numbers for previously published data sets are provided in Supplementary Table 1. Our BisSNP analyses utilized publicly available SNP calls for Arabidopsis accessions downloaded from the 1001 Genomes Project (http://1001genomes.org/data/GMI-MPI/releases/v3.1/). An R Shiny app for simulating bisulfite sequencing data and performing power analysis is available at www.tung-lab.org/protocols-and-software.html.
Author contributions
AJL and JT conceived the study; AJL, TPV, and PAPD analyzed previously published and simulated data; TPV wrote the R Shiny app; and AJL and JT wrote the manuscript, with input from all co-authors. All authors gave final approval for publication.
Conflict of interest
We declare no conflict of interest.
References
- 1.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 2.Jones P. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 3.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 4.Seymour DK, Becker C. The causes and consequences of DNA methylome variation in plants. Curr. Opin. Plant Biol. 2017;36:56–63. doi: 10.1016/j.pbi.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010;185:1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Adaptive methylation regulation of p53 pathway in sympatric speciation of blind mole rats, Spalax. Proc. Natl. Acad. Sci. 2016;113:2146–2151. doi: 10.1073/pnas.1522658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C. Rapid establishment of genetic incompatibility through natural epigenetic variation. Current Biology. 2012;22:326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 8.Hernando-Herraez I, et al. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9:e1003763. doi: 10.1371/journal.pgen.1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernando-Herraez I, Garcia-Perez R, Sharp AJ, Marques-Bonet T. DNA Methylation: Insights into Human Evolution. PLoS Genet. 2015;11:1–12. doi: 10.1371/journal.pgen.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snell-Rood E. The importance of epigenetics for behavioral ecologists (and vice versa) Behav. Ecol. 2012;19:2012. [Google Scholar]
- 11.Ledon-Rettig CC, Richards CL, Martin LB. Epigenetics for behavioral ecologists. Behav. Ecol. 2012:1–14. doi: 10.1093/beheco/ars145. [DOI] [Google Scholar]
- 12.Glastad KM, Hunt BG, Goodisman MA. Evolutionary insights into DNA methylation in insects. Curr. Opin. Insect Sci. 2014;1:25–30. doi: 10.1016/j.cois.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013 doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz RJ, et al. Transgenerational Epigenetic Instability Is a Source of Novel Methylation Variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortijo S, et al. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–8. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 17.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 2011;6:468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 18.Lister R, Pelizzola M, Dowen R, Hawkins R. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462 doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolzhenko E, Smith AD. Using beta-binomial regression for high-precision differential methylation analysis in multifactor whole-genome bisulfite sequencing experiments. BMC Bioinformatics. 2014;15:215. doi: 10.1186/1471-2105-15-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun D, et al. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 2014;15:R38. doi: 10.1186/gb-2014-15-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng H, Conneely KN, Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42:1–11. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen K, Langmead B, Irizarry R. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai PC, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol. 2015;44:1429–1441. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods. 2014;12:2–5. doi: 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakyan VK, Down Ta, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacis A, et al. Bacterial Infection Remodels the DNA Methylation Landscape of Human Dendritic Cells. Genome Res. 2015 doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 31.Takuno S, Ran J-H, Gaut BS. Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants. 2016;2:15222. doi: 10.1038/nplants.2015.222. [DOI] [PubMed] [Google Scholar]
- 32.Klughammer J, et al. Differential DNA Methylation Analysis without a Reference Genome. Cell Rep. 2015;13:2621–2633. doi: 10.1016/j.celrep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhoeven KJF, VonHoldt BM, Sork VL. Epigenetics in ecology and evolution: what we know and what we need to know. Mol. Ecol. 2016;25:1631–1638. doi: 10.1111/mec.13617. [DOI] [PubMed] [Google Scholar]
- 34.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–9. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–73. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lea A, Tung J, Zhou X. A flexible, efficient binomial mixed model for identifying differential DNA methylation in bisulfite sequencing data. PLoS Genet. 2015;11:e1005650. doi: 10.1371/journal.pgen.1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea AJ, Altmann J, Alberts SC, Tung J. Resource base influences genome-wide DNA methylation levels in wild baboons (Papio cynocephalus) Mol. Ecol. 2016;25:1681–1696. doi: 10.1111/mec.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung J, et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc. Natl. Acad. Sci. 2012;109:6490–5. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banovich NE, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:1–12. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, et al. Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Libbrecht R, Oxley PR, Keller L, Kronauer DJC. Robust DNA Methylation in the Clonal Raider Ant Brain. Curr. Biol. 2016;26:1–5. doi: 10.1016/j.cub.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle P, Clement K, Gu H, Smith Z. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krueger F. Trim Galore! 2015 [Google Scholar]
- 44.Murgatroyd C, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009;12:1559–66. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 45.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the CRF gene in adult mice. Nat. Neurosci. 2010;13:1351–3. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 46.Tobi EW, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubin MJ, et al. DNA methylation variation in Arabidopsis has a genetic basis and appears to be involved in local adaptation. eLife. 2015;4:e05255. doi: 10.7554/eLife.05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernando-Herraez I, et al. The interplay between DNA methylation and sequence divergence in recent human evolution. Nucleic Acids Res. 2015;43:8204–8214. doi: 10.1093/nar/gkv693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janowitz Koch I, et al. The concerted impact of domestication and transposon insertions on methylation patterns between dogs and grey wolves. Mol. Ecol. 2016;25:1838–1855. doi: 10.1111/mec.13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taudt A, Colomé-Tatché M, Johannes F. Genetic sources of population epigenomic variation. Nat. Rev. Genet. 2016;17:319–332. doi: 10.1038/nrg.2016.45. [DOI] [PubMed] [Google Scholar]
- 51.Gugger PF, Fitz-Gibbon S, Pellegrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and its association with climate gradients. Mol. Ecol. 2016;25:1665–1680. doi: 10.1111/mec.13563. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–23. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–8. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 55.Lippert C, et al. FaST linear mixed models for genome-wide association studies. Nat. Methods. 2011;8 doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13:R61. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao S, et al. BS-SNPer: SNP calling in bisulfite-seq data. Bioinformatics. 2015;31:4006–4008. doi: 10.1093/bioinformatics/btv507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 2009;84:131–76. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 59.Heard E, Martienssen Ra. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. Evolution of DNA methylation across insects. Mol. Biol. Evol. 2016;34 doi: 10.1093/molbev/msw264. msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants camponotus floridanus and harpegnathos saltator. Curr. Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Fan C. A neutrality test for detecting selection on DNA methylation using single methylation polymorphism frequency spectrum. Genome Biol. Evol. 2014;7:154–171. doi: 10.1093/gbe/evu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidalis A, et al. Methylome evolution in plants. Genome Biol. 2016;17:264. doi: 10.1186/s13059-016-1127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah S, et al. Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Res. 2014 doi: 10.1101/gr.176933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McRae AF, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15:R73. doi: 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weigel D, Colot V. Epialleles in plant evolution. Genome Biol. 2012;13:249. doi: 10.1186/gb-2012-13-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heard E, Martienssen RA. Transgenerational Epigenetic Inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen KD, et al. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13:R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charlesworth B, Jain K. Purifying selection, drift, and reversible mutation with arbitrarily high mutation rates. Genetics . 2014;198:1587–1602. doi: 10.1534/genetics.114.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaffe AE, Irizarry Ra. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beldomenico PM, et al. Poor condition and infection: a vicious circle in natural populations. Proc. R. Soc. B-Biological Sci. 2008;275:1753–9. doi: 10.1098/rspb.2008.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Charruau P, et al. Pervasive Effects of Aging on Gene Expression in Wild Wolves. Mol. Biol. Evol. 2016:1–29. doi: 10.1093/molbev/msw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merino S, Moreno J, Sanz JJ, Arriero E. Are avian blood parasites pathogenic in the wild? A medication experiment in blue tits (Parus caeruleus) Proc. R. Soc. B-Biological Sci. 2000;267:2507–2510. doi: 10.1098/rspb.2000.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ots I, Murumägi A, Hõrak P. Haematological health state indices of reproducing Great Tits: Methodology and sources of natural variation. Funct. Ecol. 1998;12:700–707. [Google Scholar]
- 76.Watkins Na, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants. 2016:16058. doi: 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hattab MW, et al. Correcting for cell-type effects in DNA methylation studies: reference-based method outperforms latent variable approaches in empirical studies. Genome Biol. 2017;18:24. doi: 10.1186/s13059-017-1148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng SC, et al. Correcting for cell-type heterogeneity in epigenome-wide association studies: revisiting previous analyses. Nat. Methods. 2017;14:216–217. doi: 10.1038/nmeth.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat. Methods. 2014;11:309–11. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
- 82.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2009;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bell JT, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klein HU, Hebestreit K. An evaluation of methods to test predefined genomic regions for differential methylation in bisulfite sequencing data. Brief. Bioinform. 2016;17:796–807. doi: 10.1093/bib/bbv095. [DOI] [PubMed] [Google Scholar]
- 87.Akalin A, Kormaksson M. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaffe AE, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 90.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jühling F, et al. Metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256–262. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li S, et al. An optimized algorithm for detecting and annotating regional differential methylation. BMC Bioinformatics. 2013;14(Suppl 5):S10. doi: 10.1186/1471-2105-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hebestreit K, Dugas M, Klein HU. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics. 2013;29:1647–1653. doi: 10.1093/bioinformatics/btt263. [DOI] [PubMed] [Google Scholar]
- 95.Virdi KS, et al. Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat. Commun. 2015;6:6386. doi: 10.1038/ncomms7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rockman MV. The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution. 2012;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klug M, Rehli M. Functional Analysis of Promoter CpG Methylation Using a CpG-Free Luciferase Reporter Vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 98.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016:1–14. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu C, DeWan A, Hoh J, Wang Z. A comparison of association methods correcting for population stratification in case-control studies. Ann. Hum. Genet. 2011;75:418–27. doi: 10.1111/j.1469-1809.2010.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perry G, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Piskol R, Ramaswami G, Li JB. Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet. 2013;93:641–651. doi: 10.1016/j.ajhg.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.