Abstract
Membrane proteins present a challenge for structural biology. In this article, we review some of the recent developments that advance the application of NMR to membrane proteins, with emphasis on structural studies in detergent-free, lipid bilayer samples that resemble the native environment. NMR spectroscopy is not only ideally suited for structure determination of membrane proteins in hydrated lipid bilayer membranes, but also highly complementary to the other principal techniques based on X-ray and electron diffraction. Recent advances in NMR instrumentation, spectroscopic methods, computational methods, and sample preparations are driving exciting new efforts in membrane protein structural biology.
Keywords: NMR, membrane protein, lipid, bilayer, solid-state NMR
Graphical Abstract

Introduction
Membrane proteins are encoded by 30–40% of all expressed genes, and are essential for both cellular life and human health. Due to their importance, membrane proteins are major targets of biomedical research and drug discovery efforts aimed at understanding their biological functions and harnessing their therapeutic potential. Molecular structure determination is essential for achieving both goals.
Structural biology of membrane proteins has advanced considerably in recent years. X-ray crystallography, electron microscopy (EM) and nuclear magnetic resonance (NMR) have all contributed important and complementary structural data [1–4], each with distinct advantages and unique challenges. X-ray crystallography has made important contributions to membrane protein structural biology, with notable recent successes in the area of G protein coupled receptors (GPCRs) [5–7]. EM has long been used to determine the structures of membrane proteins in proteolipid two-dimensional (2D) crystals [8], and the recent development of single-particle cryo-EM [9–11] is enabling higher resolution structures of membrane proteins to be obtained without the need to prepare large, well-ordered crystalline samples [2,12]. NMR has a long history as a key technology in advancing our understanding of the structural, chemical, and dynamic properties of lipid bilayer membranes.
Early NMR studies provided fundamental information about the structures and dynamics of phospholipid assemblies, and the effects of membrane proteins and various other membrane components on lipid bilayer membranes [13–16]. NMR, however, also plays a central role in membrane protein structural biology, providing information that is both unique and complementary to that derived from X-ray diffraction and EM. NMR methods are available for studying membrane proteins in a wide variety of samples, including soluble detergent micelles, detergent-free lipid bilayer membranes, and native cell envelope preparations [4,17–20]. The range of sample types reflects the versatility of NMR as a tool for characterizing the structures, dynamics, and functional interactions of biomolecules. NMR is also adept at characterizing intrinsically disordered regions of proteins [21]. Moreover, since NMR signals are highly sensitive to the local environment, they are extremely useful for characterizing even weak ligand binding through chemical shift changes, enabling structure activity correlations to be made for binding events or conformational changes [22].
The principal advantage of NMR as a technique for structural analysis is that is compatible with detergent-free membrane samples that are similar to the physiological protein environment [23]. This is in contrast to X-ray and single particle cryo-EM studies, which typically require non-lipid amphiphiles, such as detergents or amphiphilic polymers, for protein solubilization, and often involve extensive sample engineering, such as antibody stabilization and protein mutations, truncations, insertions and modifications [2,12,24–28]. These factors, combined with the cryogenic temperatures of the samples, all work to stabilize a single molecular conformation, thus dampening the structural plasticity that is often integral to biological function. Furthermore, in the case of EM, three-dimensional (3D) reconstruction remains challenging for proteins smaller than ∼100 kDa, and while signals can be enhanced by binding antibody fragments [29], shape asymmetry and relatively small size [30] pose impediments for high-resolution structure characterization.
Recent advances in NMR structural studies of membrane proteins reflect exciting developments in the areas of recombinant protein expression, sample preparation, pulse sequences for high-resolution spectroscopy, radio-frequency probes, high-field magnets and computational methods, which enable single atomic sites of membrane proteins to be probed with ever greater accuracy. Here we describe recent results with focus on studies in detergent-free lipid bilayers, where NMR is poised to make the most substantial contributions.
Solution NMR of membrane proteins in detergents
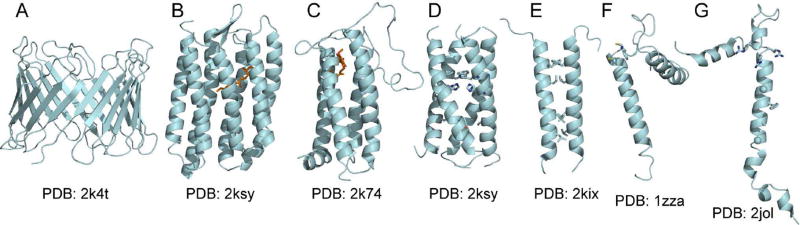
Solution NMR methods can be used for structure determination of membrane proteins in detergent micelles or detergent/lipid mixed micelles [17,18]. The protein data bank (PDB) structures of the outer mitochondrial voltage dependent anion channel VDAC-1 [31,32], the archaeal phototaxis receptor sensory rhodopsin II pSRII [33], and the bacterial inner membrane protein DsbB [34], illustrate the range of structural complexity that can be elucidated in atomic detail by solution NMR (Fig. 1). Human mitochondrial VDAC-1 forms a 19-stranded β-barrel held together by a network of hydrogen bonds (Fig. 1A). NMR chemical shift mapping experiments revealed several perturbations in the presence of protein and small molecule ligands, which were interpreted to reflect specific binding sites. Microbial pSRII, from Natronomonas pharaoni, forms a seven-transmembrane helix bundle, reminiscent of GPCRs (Fig. 1B). The structure, determined with excellent precision (0.48 Å backbone RMSD), represents the potential of solution NMR structure determination for other membrane proteins with this topology. Bacterial DsbB forms a four-helix bundle and a shorter fifth helix whose position suggests association with the membrane surface (Fig. 1C). The protein functions in disulfide bond formation and NMR analysis of its structure and dynamics helps explain how it mediates the flow of electrons together with its quinone cofactor. All three structures agree with their counterparts determined by x-ray crystallography, with no major conformational differences observed between crystalline and micelle samples.
Figure 1. Examples of membrane protein structures determined in detergent micelles by solution NMR.
The PDB file numbers are listed below each structure. (A) Mitochondrial voltage dependent anion channel VDAC [31]. (B) Archaeal photo sensory rhodopsin pSRII with bound retinal (orange) [33]. (C) Bacterial inner membrane protein DsbB with bound quinone (orange) [34]. (D) Channel domain of influenza BM2, showing His, Trp and Ser side chains (sticks) lining the pore [40]. (E) Transmembrane domain ζ-ζ dimer of the T cell receptor CD3, showing side chains (sticks) that mediate dimerization [37]. (F) Mitochondrial stannin showing the metal-binding Cys-Trp-Cys motif (sticks) at the membrane surface [36] (G) Human Na,K-ATPase regulator FXYD1 showing side chains (sticks) and Gly CA atoms (spheres) that mediate intra-membrane association with the Na,K-ATPase α subunit [38].
Solution NMR has also made significant contributions to structure determination of membrane proteins with a single transmembrane helix [35–42]. These single-pass membrane proteins have important functions in biology. They often rely on intra-membrane, helix-helix interactions for establishing their functionalities, and are often too small to form the crystallographic contacts necessary for analysis by x-ray diffraction. NMR spectroscopy, however, is not limited by this constraint and can yield precise structural information, provided that sample conditions can be established to maintain structural integrity, as illustrated by several structures determined in micelles with solution NMR (Fig. 1D–G).
Solution NMR studies of GPCRs in detergents have also provided insights about receptor-ligand binding [43–53]. Detergents, however, can perturb or disrupt receptor-ligand interactions [54,55] and influence conformational exchange between GPCR functional states [47]. Similar effects are also seen for other membrane proteins, where detergents can cause structural distortions, loss of native functionality, or gain of non-native activity due to the unmasking of adventitious ligand binding sites [23].
Amphiphilic polymers (amphipols) have been proposed as a membrane mimetic alternative to detergents [56]. Amphipol-solubilized membrane proteins can yield solution NMR spectra that appear suitable for structural studies [57]. In one notable case, amphipols have been shown to stabilize the trimeric, Chlamydia native major outer membrane protein, nMOMP, and enhance its protective ability as a vaccine [58].
Solution NMR of membrane proteins in lipid bilayer nanodiscs
The long correlation times of large protein-lipid assemblies pose a limitation on the types of membrane proteins that can be studied by solution NMR. Nevertheless, advances are being made in the development of experimental methods [59] and the preparation of specialized detergent-free lipid bilayer nanodiscs [60–62] suitable for solution NMR studies. Nanodisc samples have been used very effectively for structure/activity correlation studies of a variety of membrane proteins [60–66].
The combined use of small nanodiscs, extensive 2H-labeling of both protein and lipids, TROSY (transverse relaxation-optimized spectroscopy) sequences [67] at high magnetic fields, and non-uniform sampling schemes for signal acquisition [68–71], has enabled solution NMR structure determination of the bacterial outer membrane proteins OmpX, OmpA, and Ail [60,63,72]. These eight-stranded β-barrel proteins fall in a class that is highly amenable to NMR studies since their 3D fold can be established by measuring distance restraints across the extensive, inter-strand network of amide backbone hydrogen bonds. By contrast, the hydrogen bonds of helical proteins are intra-helical, and, while valuable, do not provide information about 3D fold. Thus, structure determination of helical proteins relies much more significantly on distance restraints measured between side chain atoms, a task that requires side chain resonance assignments.
In nanodiscs, the absence of detergent ensures that structural studies are conducted for functional protein states. Interestingly, even though a number of membrane proteins have been shown to adopt the same overall structure in lipid micelles, detergent-free nanodiscs, and 3D crystalline samples [42,60,62–64,73], the presence of detergent can have a dramatic effect on activity. For example, the solution NMR structure of the Yersinia pestis outer membrane protein Ail, has been determined in decyl-phosphocholine (DePC) detergent micelles [74] and agrees very well with its crystal structure [75]. Ail forms an eight-stranded β-barrel with four extracellular loops and three intracellular turns (Fig. 2A). The same structure is also observed in nanodiscs [72], and the NMR spectra of the protein in micelles and nanodiscs are very similar (Fig. 2B). Nevertheless, the ligand binding activity of the protein is abolished in detergent (Fig. 2C) [73], precluding structure-activity studies. Interestingly, NMR analysis indicates that the addition of DePC to Ail nanodiscs perturbs signals from the extracellular loops, suggesting that detergent molecules associating with these key sites affect activity.
Figure 2. Structure of Y. pestis outer membrane protein Ail and effect of detergent on its ligand binding activity.
(A) Solution NMR structure of Ail, determined in DePC micelles, showing Arg and Lys side chains (yellow) at the membrane surface [74]. (B) Solution NMR 1H/15N TROSY spectra of Ail in 170 mM DePC (black) or nanodiscs (red). (C) Fibronectin binding activity of Ail analyzed by enzyme linked immunosorbent assay, where increasing concentrations of Ail are added to fibronectin-coated plates. Adapted from [73].
Notably, while the solution NMR spectra of native GPCRs in detergents are typically dominated by broad, overlapped resonances, except for a few narrow signals from the disordered termini [76,77], selective truncations and mutagenesis based on directed evolution techniques [78,79] have been shown to generate GPCR sequences with conformational and dynamic properties amenable to solution NMR structural studies. For example, high resolution 1H/15N spectra were reported, recently [62], for a direct-evolved and truncated sequence of the neurotensin receptor NTR1, reconstituted in lipid nanodiscs prepared with a short and covalently circularized version of the membrane scaffold protein. These circularized nanodiscs have greater stability and size homogeneity than the non-circularized versions, and thus help enhance NMR spectral resolution.
Solid-state NMR of membrane proteins in lipids
Solid-state NMR methods are compatible with detergent-free lipid samples and are not limited by molecular size. In recent years, the structures of a number of membrane proteins have been determined in phospholipids using oriented sample (OS) and magic angle spinning (MAS) solid-state NMR approaches. Structural studies have relied primarily on experiments based on detection of dilute nuclei (15N, 13C), which can be readily introduced in a protein's amino acid sequence by growing the expression vector in isotopically defined minimal media. A wide range of experiments [4,80–83] can be performed to obtain site-specific resonance assignments and to measure restraints for structure determination. These are facilitated by the use of high magnetic fields and efficient radiofrequency probes that do not dissipate heat energy [84–86]. Isotropic chemical shifts and spin exchange signals, measured in MAS experiments, can be converted to torsion angles and inter-atomic distances, while dipolar couplings and anisotropic chemical shifts, measured using either MAS or static probes with uniaxially ordered samples, can be converted to bond orientation restraints or used to obtain information about protein dynamics [87,88].
In addition to structures, solid-state NMR studies are providing fundamental insights about the molecular mechanisms underpinning membrane protein functionalities, including: ion channel conduction [89–92], multidrug resistance [93]; antimicrobial peptide activity at membranes [94–96]; bioenergetics [97]; as well as conformation, dynamics and ligand binding of intact GPCRs [76,98–105]. The ability to examine structure, dynamics, and ligand binding in native-like samples similar to those used to assay biological functions is an important advantage of NMR spectroscopy. Solid-state NMR is making particularly exciting advances as a tool for examining the structural and dynamic properties of membrane proteins, peptides and cytoskeletal envelope components in native cell preparations [20,106–110].
Structures of membrane proteins determined by solid-state NMR (Fig. 3) include: sensory rhodopsin from Anabaena [111]; the human chemokine receptor, CXCR1 [112]; the M2 1H channel from influenza virus [113,114]; the bacterial inner membrane protein DsbB [115,116]; the Mycobacterium tuberculosis cell division protein, CrgA [117]; the membrane-inserted form of the fd bacteriophage major coat protein [118]; the bacterial mercury transporter, MerF [119]; the bacterial trimeric autotransporter YadA [120]; and the human PLB pentamer [121]. The structures of DsbB and PLB were determined with hybrid refinement approaches based on restraints from solid-state NMR and crystallography (DsbB), or on solid-state NMR and solution NMR (PLB), to enhance structural precision. Such hybrid approaches can be very valuable but should be used with caution as they may be complicated by sample-specific structural differences. Notably, the structures of influenza M2 and PLB illustrate the importance of the membrane environment. The solid-state NMR structures determined in detergent-free lipid samples are distinctly different from those determined using solution NMR or crystallography, where detergents caused distortions incompatible with membrane insertion and protein functionality [23].
Figure 3. Examples of membrane protein structures determined in detergent-free phospholipids by solid-state NMR.
The PDB file numbers are listed below each structure. (A) Anabaena sensory rhodopsin with bound retinal (yellow) [111]. (B) human chemokine receptor, CXCR1 [112]. (C) M2 1H channel from influenza virus showing His and Trp side chains (sticks) lining the channel pore [113]. (D) Bacterial inner membrane protein DsbB [115,116]. (E) Mycobacterium cell division protein, CrgA showing side chains (sticks) that mediate intra-membrane helix-helix association [117]. (F) Membrane-inserted form of the fd bacteriophage coat protein showing polar side chains (sticks) in the N-terminal helix exposed to bulk water [118].
OS Solid-state NMR
At present, the majority of solid-state NMR structures of membrane proteins available in the PDB are based on bond orientation restraints, measured in the NMR spectra of proteins in uniaxially aligned lipid bilayers. Orientation restraints are very valuable because they contain information about protein 3D structure, protein orientation in the membrane, and protein dynamics [122]. Moreover, they provide a powerful, independent method for validating structural accuracy [123]. Experimental approaches are available for obtaining resonance assignments in the NMR spectra of oriented samples, including, methods based on the direct relationship between the global membrane-integrated structure and the frequencies of the NMR signals [118,124–128], and spectroscopic methods based on spin diffusion between atomic sites [83,129–131].
Uniaxially aligned samples of membrane proteins in bilayers can yield high resolution NMR spectra that directly reflect the protein's conformational properties. For example, the 1H/15N PISEMA (polarization inversion with spin exchange and the magic angle) spectra of the human Na,K-ATPase regulatory protein FXYD2 in oriented bilayers [132] show signals from amide sites in a wheel-like pattern that reflects the ∼20° orientation of the transmembrane helix axis relative to the membrane normal (Fig. 4A, C). Spectra from bilayers aligned perpendicular to magnetic field have 15N frequencies in the range of 150–200 ppm (Fig. 4A, B), while the spectra from parallel bilayers have 15N frequencies in the range of 85–100 ppm (Fig. 4C, D). For both bilayer orientations, 15N signals at 100–130 ppm are from sites with sufficient mobility to cause isotropic averaging of the 15N chemical shift and 1H-15N dipolar coupling, as might be expected for the non-helical regions of the protein. Measureable dipolar couplings from the Arg side chains are also observed, indicating that these sites, positioned at the lipid-water interface, do not undergo rapid isotropic reorientation. Notably, the 1H-15N dipolar couplings and the 15N chemical shift frequencies of the Arg peaks also depend on the alignment of the lipid bilayer membrane in the magnetic field, indicating that these side chains have specific orientations in the context of the lipid bilayer.
Figure 4. Solid-state NMR spectra of uniformly 15N labeled FXYD2 in magnetically oriented phospholipid bilayers.
(A–D) 2D 1H/15N PISEMA) and 1D 15N OS solid-state NMR spectra of FXYD2 in lipid bilayers aligned with the membrane perpendicular (A, B) or parallel (C, D) to the magnetic field. Peaks from the transmembrane helix (TM) trace wheel-like patterns (red circles). Peaks assigned to Arg guanidinium NH groups are prominent (blue boxes). (E) Solution NMR structure of FXYD2 determined in micelles, showing the bundle of ten lowest energy structures. The positions of amide N atoms (blue spheres) were restricted by experimental plane distance restraints as described [132]. Adapted from [132].
Accurate orientation restraints can also be extracted from the powder patterns measured for, unoriented liposome samples [87,112,119]; in this case, rapid rotational diffusion of the protein around the axis perpendicular to the lipid bilayer plane is exploited to generate uniaxial order. Furthermore, chemical shift tensors themselves can provide effective restraints for structure determination and refinement [133].
MAS Solid-state NMR
High resolution MAS solid-state NMR spectra have been reported for membrane proteins in proteolipid 2D crystals, precipitates, and microcrystals [114,134–139]. These samples have the advantage of maximizing protein concentration in the NMR spectrometer, but they also have lipid to protein ratios that are lower than most biological membranes [140,141]. Low lipid content can compromise protein stability and induce the formation of non-native contacts between protein molecules with opposite transmembrane orientations [114,142,143]. Moreover, 2D crystals can be structurally heterogeneous [143], a factor that poses a unique challenge for NMR, where signals reflect ensemble properties of the bulk sample, and heterogeneity manifests as broader line widths and diminished spectral resolution. Fortunately, the only requirement for reducing inhomogeneous line broadening and obtaining single, narrow NMR lines, is that the protein adopts a homogeneous conformational ensemble state on the timescale determined by the frequency span of spin interactions. For solid-state NMR studies of membrane proteins, this can often be achieved with proteoliposomes prepared with sufficient lipid to prevent the formation of non-native conformations.
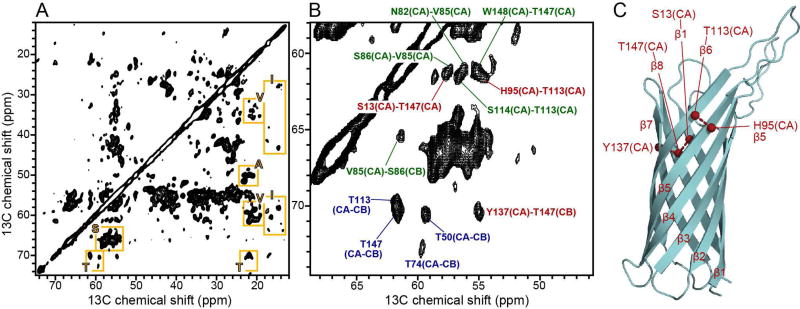
The 2D 13C/13C correlation spectrum of Ail in liposomes (Fig. 5) illustrates the spectral resolution attainable for homogeneous liposome preparations [144]. In this sample, the protein was uniformly labeled with 15N and 13C, and fractionally (∼70%) 2H labeled, with amides back exchanged to 1H during protein purification, to enable NMR detection. Deuteration dilutes the 1H concentration and suppresses the strong homonuclear 1H-1H dipolar couplings, resulting in longer lifetimes of the 13C and 15N coherences, and leading to enhanced sensitivity and resolution.
Figure 5. 2D 13C-13C spectrum of (15N, 13C, 2H) Ail in liposomes.
(A) Fingerprint region of the DARR (Dipolar Assisted Rotational Resonance) spectrum showing examples of resolved signals from Ala, Ile, Ser, Thr, Val residues (yellow boxes). (B) Expanded spectral region showing examples of short-range intra-residue correlations (blue), short-range inter-residue correlations (green), and long-range inter-residue correlations (red). (C) Structure of Ail in micelles (PDB: 2N2L) showing inter-strand connections (red) assigned in the DARR spectrum. The spectrum was obtained at 900 MHz, at 10°C, with 200 ms DARR mixing time, and 144 scans per increment. Adapted from [144].
The 13C/13C correlation spectrum together with additional 2D and 3D spectra that correlate N, CA, CO and side chain atomic sites [145–148], enable resonance assignments and the measurement of distance restraints for structure determination. Signals from Thr, Ser, Ile and Ala sites can be readily identified based on their characteristic chemical shifts, which reflect β-strand conformation. Moreover, several cross peaks observed in the 13C-13C spectrum (Fig. 5B) reflect intra-residue (Fig. 5B, blue) or inter-residue (Fig. 5B, green, red) connections, including long-range (Fig. 5B, red) connections between neighboring β-strands (Fig. 5C) that are essential for structure determination.
Recently, NMR experiments based on 1H-detection have become an important technique for enhancing sensitivity and resolution in solid-state NMR spectroscopy. These experiments are facilitated by very fast spinning rates (>40 kHz), high static magnetic fields, and 2H-labeling of the protein, to reduce homogeneous line broadening by suppressing and diluting the large network of strong homonuclear 1H-1H dipolar couplings [114,135,138,149–156]. NMR experiments of 2H-labeled proteins, based on 1H detection, require back exchange of backbone amides from 2H to 1H. Back exchange is very readily obtained for proteins that are refolded from inclusion bodies, like many b-barrels, but can be less efficient for proteins that are purified as folded entities, where strong hydrogen bond networks are established before purification. Thus, extensive deuteration may not always be a feasible approach. Nevertheless, the availability of radiofrequency probes with spinning rates greater than 100 kHz [157] is removing this obstacle by enabling NMR studies of fully protonated samples, including membrane proteins.
Solid-state NMR experiments based on 1H detection with fast spinning rates are enabling high resolution NMR spectroscopy of membrane proteins in lipids. A number of structural studies have been reported for both β-barrel and α-helical membrane proteins in liposomes and 2D crystal preparations [138,139,144,155,158,159]. For example, the 1H detected 1H/15N CP-HSQC (Cross Polarization - Heteronuclear Single Quantum Correlation) spectrum (Fig. 6A) of the Y. pestis outer membrane protein Ail, in liposomes, displays high resolution [144]. The spectrum was obtained at a magnetic field of 900 MHz, and a temperature, 30°C, where the lipid bilayer is fully liquid crystalline. Resonance line widths are in the range of 0.11–0.15 ppm (107–138 Hz) for 1H, and 0.46–0.64 ppm (42–58 Hz) for 15N, consistent with a high level of sample homogeneity, and comparable with those observed in the 1H-detected spectra of membrane proteins in 2D crystals [138,155]. Notably, the solid-state NMR spectrum overlaps significantly with the solution NMR 1H/15N TROSY spectrum (Fig. 6B) measured for Ail in nanodiscs with the same lipid composition [73], illustrating the potential of performing structural studies on essentially the same, detergent-free sample, over the wider, combined experimental time scale of solution and solid-state NMR spectroscopy.
Figure 6. 2D 1H-15N correlation spectra of Ail in phospholipid bilayers.
(A) Solid-state NMR 1H-detected CP-HSQC spectrum of (15N, 13C, 2H) labeled Ail in liposomes, recorded at 900 MHz, 30°C, with 160 scans and a MAS rate of 60 kHz. (B) Solution NMR 1H-detected TROSY spectrum of (15N,13C,2H) labeled Ail in nanodiscs prepared with 2H labeled lipids, recorded at 800 MHz, 45°C, with 128 scans. Adapted from [144].
NMR structure calculations of membrane proteins
The quality of NMR structures depends on both the accuracy and quantity of the experimental data as well as on the computational methods used in the structure calculations. Methods that facilitate NMR-restrained protein structure calculations in a physically realistic energy landscape, significantly improve structure precision, accuracy and quality.
All atom force fields, with complete chemical energy functions and explicit atomic representation of the protein, water, and lipid molecules, can be used in NMR-restrained molecular dynamics (MD) simulations of membrane proteins [113,160–162]. Recent advances are providing optimized force fields [163,164] and are enabling long MD simulations. Nevertheless, because these simulations rely on a reasonably accurate input structure, they are best suited for the final stages of refinement and remain impractical for routine de novo calculations that start from fully extended polypeptides.
Geometric restraining terms [38,165] or empirical models of membrane insertion depth [166] can be used to impose virtual water-membrane boundaries during NMR-restrained simulated annealing calculations of membrane proteins, but do not provide realistic atomic-level representations of the protein in its environment. In addition, implicit solvation models have been developed specifically for NMR-restrained MD [167–170] or simulated annealing [171–173] protocols. Among these, the implicit solvation potential EEFx (effective energy function for Xplor-NIH) is highly effective for NMR-restrained, simulated annealing calculations of both soluble and membrane proteins from unfolded templates [74,171–176]. EEFx is based on the implicit solvation energy functions developed for CHARMM [177,178] and is available with the Xplor-NIH package [179,180]. It includes an implicit membrane model that provides a physically realistic, anisotropic water-lipid environment and supports the native structures of membrane proteins.
EEFx is designed to work with NMR restraints measured in detergent-free lipid bilayer samples, but is also compatible with restraints measured in micelles [74], and with a wide range of experimental restraints measured by techniques other than NMR, to impose boundaries between hydrophobic and polar environments, establish proper protein topology and prevent water-exposed loops of side chains from folding back against protein regions that are membrane-embedded. EEFx is very effective at guiding structure calculations towards the native state, even in the absence of large numbers of experimental measurements, and yields significant improvements in structural quality, accuracy and precision, as illustrated for the structure of Anabaena sensory rhodopsin (Fig. 7).
Figure 7. The EEFx potential improves structure calculations of Anabaena sensory rhodopsin in lipid bilayers.
Structures of monomeric ASR were calculated using solid-state NMR distance and dihedral angle restraints. Bar plots represent results for: the crystal structure (PDB 1xio; pink) [181]; the average for ten models in the ensembles of the deposited solid-state NMR structure (PDB 2m3g; red) [111]; the ensemble calculated with the standard simple repulsive potential of Xplor-NIH (gray); or the ensemble calculated with the EEFx potential of Xplor-NIH (blue) [173]. (A) Agreement with the PDB crystal structure (PDB 1xio) evaluated as average pairwise RMSD of atomic coordinates. (B) Precision evaluated as average pairwise RMSD of atomic coordinates in each ensemble. (C) MolProbity score evaluation of the four structures; note this is a cost: the lower the better. (D) Comparisons of the crystal structure (PDB 1zxio, pink) with the lowest energy structure generated with EEFx (blue). The horizontal lines depict the boundaries of the 25 Å thick EEFx membrane. Adapted from [173].
Conclusions
Recent technological advances in NMR spectroscopy are opening a new chapter in membrane protein structural biology. The studies reviewed in this article, and the many other exciting efforts in laboratories around the world, reflect the breadth of capabilities of modern NMR. The ability to probe membrane proteins in detergent-free membranes, ever closer in composition to the native environment, is particularly exciting. The high resolution and high sensitivity seen in the NMR spectra reported in recent studies will significantly benefit from the additional enhancements made possible through the use of high magnetic fields, non-uniform sampling schemes that reduce experimental times or increase signal, efficient radiofrequency probes that do not dissipate heat energy, and fast MAS probes that enable 1H detection experiments.
HIGHLIGHTS.
Membrane proteins are major targets of biomedical research and drug discovery.
Molecular structure determination is essential for achieving both of these goals.
NMR is ideally suited for structural studies of membrane proteins in hydrated lipid bilayers.
Recent advances in NMR are opening a new chapter in membrane protein structural biology.
Exciting new efforts in this area reflect the breadth of capabilities of modern NMR.
Acknowledgments
Supported by grants from the National Institutes of Health (GM 118186, GM 099986, and GM 066978) and by the Biotechnology Resource for Molecular Imaging of Proteins at UCSD supported by the National Institutes of Health (P41 EB 002031).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moraes I, Evans G, Sanchez-Weatherby J, Newstead S, Stewart PD. Membrane protein structure determination - the next generation. Biochim Biophys Acta. 2014;1838:78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Zorzi R, Mi W, Liao M, Walz T. Single-particle electron microscopy in the study of membrane protein structure. Microscopy. 2016;65:81–96. doi: 10.1093/jmicro/dfv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maslennikov I, Choe S. Advances in NMR structures of integral membrane proteins. Curr Opin Struct Biol. 2013;23:555–562. doi: 10.1016/j.sbi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross TA, Ekanayake V, Paulino J, Wright A. Solidstate NMR: The essential technology for helical membrane protein structural characterization. J Magn Reson. 2014;239:100–109. doi: 10.1016/j.jmr.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lodowski DT, Angel TE, Palczewski K. Comparative analysis of GPCR crystal structures. Photochemistry and photobiology. 2009;85:425–430. doi: 10.1111/j.1751-1097.2008.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raunser S, Walz T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys. 2009;38:89–105. doi: 10.1146/annurev.biophys.050708.133649. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigorieff N. Direct detection pays off for electron cryo-microscopy. Elife. 2013;2:e00573. doi: 10.7554/eLife.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin AC, Cullis PR, Hemminga MA, Hoult DI, Radda GK, Ritchie GA, Seeley PJ, Richards RE. Application of 31P NMR to model and biological membrane systems. FEBS Lett. 1975;57:213–218. doi: 10.1016/0014-5793(75)80719-8. [DOI] [PubMed] [Google Scholar]
- 14.Griffin RG. Letter: Observation of the effect of water on the 31P nuclear magnetic resonance spectra of dipalmitoyllecithin. J Am Chem Soc. 1976;98:851–853. doi: 10.1021/ja00419a044. [DOI] [PubMed] [Google Scholar]
- 15.Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q Rev Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald PM. Deuterium NMR and the Topography of Surface Electrostatic Charge. Accounts of Chemical Research. 1997;30:196–203. [Google Scholar]
- 17.Nietlispach D, Gautier A. Solution NMR studies of polytopic alpha-helical membrane proteins. Curr Opin Struct Biol. 2011;21:497–508. doi: 10.1016/j.sbi.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Hiller S, Wagner G. The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. Curr Opin Struct Biol. 2009;19:396–401. doi: 10.1016/j.sbi.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Ladizhansky V. Recent advances in magic angle spinning solid state NMR of membrane proteins. Prog Nucl Magn Reson Spectrosc. 2014;82:1–26. doi: 10.1016/j.pnmrs.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan M, Pinto C, Houben K, Baldus M. Nuclear magnetic resonance (NMR) applied to membrane-protein complexes. Q Rev Biophys. 2016;49:e15. doi: 10.1017/S003358351600010X. [DOI] [PubMed] [Google Scholar]
- 21.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 22.Yao Y, Fujimoto LM, Hirshman N, Bobkov AA, Antignani A, Youle RJ, Marassi FM. Conformation of BCL-XL upon Membrane Integration. J Mol Biol. 2015;427:2262–2270. doi: 10.1016/j.jmb.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loll PJ. Membrane proteins, detergents and crystals: what is the state of the art? Acta Crystallogr F Struct Biol Commun. 2014;70:1576–1583. doi: 10.1107/S2053230X14025035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F Struct Biol Commun. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate CG, Schertler GF. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Avila-Sakar A, Kim J, Booth DS, Greenberg CH, Rossi A, Liao M, Li X, Alian A, Griner SL, et al. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaremko M, Jaremko L, Villinger S, Schmidt CD, Griesinger C, Becker S, Zweckstetter M. High-Resolution NMR Determination of the Dynamic Structure of Membrane Proteins. Angew Chem Int Ed Engl. 2016;55:10518–10521. doi: 10.1002/anie.201602639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat Struct Mol Biol. 2010;17:768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Cierpicki T, Jimenez RH, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 36.Buck-Koehntop BA, Mascioni A, Buffy JJ, Veglia G. Structure dynamics membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride. Journal of Molecular Biology. 2005;354:652–665. doi: 10.1016/j.jmb.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 37.Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teriete P, Franzin CM, Choi J, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles. Biochemistry. 2007;46:6774–6783. doi: 10.1021/bi700391b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Pielak RM, McClintock MA, Chou JJ. Solution structure and functional analysis of the influenza B proton channel. Nat Struct Mol Biol. 2009;16:1267–1271. doi: 10.1038/nsmb.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trenker R, Call MJ, Call ME. Progress and prospects for structural studies of transmembrane interactions in single-spanning receptors. Curr Opin Struct Biol. 2016;39:115–123. doi: 10.1016/j.sbi.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didenko T, Liu JJ, Horst R, Stevens RC, Wuthrich K. Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr Opin Struct Biol. 2013;23:740–747. doi: 10.1016/j.sbi.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horst R, Liu JJ, Stevens RC, Wuthrich K. beta(2)-adrenergic receptor activation by agonists studied with (1)(9)F NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52:10762–10765. doi: 10.1002/anie.201305286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F–NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung KY, Kim TH, Manglik A, Alvares R, Kobilka BK, Prosser RS. Role of detergents in conformational exchange of a G protein-coupled receptor. J Biol Chem. 2012;287:36305–36311. doi: 10.1074/jbc.M112.406371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim TH, Chung KY, Manglik A, Hansen AL, Dror RO, Mildorf TJ, Shaw DE, Kobilka BK, Prosser RS. The role of ligands on the equilibria between functional states of a G protein-coupled receptor. J Am Chem Soc. 2013;135:9465–9474. doi: 10.1021/ja404305k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Connor C, White KL, Doncescu N, Didenko T, Roth BL, Czaplicki G, Stevens RC, Wuthrich K, Milon A. NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc Natl Acad Sci U S A. 2015;112:11852–11857. doi: 10.1073/pnas.1510117112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang F, Yu X, Liu C, Qu CX, Gong Z, Liu HD, Li FH, Wang HM, He DF, Yi F, et al. Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and (19)F-NMR. Nat Commun. 2015;6:8202. doi: 10.1038/ncomms9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eddy MT, Didenko T, Stevens RC, Wuthrich K. beta2-Adrenergic Receptor Conformational Response to Fusion Protein in the Third Intracellular Loop. Structure. 2016;24:2190–2197. doi: 10.1016/j.str.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langelaan DN, Bebbington EM, Reddy T, Rainey JK. Structural insight into G-protein coupled receptor binding by apelin. Biochemistry. 2009;48:537–548. doi: 10.1021/bi801864b. [DOI] [PubMed] [Google Scholar]
- 54.Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nasr ML, Singh SK. Radioligand binding to nanodisc-reconstituted membrane transporters assessed by the scintillation proximity assay. Biochemistry. 2014;53:4–6. doi: 10.1021/bi401412e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. Amphipols from A to Z. Annu Rev Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 57.Etzkorn M, Zoonens M, Catoire LJ, Popot JL, Hiller S. How amphipols embed membrane proteins: global solvent accessibility and interaction with a flexible protein terminus. J Membr Biol. 2014;247:965–970. doi: 10.1007/s00232-014-9657-9. [DOI] [PubMed] [Google Scholar]
- 58.Tifrea DF, Sun G, Pal S, Zardeneta G, Cocco MJ, Popot JL, de la Maza LM. Amphipols stabilize the Chlamydia major outer membrane protein and enhance its protective ability as a vaccine. Vaccine. 2011;29:4623–4631. doi: 10.1016/j.vaccine.2011.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi K, Arthanari H, Shimada I, Wagner G. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J Biomol NMR. 2015;63:323–331. doi: 10.1007/s10858-015-9991-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dorr JM, Koorengevel MC, Schafer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA, Dafforn TR, Baldus M, Killian JA. Detergent-freeisolation, characterization, functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc Natl Acad Sci U S A. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Susac L, Horst R, Wuthrich K. Solution-NMR Characterization of Outer-Membrane Protein A from E. coli in Lipid Bilayer Nanodiscs and Detergent Micelles. Chembiochem. 2014;15:995–1000. doi: 10.1002/cbic.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shenkarev ZO, Lyukmanova EN, Butenko IO, Petrovskaya LE, Paramonov AS, Shulepko MA, Nekrasova OV, Kirpichnikov MP, Arseniev AS. Lipid-protein nanodiscs promote in vitro folding of transmembrane domains of multi-helical and multimeric membrane proteins. Biochim Biophys Acta. 2013;1828:776–784. doi: 10.1016/j.bbamem.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A. Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy. Angew Chem Int Ed Engl. 2016;55:4497–4499. doi: 10.1002/anie.201600073. [DOI] [PubMed] [Google Scholar]
- 66.Viegas A, Viennet T, Etzkorn M. The power, pitfalls and potential of the nanodisc system for NMR-based studies. Biol Chem. 2016;397:1335–1354. doi: 10.1515/hsz-2016-0224. [DOI] [PubMed] [Google Scholar]
- 67.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyberts SG, Arthanari H, Robson SA, Wagner G. Perspectives in magnetic resonance: NMR in the post-FFT era. J Magn Reson. 2014;241:60–73. doi: 10.1016/j.jmr.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin EC, Opella SJ. Covariance spectroscopy in high-resolution multi-dimensional solid-state NMR. J Magn Reson. 2014;239:57–60. doi: 10.1016/j.jmr.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suiter CL, Paramasivam S, Hou G, Sun S, Rice D, Hoch JC, Rovnyak D, Polenova T. Sensitivity gains, linearity, and spectral reproducibility in nonuniformly sampled multidimensional MAS NMR spectra of high dynamic range. J Biomol NMR. 2014;59:57–73. doi: 10.1007/s10858-014-9824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palmer MR, Suiter CL, Henry GE, Rovnyak J, Hoch JC, Polenova T, Rovnyak D. Sensitivity of Nonuniform Sampling NMR. J Phys Chem B. 2015 doi: 10.1021/jp5126415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dutta SK, Yao Y, Marassi FM. Structure and dynamics of the Yersinia pestis outer membrane protein Ail in lipid bilayers revealed by NMR. J Phys Chem B. 2017 doi: 10.1021/acs.jpcb.7b03941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding Y, Fujimoto LM, Yao Y, Plano GV, Marassi FM. Influence of the lipid membrane environment on structure and activity of the outer membrane protein Ail from Yersinia pestis. Biochim Biophys Acta. 2015;1848:712–720. doi: 10.1016/j.bbamem.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marassi FM, Ding Y, Schwieters CD, Tian Y, Yao Y. Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation. J Biomol NMR. 2015;63:59–65. doi: 10.1007/s10858-015-9963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamashita S, Lukacik P, Barnard TJ, Noinaj N, Felek S, Tsang TM, Krukonis ES, Hinnebusch BJ, Buchanan SK. Structural insights into Ail-mediated adhesion in Yersinia pestis. Structure. 2011;19:1672–1682. doi: 10.1016/j.str.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park SH, Casagrande F, Das BB, Albrecht L, Chu M, Opella SJ. Local and global dynamics of the G protein-coupled receptor CXCR1. Biochemistry. 2011;50:2371–2380. doi: 10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein-Seetharaman J, Reeves PJ, Loewen MC, Getmanova EV, Chung J, Schwalbe H, Wright PE, Khorana HG. Solution NMR spectroscopy of [alpha −15N]lysine-labeled rhodopsin: The single peak observed in both conventional and TROSY-type HSQC spectra is ascribed to Lys-339 in the carboxyl-terminal peptide sequence. Proc Natl Acad Sci U S A. 2002;99:3452–3457. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dodevski I, Pluckthun A. Evolution of three human GPCRs for higher expression and stability. J Mol Biol. 2011;408:599–615. doi: 10.1016/j.jmb.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 79.Schutz M, Schoppe J, Sedlak E, Hillenbrand M, Nagy-Davidescu G, Ehrenmann J, Klenk C, Egloff P, Kummer L, Pluckthun A. Directed evolution of G protein-coupled receptors in yeast for higher functional production in eukaryotic expression hosts. Sci Rep. 2016;6:21508. doi: 10.1038/srep21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baker LA, Folkers GE, Sinnige T, Houben K, Kaplan M, van der Cruijsen EA, Baldus M. Magic-angle-spinning solid-state NMR of membrane proteins. Methods Enzymol. 2015;557:307–328. doi: 10.1016/bs.mie.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 81.Brown LS, Ladizhansky V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Sci. 2015;24:1333–1346. doi: 10.1002/pro.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang W, Knox RW, Nevzorov AA. A spectroscopic assignment technique for membrane proteins reconstituted in magnetically aligned bicelles. J Biomol NMR. 2012;54:307–316. doi: 10.1007/s10858-012-9673-y. [DOI] [PubMed] [Google Scholar]
- 83.Koroloff SN, Nevzorov AA. Selective excitation for spectral editing and assignment in separated local field experiments of oriented membrane proteins. J Magn Reson. 2017;274:7–12. doi: 10.1016/j.jmr.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 84.Fu R, Brey WW, Shetty K, Gor’kov P, Saha S, Long JR, Grant SC, Chekmenev EY, Hu J, Gan Z, et al. Ultra-wide bore 900 MHz high-resolution NMR at the National High Magnetic Field Laboratory. Journal of Magnetic Resonance. 2005;177:1–8. doi: 10.1016/j.jmr.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 85.Li C, Mo Y, Hu J, Chekmenev E, Tian C, Gao FP, Fu R, Gor’kov P, Brey W, Cross TA. Analysis of RF heating and sample stability in aligned static solid-state NMR spectroscopy. J Magn Reson. 2006;180:51–57. doi: 10.1016/j.jmr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 86.Grant CV, Wu CH, Opella SJ. Probes for high field solid-state NMR of lossy biological samples. Journal of Magnetic Resonance. 2010;204:180–188. doi: 10.1016/j.jmr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian F, Song Z, Cross TA. Orientational constraints derived from hydrated powder samples by two-dimensional PISEMA. J Magn Reson. 1998;135:227–231. doi: 10.1006/jmre.1998.1544. [DOI] [PubMed] [Google Scholar]
- 88.Park SH, Das BB, De Angelis AA, Scrima M, Opella SJ. Mechanically, magnetically, and “rotationally aligned” membrane proteins in phospholipid bilayers give equivalent angular constraints for NMR structure determination. J Phys Chem B. 2010;114:13995–14003. doi: 10.1021/jp106043w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhate MP, Wylie BJ, Tian L, McDermott AE. Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J Mol Biol. 2010;401:155–166. doi: 10.1016/j.jmb.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Medeiros-Silva J, Mance D, Daniels M, Jekhmane S, Houben K, Baldus M, Weingarth M. 1 H-Detected Solid-State NMR Studies of Water-Inaccessible Proteins In Vitro and In Situ. Angew Chem Int Ed Engl. 2016;55:13606–13610. doi: 10.1002/anie.201606594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu F, Schmidt-Rohr K, Hong M. NMR Detection of pH-Dependent Histidine-Water Proton Exchange Reveals the Conduction Mechanism of a Transmembrane Proton Channel. J Am Chem Soc. 2011 doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fu R, Miao Y, Qin H, Cross TA. Probing Hydronium Ion Histidine NH Exchange Rate Constants in the M2 Channel via Indirect Observation of Dipolar-Dephased 15N Signals in Magic-Angle-Spinning NMR. J Am Chem Soc. 2016;138:15801–15804. doi: 10.1021/jacs.6b08376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gayen A, Banigan JR, Traaseth NJ. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52:10321–10324. doi: 10.1002/anie.201303091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hong M, Su Y. Structure and dynamics of cationic membrane peptides and proteins: insights from solid-state NMR. Protein Sci. 2011;20:641–655. doi: 10.1002/pro.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strandberg E, Zerweck J, Wadhwani P, Ulrich AS. Synergistic insertion of antimicrobial magainin-family peptides in membranes depends on the lipid spontaneous curvature. Biophysical Journal. 2013 doi: 10.1016/j.bpj.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perrin BS, Jr, Tian Y, Fu R, Grant CV, Chekmenev EY, Wieczorek WE, Dao AE, Hayden RM, Burzynski CM, Venable RM, et al. High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion. J Am Chem Soc. 2014;136:3491–3504. doi: 10.1021/ja411119m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang R, Yamamoto K, Zhang M, Popovych N, Hung I, Im SC, Gan Z, Waskell L, Ramamoorthy A. Probing the transmembrane structure and dynamics of microsomal NADPH-cytochrome P450 oxidoreductase by solid-state NMR. Biophys J. 2014;106:2126–2133. doi: 10.1016/j.bpj.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown MF, Heyn MP, Job C, Kim S, Moltke S, Nakanishi K, Nevzorov AA, Struts AV, Salgado GF, Wallat I. Solid-state 2H NMR spectroscopy of retinal proteins in aligned membranes. Biochim Biophys Acta. 2007;1768:2979–3000. doi: 10.1016/j.bbamem.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith SO. Structure and activation of the visual pigment rhodopsin. Annual review of biophysics. 2010;39:309–328. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 100.Struts AV, Salgado GF, Brown MF. Solid-state 2H NMR relaxation illuminates functional dynamics of retinal cofactor in membrane activation of rhodopsin. Proc Natl Acad Sci U S A. 2011;108:8263–8268. doi: 10.1073/pnas.1014692108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tapaneeyakorn S, Goddard AD, Oates J, Willis CL, Watts A. Solution- and solid-state NMR studies of GPCRs and their ligands. Biochim. Biophys. Acta. 2011;1808:1462–1475. doi: 10.1016/j.bbamem.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 102.Goncalves J, Eilers M, South K, Opefi CA, Laissue P, Reeves PJ, Smith SO. Magic angle spinning nuclear magnetic resonance spectroscopy of G protein-coupled receptors. Methods Enzymol. 2013;522:365–389. doi: 10.1016/B978-0-12-407865-9.00017-0. [DOI] [PubMed] [Google Scholar]
- 103.Kimura T, Vukoti K, Lynch DL, Hurst DP, Grossfield A, Pitman MC, Reggio PH, Yeliseev AA, Gawrisch K. Global fold of human cannabinoid type 2 receptor probed by solid-state 13C-, 15N–MAS NMR and molecular dynamics simulations. Proteins. 2014;82:452–465. doi: 10.1002/prot.24411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-resolution NMR spectroscopy of a GPCR in aligned bicelles. Journal of the American Chemical Society. 2006;128:7402–7403. doi: 10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J Mol Biol. 2011;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharif S, Kim SJ, Labischinski H, Schaefer J. Characterization of peptidoglycan in fem-deletion mutants of methicillin-resistant Staphylococcus aureus by solid-state NMR. B iochemistry. 2009;48:3100–3108. doi: 10.1021/bi801750u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koch K, Afonin S, Ieronimo M, Berditsch M, Ulrich A. Solid-State 19F–NMR of Peptides in Native Membranes. In: JCC Chan., editor. Solid State NMR. Springer Berlin Heidelberg; 2012. pp. 89–118. Topics in Current Chemistry, vol 306. [DOI] [PubMed] [Google Scholar]
- 108.Miao Y, Qin H, Fu R, Sharma M, Can TV, Hung I, Luca S, Gor’kov PL, Brey WW, Cross TA. M2 proton channel structural validation from full-length protein samples in synthetic bilayers and E. coli membranes. Angew Chem Int Ed Engl. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2012;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaplan M, Cukkemane A, van Zundert GC, Narasimhan S, Daniels M, Mance D, Waksman G, Bonvin AM, Fronzes R, Folkers GE, et al. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat Methods. 2015;12:649–652. doi: 10.1038/nmeth.3406. [DOI] [PubMed] [Google Scholar]
- 111.Wang S, Munro RA, Shi L, Kawamura I, Okitsu T, Wada A, Kim SY, Jung KH, Brown LS, Ladizhansky V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods. 2013;10:1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- 112.Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis A, Marassi FM, et al. Structure of the Chemokine Receptor CXCR1 in Phospholipid Bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, Miller EA, Emsley L, Pintacuda G, Chou JJ, Griffin RG. Structure and Mechanism of the Influenza A M218-60 Dimer of Dimers. J Am Chem Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang M, Sperling LJ, Berthold DA, Schwieters CD, Nesbitt AE, Nieuwkoop AJ, Gennis RB, Rienstra CM. High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data. J Biomol NMR. 2011;51:227–233. doi: 10.1007/s10858-011-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tang M, Nesbitt AE, Sperling LJ, Berthold DA, Schwieters CD, Gennis RB, Rienstra CM. Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer. J Mol Biol. 2013;425:1670–1682. doi: 10.1016/j.jmb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Das N, Dai J, Hung I, Rajagopalan MR, Zhou HX, Cross TA. Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers. Proc Natl Acad Sci U S A. 2015;112:E119–126. doi: 10.1073/pnas.1415908112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marassi FM, Opella SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Science. 2003;12:403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tian Y, Lu GJ, Marassi FM, Opella SJ. Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints. J Biomol NMR. 2014;60:67–71. doi: 10.1007/s10858-014-9852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nat Methods. 2012;9:1212–1217. doi: 10.1038/nmeth.2248. [DOI] [PubMed] [Google Scholar]
- 121.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ding Y, Yao Y, Marassi FM. Membrane protein structure determination in membrana. Acc Chem Res. 2013;46:2182–2190. doi: 10.1021/ar400041a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Murray DT, Li C, Gao FP, Qin H, Cross TA. Membrane protein structural validation by oriented sample solid-state NMR: diacylglycerol kinase. Biophys J. 2014;106:1559–1569. doi: 10.1016/j.bpj.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, et al. Imaging membrane protein helical wheels. J Magn Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 126.Marassi FM. A simple approach to membrane protein secondary structure and topology based on NMR spectroscopy. Biophys J. 2001;80:994–1003. doi: 10.1016/S0006-3495(01)76078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Angelis AA, Howell SC, Opella SJ. Assigning solid-state NMR spectra of aligned proteins using isotropic chemical shifts. Journal of Magnetic Resonance. 2006;183:329–332. doi: 10.1016/j.jmr.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 128.Tian Y, Schwieters CD, Opella SJ, Marassi FM. AssignFit: A program for simultaneous assignment and structure refinement from solid-state NMR spectra. J Magn Reson. 2012;214:42–50. doi: 10.1016/j.jmr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M, Opella SJ. Dilute spin-exchange assignment of solid-state NMR spectra of oriented proteins: acetylcholine M2 in bilayers. Journal of Biomolecular NMR. 1999;14:141–148. doi: 10.1023/a:1008391823293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Knox RW, Lu GJ, Opella SJ, Nevzorov AA. A resonance assignment method for oriented-sample solid-state NMR of proteins. Journal of the American Chemical Society. 2010;132:8255–8257. doi: 10.1021/ja102932n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gong XM, Ding Y, Yu J, Yao Y, Marassi FM. Structure of the Na,K-ATPase regulatory protein FXYD2b in micelles: implications for membrane-water interfacial arginines. Biochim Biophys Acta. 2015;1848:299–306. doi: 10.1016/j.bbamem.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wylie BJ, Schwieters CD, Oldfield E, Rienstra CM. Protein structure refinement using 13C alpha chemical shift tensors. J Am Chem Soc. 2009;131:985–992. doi: 10.1021/ja804041p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. Partial (13)C and (15)N chemical-shift assignments of the disulfide-bond-forming enzyme DsbB by 3D magic-angle spinning NMR spectroscopy. Chembiochem. 2007;8:434–442. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- 135.Linser R, Dasari M, Hiller M, Higman V, Fink U, Lopez del Amo JM, Markovic S, Handel L, Kessler B, Schmieder P, et al. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angew Chem Int Ed Engl. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- 136.Eddy MT, Ong TC, Clark L, Teijido O, van der Wel PC, Garces R, Wagner G, Rostovtseva TK, Griffin RG. Lipid dynamics and protein-lipid interactions in 2D crystals formed with the beta-barrel integral membrane protein VDAC1. J Am Chem Soc. 2012;134:6375–6387. doi: 10.1021/ja300347v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shahid SA, Markovic S, Linke D, van Rossum BJ. Assignment and secondary structure of the YadA membrane protein by solid-state MAS NMR. Sci Rep. 2012;2:803. doi: 10.1038/srep00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barbet-Massin E, Pell AJ, Retel JS, Andreas LB, Jaudzems K, Franks WT, Nieuwkoop AJ, Hiller M, Higman V, Guerry P, et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J Am Chem Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saurel O, Iordanov I, Nars G, Demange P, Le Marchand T, Andreas LB, Pintacuda G, Milon A. Local and Global Dynamics in Klebsiella pneumoniae Outer Membrane Protein a in Lipid Bilayers Probed at Atomic Resolution. J Am Chem Soc. 2017 doi: 10.1021/jacs.6b11565. [DOI] [PubMed] [Google Scholar]
- 140.Gennis RB. Biomembranes : molecular structure and function. New York: Springer-Verlag; 1989. [Google Scholar]
- 141.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 142.Dolder M, Zeth K, Tittmann P, Gross H, Welte W, Wallimann T. Crystallization of the human, mitochondrial voltage-dependent anion-selective channel in the presence of phospholipids. J Struct Biol. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- 143.Behlau M, Mills DJ, Quader H, Kuhlbrandt W, Vonck J. Projection structure of the monomeric porin OmpG at 6 A resolution. J Mol Biol. 2001;305:71–77. doi: 10.1006/jmbi.2000.4284. [DOI] [PubMed] [Google Scholar]
- 144.Yao Y, Dutta S, Park SH, Rai R, Fujimoto LM, Bobkov AA, Opella SJ, Marassi FM. High resolution solid-state NMR spectroscopy of the Yersinia pestis outer membrane protein Ail in lipid membranes. J Biomol NMR. 2017 doi: 10.1007/s10858-017-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baldus M, Petkova A, Herzfield J, Griffin R. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 146.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13C and 15N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 Tesla. Chembiochem. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 147.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters. 2001;344:631–637. [Google Scholar]
- 148.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-driven 13C-13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. The Journal of Chemical Physics. 2003;118:2325–2341. [Google Scholar]
- 149.Ishii Y, Tycko R. Sensitivity enhancement in solid state (15)N NMR by indirect detection with high-speed magic angle spinning. J Magn Reson. 2000;142:199–204. doi: 10.1006/jmre.1999.1976. [DOI] [PubMed] [Google Scholar]
- 150.Ishii Y, Yesinowski JP, Tycko R. Sensitivity enhancement in solid-state (13)C NMR of synthetic polymers and biopolymers by (1)H NMR detection with high-speed magic angle spinning. J Am Chem Soc. 2001;123:2921–2922. doi: 10.1021/ja015505j. [DOI] [PubMed] [Google Scholar]
- 151.Reif B, Griffin RG. 1H detected 1H,15N correlation spectroscopy in rotating solids. J Magn Reson. 2003;160:78–83. doi: 10.1016/s1090-7807(02)00035-6. [DOI] [PubMed] [Google Scholar]
- 152.Chevelkov V, Rehbein K, Diehl A, Reif B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew Chem Int Ed Engl. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- 153.Park SH, Yang C, Opella SJ, Mueller LJ. Resolution and measurement of heteronuclear dipolar couplings of a noncrystalline protein immobilized in a biological supramolecular assembly by proton-detected MAS solid-state NMR spectroscopy. J Magn Reson. 2013;237:164–168. doi: 10.1016/j.jmr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bockmann A, Ernst M, Meier BH. Spinning proteins, the faster, the better? J Magn Reson. 2015;253:71–79. doi: 10.1016/j.jmr.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 155.Eddy MT, Su Y, Silvers R, Andreas L, Clark L, Wagner G, Pintacuda G, Emsley L, Griffin RG. Lipid bilayer-bound conformation of an integral membrane beta barrel protein by multidimensional MAS NMR. J Biomol NMR. 2015;61:299–310. doi: 10.1007/s10858-015-9903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang R, Mroue KH, Ramamoorthy A. Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy. Acc Chem Res. 2017 doi: 10.1021/acs.accounts.7b00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Agarwal V, Penzel S, Szekely K, Cadalbert R, Testori E, Oss A, Past J, Samoson A, Ernst M, Bockmann A, et al. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew Chem Int Ed Engl. 2014;53:12253–12256. doi: 10.1002/anie.201405730. [DOI] [PubMed] [Google Scholar]
- 158.Ward ME, Ritz E, Ahmed MA, Bamm VV, Harauz G, Brown LS, Ladizhansky V. Proton detection for signal enhancement in solid-state NMR experiments on mobile species in membrane proteins. J Biomol NMR. 2015;63:375–388. doi: 10.1007/s10858-015-9997-5. [DOI] [PubMed] [Google Scholar]
- 159.Weingarth M, van der Cruijsen EA, Ostmeyer J, Lievestro S, Roux B, Baldus M. Quantitative analysis of the water occupancy around the selectivity filter of a K+ channel in different gating modes. J Am Chem Soc. 2014;136:2000–2007. doi: 10.1021/ja411450y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheng X, Im W. NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophys J. 2012;102:L27–29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheng X, Jo S, Marassi FM, Im W. NMR-based simulation studies of Pf1 coat protein in explicit membranes. Biophys J. 2013;105:691–698. doi: 10.1016/j.bpj.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cheng X, Jo S, Qi Y, Marassi FM, Im W. Solid-State NMR-Restrained Ensemble Dynamics of a Membrane Protein in Explicit Membranes. Biophys J. 2015;108:1954–1962. doi: 10.1016/j.bpj.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vanommeslaeghe K, MacKerell AD., Jr CHARMM additive and polarizable force fields for biophysics and computer-aided drug design. Biochim Biophys Acta. 2015;1850:861–871. doi: 10.1016/j.bbagen.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 165.Xu C, Gagnon E, Call ME, Schnell JR, Schwieters CD, Carman CV, Chou JJ, Wucherpfennig KW. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shi L, Traaseth NJ, Verardi R, Cembran A, Gao J, Veglia G. A refinement protocol to determine structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state NMR restraints. J Biomol NMR. 2009;44:195–205. doi: 10.1007/s10858-009-9328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Xia B, Tsui V, Case DA, Dyson HJ, Wright PE. Comparison of protein solution structures refined by molecular dynamics simulation in vacuum, with a generalized Born model, and with explicit water. J Biomol NMR. 2002;22:317–331. doi: 10.1023/a:1014929925008. [DOI] [PubMed] [Google Scholar]
- 168.Chen J, Im W, Brooks CL., 3rd Refinement of NMR structures using implicit solvent and advanced sampling techniques. J Am Chem Soc. 2004;126:16038–16047. doi: 10.1021/ja047624f. [DOI] [PubMed] [Google Scholar]
- 169.Chen J, Won HS, Im W, Dyson HJ, Brooks CL., 3rd Generation of native-like protein structures from limited NMR data, modern force fields and advanced conformational sampling. J Biomol NMR. 2005;31:59–64. doi: 10.1007/s10858-004-6056-z. [DOI] [PubMed] [Google Scholar]
- 170.Im W, Brooks CL., 3rd De novo folding of membrane proteins: an exploration of the structure and NMR properties of the fd coat protein. J Mol Biol. 2004;337:513–519. doi: 10.1016/j.jmb.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 171.Tian Y, Schwieters CD, Opella SJ, Marassi FM. A practical implicit solvent potential for NMR structure calculation. J Magn Reson. 2014;243:54–64. doi: 10.1016/j.jmr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tian Y, Schwieters CD, Opella SJ, Marassi FM. A Practical Implicit Membrane Potential for NMR Structure Calculations of Membrane Proteins. Biophys J. 2015;109:574–585. doi: 10.1016/j.bpj.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tian Y, Schwieters CD, Opella SJ, Marassi FM. High quality NMR structures: a new force field with implicit water and membrane solvation for Xplor-NIH. J Biomol NMR. 2017;67:35–49. doi: 10.1007/s10858-016-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jureka AS, Kleinpeter AB, Cornilescu G, Cornilescu CC, Petit CM. Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I. Structure. 2015;23:2001–2010. doi: 10.1016/j.str.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cornilescu G, Didychuk AL, Rodgers ML, Michael LA, Burke JE, Montemayor EJ, Hoskins AA, Butcher SE. Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. J Mol Biol. 2016;428:777–789. doi: 10.1016/j.jmb.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee W, Petit CM, Cornilescu G, Stark JL, Markley JL. The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data. J Biomol NMR. 2016;65:51–57. doi: 10.1007/s10858-016-0036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 178.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 179.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 180.Schwieters CD, Kuszewski JJ, Marius Clore G. Using Xplor,ÄìNIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 181.Vogeley L, Sineshchekov OA, Trivedi VD, Sasaki J, Spudich JL, Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]