Abstract
An extensive proteostatic network comprised of molecular chaperones and protein clearance mechanisms functions collectively to preserve the integrity and resiliency of the proteome. The efficacy of this network deteriorates during aging, coinciding with many clinical manifestations, including protein aggregation diseases of the nervous system. A decline in proteostasis can be delayed through the activation of cytoprotective transcriptional responses, which are sensitive to environmental stress and internal metabolic and physiological cues. The homeodomain-interacting protein kinase (hipk) family members are conserved transcriptional co-factors that have been implicated in both genotoxic and metabolic stress responses from yeast to mammals. We demonstrate that constitutive expression of the sole Caenorhabditis elegans Hipk homolog, hpk-1, is sufficient to delay aging, preserve proteostasis, and promote stress resistance, while loss of hpk-1 is deleterious to these phenotypes. We show that HPK-1 preserves proteostasis and extends longevity through distinct but complementary genetic pathways defined by the heat shock transcription factor (HSF-1), and the target of rapamycin complex 1 (TORC1). We demonstrate that HPK-1 antagonizes sumoylation of HSF-1, a post-translational modification associated with reduced transcriptional activity in mammals. We show that inhibition of sumoylation by RNAi enhances HSF-1-dependent transcriptional induction of chaperones in response to heat shock. We find that hpk-1 is required for HSF-1 to induce molecular chaperones after thermal stress and enhances hormetic extension of longevity. We also show that HPK-1 is required in conjunction with HSF-1 for maintenance of proteostasis in the absence of thermal stress, protecting against the formation of polyglutamine (Q35::YFP) protein aggregates and associated locomotory toxicity. These functions of HPK-1/HSF-1 undergo rapid down-regulation once animals reach reproductive maturity. We show that HPK-1 fortifies proteostasis and extends longevity by an additional independent mechanism: induction of autophagy. HPK-1 is necessary for induction of autophagosome formation and autophagy gene expression in response to dietary restriction (DR) or inactivation of TORC1. The autophagy-stimulating transcription factors pha-4/FoxA and mxl-2/Mlx, but not hlh-30/TFEB or the nuclear hormone receptor nhr-62, are necessary for extended longevity resulting from HPK-1 overexpression. HPK-1 expression is itself induced by transcriptional mechanisms after nutritional stress, and post-transcriptional mechanisms in response to thermal stress. Collectively our results position HPK-1 at a central regulatory node upstream of the greater proteostatic network, acting at the transcriptional level by promoting protein folding via chaperone expression, and protein turnover via expression of autophagy genes. HPK-1 therefore provides a promising intervention point for pharmacological agents targeting the protein homeostasis system as a means of preserving robust longevity.
Author summary
Aging is the gradual and progressive decline of vitality. A hallmark of aging is the decay of protective mechanisms that normally preserve the robustness and resiliency of cells and tissues. Proteostasis is the term that applies specifically to those mechanisms that promote stability of the proteome, the collection of polypeptides that cells produce, by a combination of chaperone-assisted folding and degradation of misfolded or extraneous proteins. We have identified hpk-1 (encoding a homeodomain-interacting protein kinase) in the nematode C. elegans as an important transcriptional regulatory component of the proteostasis machinery. HPK-1 promotes proteostasis by linking two distinct mechanisms: first by stimulating chaperone gene expression via the heat shock transcription factor (HSF-1), and second by stimulating autophagy gene expression in opposition to the target of rapamycin (TOR) kinase signaling pathway. HPK-1 therefore provides an attractive target for interventions to preserve physiological resiliency during aging by preserving the overall health of the proteome.
Introduction
Aging is sensitive to both internal and environmental stimuli, and is tuned by multiple emergent genetic circuits. External signals include the nutritive value and quantity of the food supply; internal signals originate from discrete cellular sources, such as mitochondria or ribosomes, and discrete tissue sources, such as the reproductive system[1–7].
An overarching theme currently emerging in the aging field is one of homeostasis- homeostasis at the level of genome maintenance and gene expression[8–10], and homeostasis at the level of proteome folding and stability[11, 12]. The gradual loss of homeostasis, from precision of gene expression to protein folding and degradation is a common hallmark of aging organisms. Therefore, longevity is often extendable by manipulations that increase overall stress resistance, such as thermal shock or hypoxia, by a phenomenon known as hormesis. It is generally believed that hormesis extends longevity by bolstering organismal and cellular stress response pathways, which subsequently offsets aging-related decline in these pathways[13].
A major aim in aging research is to improve quality-of-life with advancing age (often referred to as healthspan) by reinforcement of those maintenance pathways that ensure the integrity of biological processes[6]. Indeed, a number of aging-related diseases, such as Alzheimer’s and Parkinson’s dementias, are believed to arise from the decline in the systems that maintain proteome stability and plasticity, by injury to or defects in the cellular processes that promote accurate protein folding and elimination of misfolded and damaged proteins[14]. Maintaining protein homeostasis (proteostasis) is the collective process that preserves a robust and functional proteome; an equation balanced by rates of protein synthesis, protein folding, and protein turnover. Protein synthesis places stress on the proteome by increasing the total concentration of cellular protein. Protein concentrations within the cell can approach saturation levels achieved within crystals[15]. Thus, a major challenge in maintaining proteostasis is an issue of solubility, which is managed by molecular chaperones. Chaperones play an integral role both in assisting in the correct maturation of nascent polypeptides, and in the elimination of proteins through chaperone-mediated degradative pathways. Cells eliminate misfolded, damaged or unneeded polypeptides by ubiquitin-mediated proteosomal degradation as well as by macroautophagy (hereafter referred to as autophagy) at the lysosome. Yet, chaperones have a limited buffering capacity to maintain proper folding under different forms of cellular stress. Thus potent stress response mechanisms act to resolve both acute and chronic stress to the proteome, through refolding, degradation, and sequestration.
Protein homeostatic mechanisms are regulated at the transcriptional and post-transcriptional levels. For instance, the heat shock transcription factor HSF-1 activates transcription of the chaperone genetic network in response to a wide range of stresses, the most well-known being acute thermal stress[16]. A myriad of transcription factors in C. elegans have been shown to promote autophagy at the level of gene expression and autophagosome formation in response to various environmental stressors[17]; including FOXA (PHA-4)[18], TFEB (HLH-30)[19, 20], Mondo/Mlx (MML-1/MXL-2)[21], the HNF4-related nuclear hormone receptor (NHR-62)[22], and several transcription factors necessary for ER and mitochondrial unfolded protein responses[23–25]. There is a growing body of evidence that demonstrates that the loss of autophagy and the decline of proteostasis are conserved hallmarks of normal aging[12, 26, 27]. Consistently, during aging there is a general deterioration in the ability of cells to activate these transcriptional responses to proteotoxic stress. However, there is evidence in C. elegans suggesting that normal cell non-autonomous signals impair proteostasis. For example, under normal conditions, thermosensory neurons inhibit the ability of distal tissues to resolve proteotoxic stress mediated by polyglutamine expression[28], and the onset of reproduction triggers both a rapid decline in protein quality control in the soma[29] and chromatin silencing at stress response genes limits the somatic heat shock response[30]. In mammals, the autophagy system antagonizes the progression of multiple neurodegenerative disorders[31]. Thus, identifying signals that either positively or negatively impact the inducibility of proteostatic mechanisms, as well as how they are regulated and coordinated will be essential for the treatment of age-associated proteotoxic disease and to maximize healthy aging.
In this study, we describe the C. elegans homolog of the HIPK homeodomain-interacting protein kinase, or HPK-1, as an essential co-factor of multiple transcriptional responses that collectively preserve proteostasis. The Hipk gene family encodes a set of conserved kinases that act as transcriptional co-factors important for the regulation of cell growth, development, differentiation and apoptosis[32, 33]. Hipks are activated by metabolic and genotoxic stressors from yeast to mammals[34–37]. In a previous study we found that hpk-1 is necessary for wild-type lifespan and the extended longevity of insulin signaling mutants[38]. Here we show that HPK-1 reinforces proteostasis in C. elegans in response to both thermal stress and dietary restriction by transcriptional activation of chaperone and autophagy pathways, respectively. We show that HPK-1 is necessary for chaperone induction by HSF-1 in response to heat stress and opposes sumoylation of HSF-1, an inhibitory post-translational modification in mammals[39–41]. Consistently, we show that RNAi of the SUMO moiety encoded by smo-1 enhances HSF-1 dependent chaperone induction after heat stress. Thus, we propose that HPK-1 preserves HSF-1 activity in C. elegans by inhibiting sumoylation. As a separate output of HPK-1, we showed that HPK-1 (but not HSF-1) is necessary for induction of autophagosome formation and autophagy gene expression in response to dietary deprivation and reduced TORC1 signaling. HPK-1 overexpression both extended longevity and conferred protection against polyQ protein aggregate formation and toxicity. The ability of hpk-1 to increase lifespan and prevent protein aggregation relies on PHA-4(FoxA) and MXL-2(Mlx), Thus, HPK-1 functions as a regulatory hub for multiple transcription factors with proteostasis-preserving activities.
Results
The homeodomain interacting protein kinase HPK-1 extends longevity
We initially identified the hpk-1 gene in an RNAi screen aimed at identifying genes necessary for the extension of longevity in daf-2(e1370) mutant animals. The hpk-1 gene represented an attractive target for subsequent investigation because (1) members of the HIPK gene class are known to have broad physiological roles in transcription factor regulation in response to nutrient availability and other environmental cues, collectively suggesting a central role for HIPKs in stress response pathways across eukaryotes, and (2) initial characterization in our laboratory revealed that hpk-1 promotes the global maintenance of proteostasis by protecting animals from the formation of age-associated polyglutamine protein aggregates, and one of the phenotypic hallmarks of aging organisms is a gradual and progressive decline in proteostasis.
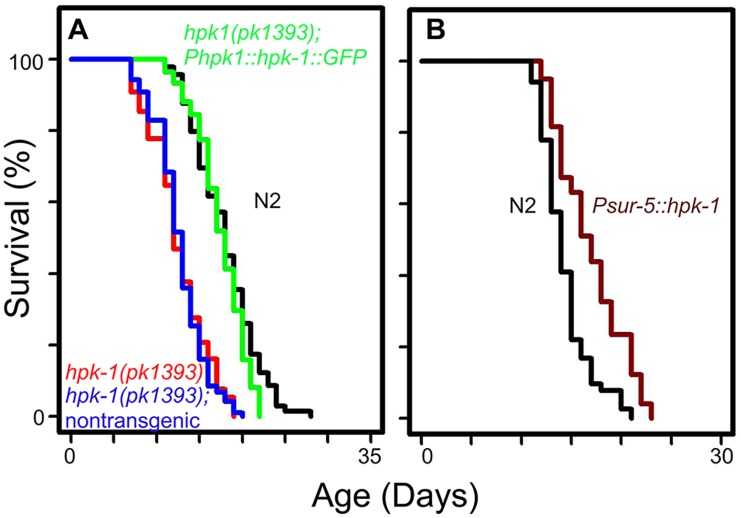
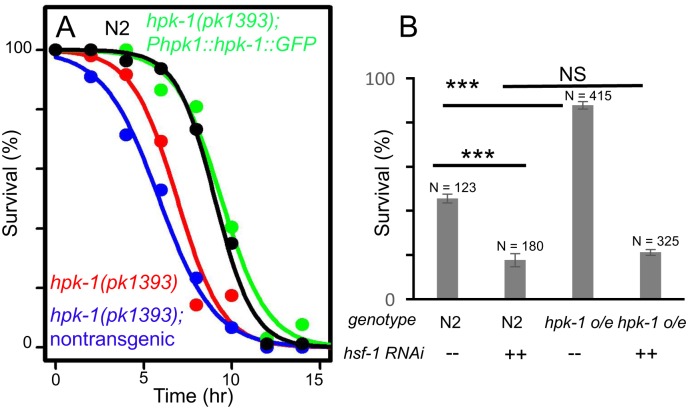
In order to verify our previous observations that hpk-1 RNAi produces a progeric phenotype, we obtained an hpk-1(pk1393) deletion mutant strain that lacks most of the kinase domain and tested whether hpk-1 was essential for normal lifespan. Loss of hpk-1 shortened mean lifespan approximately 30% from 18–21 to 12–14 days (Fig 1A, p<0.0001, S1 Table), in line with a previous study[42]. To verify that the shortened lifespan displayed by hpk-1(pk1393) animals was the result of the hpk-1 deletion and not an independent mutation, we created transgenic animals expressing an HPK-1::GFP translational fusion under control of its own promoter. Inheritance of the Phpk-1::HPK-1::GFP transgene rescued the progeric phenotype of hpk-1(pk1393), consistent with previous reports[42]. In one trial we found that hpk-1 overexpression with the endogenous promoter slightly increased lifespan (S1 Table), contrary to a previous study[42]. Non-transgenic hpk-1(pk1393) siblings remained short-lived (Fig 1A, S1 Table).
Fig 1. hpk-1 activity is necessary and sufficient to regulate lifespan.
(A) hpk-1(pk1393) null mutant animals (red) are short-lived relative to wild-type N2 (black). Phpk-1::hpk-1::GFP largely restores wild-type lifespan to hpk-1(pk1393) mutant animals (green). Lifespan of non-transgenic hpk-1(pk1393) siblings (blue) is similar to hpk-1(pk1393). (B) Overexpression of hpk-1 using the heterologous sur-5 promoter increases lifespan compared to N2 (dark red versus black). Each graph is representative of at least 3 independent lifespan experiments. Full lifespan data can be found in S1 Table.
One concern when studying mutants or gene inactivations that shorten lifespan is that such genes are essential for viability and that their disruption produces a non-specific and overall “sickly” phenotype. However, overexpression of such genes would not be predicted to extend longevity unless they exert broad regulatory control over essential processes or are themselves “rate-limiting” for lifespan (like the heat shock transcription factor hsf-1 and the daf-16/FOXO transcription factor). To determine if hpk-1 is such a gene, we tested whether constitutive overexpression of hpk-1 could increase lifespan by placing it under the control of a strong ubiquitously-expressed promoter (Psur-5). Overexpression of hpk-1 (Psur-5::HPK-1::CFP) increased mean lifespan between 7–16% (Fig 1B, p<0.0001 and S1 Table).
HPK-1 provides protection against polyglutamine aggregate formation and toxicity
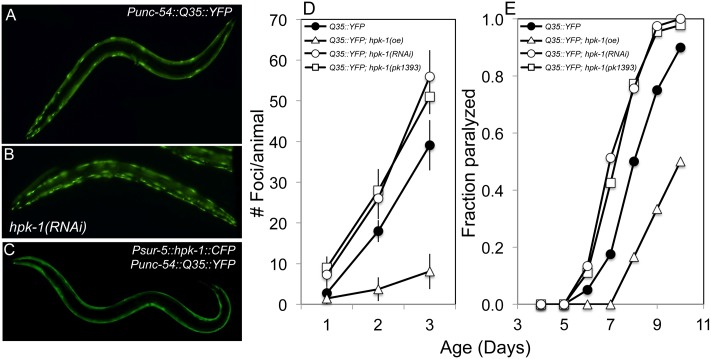
We next tested whether hpk-1 plays a cytoprotective role in maintaining protein homeostasis. Age-associated decline in protein homeostasis can be measured in C. elegans through the visualization of in vivo polyglutamine aggregate formation in muscle cells harboring the Punc-54::Q35::YFP transgene, or later in life as aggregate formation overwhelms the chaperone network and locomotory paralysis ensues[43]. Loss of hpk-1 conferred either by RNAi or the pk1393 deletion resulted in the premature accumulation of fluorescently-labeled polyglutamine Q35::YFP aggregates (Fig 2A–2C, S3 Table p <0.0001 for both comparisons). In the representative trial displayed in Fig 2, on day 2 of adulthood, wild-type animals displayed 18.0+/-2.7 aggregates while the hpk-1(pk1393) null mutant and hpk-1 RNAi-treated Q35::YFP animals averaged 28+/-5.3 and 26.0+/-5.1 aggregates, respectively (Fig 2D, S3 Table). Similarly, by day 8 of adulthood, 77–78% of hpk-1(RNAi) and hpk-1(pk1393) animals were paralyzed while 50% of control Q35::YFP animals were paralyzed (Fig 2E, S3 Table). We next tested whether overexpression of hpk-1 was capable of conferring a cytoprotective phenotype in protein aggregation assays. Overexpression of hpk-1 reduced both Q35::YFP foci accumulation (Fig 2D, S3 Table) and protected aging animals from Q35::YFP-associated paralysis (Fig 2E, S3 Table). Further experiments with additional transgenic lines were largely consistent with these results (S3 Table). Thus, hpk-1 is vital for preserving protein solubility and protecting against aggregate toxicity in adult animals as they age.
Fig 2. HPK-1 promotes protein homeostasis.
(A-C) hpk-1 activity affects the accumulation of Q35::YFP foci in muscle cells. Shown are representative images of Punc-54::polyQ::YFP animals treated with (A) control RNAi or (B) hpk-1 RNAi, and (C) transgenic animals overexpressing hpk-1 (Psur-5::HPK-1::CFP). (D) Time course of polyQ::YFP foci accumulation in conjunction with: treatment with control RNAi (black circles), hpk-1 RNAi (white circles), hpk-1(pk1393) (white squares), or hpk-1 overexpression (open triangles). Data points display the mean +/- standard deviation (S.D.) of at least 15 animals per biological replicate; at least 5 independent experiments were performed. P-values are provided in Results and in S3 Table. (E) Time course of paralysis of Punc-54::polyQ::YFP animals in conjunction with: treatment with control RNAi (black circles), hpk-1 RNAi (white circles), hpk-1(pk1393) (white squares), or hpk-1 overexpression (open triangles). Plotted data display the results for a single biological replicate. P-values and data from all trials are provided in S3 Table.
HPK-1 functions in multiple tissues and developmental stages to control longevity
In order to visualize the spatiotemporal pattern of hpk-1 expression, we analyzed the expression of a Phpk-1::hpk-1::GFP transgene. hpk-1 is expressed broadly during embryogenesis, but becomes more restricted in expression during larval development (S1A Fig). L3-stage larvae display robust expression of the GFP fusion in many head and motor neurons, and lower levels of expression in the intestine and the seam cells of the hypodermis. By late L4 stage, GFP expression is largely restricted to neurons, and is maintained in nerve cells of the head and nerve cord during adulthood, congruent with a previous study[44]. Localization of HPK-1::GFP protein is most concentrated in the nucleus often within distinct sub-nuclear sites (S1B Fig), consistent findings in mammals[45] and the predicted function of HPK-1 as a transcriptional regulator.
Identifying spatiotemporal requirements in longevity control is necessary for understanding how age-associated decline in individual tissues contributes to the larger gestalt of overall animal viability. Thus, we sought to discover where anatomically and when chronologically HPK-1 was essential for a normal lifespan. We first used stage-specific RNAi feeding to test whether the requirement of hpk-1 for normal longevity was restricted to a particular life stage. To assess whether larval-specific activities of hpk-1 are critical for normal adult lifespan, animals were raised on hpk-1 RNAi during development, and were transferred to dcr-1 RNAi at the late L4 stage in order to terminate continued silencing of hpk-1 by RNAi. Animals raised on RNAi bacteria targeting hpk-1 during larval development exhibited a shortened lifespan similar to lifelong inactivation of hpk-1 (S2A and S2B Fig), while adult-restricted inactivation of hpk-1 displayed a weaker progeric phenotype (S1C Fig). Interestingly, these temporal requirements are essentially identical to those previously described for hsf-1[46]. Given the broad developmental expression of hpk-1, we next sought to define the tissues where hpk-1 acts to promote normal longevity. Tissue-restricted RNAi of hpk-1 in the intestine or the hypodermis both caused a significant progeric phenotype (S2D and S2E Fig), consistent with intestinal and hypodermal expression being limited to larval developmental stages. In contrast, inactivation of hpk-1 in muscle cells had little effect on lifespan (S2F Fig), consistent with the absence of HPK-1 expression in these cells as determined with our fluorescent reporter (S1A Fig, and S1 File).
We next tested whether neuronal hpk-1 function was necessary for normal lifespan using an enhanced neuronal RNAi (RNAi(en)) strain, as RNAi efficiency in neurons is low in wild-type animals. Neuronal inactivation of hpk-1 showed reduced lifespan to an extent comparable to inactivation of hpk-1 by systemic RNAi (S2G Fig) and hpk-1 null mutant animals (Fig 1A). As a positive control to confirm RNAi(en) activity, daf-2(RNAi) significantly increased lifespan in the RNAi(en) strain (S2H Fig) while a control strain lacking the dsRNA channel sid-1 and enhanced neuronal RNAi did not (S2I Fig), consistent with previous reports[47, 48]. Thus hpk-1 is required across all of the tissues in which we have observed its expression during the larval stages of development to ensure wild-type lifespan. Because HPK-1::GFP expression is restricted to neurons in adult animals (S1A Fig), we interpret the longevity-extending activity of HPK-1 observed in the intestine and hypodermal seam cells (S2D and S2E Fig) to arise largely from larval-stage functions of HPK-1 in those tissues, although hpk-1 does have modest longevity-extending effects in adulthood as well (S2C Fig).
HPK-1 is a member of the HSF-1 longevity pathway
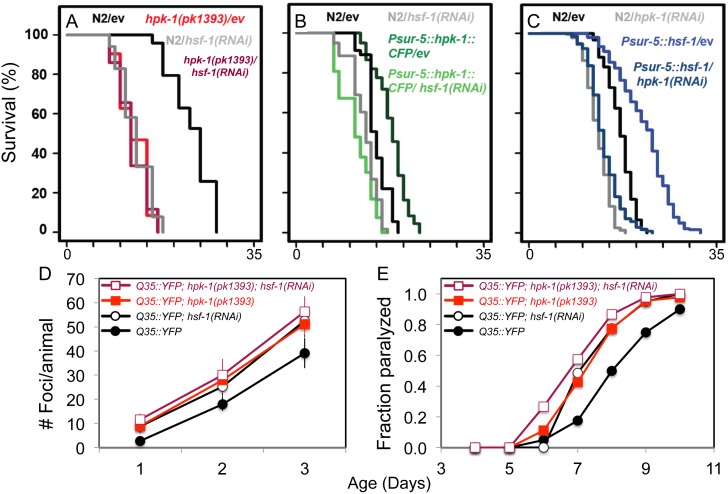
Based on a pilot screen to identify putative genetic interactions between known longevity genes and hsf-1 loss of function, we investigated the extent to which hpk-1 exerts its effects on overall longevity and proteostasis via hsf-1. We measured the extent to which hpk-1 exerts its effects on overall longevity and proteostasis via the HSF-1 pathway by examining whether their loss-of-function phenotypes were additive. We observed that hsf-1 inactivation in hpk-1(pk1393) null mutant animals did not result in a meaningful additional decrease in lifespan (Fig 3A). Further, inactivation of hsf-1 by RNAi was sufficient to suppress the increased lifespan of the long-lived hpk-1 overexpression line (Fig 3B). Conversely, hpk-1 was necessary for the extended longevity observed in hsf-1-overexpressing animals (Fig 3C). The reciprocal requirement we observed between hpk-1 and hsf-1 for animal longevity is evidence of a genetic interaction between these factors. Next we tested whether hpk-1 and hsf-1 function together or in separable pathways to protect animals from Q35::YFP foci formation by comparing animals lacking both hpk-1 and hsf-1 to animals deficient in only one of these genes. As expected, loss of hpk-1 or hsf-1 alone resulted in the premature accumulation of protein aggregates and onset of paralysis (Fig 3D and 3E). Inactivation of hsf-1 by RNAi in the absence of hpk-1 failed to produce a statistically detectable increase in the accumulation of foci or onset of paralysis over time (Fig 3D and 3E). Additional experiments corroborated these results: hpk-1 RNAi-treatment alone had as great or a greater negative impact on proteostasis when compared to hsf-1 RNAi treatment (S3 Table). These results are consistent with the notion that hpk-1 and hsf-1 function to maintain protein homeostasis and delay the progression of aging through a shared mechanism.
Fig 3. hpk-1 and hsf-1 have overlapping functions in longevity control and the preservation of proteostasis.
(A) hpk-1(pk1393) null mutant animals are short-lived compared to wild-type controls (red versus black) and hsf-1(RNAi) does not further shorten hpk-1(pk1393) lifespan (maroon versus gray). (B) Overexpression of hpk-1 increases lifespan (dark green versus black) and is hsf-1 dependent (light green). (C) Constitutive hsf-1 overexpression increases lifespan (blue versus black) and is hpk-1 dependent (light blue). Each graph is representative of at least 3 independent lifespan experiments. Full lifespan data can be found in S1 Table. (D-E) Simultaneous loss of hpk-1 and hsf-1 does not produce additive detrimental effects on protein homeostasis (maroon versus red). (D) Time course of Punc-54::Q35::YFP foci accumulation in conjunction with: treatment with control RNAi (filled circles/squares), hsf-1 RNAi (white circles/squares) in either wild-type (black traces) or the hpk-1(pk1393) null mutant background (colored traces). Data are the mean +/- S.D. of at least 15 animals from one representative trial; at least 5 independent experiments were performed. P-values are provided in Results and S3 Table. (E) Time course of paralysis of Punc-54::polyQ::YFP animals after treatment with control RNAi (filled circles/squares) or hsf-1 RNAi (white circles/squares) in either wild-type (black traces) or the hpk-1(pk1393) null mutant background (colored traces). Data is representative of one biological replicate, with at least 5 independent replicates performed. P-values and data from all trials are provided in S3 Table.
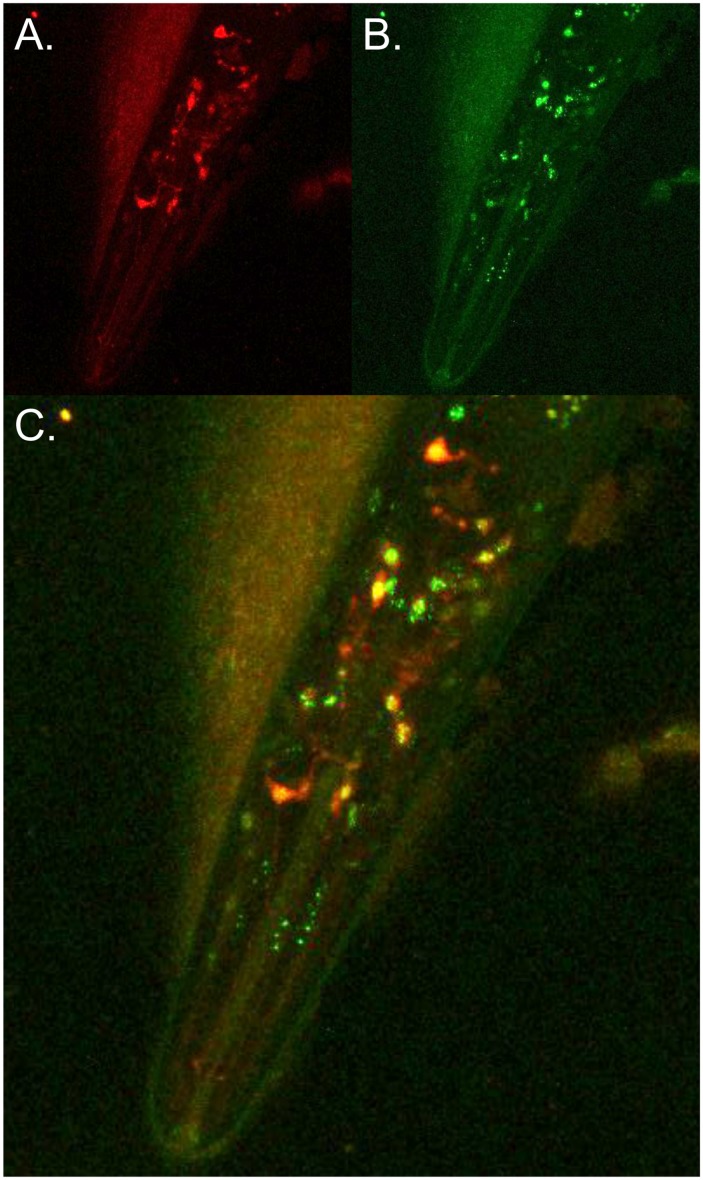
That homeodomain interacting protein kinases function as direct regulators of transcription factor activity suggested that the interaction between HPK-1 and HSF-1 may be direct. To begin to explore this possibility, we examined whether HPK-1 co-localizes with HSF-1 at the subcellular level by comparing localization of a Phsf-1::hsf-1::GFP transgene to a translational fusion between hpk-1 and the fluorescent tdtomato protein (Phpk-1::hpk-1::tdtomato) using confocal microscopy (Fig 4). Though not perfectly overlapping, HPK-1 and HSF-1 localization were often coincident with each other.
Fig 4. HPK-1 colocalizes with HSF-1 in C. elegans neurons.
(A-C) HPK-1 and HSF-1 colocalize in neurons under basal conditions. Representative image of transgenic animal co-expressing Phpk-1::HPK-1::tdtomato,Phsf-1::HSF-1::GFP: (A) red fluorescence, (B) green fluorescence, and (C) overlay.
We next sought to determine if hpk-1 shares functionality with HSF-1 with respect to the heat shock stress response by examining its role in regulating the transcriptional induction of chaperone gene expression after exposure to heat. We initially tested whether hpk-1 is important for preserving thermotolerance in response to heat shock. We observed that survival at 35°C was significantly reduced in hpk-1(pk1393) mutant animals, and that thermotolerance could be restored to wild type levels by the rescuing hpk-1 transgene (Phpk-1::hpk-1::GFP) (Fig 5A). Furthermore, we observed that the long-lived hpk-1 overexpression line (Psur-5::hpk-1::CFP) increased thermotolerance survival from ~50% in wild type animals to ~75%, and that this increase in thermotolerance was fully dependent on hsf-1 (Fig 5B).
Fig 5. hpk-1 regulates thermotolerance.
(A) hpk-1 is required for normal thermotolerance, as hpk-1(pk1393) has impaired survival at 35°C (red vs. black), which is rescued by transgenic Phpk-1::hpk-1::GFP (green). Graph represents survival data from one representative trial of three independent replica set experiments. Additional trials can be found in S2 Table; see methods and [38, 72] for additional details and statistical analysis. (B) hpk-1 overexpression increases survival to thermal stress (column 3 versus 1), dependent on hsf-1 (column 4 versus 2). Plotted data show mean and S.E.M. of a representative trial (trial 1) with 3–4 technical replicates per condition; # above each column represents total number of animals scored per condition. See S2 Table for additional details. *** indicates p-values of <0.001. P-values were calculated using ANOVA with Tukey’s HSD post-hoc, and were corrected to account for multiple testing. See S2 Table for additional trial data.
HPK-1 prevents sumoylation of HSF-1
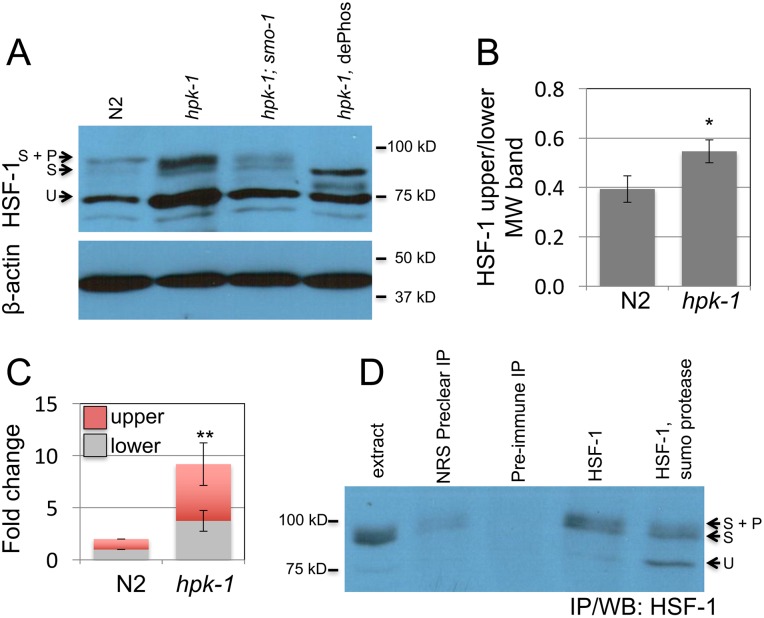
HSF-1 transcriptional activity is regulated in mammals through a complex array of post-translational modifications including phosphorylation, acetylation, and sumoylation (reviewed in[41, 49]). We sought to determine whether loss of hpk-1 altered either expression levels and/or post-translational modifications to the HSF-1 protein. Unmodified HSF-1 displayed the predicted mobility of a ~75 kD protein[50]. We additionally observed in wild-type animals two higher molecular weight isoforms between ~90 and 95 kD (Figs 6A, S3). Loss of hpk-1 resulted in an increase in the ratio of higher molecular weight isoforms of HSF-1 to the unmodified 75 kD species (Figs 6B, S3B) and an increase in overall levels of HSF-1 protein relative to the β-actin control (Figs 6A and 6C, S3C).
Fig 6. hpk-1 prevents sumoylation of HSF-1.
(A) Changes in HSF-1 post-translational modifications between early L4 wild-type and hpk-1(pk1393) animals were examined by western blot to HSF-1; smo-1(RNAi), which targets C. elegans SUMO, was used to block sumoylation, dePhos is lambda protein phosphatase treatment (other samples were mock treated). Beta-actin serves as a loading control. The ratio of modified to unmodified HSF-1 is 0.35, 0.51, and 0.35 for N2/ev, hpk-1(pk1393)/ev, and hpk-1(pk1393)/smo-1(RNAi), respectively (see S3A Fig for additional data). (B) hpk-1 prevents sumoylation of HSF-1. Ratio of HSF-1 unmodified (75kD) to modified (90-95kD, sumoylated and sumoylated plus phosphorylated). The S.E.M. from Image J quantification is shown for seven N2 and hpk-1(pk1393) replicates (* p<0.02, Student’s t-test, see S3B and S3C Fig for additional data). (C) Loss of hpk-1 induces HSF-1 in L4 animals. Protein levels of unmodified (75kD, grey), modified (90-95kD, red) HSF-1, and beta-actin were quantified in ImageJ for four replicates (see S3C Fig for additional data). (For total, upper, and lower MW fold change (respectively): ** p<0.01, <0.01, and <0.02, Student’s t-test). (D) Immunoprecipitation of HSF-1 from hpk-1(pk1393) animals followed by SUMO protease or mock treatment. First lane is protein extract prior to immunoprecipitation. Lysate was precleared of nonspecific interactions by treatment with normal rabbit serum (Invitrogen #01–6101) that was then immunoprecipitated (lane 2). Precleared lysate was then evenly divided and treated with either preimmunized rabbit serum (i.e. serum from the rabbit in which C. elegans HSF-1 antibodies were generated, prior to immunization, lane 3) or HSF-1 antiserum, followed by immunoprecipitation. Immunoprecipitated HSF-1 was then either mock treated or treated with SUMO protease (lanes 4 and 5). See methods for additional details.
Sumoylation typically results in an electrophoretic mobility upshift of ~15 kD[51]. Mammalian HSF-1 has been shown to be sumoylated[40]. Consequently, we hypothesized that the two higher MW isoforms of HSF-1 might represent a sumoylated product (S isoform) (Figs 6A and S3, lower ~90 kD MW band), and SUMO plus phosphorylation (S+P isoform) (Figs 6A and S3, upper ~95 kD MW band). Consistent with this hypothesis, lambda protein phosphatase treatment of hpk-1 null extracts resulted in the loss of the highest MW isoform of HSF-1 but not the ~90 kD isoform (Figs 6A, S3A), suggesting that the 90 kD isoform is not a result of phosphorylation events. This result confirms the hypothesis that HSF-1 phosphorylation is a modification of the 90 kD isoform. However, the 95 kD band is still observed in an hpk-1 mutant, indicating that HPK-1 is not the kinase responsible for this phosphorylation event. We determined that the higher molecular weight isoforms likely represent sumoylated forms of HSF-1 by two approaches. First, hpk-1 null mutant animals were grown on smo-1 RNAi, which reduces expression of the C. elegans SUMO gene that produces the SUMO moiety. smo-1 RNAi of hpk-1 mutant animals resulted in a decrease in the ratio of the pair of high MW bands to unmodified HSF-1 at 75 kD. Second, HSF-1 was immunoprecipitated and treated with SUMO protease, which resulted in a relative increase in the 75 kD (unmodified) HSF-1 isoform and a relative decrease in the 90–95 kD (sumoylated) bands of HSF-1 (Fig 6D). While the loss of the sumoylated species was incomplete, each result is consistent with our prediction that the higher molecular weight isoforms of HSF-1 are the result of sumoylation. Thus, HPK-1 acts directly or indirectly to oppose HSF-1 sumoylation, either by blocking sumoylation or promoting de-sumoylation.
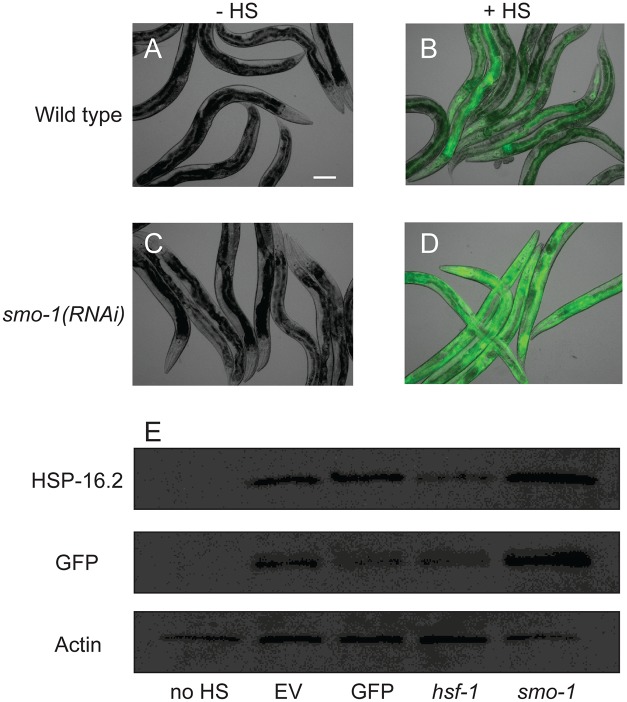
We then tested our hypothesis that sumoylation is an inhibitory modification on HSF-1 by analyzing induction of the HSF-1 transcriptional program in response to heat shock in animals grown on smo-1 RNAi (Fig 7). In response to thermal stress, smo-1 RNAi-treated animals harboring the Phsp-16.2::GFP transgene displayed enhanced induction of GFP relative to empty vector control animals (Fig 7A–7D). It is worth noting that in the absence of heat shock, the Phsp-16.2::GFP is not induced, indicating that loss of sumoylation is not sufficient of itself to activate the HSF-1 transcriptional response. In a complementary experiment, we analyzed protein induction of GFP expressed from the hsp-16.2 promoter as well as the endogenous HSP-16.2 protein in response to heat shock (Fig 7E). Prior to heat shock, animals were raised on RNAi targeting GFP, hsf-1, smo-1 or the empty vector control. In response to heat shock, both GFP and HSP-16.2 were induced. GFP but not HSP-16.2 induction was blocked by GFP(RNAi). Both GFP and HSP-16.2 induction were significantly reduced by hsf-1(RNAi), confirming that induction of these proteins was dependent on the presence of HSF-1. Finally, smo-1(RNAi) increased the level of GFP and HSP-16.2 protein when compared to the EV control, supporting our hypothesis that HSF-1 sumoylation is a modification that is likely to inhibit the HSF-1 transcriptional program in response to heat shock.
Fig 7. Heat shock induction of hsp-16.2 is enhanced by smo-1(RNAi).
(A-D) DIC and GFP overlay for Phsp-16.2::GFP worms on empty vector (A, B) or smo-1(RNAi) (C, D) with (+HS) and without (-HS) heat shock. Scale bar = 100μm. (E) Western blot for HSP-16.2, GFP and β-actin from hsp-16.2p::GFP worms grown on empty vector (EV) without heat shock (no HS) or with heat shock (EV), GFP(RNAi), hsf-1(RNAi) or smo-1(RNAi). Fold-increases for HSP-16.2/actin on smo-1(RNAi) relative to EV in three independent replicates were 2.4, 4.7, and 1.8.
Hormetic extension of natural longevity requires both hsf-1 and hpk-1
In general, the sustained hardship of chronic exposure to an environmental stressor, or application of a stressor to a level above a physiologically tolerable threshold typically compromises organism viability (and consequently limits organismal lifespan). Hormesis is the initially counterintuitive but now well-validated and generalizable phenomenon produced by acute exposure to an environmental stressor. Brief intervals of exposure to stressors (e.g. heat, ROS, etc.) can confer a “hormetic effect” characterized by a significant extension of longevity accompanied by sustained resistance to physiologic stress. For instance, a transient 30°C pulse during development and/or early adulthood increases lifespan in C. elegans, while sustained growth at 25°C hastens the onset of animal morbidity[52]. It is generally postulated that the mechanism of extended longevity following heat shock is derived from upregulation of chaperone systems and turnover of misfolded proteins, which consequently promotes long term proteostatic robustness/resiliency once heat stress is removed[53]. HSF-1 is necessary for the hormetic effect of transient heat exposure on lifespan[54].
We tested whether hpk-1 was required for heat-induced hormetic extension of longevity. While defining experimental conditions for heat-induced hormesis, we found that growth at 25°C for the first 3 days of adulthood is a form of mild heat stress that to our knowledge has not been previously documented in C. elegans (S4A–S4F Fig). We subsequently observed that transient growth at 25°C during this interval in early adulthood (when heat-resistance mechanisms at 20°C normally undergo a dramatic downregulation[29, 30, 55]) was sufficient to increase lifespan only in the presence of hsf-1 and hpk-1 (S4G and S4H Fig, p<0.001). Thus hpk-1, like hsf-1, is necessary for hormetic extension of longevity in response to heat stress, a result consistent with a positive regulatory function of HPK-1 over HSF-1.
HPK-1 is an essential component of the heat shock response
hipk family members canonically function as positive regulators of transcriptional co-activators. If the interaction between HPK-1 and HSF-1 is direct, hpk-1 could be promoting HSF-1 activity at various regulatory points in the chain of events beginning with newly translated HPK-1 and ending with induction of gene transcription by HSF-1. For example, post-translational modification of HSF-1 by HPK-1 could affect its stability, subcellular localization, DNA binding or transactivation activity at the level of recruitment of RNApol II, additional transcription factors, or chromatin modifying factors. We undertook several experiments to identify mechanisms of regulatory control.
Immediately following thermal stress, acute cytoplasmic misfolding challenges the chaperone network to free HSF-1 from Hsp90 cytoplasmic sequestration, allowing HSF-1 to undergo translocation to the nucleus, where it becomes concentrated in stress granule-like subnuclear speckles that colocalize with markers of active transcription[56]. Re-allocation and compartmentalization of HSF-1 to nuclear speckles occurs within minutes of initiating thermal stress ([57] and this study). When we tested whether hpk-1 was necessary for HSF-1 re-localization or compartmentalization in response to heat shock, we found that these early events of HSF-1 activation are hpk-1 independent, as hpk-1(RNAi) had no effect on either readout of HSF-1 activation (S5 Fig). Therefore hpk-1 is likely to play a role in subsequent events during activation and/or establishment of the HSF-1 transcriptional response.
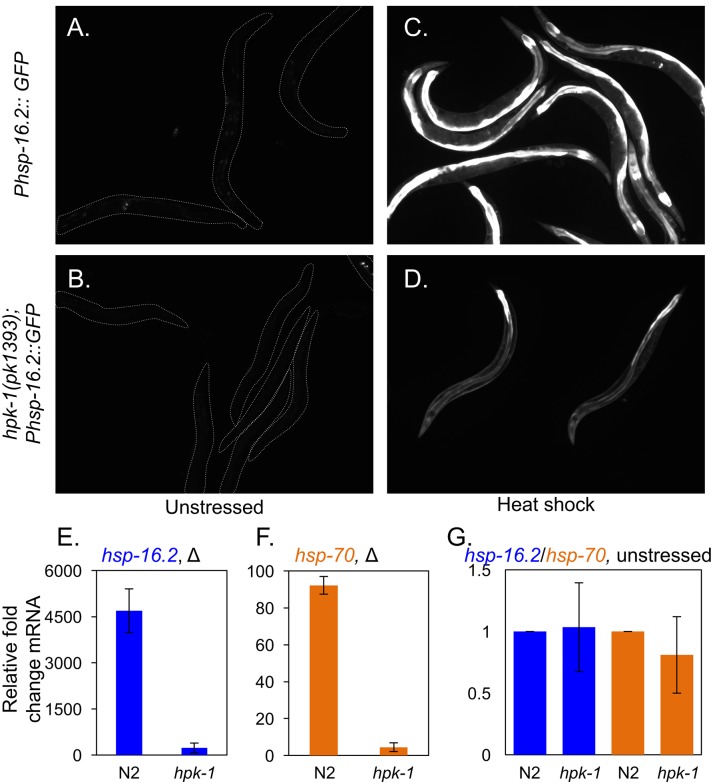
We tested whether hpk-1 was necessary for the transactivation of HSF-1 by determining whether the induction of chaperone target genes in response to thermal stress was compromised in the absence of hpk-1. We first analyzed whether hpk-1 was required for induction of the Phsp-16.2::GFP reporter. hsp-16 encodes a small chaperone that is induced by heat shock in a manner requiring hsf-1[58]. We found that hpk-1 was also necessary for hsp-16-2::GFP induction in response to transient heat shock (Fig 8A–8D) as previously shown[42]. In addition, we found that hsf-1-dependent transcriptional induction of the endogenous chaperones encoded by hsp-16.2 and hsp-70 also required the presence of hpk-1 for heat shock inducibility (Fig 8E and 8F), which unexpectedly differs from a previous report[42]. HPK-1 regulation of chaperone gene expression is dependent on heat stress, as loss of hpk-1 did not significantly alter basal expression levels of hsp-16.2 and hsp-70 (Fig 8G). In contrast, hsf-1 inactivation has been reported to reduce endogenous levels of hsp-16.2 and hsp-70 mRNA by ~40%[50], suggesting some basal HSF-1 activity remains in the absence of hpk-1.
Fig 8. hpk-1 regulates induction of the heat shock response.
(A-D) Representative images of at least 30 Phsp-16.2::GFP animals in wild type (A, C) and hpk-1(pk1393) animals (B, D) under basal conditions (A, B) or after heat shock (C, D). Animals are outlined in panels A-B. (E-F) Induction of endogenous hsp-16.2 (E) and hsp-70 (C12C8.1) (F), in N2 and hpk-1(pk1393) animals as measured by qRT-PCR. (G) Loss of hpk-1 did not alter endogenous expression of hsp-16.2 (blue) or hsp-70 (orange). Values in (E-G) are normalized to expression of act-1 and the mean fold change relative to wild-type animals, and the S.E.M. between technical replicates is shown. In total three independent experiments were performed with similar results. P-values for (E) and (F) are <0.05 and <0.01, respectively (Student’s t-test).
We also considered the possibility that HSF-1 functions upstream rather than downstream of HPK-1, or as part of a feedback loop with HSF-1. We examined HPK-1 expression in response to multiple conditions of stress including heat shock, and tested whether HPK-1 expression is regulated by HSF-1. Under basal conditions, transgenic animals that express a GFP translational fusion that includes the hpk-1 open reading frame (Phpk-1::hpk-1::GFP) displayed a broad pattern of developmental expression in the intestine, hypodermal seam cells, and neurons. The expression pattern of this transgene became restricted to neurons as animals transitioned to adulthood (Figs 9A, S1A). We next tested whether the pattern of hpk-1 expression is regulated by thermal stress. We observed robust induction of hpk-1 expression in transgenic animals expressing the translational reporter after heat shock (Fig 9B). Induction of HPK-1 was greatest in hypodermal seam cells, neurons, and to a much lesser extent within intestinal cells (Figs 9B, S5), and this induction did not require hsf-1 (Fig 9C).
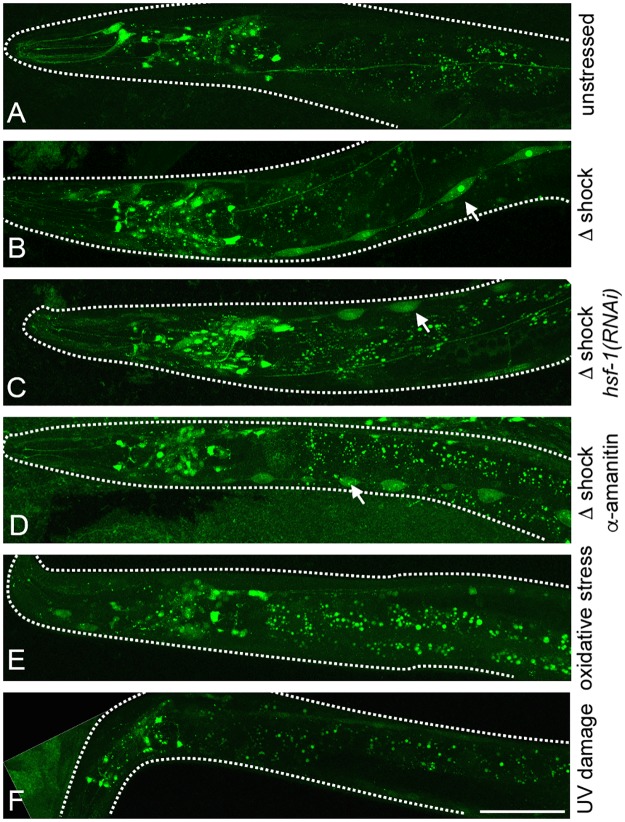
Fig 9. hpk-1 protein expression is stimulated by heat shock.
(A) Expression of HPK-1 protein (Phpk-1::HPK-1::GFP) under basal conditions is primarily restricted to neurons. Fluorescent intestinal speckles are non-specific gut granules[111]. (B-D) Heat shock induces HPK-1 protein (Phpk-1::HPK-1::GFP) levels most strongly within hypodermal seam cells (indicated by arrows) independent of hsf-1 (C) and transcription (D). Phpk-1::HPK-1::GFP animals after heat shock with (B) empty vector HT115, (C) hsf-1(RNAi), and (D) α-amanitin treatment. Increased HPK-1 expression within neurons and hypodermal seam cells is specific to heat stress as neither oxidative stress (E) or UV damage (F) altered expression. White space was artificially filled for some images and animals are outlined. GFP quantification and further analysis can be found in S6 Fig.
We asked whether HPK-1 induction was transcriptional or post-transcriptional, as early events in many stress response pathways including the heat shock response do not require active transcription. Consistently, we could not discern an increase in fluorescence in transgenic animals expressing a transcriptional fusion of the hpk-1 promoter to GFP (Phpk-1::GFP) (S7A and S7B Fig). However, we did notice that hypodermal seam cells appeared much larger and swollen compared to unstressed controls (S7A and S7B Fig, S1 File). As extrachromosomal transgenic reporter lines lack both normal gene copy number and the context of endogenous chromatin, we further examined hpk-1 mRNA levels in wild-type animals by qRT-PCR but found no significant difference in mRNA expression as a function of heat shock (S7C Fig). Importantly, endogenous hsp-16.2 mRNA was induced in the heat stressed sample (S7D Fig). Because hsf-1 is a global activator of chaperones and other heat shock response genes, we asked whether the thermal inducibility of HPK-1 translation requires hsf-1. Consistent with a mode of post-transcriptional regulation of hpk-1 in response to heat shock, hsf-1(RNAi) had no effect on the induction of the translational Phpk-1::hpk-1::GFP reporter after heat shock (Fig 9C) and induction was not blocked by pre-treatment with the RNA polymerase inhibitor α-amanitin (Fig 9D). In contrast, α-amanitin pre-treatment completely blocked the induction of the known transcriptional Phsp-16.2::GFP reporter in response to heat shock (S8 Fig). Induction of HPK-1 protein is specific to thermal stress, as oxidative damage (by tert-butyl hydroperoxide) (Fig 9E) and DNA damage (by UV) (Fig 9F) failed to alter Phpk-1::HPK-1::GFP levels or its pattern of expression. Thus, HPK-1 is specifically induced by thermal stress and this induction is post-transcriptional.
HPK-1 is inhibited by TORC1 and is necessary for TORC1-associated longevity
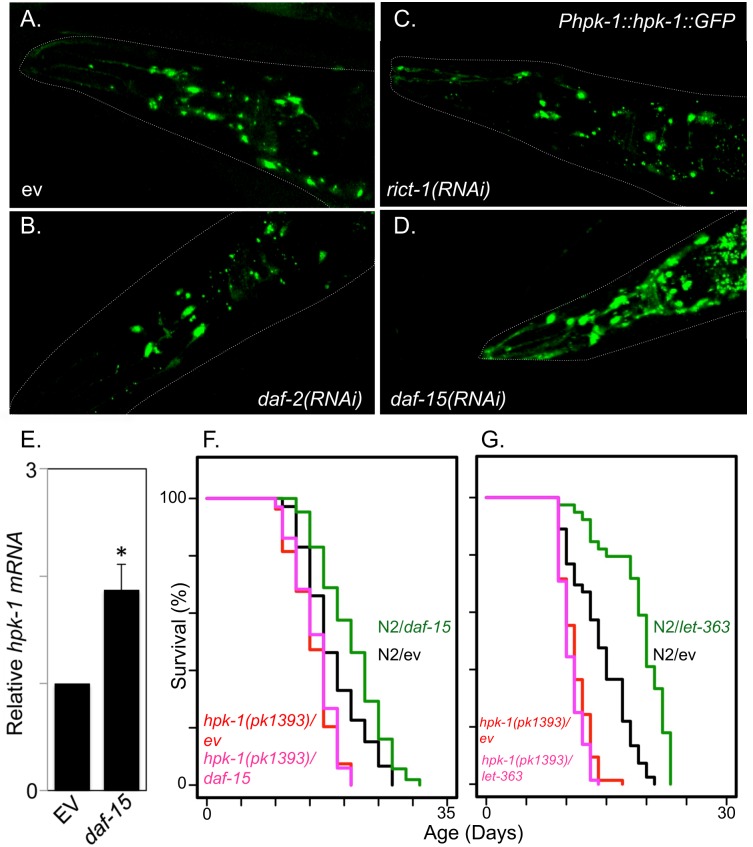
In addition to heat shock, DNA damage, and oxidative stress, another type of stress that contributes to longevity is metabolic or nutritional stress. Many longevity transcription factors linked to environmental stress responses are also responsive to changes in nutritional status. For example, HIPK2 induction under conditions of glucose deprivation has been described in mammalian cell culture[59]. This suggested that HPK-1 expression might be inhibited by the insulin-like receptor (daf-2) and/or the TOR signaling complex, both of which control key nutritional responses in C. elegans and extend longevity when disrupted. We tested whether RNAi inactivation of daf-2, daf-15 (corresponding to raptor, a TORC1 subunit), or rict-1 (corresponding to rictor, a TORC2 subunit) altered HPK-1 expression. We observed no change in expression of the Phpk-1::HPK-1::GFP reporter in response to daf-2 or rict-1 inactivation, but we did observe a significant increase in HPK-1::GFP protein expression in head neurons in daf-15(RNAi) animals (Figs 10A–10D, S8A). HPK-1 induction was distinct mechanistically from heat shock induction of HPK-1 in two ways: 1) hpk-1 mRNA levels were increased by inactivation of daf-15 (Fig 10E) but not heat shock (S7C Fig), and 2) HPK-1 induction by daf-15 RNAi treatment was restricted to neurons whereas heat shock induced a broader expression pattern (S9B–S9D Fig). daf-15(RNAi) and let-363(RNAi) (TOR kinase) have been shown to increase lifespan in wild-type animals[60, 61]. We subsequently asked whether hpk-1 was necessary for the enhanced longevity of either daf-15(RNAi) or let-363(RNAi)-treated animals. We observed suppression of the extended longevity phenotype of daf-15(RNAi) or let-363(RNAi)-treated animals to the lifespan observed in hpk-1 mutant animals alone (Fig 10F and 10G respectively), suggesting that hpk-1 may be an inhibitory target of TORC1 that is critical for changes in longevity mediated by altered TOR signaling.
Fig 10. Decreased TORC1 activates HPK-1 to extend longevity.
(A-D) Decreased TORC1 induces neuronal expression of HPK-1. Representative images of Phpk-1::HPK-1::GFP neuronal expression in day 3 adult animals after treatment with control (A), daf-2 (B), rict-1 (C) and daf-15 RNAi (D). Outlines of animals are traced in white. White space was artificially filled for (D). Additional images and quantification can found in S9 Fig. (E) Induction of endogenous hpk-1 after daf-15 inactivation as measured by qRT-PCR. Values are mean fold change and S.E.M. Three independent experiments were performed and normalized to cdc-42. * indicates a p-value <0.05 (Student’s t-test). (F, G) hpk-1 is essential for increased lifespan of daf-15(RNAi) and let-363(RNAi)-treated animals, respectively. Tabulated lifespan is provided in S1 Table.
HPK-1 promotes autophagy in response to dietary restriction and inactivation of TOR
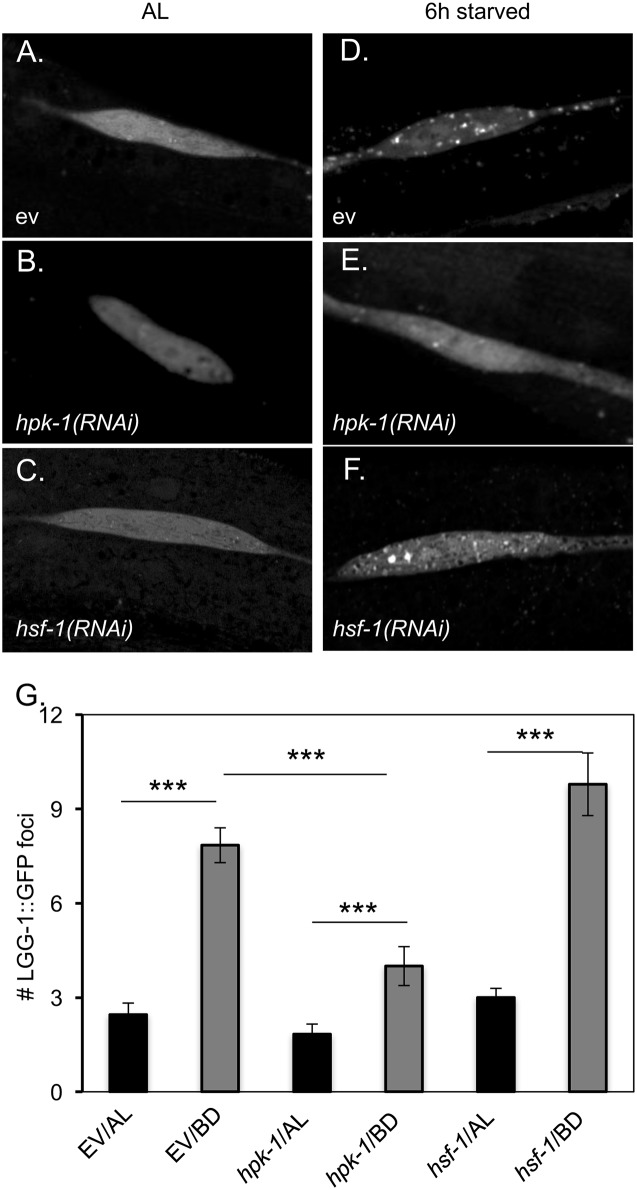
Under nutrient rich conditions, TOR promotes cellular growth by activating protein translation (e.g. transcription of translation components) while inhibiting protein turnover (e.g. transcription of chaperones[62, 63] and autophagy genes[64], and the initiation of autophagy[65]). TOR inhibition, or genetic activation of any of these targets of TOR inhibition, results in extension of longevity[18, 60]. We tested whether regulation of any of these cellular processes was dependent on hpk-1. Autophagy is induced in response to fasting across many species, and can be visualized in C. elegans using the LGG-1::GFP reporter, in which GFP is C-terminally fused to the autophagosome component LGG-1 (i.e. LC3/Atg8 in mammals and yeast, respectively). Stimulation of autophagy is observed in epidermal seam cells during fasting as the formation of discrete LGG-1::GFP puncta[18]. We tested whether hpk-1 was necessary for autophagosome formation following six hours of bacterial deprivation (BD) relative to replete, or ad libitum (AL) conditions. Consistent with published reports, LGG-1::GFP foci were rarely observed under ad libitum conditions (Fig 11A–11C), but were readily observable in seam cells upon bacterial deprivation (Fig 11D). We observed a near total loss of LGG-1::GFP foci formation in response to BD in hpk-1(RNAi)-treated animals (Fig 11E). Interestingly, LGG-1::GFP foci formation following bacterial deprivation did not require hsf-1 (Fig 11F and 11G). This result reveals that autophagosome formation in hypodermal seam cells constitutes a biological function for HPK-1 that is separable from its role in regulating HSF-1 activity and HSF-1-dependent proteostatic outputs.
Fig 11. HPK-1 but not HSF-1 is essential for autophagosome formation.
(A-F) Inactivation of hpk-1 but not hsf-1 disrupts autophagosome formation after bacterial deprivation (BD) as visualized by puncta formation for the autophagosomal reporter Plgg-1::LGG-1::GFP (Atg8p/MAP-LC3) [18]. (G) Quantification of LGG-1:GFP foci in L3 stage animals under ad libitum (AL) and bacterial deprivation (BD) conditions. BD was imposed by removal from bacterial food for 6 hours prior to scoring puncta formation. Plotted are the mean number of LGG-1::GFP puncta/seam cell visualized +/-S.D. *** indicates a p-value of <0.001 (Student’s t-test). Summary data provided in S4 Table.
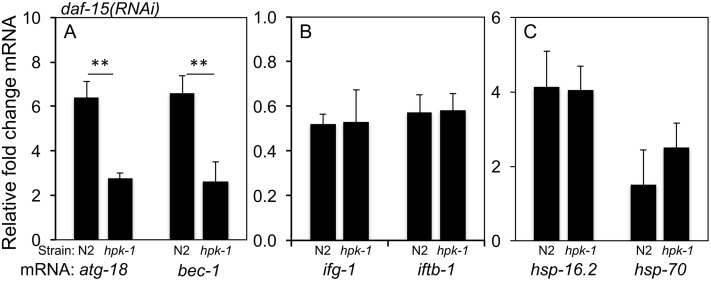
Because HIPKs are transcription factor regulators, we tested whether any of the genes negatively regulated by TOR require hpk-1 for their expression. We observed that hpk-1 is necessary for the induction of two autophagy genes (atg-18 and bec-1) previously reported to be induced in response to daf-15 inactivation and that are essential for autophagosome production (Fig 12A)[18, 20, 64, 66]. In contrast, loss of hpk-1 had no effect on daf-15(RNAi)-induced down regulation of ifg-1 and iftb-1 (eIF-4G and eIF2beta, respectively), two translation initiation genes whose transcription is induced by active TORC1 (Fig 12B). Similarly, loss of hpk-1 had no effect on the modest induction of molecular chaperones that occurs with TORC1 inactivation (Fig 12C), an induction known to be hsf-1 dependent[63]. Therefore, a previously described interaction between TORC1 and HSF-1 leading to chaperone transcriptional induction is hpk-1-independent. Conversely, HPK-1 functions specifically in the autophagy axis of TORC1 signaling while hsf-1 does not. Therefore, HPK-1 and HSF-1 must each have cellular functions that are distinct from each other in addition to their shared control of heat shock responses.
Fig 12. HPK-1 is essential for the transcriptional activation of autophagy.
(A) hpk-1 is necessary for the induction the autophagy genes atg-18 and bec-1 (Beclin1) in response to inactivation of TORC1 by daf-15(RNAi) (** indicates p<0.01, Student’s t-test). (B) In contrast, decreased TORC1 signaling represses the expression of the translation initiation factor genes ifg-1 and iftb-1 independently from hpk-1. (C) Similarly, TORC1 inhibition mildly induces hsp-16.2 and hsp-70 independently from hpk-1. Columns labeled hpk-1 indicate hpk-1(pk1393). Expression levels are presented as fold change +/- S.D. normalized to cdc-42 and averaged across four independent experiments.
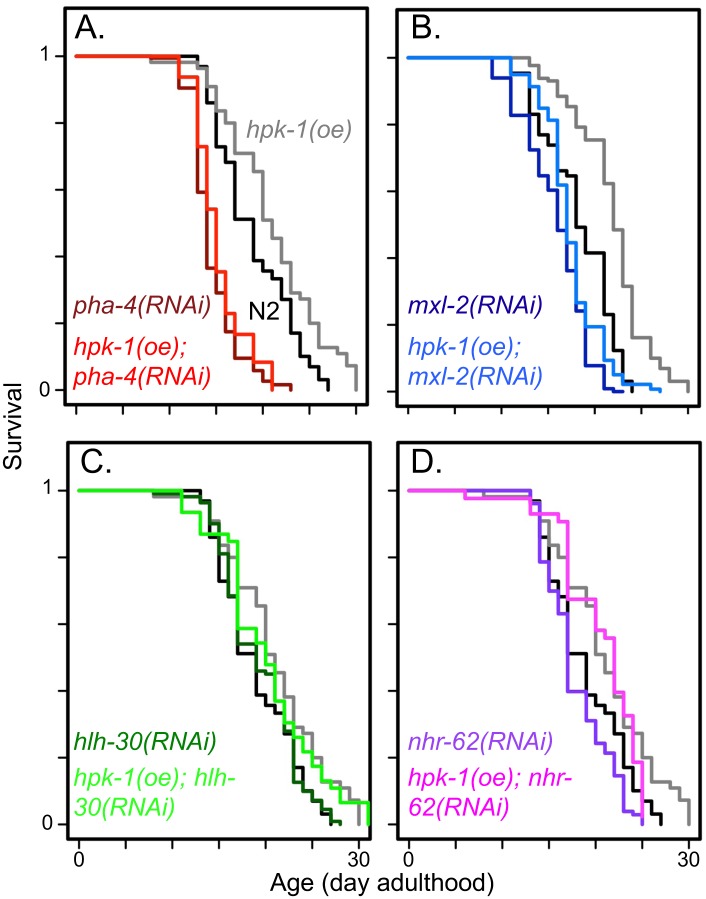
Our result that HPK-1 promotes autophagy through a mechanism independent of HSF-1 suggests that HPK-1 regulates at least one additional transcription factor that is necessary for autophagy induction in response to DR or TORC1 inactivation. PHA-4/FoxA, HLH-30/TFEB, NHR-62/HNF4-related nuclear hormone receptor are transcription factors known stimulate autophagosome assembly[18, 22, 64, 66–68]. MXL-2/Mlx (part of the MML-1/(MondoA/ChREBP) complex) has been implicated in autophagy regulation because it is the binding partner of MML-1, which promotes autophagy gene expression upon inactivation of TOR[21]. As we are primarily concerned with those functions connecting TOR and HPK-1 activity to lifespan, we examined whether the increased lifespan arising from HPK-1 overexpression (Fig 1B) was suppressed by inactivation of any of the above transcription factors. Of these, we found that inactivation of pha-4 or mxl-2 is epistatic to hpk-1 overexpression, suppressing the increased lifespan conferred by hpk-1 overexpression (Fig 13A and 13B). In contrast, animals overexpressing hpk-1 were still long-lived after inactivation of hlh-30 or nhr-62 (Fig 13C and 13D). Thus, hpk-1 overexpression extends longevity in a manner dependent on pha-4 and mxl-2, but not nhr-62 or hlh-30.
Fig 13. pha-4 and mxl-2 are required for hpk-1 to promote longevity.
(A) Overexpression of hpk-1 increases lifespan (grey versus black) and is pha-4 dependent (red traces). (B) Overexpression of hpk-1 increases lifespan dependent on mxl-2 (blue traces). (C) Loss of hlh-30 (green traces) partially suppresses the increased lifespan of hpk-1, consistent with parallel signaling or independence. (D) Loss of nhr-62 (pink/purple traces) has a minimal negative effect on both normal and the increased lifespan conferred by hpk-1 overexpression. In all cases, black traces are N2 and grey traces are Psur-5::HPK-1::CFP animals treated on control RNAi. For each panel, darker colored traces are respective RNAi-treatment of N2 animals and lighter colored traces are RNAi treatment of Psur-5::HPK-1::CFP animals. In some cases, experiments shown within this figure were performed simultaneously and split into multiple figures for readability. Full lifespan data can be found in S1 Table.
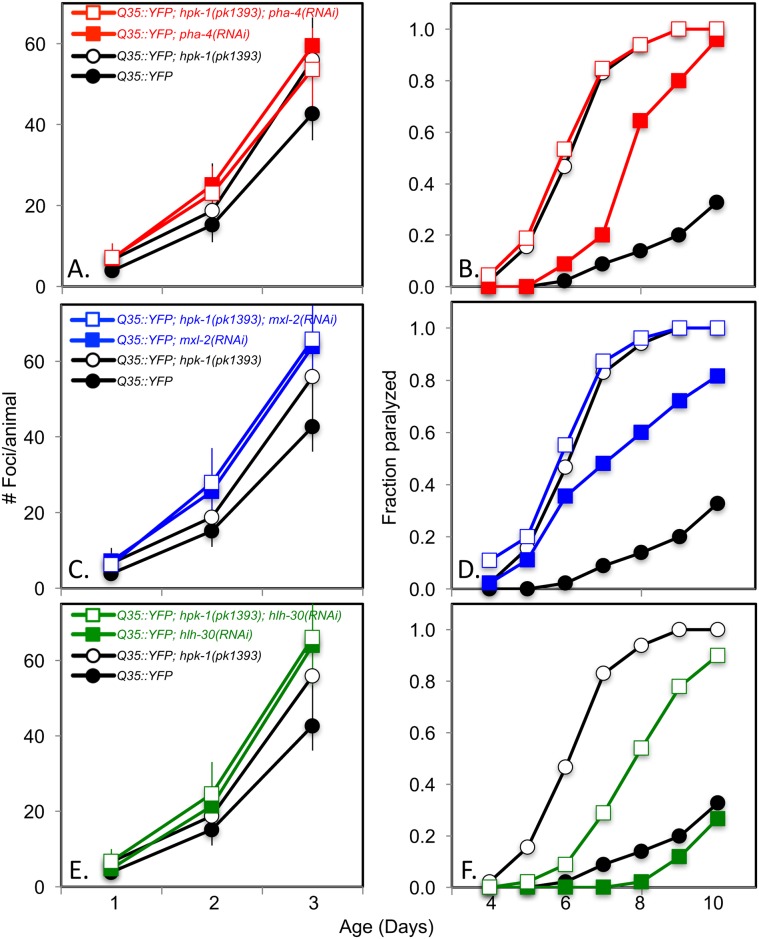
Autophagy has been shown to ameliorate aggregate formation in response to polyglutamine tracts in C. elegans, as well as other systems[69–71]. We therefore tested whether these autophagy-inducing transcription factors mitigate polyQ aggregate formation and/or toxicity in muscle cells, as has been reported in two other studies[55, 72], and whether their ability to do so requires hpk-1. Inactivating pha-4 or mxl-2 in otherwise wild-type Q35::YFP animals resulted in both accelerated protein aggregate formation and early onset of paralysis (Fig 14A–14D), consistent with previous findings. Curiously, inactivation of hlh-30 also accelerated Q35 aggregate formation without enhancing its associated locomotory toxicity.
Fig 14. pha-4 and mxl-2 intersect with hpk-1 in the maintenance of proteostasis.
(A-B) Loss of pha-4 (red traces) does not increase foci formation (A) or the onset of paralysis (B) in the absence of hpk-1 (open circles/squares). (C-D) Loss of mxl-2 (blue traces) does not increase foci formation (C) or the onset of paralysis (D) in the absence of hpk-1 (open circles/squares). (E-F) Loss of hlh-30 (green traces) does not increase foci formation (E) but delays the onset of paralysis in the absence of hpk-1 (F) (open circles/squares). For foci formation, data are the mean and standard error of the mean (S.E.M.) of at least 15 animals from one representative trial; three independent experiments were performed. ***, **, and * indicate p-values of <0.001, <0.01, and <0.05, respectively. For paralysis, data is representative of one of two trials performed with the same conditions. See S3 Table for additional details.
Next, we wished to determine if these transcription factors act in parallel or as part of an hpk-1 regulatory network that promotes protective proteostatic responses to oppose the aggregation-associated phenotypes of polyglutamine repeats. Loss of either pha-4 or mxl-2 did not worsen the accelerated polyglutamine phenotypes of an hpk-1(pk1393) mutant, suggesting that pha-4 and mxl-2 protect against aggregate formation in conjunction with hpk-1, possibly as direct transcription factor targets. In contrast, the hlh-30 results were equivocal: while hlh-30 RNAi did not accelerate the formation of Q35::YFP aggregates in the absence of hpk-1, hlh-30 RNAi surprisingly resulted in partial restoration of the premature aggregate-associated motility defects of hpk-1(pk1393). The apparent contradiction that loss of hlh-30 ameliorates the proteotoxicity of an hpk-1(pk1393) mutant will require additional experimentation to determine the regulatory relationships between hlh-30, hpk-1, mxl-2, and pha-4. Nevertheless, our results- which tentatively place hpk-1 in a regulatory network with mxl-2/pha-4 but not hlh-30- are consistent with previous reports that have found hlh-30 and pha-4 have distinct pro-longevity functions[19] and pha-4 and mxl-2 have similar pro-longevity functions[72]. It is tempting to speculate that loss of hlh-30 may be compensated by upregulation of MXL-2 or PHA-4, which partially rescues the proteotoxicity that results from loss of hpk-1.
Bacterial deprivation is known to activate autophagy and the nuclear translocation of DAF-16/FoxO and HLH-30/TFEB[19, 73]. Since hpk-1 is required for autophagosome formation after bacterial deprivation, we tested whether hpk-1 was also required for DAF-16 or HLH-30 activation. Loss of hpk-1 did not impair cytoplasm-to-nuclear translocation of HLH-30::GFP or DAF-16::GFP after bacterial deprivation (S10 and S11 Figs). This is consistent with the notion that hpk-1 and daf-16/hlh-30 have separable functions in nutrient sensing. Collectively, our results support a model in which PHA-4 and MXL-2 represent a nutritionally-responsive arm of HPK-1 regulation in addition to its role in HSF-1 activation in response to thermal stress and in preserving the integrity of the proteome under unstressed conditions.
Discussion
HPK-1 belongs to a conserved family of homeodomain-interacting kinases that increase resistance to thermal and nutritional stresses
In this study we describe HPK-1 as a transcriptional regulator of proteostasis and longevity in C. elegans. HPK-1 is the lone representative of the homeodomain-interacting kinase gene family in C. elegans, a relatively understudied family of kinases. hpk-1 is most closely related to yak1 in Saccharomyces cerevisiae and to the HIPKs and DYRK kinases in mammals. In mammals a total of four HIPK orthologues respond to a number of external cues including the DNA damage response, hypoxia response, reactive oxygen species (ROS), glucose availability, and viral infection[32, 45, 74, 75]. In general HIPK family members regulate the activity of transcription factors, chromatin modifiers, signaling molecules and scaffolding proteins in response to cellular stress. For example, genotoxic damage induces mammalian Hipk2, and HIPK2 potentiates p53 pro-apoptotic activity through direct phosphorylation[34].
We originally identified hpk-1 in an RNAi screen for genes acting in the daf-2/insulin signaling pathway (i.e. genes necessary for the increased lifespan of daf-2 mutants). A subset of the gene inactivations that shorten daf-2(e1370) lifespan also confer no additional lifespan shortening effect in a daf-2;daf-16 genetic background[38], implying function specifically within the ILS pathway. hpk-1(RNAi) met these phenotypic criteria, but we ultimately concluded that hpk-1 was unlikely to be component of the canonical DAF-2(insulin/IGF1R)-AGE-1(PI3K)-AKT-DAF-16(FoxO) signaling pathway, because loss of hpk-1 only modestly suppressed daf-2 lifespan, and to a degree that was not proportionally greater than the reduction of lifespan observed in response to hpk-1 inactivation in wild-type animals[38]. Additionally, induction of the DAF-16 target gene sod-3 under conditions of decreased ILS does not require hpk-1[38]. Our conclusion has since been supported by a separate study reporting that only eight of 259 DAF-16/ILS regulated genes showed decreased expression in animals lacking hpk-1[42]. Lastly, in this manuscript we show that decreased ILS does not alter hpk-1 expression (Fig 10B). Collectively these findings are more consistent with a model where HPK-1 functions not within but parallel to the canonical ILS pathway.
HPK-1 extends longevity, thermotolerance and preservation of proteostasis by activation of HSF-1
We show that overexpression of hpk-1 extends natural longevity, which suggests that HPK-1 exerts a regulatory function on longevity pathways, rather than simply being required for some essential physiological function. We show that a translational HPK-1::GFP reporter is expressed broadly during C. elegans embryogenesis and larval development (in intestine, hypodermal seam cells and neurons), but its expression pattern becomes restricted to neurons in adults. These data are consistent with reported patterns of HIPK expression in mammals and C. elegans [76, 77]. Because stress response pathways are intimately tied to longevity, we tested whether hpk-1 expression was induced by oxidative stress, DNA damage or heat shock. We found that heat shock induces HPK-1 in the same tissues where it is normally expressed during development: hypodermis and neurons, and to a lesser extent, the intestine. We showed that heat shock induction was at the protein level, as it occurred in the presence of α-amanitin. A role for HPK-1 in heat shock responses is likely to be evolutionarily conserved, as yak1 is also induced by heat stress and is required for normal thermal stress survival in S. cerevisiae [78, 79]. In contrast, we observed no induction of HPK-1 in response to oxidative or DNA damage.
We investigated whether hpk-1 functions as part of the well-known HSF-1 heat shock response pathway. We found that hpk-1 was required for HSF-1 mediated thermotolerance, the hormetic extension of longevity, and transcriptional induction of two key HSF-1 target genes, the chaperones encoded by hsp-16.2 and hsp-70. hsf-1 and hpk-1 are mutually required for the increased lifespan arising from overexpression of the other. We observed that an HPK-1::GFP fusion protein was localized to subcellular structures coincident with HSF-1 localization, an observation consistent with a model in which HPK-1 and HSF-1 reside within a regulatory complex. We believe that HPK-1 is likely to be a positive regulator of HSF-1 activity because of its homology to kinases that activate transcription factors by phosphorylation. The homology of HPK-1 to the homeodomain-interacting kinase family, together with our result that HPK-1 induction was not blocked by global inhibition of transcription meant that we were unsurprised to discover that the heat shock inducibility of HPK-1 was also HSF-1-independent,
A separate study has recently reported that C. elegans HPK-1 is induced by thermal stress. However the authors concluded that hpk-1 was not part of the HSF-1 heat shock response because they saw no dependence on hpk-1 for induction of endogenous molecular chaperones after heat shock[42]. We believe we can reconcile this discrepancy as a consequence of the timing at which chaperone induction was measured. We heat shocked animals and examined endogenous chaperone induction during late larval development, while Berber et al. tested for chaperone induction after the onset of reproduction. However, the onset of reproductive maturity in C. elegans is characterized by an extreme downshift in the ability of animals to respond to heat shock. Within 4 to 8 hours of the onset of reproduction, repressive chromatin marks are laid down at stress loci; these repressive chromatin modifications severely curtail the ability of animals to respond to heat shock[30]. This seems a likely explanation for why Berber et al. observed only low-level induction of hsp-70 and hsp-16 in response to heat shock, and why they were unable to detect a dependency on hpk-1. In contrast, we conducted our experiments prior to the timing of chromatin repression at heat shock loci. Resultantly, we observed a far larger transcriptional induction for hsp-70 and hsp-16.2 compared to Berber et al. (~100 and 4500-fold, respectively, versus ~6–8 fold) and nearly all of this induction required the presence of hpk-1. This suggests that hpk-1 is essential for the activation of the heat shock response prior to chromatin silencing at stress loci, but after the heat shock response is compromised through chromatin remodeling at stress loci, hpk-1 is no longer essential for the limited transcriptional activation of heat shock genes. As Berber et al. did not report whether the residual expression of chaperones they observed requires hsf-1 itself, it is impossible to say whether this modest transcriptional induction is HSF-1 mediated or occurs through some other mechanism.
HSF-1 promotes global proteostasis even in the absence of heat stress by regulating the transcription of chaperones, which maintain stability and solubility of the proteome both by assisting in the folding of newly translated polypeptides and by directing chronically misfolded proteins to the proteasome for degradation. The protective functions of the chaperone system are particularly relevant to diseases of the nervous system caused by inappropriate protein aggregation and resulting neural toxicity. Expression of aggregation-prone polyQ transgenic constructs in C. elegans provides both a method for detecting the proteostatic stress level of tissues and a means of identifying protective and risk factors for aggregation diseases. Inactivation of hsf-1, for instance, causes a premature accumulation of polyglutamine-YFP puncta in muscle cells[80].
When we tested whether HPK-1 displays a similar protective effect on polyQ aggregate formation, we found that HPK-1 is as, if not slightly more important than, HSF-1 in its ability to delay insoluble aggregate formation and associated paralysis in Q35:YFP animals (see S3 Table). Moreover, hsf-1 RNAi did not increase the number or toxicity of polyQ aggregates when combined with the hpk-1(pk1393) mutation, showing that HSF-1 confers its protective effects entirely under the regulatory umbrella of HPK-1. Strikingly, HPK-1 overexpression exerted a potent protective effect against polyQ proteotoxicity by dramatically reducing the rate of foci formation and paralysis.
HPK-1 opposes inhibitory sumoylation of HSF-1
There has been significant effort invested in defining the mechanism(s) of activation of the heat shock transcription factor HSF-1, an effort complicated by the large number of post-translational modifications on HSF-1. Biochemical analysis of HSF-1 regulation is an extremely active area of research from yeast to mammals. Most studies of HSF-1 activation have been performed in tissue culture and ex vivo models, but there is very little information about the regulation of HSF-1 by post-translational modification in living animals. In this study, we describe a pair of HSF-1 modifications in C. elegans that correlate with reduced transcriptional activity of HSF-1 during aging, and in a manner dependent on hpk-1.
Since HPK-1 is a protein kinase that co-localizes into subnuclear foci with HSF-1, the simplest model is that HPK-1 stimulates HSF-1 activity through direct phosphorylation. There are multiple examples in mammals that phosphorylation of a transcription factor/co-factor directly prevents subsequent sumoylation, thereby increasing the activation potential of that transcription factor; examples include PML protein, p53, and c-Jun[81, 82]. While we were unable to resolve a phospho-isoform of HSF-1 attributable to HPK-1 activity, not all phosphorylation events produce a mobility shift. Alternatively, our antibody may not recognize the relevant phospho-isoform of HSF-1.
It would also be informative to test whether HSF-1 regulation requires an active HPK-1 kinase domain. There are at least 19 phosphorylation sites on human HSF1, a subset of which are conserved in C. elegans (S5 Table and[41]). Consistent with the possibility of direct interaction between HPK-1 and HSF-1, S. cerevisiae Yak1 directly phosphorylates Hsf1 in response to glucose stress, albeit within a region not conserved in multicellular eukaryotes [36]. Alternatively, HPK-1 may act indirectly to prevent HSF-1 sumoylation. In mammals both sumoylation and acetylation occurs at HSF-1 K298[49], the latter through the opposing acetyltransferase activities of p300 and SIRT1[41]. Consistent with the possibility of indirect interaction, mammalian HIPK2 directly phosphorylates SIRT1 to restrict activity after DNA damage[83]. Given the complexity of post-translational modification to HSF-1, determination of whether HPK-1 directly phosphorylates HSF-1, and identification of the relevant residue(s) will require analysis using mass spectrometry.
Sumoylation of transcription factors is often associated with reduced transactivation activity [84]. Specifically, sumoylation of HSF family members has previously been reported in mammals, and these isoforms possess decreased transcriptional activity[39, 85]. Our findings that smo-1(RNAi) leads to an increase in HSF-1 chaperone induction in response to heat shock support a hypothesis in which sumoylation of HSF-1 is inhibitory in C. elegans as well. Moreover, our data support a role for HPK-1 in preventing HSF-1 sumoylation (and its coupled phosphorylation event) through the end of development, at which point HPK-1 becomes restricted to low level expression in neurons. This timing for the loss of systemic HPK-1 expression correlates with the onset of transcriptional silencing at stress loci[30] and a decline in multiple additional protein quality control mechanisms (reviewed in[29]). Opposition of sumoylation by HPK-1 may provide a molecular mechanism for a long-standing question in the aging field concerning the decline of HSF-1 activity in aging animals. In mammals, HSF1 DNA binding activity and chaperone expression levels both decline with aging, while the abundance of HSF1 protein does not[41]. We propose that sumoylation of HSF-1 is inactivating, and that increased accumulation of sumoylated HSF-1 with reproductive age explains the decline of basal HSF-1 activity and the resulting decay of proteostasis in aging animals. We also propose that a critical longevity function of HPK-1 is to protect HSF-1 against age-dependent inactivation—by delaying/preventing sumoylation or by driving de-sumoylation.
HPK-1 stimulates transcriptional induction of autophagy in response to dietary restriction. TORC1 blocks autophagy by inhibition of HPK-1 expression in the nervous system
We describe earlier in the discussion how we initially identified a role for hpk-1 in longevity control, and provide the rationale for our conclusion that the pro-longevity functions of hpk-1 belong to pathway(s) acting in parallel to the ILS/FOXO signaling pathway. We have shown that hpk-1 promotes HSF-1 mediated transcription—within the context of the heat shock pathway and basal unstressed conditions—to promote global proteostasis via chaperone systems. We also wondered if hpk-1 might function more broadly to regulate stress-responsive transcription factors. We suspected that hpk-1 might modulate nutritional stress responses controlled by the TOR signaling pathway. First, a wealth of data in mammals and C. elegans places daf-15 (Raptor, a TORC1 subunit) in a pathway parallel to ILS based on distinct activities within each pathway that modulate longevity[60]. However, daf-2/ILS and daf-15/TOR do not always function in isolated linearity with respect to each other, and under certain conditions these kinases converge on common transcription factors necessary for extension of longevity, including SKN-1(Nrf2), HSF-1, MXL-2, and DAF-16[21, 50, 63, 72, 73, 86–88]. Only a subset of transcriptional targets is shared. For example, TORC1 inactivation does not stimulate DAF-16 nuclear translocation and activation (as daf-2 inactivation does)[73]. Though the importance of HIPKs in orchestrating stress responses has only begun to emerge, several pieces of evidence suggest that HIPKs may reside at the crossroads of ILS and TOR signaling. In mammals, multiple mechanisms link insulin/IGF (insulin-like growth factor) levels to activation of mTORC1[89]. Knockdown of HIPK1, 2 or 3 attenuates insulin secretion in response to glucose and HIPKs bind to insulin promoters, suggesting that HIPKs may activate insulin expression[37]. Glucose deprivation can also activate Hipk1[75, 90]. In budding yeast, the HIPK homolog Yak1 is activated by rapamycin treatment[91] and by glucose depletion[92]. For these reasons, it seemed worth testing if hpk-1 might function within the TOR pathway, potentially as a point of regulation between TOR growth signaling and the nutrient deprivation transcriptional programs repressed by TOR.
First, we found that inactivation of daf-15 stimulated neuronal expression of HPK-1, while inactivation of daf-2 did not. HPK-1 induction in response to daf-15 RNAi was restricted to the nervous system, in contrast to the broader induction of HPK-1 in response to heat shock that also included HPK-1 expression in seam cells and the intestine (weakly). When we examined the transcriptional programs controlled by TOR, we found that HPK-1 was necessary for the induction of autophagy genes bec-1 and atg-8 following TOR inactivation, but that hpk-1 had no effect on the transcription of translation initiation factor genes induced by TOR activation. In further support that HPK-1 can stimulate autophagy, we observed that the induction of autophagosome formation (LGG-1::GFP) in response to DR required hpk-1 while interestingly, hsf-1 was not required. Stimulation of autophagy by hpk-1 in response to DR must therefore involve the activation of other transcription factor(s) that are not HSF-1.
Autophagy is thought to occur in at least two separate phases: a rapid increase in autophagic flux that occurs entirely through post-translational protein modifications within minutes to hours after stress, which is followed by a subsequent extended phase reliant on the activation of transcriptional programs[17, 64]. Dietary restriction and TOR inactivation induce protein turnover in C. elegans by stimulating autophagy, which requires multiple DR-responsive transcription factors, including: pha-4(FoxA), mxl-2(MLX), hlh-30(TFEB) and nhr-62(NHR4α). In addition to transcriptional controls, the TORC1 complex inhibits autophagy directly by inhibitory phosphorylation of autophagy components necessary for initiation of autophagy. Because hpk-1 functions biologically by activation of transcription factors, it seems likely that HPK-1 acts during the “extended phase” of autophagy induction by activating one or more of the transcription factors required for autophagy induction in response to DR.
Distinct roles for HSF-1 and HPK-1 in autophagy induction in response to thermal and nutritional stress
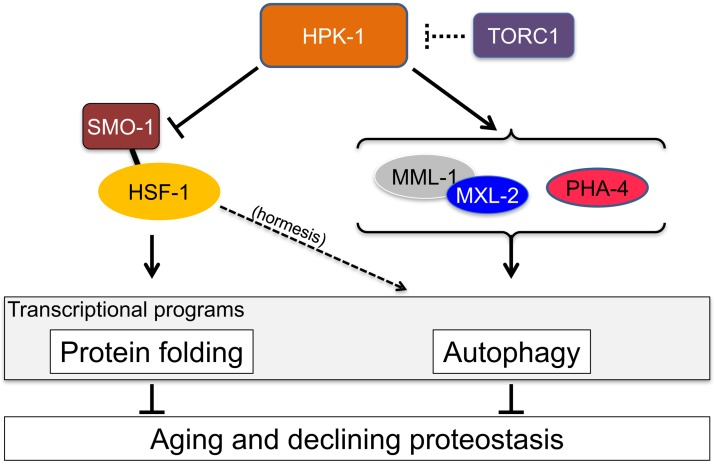
Our results are consistent with a general model in which HPK-1 promotes protein homeostasis by two separate mechanisms, each of which can function under basal as well as stressed conditions. HPK-1 stimulates transcription of molecular chaperones via HSF-1, which decreases the physiological load of misfolded proteins in vivo. In parallel, HPK-1 stimulates autophagy via PHA-4 and MXL-2, allowing existing proteins to be catabolized into useable metabolites, also reducing the physiological protein load. Under conditions of heat shock, the requirement for HPK-1 becomes more pronounced as the need for a proteostasis compensation mechanism is dramatically increased in response to protein unfolding. Under conditions of nutrient stress, proteostasis is not directly compromised, but the demand for metabolic building blocks must be met using existing cellular resources. In this case, protein turnover by autophagy may be an effective way to supply essential metabolites; at the same time that it improves proteostasis by clearing protein aggregates.
A recent study identified a role for HSF-1 in the induction of autophagy after hormetic heat shock or ectopic over-expression of hsf-1[70]. Our finding that hpk-1 is essential for the beneficial effect of hormetic heat shock on lifespan (S4G Fig) would be consistent with the notion that hpk-1 may also be essential for the induction of autophagy gene expression conferred by either hsf-1 overexpression or hormetic heat shock, as well as after nutrient stress. Induction of autophagosome formation may also be regulated differently in different tissues: we find that hpk-1 but not hsf-1 is required for autophagosome formation in hypodermal seam cells, while heat mediated autophagosome formation in muscle, intestinal, and cells within the nerve ring require hsf-1[70].
Extension of longevity and proteome maintenance by HPK-1 requires the autophagy-inducing transcription factors PHA-4 and MXL-2
Upon nutrient deprivation, hlh-30 (TFEB), pha-4 (FoxA), the nuclear hormone receptor nhr-62, and the Myc family transcription factors mml-1 (Mondo A/ChREBP) and mxl-2 (Mlx) have all been shown independently to be essential for at least one aspect of the induction of autophagy[21, 22, 64]. TORC1 inactivation is thought to increase lifespan by inducing autophagy and by decreasing global protein translation (reviewed in[65]). Dietary restriction reduces TOR signaling[93]. We and others have shown that either conditions of dietary restriction or inactivation of TORC1 depend upon hpk-1, pha-4 and mxl-2 to extend longevity (this study and[21, 67, 72, 73]). Similarly, we show that the extension of longevity conferred by overexpression of hpk-1 is suppressed by pha-4 and mxl-2, but not hlh-30 or nhr-62 (Fig 13), suggesting that autophagy contributes to the extended longevity of animals overexpressing HPK-1 in addition to the induction of chaperone genes driven by HSF-1. HPK-1 could represent the crucial regulation point between TORC1 and its target transcription factors PHA-4 and MXL-2. We have demonstrated that TOR inhibits expression of hpk-1 mRNA (Fig 10E) and protein levels of HPK-1 (Figs 10D, S8). Mammalian Hipks are unstable and HPK-1 could be inhibited by TOR post-translationally through regulation of its stability. A particularly exciting possibility is that HPK-1 acts as a nutritional switch operated by TOR. For instance, phosphorylation of HPK-1 by TOR may destabilize HPK-1 and/or target it for degradation. Under replete conditions, autophagy gene transcription would be OFF because HPK-1 protein levels are being maintained at only low levels by TOR kinase. Inactivation of TOR kinase by dietary restriction would lead to stabilization of HPK-1, allowing protein levels to accumulate. HPK-1 could in turn phosphorylate and activate downstream transcription factors (PHA-4 and MXL-2) that stimulate expression of genes necessary for autophagosome biogenesis.
A complication of this model is that it only provides a sequence of action for TOR, HPK-1 and PHA-4/MXL-2 when they are functioning within the same tissue. We have shown that TORC1(RNAi) stimulates HPK-1 expression in neurons, while the Q35::YFP transgene is only expressed in muscle. Therefore, it is likely that endocrine signals arise downstream of HPK-1 in order to regulate cellular functions like autophagy in non-neuronal tissues. Precedent for the neuroendocrine relay of stress responses has arisen in recent years, including the unfolded protein responses of the ER UPRER, mitochondria (UPRmito), and the heat shock response[25, 94–96]. Activation of either of these stress response pathways in neurons stimulates activation of the self-same UPR in non-neuronal tissues, and a recent report suggests that the transcription factor actors of these responses are required both “upstream” (in neurons) and “downstream” (in non-neuronal tissues) in the neuroendocrine circuit. A specific example is stimulation of the ERUPR in non-neuronal tissues by activation of the UPRER in neurons. XBP-1, a cell autonomously acting transcription factor is required in both neurons and non-neuronal tissues in order for the UPRER activation in neurons to elicit the UPRER in distal tissues. Aside from evidence that serotonergic signaling is involved, the mechanism(s) by which specific cellular stress response pathways send or receive endocrine signals between tissues in order to generate systemic responses has yet to determined[97, 98].
Activation of PHA-4 and MXL-2 transcriptional programs by HPK-1 protects animals from polyglutamine aggregate formation
In addition to evaluating the requirement for autophagy transcription factors in mediating the increased lifespan arising from HPK-1 overexpression, we tested the possibility that PHA-4, MXL-2 and HLH-30 were targets of HPK-1 regulation in the proteostatic context of polyQ aggregation. First we quantitated the dynamics of Q35::YFP aggregate formation and toxicity in the presence or absence of pha-4, mxl-2 and hlh-30 using RNAi. We found that inactivation of all three transcription factors significantly accelerated the accumulation of Q35::YFP aggregates. Inactivation of pha-4 and mxl-2 accelerated the rates of toxicity-induced paralysis. Curiously, hlh-30 RNAi partially rescued the paralysis phenotype of the hpk-1(pk1393) mutant. In the Q35::YFP; hpk-1(pk1393) background, inactivation of pha-4, mxl-2 or hlh-30 did not increase aggregate formation above the already elevated level caused by the absence of hpk-1. Similarly, pha-4(RNAi) and mxl-2(RNAi) had no increased effect on the accelerated rates of paralysis observed in the hpk-1(pk1393) mutant strain relative to wild type. Our finding that inactivation of pha-4 and mxl-2 causes an increase in aggregate formation suggests that autophagy can in fact mitigate the formation of toxic Q35::YFP aggregates in muscle cells. This supports our model: extension of longevity depends specifically on the stimulation of autophagy by hpk-1—via activation of the pha-4 and mxl-2 transcription factors—and suggests that HPK-1, PHA-4, and MXL-2 may comprise a larger transcriptional circuit to regulate autophagy gene expression under specific conditions of nutrient stress.
Proteostatic robustness is determined by the mutual opposition of TOR-mediated growth signaling and HPK-1-mediated stress responses
Aging is highly dependent upon the cellular processes that maintain proteostasis[99, 100], and diverse age-related neurodegenerative diseases (e.g. Alzheimer, Parkinson and Huntington diseases) are linked to compromised autophagy and the decline of proteostasis[99–102]. Proteostasis is achieved by the careful balancing of rates of protein synthesis with chaperone-mediated protein folding and turnover of unfolded polypeptides via proteasome- and autophagy-mediated degradation pathways. When proteostability is compromised, HSF-1 promotes restoration of proteostasis by induction of molecular chaperones and proteosomal degradation of misfolded proteins[49]. In contrast, nutrient rich conditions challenge proteome stability by activation of the TOR pathway, which stimulates protein translation and inhibits protein turnover. When nutrients are limited, TOR kinase is inactivated, prompting protein degradation through autophagy into constituent metabolites, which serves the dual function of reducing the overall concentration of proteins that must remain soluble while providing biosynthetic building blocks that can be remobilized towards pro-survival processes[103]. It is worthy of notice that HPK-1 and TOR act in physiological opposition; HPK-1 promotes catabolic processes that decrease proteotoxic stress, while TOR promotes an anabolic physiological state represented by an overall increase in cellular protein. Overexpression of HPK-1 or inhibition of the TOR pathway promotes proteome re-stabilization and concomitant extension of longevity.
Significance: Proteostasis in the context of aggregation diseases
Most neurodegenerative diseases are characterized by an age-dependent onset, which share the common molecular feature of abnormal protein aggregation leading to neuronal cell death. A number of studies have linked loss of either HSF1 or autophagy to the promotion of neurodegenerative diseases[104, 105]. We have discovered that HPK-1 functions as a central regulatory node linking these two complementary proteostatic mechanisms that act to preserve the proteome (Fig 15), and justifies consideration of HPK-1 as an attractive intervention point for the improvement of healthspan and protection against proteostatic diseases.
Fig 15. HPK-1 delays aging and maintains proteostasis by potentiating TORC1 mediated autophagy and blocking HSF-1 inactivation through sumoylation.
Model of HPK-1 functions in longevity control: HPK-1 functions as a central hub to maintain proteostasis by preventing sumoylation and inactivation of HSF-1 and by stimulating the expression of autophagy genes by pha-4 and mxl-2. TORC1 inhibits hpk-1 expression to limit the induction of autophagy genes under basal conditions. Under nutrient stress, TORC1 is inactivated resulting in increased hpk-1 expression, which promotes autophagy gene expression through PHA-4/FoxA and MXL-2/Mlx. Thermal stress increases HPK-1 protein levels to reduce the threshold of activation of the heat shock response, and HPK-1 promotes longevity through modulation of HSF-1 activity under normal growth conditions.
Experimental procedures
C. elegans strains and maintenance, RNAi, and nematode culture
Strains were maintained at 20°C on standard NGM plates with OP50. Strains and genotypes used in this study are:
N2 wild type (CGCM)
AVS392: hpk-1(pk1393) (received 6X backcrossed from CGC and backcrossed 5 additional times)
AVS84/TJ375: gpIs1[Phsp-16.2::GFP]
AVS85/AM140: rmIs132[Punc-54::Q35::YFP]
AVS253/DA2123: adIs2122[lgg-1::GFP + pRF4 (rol-6(su1006))]
AVS393-394: artEx11-12[Phpk-1::GFP+ pRF4(rol-6(su1006))] (two independent lines)
AVS181/KWN147: pha-1(e2123ts) III; rde-1(ne219) V; rnyEx078[Pnhx-2::RDE-1; Pnhx-2::pHluorin; pCL1(pha-1+)] (intestinal RNAi strain)
AVS206/NR222: rde-1(ne219) V; kzIs9 [pKK1260(lin-26p::nls::GFP) + pKK1253(lin-26p::rde-1) + pRF6(rol-6(su1006)] (hypodermal RNAi strain)
AVS454/NR350: rde-1(ne219) V; kzIs20[pDM#715(hlh-1p::rde-1) + pTG95(sur-5p::NLS::GFP)] (muscle RNAi strain)
AVS216/NL3321: sid-1(pk3321) V
AVS265/TU3401: sid-1(pk3321) V; uIs69[punc-119::sid-1;pmyo-2::mCherry] V.
AVS309, 310 and 396: artEx26-28[Phpk-1::hpk-1::GFP+ pRF4(rol-6 (su1006))] (Three independent lines)
AVS413: hpk-1(pk1393); artEx29[Phpk-1::hpk-1::GFP + pRF4(rol-6 (su1006))]
AVS411: artEx30[Phpk-1::hpk-1::tdtomato + pAH71(Phsf-1::hsf-1::GFP) + pRF4(rol-6(su1006)]
AVS408, 409: artEx31-32[Psur-5::hpk-1::CFP + pCFJ90(Pmyo-2::m-cherry)]
AVS397, 398: gpIs1[Phsp-16.2::GFP]; artEx35-36[Psur-5::hpk-1::CFP + pCFJ90(Pmyo-2::m-cherry)] (two independent lines)
AVS401/AGD710: uthIs235[Psur-5p::hsf-1 FL, + Pmyo-2::tdTomato]
AVS403, 404: rmIs132[unc-54p::Q35::YFP]; artEx37-38[Psur-5::hpk-1::CFP + pCFJ90 (Pmyo-2::m-cherry)] (two independent lines)
AVS438/EQ87: [Phsf-1::hsf-1::GFP + pRF4(rol-6(su1006))]
AVS405/OG497: drSi13[hsf-1p::hsf-1::GFP::unc-54 3'UTR + Cbr-unc-119(+)]
AVS469/TJ356: zIs356[daf-16p::daf-16a/b::GFP pRF4(rol-6(su1006))]
AVS474/MAH240: sqIs17[hlh-30p::hlh-30::GFP + pRF4(rol-6(su1006))]
AVS464: hpk-1(pk1393); rmIs132[Punc-54::Q35::YFP]
AVS471: rmIs132[Punc-54::Q35::YFP]; artEx32[Psur-5::hpk-1::CFP + pCFJ90 (Pmyo-2::m-cherry)]
Multiple strain designations per genotype indicate independently generated isolates of identical genotype. In cases where both an AVS and an outside laboratory designation are given, the latter indicates the source strain we received and the former our in-house laboratory designation.
RNAi
RNAi clones originated from early copies of E. coli glycerol stocks of the comprehensive RNAi libraries generated in the Ahringer and Vidal laboratories. Each RNAi colony was grown overnight in Luria broth with 50 μg ml–1 ampicillin and then seeded onto 24-well RNAi agar plates containing 5 mM isopropylthiogalactoside to induce dsRNA expression overnight at room temperature. All of the RNAi clones used in this study were verified by DNA sequencing and subsequent BLAST analysis to confirm their identity.
Generation of transgenic strains
Fusion of promoter and gene sequences to reporters were carried about using the SOEing technique [106]. Pooled PCR products were microinjected to generate transgenic lines as follows.
Phpk-1::GFP: 2kb upstream of ATG start for the longest isoform of hpk-1 was amplified from fosmid WRM066aH12 and fused to GFP (from pPD95.75) (Addgene) using the following primers:
Phpk-1 Primer A 5’- AATTTTCAAGATAGGGCCGCCG
Phpk-1 Primer B (nested to GFP) 5’-AGTCGACCTGCAGGCATGCAAGCTCGCCACCCAATCAAACAATCG
Phpk-1 Primer C 5’- AGCTTGCATGCCTGCAGGTCG
Phpk-1 Primer D 5’- AAGGGCCCGTACGGCCGACTA
Phpk-1 Primer A’- 5’-CCTCCTTCGCCAAGTTTCGA-3’GFP
GFP Primer D* (nested)- 5’- GGAAACAGTTATGTTTGGTATA-3’
30ng of fused PCR product was injected with 120ng of pRF4 (rol-6) as a coinjection marker.
Phpk-1::hpk-1::GFP: C-terminal fusion of full-length hpk-1 to GFP was generated using the fosmid template WRM066aH12 and pPD95.75 as above with following primers:
hpk-1 Primer A 5’- AATTTTCAAGATAGGGCCGCCG
hpk-1 Primer B nested to GFP with mutated STOP codon 5’-AGTCGACCTGCAGGCATGCAAGCTTATGCGAGTGCAATGAAGTGATGAGT
hpk-1 Primer A’ 5’-CCTCCTTCGCCAAGTTTCGA
GFP Primer D* (nested) 5’-GGAAACAGTTATGTTTGGTATA
5ng of PCR product was injected into wild type and hpk-1(pk1393) animals with 150ng of pRF4 as coinjection marker.
Phpk-1::hpk-1::tdtomato C-terminal fusion of full-length hpk-1 to GFP was generated using the fosmid WRM066aH12 and pDS9 (tdTomato) as templates
hpk-1 Primer A 5’- AATTTTCAAGATAGGGCCGCCG -3’
hpk-1 PrimerB nested to tdtomato 5’-AGTCGACCTGCAGGCATGCAAGCTTTATGCGAGTGCAATGAAGTGATGAGT-3’
Primer C tdTomato 5’-AAGCTTGCATGCCTGCAGGTCG-3’
Primer D tdTomato 5’-GGAAACAGTTATGTTTGGTATA-3’
For visualizing co-localization of HSF-1::GFP with HPK-1::tdTomato, pAH71 (P-hsf-1::hsf-1::GFP) was obtained from Dr. Hsu (University of Michigan, Ann Arbor) and injected at a concentration of 15ng/ul either with 5ng/ul or 30ng/ul of P-hpk-1::hpk-1::tdtomato.
Psur-5::hpk-1::CFP: 1.5kb promoter region of sur-5 amplified from genomic DNA was fused to hpk-1 genomic fragment and CFP at the C-terminus in a nested PCR reaction. CFP was amplified from pPD134.96.
Psur-5 PrimerA cattcgggctggaaatctgaatg
Psur-5 PrimerA' gagtcccaccatgcttgtcatc
Psur-5:: hpk1 PrimerB CGGAGTTCTTACGCTTTGGCATgcgatatcaccacttctaggcgt
Primer D CFP CTTTTGGGCCCAAGCGAGG
5ng/ul of the PCR fusion construct, 5ng/ul of Pmyo-2::m-cherry (pCFJ90), and Log2 DNA ladder (NEB N3200S) was included as carrier DNA.
Lifespan assays
Lifespan assays were performed essentially as described [107]. Synchronized L1 animals were seeded onto plates and allowed to develop to L4 stage at 20°C, with two exceptions: for the hormesis experiments (S5G and S5H Fig) animals were shifted to 25°C at L4 and maintained at this elevated temperature until day 3 of adulthood and then shifted back to 20°C; and for the intestinal specific RNAi (S2D Fig) animals were maintained at 25°C during development (this strain is a pha-1 rescue strain that must be maintained at 25°C during development to ensure maintenance of the extrachromosomal transgene[108]). In all cases, at L4 2’ fluoro– 5’deoxyuridine (FUDR) was added to a final concentration of 400 uM to prevent progeny production (defined as day 0 adulthood). Viability was scored every day or every other day as indicated in each figure. Survival analysis was performed for all the experiments using the Kaplan-Meier estimator and Peto & Peto’s generalized Wilcoxon test, both as implemented in the R package survival version 2.38 (http://CRAN.R-project.org/package=survival). P-values were corrected for multiple testing with the Benjamini-Hochberg FDR approach[109].
To determine temporal requirements in longevity control (S2A–S2C Fig), synchronized L1 animals were seeded on control or hpk-1 RNAi according to the following conditions: for constitutive RNAi L1 animals exposed to each of the RNAi conditions throughout lifespan, for developmental gene inactivation animals were fed E. coli expressing dsRNA to hpk-1 or control (empty vector) from L1-L4 after which animals were moved to dcr-1 (dicer-1) RNAi, and for adult inactivation animals were maintained on control RNAi plates until L4 and then moved to either control, hsf-1 or hpk-1 RNAi plates.
Detailed information on all lifespan trials, including number of animals scored can be found in S1 Table.
Thermotolerance assays
Time course survival assays at high temperature were conducted using the replica set method as previously described [72]. In brief, synchronized L1 animals were allowed to develop at 20°C; FUDR was added at the L4 stage and at day three adulthood animals were moved to 35°C for the indicated time period, allowed to recover for 1–2 hours at 20°C, and viability was scored every 2 hours for up to 14 hours. Significance of pairwise comparisons between genotypes was determined by fitting the data to logit curves with pooled data from all three trials and randomized permutation testing with 100,000 iterations to determine p-values, as previously reported in [38, 72]. For single time point thermotolerance assays, synchronized day 1 adult animals were heat shocked at 35°C for 9–10 hours and viability was scored the next day. 3–4 technical replicates were included in each of two independent trials. Statistical testing for single time point assays was performed with ANOVA followed by Tukey’s HSD post-hoc test, and p-values were corrected to account for multiple testing.
Fluorescent analysis of transgenic lines
Induction of Phpk-1::GFP and Phpk-1::hpk-1::GFP after heat shock
To assess changes of hpk-1 expression after heat shock, late L4 transgenic animals expressing either the transcriptional (Phpk-1::GFP) or translational (Phpk-1::hpk-1::GFP) reporter were heat shocked at 37°C for 1 hour and allowed to recover for at least 15 minutes. Animals were then immobilized on 2% agarose pads with 1mM tetramisole hydrochloride and imaged using Leica SP5 confocal microscope at 40X magnification. Z-stack imaging was performed and 3-D projection was used to create composite images. Three independent trials were performed with a minimum of 20 animals per condition.
Induction of Phpk-1::hpk-1::GFP in response to metabolic stress
To assess changes of hpk-1 expression after altered metabolic signaling, L1 Phpk-1::hpk-1::GFP transgenic animals were seeded on control, daf-15, rictor-1 or daf-2 RNAi. 400 μM of FUDR was added at the L4 stage and animals were visualized on day 4 of adulthood using confocal microscope Leica SP5. Z-stack imaging was performed for each image that was combined using 3D projection on Leica software LAS to give a composite image. At least three independent trials visualizing a minimum of 15–20 animals each were performed.
Induction of Phpk-1::hpk-1::GFP after UV and oxidative stress
UV damage was induced in late L4 transgenic animals by exposure to 300 μjoules (Stratalinker). Oxidative stress was induced by 1hr of exposure to 7.5mM tert-butyl hydrogen peroxide (as in [89]). Animals were allowed to recover for at least fifteen minutes prior to imaging on confocal microscope Leica SP5 at 40X. At least 15–20 animals were visualized for each condition.
Induction of Phsp-16.2::GFP after heat shock
The Phsp-16.2::GFP animals imaged in Fig 7A–7D were heat shocked on day 1 of adulthood, for 3 hours at 35°C and imaged after 3 hours of recovery. Phsp-16.2::GFP transgenic animals imaged in Fig 8A–8D were heat shocked at 35°C for 1 hour on day 3 of adulthood, allowed to recover for 7–8 hrs and imaged using a Zeiss Axio Imager M2m using QCapturePro v6.0 software. Four independent trials with a minimum of 30 animals per experiment were performed.
Measurement of autophagosome formation using the lgg-1::GFP reporter
LGG-1::GFP foci formation was visualized in L3 stage animals that were raised on either control (EV), hsf-1 or hpk-1 RNAi from the preceding parental generation and imaged using confocal Leica software at 63X magnification. 15–20 animals were scored for seam cell LGG-1::GFP puncta accumulation after 6 hrs of bacterial deprivation (BD) or ad libitum (AL).
Measurement of HLH-30::GFP and DAF-16::GFP subcellular localization
HLH-30::GFP and DAF-16::GFP transgenic animals were treated with empty vector or hpk-1(RNAi) starting at the L1 stage, FUDR was added at L4, and at day 3 of adulthood animals were either subjected to bacterial deprivation or allowed to continue to feed ad libitum for 24 hours. For each condition, immediately prior to visualization 10 animals were moved to a fresh plate with sodium azide to prevent movement. GFP localization was then imaged on a Zeiss Axio Imager M2m using QCapturePro v6.0 software. At least 30 animals were visualized for each condition.
Polyglutamine aggregation and locomotory analyses
Synchronized Punc-54::polyQ35::YFP L1 progeny were treated with the indicated RNAi and FUDR was added at L4 stage to prevent progeny production. To quantify fluorescent foci formation, between 15–50 animals per technical replicate were scored blind daily from days one to three of adulthood. To assess paralysis, prodded animals that responded with head movement (and were therefore still alive) but no backward or repulsive motion were scored as paralyzed (as described in [72]). Statistical testing between pairs of conditions for differences in paralysis rates was performed with Cox modeling and the Wald test.
Generation of HSF-1 antibodies
Rabbit polyclonal antibodies to C. elegans HSF-1 were generated using purified, recombinant C. elegans HSF-1, which was recovered from E. coli expressing His6-HSF-1 from a modified version of the pET-28a(+) vector by affinity purification and subsequent on column thrombin cleavage to remove the His6 tag. Polyclonal rabbit antibodies to the full-length, untagged protein were generated by Covance (http://www.covance.com/services/lead-optimization/immunology/custom-antibody-development.html) through injection of two rabbits, of which the one with the least background immunogenicity was chosen.
Immunoblotting
Approximately 1000 synchronized animals were collected with M9, washed, and pelleted animals were snap frozen in liquid nitrogen. Pellets were lysed in RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0)) with 40 mM N-Ethylmaleimide (NEM, Thermo #PI23030); per 1 mL RIPA, 40 μL of 100x Halt Protease & Phosphatase Inhibitor (Thermo #78446), and 40 μL 100x EDTA (0.5 M) was added. Briefly, extracts were vortexed with Zirconin 2.0 mm beads (#11079124ZX, BioSpec) and lysate was separated from pellet by centrifugation. Samples were normalized using a Bradford assay and was resolved by 6% SDS-PAGE. The antibodies used to probe membranes of the immunoblots were anti-HSF-1 (1:500), and anti-beta-actin (1:1000) (Cell Signaling #4967).
The ratio of HSF-1 unmodified (75kD) to modified (90-95kD, sumoylated and sumoylated plus phosphorylated) was determined based on the quantification of band intensity in Image J. Protein levels of unmodified (75kD), modified (90-95kD) HSF-1, and beta-actin were also quantified in ImageJ from western blots. HSF-1 expression was first normalized to matched beta-actin levels and then relative fold-change compared to N2 was calculated. The S.E.M. from quantified extracts/western blots was calculated and in both cases significance was determined by Student’s t-test.
For detection of HSP-16.2 protein and GFP reporter expression on RNAi, 100 worms grown at 16°C on RNAi were collected in 25 μL dH2O. An equal volume of 2x SDS-PAGE loading dye was added followed by 15 min boiling. A 10–20% Tris-HCl gel (Bio-Rad) was loaded with 15μL (~30 worms) after another 15 min boiling, run, transferred to nitrocellulose, and blocked in 2% milk in 1X TTBS (1M Tris, 150 mM NaCl, 0.1%Tween 20). Membrane was probed simultaneously for HSP-16.2 (1:5000, rabbit, #5506 R120; kind gift of Chris Link, UC Boulder), GFP (1:1000, mouse, Roche 7.1 and 13.1) and β-actin (1:2000, mouse, Sigma AC-15) at 4°C overnight. Blots were washed in 0.1% milk 1X TTBS three times. Secondary antibodies were anti-mouse HRP and anti-rabbit HRP (1:5882 dilution both, Amersham). Blots were visualized with Amersham ECL Western Blotting System (RPN2108).
Immunoprecipitation
For HSF-1 immunoprecipitations, L4 C. elegans pellets were collected as per sample collection for immunoblotting. C. elegans pellets were lysed in 20 mM Tris-HCl (pH 8.0), 170 mM KCl, 0.5% NP-40 (IGEPAL CA-360), 2 mM EDTA, with 40 μL of 100x Halt Protease & Phosphatase Inhibitor, 40 μL 100x EDTA (0.5 M) (Thermo #78446) per mL of lysis buffer. Lysates were prepared and quantified as above, and then 1.5 mg of lysate was precleared by the addition of 10 μL of normal rabbit serum (Invitrogen # 016101) and incubated with rotation for 1 h at 4°C, at which time the equivalent of 120 μL of drained Protein-A-Sepharose was added to immunoprecipitate non-specific interactions and incubation continued an additional 2 h. The proteins were eluted from the nonspecific pellet fraction with the addition of equal volume 2x SDS reducing buffer (1x SDS reducing buffer: 62.5 mM Tris (pH 6.8), 10% glycerol, 0.02% SDS, and 5% β-mercaptoethanol) and boiled for 5 min. Precleared lysate was evenly divided, antibodies were added, and lysates were incubated with rotation for 2 h at 4°C. The antibodies used in various immunoprecipitations were as follows: for HSF-1 1 μL of HSF-1 immune serum, and for negative controls either 1 μL of commercially available NRS (Invitrogen # 016101) or pre-immune serum from the rabbit prior to injection with recombinant C. elegans recombinant HSF-1. The equivalent of 50 μL of drained beads of Protein A-Sepharose was added, and lysates were incubated with rotation for an additional 2 h at 4°C. Immunoprecipitates were washed three times with lysis buffer, resuspended in 2x SDS reducing buffer, boiled for 5 min, and resolved by 6% SDS-PAGE. Subsequent immunoblotting for HSF-1 was carried out as described above.
SUMO protease treatment of immunoprecipitation
Prior to elution after immunoprecipitation, beads of Protein A-Sepharose were washed three times with 1x SUMO Protease Buffer (50 mM Tris-HCl (pH8.0), 0.2% Igepal (NP-40), 150 mM NaCl, 1 mM DTT), and resuspended in 100 μL of 1x SUMO protease buffer with or without 5 μL of SUMO protease (ULP1; Invitrogen #12588–018). Reactions were performed at 20°C for 1 hour with nutating. Immunoprecipitate was eluted with the addition of equal volume 2x SDS reducing buffer and boiled for 5 min.
Phosphatase treatment of protein extracts
Extracts were prepared as above but with HALT complete protease inhibitor cocktail (Thermo #87785) without EDTA. Protein dephosphorylation was carried out with lambda Protein Phosphatase (λPP, New England Biolabs #P0753S) treatment. For λPP treatment 25 μg total protein was incubated for 1 h at 30°C in 1x NEBuffer for MetalloPhosphatases (PMP), 1 mM MnCl2, and 400U of λPP. As a negative control mock λPP treatments used enzyme that had been inactivated by 10 min of boiling prior to addition and samples contained Halt Protease & Phosphatase Inhibitor (Thermo #78446). In all cases, after 1 h of incubation SDS reducing buffer was added and samples were boiled 5 minutes.
Quantitative real time PCR
For measurement of hsp gene induction in response to heat shock, late L4 animals were heat shocked at 37°C for one hour and allowed to recover for two hours before harvesting in M9 buffer. For measurement of atg-18, bec-1, ifg-1, iftb-1, hsp-16.2, and hsp-70 mRNA levels after daf-15 inactivation, synchronized populations were grown until L4 at 20°C on RNAi plates and animals were harvested on day 1 of adulthood. RNA was isolated using Trizol reagent (Life Technologies). RNA concentration was measured using a Nanodrop, and RNA preparations were reverse transcribed into cDNA using the BioRad cDNA synthesis kit (#1708890) as per manufacturer’s protocol. For all qRT-PCR primer pairs, (-) RT test reactions were run to confirm mRNA purity and target specificity. Quantitative real time PCR was performed using SYBR green supermix (Biorad) with three biological and two technical replicates for each condition. Primer sets with at least one primer spanning the exon were used to amplify the gene of interest. cdc-42 mRNA levels were used for normalization of autophagy and translation gene expression. act-1 mRNA levels were used for normalization of hsp gene expression. Fold change in mRNA levels was determined using ΔΔ Ct method [110]. Primer sequences used were as follows:
act-1F 5’-CCATCATGAAGTGCGACATTG
act-1R 5’-CATGGTTGATGGGGCAAGAG
hsp-16.2F 5’-CTCCATCTGAGTCTTCTGAGATTGT
hsp-16.2R 5’-CTCCTTGGATTGATAGCGTACGA
hsp-70F 5’-GATGAAGTTGTCTTGGTTGG
hsp-70R 5’-CAGTTGAGGTCCTTCCCATTG
atg-18F 5’-ACTTGAGAAAACGGAAGGTGTT
atg-18R 5’-TGATAGCATCGAACCATCCA
cdc-42F 5’-AGCCATTCTGGCCGCTCTCG
cdc-42R 5’-GCAACCGCTTCTCGTTTGGC
bec-1F 5’-TTTTGTTGAAAGAGCTCAAGGA
bec-1R 5’-’CAACCAGTGAATCAGCATGAA
ifg-1F 5’-GCTAGCTGATTTCGGATTGG
ifg-1R 5’-CACGGTATGGAACCTGCTG
iftb-1F 5’-ATGGCACATTACGTACTCCTTG
iftb-1R 5’-CGGAAGTAACTGTTTGATTGTGAA
hpk-1F 5’-AGTATGCACAGCTCCATCAC
hpk-1R 5’-CCATTATTGGGACCGGAACA
Ethics statement
This study did not employ Human Subject Research or Animal Research that would require IACUC oversight or approval.
Supporting information
(A) HPK-1 is broadly expressed during development, with limited expression by adulthood. (A, Top left) Expression of Phpk-1::HPK-1::GFP during embryogenesis. (A, top right) Phpk-1::HPK-1::GFP expression becomes increasingly restricted by the L3 stage of development to neurons (arrow with N), intestinal cells (arrow with I), and the hypodermis (arrow with H). (A bottom panel) HPK-1 expression is limited to neuronal cells by the L4 stage of development and in adults (not shown). Brightfield insets for each image and white dotted lines trace animals. In all cases, representative images are shown of at least fifteen animals from two independently derived lines. (B) HPK-1 is localized in the nucleus. Phpk-1::HPK-1::GFP expression with the overlay of DIC image. Arrows indicate examples of localized expression within nuclei.
(TIF)
(A) Constitutive inactivation of hpk-1 (red) from the L1 stage by feeding-based RNAi in wild-type N2 worms decreases lifespan compared to control (EV) (black) RNAi. (B) hpk-1 RNAi only during development (red) results in a decrease in lifespan comparable to constitutive hpk-1 RNAi. Animals were moved to dcr-1(RNAi) at L4. (C) hpk-1 RNAi (red) initiated at L4 decreases lifespan to a smaller, but significant extent. HPK-1 spatial requirements mirror tissues of expression. (D) Intestinal or (E) hypodermal specific gene inactivation of hpk-1 (red) reduces lifespan. In contrast, (F) muscle specific gene inactivation of hpk-1 has a minimal effect on lifespan, consistent with a lack of muscle expression during larval development (S1 File). (G) Neuronal inactivation of hpk-1 (pink) reduces lifespan. (H) Neuronal inactivation of daf-2 increases lifespan (orange), consistent with previous reports[47, 48]. (I) sid-1(pk3321) loss of function animals have impaired RNAi: hpk-1(RNAi) (pink) and daf-2(RNAi)-treatment (red), respectively. Full lifespan data can be found in S1 Table.
(TIF)
(A) Inactivation of the SUMO moiety smo-1 by RNAi prevents formation of higher molecular weight isoforms of HSF-1. Data supports Fig 6A. RNAi knockdown of smo-1 showed some variability in the degree of effectiveness, but generally decreased the relative proportion of higher molecular weight isoforms of HSF-1. Relative proportion of modified HSF-1 shown in the lower panel is 0.49, 0.35, and 0.77 for N2/ev, hpk-1(pk1393)/smo-1, and hpk-1(pk1393)/ev, respectively. ev = empty vector RNAi treatment (L4440). dePhos is λ protein phosphatase treatment. β-actin serves as a loading control. (B, C) Additional replicates for quantification of the ratio of modified to unmodified HSF-1 are shown in Fig 6B. For (B) no β-actin control was available, which precluded quantifying differences in overall expression levels between samples. (C) Additional replicates for quantifying differences in overall HSF-1 levels between N2 and hpk-1(pk1393) shown in Fig 6C. β-actin serves as a loading control. In all cases U = unmodified (75 kD), S = sumoylated (~90 kD), and S+P = sumoylated and phosphorylated (~95 kD) isoforms of HSF-1.
(TIF)
(A, B) Phsp-16.2::GFP is not expressed at 20°C (basal conditions) in day one adult animals. Outlines of animals are traced in white. (C, D, E, F) Phsp-16.2::GFP is induced heterogeneously across tissues after continued exposure to mild temperature stress of 25°C in day one adult animals. (G, H) Transient hormetic exposure to 25°C increases mean lifespan (gray versus black), which is dependent on both hpk-1 (G, red) and hsf-1 (H, blue). See S1 Table for comprehensive lifespan data.
(TIF)
(A) Strain expressing a single copy of HSF-1::GFP is diffusely localized in the nucleus under basal conditions. (B) Strain expressing a single copy of HSF-1::GFP forms nuclear stress granules after heat shock. (C-D) Strain expressing a single copy of HSF-1::GFP treated with hpk-1(RNAi) does not alter HSF-1 basal localization (C) or formation of stress granules after heat stress (D). Results were consistent across three independent trials; approximately 20 animals were visualized per trial and all animals produced nuclear stress granules regardless of hpk-1 status. (E) Strain with low copy HSF-1::GFP shows increased nuclear accumulation after heat stress (column 1 to 2), which is not altered in the absence of hpk-1 (compare 3 to 4). Two independent trials produced similar results, with 10 animals per condition in each trial. Graph includes results from one trial (*** p <0.001, Student’s t-test).
(TIF)
(A) Mean GFP intensity was measured in ImageJ for: head neurons, 2–4 hypodermal seam cells, and 4 spots of internal non-neuronal, non-seam cell background fluorescence for the indicated number of animals (n). Head and seam cell GFP intensity was normalized to average background fluorescence, and the contribution of background fluorescence was subtracted. Mean and standard deviation of head neuron and seam cell fluorescence in unstressed and heat shocked animals was calculated, and normalized to unstressed animals. * indicates p<0.0005, ** p<0.0002 (Student’s t-test). (B) As an independent verification of increased fluorescence within head neurons, we applied a matrix for assessing GFP levels via double-blind visualization, which allowed us to score a larger cohort of animals +/- heat shock. GFP expression was scored as “strong”, “moderate”, or “weak”. * p<0.03 (Mann-Whitney).
(TIF)
(A-B). Phpk-1::GFP expression in L4 animals under unstressed conditions (A) or after 1 hour heat shock (B). For additional images see S1 File. (C) Endogenous hpk-1 mRNA levels of L4 wild-type unstressed animals or following a 15-minute recovery after 1 hour of heat shock at 35°C. (D) Endogenous hsp-16.2 mRNA levels are induced following a 15-minute recovery after 1 hour of heat shock (p<0.05, Student’s t-test). Fold change is relative to unstressed N2. Note: mRNA from the same extracts was used in (C) and (D). In all cases expression is normalized to act-1 mRNA and error bars represent the standard deviation of a minimum of three replicates in each of two independent trials.
(TIF)
(A-C) Expression of Phsp-16.2::GFP animals under the following conditions: unstressed (A), 2 hours of recovery following heat shock for 1 hour at 35°C heat shock (B), and heat shock as in (B) but with prior treatment with 100 ug/mL α-amanitin treatment (C) (as described in [112]). Note, images in (B) and (C) were taken at a similar magnification, which was higher than image (A). Outlines of animals are traced in white.
(TIF)
(A) Mean GFP intensity within head neurons was measured in ImageJ for Phpk-1::HPK-1::GFP expressing animals after empty vector or daf-15(RNAi)-treatment. Change in expression was normalized to empty vector; error bars represent the standard deviation of six animals. * p<0.03 (Student’s t-test). (B-D) Additional images of Phpk-1::HPK-1::GFP expression after empty vector control or daf-15(RNAi) treatment. Expression of Phpk-1::HPK-1::GFP is only induced within neurons after daf-15 (Raptor) inactivation compared to control. While expression within hypodermal seam cells varied between animals, no differences in expression between empty vector and daf-15 RNAi could be discerned, which is in contrast to heat shock. Images in (C) and (D) were stitched together from several overlapping high-resolution images and white space was artificially filled.
(TIF)
Phlh-30::HLH-30::GFP animals were treated with either empty vector (A, C) or hpk-1(RNAi) (B, D) and then were placed under ad libitum or bacterial deprivation conditions for 24 hours and HLH-30::GFP subcellular localization was assessed.
(TIF)
Pdaf-16::DAF-16::GFP animals were treated with either empty vector (A, C) or hpk-1(RNAi) (B, D) and then were placed under ad libitum or bacterial deprivation conditions for 24 hours and DAF-16::GFP subcellular localization was assessed.
(TIF)
This dataset contains all lifespan data discussed within this manuscript, as well as data from other experiments that support lifespan data represented in in-text and supplemental figures. This dataset also contains select statistical analyses to support the significance of the lifespan differences described in the text.
(XLSX)
This dataset contains all data pertaining to Fig 5: including median lifespan, p-values, and total number of animals examined for thermotolerance assays.
(XLSX)
This dataset contains all data pertaining to Figs 2D and 2E, 3D and 3E and 13: including: total number of animals examined for polyQ foci formation and paralysis, quantification, and statistical analysis. Supporting experiments with additional independently derived transgenic lines are also reported.
(XLSX)
This dataset contains all data pertaining to Fig 10: including total number of cells examined for autophagosome formation, average number of foci per cell, and S.E.M.
(XLS)
(XLSX)
(PDF)
Acknowledgments
We would like to thank Dr. Gary Ruvkun at Massachusetts General Hospital and Harvard Medical School and Dr. Benoit Biteau at the University of Rochester Medical Center for critical reading of the manuscript. Some strains were provided by the CGC (Caenorhabditis Genetics Center), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). A plasmid containing HSF-1::GFP and the strain EQ87 were generously provided by Dr. Hsu at The University of Michigan. Dr. Keith Nehrke at the University of Rochester Medical Center kindly provided strain KWN147. HSF-1 antibodies were generated by Dr. Peter Douglas and Dr. Michael French, while they were in the laboratories of Dr. Andrew Dillin at The University of California, Berkeley and Dr. Anthony Hunter at The Salk Institute for Biological Studies (respectfully); and we sincerely thank them for sharing this valuable resource. We thank Dr. Malene Hansen (Sanford Burnham Prebys Medical Discovery Institute) and Dr. Keith Nehrke for assisting with image analysis and helpful intellectual discussions. We thank Dr. Alison Frand (The University of California, Los Angeles) for discussions of the publication and review process.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The Ellison Medical Foundation New Scholars in Aging Fellowship (AG-NS-0681-10) (http://www.ellisonfoundation.org) provided support for salaries and supplies. The Glenn Foundation for Medical Research (Glenn Award for Research in Biological Mechanisms of Aging PS#: 213633) (http://glennfoundation.org) provided support for equipment and supplies. ARRA funding for faculty recruitment in Aging and Metabolism Research (P30 AG036489-01) (www.nih.gov) provided support for equipment, salaries, and supplies. EAM and TL were supported by the National Institutes of Health (R01GM105655). The National Institute On Aging of the National Institutes of Health (R01AG043421) provides current salary support for AVS and JAM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–12. doi: 10.1038/nature08980 . [DOI] [PubMed] [Google Scholar]
- 2.Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 2015;88(Pt B):290–301. doi: 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiLoreto R, Murphy CT. The cell biology of aging. Mol Biol Cell. 2015;26(25):4524–31. doi: 10.1091/mbc.E14-06-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–54. Epub 2008/04/01. doi: 10.1146/annurev.biochem.77.061206.171059 . [DOI] [PubMed] [Google Scholar]
- 5.Riera CE, Dillin A. Emerging Role of Sensory Perception in Aging and Metabolism. Trends Endocrinol Metab. 2016;27(5):294–303. doi: 10.1016/j.tem.2016.03.007 . [DOI] [PubMed] [Google Scholar]
- 6.Shore DE, Ruvkun G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 2013;23(9):409–20. Epub 2013/06/04. doi: 10.1016/j.tcb.2013.04.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447(7144):545–9. doi: 10.1038/nature05904 . [DOI] [PubMed] [Google Scholar]
- 8.Lee SS, Tyler JK. Physiology: Stressed-out chromatin promotes longevity. Nature. 2016;534(7609):625–6. doi: 10.1038/534625a . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pu M, Ni Z, Wang M, Wang X, Wood JG, Helfand SL, et al. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29(7):718–31. doi: 10.1101/gad.254144.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A, Kumar N, Matai L, Jain V, Garg A, Mukhopadhyay A. A chromatin modifier integrates insulin/IGF-1 signalling and dietary restriction to regulate longevity. Aging Cell. 2016;15(4):694–705. doi: 10.1111/acel.12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–38. doi: 10.1101/gad.1657108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64(2):167–70. doi: 10.1093/gerona/gln071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epel ES, Lithgow GJ. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J Gerontol A Biol Sci Med Sci. 2014;69 Suppl 1:S10–6. doi: 10.1093/gerona/glu055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009;19(2):52–61. doi: 10.1016/j.tcb.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157(1):52–64. doi: 10.1016/j.cell.2014.03.007 . [DOI] [PubMed] [Google Scholar]
- 16.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11(8):545–55. doi: 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, et al. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol. 2013;23(5):310–22. doi: 10.1016/j.semcancer.2013.05.008 . [DOI] [PubMed] [Google Scholar]
- 18.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4(2):e24 doi: 10.1371/journal.pgen.0040024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15(6):668–76. doi: 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nature communications. 2013;4:2267 doi: 10.1038/ncomms3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura S, Karalay O, Jager PS, Horikawa M, Klein C, Nakamura K, et al. Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nature communications. 2016;7:10944 doi: 10.1038/ncomms10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, et al. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet. 2013;9(7):e1003651 doi: 10.1371/journal.pgen.1003651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo B, Huang X, Zhang P, Qi L, Liang Q, Zhang X, et al. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO reports. 2014;15(6):705–13. doi: 10.1002/embr.201338310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haroon S, Vermulst M. Linking mitochondrial dynamics to mitochondrial protein quality control. Curr Opin Genet Dev. 2016;38:68–74. doi: 10.1016/j.gde.2016.04.004 . [DOI] [PubMed] [Google Scholar]
- 25.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320(5877):811–4. doi: 10.1126/science.1156093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106(35):14914–9. doi: 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vellai T, Takacs-Vellai K, Sass M, Klionsky DJ. The regulation of aging: does autophagy underlie longevity? Trends Cell Biol. 2009;19(10):487–94. doi: 10.1016/j.tcb.2009.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci U S A. 2011;108(34):14204–9. doi: 10.1073/pnas.1106557108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shai N, Shemesh N, Ben-Zvi A. Remodeling of Proteostasis Upon Transition to Adulthood is Linked to Reproduction Onset. Current genomics. 2014;15(2):122–9. doi: 10.2174/1389202915666140221005023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbadia J, Morimoto RI. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol Cell. 2015;59(4):639–50. doi: 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. The Journal of clinical investigation. 2015;125(1):65–74. doi: 10.1172/JCI73944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calzado MA, Renner F, Roscic A, Schmitz ML. HIPK2: a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle. 2007;6(2):139–43. doi: 10.4161/cc.6.2.3788 . [DOI] [PubMed] [Google Scholar]
- 33.Rinaldo C, Siepi F, Prodosmo A, Soddu S. HIPKs: Jack of all trades in basic nuclear activities. Biochim Biophys Acta. 2008;1783(11):2124–9. doi: 10.1016/j.bbamcr.2008.06.006 . [DOI] [PubMed] [Google Scholar]
- 34.Hofmann TG, Stollberg N, Schmitz ML, Will H. HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res. 2003;63(23):8271–7. . [PubMed] [Google Scholar]
- 35.Kim EA, Noh YT, Ryu MJ, Kim HT, Lee SE, Kim CH, et al. Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J Biol Chem. 2006;281(11):7489–97. doi: 10.1074/jbc.M507227200 . [DOI] [PubMed] [Google Scholar]
- 36.Lee P, Cho BR, Joo HS, Hahn JS. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol Microbiol. 2008;70(4):882–95. doi: 10.1111/j.1365-2958.2008.06450.x . [DOI] [PubMed] [Google Scholar]
- 37.Shojima N, Hara K, Fujita H, Horikoshi M, Takahashi N, Takamoto I, et al. Depletion of homeodomain-interacting protein kinase 3 impairs insulin secretion and glucose tolerance in mice. Diabetologia. 2012;55(12):3318–30. doi: 10.1007/s00125-012-2711-1 . [DOI] [PubMed] [Google Scholar]
- 38.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21(22):2976–94. doi: 10.1101/gad.1588907 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103(1):45–50. doi: 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakai A. Molecular basis of HSF regulation. Nature structural & molecular biology. 2016;23(2):93–5. doi: 10.1038/nsmb.3165 . [DOI] [PubMed] [Google Scholar]
- 41.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–115. doi: 10.1146/annurev-biochem-060809-095203 . [DOI] [PubMed] [Google Scholar]
- 42.Berber S, Wood M, Llamosas E, Thaivalappil P, Lee K, Liao BM, et al. Homeodomain-Interacting Protein Kinase (HPK-1) regulates stress responses and ageing in C. elegans. Scientific reports. 2016;6:19582 doi: 10.1038/srep19582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99(16):10417–22. doi: 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berber S, Llamosas E, Thaivalappil P, Boag PR, Crossley M, Nicholas HR. Homeodomain interacting protein kinase (HPK-1) is required in the soma for robust germline proliferation in C. elegans. Developmental dynamics: an official publication of the American Association of Anatomists. 2013;242(11):1250–61. doi: 10.1002/dvdy.24023 . [DOI] [PubMed] [Google Scholar]
- 45.de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell. 2012;46(4):472–83. doi: 10.1016/j.molcel.2012.03.003 . [DOI] [PubMed] [Google Scholar]
- 46.Volovik Y, Maman M, Dubnikov T, Bejerano-Sagie M, Joyce D, Kapernick EA, et al. Temporal requirements of heat shock factor-1 for longevity assurance. Aging Cell. 2012;11(3):491–9. doi: 10.1111/j.1474-9726.2012.00811.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95(2):199–210. [DOI] [PubMed] [Google Scholar]
- 48.Wolkow CA. Life span: getting the signal from the nervous system. Trends in neurosciences. 2002;25(4):212–6. . [DOI] [PubMed] [Google Scholar]
- 49.Vihervaara A, Sistonen L. HSF1 at a glance. J Cell Sci. 2014;127(Pt 2):261–6. doi: 10.1242/jcs.132605 . [DOI] [PubMed] [Google Scholar]
- 50.Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148(1–2):322–34. doi: 10.1016/j.cell.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilgarth RS, Sarge KD. Detection of sumoylated proteins. Methods in molecular biology (Clifton, NJ. 2005;301:329–38. doi: 10.1385/1-59259-895-1:329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6(6):413–29. [DOI] [PubMed] [Google Scholar]
- 53.Cypser JR, Johnson TE. Multiple stressors in Caenorhabditis elegans induce stress hormesis and extended longevity. J Gerontol A Biol Sci Med Sci. 2002;57(3):B109–14. . [DOI] [PubMed] [Google Scholar]
- 54.Cypser JR, Tedesco P, Johnson TE. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41(10):935–9. doi: 10.1016/j.exger.2006.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shemesh N, Shai N, Ben-Zvi A. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell. 2013;12(5):814–22. doi: 10.1111/acel.12110 . [DOI] [PubMed] [Google Scholar]
- 56.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–80. . [DOI] [PubMed] [Google Scholar]
- 57.Morton EA, Lamitina T. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell. 2013;12(1):112–20. doi: 10.1111/acel.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168(4):1937–49. doi: 10.1534/genetics.104.028423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garufi A, Ricci A, Trisciuoglio D, Iorio E, Carpinelli G, Pistritto G, et al. Glucose restriction induces cell death in parental but not in homeodomain-interacting protein kinase 2-depleted RKO colon cancer cells: molecular mechanisms and implications for tumor therapy. Cell death & disease. 2013;4:e639 doi: 10.1038/cddis.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131(16):3897–906. doi: 10.1242/dev.01255 . [DOI] [PubMed] [Google Scholar]
- 61.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426(6967):620 doi: 10.1038/426620a . [DOI] [PubMed] [Google Scholar]
- 62.McCormick MA, Tsai SY, Kennedy BK. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci. 2011;366(1561):17–27. doi: 10.1098/rstb.2010.0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo K, Choi E, Lee D, Jeong DE, Jang SK, Lee SJ. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell. 2013;12(6):1073–81. doi: 10.1111/acel.12140 . [DOI] [PubMed] [Google Scholar]
- 64.Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy. 2015;11(6):867–80. doi: 10.1080/15548627.2015.1034410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–45. doi: 10.1038/nature11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21(18):1507–14. doi: 10.1016/j.cub.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447(7144):550–5. doi: 10.1038/nature05837 . [DOI] [PubMed] [Google Scholar]
- 68.Gelino S, Chang JT, Kumsta C, She X, Davis A, Nguyen C, et al. Intestinal Autophagy Improves Healthspan and Longevity in C. elegans during Dietary Restriction. PLoS Genet. 2016;12(7):e1006135 doi: 10.1371/journal.pgen.1006135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3(1):21–5. . [DOI] [PubMed] [Google Scholar]
- 70.Kumsta C, Chang JT, Schmalz J, Hansen M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nature communications. 2017;8:14337 doi: 10.1038/ncomms14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–63. doi: 10.1016/j.cell.2014.05.048 . [DOI] [PubMed] [Google Scholar]
- 72.Johnson DW, Llop JR, Farrell SF, Yuan J, Stolzenburg LR, Samuelson AV. The Caenorhabditis elegans Myc-Mondo/Mad complexes integrate diverse longevity signals. PLoS Genet. 2014;10(4):e1004278 doi: 10.1371/journal.pgen.1004278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18(18):1355–64. doi: 10.1016/j.cub.2008.07.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song JJ, Lee YJ. Effect of glucose concentration on activation of the ASK1-SEK1-JNK1 signal transduction pathway. J Cell Biochem. 2003;89(4):653–62. doi: 10.1002/jcb.10541 . [DOI] [PubMed] [Google Scholar]
- 75.Song JJ, Lee YJ. Role of the ASK1-SEK1-JNK1-HIPK1 signal in Daxx trafficking and ASK1 oligomerization. J Biol Chem. 2003;278(47):47245–52. doi: 10.1074/jbc.M213201200 . [DOI] [PubMed] [Google Scholar]
- 76.Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, et al. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol Cell Biol. 2006;26(7):2758–71. doi: 10.1128/MCB.26.7.2758-2771.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raich WB, Moorman C, Lacefield CO, Lehrer J, Bartsch D, Plasterk RH, et al. Characterization of Caenorhabditis elegans homologs of the Down syndrome candidate gene DYRK1A. Genetics. 2003;163(2):571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11(8):4045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hartley AD, Ward MP, Garrett S. The Yak1 protein kinase of Saccharomyces cerevisiae moderates thermotolerance and inhibits growth by an Sch9 protein kinase-independent mechanism. Genetics. 1994;136(2):465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–5. doi: 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- 81.Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem. 2000;275(18):13321–9. . [DOI] [PubMed] [Google Scholar]
- 82.Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17(1):61–70. doi: 10.1093/emboj/17.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conrad E, Polonio-Vallon T, Meister M, Matt S, Bitomsky N, Herbel C, et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 2016;23(1):110–22. doi: 10.1038/cdd.2015.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18(1):1–12. doi: 10.1016/j.molcel.2005.03.012 . [DOI] [PubMed] [Google Scholar]
- 85.Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol. 2006;26(3):955–64. doi: 10.1128/MCB.26.3.955-964.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132(6):1025–38. doi: 10.1016/j.cell.2008.01.030 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6(8). doi: 10.1371/journal.pgen.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, et al. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell metabolism. 2012;16(4):526–37. doi: 10.1016/j.cmet.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang K, Fingar DC. Growing knowledge of the mTOR signaling network. Seminars in cell & developmental biology. 2014;36:79–90. doi: 10.1016/j.semcdb.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ecsedy JA, Michaelson JS, Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol Cell Biol. 2003;23(3):950–60. doi: 10.1128/MCB.23.3.950-960.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119(7):969–79. doi: 10.1016/j.cell.2004.11.047 . [DOI] [PubMed] [Google Scholar]
- 92.Moriya H, Shimizu-Yoshida Y, Omori A, Iwashita S, Katoh M, Sakai A. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 2001;15(10):1217–28. doi: 10.1101/gad.884001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaeberlein M, Kennedy BK. Hot topics in aging research: protein translation and TOR signaling, 2010. Aging Cell. 2011;10(2):185–90. doi: 10.1111/j.1474-9726.2010.00665.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fawcett TW, Sylvester SL, Sarge KD, Morimoto RI, Holbrook NJ. Effects of neurohormonal stress and aging on the activation of mammalian heat shock factor 1. J Biol Chem. 1994;269(51):32272–8. . [PubMed] [Google Scholar]
- 96.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153(7):1435–47. doi: 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berendzen KM, Durieux J, Shao LW, Tian Y, Kim HE, Wolff S, et al. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell. 2016;166(6):1553–63 e10. doi: 10.1016/j.cell.2016.08.042 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tatum MC, Ooi FK, Chikka MR, Chauve L, Martinez-Velazquez LA, Steinbusch HW, et al. Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr Biol. 2015;25(2):163–74. doi: 10.1016/j.cub.2014.11.040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harbor perspectives in biology. 2011;3(5). doi: 10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kikis EA, Gidalevitz T, Morimoto RI. Protein homeostasis in models of aging and age-related conformational disease. Advances in experimental medicine and biology. 2010;694:138–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–95. doi: 10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naidu SD, Dinkova-Kostova AT. Regulation of the mammalian heat shock factor 1. Febs Journal. 2017;284(11):1606–27. doi: 10.1111/febs.13999 [DOI] [PubMed] [Google Scholar]
- 105.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nature reviews. 2015;16(6):345–57. doi: 10.1038/nrn3961 . [DOI] [PubMed] [Google Scholar]
- 106.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques. 2002;32(4):728–30. . [DOI] [PubMed] [Google Scholar]
- 107.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300(5619):644–7. doi: 10.1126/science.1083614 [DOI] [PubMed] [Google Scholar]
- 108.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22(9):1762–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological). 1995:289–300. [Google Scholar]
- 110.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 111.Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, et al. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16(7):3273–88. doi: 10.1091/mbc.E05-01-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell metabolism. 2010;12(3):260–72. doi: 10.1016/j.cmet.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) HPK-1 is broadly expressed during development, with limited expression by adulthood. (A, Top left) Expression of Phpk-1::HPK-1::GFP during embryogenesis. (A, top right) Phpk-1::HPK-1::GFP expression becomes increasingly restricted by the L3 stage of development to neurons (arrow with N), intestinal cells (arrow with I), and the hypodermis (arrow with H). (A bottom panel) HPK-1 expression is limited to neuronal cells by the L4 stage of development and in adults (not shown). Brightfield insets for each image and white dotted lines trace animals. In all cases, representative images are shown of at least fifteen animals from two independently derived lines. (B) HPK-1 is localized in the nucleus. Phpk-1::HPK-1::GFP expression with the overlay of DIC image. Arrows indicate examples of localized expression within nuclei.
(TIF)
(A) Constitutive inactivation of hpk-1 (red) from the L1 stage by feeding-based RNAi in wild-type N2 worms decreases lifespan compared to control (EV) (black) RNAi. (B) hpk-1 RNAi only during development (red) results in a decrease in lifespan comparable to constitutive hpk-1 RNAi. Animals were moved to dcr-1(RNAi) at L4. (C) hpk-1 RNAi (red) initiated at L4 decreases lifespan to a smaller, but significant extent. HPK-1 spatial requirements mirror tissues of expression. (D) Intestinal or (E) hypodermal specific gene inactivation of hpk-1 (red) reduces lifespan. In contrast, (F) muscle specific gene inactivation of hpk-1 has a minimal effect on lifespan, consistent with a lack of muscle expression during larval development (S1 File). (G) Neuronal inactivation of hpk-1 (pink) reduces lifespan. (H) Neuronal inactivation of daf-2 increases lifespan (orange), consistent with previous reports[47, 48]. (I) sid-1(pk3321) loss of function animals have impaired RNAi: hpk-1(RNAi) (pink) and daf-2(RNAi)-treatment (red), respectively. Full lifespan data can be found in S1 Table.
(TIF)
(A) Inactivation of the SUMO moiety smo-1 by RNAi prevents formation of higher molecular weight isoforms of HSF-1. Data supports Fig 6A. RNAi knockdown of smo-1 showed some variability in the degree of effectiveness, but generally decreased the relative proportion of higher molecular weight isoforms of HSF-1. Relative proportion of modified HSF-1 shown in the lower panel is 0.49, 0.35, and 0.77 for N2/ev, hpk-1(pk1393)/smo-1, and hpk-1(pk1393)/ev, respectively. ev = empty vector RNAi treatment (L4440). dePhos is λ protein phosphatase treatment. β-actin serves as a loading control. (B, C) Additional replicates for quantification of the ratio of modified to unmodified HSF-1 are shown in Fig 6B. For (B) no β-actin control was available, which precluded quantifying differences in overall expression levels between samples. (C) Additional replicates for quantifying differences in overall HSF-1 levels between N2 and hpk-1(pk1393) shown in Fig 6C. β-actin serves as a loading control. In all cases U = unmodified (75 kD), S = sumoylated (~90 kD), and S+P = sumoylated and phosphorylated (~95 kD) isoforms of HSF-1.
(TIF)
(A, B) Phsp-16.2::GFP is not expressed at 20°C (basal conditions) in day one adult animals. Outlines of animals are traced in white. (C, D, E, F) Phsp-16.2::GFP is induced heterogeneously across tissues after continued exposure to mild temperature stress of 25°C in day one adult animals. (G, H) Transient hormetic exposure to 25°C increases mean lifespan (gray versus black), which is dependent on both hpk-1 (G, red) and hsf-1 (H, blue). See S1 Table for comprehensive lifespan data.
(TIF)
(A) Strain expressing a single copy of HSF-1::GFP is diffusely localized in the nucleus under basal conditions. (B) Strain expressing a single copy of HSF-1::GFP forms nuclear stress granules after heat shock. (C-D) Strain expressing a single copy of HSF-1::GFP treated with hpk-1(RNAi) does not alter HSF-1 basal localization (C) or formation of stress granules after heat stress (D). Results were consistent across three independent trials; approximately 20 animals were visualized per trial and all animals produced nuclear stress granules regardless of hpk-1 status. (E) Strain with low copy HSF-1::GFP shows increased nuclear accumulation after heat stress (column 1 to 2), which is not altered in the absence of hpk-1 (compare 3 to 4). Two independent trials produced similar results, with 10 animals per condition in each trial. Graph includes results from one trial (*** p <0.001, Student’s t-test).
(TIF)
(A) Mean GFP intensity was measured in ImageJ for: head neurons, 2–4 hypodermal seam cells, and 4 spots of internal non-neuronal, non-seam cell background fluorescence for the indicated number of animals (n). Head and seam cell GFP intensity was normalized to average background fluorescence, and the contribution of background fluorescence was subtracted. Mean and standard deviation of head neuron and seam cell fluorescence in unstressed and heat shocked animals was calculated, and normalized to unstressed animals. * indicates p<0.0005, ** p<0.0002 (Student’s t-test). (B) As an independent verification of increased fluorescence within head neurons, we applied a matrix for assessing GFP levels via double-blind visualization, which allowed us to score a larger cohort of animals +/- heat shock. GFP expression was scored as “strong”, “moderate”, or “weak”. * p<0.03 (Mann-Whitney).
(TIF)
(A-B). Phpk-1::GFP expression in L4 animals under unstressed conditions (A) or after 1 hour heat shock (B). For additional images see S1 File. (C) Endogenous hpk-1 mRNA levels of L4 wild-type unstressed animals or following a 15-minute recovery after 1 hour of heat shock at 35°C. (D) Endogenous hsp-16.2 mRNA levels are induced following a 15-minute recovery after 1 hour of heat shock (p<0.05, Student’s t-test). Fold change is relative to unstressed N2. Note: mRNA from the same extracts was used in (C) and (D). In all cases expression is normalized to act-1 mRNA and error bars represent the standard deviation of a minimum of three replicates in each of two independent trials.
(TIF)
(A-C) Expression of Phsp-16.2::GFP animals under the following conditions: unstressed (A), 2 hours of recovery following heat shock for 1 hour at 35°C heat shock (B), and heat shock as in (B) but with prior treatment with 100 ug/mL α-amanitin treatment (C) (as described in [112]). Note, images in (B) and (C) were taken at a similar magnification, which was higher than image (A). Outlines of animals are traced in white.
(TIF)
(A) Mean GFP intensity within head neurons was measured in ImageJ for Phpk-1::HPK-1::GFP expressing animals after empty vector or daf-15(RNAi)-treatment. Change in expression was normalized to empty vector; error bars represent the standard deviation of six animals. * p<0.03 (Student’s t-test). (B-D) Additional images of Phpk-1::HPK-1::GFP expression after empty vector control or daf-15(RNAi) treatment. Expression of Phpk-1::HPK-1::GFP is only induced within neurons after daf-15 (Raptor) inactivation compared to control. While expression within hypodermal seam cells varied between animals, no differences in expression between empty vector and daf-15 RNAi could be discerned, which is in contrast to heat shock. Images in (C) and (D) were stitched together from several overlapping high-resolution images and white space was artificially filled.
(TIF)
Phlh-30::HLH-30::GFP animals were treated with either empty vector (A, C) or hpk-1(RNAi) (B, D) and then were placed under ad libitum or bacterial deprivation conditions for 24 hours and HLH-30::GFP subcellular localization was assessed.
(TIF)
Pdaf-16::DAF-16::GFP animals were treated with either empty vector (A, C) or hpk-1(RNAi) (B, D) and then were placed under ad libitum or bacterial deprivation conditions for 24 hours and DAF-16::GFP subcellular localization was assessed.
(TIF)
This dataset contains all lifespan data discussed within this manuscript, as well as data from other experiments that support lifespan data represented in in-text and supplemental figures. This dataset also contains select statistical analyses to support the significance of the lifespan differences described in the text.
(XLSX)
This dataset contains all data pertaining to Fig 5: including median lifespan, p-values, and total number of animals examined for thermotolerance assays.
(XLSX)
This dataset contains all data pertaining to Figs 2D and 2E, 3D and 3E and 13: including: total number of animals examined for polyQ foci formation and paralysis, quantification, and statistical analysis. Supporting experiments with additional independently derived transgenic lines are also reported.
(XLSX)
This dataset contains all data pertaining to Fig 10: including total number of cells examined for autophagosome formation, average number of foci per cell, and S.E.M.
(XLS)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.