Abstract
Cellular heterogeneity is a common feature in breast cancer, yet an understanding of the coexistence and regulation of various tumor cell subpopulations remains a significant challenge in cancer biology. In the current study, we approached tumor cell heterogeneity from the perspective of Wnt pathway biology to address how different modes of Wnt signaling shape the behaviors of diverse cell populations within a heterogeneous tumor landscape. Using a syngeneic TP53-null mouse model of breast cancer, we identified distinctions in the topology of canonical Wnt β-catenin-dependent signaling activity and non-canonical β-catenin-independent Ror2-mediated Wnt signaling across subtypes and within tumor cell subpopulations in vivo. We further discovered an antagonistic role for Ror2 in regulating canonical Wnt/β-catenin activity in vivo, where lentiviral shRNA depletion of Ror2 expression augmented canonical Wnt/β-catenin signaling activity across multiple basal-like models. Depletion of Ror2 expression yielded distinct phenotypic outcomes and divergent alterations in gene expression programs among different tumors, despite all sharing basal-like features. Notably, we uncovered cell state plasticity and adhesion dynamics regulated by Ror2, which influenced Ras Homology Family Member A (RhoA) and Rho-Associated Coiled-Coil Kinase 1 (ROCK1) activity downstream of Dishevelled-2 (Dvl2). Collectively, these studies illustrate the integration and collaboration of Wnt pathways in basal-like breast cancer, where Ror2 provides a spatiotemporal function to regulate the balance of Wnt signaling and cellular heterogeneity during tumor progression.
Introduction
The advent of high-throughput molecular profiling has helped classify breast cancers into distinct subtypes by global gene expression-based approaches. These ‘intrinsic’ subtypes include Luminal A, Luminal B, Her2-enriched, basal-like, claudin-low and normal-like, and complement classical pathology to reveal distinctions in breast cancer incidence, survival and treatment responses.1, 2, 3 Triple negative breast cancers lack expression of estrogen and progesterone receptors (ER and/or PR) and amplification of epidermal growth factor receptor 2 (Her2), and are further divided into the basal and claudin-low subtypes.4, 5 Notably, breast cancer subtypes reflect differentiation states of the normal epithelial hierarchy, spanning cell states that reflect the multipotent stem cell down to fully differentiated ER+ luminal cells.6 The basal-like and claudin-low subtypes are suggested to reflect the less-differentiated luminal progenitor and mammary stem cell states, respectively, with claudin-low tumors exhibiting more mesenchymal-like characteristics.7 Considerable evidence now suggests that while global gene expression-based approaches capture the intertumoral heterogeneity between individual tumors and subtypes, these approaches fail to resolve the cellular diversity present within individual tumors. Indeed, more refined molecular analyses now demonstrate considerable genetic and phenotypic variations within tumors, where vast intratumoral cellular hierarchies exist.8
Wnt signals are categorized by their ability to either stabilize β-catenin in the nucleus (canonical/β-catenin-dependent) or elicit alternative routes of intracellular signaling independent of β-catenin stabilization (non-canonical/β-catenin-independent).9 Canonical Wnt signaling controls numerous biological processes that include cell fate determination, self-renewal and tissue homeostasis.10 Non-canonical Wnt signals regulate organized cellular movements, cell shape and the orientation of proper cell polarity in development.11, 12 A canonical or non-canonical Wnt signaling outcome within a cell depends, in part, on the receptor context.12, 13, 14 Canonical Wnt signaling utilizes Frizzled receptors (Fzd) together with co-receptors low-density lipoprotein-related receptor proteins 5/6 (Lrp5/6) to stabilize intracellular β-catenin and transcriptional activation of T-cell factor/Lymphoid enhancer-binding factor (Tcf/Lef)-dependent outputs, while non-canonical Wnt signals require Fzd receptors independent of Lrp5/6 and/or utilize the alternative Wnt receptors Ror1, Ror2 or Ryk. Alternative Wnt receptors are implicated in conveying a Wnt5a signal; however, Ror2 represents a favored receptor candidate of non-canonical Wnt5a signaling due to similarities in spatiotemporal expression across tissues and overlap between loss-of-function mutants.15, 16, 17, 18 While less defined, alternative Wnt outputs downstream of Ror2 include Rho and Rac GTPases, c-Jun N-terminal kinase (JNK), the calcium-sensitive kinases calmodulin-dependent protein kinase (CAMKII) and protein kinase C (PKC), and nuclear factor of activated T cells (NFAT).9, 11 The ability of non-canonical Wnt signaling to inhibit the canonical Wnt pathway in some contexts emphasizes the cross talk and lack of linearity between pathways. Deciphering this integration remains an unresolved task both in development and disease.
Activating mutations within the Wnt pathway are infrequent in breast cancer;19 however, hyperactivated levels of canonical Wnt signaling exist within basal-like breast cancers.20, 21, 22 Studies also demonstrate a role for non-canonical Wnt signaling in breast cancer, with either negative or positive correlations regarding clinical outcome, involving a variety of alternative Wnt signaling constituents that include Vangl,23 Fzds,24 Rors25, 26 and individual Wnts.27, 28 The broad spectrum of non-canonical Wnt signaling mechanisms and diverse receptor repertoires complicate the assignment of generalized roles for alternative Wnt signaling in breast cancer. An outstanding challenge is to decipher how inter- and intratumoral heterogeneity factor into the equation when considering the Wnt signaling landscape and the integration of multiple arms of Wnt signaling within a tumor.
In the current study, we utilize a transplantable syngeneic TP53-null genetically engineered mouse model of breast cancer to interrogate the function of the alternative Wnt receptor Ror2 in breast cancer. We establish the integration of Wnt pathways within basal-like TP53-null tumors and an inverse correlation between Wnt β-catenin-dependent and -independent pathways among tumor cell subpopulations. Furthermore, we identify context-dependent functions for Ror2 with respect to tumor heterogeneity, and uncover unique nodes of signaling downstream of Ror2 which regulate plasticity, cell fate and cell–cell interactions within the tumor landscape.
Results
Assessment of Wnt/β-catenin-dependent activity and expression of the alternative Wnt receptor Ror2 uncovers the presence of inter- and intratumoral heterogeneity within the TP53-null mammary tumor model
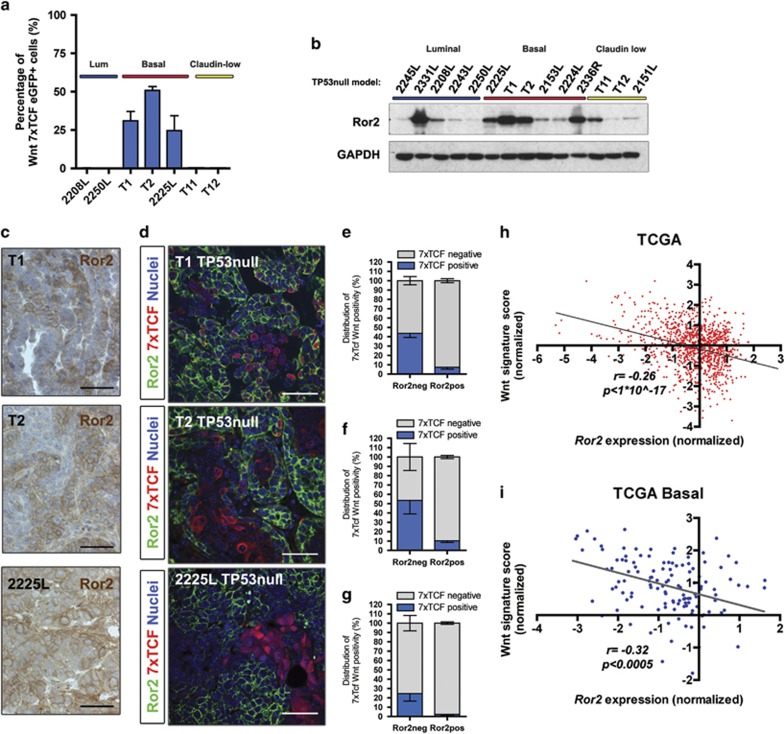
We utilized a collection of transplantable mammary tumors representing characteristics of human breast cancer with respect to histology, gene expression programs and subtype, derived from the TP53-null genetically engineered mouse model,29, 30, 31 to assess the integration of Wnt/β-catenin-dependent and -independent pathways in vivo. Using a subset of these models representing luminal, basal-like and claudin-low subtypes, we first examined the Wnt pathway landscape across heterogeneous TP53-null models. By transduction of tumor cells with a lentiviral reporter harboring a 7xTCF/Lef responsive promoter upstream of eGFP,32 we observed that only basal-like TP53-null tumors (T1, T2, 2225L) harbored canonical Wnt-active cell populations relative to luminal and claudin-low tumors, similar to that observed in human breast cancers (Figure 1a).21, 22, 33 Within TP53-null tumors with intrinsic Wnt activity, we also detected expression of the alternative Wnt receptor Ror2, a mediator of non-canonical Wnt signaling (Figures 1b–g). Although Wnt/β-catenin-dependent pathway activity was restricted to basal-like tumors, we observed varying levels of Ror2 expression in all subtypes of the TP53-null model, supporting context-dependent functions for Wnt/β-catenin-independent signaling within specific subtypes (Figure 1b). Upon closer examination of basal-like tumors, we identified prevalent expression of Ror2, with a heterogeneous and graded expression pattern, often distinct from cell populations harboring an active canonical Wnt/β-catenin-dependent signal based on 7xTCF-eGFP reporter activity in vivo (Figures 1c and d). As opposed to the normal mammary epithelium,34 some overlap of Ror2 expression with Wnt/β-catenin-dependent activity was detected within the TP53-null tumors, demonstrating lack of complete mutual exclusivity (Figures 1d–g). Interestingly, we identified a negative correlation between Ror2 expression and an active canonical Wnt signature across human breast tumors in the TCGA database (Figure 1h). These results were further validated in human breast tumors of the basal-like subset (Figure 1i), suggesting that Wnt/β-catenin-independent signaling through Ror2 may function as a cellular rheostat to control levels of Wnt/β-catenin-dependent activity within the tumor landscape.
Figure 1.
Assessment of canonical Wnt activity and Ror2 expression uncovers the complex coexistence of Wnt pathways in TP53-null mouse models and human breast cancer. (a) The percentage of canonical Wnt-active cells in basal-like TP53-null tumors relative to luminal and claudin-low, based on fluorescence-activated cell sorting (FACS) analysis of tumors harboring a lentiviral Wnt reporter with a 7xTCF responsive promoter upstream of eGFP. FACS percentages for basal-like tumors: T1 (31.0±6.1%, n=3 tumors), T2 (50.9±2.5%, n=3 tumors) and 2225L (24.6±9.8%, n=3 tumors). (b) Western blot for Ror2 in a panel of TP53-null tumors representing luminal, basal-like and claudin-low subtypes. (c) Immunohistochemistry for Ror2 within T1, T2 and 2225L basal-like TP53-null models depicting a spectrum of Ror2 positivity within each basal-like model. Scale 50 μm. (d) Co-immunofluorescence for Ror2 (green) and 7xTCF-mCherry (red), demarcating active canonical Wnt activity, across three TP53-null basal-like models. Scale 50 μm. (e–g) Quantitation of immunofluorescence staining of mCherry positive (7xTCF responsive) Wnt populations within Ror2-negative and Ror2-positive populations within basal-like tumors (e) T1 (43.7±4.4% in Ror2-neg vs 7.3±2.1% in Ror2-pos tumor cells, n=6 tumors), (f) T2 (53.5±14.14% in Ror2-neg vs 10.2±1.7% in Ror2-pos tumor cells, n=6 tumors) and (g) 2225L (24±8.2% in Ror2-neg vs 2.2±1.3% in Ror2-pos tumor cells). (h) Scatter plot of individual human breast tumors within the TCGA database illustrating an inverse correlation between Ror2 expression and an active canonical Wnt signature (Spearman rank correlation, n=1095 cases, correlation coefficient: −0.26, one-sided P<1 × 10−17). (i) Scatter plot of human breast tumors of the basal-like subtype within TCGA demonstrating an inverse correlation between Ror2 expression and an active canonical Wnt signature (Spearman rank correlation, 122 cases, correlation coefficient: −0.32, one-sided P<0.0005).
Depletion of Ror2 expression in basal-like TP53-null models impairs tumor growth and enhances canonical Wnt signaling across basal-like TP53-null models despite distinctions in both histopathology and gene expression outcomes
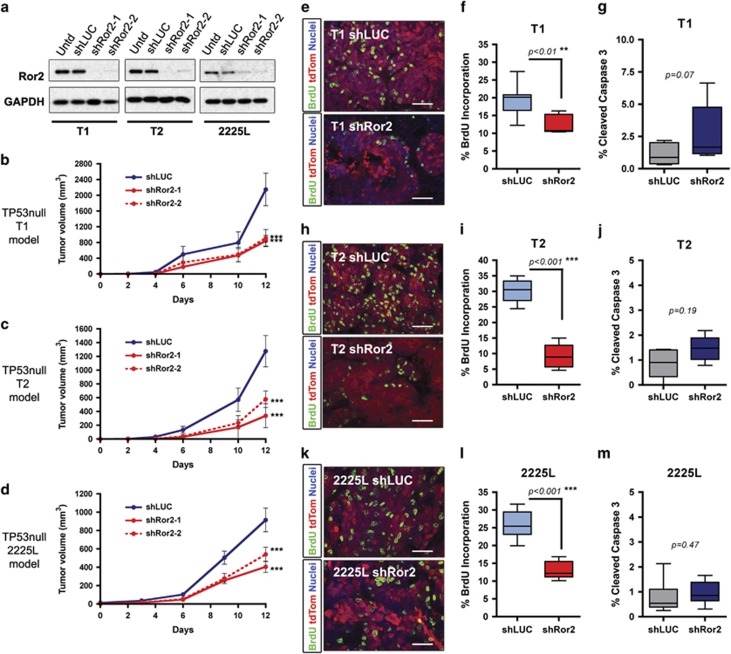
Given the co-occurrence of Ror2 expression with decreased Wnt/β-catenin signaling activity in basal-like TP53-null models, we used a lentiviral shRNA approach to deplete Ror2 in vivo and investigate effects on both the canonical Wnt landscape and tumor progression. Interestingly, Ror2 knockdown (Figure 2a) impaired tumor growth relative to controls that harbored a nonspecific hairpin (Figures 2b–d). Importantly, expression of Ror1, a second Ror family member possessing 56% amino acid homology with Ror2,35 was not impacted upon Ror2 knockdown (Supplementary Figures S1a and b). Ror2-depleted tumors exhibited a decrease in proliferation assessed by BrdU incorporation across T1 (Figures 2e and f), T2 (Figures 2h and i) and 2225L (Figures 2k and l) models, with no statistically significant changes observed in apoptosis (Figures 2g, j and m).
Figure 2.
Lentiviral silencing of Ror2 expression in basal-like TP53-null models impairs tumor growth. (a) Western blots for Ror2 illustrating shRNA depletion of Ror2 protein levels with two independent hairpins in T1, T2 and 2225L TP53-null basal-like models. GAPDH protein depicts equal loading between samples. (b–d) Tumor growth curves representing changes in tumor volume over time (days) for (b) T1, (c) T2 and (d) 2225L, comparing changes in tumor volume between shLUC control tumors and shRor2 tumors (n=6 tumors within each group, ***P<0.001). Volumes were calculated using the formula, volume=(length × width2)/2. (e,f,h,i,k,l) Proliferation of shLUC vs shRor2 tumors based on BrdU incorporation. (e,h,k) Representative immunofluorescence images of BrdU-positive cells (green) among transduced tumor cells (red) between shLUC and shRor2 groups within (e) T1, (h) T2 and (k) 2225L. Scale 50 μm. (f,i,l) Quantitation of BrdU-positive cells (green) among transduced tumors cells (red) between shLUC and shRor2 groups among (f) T1 (shLUC 19.4±4.2% vs shRor2 12.6±2.5%, n=6 **P<0.01), (i) T2 (shLUC 30.1±3.6% vs shRor2 9.1±3.8%, n=6, ***P<0.001), (l) 2225L (shLUC 26.1±3.9% vs shRor2 13.0±2.4%, n=6, ***P<0.001). (g,j,m) Quantitation of apoptosis, CC3 positivity, among transduced tumor cells between shLUC and shRor2 groups among (g) T1 (shLUC 1.0±0.4% vs shRor2 3.5±3.2%, n=6), (j) T2 (shLUC 0.90±0.52% vs shRor2 1.5±0.50%, n=6) and (m) 2225L (shLUC 0.80±0.65% vs shRor2 0.96±0.47%, n=6).
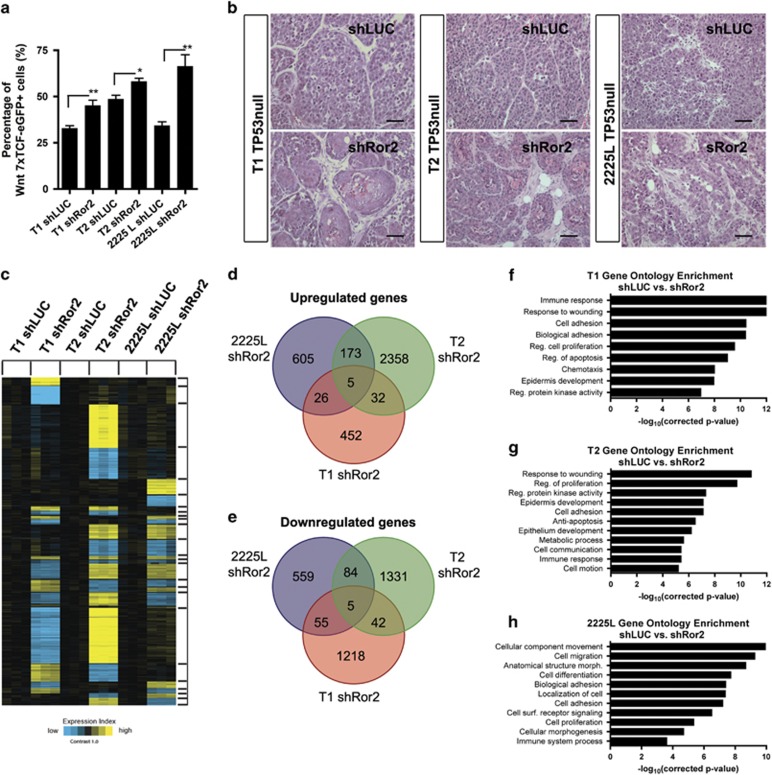
Co-transduction of tumors with the 7xTCF-eGFP Wnt reporter revealed that Ror2-depleted tumors displayed an increase in the percentage of cells harboring canonical Wnt/β-catenin-dependent activity, suggesting that depletion of Ror2 abrogated the inhibition of canonical Wnt signaling in vivo (Figure 3a). Conversely, lentiviral overexpression of Ror2 above endogenous levels enhanced the antagonism of canonical Wnt signaling in vivo (Supplementary Figures S2a–c). Although Ror2 loss enhanced canonical Wnt signaling across all basal-like tumor models (Figure 3a), other phenotypic outcomes were distinct. For example, histological examination of tumors revealed squamous differentiation present in T1 and T2 models upon Ror2 loss, previously observed in other GEM and PDX models with hyperactive Wnt signaling (Figure 3b and Supplementary Figure S1c).36, 37, 38, 39 Loricrin, a late epidermal differentiation marker, was expressed within areas exhibiting elevated 7xTCF-eGFP activity and squamous features within T1 (Supplementary Figures S1d and e). However, depletion of Ror2 in 2225L yielded a more disorganized tumor architecture and increase in stroma, with no squamous differentiation evident (Figure 3b and Supplementary Figure S1c).
Figure 3.
Depletion of Ror2 expression enhances canonical Wnt signaling across basal-like TP53-null models despite distinctions in both histopathology and gene expression outcomes. (a) Elevated Wnt/β-catenin signaling occurs in the absence of Ror2 in all three basal-like TP53-null models. Quantitation of fluorescence-activated cell sorting analysis of 7xTCF-eGFP positivity within shLUC and shRor2 tumors within T1, T2 and 2225L models (n=3 shLUC tumors and n=3 shRor2 tumors per basal-like model, *P<0.05, **P<0.001). (b) H&E staining of shLUC and shRor2 tumors showing squamous differentiation in T1 and T2 and disorganization in 2225L upon Ror2 loss. Scale 50 μm. (c) Supervised clustering and heat map showing gene expression changes (P<0.01 by t-test, fold change>1.4) across TP53-null basal-like models T1, T2 and 2225L, harboring shLUC vs shRor2 hairpins (derived from sorted transduced tdTomato-positive tumor cells). Within each model (T1, T2, 2225L), expression values were centered on the corresponding control. Bright yellow/blue represents minimum of two-fold change from the corresponding control. (d, e) Venn diagrams of (d) upregulated and (e) downregulated genes represented in shLUC and shRor2 groups within T1, T2 and 2225L, illustrating gene expression overlap between basal-like TP53-null models in response to Ror2 depletion. (f–h) Gene ontology analysis illustrating the enrichment of particular gene expression programs within (f) T1, (g) T2 and (h) 2225L.
Differences in histological outcomes despite a common trend in elevated Wnt/β-catenin-dependent activity upon Ror2 loss prompted a global assessment of gene expression alterations by microarray between shLUC and shRor2 tumors among basal-like models. Microarray analysis of sorted, lineage-depleted tdTomato-positive shLUC and shRor2 tumor cell populations failed to resolve a prominent Ror2 gene signature across models, but revealed distinct gene expression signatures in response to Ror2 depletion (Figure 3c). These differences were confirmed when assessing the overlap in up- and downregulated genes shared between models upon Ror2 depletion, with very few genes similarly changed across all three models (Figures 3d and e and Supplementary Figures S3a and b). Gene ontology (GO) enrichment analysis revealed cellular motifs including immune response, cell adhesion and epidermis development for T1 (Figure 3f); response to wounding, epidermis development, metabolism and cell adhesion for T2 (Figure 3g); and cell migration, biological adhesion and cellular morphogenesis for 2225L (Figure 3h). These data, therefore, uncovered unexpected context-dependent functions for Ror2 during mammary tumor progression, despite all models being designated as basal-like.
Loss of Ror2 expression in vivo results in changes in tumor cell plasticity between basal-like and claudin-low states
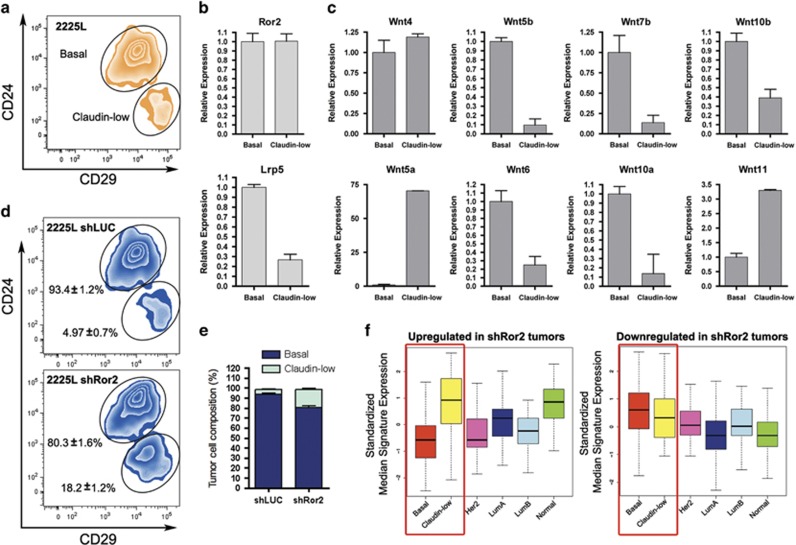
The presence of a strong cell adhesion and migration GO signature upon Ror2 depletion in 2225L prompted a more detailed investigation of Ror2 function within this model, given the role of non-canonical Wnt signaling in mediating cell adhesion, morphogenesis and polarity. Although gene expression profiling of the bulk 2225L TP53-null tumors categorized them as basal-like, further refinement of this model using flow cytometry for cell surface markers CD24 and CD29 resolved two distinct cell populations: a bulk CD24+CD29+ population and minor CD24negCD29+ population comprising ~5% of the tumor (Figure 4a). By gene expression profiling, these two populations clustered into distinct CD24+CD29+ basal-like and CD24negCD29+ claudin-low populations, similar to that observed in human cell lines.40 Intriguingly, when assessing expression of Ror2 and other Wnt receptors and ligands, we discovered that the basal-like and claudin-low populations harbored differences in Wnt pathway components. Of note, Ror2 expression was similar across the two populations (Figure 4b). Conversely, the Lrp5 co-receptor for Wnt/β-catenin-dependent signaling showed enrichment within the basal-like relative to claudin-low populations (Figure 4b), while some Wnt ligands showed enrichment within either basal-like (Wnt5b, Wnt6, Wnt7b, Wnt10a, Wnt10b) or claudin-low (Wnt5a, Wnt11) subpopulations, with neutral expression patterns present as well (Wnt4) (Figure 4c). These data suggest distinct subpopulations of tumor cells likely utilize Wnt signals in a refined manner to shape-specific cellular interactions and outcomes related to cell fate and coordinated movements, much like the normal mammary gland.
Figure 4.
shRNA depletion of Ror2 expression in vivo results in changes in tumor cell plasticity between basal-like and claudin-low subpopulations in the 2225L TP53-null model. (a) Representative fluorescence-activated cell sorting (FACS) plot depicting the segregation of 2225L tumors into two distinct populations based on CD24 and CD29 cell surface marker staining. (b) Quantitative real-time PCR of Ror2 and Lrp5 within sorted basal-like and claudin-low fractions from 2225L. (c) Quantitative real-time PCR of Wnt ligands within sorted basal-like and claudin-low subpopulations within 2225L. (d) FACS plots of shLUC and shRor2 tumors stained with CD24 and CD29, illustrating the shift in tumor cell populations upon Ror2 loss, from basal-like to claudin-low. (e) Quantitation of basal-like and claudin-low populations within shLUC and shRor2 2225L TP53-null tumors (shLUC basal-like 93.4±1.2% claudin-low 4.97±0.7% vs shRor2 basal-like 80.3±1.6% claudin-low 18.2±1.2%, n=5). (f) Box plots depicting gene expression signatures across subtypes represented in shRor2 vs shLUC tumors. Genes upregulated in shRor2 2225L tumors are associated with claudin-low features. Genes downregulated are associated with basal-like features (red boxes).
Interestingly, loss of Ror2 within 2225L resulted in a four-fold expansion of the claudin-low CD24negCD29+ population, revealing a shift in the cellular composition within the tumors (Figures 4d and e). This increased proportion of claudin-low to basal tumor cells was additionally reflected by gene expression profiling of shLUC and shRor2 tumors, where a claudin-low gene signature was enriched upon Ror2 loss (Figure 4f). Specifically, expression analysis of control versus Ror2-depleted tumors confirmed that genes upregulated in shRor2 tumors were associated with claudin-low features, while genes downregulated were associated with basal-like features (Figure 4f), reflecting the shift in balance of tumor cell subsets and change in Wnt signaling landscape across tumors.
Impaired Ror2 signaling enhances tumor cell migration by altering cytoskeletal and adhesion dynamics
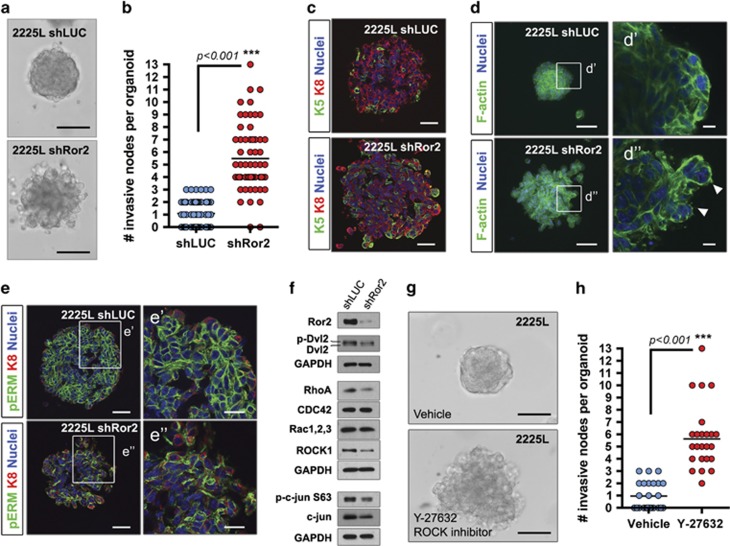
The fact that claudin-low breast cancers lack tight junctions and possess features of an epithelial-to-mesenchymal transition was intriguing when considering certain features identified in shRor2 2225L tumors, particularly the enriched cell adhesion and migration gene signature and histological features that indicated a less organized tumor landscape. As a surrogate assay, we developed three-dimensional models of lentivirus-infected tumor organoids for short-term phenotypic assessment, harboring shLUC and shRor2 hairpins, to recapitulate Ror2-intact versus -depleted tumors. Within the 2225L TP53-null basal-like organoid model, depletion of Ror2 conferred a pro-migratory phenotype, accompanied by more protrusive cellular fronts along with disseminated clusters of cells into the surrounding matrix (Figures 5a and b). T1 and T2 tumor organoids, on the other hand, lacked any evidence of invasive outcome in vitro and recapitulated the squamous phenotype upon Ror2 loss, marked by the presence of keratin pearls within the centers of shRor2 organoids (Supplementary Figures S4a and b). Within 2225L organoids, Keratin-8 (K8) and -5 (K5) cells, demarcating luminal and basal cell identities, respectively, were similarly heterogeneous between shLUC and shRor2 groups (Figure 5c). Since cytoskeletal defects were evident upon Ror2 loss in normal mammary development, we examined the distribution of F-actin in 2225L organoids by phalloidin staining. Of note, changes in cortical F-actin patterns were evident in shRor2 organoids relative to controls, where irregularities in cortical F-actin continuity were observed along with F-actin projections into the surrounding matrix (Figure 5d, shLUC magnified d′ and shRor2 d″). Additionally, the distribution of phosphorylated-Ezrin/Radixin/Moesin (pERM), a protein family necessary in crosslinking the actin cytoskeleton and plasma membrane, was discontinuous between cell–cell junctions in shRor2 organoids versus shLUC organoids, where a continuous pattern of pERM staining was evident (Figure 5e, magnified shLUC e′ and shRor2 e″). Indeed, other genes associated with junction integrity, such as desmocolin and desmoglein, were downregulated within shRor2 tumors. Collectively, these data suggest that alterations in cytoskeleton dynamics and a loss of cellular adhesion occur upon Ror2 loss in 2225L tumors.
Figure 5.
Three-dimensional modeling of tumor organoids reveals alterations in cytoskeletal and adhesion dynamics upon Ror2 loss. (a) Brightfield DIC images showing enhanced migration of shRor2 organoids into the surrounding matrix. Scale 50 μm. (b) Quantitation of cellular protrusions emanating into the surrounding matrix in shLUC vs shRor2 organoids (***P<0.001, n=63 organoids per shLUC and shRor2 group). (c) Immunofluorescence of shLUC and shRor2 organoids for K8 (red) and K5 (green). Scale 50 μm. (d) Phalloidin staining and Maximum Intensity Projection of shLUC and shRor2 organoids demonstrating alterations in F-actin dynamics upon Ror2 loss. Scale 50 μm. (d′) Magnified slice of shLUC organoid within (d) depicting cortical F-actin staining at cell junctions in shLUC 2225L organoids. (d″) Magnified slice of shRor2 organoid within (d) illustrating the change in patterning of F-actin and presence of F-actin projections within protrusive nodes of shRor2 organoids (arrows). Scale 10 μm. (e) Immunofluorescence of pERM (green), K8 (red) and nuclei (blue), showing changes in pERM upon Ror2 loss. Scale 50 μm. (e′) Magnified shLUC pERM staining. (e″) Magnified shRor2 pERM staining. Scale 25 μm. (f) Western blot of 2225L shLUC vs shRor2 organoids for Ror2, p-Dvl2, Dvl2, RhoA, CDC42, Rac1/2/3, ROCK1, p-c-jun S63 and c-jun. (g) Representative brightfield DIC images of shLUC vs shRor2 organoids after administration of Y-27632, a ROCK inhibitor (50 μm). Scale 50 μm. (h) Quantitation of the number of invasive nodes per organoids between control and ROCK inhibitor-treated organoids after 3 days (***P<0.001, n=30 organoids per shLUC and shRor2 group).
The Rho family of GTPases, a key mediator of actin cytoskeletal rearrangements and cell adhesion/migration cues, represents an important signaling hub downstream of Wnt/β-catenin-independent signaling in many developmental contexts responsible for orientation of cell polarity and morphogenesis.41 Interestingly, RhoA and Rho-Associated Coiled-Coil Containing Protein Kinase 1 (ROCK1) protein expression were downregulated in shRor2 2225L organoids relative to shLUC organoids, while CDC42 and Rac1/2/3 levels were unchanged (Figure 5f). Additionally, expression of p-c-Jun (Ser63) was reduced upon Ror2 depletion, suggesting a potential RhoA→ROCK1→JNK→c-Jun signaling cascade mediated by Ror2 to control tumor intercellular interactions and adhesive states during tumor progression (Figure 5f). Administration of a ROCK inhibitor, Y-27632, to 2225L organoids similarly resulted in organoids that exhibited disorganization and invasive fronts into the surrounding matrix, partially phenocopying Ror2-depleted organoids (Figures 5g and h). Of note, neither T1 nor T2 organoids showed changes in the expression of proteins within the Rho-GTPase and JNK pathways, suggesting divergent signaling outputs downstream of Ror2 across basal-like models (Supplementary Figures S4c and d).
This divergence likely occurs either at the same level, or downstream, of the molecular hub of Wnt signaling, Dishevelled (Dvl). Specifically, we identified a decrease in the phosphorylated form of Dvl2 in shRor2 2225L organoids evident as the slower mobility band in western blot (Figure 5f). Impaired Ror2 function in T2 organoids similarly resulted in a decline in p-Dvl2 (Supplementary Figure S4d); however, T2-associated phenotypes were likely from alternative modes of signaling downstream of Dvl2, given the lack of apparent change in Rho-GTPase and JNK pathways as in 2225L. Moreover, Ror2-depleted T1 organoids exhibited a Dvl2-independent phenotype, with no change in p-Dvl2 (Supplementary Figure S4c). Collectively, these results implicate the Rho-GTPase and JNK pathways downstream of Ror2 in 2225L TP53-null tumors in mediating appropriate cytoskeleton and adhesive cues among tumor cells. The presence of intertumoral heterogeneity, as seen for other basal-like models T1 and T2, highlight the importance of cellular context in interpreting broad functions for alternative Ror2 signaling in breast cancer. Furthermore, these studies, using three independent TP53-null genetically engineered mouse models, illustrate the considerable heterogeneity reported for basal-like breast cancers within subgroups5 and the complex coexistence of tumor cell subpopulations during tumor progression.42, 43
Discussion
Our results revealed that tumor heterogeneity and cellular context considerably influence the Wnt pathway output during tumor progression. In particular, we demonstrated that expression of the alternative Wnt receptor Ror2 resides in multiple TP53-null breast tumor models across luminal, basal-like and claudin-low subtypes, whereas Wnt/β-catenin-dependent signaling was preferentially restricted to the basal-like TP53-null subtype. While it is appreciated that a hyperactivated Wnt pathway exists in basal-like breast cancers,20, 21, 22 we discovered that Ror2 expression is negatively correlated with Wnt/β-catenin-dependent pathway activity within the intratumoral landscape of basal-like TP53-null tumor models and across human breast tumor samples within the TCGA. We further showed that Ror2 expression modulation by lentiviral-mediated shRNA silencing influences the level of Wnt/β-catenin-dependent signaling in vivo by diminishing this antagonism. Thus, in the context of tumor heterogeneity, a tumor cell’s expression of Ror2 regulates the spatial distribution, duration and amplitude of canonical Wnt signaling within a tumor, depending on the repertoire of Wnt signaling components present. In hematopoiesis and other developmental systems, the particular dosage of canonical Wnt signaling is critical with respect to dictating lineage specification and cellular output.44, 45 In the current study, the balance of Ror2 expression coupled with Wnt/β-catenin-dependent pathway activity within the tumor represents an important factor in tumor progression. Whether these gradients of expression denote a hierarchy of cellular differentiation within the tumor or influence cellular plasticity remains to be determined; however, breast cancers likely harbor Wnt signaling gradients that supply spatiotemporal and contextual cues for tumor cells during progression and metastasis, as has been observed in many developmental contexts.46, 47, 48
Although canonical Wnt signaling was increased upon Ror2 loss, the phenotypic outcomes of Ror2 depletion were not equivalent across basal-like TP53-null models. The lack of a consensus Ror2 signature derived from in vivo knockdown experiments reflects the intertumoral heterogeneity observed between models, despite the common loss of TP53 and their clustering as basal-like. Nonetheless, these studies reveal unique functions for Ror2 depending on cell context and evoke a more complicated view of Wnt pathway integration and signaling downstream of Ror2 in breast cancer. We suspect that Ror2 loss differentially impacts the net balance of Wnt signaling within basal-like models to varying extents, likely dependent on the existing repertoire of Wnt ligands, receptors, modulators and downstream signaling constituents. In the case of the 2225L TP53-null model, we identified interesting differences in Wnt receptor and ligand expression across basal-like and claudin-low subpopulations. Loss of Ror2 expression, if coupled to other receptors, could release Wnt receptor availability of Fzd to engage other co-receptors, allowing for alterations in signaling output.49 Additionally, changes in Wnt receptor availability may alter the output of a single Wnt, as has been shown for Wnt5a.14 Wnt5a, known to signal through Ror2, exhibits both tumor suppressive and promoting roles in breast cancer.24, 50, 51, 52, 53, 54 The combination of Wnt receptors and presence/absence of Ror2 could provide an explanation for the contrasting functions previously proposed for Wnt5a. Deciphering these intricacies is essential to unraveling subtype-specific and more refined functions for alternative Wnt signaling in breast cancer progression.
In spite of the absence of a unique gene signature downstream of Ror2, GO enrichment analysis identified a consistent cell adhesion and migration program impacted in all three models, most pronounced within 2225L compared with either T1 or T2. The presence of altered cell adhesion programs was intriguing, particularly within 2225L, given the role of non-canonical Wnt signaling in directing cell movements and polarity decisions.11 Lentiviral silencing of Ror2 in three-dimensional cultures disrupted cytoskeletal F-actin localization along with proper patterning of pERM, a linker between the actin cytoskeleton and the plasma membrane.55, 56 The fact that ERM proteins interact with membrane proteins and the actin cytoskeleton to organize membrane-cytoskeletal-associated complexes suggests Ror2 may be instrumental in regulating cytoskeleton remodeling and intercellular adhesions during tumor progression. Indeed, the pERM landscape within shRor2 tumors of 2225L was drastically changed (Supplementary Figures S5a and b), suggesting that non-canonical Wnt/β-catenin-independent signaling through Ror2 is integral for proper cytoskeletal and intercellular interactions among tumor cells.57 Moreover, ERM proteins have been strongly implicated in orchestrating metastatic potential in multiple cancers.58, 59 In three-dimensional culture, we identified an impaired Dvl2→RhoA→ROCK1→JNK pathway downstream of Ror2 silencing, accompanied by disruption in organoid integrity and enhanced migratory capability. ROCK1 inhibition phenocopied the loss of Ror2 in vitro, implicating ROCK signaling downstream of Ror2 in guiding cellular adhesions in vivo. Several studies now demonstrate the importance of the Rho pathway in mediating adhesion and migration cues within the tumor microenvironment, likely cancer- and cell-type dependent.60
Akin to their normal tissue counterpart, breast cancers harbor subpopulations of cells composed of a hierarchy, representing various states of cellular differentiation.6, 61 These states of cellular differentiation are reflected within and across breast cancer intrinsic subtypes and offer significant insights into the cellular composition and molecular makeup of a given tumor. Within these cellular landscapes, the coexistence of subpopulations likely involves paracrine and collaborative interactions that provide a supportive function during tumor progression.62, 63 Here, we identified a bulk basal-like and minor claudin-low subpopulation within the genomically classified basal-like TP53-null model, 2225L, where Ror2 regulated the proportion of these subpopulations in vivo. Of note, basal-like and claudin-low subtypes possess very similar genomic features that mirror luminal progenitor and bi-potent mammary stem cells, respectively, within the normal breast hierarchy.6 The specific increase in the claudin-low subpopulation by CD24/CD29 cell surface expression and enrichment in claudin-low-associated genes upon Ror2 loss is intriguing, given the cell adhesion defects and enhanced migration capacity of Ror2-depleted tumor cells observed in three-dimensional organoids. Claudin-low breast cancers possess mesenchymal and stem cell-associated features and lack tight junction proteins,4, 7, 40 properties associated with conveying metastatic potential within breast and other solid tumors. Moreover, claudin-low signatures have been identified in residual tumor tissue after either endocrine or chemotherapy.64 Although triple negative breast cancers exhibit a pathological complete response (pCR) of 30–40%,65, 66 plasticity from basal-like to claudin-low states, as observed upon Ror2 depletion, may be an important factor with regard to clinical response and treatment resistance.
Thus, our data couple cell adhesion defects with alterations in cell state plasticity within 2225L breast tumor models downstream of Ror2 loss. The alternative Wnt receptor, Ror2, therefore, might not only provide a spatial constraint for canonical Wnt signaling topology, but may also restrain the potential of tumor cells to migrate and intravasate within the context of a heterogeneous tumor landscape. Computational models now show the impact of short-range dispersal and cell turnover on cell mixing within a tumor, responsible for the regulation of intratumoral heterogeneity, metastatic potential and treatment resistance.67 The identification of these subpopulations and the regulation of their coexistence within the tumor, beyond Wnt signaling, are therefore both biologically and clinically important.
Materials and methods
Mouse strains, TP53null transplant lines and maintenance
This study was performed in accordance with the rules of the Guide for the Care and Use of Laboratory Animals of the NIH. All mice were maintained and killed according to guidelines of our institutional IACUC (Protocol AN-504). Balb/c inbred female mice (strain #047, 3–4 weeks of age) were purchased from ENVIGO (Houston, TX, USA) and used to propagate the TP53-null tumors. TP53-null mammary tumors were previously generated by transplantation of donor mammary epithelium from Balb/c mice, where TP53 was deleted from the germline, into syngeneic hosts for derivation of TP53-null mammary tumors.30, 31 Subsequent mammary tumors were maintained and propagated in the mouse as a transplantable bank of TP53-null tumors representative of breast cancer subtypes.
Lentiviral plasmids
Lentiviral LeGO plasmids68 and shRNA and ORF sequences for Ror2 were derived and validated as described.34 Wnt pathway reporters32 (Addgene, plasmids 7TGC, 7TG and 7TF #24304, #24314 and #24306, Cambridge, MA, USA) were validated in mammary epithelial cells as described.34
Tumor cell isolation
Primary TP53-null tumor cells were isolated similarly to normal primary mammary epithelial cells.34 Digests were subjected to a series of short centrifugation steps (8 s × 1500 r.p.m.) to enrich for tumor organoids. Organoids were subjected to 0.05% trypsin at 37 °C for 5 min, washed and filtered through a 0.40-μm cell strainer to obtain single cells. Single cells were washed 2 × with phosphate-buffered saline.
Lentiviral transduction of primary TP53-null tumor cells
Transduction of primary TP53-null tumor cells was performed as described,34 with modifications. Single tumor cells were seeded (500 000 cells/well) in a 24-well plate and infected at an MOI of 30 with lentivirus in growth media. Cells were infected overnight and were then washed with phosphate-buffered saline and resuspended in a 1:1 ratio of growth factor-reduced Matrigel/HBSS at 25 000 cells/10 μl injection volume.
Fluorescence-activated cell sorting staining and analysis
Lineage-positive cells were depleted from single-cell preparations using the EasySep Mouse Mammary Stem Cell Enrichment kit (#19757; Stemcell Technologies, Vancouver, BC, Canada). For transduced tumors, tdTomato fluorescence was used to positively select Lin− tumor cells for analysis and sorting. Single-cell preparations were resuspended (107 cells/ml in HBSS+ (containing 2% fetal bovine serum with 10 mm HEPES buffer)) for antibody staining. Antibody incubations were performed on ice for 30 min. Cells were stained with anti-mouse CD24-Pacific Blue and anti-mouse CD29-APC (1:100; BioLegend, San Diego, CA, USA). Cells were either fluorescence-activated cell sorting analyzed (LSR Fortessa; BD Biosciences, Franklin Lakes, NJ, USA) or sorted (Aria II; BD Biosciences). Data analysis was performed using FlowJo, version 9.5.3 (Tree Star, Ashland, OR, USA).
Tumor organoids assays
Lineage-depleted tumor cells were transduced as described above. Eight-well chamber slides were coated with growth factor-reduced Matrigel (5 μl). Aggregated organoids were washed in phosphate-buffered saline before suspending in growth factor-reduced Matrigel at 50 000 cells/40 μl. Within each chamber, 40 μl of cell suspension was plated and solidified at 37 °C for 30 min. Cultures were overlaid with 500 μl of growth media. For western blots, organoids were retrieved with matrigel recovery solution (#354253; Corning) and washed prior to lysis with RIPA buffer.
RNA isolation and gene expression profiling
Total RNA was purified from 20 to 30 mg of mouse mammary tumors or sorted samples (tdTomato+) following the manufacturer's protocol (#74104; Qiagen, Germantown, MD, USA). The BCM Genomic and RNA Profiling Core performed sample quality checks using the Nanodrop ND-1000 (ThermoFisher Scientific, Waltham, MA, USA) and a Bioanalyzer Nano chip (Agilent Technologies, Santa Clara, CA, USA). Murine TP53-null mammary tumors were microarray profiled as previously described69 and uploaded to the gene expression omnibus (GEO) under accession number GSE93815. RNA was reverse transcribed and labeled with cyanine-5 (Cy5) dye for experimental samples and cyanine-3 (Cy3) dye for mouse reference samples70 using Agilent Low RNA Input Fluorescent Linear Amplification Kit. Equal quantities of labeled mouse reference and tumor RNA were co-hybridized overnight to custom Agilent microarrays, washed, scanned and signal intensities determined. Top differential genes found for any of the three models (P<0.01 by t-test using log-transformed data, fold change >1.4) were clustered, using a supervised approach.71 Breast cancer gene expression profiles from TCGA were scored for a previously reported Wnt signature,72 by first normalizing expression values to standard deviations from the median across tumor profiles, then applying our previously described t-score metric.73 Enriched GO terms within gene sets was assessed using Sigterms.74 Gene expression signatures were created by performing a two-class (shRor2 vs shLuc) Significance Analysis of Microarrays (SAM) analysis on the microarray data set.75 Signatures in Figure 4f were defined as all genes either highly or lowly expressed with a false discovery rate <5%. Expression scores for each gene signature were determined by calculating the standardized median expression of the signature within each sample of the UNC308 human breast cancer data set.7
Quantitative real-time PCR
Quantitative real-time PCR was performed using SYBR green methodology (Applied Biosystems, Foster City, CA, USA). Primer sequences (Supplementary Table S1) were designed using the Roche Universal Probe Library. Relative gene expression changes were determined after normalizing to β-actin and calculating the ΔΔCT. s.d. calculations were performed on fold changes observed among biological replicates, derived from the calculated 2−ΔΔCt.
Processing of tumors tissue and organoids cultures
Mice were administered 60 μg/g body weight BrdU via intraperitoneal injection 2 h before killing. Tumors were dissected and fixed in 4% paraformaldehyde overnight at 4 °C before processing to paraffin blocks. Organoid cultures were washed with phosphate-buffered saline and fixed in 4% paraformaldehyde (10 min) before processing to paraffin block or staining with Phalloidin (#A12379; ThermoFisher Scientific).
Immunostaining and western blotting
Tissue and organoid sections were deparaffinized, rehydrated and subjected to Tris-EDTA antigen retrieval for 20 min in a microwave. Primary antibodies were applied overnight at 4 °C. Antibodies and concentrations were: Ror2 (1:100; Nt 2535-2835, Ror2-s; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), eGFP (1:250; ab290; Abcam, Cambridge, MA, USA), K8 (1:250; TROMA-1; Developmental Studies Hybridoma Bank), K5 (1:5,000; PRB-160P; Covance, Princeton, NJ, USA), BrdU (1:250; ab6326; Abcam), CC3 (1:200; 9661; Cell Signaling Technology (CST), Danvers, MA, USA) and pERM (1:500; CST). Tyramide amplification was performed for Ror2 detection according to the manufacturer's instructions (NEL701A001KT; PerkinElmer, Waltham, MA, USA). For western blotting, the following antibodies were used: Ror1 (1:1000, #4102; CST), Dvl2 (1:1000, #3216; CST), RhoA (1:1000, #2117; CST), Rac1/2/3 (1:1000, #2465; CST), CDC42 (1:1000, #2466; CST), c-jun (1:1000, #9265; CST), p-c-jun s63 (1:1000, #9261; CST).
Microscope image acquisition
Sample preparation and microscope image acquisition were carried out as previously described.34
Statistical analysis
Unpaired two-tailed Student’s t-tests were performed on analyses involving two-group comparisons unless otherwise noted. Quantitative measurements were performed in ImageJ or Excel (Microsoft; *P<0.05; **P<0.01; ***P<0.001). Plotted values represent means±s.d. In all cases, experiments were reproduced across multiple (3) experiments.
Acknowledgments
This work was conducted with the help of the Baylor College of Medicine Cytometry and Cell Sorting Core (National Institutes of Health [NIH] P30 AI036211 and S10 RR024574), the Genomics and RNA Profiling Core, the Integrated Microscopy Core (NIH HD007495, DK56338 and CA125123), with funding from the Dan L. Duncan Cancer Center, the John S. Dunn Gulf Coast Consortium for Chemical Genomics and the Lester and Sue Smith Breast Center Pathology Core. We would also like to thank Yiqun Zhang for help and expertise with the microarray analysis. This work was funded by a National Cancer Institute Award NCI R01 CA016303-40 to JMR, NIH R01 CA195754 to CMP and JMR, a Department of Defense Award W81XWH-10-0356 to KPR and NIH CA125123 to CJC.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003; 100: 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol 2011; 5: 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou CM. Mammary development meets cancer genomics. Nat Med 2009; 15: 842–844. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest 2011; 121: 3810–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol 2009; 10: 468–477. [DOI] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014; 346: 1248012. [DOI] [PubMed] [Google Scholar]
- van Amerongen R. Alternative Wnt pathways and receptors. Cold Spring Harb Perspect Biol 2012; 4: a007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development 2009; 136: 3205–3214. [DOI] [PubMed] [Google Scholar]
- Beermann A, Pruhs R, Lutz R, Schroder R. A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development 2011; 138: 2793–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 2006; 4: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L et al. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci USA 2012; 109: 4044–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 1999; 126: 1211–1223. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Takeda K, Oishi I, Nomi M, Ikeya M, Itoh K et al. Mouse Ror2 receptor tyrosine kinase is required for the heart development and limb formation. Genes Cells 2000; 5: 71–78. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Kimble RB, Poueymirou WT, Rojas J, Masiakowski P, Valenzuela DM et al. Ror2, encoding a receptor-like tyrosine kinase, is required for cartilage and growth plate development. Nat Genet 2000; 24: 271–274. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol 2012; 4: a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol 2014; 16: 1004–1015, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A et al. Beta-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol 2011; 24: 209–231. [DOI] [PubMed] [Google Scholar]
- Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol 2010; 176: 2911–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvirajesinghe TM, Bertucci F, Jain A, Scerbo P, Belotti E, Audebert S et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat Commun 2016; 7: 10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014; 159: 844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One 2012; 7: e31127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Quadir A, Hawkins NJ, Jary E, Llamosas E, Kumar D et al. Expression of the novel Wnt receptor ROR2 is increased in breast cancer and may regulate both beta-catenin dependent and independent Wnt signalling. J Cancer Res Clin Oncol 2015; 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson M, Dejmek J, Bendahl PO, Andersson T. Loss of Wnt-5a protein is associated with early relapse in invasive ductal breast carcinomas. Cancer Res 2002; 62: 409–416. [PubMed] [Google Scholar]
- Lejeune S, Huguet EL, Hamby A, Poulsom R, Harris AL. Wnt5a cloning, expression, and up-regulation in human primary breast cancers. Clin Cancer Res 1995; 1: 215–222. [PubMed] [Google Scholar]
- Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci USA 2012; 109: 2778–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene 2000; 19: 1052–1058. [DOI] [PubMed] [Google Scholar]
- Medina D, Kittrell FS, Shepard A, Stephens LC, Jiang C, Lu J et al. Biological and genetic properties of the p53 null preneoplastic mammary epithelium. FASEB J 2002; 16: 881–883. [DOI] [PubMed] [Google Scholar]
- Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE 2010; 5: e9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci USA 2010; 107: 3522–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roarty K, Shore AN, Creighton CJ, Rosen JM. Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J Cell Biol 2015; 208: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Takeuchi S, Hashimoto R, Nagabukuro A, Ueda T, Liu ZJ et al. Spatio-temporally regulated expression of receptor tyrosine kinases, mRor1, mRor2, during mouse development: implications in development and function of the nervous system. Genes Cells 1999; 4: 41–56. [DOI] [PubMed] [Google Scholar]
- Green JL, La J, Yum KW, Desai P, Rodewald LW, Zhang X et al. Paracrine Wnt signaling both promotes and inhibits human breast tumor growth. Proc Natl Acad Sci USA 2013; 110: 6991–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowy J, Adissu HA, Liu JC, Zacksenhaus E, Woodgett JR. Effect of glycogen synthase kinase-3 inactivation on mouse mammary gland development and oncogenesis. Oncogene 2015; 34: 3514–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Rosner A, Nozawa M, Byrd C, Morgan F, Landesman-Bollag E et al. Activation of different Wnt/beta-catenin signaling components in mammary epithelium induces transdifferentiation and the formation of pilar tumors. Oncogene 2002; 21: 5548–5556. [DOI] [PubMed] [Google Scholar]
- Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR et al. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol 2002; 161: 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat 2013; 142: 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev 2009; 23: 265–277. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Ng CK, Piscuoglio S, Weigelt B, Reis-Filho JS. Breast cancer intra-tumor heterogeneity. Breast Cancer Res 2013; 16: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CK, Pemberton HN, Reis-Filho JS. Breast cancer intratumor genetic heterogeneity: causes and implications. Expert Rev Anticancer Ther 2012; 12: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Brechbuhl HM, Ghosh M, Smith MK, Smith RW, Li B, Hicks DA et al. Beta-catenin dosage is a critical determinant of tracheal basal cell fate determination. Am J Pathol 2011; 179: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 2011; 9: 345–356. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M et al. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol 2008; 10: 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell 2011; 20: 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sinha T, Wynshaw-Boris A. Wnt signaling in mammalian development: lessons from mouse genetics. Cold Spring Harb Perspect Biol 2012; 4: a007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev 2010; 24: 2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherding N, Kusner D, Kolb R, Xie Q, Li W, Yuan F et al. Paracrine WNT5A signaling inhibits expansion of tumor-initiating cells. Cancer Res 2015; 75: 1972–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass RA, Sarshad AA, Carson BB, Feenstra JM, Kaur A, Obrdlik A et al. Wnt5a signals through DVL1 to repress ribosomal DNA transcription by RNA polymerase I. PLoS Genet 2016; 12: e1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Crossman DK, Mitchell EH, Sohn P, Crowley MR, Serra R. WNT5A inhibits metastasis and alters splicing of Cd44 in breast cancer cells. PLoS ONE 2013; 8: e58329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roarty K, Baxley SE, Crowley MR, Frost AR, Serra R. Loss of TGF-beta or Wnt5a results in an increase in Wnt/beta-catenin activity and redirects mammary tumour phenotype. Breast Cancer Res 2009; 11: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 2014; 25: 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 2010; 11: 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Fehon RG. Ezrin, Radixin and Moesin: key regulators of membrane-cortex interactions and signaling. Curr Opin Cell Biol 2011; 23: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin M, Chirivino D, Naba A, Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr 2011; 5: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci 2014; 127(Pt 2): 267–275. [DOI] [PubMed] [Google Scholar]
- Ren L, Hong SH, Cassavaugh J, Osborne T, Chou AJ, Kim SY et al. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene 2009; 28: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CD, Fan C, Mitin N, Baker NM, George SD, Graham DM et al. Rho GTPase transcriptome analysis reveals oncogenic roles for Rho GTPase-activating proteins in basal-like breast cancers. Cancer Res 2016; 76: 3826–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 2009; 15: 907–913. [DOI] [PubMed] [Google Scholar]
- Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014; 508: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Tsimelzon A, Chang CH, Fan C, Wolff A, Perou CM et al. Intratumoral heterogeneity in a Trp53-null mouse model of human breast cancer. Cancer Discov 2015; 5: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 2009; 106: 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck S, Ross JS, Royce M, McKenna Jr EF, Perou CM, Avisar E et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/- trastuzumab. Breast Cancer Res Treat 2012; 132: 781–791. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008; 26: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 2015; 525: 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral 'gene ontology' (LeGO) vectors for functional gene analysis. Mol Ther 2008; 16: 698–706. [DOI] [PubMed] [Google Scholar]
- Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol 2013; 14: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol 2008; 26: 4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li Y, Chen Y, Podsypanina K, Chamorro M, Olshen AB et al. Changes in gene expression during the development of mammary tumors in MMTV-Wnt-1 transgenic mice. Genome Biol 2005; 6: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One 2012; 7: e34546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA 2008; 14: 2290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.