Summary
Chemokines are key signals in the immune system and play an important role as proinflammatory mediators in the pathology of inflammatory bowel disease and colorectal cancer, making them an important target for therapy. Recombinant lactic acid bacteria (LAB) were engineered to bind CC and CXC chemokines by displaying chemokine‐binding proteins evasin‐1, evasin‐3 and evasin‐4 on their surface. Evasin genes were cloned into lactococcal surface display vector and overexpressed in L. lactis NZ9000 and NZ9000ΔhtrA in fusion with secretion signal and surface anchor. Evasin‐displaying bacteria removed from 15% to 90% of 11 different chemokines from the solution as determined with ELISA and Luminex multiplexing assays, whereby L. lactis NZ9000ΔhtrA proved more efficient. Lactobacillus salivarius ATCC 11741 was coated with L. . lactis‐expressed evasin fusion protein, and its ability to bind chemokines was also confirmed. Evasin‐3‐displaying L. lactis removed 76.0% of IL‐1β‐induced CXCL8 from the supernatant of Caco‐2 epithelial cells. It also prevented secretion of CXCL8 from Caco‐2 cells in a time‐dependent manner when added before induction with IL‐1β. Evasin‐displaying LAB have the ability to bind multiple chemokines simultaneously and exert synergistic activity. This innovative treatment approach therefore has the potential for mucosal therapy of inflammatory bowel disease or colorectal cancer.
Introduction
Chemokines are small chemoattractant cytokines acting via seven transmembrane G protein‐coupled receptors (GCPRs) that selectively induce recruitment and activation of immune cells to the site of infection. They are classified into four subfamilies, C, CC, CXC and CX3C, on the basis of the number and spacing of the conserved cysteine residues in the amino‐terminus of the protein (Zlotnik and Yoshie, 2000). Historically, their name derived from their function (e.g. macrophage inflammatory protein: MIP‐1α, β), while recently, a generic nomenclature has been applied (e.g. CCL1, CCL2). Chemokines and chemokine receptors play an important role in injury, inflammation, wound repair and cancer (Proudfoot et al., 2010) and are useful as diagnostic, prognostic and therapeutic targets (Castellani et al., 2007). Local mucosal recruitment and activation of neutrophils is a fundamental event in the pathogenesis of inflammatory bowel disease (IBD) and is mediated by interaction of chemokines with chemokine receptors on their target cells (Ajuebor and Swain, 2002; Wang et al., 2009; Atreya and Neurath, 2010). Expression of several chemokines, especially CXCL8, and also of the corresponding receptors, CXCR1 and CXCR2, is constantly increased during the active phase of IBD (Raab et al., 1993). Besides CXCL8, the levels of CXC chemokines CXCL1, CXCL5 and CXCL6 have been shown to be positively correlated with severity of inflammation in IBD patients (Ina et al., 1997). Additionally, recurrent inflammation and tissue destructive lesions that are accompanied by uncontrolled activation of effector immune cells in mucosa are associated with increased risk of colorectal cancer (CRC) (Triantafillidis et al., 2009; Sebastian et al., 2014). The levels of proinflammatory chemokines, such as CCL2, CCL3, CCL4 and CCL5, and pro‐angiogenic chemokines, such as CXCL1, CXCL5 and CXCL8, are also elevated in human colon tumour tissues as compared to the matched normal tissues, indicating that these chemokines and their receptors play an important role in regulating colon tumour progression, angiogenesis and metastasis (Baier et al., 2005). The development of chemokine‐binding proteins as potential therapeutic agents in IBD, or as preventive agents in inflammation‐associated colorectal cancer, is therefore of great interest (Triantafillidis et al., 2011).
Many pathogenic organisms have developed chemokine and cytokine mediators with the ability to interfere with the host chemokine network and escape host detection and defence systems (Proudfoot et al., 2015). Evasins belong to a family of small chemokine‐binding proteins (CKBPs). They have been identified in the salivary gland of the brown tick Rhipicephalus sanguineus and are probably used by the tick to inhibit the chemokine‐mediated recruitment of leucocytes to the bite site (Frauenschuh et al., 2007). CKBPs have also been identified in several other organisms, including viruses, worms and arthropods (Seet and McFadden, 2002; Gonzalez‐Motos et al., 2016). Unlike other CKBPs, evasins are smaller proteins (7–11 kDa) and display a more selective profile and a unique mechanism of chemokine binding (Deruaz et al., 2008; Bonvin et al., 2014). Evasins recognize and interact with chemokines with various degrees of selectivity and inhibit the binding of chemokines to their receptors and/or cell surface glycosaminoglycans. Evasin‐1 binds CCL3, CCL4 and CCL18 (Frauenschuh et al., 2007), evasin‐3 binds CXCL1 and CXCL8 (Deruaz et al., 2008), and evasin‐4 is able to interact with almost 20 chemokines of the CC subfamily, particularly with CCL3, CCL5, CCL18 and CCL21 (Deruaz et al., 2013). Evasins have inhibited cellular recruitment in different murine models of disease and demonstrated potent anti‐inflammatory properties (Deruaz et al., 2008; Castor et al., 2010; Russo et al., 2011; Braunersreuther et al., 2013; Copin et al., 2013; Montecucco et al., 2014; Bonvin et al., 2016).
Various lactic acid bacteria and other microorganisms have been suggested as a possible therapeutic approach for IBD treatment (Ng et al., 2009). Their therapeutic efficiency can be further improved by genetic modification of LAB (Kleerebezem and de Vos, 2011; Berlec et al., 2012; De Moreno de LeBlanc et al., 2015). Several strategies have been described including the production of antioxidant enzymes (Han et al., 2006), anti‐inflammatory cytokine IL‐10 (Schotte et al., 2000) or other anti‐inflammatory compounds (Vandenbroucke et al., 2010). A somewhat different strategy included the display of cytokine‐binding proteins on the surface of LAB (Ravnikar et al., 2010; Zadravec et al., 2015a,b; Berlec et al., 2016; Kosler et al., 2017), with the goal of preventing cytokine proinflammatory action. In the present research, novel therapeutic targets, chemokines, have been addressed in a similar fashion – chemokine‐binding proteins (CKBPs), evasins, were displayed on the surface of LAB and their ability to bind different chemokines was demonstrated. Engineered LAB constitute an innovative approach for the treatment of IBD or CRC.
Results
Expression and surface display of evasins on L. lactis NZ9000 and L. lactis NZ9000ΔhtrA
Synthetic evasin genes (Frauenschuh et al., 2007; Deruaz et al., 2008) with L. lactis‐optimized codon usage were cloned into the plasmid for surface display (pSDBA3b) (Ravnikar et al., 2010). Evasin gene constructs (Fig. 1) were expressed as fusion proteins under the control of NisA promotor. Evasin_B domain fusion proteins were composed of four functional parts, including a signal sequence for secretion to the growth medium [derived from the Usp45 (van Asseldonk et al., 1990)], the gene for chemokine‐binding protein (evasin‐1, evasin‐3 or evasin‐4), the gene for reporter protein B domain (IgG‐binding B domain of staphylococcal protein A) and the gene for peptidoglycan‐binding domain of AcmA (Buist et al., 1995) for surface attachment. Evasin fusion proteins (Fig. 1) are similar to evasin_B domain fusion proteins, lacking only the B domain reporter protein.
Figure 1.
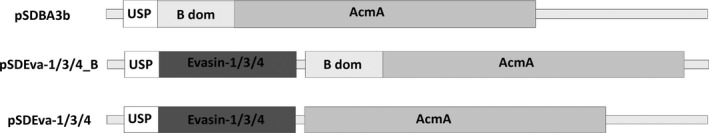
Gene constructs for lactococcal surface display. USP: gene for Usp45 signal peptide for secretion to the growth medium (84 bp). B dom: gene for reporter protein B domain of Staphylococcal protein A (174 bp). Evasin‐1/3/4: genes for chemokine‐binding evasin‐1 (327 bp), evasin‐3 (243 bp) and evasin‐4 (312 bp) respectively. AcmA: gene for C‐terminal part of AcmA protein‐containing 3 LysM repeats for surface anchoring (642 bp).
Expression of evasin_B domain fusion proteins was evaluated with specific antiprotein A antibody (recognizing B domain) using Western blot (Fig. 2). When fusion proteins were expressed in L. lactis NZ9000 (Fig. 2A), the bands with the highest molecular weight corresponded to the weight of the full‐length proteins (Eva‐1_B 42.2 kDa, Eva‐3_B 39.8 kDa, Eva‐4_B 43.2 kDa and B dom 32.9 kDa). The bands of lower molecular weight corresponded to degradation products. The expression of evasin_B domain fusion proteins in L. lactis NZ9000ΔhtrA (Fig. 2B) yielded bands of similar molecular weights; however, the extent of degradation was significantly lower (Fig. 2B). No expression was detected in negative control Fig 2A and B) or without nisin induction (not shown).
Figure 2.
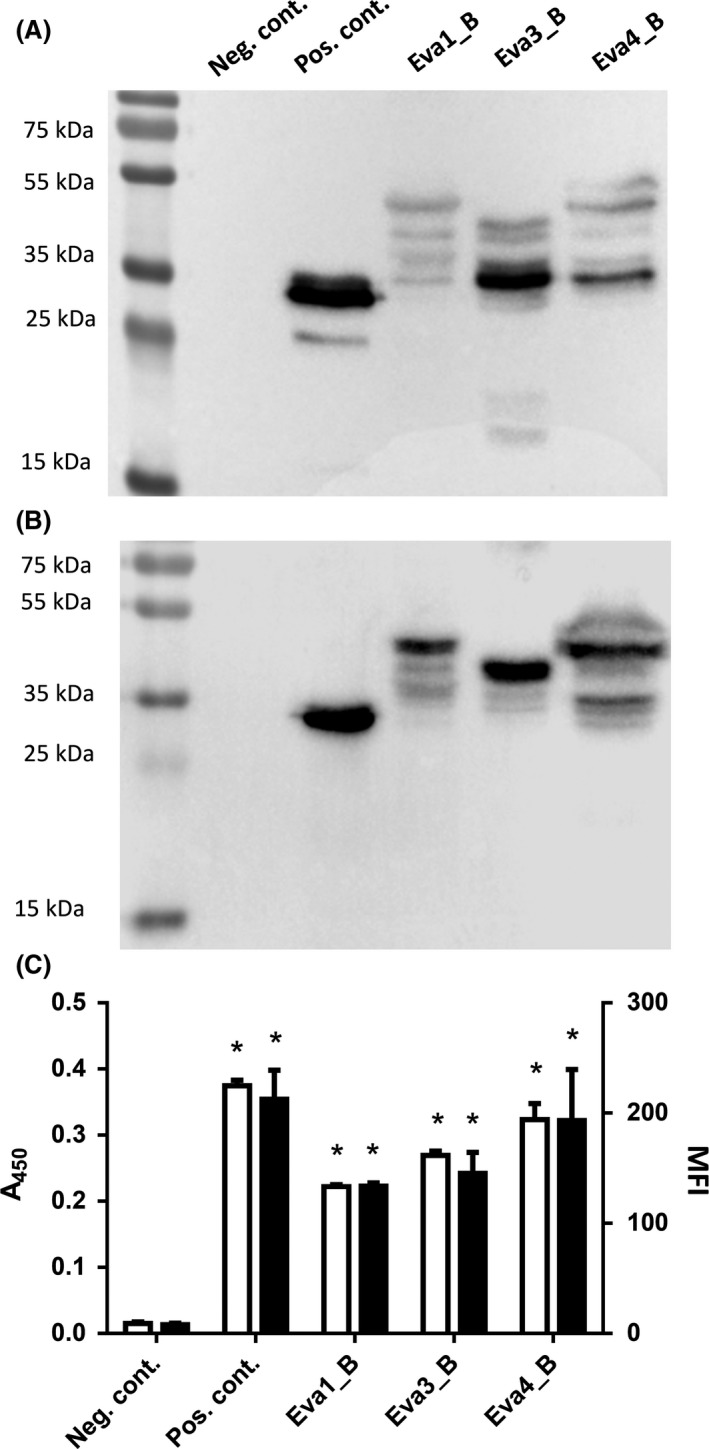
Western blot of (A) L. lactis NZ9000 and (B) L. lactis NZ9000ΔhtrA cells expressing evasin‐1, evasin‐3 and evasin‐4 in fusion with Usp45 secretion signal, B domain reporter protein and LysM‐containing AcmA domain. Whole‐cell ELISA (C, white bars) and flow cytometric (C, black bars) analysis of L. lactis NZ9000ΔhtrA cells expressing evasin‐1, evasin‐3 and evasin‐4 in fusion with B domain. Neg. cont.: negative control‐containing empty plasmid pNZ8148. Pos. cont.: positive control‐containing plasmid pSDBA3b (display of B domain). The results were from three independent experiments performed in triplicate and are expressed as mean ± SD. Significant difference (*P < 0.01) between evasin‐expressing cells and negative control (empty plasmid) is marked with an asterisk.
Surface display of evasin_B domain fusion proteins on L. lactis NZ9000 and L. lactis NZ9000ΔhtrA was confirmed and quantified with whole‐cell ELISA and flow cytometry (Fig. 2C), using antibodies against B domain. The extent of surface display on L. lactis NZ9000ΔhtrA is shown as it was higher than that achieved on L. lactis NZ9000. Statistically significant (P < 0.01; t‐test) display of evasin fusion proteins on L. lactis cell surface as compared to the negative control was observed with both methods. Flow cytometry of evasin_B domain fusion protein‐displaying cells showed a shift in mean fluorescence intensity (MFI) (Fig. S1) in comparison with the negative control (pNZ8148). The extent of displayed evasin_B domain fusion proteins was lower than that of displayed B domain fusion protein without evasins in the positive control. The highest extent of surface display was observed with evasin‐4_B fusion protein with both methods.
Evaluation of chemokine binding by L. lactis NZ9000 and L. lactis NZ9000ΔhtrA with surface‐displayed evasins
Preliminary evaluation of chemokine‐binding ability was performed with L. lactis NZ9000, and the results are shown in Table 1. The chemokine selectivity of evasin‐1‐ and evasin‐4‐displaying L. lactis NZ9000 for CC chemokines was determined by analysing the binding of CCL3, CCL4 and CCL5 by ELISA. 2 × 109 L. lactis NZ9000 cells with surface‐displayed evasin‐1 fusion protein bound 23% of CCL3 from the solution, but could not bind CCL5 (Table 1). 2 × 109 evasin‐4‐expressing L. lactis NZ9000 cells removed around 40% of CCL5 from the solution, but did not bind CCL3. None of the cells bound CCL4.
Table 1.
The percentage of CC and CXC chemokines removed from solution after incubation with 2 × 109 or 1 × 1010 L. lactis NZ9000 cells that displayed on their surface evasin‐1 (pSDEva1), evasin‐4 (pSDEva4) or evasin‐3 (pSDEva3), relative to empty plasmid‐containing control cells. Chemokine concentration was determined with ELISA and multiplexing Luminex assay system MAGPIX
| L. lactis NZ9000 | Determination methods | Number of cells | ||
|---|---|---|---|---|
| pSDEva1 | pSDEva4 | |||
| CCL3 (MIP‐1α) | 23.2 ± 5.1 | n.b. | ELISA | 2 × 109 |
| CCL4 (MIP‐1β) | n.b | n.b. | ||
| CCL5 (RANTES) | n.b | 38.7 ± 9.6 | ||
| CCL3 (MIP‐1α) | 65.8 | 48.5 | LUMINEX | 1 × 1010 |
| CCL11 (Eotaxin) | n.b. | 23.2 | ||
| CCL18 (PARC) | n.b. | 19.5 | ||
| CCL24 (Eotaxin‐2) | n.b. | n.b. | ||
| CCL25 (TECK) | n.b. | 29.3 | ||
| pSDEva3 | ||||
| mCXCL1 (KC) | 56.7 ± 7.0 | ELISA | 2 × 109 | |
| CXCL2 (Gro‐β) | 44.1 ± 1.5 | |||
| mCXCL2 (MIP‐2) | 67.0 ± 6.6 | |||
| CXCL8 (IL‐8) | 54.5 ± 2.1 | |||
| CXCL1 (Gro‐α) | n.b. | LUMINEX | 2 × 109 | |
| CXCL4 (PF4) | n.b. | |||
| CXCL5 (ENA78) | 16.7 ± 1.4 | |||
| CXCL6 (GCP‐2) | 20.0 ± 4.0 | |||
| CXCL16 | n.b. | |||
n.b., no statistically significant binding. Murine chemokines are denoted by m.
Apart from using ELISA, binding of chemokines CCL3 (MIP‐1α), CCL11 (Eotaxin), CCL18 (PARC), CCL24 (Eotaxin‐2) and CCL25 (TECK) was also measured simultaneously with the 5‐plex magnetic assay system MAGPIX (Table 1). 1 × 1010 L. lactis NZ9000 cells with surface‐displayed evasin‐1 bound only CCL3 from the mixture of five different chemokines. On the other hand, 1 × 1010 L. lactis NZ9000 cells with surface‐displayed evasin‐4 bound CCL3, CCL11, CCL18 and CCL25 from the mixture. No binding of CCL24 by either evasin‐1‐ or evasin‐4‐displaying L. lactis was observed. The presence of B domain had little effect on binding by evasin‐1‐ and evasin‐4‐displaying L. lactis, and the results are therefore not shown.
L. lactis NZ9000 cells with surface‐displayed evasin‐3 bound and removed various portions of CXC chemokines murine CXCL1 (KC), human CXCL2, murine CXCL2 (MIP‐2) and human CXCL8 (IL‐8) from the solution, as evaluated by ELISA (Table 1). The extent of chemokine removal was, in general, lower with B domain‐containing fusion protein (not shown). Human chemokines CXCL1 (Gro‐α), CXCL4 (PF4), CXCL5 (ENA78), CXCL6 (GCP‐2) and CXCL16 were evaluated simultaneously with a 5‐plex screening assay using the Luminex system. 2 × 109 L. lactis NZ9000 cells with surface‐displayed evasin‐3 removed portions of CXCL5 and CXCL6 from the solution, while no binding of CXCL1, CXCL4 and CXCL16 was observed (Table 1).
Binding of CCL3, CCL5 and CXCL8 was studied in more detail by taking into account the influence of bacterial cell number and bacterial strain. The CCL3 and CXCL8 binding with L. lactis NZ9000ΔhtrA (Fig. 3) was 1.3‐ and 2.0‐fold better, respectively, than that achieved with L. lactis NZ9000 (Fig. S2), while for CCL5, the difference was less pronounced. Evasin‐1‐displaying L. lactis NZ9000ΔhtrA in concentration of 6 × 109, 3 × 109 and 6 × 108 cells ml−1 removed 54.8%, 50.5% and 27.8% of CCL3 from the solution, respectively, relative to that of the control bacteria (Fig. 3). The extent of CCL5 removal by evasin‐4‐displaying L. lactis NZ9000ΔhtrA also depended on the bacterial cell number and was decreased by lowering the number of cells. Evasin‐4‐displaying L. lactis NZ9000ΔhtrA in concentration of 6 × 109, 3 × 109 and 6 × 108 cells ml−1 removed 59.7%, 42.3% and 13.0% of CCL5 from the solution, respectively, relative to the control bacteria. Evasin‐3‐displaying L. lactis NZ9000ΔhtrA cells at 6 × 109, 3 × 109 and 6 × 108 cells ml−1 removed 94.0%, 90.4% and 83.2%, of CXCL8, relative to the control bacteria (Fig. 3).
Figure 3.
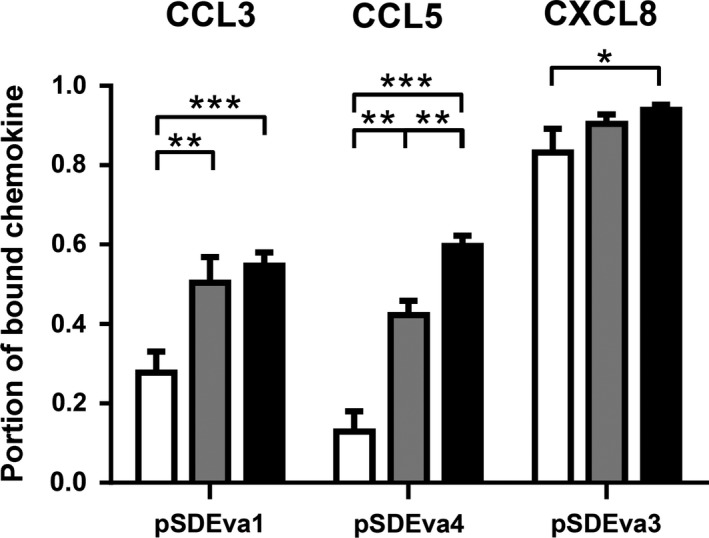
ELISA‐determined percentage of CCL3, CCL5 and CXCL8 removed after incubation with 6 × 108 (white bars), 3 × 109 (grey bars) or 6 × 109 (black bars) cells mL−1 of recombinant L. lactis NZ9000ΔhtrA cells that displayed evasin‐1 (pSDEva1), evasin‐4 (pSDEva4) or evasin‐3 (pSDEva3). The results were from three independent experiments performed in triplicate and are expressed as mean ± SD. Significant differences (*P < 0.05; **P < 0.01; ***P < 0.001) are marked with asterisks.
Chemokine binding by Lb. salivarius heterologously coated with evasin fusion proteins
2 × 109 Lb. salivarius cells were incubated with different volumes of conditioned sterile growth medium of L. lactis NZ9000ΔhtrA that contained evasin‐1, evasin‐3 and evasin‐4 fusion proteins. This resulted in heterologous non‐covalent attachment of evasin fusion proteins to the surface of Lb. salivarius via the peptidoglycan‐binding domain of AcmA. Binding of CCL3, CCL5 and CXCL8 was achieved by coating Lb. salivarius with evasin‐1, evasin‐4 and evasin‐3 fusion proteins respectively (Table 2). Decreasing the volume of the lactococcal growth medium (20, 10, 5 ml) resulted in slight, but statistically significant decrease in chemokine binding (Fig. S3).
Table 2.
The percentage of chemokines CCL3, CCL5 and CXCL8 removed from solution after incubation with 2 × 109 Lb. salivarius cells that were previously coated with evasin‐1 (pSDEva1), evasin‐3 (pSDEva3) and evasin‐4 (pSDEva4) fusion proteins, respectively, relative to control Lb. salivarius cells. Growth media of recombinant L. lactis NZ9000ΔhtrA served as a source of fusion proteins. The extent of binding was normalized relative to the control Lb. salivarius cells incubated with the growth medium of L. lactis harbouring empty plasmid pNZ8148
| Lactobacillus salivarius coated with growth medium of. L. lactis NZ9000ΔhtrA containing | |||
|---|---|---|---|
| pSDEva1 | pSDEva3 | pSDEva4 | |
| CCL3 | 86.6 ± 1.9 | n.b. | 47.0 ± 1.2 |
| CCL5 | n.b. | n.b. | 49.0 ± 5.5 |
| CXCL8 | n.b. | 79.7 ± 2.3 | n.b. |
n.b., no statistically significant binding.
Evasin‐3‐displaying L. lactis and Lb. salivarius removed CXCL8 secreted by IL‐1β‐induced intestinal epithelial cells
Basal secretion of CXCL8 to the culture medium by untreated Caco‐2 cells was determined to be 9.5 pg ml−1. Treatment of Caco‐2 cells with IL‐1β resulted in increased CXCL8 production. The dose–response curve of CXCL8 secretion, following treatment of Caco‐2 cells with varying concentrations of IL‐1β (0.25–250 ng ml−1), showed a peak in CXCL8 production (~600 pg ml−1) at a concentration of 25 ng ml−1 IL‐1β (Fig. S4A), resulting in more than 60‐fold increase. CXCL8 secretion by Caco‐2 cells increased rapidly during first 6 h after addition of 25 ng ml−1 of IL‐1β (Fig. S4B).
The effect of evasin‐3‐displaying L. lactis NZ9000 on the basal secretion of CXCL8 by Caco‐2 cells (without IL‐1β addition) was compared to that of control L. lactis NZ9000 (containing empty plasmid pNZ8148) or wild‐type E. coli DH5α. 2 × 109 bacterial cells ml−1 were used, representing a multiplicity of infection (moi) of 2000:1 (bacterial: epithelial cells). L. lactis NZ9000 with surface‐displayed evasin‐3 decreased CXCL8 concentration in the supernatant of Caco‐2 cells for 26.8% in comparison with empty plasmid control‐containing L. lactis NZ9000 (Fig. 4A), while, on the contrary, incubation with E. coli cells increased the CXCL8 secretion for 53.3%. Induction of CXCL8 production in Caco‐2 cells by adding 25 ng ml−1 of IL‐1β for 6 h, followed by 2‐h incubation with bacteria, resulted in much higher total concentrations of CXCL8. However, similar effects of bacteria were observed: control L. lactis NZ9000 (containing empty plasmid pNZ8148) had no effect on CXCL8 production, while evasin‐3‐displaying L. lactis NZ9000 caused 26.3% reduction in CXCL8 in the supernatant of Caco‐2 cells (Fig. 4A).
Figure 4.
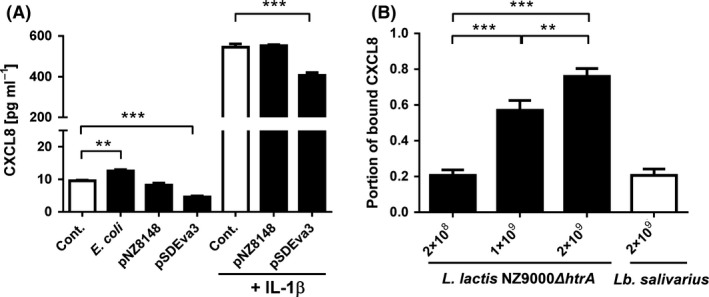
ELISA‐determined concentration (A) or portion of bound (B) CXCL8 in the supernatant of Caco‐2 cells.
A. Caco‐2 cells with or without IL‐1β induction were untreated (Cont.) or were exposed to 2 × 109 cells ml−1 of bacteria (E. coli DH5α, L. lactis NZ9000 containing empty plasmid control pNZ8148 or plasmid pSDEva3).
B. Removal of CXCL8 from the supernatant of IL‐1β‐induced Caco‐2 cells by 2 × 108, 1 ×109 or 2 × 109 cells ml−1 of L. lactis NZ9000 ΔhtrA (white bars) or 2 × 109 cells ml−1 of Lb. salivarius cells coated with growth medium of L. lactis NZ9000ΔhtrA (black bars) displaying evasin‐3. The results were from three independent experiments performed in triplicate and are expressed as mean ± SD. Significant differences (t‐test) are marked with asterisks. (** < 0.01; ***P < 0.001).
The bacterial removal of CXCL8 secreted from IL‐1β‐induced Caco‐2 cells was optimized by varying the number of bacterial cells and bacterial strain. Significant increase in the extent of CXCL8 removal was observed using L. lactis NZ9000ΔhtrA strain (Fig. 4B). 2 × 109 evasin‐3‐expressing L. lactis NZ9000ΔhtrA reduced the concentration of CXCL8 by 76.0%, 1 × 109 cells reduced the concentration of CXCL8 by 57.0%, and 2 × 108 cells still reduced the concentration by 20.7% (Fig. 4B). 2 × 109 Lb. salivarius cells coated with evasin‐3 fusion protein‐containing growth medium of L. lactis NZ9000ΔhtrA were less effective and removed only 20.7% of CXCL8 (Fig. 4B). Viability of Caco‐2 cells after incubation with bacteria was above 90% (Fig. S5A).
Evasin‐3‐displaying L. lactis and Lb. salivarius prevented IL‐1β‐induced secretion of CXCL8 from intestinal epithelial cells
The preventive effect of evasin‐3‐displaying bacteria on CXCL8 secretion was evaluated by pre‐incubating Caco‐2 cells with bacterial cells for 1 h, prior to addition of IL‐1β. The concentration of CXCL8 was monitored over 6 h (Fig. 5). 1 × 109 evasin‐3‐displaying L. lactis NZ9000ΔhtrA cells significantly impeded the time‐dependent increase in CXCL8 concentration in comparison with control cells (harbouring pNZ8148) or absence of bacteria. The final CXCL8 concentration after 6 h of monitoring was for 62% lower in the supernatant of Caco‐2 cells pre‐treated with evasin‐3‐displaying L. lactis NZ9000ΔhtrA cells than in the supernatant without bacterial cells. Lb. salivarius cells coated with evasin‐3 fusion protein from growth medium of L. lactis NZ9000ΔhtrA were less effective. They reduced the final CXCL8 concentration in Caco‐2 supernatant by 27% in comparison with the Caco‐2 supernatant without bacteria after 6 h of monitoring.
Figure 5.
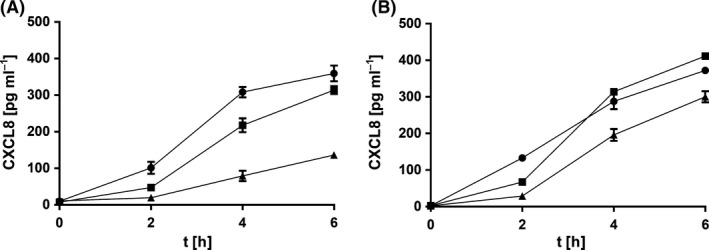
Time‐dependent secretion of CXCL8 by Caco‐2 cells after the preventive 1‐h treatment with evasin‐3‐displaying bacteria (triangles) L. lactis NZ9000ΔhtrA (A) or Lb. salivarius coated with evasin‐3‐fusion protein‐containing growth medium of L. lactis NZ9000ΔhtrA (B) that was followed by the addition of 25 ng ml−1 IL‐1β (at time 0). CXCL8 secretion by Caco‐2 cells was compared to that after incubation with control bacteria (squares; L. lactis NZ9000ΔhtrA harbouring pNZ8148 (A) or Lb. salivarius coated with growth medium of L. lactis NZ9000ΔhtrA cells harbouring pNZ8148 (B) or to that without bacteria (circles).
Discussion
Engineered evasin‐displaying bacteria were able to bind and remove 11 different recombinant chemokines of CC and CXC subfamilies. Synergistic targeting of different chemokines and prevention of the secretion of CXCL8 by the Caco‐2 intestinal cell line suggest a significant potential of engineered bacteria in the treatment of IBD or CRC by addressing the relatively unexplored option of oral neutralization of chemokines, coupled by beneficial effects of probiotics.
Surface display of evasins on L. lactis was achieved using nisin‐controlled expression (Mierau and Kleerebezem, 2005) of fusion proteins consisting of chemokine‐binding proteins evasins (evasin‐1, evasin‐3 and evasin‐4) (Deruaz et al., 2008), C‐terminal peptidoglycan‐binding domain of AcmA (Steen et al., 2003) and Usp45 secretion signal (van Asseldonk et al., 1990; Ravnikar et al., 2010; Zadravec et al., 2015a,b, 2016). Genes for evasins were codon optimized for L. lactis to favour higher protein yield. A 15‐amino acid peptide linker (Gly4Ser)3 (Trinh et al., 2004) was included between the evasin and the peptidoglycan‐binding domain in evasin‐1 and evasin‐3 gene constructs to enable mobility and flexibility of surface‐displayed evasins. The B domain of staphylococcal protein A was included as a part of fusion proteins to serve as a reporter protein, because its effective expression and surface display in L. lactis has already been demonstrated (Ravnikar et al., 2010). Expression of evasin fusion proteins containing B domain was confirmed by Western blot. The multiple bands of lower molecular weight were a consequence of AcmA degradation (Poquet et al., 2000). Expression of fusion proteins in the L. lactis NZ9000ΔhtrA strain that lacks extracellular protease HtrA (Lindholm et al., 2004; Cortes‐Perez et al., 2006) resulted in significant reduction of proteolytic degradation by HtrA and higher recombinant protein yields.
Surface display of evasin fusion proteins was confirmed and its extent quantified with whole‐cell ELISA and flow cytometry by detecting B domain reporter. The extent of surface display of evasin‐B domain fusion proteins was lower than that of fusion proteins containing only B domain. This may be the consequence of lower protein yield, larger size of the fusion protein, hindered availability of B domain in the displayed fusion protein or higher proteolytic susceptibility. The level of surface display was highest with evasin‐4 fusion protein, the only fusion protein in which (Gly4Ser)3 linker was not included due to cloning issues. Although surprising, this is in agreement with our previous report in which the linker did not contribute to the extent of surface display (Zadravec et al., 2014). The lower level of fusion protein degradation in NZ9000ΔhtrA resulted in the increased extent of surface display. Although this is in accordance with expectations, it was not observed in our previous study in which only B domain was displayed (Ravnikar et al., 2010), possibly pointing to the susceptibility of evasins or linker to degradation. Taken together, these results demonstrate that L. lactis NZ9000ΔhtrA strain stabilizes surface‐associated proteins and is more appropriate as an expression host for evasin fusion proteins.
The functionality of all three evasins on the bacterial surface was confirmed indirectly, via the greater ability of evasin‐displaying bacterial cells to remove chemokines from the solution than that of control cells. The functional expression is noteworthy because of the presence of intramolecular disulfide bonds which are less common in proteins of Gram‐positive LAB (Davey et al., 2016). Among recombinant proteins, disulfide bond‐containing bacteriocins have been produced in L. lactis, and evasins may share the same mechanisms of disulfide bond formation (Back et al., 2016; Davey et al., 2016).
Using different evasins‐displaying bacteria, the binding of 11 different chemokines: five human CC chemokines (CCL3, CCL5, CCL11, CCL18 and CCL25), four human CXC chemokines (CXCL2, CXCL8, CXCL5 and CXCL6) and two murine CXC chemokines (CXCL1 and CXCL2) has been demonstrated. The binding specificity of evasins was in accordance with that published. The extent of chemokine removal varied from app. 15% (CXCL5) to more than 90% (CXCL8) and was largely dependent on bacterial concentration. As seen with the expression and surface display, the binding of chemokines was improved when fusion proteins were expressed in L. lactis NZ9000ΔhtrA. The presence of B domain in fusion proteins had minor, usually detrimental, effect on binding, although positive effects were also observed, pointing to the importance of steric positioning of evasins for binding of different chemokines.
CC chemokines were removed by evasin‐1‐ and evasin‐4‐displaying bacteria. Evasin‐1‐displaying bacteria were capable of binding CCL3, but not the closely related CCL5, although binding of CCL5 by evasin‐1 was previously reported (Frauenschuh et al., 2007). This may be the consequence of insufficient concentration of cells, or incorporation of evasin‐1 into the fusion protein via its C‐terminus, as it was shown that C‐terminus was involved in chemokine binding (in contrast to evasin‐4 that binds chemokines via its N‐terminus) (Deruaz et al., 2013). Evasin‐4 has a broader selectivity pattern than evasin‐1 and evasin‐3 and was previously shown to bind almost 20 chemokines of the CC subfamily, among them CCL5 with the highest affinity (Deruaz et al., 2013). Removal of CCL5 by evasin‐4‐displaying L. lactis cells was indeed very effective (42.3%). Apart from CCL5, evasin‐4‐displaying cells also bound CCL3, CCL11, CCL18 and CCL25 in a multiplex Luminex assay.
CXC chemokines (human CXCL2, CXCL5, CXCL6, CXCL8 and mouse homologues CXCL1 and CXCL2) were bound by evasin‐3‐displaying L. lactis cells. Binding of human CXCL1 was not detected, possibly due to the multiplex Luminex assay format and the competition by five different chemokines. This could also be the reason for the generally lower percentage of binding detected with Luminex. Detailed characterization of binding with evasin‐3‐displaying L. lactis cells was performed with CXCL8. As observed with evasin‐1‐ or evasin‐4‐displaying cells, binding of CXCL8 was more effective with higher number of evasin‐3‐displaying bacterial cells.
Lactobacillus salivarius ATCC 11741 has been established earlier as a suitable host for heterologous surface display of proteins containing the peptidoglycan‐binding domain of AcmA (Zadravec et al., 2015a,b; Kosler et al., 2017). It was shown that not all AcmA‐containing fusion protein, produced and secreted by recombinant L. lactis, binds to the producer cells. The fusion protein‐containing growth medium can therefore be used for heterologous coating of non‐recombinant bacterial cells (Bosma et al., 2006), representing a non‐GMO alternative (Zadravec et al., 2015a,b) or a surface display option for hard to transform bacteria. Lb. salivarius was coated with evasin fusion proteins produced by L. lactis, and its ability to bind chemokines was confirmed.
Inflammation of the intestinal epithelial cells is usually driven by inflammatory cytokines from the immune cells of the lamina propria. This can be recreated in the Caco‐2 epithelial cell model by stimulation with IL‐1β which triggers strong secretion of CXCL8 (Luerce et al., 2014) that can accumulate in the growth medium for several hours. Evasin‐3‐displaying L. lactis significantly reduced CXCL8 concentration (by up to 76.0%) in Caco‐2 cell supernatant in comparison with untransformed L. lactis or E. coli. Evasin‐3‐coated Lb. salivarius was less effective in binding Caco‐2‐produced CXCL8 than evasin‐3‐displaying L. lactis, despite their similar binding abilities as observed with ELISA (Tables 1 and 2). This might be due to the intrinsic slightly proinflammatory phenotype of Lb. salivarius that was shown to be able to induce TNFα production in THP‐1 cells (Diaz et al., 2013). However, Lb. salivarius coated with evasin‐3 fusion protein‐containing NZ9000ΔhtrA growth medium could still remove around 20% of CXCL8 from the Caco‐2 supernatant.
Preventive addition of evasin‐3‐displaying L. lactis to Caco‐2 cells for 1 h before exposure to IL‐1β was shown to reduce CXCL8 secretion in a time‐dependent manner. A less intense, but still significant decrease in CXCL8 was observed with control L. lactis, in line with the intrinsic anti‐inflammatory activity of L. lactis (Berlec et al., 2016). The decrease achieved with evasin‐3‐displaying Lb. salivarius was again less pronounced than that achieved with evasin‐3‐displaying L. lactis.
The preventive addition of chemokine‐binding bacteria to the epithelial cell model resembles the intended administration of bacteria in intestinal inflammations and suggests a significant potential for mucosal therapy of inflammation occurring in IBD or colorectal cancer. This potential is strengthened by the intrinsic evasin affinity for different chemokines. The chemokine system is promiscuous, with individual chemokines binding to several receptors and individual receptors recognizing multiple chemokines (Allegretti et al., 2012; Steen et al., 2014). Targeting a single chemokine or single receptor would therefore not be sufficient to exert a significant effect in vivo (Koelink et al., 2012). However, targeting multiple chemokines with three different evasins could avoid redundancy and enable synergy (Bonvin et al., 2016). The ability of evasin‐displaying bacteria to bind multiple cytokines simultaneously has been confirmed in this study using multiplex assays.
To conclude, we present for the first time the expression of biologically active evasins and their display on the surface of LAB. We have shown that evasin‐displaying LAB are able to bind at least 11 different chemokines, some of them at the same time. Apart from binding the recombinant chemokines, the evasin‐displaying bacteria also prevented CXCL8 secretion in the epithelial cell model. The results obtained thus warrant expanding the studies on chemokine‐binding bacteria to animal models of intestinal inflammation with the aim of confirming their usefulness in the treatment of IBD and CRC.
Experimental procedures
Bacterial strains, media and growth conditions
The bacterial strains used in this study are shown in Table S1. Lactococcus lactis NZ9000 (de Ruyter et al., 1996; Kuipers et al., 1998; Mierau and Kleerebezem, 2005) and Lactococcus lactis NZ9000ΔhtrA (Lindholm et al., 2004) were grown at 30 °C in M17 medium (Sigma) supplemented with 0.5% glucose (GM‐17) without agitation or in the same medium solidified with 1.5% agar. To maintain selection pressure on transformation, 10 μg ml−1 chloramphenicol or erythromycin, or both, was added to the growth medium of L. lactis NZ9000 and L. lactis NZ9000ΔhtrA. Lactobacillus salivarius ATCC 11741 was grown in De Man, Rogosa and Sharpe (MRS) medium (Merck) at 37 °C without aeration. E. coli strain DH5α was grown at 37 °C with agitation in lysogeny broth (LB) medium supplemented with 100 μg ml−1 ampicillin.
DNA manipulation and plasmid construction
Detailed cloning procedures are described in Supporting information. Primers (IDT) and plasmids are listed in Table S1. Scheme of evasin constructs pSDEva1, pSDEva3 and pSDEva4 is shown in Fig. 1.
Expression of evasin fusion proteins in L. lactis
Overnight cultures of L. lactis NZ9000 and L. lactis NZ9000ΔhtrA harbouring plasmids pNZ8148, pSDBA3b, pSDEva1, pSDEva1_B, pSDEva3, pSDEva3_B, pSDEva4 and pSDEva4_B were diluted (1:100) in 10 ml (or 100 ml) of fresh GM‐17 medium and grown to an optical density (A600) of 0.50–0.80. Fusion protein expression was induced with 25 ng ml−1 nisin (Fluka) (de Ruyter et al., 1996; Kuipers et al., 1998; Mierau and Kleerebezem, 2005). After 3 h of incubation, 1 ml of culture was stored at 4 °C for flow cytometric analysis, and the remaining cell culture was centrifuged at 5000 g for 10 min. The cell pellet was resuspended in 400 μl of phosphate‐buffered saline (PBS, pH 7.4) and stored at −20 °C for SDS‐PAGE analysis, or resuspended in different volumes of PBS for whole‐cell ELISA and assay of the chemokine‐binding ability. The supernatant was decanted, filtered with a 0.22 μm pore size filter (Minisart; Millipore, Darmstadt, Germany), aliquoted and stored at −20 °C for testing the heterologous coating of non‐recombinant bacteria with evasin fusion proteins.
Heterologous coating of Lactobacillus salivarius with evasin fusion proteins
The coating of Lb. salivarius was performed as described previously (Zadravec et al., 2015a,b; Kosler et al., 2017). Lb. salivarius overnight culture containing 2 × 109 cells ml−1 was centrifuged (5000 g, 5 min, 4 °C) and resuspended in different volumes (10 ml in Caco‐2 cell experiments) of the conditioned growth medium of L. lactis NZ9000 or L. lactis NZ9000ΔhtrA that contained evasin fusion proteins. Bacterial suspension was shaken gently for 2 h at room temperature (RT). After centrifugation (5000 g, 5 min, 4 °C), the cells were resuspended in PBS for assaying the chemokine‐binding ability with ELISA as described below. The growth medium of empty plasmid (pNZ8148)‐containing L. lactis was used as a control and was incubated with Lb. salivarius in the same manner.
SDS‐PAGE and Western blot
Detailed procedure is described in Supporting information.
Flow cytometry
Details of flow cytometry are described in Supporting information.
Whole‐cell enzyme‐linked immunosorbent assay (ELISA)
The whole‐cell enzyme‐linked immunosorbent assay (ELISA) was performed as described previously (Lindholm et al., 2004; Zadravec et al., 2015a,b). Absorbances were read at 450 nm using an Infinite M1000 (Tecan, Salzburg, Austria). Detailed procedure is described in Supporting information.
Chemokine binding by evasin‐displaying lactic acid bacteria
Different volumes of L. lactis NZ9000 and L. lactis NZ9000ΔhtrA expressing evasin‐1, evasin‐3 or evasin‐4 and Lactobacillus salivarius ATCC 11741 coated with evasin‐1, evasin‐3 and evasin‐4 fusion proteins were centrifuged (5000 g, 5 min, 4 °C), washed twice with 500 μl PBS and finally resuspended in 200 μl of PBS containing various concentrations of chemokine standards (from ELISA kits, see below) and incubated 2 h at room temperature (RT) with gentle shaking. Cells were then removed by centrifugation (5000 g, 10 min, 4 °C) and 100 μl of the supernatant collected to examine the content of chemokines using ELISA kits (Mabtech, Nacka Strand, Sweden; R&D Systems, Minneapolis, MN, USA; and PeproTech, London, UK). Alternatively, 50 μl of the supernatant was used to examine the content of chemokines in multiplexing system (Luminex 100 or MAGPIX, 's‐Hertogenbosch, The Netherlands). The chemokine‐binding ability of evasin‐displaying bacteria was presented as the portion of cytokine removed from the solution by evasin‐displaying bacteria, in comparison with control bacteria, harbouring pNZ8148 plasmid.
ELISA for chemokine concentration determination
CXCL8 and CCL4 were measured using a Human IL‐8 (CXCL8) and Human MIP‐1β (CCL4) ELISA development kit (Mabtech) following the manufacturer's instructions. CCL3 (MIP‐1α) and CCL5 (RANTES) were measured using DuoSet ELISA (R&D Systems). CXCL2 (Gro‐β), CXCL16, murine CXCL1 (KC) and murine CXCL2 (MIP‐2) were measured using mini ELISA development kits (PeproTech). Standard curves for chemokines and detailed procedure of ELISA are described in Supporting information.
Luminex multiplexing system assays for chemokine concentration determination
Human chemokines CXCL1 (Gro‐α), CXCL4 (PF4), CXCL5 (ENA78), CXCL6 (GCP‐2), CXCL8 and CXCL16 levels were measured using Luminex screening human assay kit, the xMAP Luminex fluorescent bead‐based technology (R&D Systems) according to manufacturer's instructions, and fluorescence signal was read on a Luminex 100 System (Luminex). The trimmed median value was used to derive the standard curve and calculate sample concentration. Data from three independent experiments were considered. Human chemokines CCL3 (MIP‐1α), CCL11 (Eotaxin), CCL18 (PARC), CCL24 (Eotaxin‐2) and CCL25 (TECK) were measured using a 5‐plex magnetic Luminex screening assay (R&D Systems). The samples were assayed in MAGPIX system, and data were analysed with xPONENT software (Luminex). Standard curves for chemokines are described in Supporting information.
Caco‐2 cell culturing and incubation with bacteria
Caco‐2 cells (ATCC HTB‐37), a human colon adenocarcinoma cell line, were cultured in Dulbecco's modified Eagle's medium (DMEM, high glucose) with GlutaMAX (Gibco, Life Technologies, Paisley, UK) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, Life Technologies), 25 mM HEPES (Sigma‐Aldrich, St. Louis, MO, USA), 100 U m−1 penicillin, 100 μg ml−1 streptomycin (Gibco, Life Technologies) and 1% Eagle's minimum essential medium (MEM) non‐essential amino acid solution (Sigma‐Aldrich) in a humidified atmosphere containing 5% CO2 at 37 °C.
Caco‐2 cells were seeded at 1 × 105 cells per well in 24‐well plates and incubated at 37 °C with 5% CO2 for 24 h before treatment, as reported (Luerce et al., 2014). Secretion of CXCL8 was induced by the addition of different concentrations (250, 25, 2.5, 0.25, 0.025, 0.0025, 0.00025 ng ml−1) of recombinant human IL‐1β (Cell Genix, Freiburg, Germany). To examine the production of CXCL8, Caco‐2 cells were incubated with 25 ng ml−1 of IL‐1β for different time periods (0–48 h). 25 ng ml−1 of IL‐1β for 6 h was used when incubating with bacteria. Cell cultures were centrifuged (10 min 200 g at 4 °C and 10 min 16 000 g at 4 °C), and the supernatant was collected and stored at −80 °C until analysis. CXCL8 levels were measured using a Human IL‐8 (CXCL8) ELISA development kit (Mabtech) as described above.
Cultures of E. coli DH5α, L. lactis NZ9000 or L. lactis NZ9000ΔhtrA, harbouring plasmids pNZ8148 (control) and pSDEva3, as well as evasin‐3‐coated Lb. salivarius, were centrifuged, washed two times with PBS and finally resuspended in DMEM. In CXCL8 removal experiments, bacterial cells were added to the Caco‐2 cell culture (preincubated with IL‐1β for 6 h) at a final concentration of 2 × 108, 1 × 109 or 2 × 109 cells ml−1. CXCL8 secretion was measured after 2 h of co‐incubation. To prevent CXCL8 secretion, we incubated bacterial cells with Caco‐2 cells for 1 h before inducing with IL‐1β for another 6 h. Untreated Caco‐2 cells secreting baseline levels of CXCL8 were used as a control. Data from three independent experiments were considered. Viability of Caco‐2 cells after incubation with bacteria was tested with Trypan blue exclusion staining.
Statistical analyses
All data are presented as means ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism 6.01 software (San Diego, CA, USA). Student's t‐test was used to compare the differences. Significant differences (t‐test; *P < 0.05; **P < 0.01; ***P < 0.001) from the control are marked with an asterisk. The portion of removal of chemokine by bacteria was calculated only when the difference in chemokine concentration between evasin‐displaying and control bacteria was significant (t‐test; P < 0.05).
Conflict of interest
None declared.
Supporting information
Table S1. Strains, plasmids and primers used in the study.
Fig. S1. Shift in fluorescence intensity.
Fig. S2. ELISA‐determined percentage of CCL3, CCL5 and CXCL8 removed after incubation with 6 × 108 (white bars), 3 × 109 (gray bars), or 6 × 109 (black bars) cells/mL of recombinant L. lactis NZ9000 cells that displayed evasin‐1 (pSDEva1), evasin‐4 (pSDEva4), or evasin‐3 (pSDEva3).
Fig. S3. The portion of CCL3 removed from the solution after incubation with 2 x 109 Lb. salivarius cells coated with evasin‐1 (white bars) or evasin‐4 (black bars) fusion proteins.
Fig. S4. ELISA‐determined CXCL8 secretion by Caco‐2 cells exposed to IL‐1β.
Fig. S5. Percentage of viable Caco‐2 cells determined with trypan blue exclusion staining after 2 h of incubation with different number of L. lactis NZ9000ΔhtrA cells (2 × 108, 1 × 109 and 2 × 109; A); or after 7 h of incubation of Caco‐2 cells with bacterial cells (L. lactis NZ9000ΔhtrA and Lb. salivarius coated with conditioned medium of L. lactis NZ9000ΔhtrA; B).
Data S1. Supplemental methods.
Acknowledgements
This work was supported by the Slovenian Research Agency (grant number P4‐0127). We are grateful to Prof. Roger Pain for critical reading of the manuscript.
Microbial Biotechnology (2017) 10(6), 1732–1743
Funding Information
This work was supported by the Slovenian Research Agency (grant number J1‐5450, P4‐0127).
References
- Ajuebor, M.N. , and Swain, M.G. (2002) Role of chemokines and chemokine receptors in the gastrointestinal tract. Immunology 105: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti, M. , Cesta, M.C. , Garin, A. , and Proudfoot, A.E. (2012) Current status of chemokine receptor inhibitors in development. Immunol Lett 145: 68–78. [DOI] [PubMed] [Google Scholar]
- van Asseldonk, M. , Rutten, G. , Oteman, M. , Siezen, R.J. , de Vos, W.M. , and Simons, G. (1990) Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95: 155–160. [DOI] [PubMed] [Google Scholar]
- Atreya, R. , and Neurath, M.F. (2010) Chemokines in inflammatory bowel diseases. Dig Dis 28: 386–394. [DOI] [PubMed] [Google Scholar]
- Back, A. , Borges, F. , Mangavel, C. , Paris, C. , Rondags, E. , Kapel, R. , et al (2016) Recombinant pediocin in Lactococcus lactis: increased production by propeptide fusion and improved potency by co‐production with PedC. Microb Biotechnol 9: 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, P.K. , Eggstein, S. , Wolff‐Vorbeck, G. , Baumgartner, U. , and Hopt, U.T. (2005) Chemokines in human colorectal carcinoma. Anticancer Res 25: 3581–3584. [PubMed] [Google Scholar]
- Berlec, A. , Ravnikar, M. , and Strukelj, B. (2012) Lactic acid bacteria as oral delivery systems for biomolecules. Pharmazie 67: 891–898. [PubMed] [Google Scholar]
- Berlec, A. , Perse, M. , Ravnikar, M. , Lunder, M. , Erman, A. , Cerar, A. , and Strukelj, B. (2016) Dextran sulphate sodium colitis in C57BL/6J mice is alleviated by Lactococcus lactis and worsened by the neutralization of Tumor necrosis Factor alpha. Int Immunopharmacol 43: 219–226. [DOI] [PubMed] [Google Scholar]
- Bonvin, P. , Dunn, S.M. , Rousseau, F. , Dyer, D.P. , Shaw, J. , Power, C.A. , et al (2014) Identification of the pharmacophore of the CC chemokine‐binding proteins Evasin‐1 and ‐4 using phage display. J Biol Chem 289: 31846–31855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin, P. , Power, C.A. , and Proudfoot, A.E. (2016) Evasins: therapeutic potential of a new family of chemokine‐binding proteins from ticks. Front Immunol 7: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma, T. , Kanninga, R. , Neef, J. , Audouy, S.A. , van Roosmalen, M.L. , Steen, A. , et al (2006) Novel surface display system for proteins on non‐genetically modified gram‐positive bacteria. Appl Environ Microbiol 72: 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunersreuther, V. , Montecucco, F. , Pelli, G. , Galan, K. , Proudfoot, A.E. , Belin, A. , et al (2013) Treatment with the CC chemokine‐binding protein Evasin‐4 improves post‐infarction myocardial injury and survival in mice. Thromb Haemost 110: 807–825. [DOI] [PubMed] [Google Scholar]
- Buist, G. , Kok, J. , Leenhouts, K.J. , Dabrowska, M. , Venema, G. , and Haandrikman, A.J. (1995) Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol 177: 1554–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani, M.L. , Bhattacharya, K. , Tagen, M. , Kempuraj, D. , Perrella, A. , De Lutiis, M. , et al (2007) Anti‐chemokine therapy for inflammatory diseases. Int J Immunopathol Pharmacol 20: 447–453. [DOI] [PubMed] [Google Scholar]
- Castor, M.G. , Rezende, B. , Resende, C.B. , Alessandri, A.L. , Fagundes, C.T. , Sousa, L.P. , et al (2010) The CCL3/macrophage inflammatory protein‐1 alpha‐binding protein evasin‐1 protects from graft‐versus‐host disease but does not modify graft‐versus‐leukemia in mice. J Immunol 184: 2646–2654. [DOI] [PubMed] [Google Scholar]
- Copin, J.C. , da Silva, R.F. , Fraga‐Silva, R.A. , Capettini, L. , Quintao, S. , Lenglet, S. , et al (2013) Treatment with Evasin‐3 reduces atherosclerotic vulnerability for ischemic stroke, but not brain injury in mice. J Cereb Blood Flow Metab 33: 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes‐Perez, N.G. , Poquet, I. , Oliveira, M. , Gratadoux, J.J. , Madsen, S.M. , Miyoshi, A. , et al (2006) Construction and characterization of a Lactococcus lactis strain deficient in intracellular ClpP and extracellular HtrA proteases. Microbiol 152: 2611–2618. [DOI] [PubMed] [Google Scholar]
- Davey, L. , Halperin, S.A. , and Lee, S.F. (2016) Thiol‐disulfide exchange in Gram‐positive Firmicutes . Trends Microbiol 24: 902–915. [DOI] [PubMed] [Google Scholar]
- de Moreno de LeBlanc, A. , Del Carmen, S. , Chatel, J.M. , Miyoshi, A. , Azevedo, V. , Langella, P. , et al (2015) Current review of genetically modified lactic acid bacteria for the prevention and treatment of colitis using murine models. Gastroenterol Res Pract 2015: 146972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruaz, M. , Frauenschuh, A. , Alessandri, A.L. , Dias, J.M. , Coelho, F.M. , Russo, R.C. , et al (2008) Ticks produce highly selective chemokine binding proteins with antiinflammatory activity. J Exp Med 205: 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruaz, M. , Bonvin, P. , Severin, I.C. , Johnson, Z. , Krohn, S. , Power, C.A. , and Proudfoot, A.E. (2013) Evasin‐4, a tick‐derived chemokine‐binding protein with broad selectivity can be modified for use in preclinical disease models. FEBS J 280: 4876–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, M.A. , Bik, E.M. , Carlin, K.P. , Venn‐Watson, S.K. , Jensen, E.D. , Jones, S.E. , et al (2013) Identification of Lactobacillus strains with probiotic features from the bottlenose dolphin (Tursiops truncatus). J Appl Microbiol 115: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenschuh, A. , Power, C.A. , Deruaz, M. , Ferreira, B.R. , Silva, J.S. , Teixeira, M.M. , et al (2007) Molecular cloning and characterization of a highly selective chemokine‐binding protein from the tick Rhipicephalus sanguineus . J Biol Chem 282: 27250–27258. [DOI] [PubMed] [Google Scholar]
- Gonzalez‐Motos, V. , Kropp, K.A. , and Viejo‐Borbolla, A. (2016) Chemokine binding proteins: an immunomodulatory strategy going viral. Cytokine Growth Factor Rev 30: 71–80. [DOI] [PubMed] [Google Scholar]
- Han, W. , Mercenier, A. , Ait‐Belgnaoui, A. , Pavan, S. , Lamine, F. , Van Swam, I.I. , et al (2006) Improvement of an experimental colitis in rats by lactic acid bacteria producing superoxide dismutase. Inflamm Bowel Dis 12: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Ina, K. , Kusugami, K. , Yamaguchi, T. , Imada, A. , Hosokawa, T. , Ohsuga, M. , et al (1997) Mucosal interleukin‐8 is involved in neutrophil migration and binding to extracellular matrix in inflammatory bowel disease. Am J Gastroenterol 92: 1342–1346. [PubMed] [Google Scholar]
- Kleerebezem, M. , and de Vos, W.M. (2011) Lactic acid bacteria: life after genomics. Microb Biotechnol 4: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelink, P.J. , Overbeek, S.A. , Braber, S. , de Kruijf, P. , Folkerts, G. , Smit, M.J. , and Kraneveld, A.D. (2012) Targeting chemokine receptors in chronic inflammatory diseases: an extensive review. Pharmacol Ther 133: 1–18. [DOI] [PubMed] [Google Scholar]
- Kosler, S. , Strukelj, B. , and Berlec, A. (2017) Lactic acid bacteria with concomitant IL‐17, IL‐23 and TNFalpha‐ binding ability for the treatment of inflammatory bowel disease. Curr Pharm Biotechnol 18: 318–326. [DOI] [PubMed] [Google Scholar]
- Kuipers, O.P. , de Ruyter, P.G.G.A. , Kleerebezem, M. , and de Vos, W.M. (1998) Quorum sensing‐controlled gene expression in lactic acid bacteria. J Biotechnol 64: 15–21. [Google Scholar]
- Lindholm, A. , Smeds, A. , and Palva, A. (2004) Receptor binding domain of Escherichia coli F18 fimbrial adhesin FedF can be both efficiently secreted and surface displayed in a functional form in Lactococcus lactis . Appl Environ Microbiol 70: 2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerce, T.D. , Gomes‐Santos, A.C. , Rocha, C.S. , Moreira, T.G. , Cruz, D.N. , Lemos, L. , et al (2014) Anti‐inflammatory effects of Lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. Gut Pathog 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierau, I. , and Kleerebezem, M. (2005) 10 years of the nisin‐controlled gene expression system (NICE) in Lactococcus lactis . Appl Microbiol Biotechnol 68: 705–717. [DOI] [PubMed] [Google Scholar]
- Montecucco, F. , Mach, F. , Lenglet, S. , Vonlaufen, A. , Gomes Quindere, A.L. , Pelli, G. , et al (2014) Treatment with Evasin‐3 abrogates neutrophil‐mediated inflammation in mouse acute pancreatitis. Eur J Clin Invest 44: 940–950. [DOI] [PubMed] [Google Scholar]
- Ng, S.C. , Hart, A.L. , Kamm, M.A. , Stagg, A.J. , and Knight, S.C. (2009) Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis 15: 300–310. [DOI] [PubMed] [Google Scholar]
- Poquet, I. , Saint, V. , Seznec, E. , Simoes, N. , Bolotin, A. , and Gruss, A. (2000) HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol Microbiol 35: 1042–1051. [DOI] [PubMed] [Google Scholar]
- Proudfoot, A.E. , Power, C.A. , and Schwarz, M.K. (2010) Anti‐chemokine small molecule drugs: a promising future? Expert Opin Investig Drugs 19: 345–355. [DOI] [PubMed] [Google Scholar]
- Proudfoot, A.E. , Bonvin, P. , and Power, C.A. (2015) Targeting chemokines: pathogens can, why can't we? Cytokine 74: 259–267. [DOI] [PubMed] [Google Scholar]
- Raab, Y. , Gerdin, B. , Ahlstedt, S. , and Hallgren, R. (1993) Neutrophil mucosal involvement is accompanied by enhanced local production of interleukin‐8 in ulcerative colitis. Gut 34: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnikar, M. , Strukelj, B. , Obermajer, N. , Lunder, M. , and Berlec, A. (2010) Engineered lactic acid bacterium Lactococcus lactis capable of binding antibodies and tumor necrosis factor alpha. Appl Environ Microbiol 76: 6928–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, R.C. , Alessandri, A.L. , Garcia, C.C. , Cordeiro, B.F. , Pinho, V. , Cassali, G.D. , et al (2011) Therapeutic effects of evasin‐1, a chemokine binding protein, in bleomycin‐induced pulmonary fibrosis. Am J Respir Cell Mol Biol 45: 72–80. [DOI] [PubMed] [Google Scholar]
- de Ruyter, P.G. , Kuipers, O.P. , and de Vos, W.M. (1996) Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol 62: 3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte, L. , Steidler, L. , Vandekerckhove, J. , and Remaut, E. (2000) Secretion of biologically active murine interleukin‐10 by Lactococcus lactis . Enzyme Microb Technol 27: 761–765. [DOI] [PubMed] [Google Scholar]
- Sebastian, S. , Hernández, V. , Myrelid, P. , Kariv, R. , Tsianos, E. , Toruner, M. , et al (2014) Colorectal cancer in inflammatory bowel disease: results of the 3rd ECCO pathogenesis scientific workshop (I). J Crohn Colitis 8: 5–18. [DOI] [PubMed] [Google Scholar]
- Seet, B.T. , and McFadden, G. (2002) Viral chemokine‐binding proteins. J Leukoc Biol 72: 24–34. [PubMed] [Google Scholar]
- Steen, A. , Buist, G. , Leenhouts, K.J. , El Khattabi, M. , Grijpstra, F. , Zomer, A.L. , et al (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem 278: 23874–23881. [DOI] [PubMed] [Google Scholar]
- Steen, A. , Larsen, O. , Thiele, S. , and Rosenkilde, M.M. (2014) Biased and G protein‐independent signaling of chemokine receptors. Front Immunol 5: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafillidis, J.K. , Nasioulas, G. , and Kosmidis, P.A. (2009) Colorectal cancer and inflammatory bowel disease: epidemiology, risk factors, mechanisms of carcinogenesis and prevention strategies. Anticancer Res 29: 2727–2737. [PubMed] [Google Scholar]
- Triantafillidis, J.K. , Merikas, E. , and Georgopoulos, F. (2011) Current and emerging drugs for the treatment of inflammatory bowel disease. Drug Des Devel Ther 5: 185–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, R. , Gurbaxani, B. , Morrison, S.L. , and Seyfzadeh, M. (2004) Optimization of codon pair use within the (GGGGS)3 linker sequence results in enhanced protein expression. Mol Immunol 40: 717–722. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke, K. , de Haard, H. , Beirnaert, E. , Dreier, T. , Lauwereys, M. , Huyck, L. , et al (2010) Orally administered L. lactis secreting an anti‐TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3: 49–56. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Dubois, R.N. , and Richmond, A. (2009) The role of chemokines in intestinal inflammation and cancer. Curr Opin Pharmacol 9: 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadravec, P. , Mavric, A. , Bogovic Matijasic, B. , Strukelj, B. , and Berlec, A. (2014) Engineering BmpA as a carrier for surface display of IgG‐binding domain on Lactococcus lactis . Protein Eng Des Sel 27: 21–27. [DOI] [PubMed] [Google Scholar]
- Zadravec, P. , Strukelj, B. , and Berlec, A. (2015a) Heterologous surface display on lactic acid bacteria: non‐GMO alternative? Bioengineered 6: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadravec, P. , Strukelj, B. , and Berlec, A. (2015b) Improvement of LysM‐mediated surface display of designed ankyrin repeat proteins (DARPins) in recombinant and nonrecombinant strains of Lactococcus lactis and Lactobacillus species . Appl Environ Microbiol 81: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadravec, P. , Mareckova, L. , Petrokova, H. , Hodnik, V. , Perisic Nanut, M. , Anderluh, G. , et al (2016) Development of recombinant Lactococcus lactis displaying albumin‐binding domain variants against Shiga toxin 1 B subunit. PLoS ONE 11: e0162625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik, A. , and Yoshie, O. (2000) Chemokines: a new classification system and their role in immunity. Immunity 12: 121–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Strains, plasmids and primers used in the study.
Fig. S1. Shift in fluorescence intensity.
Fig. S2. ELISA‐determined percentage of CCL3, CCL5 and CXCL8 removed after incubation with 6 × 108 (white bars), 3 × 109 (gray bars), or 6 × 109 (black bars) cells/mL of recombinant L. lactis NZ9000 cells that displayed evasin‐1 (pSDEva1), evasin‐4 (pSDEva4), or evasin‐3 (pSDEva3).
Fig. S3. The portion of CCL3 removed from the solution after incubation with 2 x 109 Lb. salivarius cells coated with evasin‐1 (white bars) or evasin‐4 (black bars) fusion proteins.
Fig. S4. ELISA‐determined CXCL8 secretion by Caco‐2 cells exposed to IL‐1β.
Fig. S5. Percentage of viable Caco‐2 cells determined with trypan blue exclusion staining after 2 h of incubation with different number of L. lactis NZ9000ΔhtrA cells (2 × 108, 1 × 109 and 2 × 109; A); or after 7 h of incubation of Caco‐2 cells with bacterial cells (L. lactis NZ9000ΔhtrA and Lb. salivarius coated with conditioned medium of L. lactis NZ9000ΔhtrA; B).
Data S1. Supplemental methods.


