 Different starter unit and complex tailoring steps for type II polyketide synthase in trioxacarcin biosynthesis.
Different starter unit and complex tailoring steps for type II polyketide synthase in trioxacarcin biosynthesis.
Abstract
Trioxacarcins (TXNs) are highly oxygenated, polycyclic aromatic natural products with remarkable biological activity and structural complexity. Evidence from 13C-labelled precursor feeding studies demonstrated that the scaffold was biosynthesized from one unit of l-isoleucine and nine units of malonyl-CoA, which suggested a different starter unit in the biosynthesis. Genetic analysis of the biosynthetic gene cluster revealed 56 genes encoding a type II polyketide synthase (PKS), combined with a large amount of tailoring enzymes. Inactivation of seven post-PKS modification enzymes resulted in the production of a series of new TXN analogues, intermediates, and shunt products, most of which show high anti-cancer activity. Structural elucidation of these new compounds not only helps us to propose the biosynthetic pathway, featuring a type II PKS using a novel starter unit, but also set the stage for further characterization of the enzymatic reactions and combinatorial biosynthesis.
Introduction
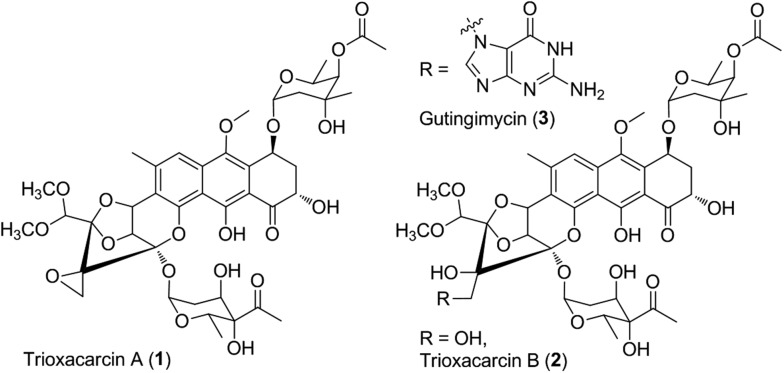
Microorganisms can produce a large variety of biologically active secondary metabolites representing a vast diversity of fascinating molecular architecture, which usually attract attention for chemical synthesis, mode of action, biosynthesis, and even drug discovery studies. As an example, trioxacarcin A (TXN-A, 1, Fig. 1) represents a special family of complex aromatic natural products, which was first isolated from Streptomyces bottropensis DO-45 (NRRL 12051) in 1981,1–3 and subsequently re-isolated from a marine Streptomyces sp. B8652 with a series of analogues in 2004.4,5 It displays extraordinary anti-bacterial, anti-malarial, and anti-tumor activity with sub-nanomolar IC70 values in various cancer cell lines.1–5 Structurally, TXN-A contains an unusual condensed polycyclic trisketal, bearing a fused spiro-epoxide, which is believed to be a “warhead” to covalently bind to DNA, followed by cleavage of the resultant TXN–DNA complex, to yield another natural product gutingimycin (3, Fig. 1) through an abstraction of the guanine.6,7 In addition, it has unique glycosylation patterns, including a rare γ-branched octose.
Fig. 1. Chemical structure of trioxacarcin (TXN) and relative natural products.
The high biological activities, especial anti-cancer activity, along with unusual and complex structural features of TXN-A distinguish it from other aromatic polyketides, thus providing an interesting but challenging target for total synthesis. Recently, Myers's group successfully established a multiply convergent, component-based route to chemically synthesize TXN-A and its structural analogues.8,9 However, the biosynthetic studies have never been explored to these structurally complex antibiotics. Herein, we describe (1) incorporation studies with 13C-labelled precursors, which elucidated the biosynthetic origin of the scaffold for the TXN family of natural products; (2) the genetic characterization of txn gene cluster, which afforded four polyketide derivates and seven TXN analogues; and (3) a proposed biosynthetic pathway, involving a different starter unit for priming type II polyketide synthase (PKS) and complex tailoring steps.
Results and discussion
Biosynthetic origin of the polycyclic scaffold of TXNs
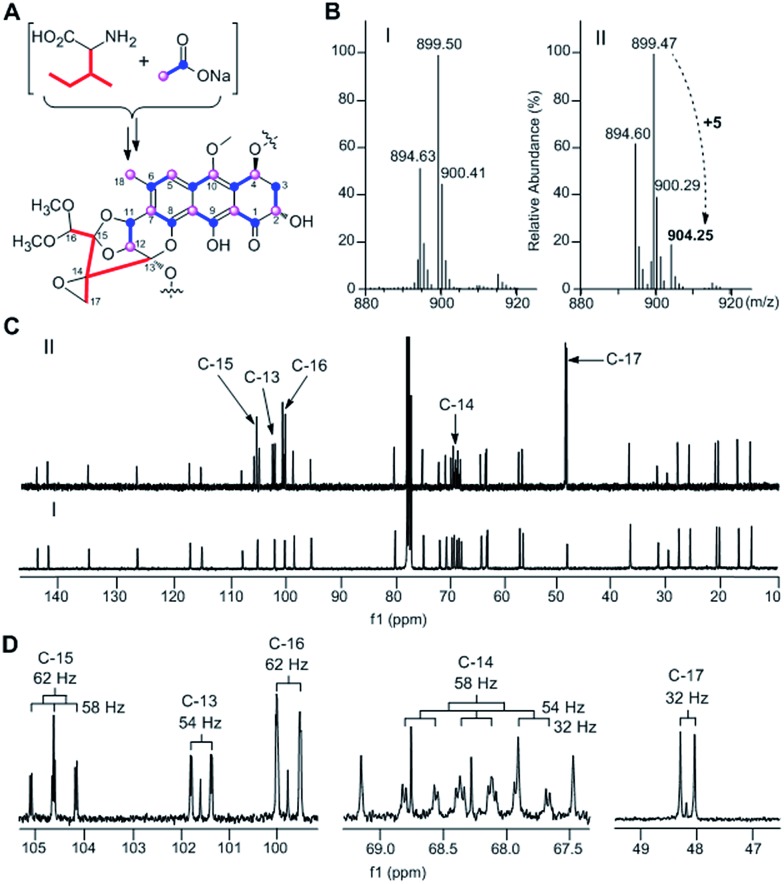
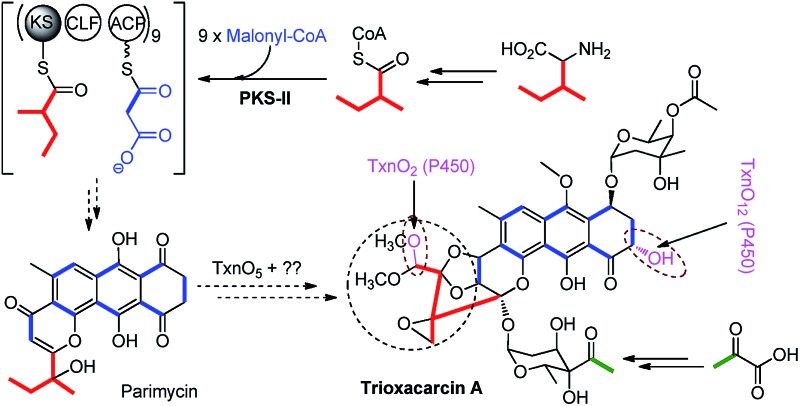
TXN-A was originally isolated from S. bottropensis DO-45 with the isolation of 20 mg from an 18 L fermentation broth;1,2 however this titer was not efficient enough for biosynthetic studies. In our early efforts to optimize the fermentation and isolation processes, we noticed that the yield of TXN-A could be significantly improved by a hundred times through the addition of the hydrophobic resin HP-20 into the fermentation medium, even up to titers of 100–200 mg L–1 in shaking flasks.10 Under this optimized condition, the precursors [1-13C]-acetate, [2-13C]-acetate, and [1,2-13C]-acetate were added to a fermentation culture (a total of 0.7 g L–1) by pulse feeding after 48, 56, 64, 72, 80, 88 h of incubation in separate incorporation experiments, and the fermentation lasted 120 h. TXN-A isolated from the feeding fermentations was subjected to 13C-NMR analysis to confirm the polyketide extender units of the scaffold (ESI, Table S1†). All the 13C abundance at each position of the TXN-A backbone could be sufficiently separated and identified (ESI, Fig. S1†). These incorporation results are summarized in Table S1† and Fig. 2A. Significant enrichment was observed at C-1, C-3, C-4a, C-6, C-8, C-9, C-10a, and C-11 in the [1-13C]-acetate labelled TXN-A, as well as C-2, C-4, C-5, C-7, C-8a, C-9a, C-10, C-12, and C-18 in the [2-13C]-acetate labelled TXN-A; both suggested the folding pattern of the polyketide chain in Fig. 2A, which was further supported by the [1,2-13C]-acetate feeding results (Fig. S1 and Table S1†). Obviously, the right ring contains three malonate-derived intact acetate units (C-9a to C-1, C-2 to C-3, and C-4 to C-4a), which suggested the folding pattern of the polyketide chain could be classified as a typical Streptomyces mode.11 In addition, the incorporation of the [2-13C]-acetate to C-18 indicated that a decarboxylation step should be involved in the formation of a fused-ring skeleton. However, the labelled pattern of the five-carbon fragment (C-13, C-14, C-15, C-16, and C-17) remains confused, which hints that this five-carbon unit may be derived from another origin. Moreover, the five-carbon unit was likely employed by the type II PKS as a non-acetate starter unit to generate the polyketide in which the decarboxylation is usually performed on the last carbon of the fully elongated polyketide chain.
Fig. 2. Characterization of the biosynthetic origin of TXNs by precursor feeding experiments. (A) Summary of feeding results with 13C-labeled sodium acetate and 13C6-l-isoleucine (Ile). (B) MS analysis of production of TXN by fermentation without (I) or with 13C6-l-Ile (II). (C) 13C-NMR spectra of TXN-A with (II) and without (I) feeding of 13C6-l-Ile. The enhanced signals of C-13, C-14, C15, C-16, and C-17 are marked. (D) The enlarged parts of 13C-NMR spectra from feeding experiment.
A five-carbon unit (C-13 to C-17), most possibly from 2-methylbutyryl-CoA, serving as the starter unit of PKS, is seldom observed in natural product biosynthesis. The only exception is involved in the biosynthesis of avermectin “a” components, which are 16-membered macrocyclic lactones generated by type I PKS through loading 2-methylbutyryl-CoA as the starter unit.12 For the type II PKS, although non-acetate starter units, including propionate, malonamate, polyketide or fatty acid, and even amino acid derivates have also been employed,13 2-methylbutyryl-CoA has never been reported as a starter unit to generate aromatic polyketides. Given the fact that 2-methylbutyryl-CoA is usually derived from l-isoleucine (Ile) through deamination and decarboxylation by transaminase and branched-chain 2-oxo acid dehydrogenase in vivo, we performed the feeding experiment with 13C6-l-Ile to validate this hypothesis (Fig. 2A). Remarkably, ESI-MS showed TXN-A from this feeding experiment was +5 m/z heavier than that without feeding (Fig. 2B), indicating the incorporation of a five-carbon unit which arose from an intact Ile. Further specific and significant signal enrichment at C-13 to C-17 (Fig. 2C) in the 13C-NMR spectra, and all the 13C–13C coupling data (Fig. 2D) are consistent with the same conclusion (JC-16/C-15 = 62 Hz, JC-15/C-14 = 58 Hz, JC-13/C-14 = 54 Hz, and JC-17/C-14 = 32 Hz), which are in agreement with this five-carbon unit originating from Ile via an intact incorporation manner. Thus, these results unambiguously demonstrated that the missing five-carbon unit, C-13 to C-17, is derived from l-Ile, which most likely follows a deamination and decarboxylation process similar to that of the avermectin “a” components biosynthesis.12
Cloning, sequencing, and identification of the biosynthetic gene cluster of TXNs
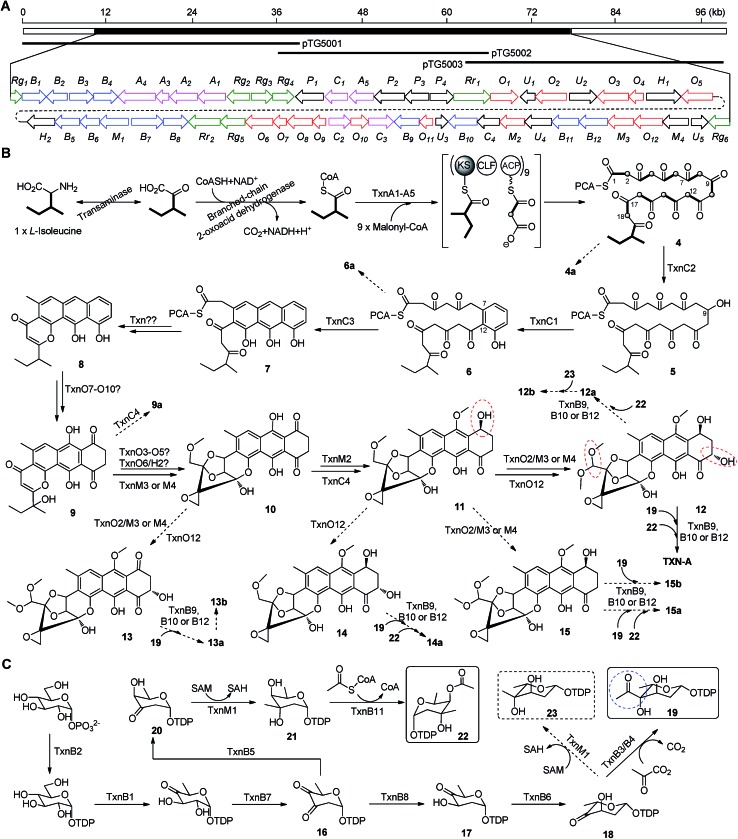
The aromatic polycyclic skeleton of TXNs and the primary 13C-labeled acetate feeding experiments suggest that a type-II PKS should be involved in the biosynthesis. Therefore, we cloned the gene cluster by the PCR approach specific for accessing the genes encoding a ketosynthase (KS)-chain length factor (CLF) heterodimer.14 By screening the genomic library and the subsequent chromosome walking, a 102 kb contiguous DNA sequence was mapped into three overlapping fosmids (pTG5001, pTG5002 and pTG5003, Fig. 3A). Sequencing and bioinformatic analysis of these fosmids revealed 91 ORFs, most of which (the txn gene cluster) are deposited in the GenBank under the accession no. KP410250.
Fig. 3. Biosynthetic gene cluster and proposed biosynthetic pathway of TXN-A and relative metabolites. (A) Localization of the cloned DNA region as represented by three overlapping fosmids and organization of the txn gene cluster. Color coding indicates the genes for the PKS and PKS associated enzymes (pink), tailoring enzymes (red), deoxysugar (blue), regulators and resistant proteins (green), and all others (black). (B) Proposed model for type II PKS and post-PKS modification. (C) Proposed biosynthetic pathway of two deoxysugar moieties. The significant points of the pathway were highlighted by colored circles.
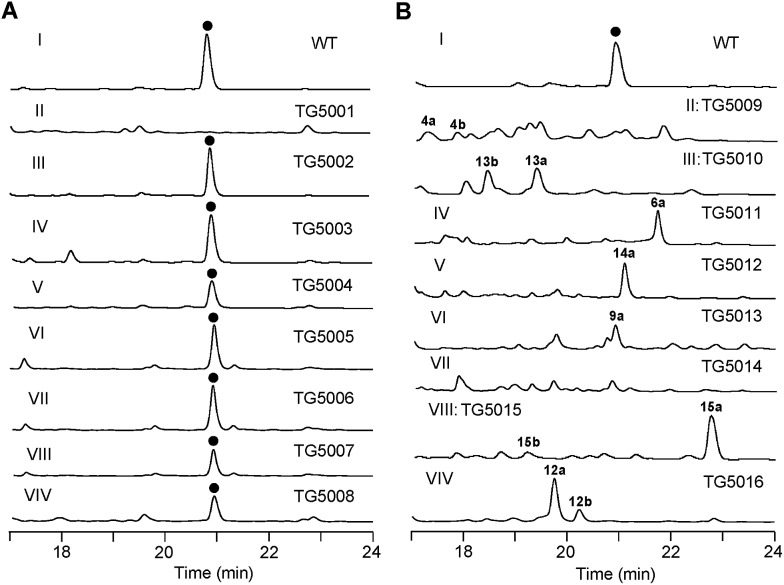
To verify that the cloned gene cluster was involved in TXNs biosynthesis, we constructed a mutant strain TG5001 in which the txnA1 gene encoding KS was inactivated by gene disruption (ESI, Fig. S2†). As expected, this mutant strain completely abolished the production of TXN-A (Fig. 4A-II), which proved the essential role of this gene cluster governing TXN biosynthesis. Next inactivation of the genes orf–2 (acyltransferase), orf–1 (unknown), orf+11 (cytochrome P450), and orf+3 (tRNA-synthetase) had no effect on TXN-A production; whereas, inactivation of txnRg1 (regulator) or txnRg6 (regulator) led to obviously decreased the yield of TXN-A (Fig. 4A-III to VIII), which suggested that the txn gene cluster may range from txnRg1 to txnRg6, encompassing 56 ORFs (Fig. 3A and Table 1).
Fig. 4. Genetic characterization of the genes for TXN biosynthesis in vivo. HPLC analysis of TXN-A and analogues production (UV at 271 nm) from S. bottropensis: (I) wild-type NRRL 12051, (A-II) mutant TG5001 (ΔtxnA1), (A-III) TG5002 (Δorf–3), (A-IV) TG5003 (Δorf–1), (A-V) TG5004 (ΔtxnRg1), (A-VI) TG5005 (Δorf+11), (A-VII) TG5006 (Δorf+3), (A-VIII) TG5007 (ΔtxnRg6), (A-VIV) TG5008 (ΔtxnA4); (B-II) TG5009 (ΔtxnC2), (B-III) TG5010 (ΔtxnC4), (B-IV) TG5011 (ΔtxnC3), (B-V) TG5012 (ΔtxnO2), (B-VI) TG5013 (ΔtxnO5), (B-VII) TG5014 (ΔtxnO6), (B-VIII) TG5015 (ΔtxnO12), (B-VIV) TG5016 (ΔtxnB4). () TXN-A. The genotypes of all the mutants were confirmed by PCR analysis, and the results were summarized in Fig. S2.† .
Table 1. Deduced functions of ORFs in txn biosynthetic gene cluster.
| Gene | AA a | Protein homolog (accession no.), origin | S/I b (%) | Proposed function |
| txnRg1 | 94 | LuxR family regulator (016578673), S. albulus | 65/55 | Regulator |
| txnB1 | 330 | ChlC2 (AAZ77689), S. antibioticus | 76/67 | dTDP-glucose 4,6-dehydratase |
| txnB2 | 290 | AclY (BAB72036), S. galilaeus | 86/74 | dTDP-glucose synthase |
| txnB3 | 327 | KstD7 (AFJ52686), Micromonospora sp. TP-A0468 | 76/66 | Pyruvate dehydrogenase-α |
| txnB4 | 345 | KstD8 (AFJ52687), Micromonospora sp. TP-A0468 | 86/79 | Pyruvate dehydrogenase-β |
| txnA4 | 561 | OxyP (AAZ78339), S. rimosus | 64/53 | MAT |
| txnA3 | 90 | SsfC (ADE34520), S. sp. SF2575 | 76/53 | ACP |
| txnA2 | 406 | Snoa2 (CAA12018), S. nogalater | 77/66 | CLF (KSβ) |
| txnA1 | 420 | PgaA (AAK57525), S. sp. PGA64 | 84/72 | KSα |
| txnRg2 | 263 | DnrI (EFL25867), S. himastatinicus ATCC 53653 | 78/63 | SARP-family regulator |
| txnRg3 | 394 | 2-Component kinase (ADO32765), S. vietnamensis | 54/40 | 2-Component kinase |
| txnRg4 | 203 | 2-Component regulator (CAA09631), S. violaceoruber | 82/69 | 2-Component regulator |
| txnP1 | 579 | RkA (ACZ65474), S. sp. 88-682 | 57/43 | ATP-dependent CoA synthetase |
| txnC1 | 318 | ORF27 (AEM44304), e-DNA | 64/50 | Aromatase |
| txnA5 | 344 | CosE (ABC00733), S. olindensis | 70/58 | KS-III |
| txnP2 | 543 | 2-Isopropylmalate synthase (ACY99077), Thermomonospora curvata DSM 43183 | 70/58 | 2-Isopropylmalate synthase |
| txnP3 | 417 | Acyl-CoA transferase/dehydratase (EIE99664), S. glauca K62 | 67/56 | Dehydratase or isomerase |
| txnP4 | 260 | Ketoreductase (EDY66493), S. pristinaespiralis ATCC 25486 | 63/46 | Short-chain dehydrogenase |
| txnRr1 | 500 | Actinorhodin transporter (EFL40860), S. griseoflavus Tu4000 | 64/48 | Transporter |
| txnO1 | 345 | Dehydrogenase (ACZ83978), Streptosporangium roseum DSM43021 | 74/61 | Dehydrogenase |
| txnU1 | 126 | Tcur_2795 (ACY98340), Thermomonospora curvata DSM 43183 | 40/33 | Unknown |
| txnO2 | 401 | P450 (CBX53644), S. platensis | 66/52 | Cytochrome P450 |
| txnU2 | 366 | O3I_28241 (EHY24336), Nocardia brasiliensis ATCC 700358 | 69/53 | Unknown |
| txnO3 | 411 | ThcD (AAC45752), Rhodococcus erythropolis | 62/48 | Ferredoxin reductase |
| txnO4 | 107 | 2Fe-2S ferredoxin (ZP_09514545), Oceanicola sp. S124 | 68/52 | Ferredoxin |
| txnH1 | 494 | Putative tripeptidylaminopeptidase (AAP85358), S. griseoruber | 68/59 | Hydrolase |
| txnO5 | 409 | ORF29 (AAP85338), S. griseoruber | 68/53 | Cytochrome P450 |
| txnH2 | 373 | Microsomal epoxide hydrolase (EHI80707), Frankia sp. CN3 | 68/56 | Epoxide hydrolase |
| txnB5 | 328 | PokS9 (ACN64856), S. diastatochromogenes | 70/60 | dNDP-hexose-4-ketoreductase |
| txnB6 | 213 | PokS7 (ACN64855), S. diastatochromogenes | 82/72 | 3,5-Epimerase |
| txnM1 | 413 | TylCIII (AAD41823), S. fradiae | 84/73 | dNDP-hexose 3-C-MT |
| txnB7 | 488 | SaqS (ACP19377), Micromonospora sp. Tu 6368 | 71/62 | dNDP-hexose 2,3-dehydratase |
| txnB8 | 321 | SaqT (ACP19378), Micromonospora sp. Tu 6368 | 70/62 | dNDP-hexose 3-ketoreductase |
| txnRr2 | 500 | EmrB/QacA (EGE43895), S. griseus XylebKG1 | 75/59 | Transporter |
| txnRg5 | 339 | DeoR regulator (ACZ87003), Streptosporangium roseum DSM43021 | 77/70 | Regulator |
| txnO6 | 406 | ORF3 (AAD28449), S. lavendulae | 63/45 | Cytochrome P450 |
| txnO7 | 175 | PokC1 (ACN64848), S. diastatochromogenes | 45/35 | Cyclase or hydroxylase |
| txnO8 | 371 | AlnT (ACI88867), S. sp. CM020 | 57/43 | Hydroxylase |
| txnO9 | 154 | CalC (AAM70338), Micromonospora echinospora | 50/36 | Cyclase or hydroxylase |
| txnC2 | 261 | HedA (AAP85364), S. griseoruber | 83/71 | Ketoreductase |
| txnO10 | 178 | AsuE2 (ADI58638), S. nodosus subsp. asukaensis | 57/43 | Flavin reductase |
| txnC3 | 304 | Gra-ORF33 (ADO32793), S. vietnamensis | 68/56 | 2,3-Cyclase |
| txnB9 | 383 | SsfS6 (ADE34512), S. sp. SF2575 | 55/38 | Glycosyl transferase |
| txnO11 | 148 | Aln2 (ACI88858), S. sp. CM020 | 52/41 | Cyclase or hydroxylase |
| txnU3 | 121 | GrhI (AAM33661), S. sp. JP95 | 46/28 | Unknown |
| txnB10 | 424 | UrdGTa1 (AAF00214), S. fradiae | 61/47 | Glycosyl transferase |
| txnC4 | 240 | RedLA2 (AAT45284), S. tubercidicus | 82/73 | Ketoreductase |
| txnM2 | 340 | MetLA2 (AAT45283), S. tubercidicus | 79/70 | O-Methyltransferase |
| txnU4 | 388 | PAI11_01900 (EHN12885), Patulibacter sp. I11 | 82/69 | Unknown |
| txnB11 | 397 | Azi15 (ABY83154), S. sahachiroi | 63/50 | O-Acyltransferase |
| txnB12 | 427 | UrdGTa1 (AAF00214), S. fradiae | 61/47 | Glycosyl transferase |
| txnM3 | 339 | DmpM (AFE08598), Corallococcus coralloides DSM 2259 | 62/45 | O-Methyltransferase |
| txnO12 | 407 | FosK (AEC13077), S. pulveraceus | 67/54 | Cytochrome P450 |
| txnM4 | 340 | DmpM (AFE08598), Corallococcus coralloides DSM 2259 | 61/44 | O-Methyltransferase |
| txnU5 | 182 | RAM_06565 (AEK39805), Amycolatopsis mediterranei S699 | 75/64 | Unknown |
| txnRg6 | 286 | SARP regulator (ACU39492), Actinosynnema mirum DSM 43827 | 53/40 | Regulator |
aAmino acid.
bSimilarity/identity.
PKS and polyketide processing enzymes in TXN-A scaffold biosynthesis
Bioinformatic analysis not only gives the expected minimal PKS encoded by txnA1 (KS), txnA2 (CLF), and txnA3 (acyl carrier protein, ACP), but also reveals a malonyl-CoA:ACP transacylase (MAT, txnA4) and a KS-III (txnA5), which are less frequently involved in the type-II PKS machinery.13,15 The inactivation of txnA4 significantly reduced the production of TXN-A with 20–30% to that of WT (Fig. 4A-VIV). This phenomenon is reasonable, for the partially functional complementation by the MAT of fatty acid biosynthesis. Deep analysis of CLF (TxnA2) revealed the gatekeeper residues as G113-L117-W195-V110-G196-M151-F134, which shows specificity towards the C-23 polyketide length.16,17 Additionally, two genes (txnC2 and txnC4) encode enzymes bearing a high sequence homology (60–75% identity) with typical ketoreductases (KRs), while TxnC2 is closer to the C-9 KRs, which are involved in the folding and cyclization of the nascent polyketide chain.18,19 TxnC1 is relatively close to the aromatase likely responsible for the C7–C12 cyclization of the first ring, followed the ketoreduction of C-9 by TxnC2, and TxnC3 shares a high sequence similarity with the 2,3-cyclase, which catalyzes the second and third cyclization steps to form the aromatic ring intermediate 7 (Fig. 3B).
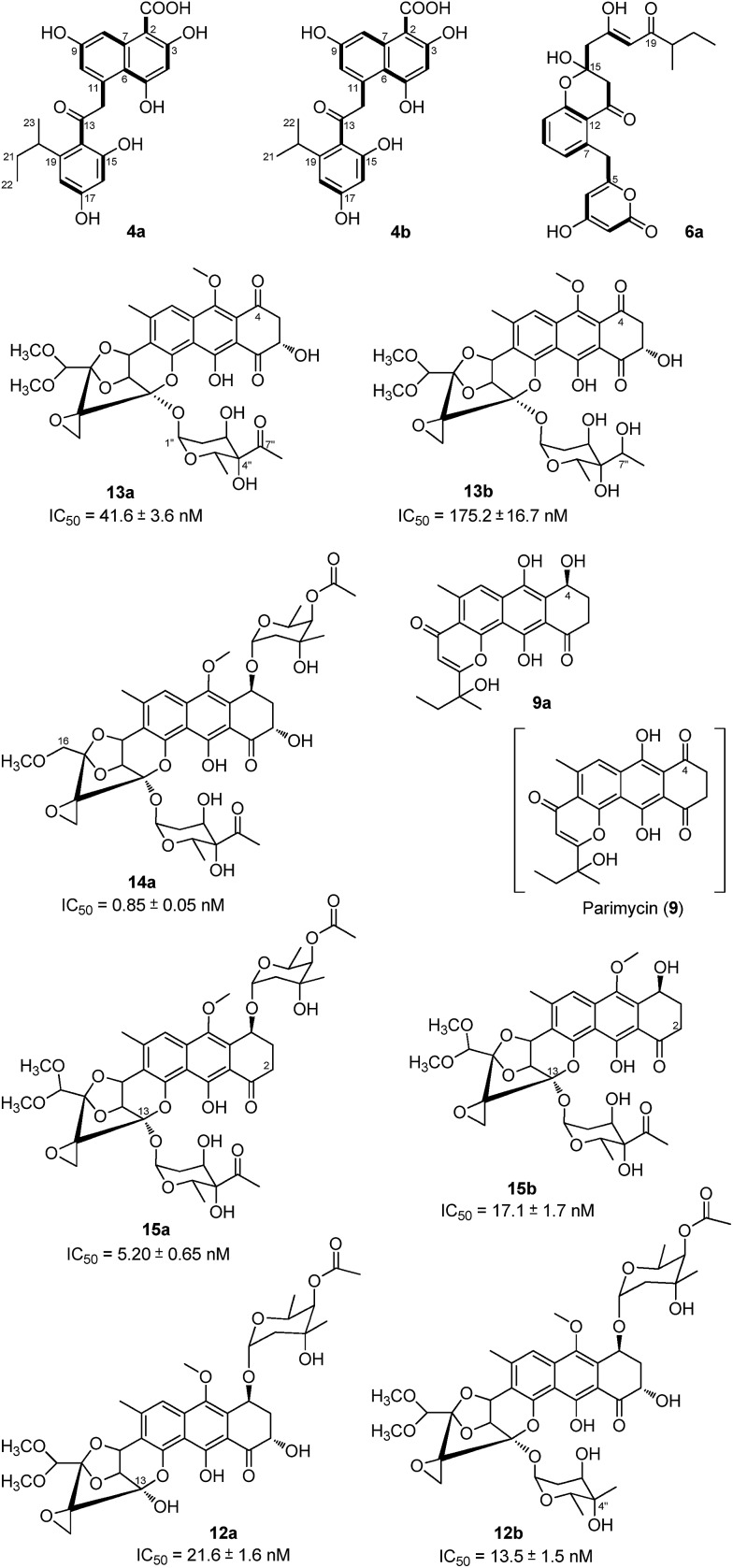
To verify the hypothetical functions of the relative genes in TXN-A biosynthesis, txnC2, txnC3 and txnC4 were inactivated separately by gene replacement with the aac(3)IV apramycin-resistance gene (ESI, Fig. S2†). The resultant mutant strains S. bottropensis TG5009 (ΔtxnC2), TG5010 (ΔtxnC4) and TG5011 (ΔtxnC3) all abolished production of TXN-A; whereas, each of the three mutants accumulated new compounds that are different from TXNs (Fig. 4B-II to IV). Following the optimized fermentation and isolation processes (including silica gel and Sephadex LH-20 column chromatography, preparative HPLC et al.), we obtained 10 mg of 4a and 6 mg of 4b from an 8 L culture of the TG5009 strain; 11 mg of 13a and 2 mg of 13b from a 4 L broth of the TG5010 strain; as well as 40 mg of 6a from a 2 L culture of the TG5011 strain. The chemical structures of these compounds were elucidated by MS, HRMS and 1D, 2D-NMR spectra (Fig. S3–S17, S28–S32 and Tables S4, S5, S8, S11†) and are summarized in Fig. 5. These results strongly support the biological function of the respective gene and the proposed biosynthetic pathway (Fig. 3B). Firstly, the production of compounds 4a and 4b by the TG5009 (ΔtxnC2) mutant verified that TxnC2 reduces the C-9 keto group of the nascent polyketide chain. More importantly, this result indicated that the reduction of C-9 is necessary for the next C7–C12 cyclization and aromatization, and a similar opinion has been widely accepted in type II PKS.19,20 Whereas, the production of a small amount of 4b is unexpected but reasonable, which could be derived from the incorporation of l-valine through deamination and decarboxylation, similar to that of the avermectin “b” components biosynthesis.12,21 Secondly, the TG5011 (ΔtxnC3) mutant affording compound 6a doubly confirmed that TxnC2/C1 catalyzes the C7–C12 first-ring cyclization and aromatization, and a similar cyclized compound SEK4 had been generated by an octaketide minimal PKS, except with a different starter unit and chain length.19,22 Thirdly, the isolation of 13a and 13b from the TG5010 (ΔtxnC4) mutant suggested that this KR catalyzes another ketoreduction, such as 10 into 11 (Fig. 3B), which affords the hydroxyl group for deoxysugar attachment. Together, the two new compounds 4a and 6a further established a different five-carbon starter unit for type-II PKS in TXN biosynthesis. Given the fact that the starter unit has been proven to be an attractive point for engineering aromatic polyketide biosynthetic machinery,21,23 the discovery of the different starter unit in TXN-A biosynthesis will also substantiate the potential for similar efforts.
Fig. 5. Chemical structures of the TXN-A analogues or relative metabolites produced by the mutants.
In a typically bacterial type II PKS system, a MAT sharing with fatty acid biosynthesis loads malonyl-CoA onto the thiol group of the 4′-phosphopantheinyl arm attached to the ACP, which is subsequently decarboxylated to generate an acetate starter unit and also used as extender units catalyzed by a KS-CLF heterodimer.13,15 Meanwhile, non-acetate starter units have been increasingly observed as alternative primers and usually involve an additional KS-III.13 Based on the precursor feeding, bioinformatic analysis and genetic characterization results, we could propose that the biosynthetic pathway of the TXN-A polyketide backbone follows the action of a special type II PKS (TxnA1–A2–A3) as illustrated in Fig. 3B. The enzymes involved in the branched-chain fatty acids catabolism, a transaminase and a branched-chain 2-oxo acid dehydrogenase catalyze the deamination and decarboxylation reactions to generate 2-methylbutyryl-CoA, which might be a direct starter unit for the KS of type II PKS primed by KS-III, TxnA5. Nine units of malonyl-CoA are subsequently incorporated into the PKS biosynthetic system by MAT (TxnA4) to form the full elongated polyketide chain 4. Next, the PKS associated enzymes KR (TxnC2), aromatase (TxnC1), and cyclase (TxnC3) are required to carry out the regioselective folding and cyclization of the nascent chain to yield the aromatic polycyclic backbone 7. Subsequently, a decarboxylation and further cyclization steps should be involved to yield the intermediate 8.
Tailoring enzymes for further modifications in TXN-A scaffold biosynthesis
The extremely complex structural features of TXN-A indicated that a large amount of unusual post-PKS modification steps should be involved to construct the framework. Indeed, the txn gene cluster encodes four methyltransferases (MTs, TxnM1–M4), twelve enzymes possibly related to oxidation–reduction (TxnO1–O12), two hydrolases (TxnH1–H2), and nine proteins with unknown functions (TxnP1–P4, TxnU1–U5). Except for the MTs, most of the tailoring enzymes could not be easily assigned physiological roles in the biosynthetic pathway.
In total, four cytochrome P450 enzymes (P450s) encoded by txnO2, O5, O6 and O12 attracted our attention because this family of oxidative hemoproteins could catalyze many different reactions for structural diversification in natural product biosynthesis.24 Therefore, we constructed the respective gene replacement mutants S. bottropensis TG5012 (ΔtxnO2), TG5013 (ΔtxnO5), TG5014 (ΔtxnO6) and TG5015 (ΔtxnO12), and analyzed the metabolites produced by HPLC and LC-MS. The results showed that each of the four mutants afforded compounds different from the wild type (Fig. 4B-V to VIII). Although attempts to isolate new compounds from the TG5014 (ΔtxnO6) mutant were unsuccessful for the low yield and instability, we finally obtained 40 mg of 14a from a 1 L culture of the TG5012 strain, 15 mg of 9a from a 4 L fermentation broth of the TG5013 mutant, as well as 20 mg of 15a and 4 mg of 15b from a 2 L culture of the TG5015 strain. Evaluation of the MS and NMR spectra and comparison with TXN-A (Fig. S18–S21, S33–S44 and Tables S6, S9–S11†) led to the successful assignment of the chemical structures of all these new compounds (Fig. 5).
Structurally, compound 9a is close to parimycin (9, Fig. 5), which was isolated from another TXN-A producing strain, marine Streptomyces sp. B8652, as a novel 2,3-dihydro-1,4-anthraquinone unrelated to TXNs.25 The isolation of 9a from a ΔtxnO5 mutant not only hinted that this P450 plays a key role in the formation of the highly oxygenated polycyclic skeleton, but also suggested that 9 or 9a should be the intermediate for the bio-generation of TXN-A (Fig. 3B). We believe that TxnO5 (P450), or/and TxnO6 (P450), TxnO4 (ferredoxin), TxnO3 (ferredoxin reductase), TxnH2 (epoxide hydrolase), and TxnM3 or M4 (MT) should be involved in the transformation of 10 from 9 (Fig. 3B and S45†), while this complex process may need more uncharacterized enzymes. In addition, the production of 14a by the ΔtxnO2 mutant and 15a/15b by the ΔtxnO12 mutant showed that the P450s catalyze hydroxylation at the C-16 and C-2 positions, respectively.
Deoxysugars pathway in TXN-A biosynthesis
Glycosylation modifications of natural products are usually important diversification steps leading to the corresponding ultimately bioactive compounds.26 TXN-A contains two deoxysugar moieties, including a rare γ-branched octose with a two-carbon side chain attached at C-4′′ position. A total of thirteen genes (txnB1–txnB12 and txnM1) in the txn gene cluster encoding enzymes are consistent with the biosynthesis of two sugar moieties and subsequently attachment to the aglycon (Fig. 3C and B). A thymine diphosphate (dTDP)-glucose synthetase (TxnB2), a dTDP-glucose 4,6-dehydratase (TxnB1) and a dNDP-hexose 2,3-dehydratase (TxnB7) catalyze the generation of 16 from glucose-1-phosphate, which possibly served as the branch point for the biosynthesis of deoxysugar donors 19 and 22 (Fig. 3C). Sequentially acted on by a 4-ketoreductase (TxnB5), a dTDP-hexose 3-C-MT (TxnM1), and an O-acyltransferase (TxnB11), the intermediate 16 could be converted into 22, a deoxysugar donor for the formation of TXN-A (Fig. 3C). On the other hand, a 3-ketoreductase (TxnB8) and a 3,5-epimerase (TxnB6) would perform the generation of 18 from 16, which could be further attached with a two-carbon side chain derived from pyruvate catalyzed by a two-component pyruvate dehydrogenase like enzyme (TxnB3/B4) to yield another deoxysugar donor 19 (Fig. 3C). A similar process was also proposed for the same deoxysugar moiety in the biosyntheses of kosinostatin,14 yersiniose A,27 and avilamycin A.28 Finally, two deoxysuger donors 19 and 22 would be installed onto the TXN scaffold catalyzed by glycosyl transferases (TxnB9, TxnB10 or TxnB12) to afford the final product TXN-A.
To obtain further insight into the deoxysugars pathway, especially the usual γ-branched octose, we inactivated the txnB4 gene, resulting in the mutant strain S. bottropensis TG5016 (ΔtxnB4). This mutation completely abolished TXN-A production, but yielded two new compounds (Fig. 4B-VIV). After fermentation and purification, we isolated 10 mg of 12a and 3 mg of 12b from a 1 L culture, and the structures are shown in Fig. 5 (Fig. S22–S27 and Tables S7, S11†). Compared with TXN-A, the major compound 12a has lost the γ-branched octose moiety at 13-OH, which means that the respective glycosyl transferase bears a relatively strict substrate specificity toward the two-carbon side chain. The production of the minor compound 12b revealed that a sugar C-MT, most likely TxnM1, catalyzes a methylation reaction to form a new sugar donor 23, which partially completed 19 to generate 12b, though it is not the perfect sugar donor for the glycosyl transferase comparable to the native 19 (Fig. 3C and B).
Bioactivity of TXN analogues and primary structure–activity relationship
With seven TXN-A analogues in hand, we subsequently performed in vitro cytotoxicity assays of these compounds using cultured Jurkat cells. As a positive control, TXN-A shows high activity, with an IC50 value of 0.78 ± 0.08 nM; and the IC50 values of these analogues were also measured and are listed below the respective structure in Fig. 5. The most potent compound 14a, exhibits excellent activity, having an IC50 value of 0.85 ± 0.05 nM, which is comparable to that of TXN-A. Another promising compound 15a (IC50 = 5.20 ± 0.65 nM), which was also chemically synthesized by Myers's group,9 suggested that the 2-OH group is changeable for further drug development. In addition, the cytotoxicity of TXN-A is higher than that of 12a or 13a, and that 15a is more active than 15b revealed that either of two deoxysugar moieties is important for the anti-cancer activity. Another interesting conclusion could be drawn; that the two-carbon side chain of the γ-branched octose is important for the biological activity of TXN-A, because 12b is more than 10-fold less potent. Furthermore, the keto group at C-7′′ of this octose side chain could also contribute to the anti-cancer activity, which was supported by the observation of the reduced potency of 13b compared to 13a.
Conclusions
Currently, the bacterial aromatic polyketides generated by type II PKSs have been well studied.13,15,29,30 However, the unusual structure of TXN-A distinguishes it from others and indicated that a unique biosynthetic machinery, including a series of sophisticated modifications should be involved in the pathway. Our feeding experiments and genetic characterization of the txn gene cluster have now revealed a novel precursor pathway for type II PKS. In addition, the TXN biosynthesis system employs extremely complex tailoring modifications, which suggested a vast array of enzymatic reactions to be explored. These findings have expanded our understanding of type II PKSs and set the stage for further combinatorial biosynthesis to yield more analogues towards drug discovery.
Supplementary Material
Acknowledgments
We are grateful for the supporting grants from the 973 (2013CB836900), NSFC (81202442 & 81373307), and STCSM (14XD1404500) Programs.
Footnotes
†Electronic supplementary information (ESI) available: The experimental procedures, strains, plasmids and PCR primers, and compound characterization. See DOI: 10.1039/c5sc00116a
References
- Tomita F., Tamaoki T., Morimoto M., Fujimoto K. J. Antibiot. 1981;34:1519–1524. doi: 10.7164/antibiotics.34.1519. [DOI] [PubMed] [Google Scholar]
- Tamaoki T., Shirahata K., Iida T., Tomita F. J. Antibiot. 1981;34:1525–1530. doi: 10.7164/antibiotics.34.1525. [DOI] [PubMed] [Google Scholar]
- Fujimoto K., Morimoto M. J. Antibiot. 1983;36:1216–1221. doi: 10.7164/antibiotics.36.1216. [DOI] [PubMed] [Google Scholar]
- Maskey R. P., Sevvana M., Usón I., Helmke E., Laatsch H. Angew. Chem., Int. Ed. 2004;43:1281–1283. doi: 10.1002/anie.200352312. [DOI] [PubMed] [Google Scholar]
- Maskey R. P., Helmke E., Kayser O., Fiebig F. H., Maier A., Busche A., Laatsch H. J. Antibiot. 2004;57:771–779. doi: 10.7164/antibiotics.57.771. [DOI] [PubMed] [Google Scholar]
- Fitzner A., Frauendrof H., Laatsch H., Diederichsen U. Anal. Bioanal. Chem. 2008;390:1139–1147. doi: 10.1007/s00216-007-1737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoh R., Laatsch H., Sheldrick G. M. Nucleic Acids Res. 2008;36:3508–3514. doi: 10.1093/nar/gkn245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Švenda J., Hill N., Myers A. G. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6709–6714. doi: 10.1073/pnas.1015257108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magauer T., Smaltz D. J., Myers A. G. Nat. Chem. 2013;5:886–893. doi: 10.1038/nchem.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.-H., Zhang M., Pan H.-X., Chen X.-D., Tang G.-L. Chin. J. Org. Chem. 2014;34:1376–1381. [Google Scholar]
- Thomas R. A. ChemBioChem. 2001;2:612–627. doi: 10.1002/1439-7633(20010903)2:9<612::AID-CBIC612>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Ōmura S. Chem. Rev. 1997;97:2591–2609. doi: 10.1021/cr960023p. [DOI] [PubMed] [Google Scholar]
- Hertweck C., Luzhetskyy A., Rebets Y., Bechthold A. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- Ma H.-M., Zhou Q., Tang Y.-M., Zhang Z., Chen Y.-S., He H.-Y., Pan H.-X., Tang M.-C., Gao J.-F., Zhao S.-Y., Igarashi Y., Tang G.-L. Chem. Biol. 2013;20:796–805. doi: 10.1016/j.chembiol.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Das A., Khosla C. Acc. Chem. Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Tsai S.-C., Khosla C. J. Am. Chem. Soc. 2003;125:12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay A. T., Maltby D. A., Medzihradszky K. F., Khosla C., Stroud R. M. Nat. Struct. Mol. Biol. 2004;11:888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- Lackner G., Schenk A., Xu Z., Reinhardt K., Yunt Z. S., Piel J., Hertweck C. J. Am. Chem. Soc. 2007;129:9306–9312. doi: 10.1021/ja0718624. [DOI] [PubMed] [Google Scholar]
- Zhou H., Li Y., Tang Y. Nat. Prod. Rep. 2010;27:839–868. doi: 10.1039/b911518h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yoon P., Yu T. W., Floss H. G., Hopwood D. A., Moore B. S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3622–3627. doi: 10.1073/pnas.96.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. S., Hertweck C. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- Fu H., Ebert-Khosla S., Hopwood D. A., Khosla C. J. Am. Chem. Soc. 1994;116:4166–4170. [Google Scholar]
- Lee T. S., Khosla C., Tang Y. J. Am. Chem. Soc. 2005;127:12254–12262. doi: 10.1021/ja051429z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust L. M., Sherman D. H. Nat. Prod. Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskey R. P., Helmke E., Fiebig H.-H., Laatsch H. J. Antibiot. 2002;55:1031–1035. doi: 10.7164/antibiotics.55.1031. [DOI] [PubMed] [Google Scholar]
- Thibodeaux C. J., Melancon III C. E., Liu H.-W. Angew. Chem., Int. Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Guo Z., Liu H.-W. J. Am. Chem. Soc. 1998;120:11796–11797. [Google Scholar]
- Treede I., Hauser G., Mühlenweg A., Hofmann C., Schmidt M., Weitnauer G., Glaser S., Bechthold A. Appl. Environ. Microbiol. 2005;71:400–406. doi: 10.1128/AEM.71.1.400-406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsä-Ketelä M., Niemi J., Mäntsälä P., Schneider G. Top. Curr. Chem. 2008;282:101–140. [Google Scholar]
- Kharel M. K., Pahari P., Shepherd M. D., Tibrewal N., Eric Nybo S., Shaaban K. A., Rohr J. Nat. Prod. Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.