Abstract
The T cell repertoire is a function of thymic V(D)J rearrangement and of peripheral selection. The mature repertoire embodies TCR sequences that are important for survival and can identify important structural aspects of the TCR. Analysis of the circulating BV19 CD8 T cell TCR repertoire showed that a majority of NDN encoded CDR3 amino acid motifs start at CDR3 position 4, well within the V region. Rearrangement at this position indicates that the DNA hairpin loop is not opened at the position adjacent to the recombination signal sequence, but rather trimmed back three or more bases. Here we show that the rearrangement frequency distribution within the V region reveals selection on CDR3 position 4. The selection is already established in SP CD8 thymocytes. Crystal structures reveal a possible basis for this selection due to the location of this residue in a bend which positions the remaining portion of the CDR3 to interact with the peptide and MHC (pMHC). Examination of other BV families also shows selection for rearrangement within the V region of a number of genes and for both CD8 and CD4 cells. The exact profile of rearrangement within the V region appears to be V gene-specific. Frequent observation of side chains associated with turn motifs at CDR3 positions 3 and 4 fits with the structural need for a bend. The data are discussed in terms of the generation of a structural turn motif, the rearrangement mechanism, and selection of the repertoire on pMHC.
INTRODUCTION
The antigen-specific receptors of the adaptive immune system are generated by a rearrangement mechanism in which the RAG genes mediate cleavage and ligation of the DNA. The resulting DNA hairpin loop is cleaved by the protein Artemis. The DNA ends thus generated can be extended by deoxynucleotide terminal transferase and the two ends trimmed and resealed during classical non-homologous end joining. For TCRBV (hereafter referred to as BV) and IGH genes, the first stage in the process of generating a receptor is the rearrangement of a Diversity sequence element next to the Joining region (D to J) followed by rearrangement of the Variable segment to the DNJ. This results in a region of the receptor that is encoded by V gene sequences followed by NDN sequences and finally J region sequences. Asymmetric hairpin cleavage can also give rise to palindromic sequences referred as P nucleotides (see Reference 1 for recent review of the rearrangement process).
The region between the conserved cysteine in the V region and the conserved phenylalanine-glycine doublet in the J region is referred to as the third complementarity determining region (CDR3) which is involved in the recognition of peptide:MHC (pMHC) complexes. The CDR3 has a component that is V region derived, one that is composed of N and/or P nucleotides, a possible D-region derived component, another N/P component, and finally a J region-derived component. We will be referring both to the amino acid numbering of the CDR3 (CDR3 a.a.) as well as the base pair (CDR3 b.p.) numbering, the former when referring to the protein structure, and the latter when referring to the actual rearrangement position.
Generation of a diverse repertoire of TCR is the first stage of generating the memory repertoire that will be responsible for maintaining the response to recurring pathogen exposures and thus the health of an individual. By middle age such memory repertoires are well established. We have analyzed the mature repertoire of CD8 T cells expressing the BV19 gene by high throughput sequencing (2, 3) and observed that the predominant site for rearrangement (i.e. initiation of the NDN region) is at CDR3 a.a. position 4. This is somewhat surprising because in current models of V(D)J joining the Artemis-mediated resolution of the DNA hairpin loop generated by RAG nuclease (4) occurs most frequently at the original site of ligation. This would yeild a symmetric cleavage regenerating the same DNA ends. Less frequently the cleavage would be asymmetric by a base or two on either side. Either case would lead to most NDN regions starting at CDR3 a.a. position 5, not position 4. Here we examine the exact frequency of rearrangement across this portion of the CDR3 for the published BV19 data and compare it with CDR3 sequences from CD8SP thymocytes, a number of BV rearrangements from CD4 T cells, IgH, and also with other TCR β-chain CDR3 sequences available at the NCBI site. We interpret our results in terms of TCRBV structures and of our current understanding of the rearrangement mechanism. We propose that rearrangement within the V gene is involved in generating a turn in the CDR3 backbone that allows the remainder of the CDR3 to contact pMHC.
METHODS
T cell isolation, cDNA synthesis, amplification of TCR, and 454 sequence analysis are described in more detail elsewhere (2, 3) including error estimation, and steps taken in cleaning the data. In brief, PBMC were collected from buffy coats from normal whole blood donations and either CD8 or CD4 cells isolated. V-gene specific PCR amplification was performed using our standard “spectratyping” BV and BC primers (5). Amplicons were analyzed on a Roche GS-FLX Genome Sequencer at the Human and Molecular Genomic Center Sequencing Facility (www.hmgc.mcw.edu) of the Medical College of Wisconsin. Sequences derived from each sample were downloaded in fasta format and analyzed using our proprietary “CDR3Reader” software, which assigns clonotype names according to the naming convention described by Yassai et al (6). Clonotype is used to refer to the unique CDR3 nucleotide sequence of the TCR β-chain gene. Data from CDR3 Reader were analyzed using Microsoft Excel.
DNA prepared from CD8 SP thymocytes isolated from thymic surgical discards as part of a previous study (7) was used to generate sequences of BV19 - BJ2.7 amplicons on the same 454 platform and analyzed in a similar manner.
Sequence data from CD4 cells isolated from PBMC isolated from three middle-aged subjects (UPN11, UPN 50 and UPN902) were also analyzed using the same platform. The data are for BV families from which some clonotypes are involved in HLA-DR1 restricted recall responses to an influenza M1-derived peptide.
Two sources of IgH sequences were used. IgH sequences from naïve B cells PBMC were provided by Dr Dunn-Walters (8). Rearrangement sites were determined using V-Quest software at the IMGT website (9). A set of IgH sequences from naïve B cells (10) was obtained from Dr Patrick Wilson, U of Chicago and rearrangement sites were also identified using V-Quest.
TCR sequences were also obtained from data published by Wang and colleagues (11) and available on the NIH Sequence Read Archive (SRA at https://www.ncbi.nlm.nih.gov/sra) website (ref 12, PRJNA79519). We also used data from another CD4 dataset available on the SRA site (13, SRP011414). Both these studies used the 454 sequencing platform. These downloaded sequences were also analyzed using CDR3 Reader and Excel.
TCR structures were obtained from data deposited in the PDB. The PDB files used for the analysis are included in Table 1. PyMol software (14) was used to select atoms within 5Ǻ of any atoms in the amino acid at position 4. This distance was chosen to be sure that possible contacts could not be missed if the actual molecules showed more flexibility than the crystallized molecules. The atoms falling within this distance were identified and are given in Table 1. The superimpose command followed by some manual manipulation was used to overlap the TCR β-chain structures for Figure 3.
RESULTS
We use a genetic definition of the CDR3 as the amino acids starting immediately after the conserved cysteine in the V region and extending to the amino acid immediately before the conserved phenylalanine-glycine in the J region. After rearrangement, the CDR3 has three genetic components, the 3’ end of V region, the D region, and the 5’ end of the J region. Flanking the D region are untemplated nucleotides added by terminal deoxynucleotidyl transferase (TdT), referred to as N-nucleotides, and by asymmetric resolution of the DNA hairpin loop resulting in palindromic insertions after a filling-in reaction (P nucleotides). For any V - J pair, the amino acids encoded by the V and J are fixed, with the diversity being a function of the sequence and position of the NDN component. Our use of the NDN term includes possible P nucleotide contribution. Thus, the portion of the CDR3 derived from the NDN generates the most diversity in the TCR. While the rearrangement site is identified at the nucleotide level, in terms of amino acid motif analysis the NDN start site is defined by the CDR3 amino acid position at which it occurs. An amino acid is considered to be NDN encoded even if the rearrangement is at the third codon position and results in a synonymous substitution.
Rearrangement in BV19 from CD8 cells
BV19 encodes four amino acids after the cysteine and the first two bases of a fifth, the latter resulting in either Asp or Glu (CASSID/E). Thus, the NDN can formally start from CDR3 amino acid position 1 to 5. As described (2), none of the 12,690 clonotypes had an NDN start site at CDR3 position 1, and slightly more than half (51%) of NDN regions start at CDR3 position 4.
Importance of CDR3 position amino acid 4 based on selection
We examined the NDN start (insertion) site at the nucleotide level to determine exactly where the rearrangement took place. The analysis was performed as a function of CDR3 lengths. When the number of clonotypes with NDN start sites at each possible CDR3 nucleotide position is plotted (Fig 1A) the results are striking in that the middle base of amino acid position 4 (bp11), there is a much lower frequency of starts than might be expected based on the trends at the surrounding two bp positions. This observation is independent of the CDR3 length. Such a decrease is an indicator that not all amino acids are permitted at this position.
Figure 1.
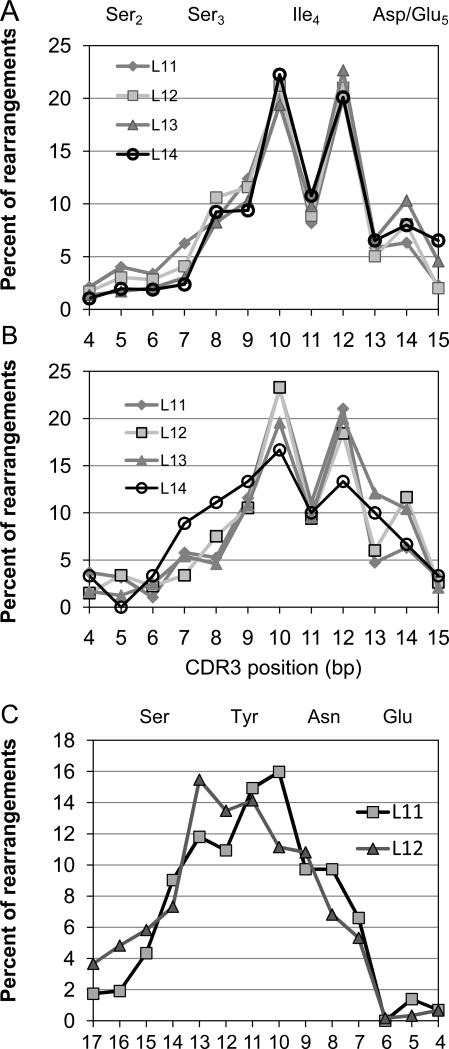
Rearrangement site frequencies along the BV19 gene at the nucleotide level. A. The percentage of NDN start sites at CDR3 nucleotide positions 4 to 15 for the circulating repertoire of UPN204 is shown. The amino acid encoded by each triplet is shown above the graph. Data is shown on the basis of the CDR3 length in amino acids as identified in the inset. B. rearrangement site data for CD8 single positive thymocytes from an HLA-A2 subject. C. J2S7 rearrangements are shown for the circulating repertoire of the same dataset as analyzed in panel A. Numbering of b.p. positions is relative to the start of the conserved FG motif. The amino acids encoded by the triplets are shown above the graph.
Because the PBMC are from an older subject the observation could be the result of the peripheral selection involved in generating mature memory. Alternatively, this result may represent the importance of CDR3 position four in generating the proper TCR structure to allow thymic selection and thus be already visible in mature thymocytes. Therefore, we undertook a similar analysis of BV19 sequence data from CD8 SP thymocytes from an HLA-A2 subject. These data (Fig 1B) showed a similar drop in number of clonotypes with rearrangements at CDR3 bp 11.
Rearrangements at the D – J joining site
At the other end of the NDN region, the D to J rearrangement distribution (NDN end site) was analyzed for a number of J regions and CDR3 lengths. The J2s7 data is shown (Fig 1C) and approaches a normal distribution as might be expected of a rearrangement process followed by equally distributed selection. J2.7 is the most commonly used J region in the BV19 dataset and provides for a robust analysis. Data for other J regions with sufficient sample numbers for the analysis were similar. It should be pointed out that not only is the distribution of rearrangement sites relatively smooth, the circulating clonotypes analyzed are predominantly the result of rearrangement within the J region.
Selection along the CDR3 of BV19
The decreased frequency of CDR3 with rearrangement at the middle base of the codon encoding position 4 could be due to selection post-rearrangement. If so, analysis of the amino acids observed at this position in comparison with neighboring positions should be informative. We therefore examined the first NDN-encoded amino acid at each CDR3 nucleotide start position. N addition due to the action of TdT is considered relatively random although the enzyme has a tendency to prefer adding G (15). Ligation of D-encoded nucleotides would add a G or whichever base is present at a nuclease-generated 5’- end of the D-region; be it exonuclease activity or endonuclease cleavage of a D-region hairpin. The effect of the base addition on the amino acid encoded will be a function of the position of rearrangement.
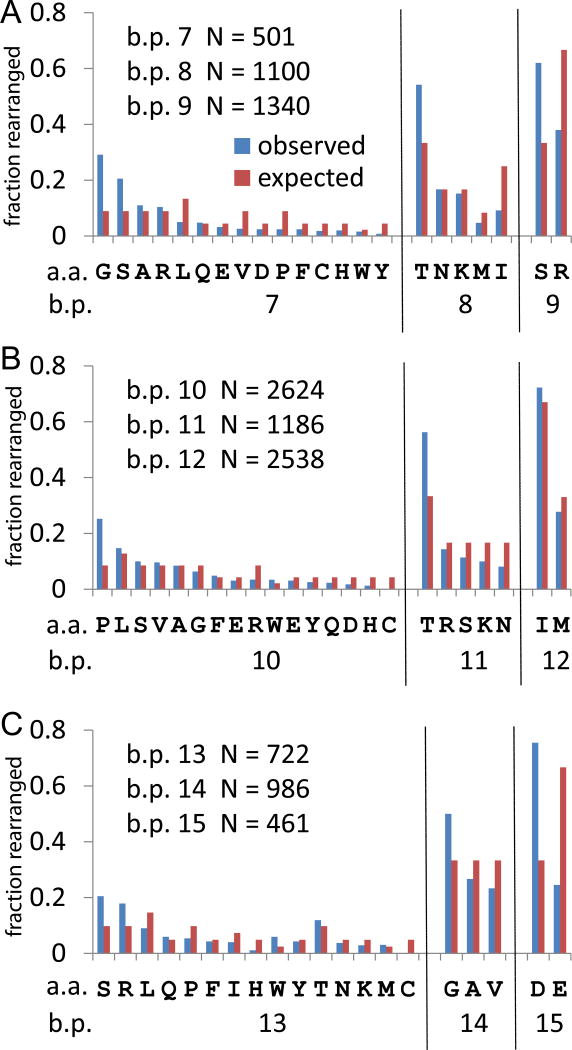
Panel A in Figure 2 shows the various rearrangements that can give rise to NDN amino acids at CDR3 a.a. position 3. If left intact, the germline sequence encodes Ser. This amino acid position corresponds to CDR3 b.p. positions 7, 8, and 9, identified at the bottom of the panel. For each CDR3 b.p. position the potential amino acids encoded are listed using the one letter code. The number of observations of each amino acid is plotted as the fraction of clonotypes which rearranged at that b.p. position. The number of clonotypes rearranged at each position is given in the upper left of the panel. The expected fraction based on number of codons encoding the a.a. is plotted as a reference. Rearrangement at bp position 7 can encode any one of fifteen amino acids. Two of these, Gly and Ser, are observed at higher than expected levels, with observed to expected ratios of 3.3 and 2.3 respectively. While the Gly codon usage shows some fluctuation in codon use, almost half (49%) of the Ser is encoded by the TCC codon. A significant codon skew is defined by an observed/expected ratio >1.5, or < 0.5. A skew in codon usage is a likely indicator of the action of a molecular mechanism associated with rearrangement. At b.p. position 8, Thr is overrepresented 1.6 fold, with 70% of the Thr encoded by the ACC codon. This is almost a threefold skew in codon usage. The other four amino acids encoded at this position show a decreasing observed to expected ratio, with Met and Ile being underrepresented. Rearrangement at CDR3 b.p. position 9 can either regenerate Ser or encode Arg. It appears that Arg is selected against as it should represent 67% of rearrangements but is observed only 38% of time.
Figure 2.
Amino acid utilization at the rearrangement site. A. Data for rearrangements at CDR3 amino acid position 3. Rearrangements can occur at CDR3 b.p. position 7, 8, 9. The number of clonotypes observed rearranging at each CDR3 b.p. position is given in the upper left of the panel. The observed (blue) and expected (red) frequencies are shown for each amino acid. The x-axis identifies the possible encodings for rearrangements at the three CDR3 b.p. positions. B. Rearrangements at CDR3 amino acid position 4. C. Rearrangements at CDR3 amino acid position 5.
Rearrangements that would affect the amino acid at CDR3 a.a. position 4 occur at CDR3 b.p. positions 10 to 12 (Figure 2B), with the number of rearrangements at position 11 being lower than at either 10 or 12 (data in upper left of panel). Rearrangement at CDR3 b.p. position 10 can encode 16 possible amino acids. Proline represents 26% of the observations which is a 2.93 fold excess over the expected frequency. The CCC codon is observed 48% of the time which represents almost a two fold increase over the expected frequency. Of the remaining 14 amino acids, seven are close to the expected frequency with W also observed at higher frequency and R, D, H, and C at lower than expected frequency. Rearrangement at bp position 11 can result in either one of 5 possible amino acids (T, R, S, K, N), with T occurring at higher than expected frequency (56% in lieu of the expected 33%). The codon utilization is skewed with the ACC codon represented 46% of the observed rearrangements. This is the same skew as observed for rearrangements at CDR3 b.p. position 8. If the contribution of the ACC codon skew is not considered, the observed to expected frequencies for T,R, S, and K are close to expected, with N (0.66) representing the low outlier. Thus, in spite of there being fewer clonotypes with rearrangement at this position than at either of the two flanking positions, there is no evidence of selection against a particular amino acid. Rearrangement at the 3rd codon position (b.p. 12) can yield Ile and Met, with two of three possibilities leading to Ile and one of three to Met. The frequency of these two amino acids was that expected by chance. Comparing position 11, at which there is a decreased frequency of rearrangements, with positions 10 or 12 does not show evidence of a major skew in amino acid usage compatible with focused selection.
All amino acids observed at CDR3 a.a. position 5 are NDN encoded as the germ-line sequence can only contribute the first two bases of the codon. Rearrangements at CDR3 bp position 13 can encode 15 different amino acids (Figure 2C). Ser is the most frequently observed amino acid at this position and the codon usage is relatively equally distributed. Arg is also observed more frequently than expected. However 43% of the observations represent the AGG codon which represents ~2.6 fold over the expected value of 17%. This observation could be explained by use of the D1 region starting at the second amino acid of reading frame 3 (DRG). Of the 55 observations of the AGG codon, 3 were associated with the full RG dimer (AGG GGG) expected with D1 region use, 12 with the partial RD dimer (AGG GGx) and an additional 12 with shorter D1 use (AGG G associated with RV, RA, RD, or RE dimer motifs). Interestingly tryptophan was observed at frequency of ~6%, but this represented over two-fold higher than expected. His and Cys represented the two amino acids that occurred at much lower than expected frequencies. Rearrangements at CDR3 position 14 can encode Gly, Ala or Val with equal probability. Gly is observed at slightly higher frequency than either Ala or Val. There is a general skewing toward Gly at most NDN positions (2). There was also a slight skew in the use of the GGG codon. There was no skew in the Val or Ala encoding. Examining addition NDN additions at bp15, which corresponds to a symmetrical cleavage of the hairpin loop at the 3’ end of the V region, should result in equivalent expected frequency of either D or E. For the 25% of rearrangements encoding E, 48% were encoded by GAA and 52% by GAG, indicating a relatively random process for generation of E. However, 73% of the D was encoded as GAT in lieu of the expected 50% (obs/exp ≈ 1.5). This is compatible with a P nucleotide addition. The examination of the codon usage for rearrangements that occur at CDR3 bp 13 to 15, representing the codon for CDR3 amino acid position 5, show evidence for random addition as well a more specific mechanisms such as skewed D region use or P nucleotide addition.
Comparison of rearrangements at CDR3 amino acid position 4 do not show any major differences from those observed at the two flanking positions, as might be expected if there were some side chains that were strongly selected against. The skewing observed could often be explained by selection of a particular codon as opposed to equal selection of all possible codons. The former observation implicates a more subtle reason for selection against rearrangements at b.p. position 11. The latter observations suggest a connection of codon skew to a particular aspect of the rearrangement mechanism.
Structural importance of CDR3 position 4
The high frequency of NDN start site at CDR3 position 4 may be due to the role of the amino acid at this position in TCR recognition of pMHC. Therefore we analyzed available crystal structures of TCR β-chains that are part of TCR-peptide-HLA-A2 structures. A number of these are available including the structure of BV19 complexed with M158–66 and HLA-A2 (16). We examined the BV19 structure as well as those of a four TRBV6-5 structures (17–20) and identified the contacts made by the CDR3 position 4 residue (Table 1). The possible contacts are defined for the various CDR of the TCR beta and alpha chains, the peptide, and the 1st domain of HLA-A2. A 5Å distance was chosen to compensate for possible small movements of the backbone. This distance insures that observed lack of contacts is valid under more stringent conditions. For all the different structures the residue at position 4 predominantly contacted other beta CDR3 residues as well a region of the beta CDR1. For the TRBV6-5 chains, two chains of different CDR3 length had the germ-line-encoded Tyr at position 4, and both also showed possible contact with the peptide. Of these two, the 2P5R TCR (L12) also showed a possible contact with the alpha chain CDR3.
Table 1.
Contacts between CDR3 position 4 and other amino acids in the TCR, peptide or MHC.
| TCR ID | M1 | A6 | A6 | B7 | 2P5E | RA14 |
|---|---|---|---|---|---|---|
| BV (reference) | BV19 (16) | BV6 - 5 (17) | BV6 - 5 (18) | BV6 - 5 (17) | BV6 - 5 (19) | BV6 - 5 (20) |
| PDB File | 1OGA | 1QRN | 3H9S | 1BD2 | 2P5E | 3GSN |
| vNDN CDR3L | assIRS L11 | asRPGLAGGRP L14 | asRPGLAGGRP L14 | assyPGGF L13 | assyLGNTGE L12 | assPVTGGIYG L13 |
| peptide source | GILGFVFTL M1 | LLFGYPVYV Tax | MLWGYLQYV Tel | LLFGYPVYV Tax | SLLMWITQC NY-ESO-1 | NLVPMVATV HCMV |
| CDR3 Position 4 | I96 | P96 | P96 | Y96 | Y96 | P95 |
| 5Ǻ Contacts | a.a. seq start | a.a. seq start | a.a. seq start | a.a. seq start | a.a. seq start | a.a. seq start |
| TCRB CDR3 | SS-RS-YE 94 | SR-GLAG-RPE 94 | SR-GLAGGRPE 94 | SS-PGGGFY 94 | SS-LGN-GE 91 | SS-VT---YG 93 |
| TCRB CDR1 | MHNEY 27 | NHEY 28 | HEY 29 | NHEY 28 | MHNEY 25 | NHEY 28 |
| TCRB CDR2 | none | none | none | none | none | none |
| TCRA CDR3 | none | none | none | none | R 93 | none |
| TCRA CDR1 | none | none | none | none | none | none |
| TCRA CDR2 | none | none | none | none | none | none |
| peptide | none | none | none | Y 8 | W - - Q 5 | none |
| HLA-A2 domain 1 | none | none | none | none | none | none |
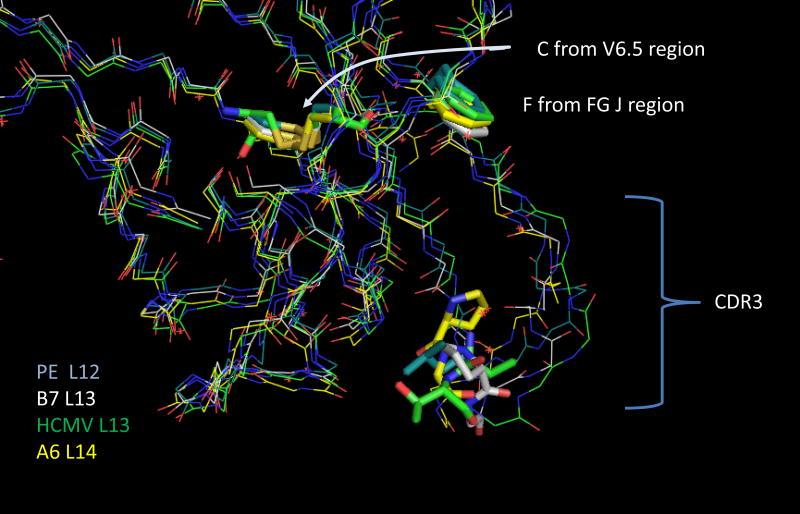
The structural basis of the importance of this position is most clearly observed by examining an alignment of the four TCR TRBV6-5 exemplars (Fig 3). The position of the conserved V-region cysteine, the conserved J-region phenylalanine-glycine, and CDR3 position 4 are shown as tubes. It is clear that the amino acid at CDR3 position 4 is involved in making a sharp bend in the structure, which allows the downstream CDR3 positions to contact the pMHC. This role helps explain the increased frequency of proline at this position as noted in Figure 2 and previously (2). Indeed two of the structures in Fig 3 have proline at this position.
Figure 3.
Alignment of four BV6 CDR3 structures. The structures are identified on the lower left by the color of the carbon atoms. The CDR3 length is also shown. The β-chain names are also in Table 1 with the publication reference and PDB ID. Tube structures identify CDR3 position 4, the conserved C’ terminal Cys of the BV6.5 chain and the conserved PheGly in the J-region. The crystal structure data were manipulated in PyMol (9).
We also examined three cases in which the structures of TCR-pMHC complexes were compared with structures solved of the TCR alone to determine if the turn might be induced during the pMHC recognition event or whether it was integral to the TCR structure in itself. The turn associated with position 4 was maintained in the unliganded TCR (Supplemental Figure 1). There were some differences in the backbone downstream of position 4.
Rearrangement site distributions show a V gene-dependence for both CD8 and CD4 T cells
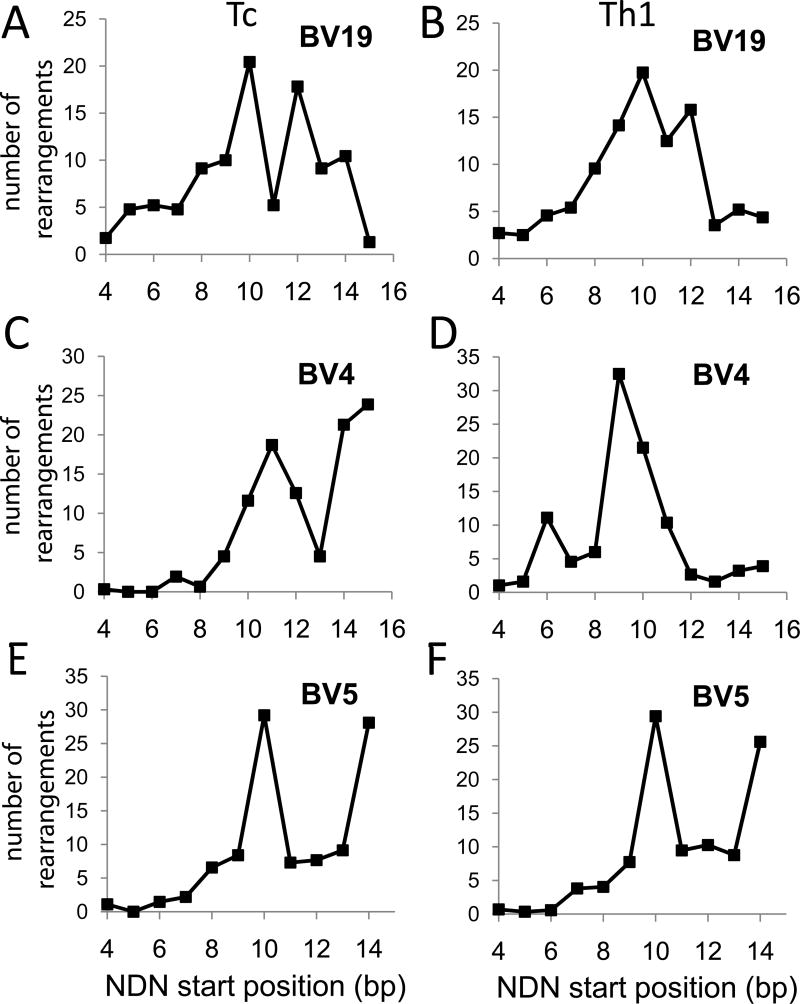
It is important to determine if the rearrangement distribution observed for BV19 in CD8 cells was specific or a general phenomenon for all TCR. The similarities in the BV18 and BV6 structures already hinted that this may be the case. We therefore examined additional high throughput sequence data available at the NIH Sequence Read Archive (SRA) web site. The data in Figure 4 are from SRX013279 and represent published data (11). We analyzed the BV19 data for CD8 T cells (Tc) and observed the decrease of rearrangements at bp position 11 similar to that seen in our data (Fig 4A). Data from the same analysis of cytotoxic T cells shows that BV4 has a mixed distribution with increased frequency at position 11 and also at the end of the V gene (Fig 4C, bp position 15). BV5 also showed a biphasic distribution (Fig 4E). Data from Th1 cells (CD4) was similar to CD8 cells for BV19 and BV5 (Fig 4B and F), but the BV4 data (Figure 4D) were missing the increase at CDR3 positions 14 and 15.
Figure 4.
Comparison of published CD8 vs CD4 rearrangement frequencies along the V gene. The Y axes count the number of clonotypes rearranged (NDN insertions) at each CDR3 b.p. position (number of rearrangements; n. o. r.). The data are from Wang et al (11). Panels A, C, E show data from Tc (CD8 cells). Panels B, D, F show data from Th1 (CD4) cells. The BV analyzed is identified above each panel.
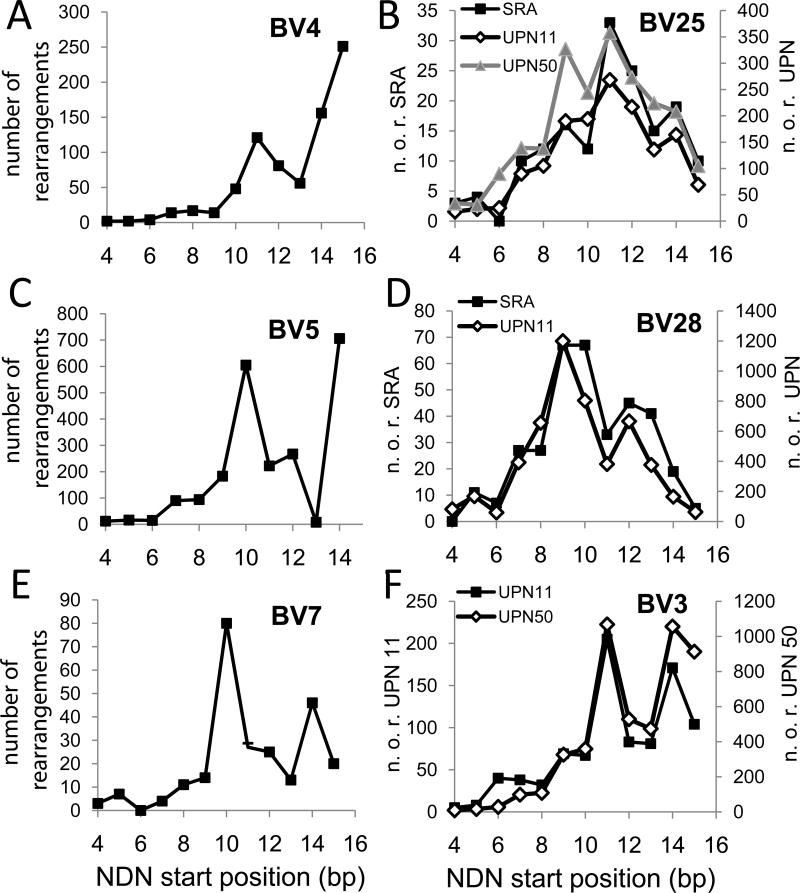
Additional CD4 data was obtained from the NIH SRA site and from our own studies (Figure 5). Data identified as SRA are from ref 13 and the data from our unpublished studies represented BV families involved in HLA-DR1-restricted responses to influenza virus epitopes. The BV4 data (Figure 5A) showed a different distribution from either the CD4 or CD8 data shown in Figure 4. The BV5 data (Fig 5C) was similar to that shown in Figure 4 with highest frequency of rearrangement at bp 10 and 14, except that rearrangement at position 13 was decreased and that at 14 is increased. BV7 (Fig 4E) shows a different pattern in that rearrangement is maximal at bp 10, but the frequency at bp 14 is higher than that at 15. The distribution of BV25 rearrangements is from three different individuals, with similar patterns for all three (Fig 5B). The distribution is relatively symmetric around a maximum at bp11 with an increase at bp9. The BV28 data (Fig 5D) is similar in the two subjects analyzed, with a generally symmetric distribution characterized by a decrease at bp11. The BV3 pattern was also reproducible between subjects (Fig 5F), and resembled BV7 in that there was a spike in rearrangements at bp14. Overall the TCR rearrangements profiles showed reproducible, V-gene specific patterns that varied between those that a high frequency towards the end of the V gene to those that were symmetrical around bp 10 or 11 within the V gene. Rearrangements involving bp 10 – 12 map to CDR3 amino acid position 4.
Figure 5.
Rearrangement frequencies along the BV gene in CD4 cells for six BV families. BV gene families are identified in each panel. The Y-axes count the number of clonotypes rearranged at each position. Data taken from SRP011414 are shown as filled squares in panels A to E. Data from our own studies are squares, triangles or diamonds and identified in the panel. In the right panels, the axes describing number of rearrangements varied by data sources (SRA or UPN) and these are identified as part of the axis title.
Rearrangement within the V region reveals increase use of amino acids associated with promoting tight turns
In light of the different rearrangement site profiles for the BV3 and BV28 genes with respect to BV19, we analyzed the NDN amino acid use for these two genes. Comparing and contrasting the data from three families is useful. The overall a.a. use was similar for the three V genes, with glycine being the most frequent, followed by S, T, R, L, P and A, whose exact ranking differed by V gene. The NDN amino acid usage for both V genes is available as Supplemental Figure 2.
To compare the amino acid at the rearrangement position in the context of a longer NDN sequence, we investigated the NDN amino acid usage from clonotypes that had at least three amino acids in the NDN region, i.e. minimum triplet motif. In Figure 6 we show the amino acid frequency along the CDR3, based on the base pair position of the rearrangement. The relative frequency of the amino acid is plotted as the ratio of the number of clonotypes with that amino acid at that position relative to all the clonotypes encoding an amino acid at that position. This data is broken down on the basis of nine possible rearrangement positions (CDR3 bp 7 to bp 15). These correspond to CDR3 a.a. positions three to five. Starting at CDR3 a.a. position 3, the C-terminal V region nucleotide sequence for BV3 is AGC CAA GA, and that for BV28 is AGT TTA TG. These would encode Ser, Gln and either Asp or Glu for BV3, and Ser, Leu and either Trp or Cys for BV28.
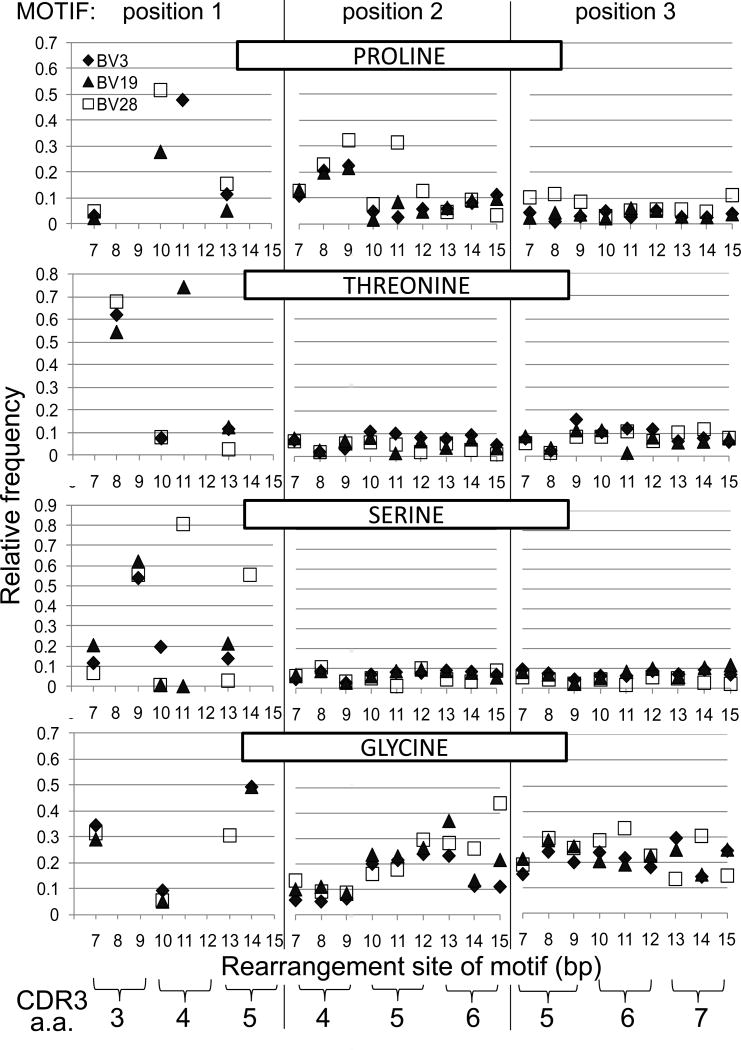
Figure 6.
Select amino acid frequencies in triplet NDN motifs based on rearrangement site for BV3 (CD4 - black diamond), BV19 (CD8 - triangle), and BV28 (CD4 - open square). Data is shown for four informative amino acids, identified across the top of each series of three panels. Each series of three panels shows the frequency of that amino acid on the basis of the b.p. of the rearrangement site. Amino acid use at position 1 of the triplet motif is dictated by the V gene sequence at which rearrangements occur. The second and third motif positions will be based on N, P or D additions. The inset in the top left identifies the V gene symbol used. The correspondence between CDR3 amino acid position and rearrangement site in b.p. is shown at the bottom. e.g. Pro at the second position of the triplet motif that rearranged at b.p. position 9, corresponds to CDR3 amino acid position 4. See text for further description.
We present data for four of the six most informative amino acids with respect to the stereochemistry of tight turns. The amino acid and its position in the triplet motif is identified in each panel. Position 1 will be the amino acid at the rearrangement site which is constrained by the V region sequence. The following two positions are a function of TTase addition, D-region use, or possible P nucleotide (NDP) and all twenty amino acids are a formal possibility. If a motif starts at a b.p. position encoding a.a. 3, then the next position of the triplet will a.a. 4 and the final a.a.5.
Proline (upper three panels) is associated with turns in the secondary structure (21). At the rearrangement position it can be encoded at b.p. 7, 10 and 13, for the case of BV19 (triangle) and 28 (square). This corresponds to the first codon position of each of the three amino acid positions which has the highest degree of freedom as 15 or 16 different amino acids can be encoded. For BV3 (diamond), proline can be encoded at positions 7 and 13, but also at position 11 where it is one of three possibilities. For all three BV, at position 7 the observed frequency is between 3 to 5% (0.03 to 0.05) which is lower than the expected 9% (Fig 6. top left panel). A striking observation is the increased frequency at position 10, which is 3 to 5 fold higher than expected for BV19 and 28 respectively. For BV3 at position 11, the observed relative frequency (48%) is ~1.4 fold higher than the expected 33%. At position 13, the observed relative frequency is lower than expected for BV19 and slightly higher for BV3 and BV28. When proline is observed at position 2 of triplet motifs (middle panel) and encoded by clonotypes that have rearranged at position 7, 8 or 9, there is another striking finding in that proline is observed more frequently than the 0.06 expected on the basis of random addition. These prolines are in the second position of rearrangements that took place at CDR3 amino acid position 3, i.e. they are at position 4. The frequency increases across these three b.p. positions to the point that for clonotypes that rearranged at position 9, there is a three to five fold increase in the observed proline frequency. Such an increase in proline observation relative to rearrangement position may have mechanistic implications. BV28 shows some sporadic increases in Pro frequency in clonotypes that rearranged at later CDR3 positions, but in general the Pro frequency is close to that expected. For BV3 and BV19, rearrangements at position 15 could result in P nucleotide additions which would skew the next amino acid to proline. The two bases at 13 and 14 are GA and a two b/p. P-nucleotide addition would add TC, resulting in GAT Cxx. The occurrence of GAT Cxx is over twice that of GAC Cxx (27 vs 10), indicating a probable role for this mechanism. Examining the third amino acid in motifs (right panel) where the rearrangement was at bp7, 8 or 9, show a small increase for BV28. These prolines would map to CDR3 amino acid 5. At the remaining positions frequencies are close to those expected.
Threonine and Ser are two amino acids that give rise to beta turns based on H-bonding between the side chain oxygen and the main chain nitrogen of the third residue downstream (S/T turns). Threonine can be encoded by rearrangements at position 8, 10 and 13 for BV3 and BV28, and at positions 8, 11 and 13 for BV19. Rearrangements at position 8 can encode five possible amino acids and the expected frequency of Thr is highest at 33% (0.33 on graph). However, for all three BV the observed frequency is ~two fold higher than expected. For rearrangements at positions 10 and 13, the expected frequency is ~8% (0.08), and the observations are close to expectations. For BV19 rearrangements at position 11, the expected Thr frequency is 33%, whereas the observed frequency is over two fold higher. Thr frequencies at the next two positions in the triplet motif are close to the expected ~6%. Ser can be encoded by rearrangements at positions 7, 9, and 13 in all thre BV genes, and at position 11 for BV19 and BV28 as well as position 14 for BV28. At position 7 the expected frequency is ~9% and only BV19 shows an increase in the observed data. At position 9 the expected frequency is ~33%, and for all three BV the observed frequency is almost two fold higher. It should be pointed out that if rearrangement occurs 3’ of b.p. 9, CDR3 amino acid 3 will be the germline encoded Ser. At position 10, Ser is expected at 14%, 9% and 4% for BV3 to 28 respectively; and is under represented in BV19 and BV28 clonotypes and over represented in BV3. At position 11, Ser is expected ~17% for BV19 and 44% for BV28. It is again under represented in BV19 clonotypes but over represented almost two fold in BV28 clonotypes. At position 13, Ser is expected at a frequency of 17% for BV3 and BV19 and 4% for BV28 and the observations match expectation. At position 14, Ser is expected at 40% for BV28, but is slightly more represented in the measurements (~55%). The occurrence of Ser at the second and third position in the triplet motifs that start at these various positions is generally lower than the 10% expected.
Glycine is a turn enabling residue because its lack of a side chain allows a tighter turn geometry. For this and for perhaps other steric reasons glycine is the most common amino acid in NDN regions. In the data examined it can be encoded when rearrangements take place at positions 7 and 10 for all three BV and at position 13 for BV28 and 14 for BV3 and BV19. The expected frequency at both position 7 and 10 is ~9%, which is observed for position 10 but at position 7 Gly is observed at three fold higher frequency than expected. The same expectation and fold increase is also the case for BV28 at position 13. For BV3 and BV19 rearrangements at position 14, the expectation is ~33% and the observed is ~50%.
Thus, these four amino acids all show at least one position at which they are both frequently observed and this frequency is higher than expected, the latter a sign of selection.
Based on the distal V region sequence, there are other amino acids that can appear at high frequency if the rearrangement site is within the V region (e.g. Arg, Phe, Gln, Ile or Leu). We did not show these in the triplet motif format because either they are observed at expected levels, or if the other encoding possibility includes Gly, Thr, Ser or Pro, they are observed less frequently than expected.
P nucleotide additions and proline frequencies
We have already reported that the increased incidence of proline in BV19 can in part be ascribed to P nucleotide additions from the 5’ end of intact D regions (3). We therefore examined the triplet motif data to determine if this is the case for the BV3 and BV28 data where the incidence of proline is observed at the second or third position after the rearrangement within the V gene. Maintenance of an intact 5’ end of either D region means that a glycine would appear after the proline; glycine is encoded at the 5’ end of both D regions in either the first or second reading frame.
The number of rearrangements at position 7 is low and the number of xPG triplets for analysis is limited. Therefore, the focus was on rearrangements at position 8 and 9, which had higher levels of proline occurring after the initial rearrangement position (Fig 6). Because of the nature of rearrangement at positions 8 and 9, TP would be the most common rearrangement at position 8 and SP or RP at position 9. We therefore examined TPG triplets from rearrangements at b.p. 8 and RPG and SPG triplet motifs at b.p.9. Table 2 lists the number of Txx, Rxx, and Sxx encoding clonotypes and then breaks down the distribution of TPx, RPx and SPx clonotypes. The number of RPG clonotypes and the number of RPG clonotypes whose codon use is compatible with P nucleotide addition is then given. The percent of each population relative to the population above it is shown.
Table 2.
Distribution of proline-containing triplet motifs rearranged at b.p. 8 and 9 with potential of being generated by P nucleotide addition.
| b.p.8 | b.p.9 | |||||||
|---|---|---|---|---|---|---|---|---|
| motif | count | % | motif | count | % | motif | count | % |
| BV3 | ||||||||
| Txx | 592 | Rxx | 217 | Sxx | 254 | |||
| TPx | 31 | 5.24 | RPx | 46 | 21.3 | SPx | 59 | 23.2 |
| TPG | 7 | 22.6 | RPG | 15 | 32.6 | SPG | 10 | 17 |
| TPG (P) | 1 | 14.3 | RPG (P) | 9 | 60 | SPG (P) | 6 | 60 |
| BV19 | ||||||||
| Txx | 592 | Rxx | 500 | Sxx | 823 | |||
| TPx | 153 | 25.8 | RPx | 62 | 12.4 | SPx | 223 | 27.1 |
| TPG | 60 | 39.2 | RPG | 17 | 27.4 | SPG | 76 | 34.1 |
| TPG (P) | 28 | 46.7 | RPG (P) | 9 | 52.9 | SPG (P) | 46 | 60.5 |
| BV28 | ||||||||
| Txx | 675 | Rxx | 817 | Sxx | 1018 | |||
| TPx | 170 | 5.24 | RPx | 197 | 24.1 | SPx | 394 | 38.7 |
| TPG | 7 | 22.6 | RPG | 52 | 26.4 | SPG | 121 | 30.7 |
| TPG (P) | 1 | 14.3 | RPG (P) | 25 | 48.1 | SPG (P) | 39 | 32.2 |
The BV3 data did not provide any evidence for increased representation of P nucleotide-compatible TPG codons. While the numbers of PG encoding clonotypes for rearrangement at b.p. position 9 were limited, ~60% of both RPG and SPG codons were compatible with P nucleotide addition.
The data for BV19 rearrangements at bp positions 8 and 9 shows a higher proportion of clonotypes encoding Pro in the second position and in all three cases the percent of codons compatible with P nucleotide addition was 46% or higher.
Analysis of BV28 rearrangements at bp position 8 and 9 also show a strong propensity for encoding proline at the second position. While only 21% of TPG clonotypes were compatible with P nucleotide addition, this is almost twice than the 12.5% expected by chance alone. For RPG and SPG clonotypes the codon usage compatible with P nucleotide addition was higher. BV28 also shows an increase of Pro at the second position of motifs that rearrange at CDR3 bp position 11. These are often associated with the increased Ser rearrangement observed within the V region at position 11 (Fig 6). Of the 69 SPG motifs that are encoded at position 11. 36% show a codon usage that can be attributed to P nucleotide addition.
For BV 28, there is also a small increase in Pro at the third position of motifs rearranging at positions 7 to 9 (Fig 6). These are associated with multiple clonotypes encoding a limited number of motifs, with only a small percentage showing possible P nucleotide origins.
Thus, while P nucleotide addition can play a major role in the increased frequency of proline residues, it does not account for many of the cases of observations of proline in the triplet motifs analyzed.
IgH V-DJ rearrangements are predominantly at the end of the V region
IgH rearrangements were examined from naïve B cell datasets (8, 10) and mapped the V to DJ rearrangement site. Only VH genes with 8bp of germline encoded V-region after the conserved 3’ cysteine were examined. These constitute ~ 97% of the families. For these genes CDR3 positions 1, 2, and 3 can be V-gene encoded, with the final amino acid being D/E for V regions ending in GA, G for those ending in GG, and H for V ending in CA. The pattern observed was different from that of the TCR, with most rearrangements occurring at the end of the V or one base into the V (Fig 7A). Therefore, it is likely that the selection observed at position 4 (bp10 – 12) in the T cells is not a general outcome of V to DJ rearrangement but is indicative of an important role for the amino acid at this position during thymic selection.
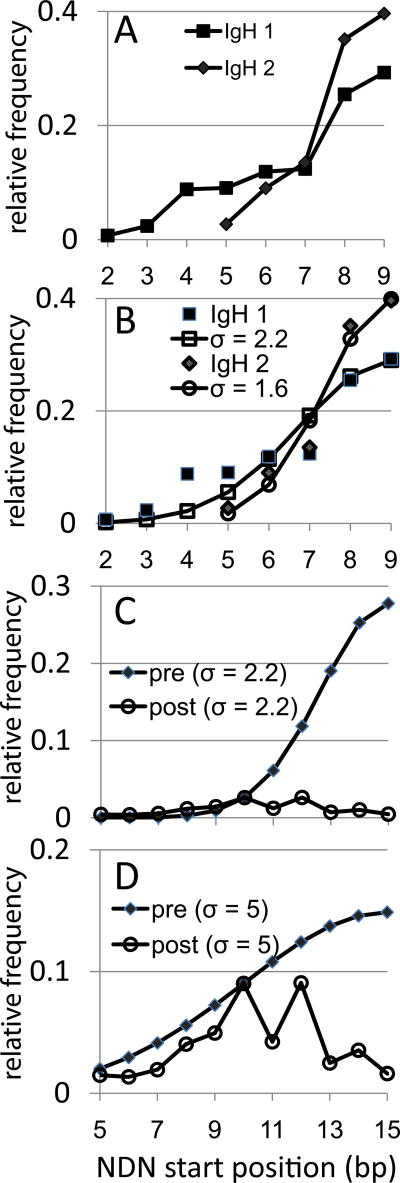
Figure 7.
IgH rearrangement along the V gene and estimating selection on BV gene rearrangements under an IgH-like rearrangement scheme. The data are shown as the relative frequency (as a fraction) of the number of rearrangements each CDR3 position. A. IgH rearrangement frequencies along the VH gene from two different sources of naïve B cells. The data are for VH families that have 8bp of germline encoded sequence 3’ of the second conserved cysteine. Rearrangements at position 9 have retained all eight of the germline bp. IgH1 squares, N=420, IgH2 diamonds, N=108. V-derived CDR3 amino acids are not identified as they would vary between different VH families. B. Fitting a half of a normal distribution to the IgH data. Normal distribution values calculated for a sigma of 2.2 (open squares) and 2.3 (open circles) are shown. The IgH values are the same as in panel A. C. Model of selection on BV19 assuming initial post-rearrangement distribution is IgH like (filled diamonds, sigma 2.2).The post- selection BV19 data (open circles) are normalized to the pre-selection distribution at position 10. D. Approximating a multi-mechanistic process (hairpin cleavage and exonuclease) by broadening the sigma to 5 (filled diamonds). BV19 data (open circles) are normalized to the pre-selection distribution at position 10.
Approximating the level of selection assuming an initial IgH-like rearrangement profile
To model the extent of selection needed if the initial distribution of rearrangements is IgH-like, we fit the IgH data to half of a normal distribution. The use of a normal distribution is based on the assumption of a random resolution of the hairpin around some optimal position, here assumed to be bp 8 of the CDR3, the end of the V gene. The NDN would therefore start at bp9, the first bp after the end of the intact V gene). The V to DJ portion would represent half of this normal distribution; with the DJ end representing the other half. Each IgH data set fit a normal distribution with sigma values of 2.2 for IgH dataset 1 (stdev = 0.03) and a sigma of 1.6 (stdev 0.03) for the IgH 2 dataset (Figure 7B). Such tight normal distributions are compatible with the action of Artemis in hairpin loop resolution (22).
We then calculated a normal distribution at sigma 2.2 for a 14 bp V region (15 bp being mean NDN start site) to generate an initial probability mass distribution for BV19 (Figure 7C). Superimposed on this is the pattern of the BV19 as we observe it, with the data being normalized at bp10, the highest frequency in the BV19 data. The post-selection BV19 mass distribution is <20% of the initial distribution. If a sigma value of 1.6 is used to generate the initial distribution, the percentage remaining is even lower. Thus ~20% retention is a generous estimate. This relation would hold even if there were prior selection events. For example, the initial distribution could already be corrected for proper in-frame selection, ~80% of which would then be negatively selected to insure the predominant use of those in-frame clonotypes that rearranged at CDR3 position 4. “Negatively” is used here to mean eliminated for structural reasons, without direct reference to positive or negative selection in the immunological sense. In Panel D we show the relation between a hypothetical model of rearrangement that generates a flatter initial distribution of rearrangement sites and the observed data. This would decrease the intensity of the required selection. This point is discussed further below.
DISCUSSION
T cell repertoires that are the culmination of a long period of adaptive responses can be source of novel information about the mechanisms that generate functional diversity. One of the observations made as part of such an analysis was the high frequency of NDN start sites at CDR3 position 4, which corresponds to the penultimate amino acid encoded by the V gene. 51% of the NDN start sites occur at CDR3 position 4, independent of CDR3 length (2). Our current understanding of rearrangement would predict that most of the NDN starts should have been at CDR3 position 5; after the ultimate or penultimate base of the V region. This is indeed the case for IgH. By examining our own and publicly available TCR data, we show that: NDN start sites are most often observed within the V region portion of the CDR3, NDN start sites are not smoothly represented at the DNA level, and the distribution of NDN start site frequencies can vary by the BV gene examined. For example BV28 rearrangements were maximal at CDR3 bp position 9, which maps to amino acid 3. The above conclusion hold for both CD4 and CD8 T cells implying a general phenomenon pertaining to recognition of both class I and class II MHC. These data raise two issues, why rearrangement within the V is so frequently observed, and is internal rearrangement an inherently wasteful process.
Crystal structures yield a relatively straightforward explanation for the importance of CDR3 amino acid position 4 in complexes where BV6 was recognizing various peptides bound to HLA-A2. Position 4 was involved in forming a bend/turn that involved minimal contact with pMHC even when the contact neighborhood was extended to 5Ǻ. The turn is required for the remainder of the CDR3 to maintain close contact with the pMHC. The turn was observed even in structures of TCR not bound to pMHC. The clear lack of contact with pMHC is in part due to the simplicity of the pMHCI structure recognized by CD8 cells. The turn is also present in TCR DR1 complex structures examined, but because of the more open fold and greater peptide accessibility, position 4 can also make contact with the peptide. This indicates that, for class II MHC, the turn may be induced in the process of docking. The requirement for an almost immediate turn in the CDR3 does not extend to IgH.
The amino acid usage at the rearrangement sites within the V gene often generate residues associated with beta turn-like structures. Ser is encoded in the V region, but Thr is also frequently observed. These can participate in S/T turns (23). Interestingly, while Asp is more frequently selected as the position 5 residue, in BV19 and BV28, two V genes where it is a possibility, Asn which might be expected as part of the Asx turn motif, is not observed at higher frequencies in NDN motifs. There is also an increase in proline, which can facilitate turns in the backbone, at the rearrangement site or at the next position in the NDN. Higher frequencies of these amino acids are compatible with the need for a turn in the CDR3 to facilitate pMHC recognition.
Glycine which is observed in high frequency throughout the NDN region can potentiate turns because the lack of a side chain allows a tighter bend in the previous linkage (24). This lack of steric hindrance can also make the NDN region more susceptible to direct interactions between a flexible main chain and side chains of the pMHC. The flexibility of poly-glycine runs has been presented in the context of TCRAV (25). Arginine, which is usually in the top four frequent amino acids in the NDN, also has an adaptable side chain capable of forming multiple H-bonds that are complementary to Ser and Thr.
There has been a long held theory of a co-evolution of MHC and TCR (26–28), which has interested us as well (29). Analysis of this co-evolution is predominantly based on CDR1 and CDR2 contributions as these are the major germ-line components that define the V region, and on which selection and thus evolution can function. We would argue that the distal V region contribution as part of the CDR3 is also important in pMHC and TCR interaction and thus selection. However, the driving force is the need to maintain a mechanism by which an almost immediate turn is generated. This drive has less to do with locus or allelic MHC polymorphism and more with the general ability to interact with the pMHC complex. There would be two components to the evolution of the V-end, the first would be the germ line structure (classical evolution) and the second would be the genetics of the mechanism driving rearrangement within the V-end (somatic evolution).
The germline V-end provides the encoding potential of this region as the rearrangement machinery trims back the germ line sequences. We observe this in the restricted nature of possibilities for the first amino acid at the rearrangement position. With respect to the distal V-end, the general rule for both human and mouse germline sequences is that the V contribution to the CDR3 is one amino acid longer for BV as compared to IgH. However, the amino acids observed after the Cys are generally Ala and Ser for both BV and IgH, with the “extra” a.a. in BV usually comprising L, Q, Y or E. The last two bases in the BV and IgH are generally GC, GA, or TC; but can be other combinations. The last two bases have an impact on the resulting repertoire. For example, the very low frequency of NDN starts at b.p. position 15 in BV28 (TG) results from the possible encodings. These are Cys, which is rarely observed in NDN sequences, the TGA termination codon, or Trp. The latter, while possibly a viable amino acid appears to be heavily selected against. The amino acid choices for BV25 (TA, rearrangement data in Fig 4) which also showed very low frequency or rearrangement at b.p. 15 are either Tyr or the ochre and amber terminators. The maintenance of poor potential “V-ends” supports the concept that rearrangements within the V region are salutary.
With respect to the mechanism driving the rearrangement within the V end, there are two major possibilities. The first is that BV rearrangement follows the same pattern as IgH, and that the level of unusable rearrangements will be very high. The second possibility would use evolution to argue selection for a more efficacious use of early thymocytes. There are data using human pre-B cell lines transfected with model rearrangement substrates (30) that show a reasonable amount of trimming with a general pattern that is equi-proportional over the four interior bp positions from the V-gene end. However, even if broader Artemis cleavage, and more nuclease trimming can flatten the distribution of rearrangement, there is still a considerable (50%) overhead of unused TCR (Fig 7D). One might also consider a mechanism by which the mean rearrangement start position is moved internal to the V gene (fixed asymmetric cleavage). In this regard it is interesting that D - J rearrangements present in the BV19 data are predominantly symmetrical within the J region.
Combinations of both mechanisms are possible. For example, primary rearrangements may be Ig-like, and any additional editing rearrangements on the same chromosome may use an altered mechanism that favors resolving hairpins in a manner that provides for rearrangement within the V region. We have previously proposed that the frequent IRSS rearrangements observed in CD8 SP thymocytes may have resulted from a direct V to J rearrangement, with the J region contributing the RS as part of an extended palindrome and the rearrangement at the V end taking place at CDR3 position 4, resulting in V-P-J rearrangements (6). Since J2.7 is the most frequent participant in such rearrangements and is the last J region, this form of rearrangement may represent a final chance at edited selection. Any such mechanism for broader or shifted rearrangement patterns may have evolved for BV rearrangement specifically.
During the generation of the repertoire one would expect the selection of a proper NDN would be a primary step. If the CDR3 cannot accommodate the pMHC the thymocyte would be a candidate for death by ignorance. β-selection has long been recognized as an important stage in thymocyte development, insuring rearrangement resulted in a protein that could bind to a surrogate alpha. This stage would also constitute a perfect occasion to select those thymocytes that have the correct turn structure to align with the proper part of the pMHC complex. This would require pMHC contact and evidence that this is indeed the case has been recently presented (31). This is probably not the only time at which the generation of a proper fit is selected as we have shown that following α-chain rearrangement there is a selection for SP thymocytes with β-chains with shorter CDR3 lengths (32). The binding of an actual α-chain could have indirect consequences on the NDN fold structure of the β-chain (33) that was not properly mimicked by the pre-Tα; thus a further selection.
We propose that an indirect (co-operative) mechanism (33) can also explain the loss of many BV19 clonotypes that rearranged at b.p. position 11. Under this scenario any of the possible position 11 a.a. (T, N, S, K, R) are present at position 4 would not function well in the context of other amino acids in the TCR. This could happen for a number of reasons ranging from protein folding to instability, and at multiple stages during thymocyte maturation, including during α-chain pairing. The final result would be SP thymocytes that show the selected rearrangement pattern.
To some extent we are only starting to grasp the complexity of the somatic evolutionary process of generating the naïve repertoire and how this evolves to become the mature repertoire. The current analysis of functionally relevant TCR β-chains indicates the existence of a complex selection process with a goal of generating pMHC recognition. It is likely that with further analyses additional mechanisms involved in the process will be identified.
Supplementary Material
Acknowledgments
We thank Dr Liz Worthy and Mike Tschannen at the Human and Molecular Genetics Center of the Medical College of Wisconsin for 454 sequencing. We thank Drs Deborah Dunn-Walters and Patrick Wilson for providing IGHV sequence data.
This work was supported by NIH Contract NO1 AI-50032 and NIAID U19 AI062627.
References
- 1.Roth DB. V(D)J Recombination: Mechanism, errors, and fidelity. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0041-2014. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yassai MB, Demos W, Janczak T, Naumova EN, Gorski J. CDR3 clonotype and amino acid motif diversity of BV19 expressing circulating human CD8 T cells. Hum Immunol. 2016;77:137–145. doi: 10.1016/j.humimm.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yassai MB, Demos W, Gorski J. CDR3 motif generation and selection in the BV19-utilizing subset of the human CD8 T cell repertoire. Mol Immunol. 2016;72:57–64. doi: 10.1016/j.molimm.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Deist F, Poinsignon C, Moshous D, Fischer A, de Villartay JP. Artemis sheds new light on V(D)J recombination. Immunol Rev. 2004;200:142–155. doi: 10.1111/j.0105-2896.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 5.Yassai MB, Naumova EN, Gorski J. Generation of TCR spectratypes by multiplex PCR for T cell repertoire analysis. In: Oksenberg JR, editor. The human antigen T cell receptor: Selected protocols and applications. Landes Bioscience; Austin TX: 1997. pp. 326–372. [Google Scholar]
- 6.Yassai MB, Naumov Y, Naumova EN, Gorski J. A clonotype nomenclature for T cell receptors. Immunogenetics. 2009;61:493–502. doi: 10.1007/s00251-009-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yassai MB, Bosenko D, Unruh M, Zacharias G, Reed E, Demos W, Ferrante A, Gorski J. Naive T cell repertoire skewing in HLA-A2 individuals by a specialized rearrangement mechanism results in public memory clonotypes. J Immunol. 2011;186:2970–2977. doi: 10.4049/jimmunol.1002764. [DOI] [PubMed] [Google Scholar]
- 8.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duty JA, Szodoray P, Zheng NY, Koelsch KA, Zhang Q, Swiatkowski M, Mathias M, Garman L, Helms C, Nakken B, Smith K, Farris AD, Wilson PC. Functional anergy in a subpopulation of naive B cells from healthy humans that express autoreactive immunoglobulin receptors. J Exp Med. 2009;206:139–151. doi: 10.1084/jem.20080611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci USA. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. doi: 10.1073/pnas.0913939107. https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP001441. [DOI] [PMC free article] [PubMed]
- 13.High throughput analysis of natural regulatory and conventional T cell receptor repertoires following human H1N1 vaccination http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP011414.
- 14.The PyMOL Molecular Graphics System, Version 1.8. Schrödinger, LLC: [Google Scholar]
- 15.Basu M, Hegde MV, Modak MJ. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- 16.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 17.Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, Wiley DC. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 18.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sami M, Rizkallah PJ, Dunn S, Molloy P, Moysey R, Vuidepot A, Baston E, Todorov P, Yi L, Gao F, Boulter JM, Jakobsen BK. Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry. Protein Eng. Des. Sel. 2007;20:397–403. doi: 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- 20.Gras S, Saulquin X, Reiser JB, Debeaupuis E, Echasserieau K, Kissenpfennig A, Legoux F, Chouquet A, Le Gorrec M, Machillot P, Neveu B, Thielens N, Malissen B, Bonneville M, Housset D. Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J. Immunol. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 21.Huber R, Steigemann W. Two cis-prolines in the Bence-Jones protein Rei and the cis-pro-bend. FEBS Lett. 1974;48:235–237. doi: 10.1016/0014-5793(74)80475-8. [DOI] [PubMed] [Google Scholar]
- 22.Lu H, Schwarz K, Lieber MA. Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res. 2007;35:6917–6923. doi: 10.1093/nar/gkm823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duddy WJ, Nissink JW, Allen FH, Milner-White EJ. Mimicry by asx- and ST-turns of the four main types of beta-turn in proteins. Protein Sci. 2004;13:3051–3055. doi: 10.1110/ps.04920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford JL, Lipscomb WN, Schellman CG. The reverse turn as a polypeptide conformation in globular proteins. Proc Natl Acad Sci U S A. 1973;70:538–542. doi: 10.1073/pnas.70.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naumov YN, Naumova EN, Yassai MB, Kota K, Welsh RM, Selin LK. Multiple glycines in TCR alpha-chains determine clonally diverse nature of human T cell memory to influenza A virus. J Immunol. 2008;181:7407–7419. doi: 10.4049/jimmunol.181.10.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerne KN. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 27.Huseby E, Kappler J, Marrack P. TCR-MHC/peptide interactions: kissing-cousins or a shotgun wedding? Eur J Immunol. 2004;34:1243–50. doi: 10.1002/eji.200425000. [DOI] [PubMed] [Google Scholar]
- 28.Sharon E, Sibener LV, Battle A, Fraser HB, Garcia KC, Pritchard JK. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battaglia M, Gorski J. Evidence for preferred MHC class II-TCR recognition independent of the source of bound peptide. Eur. J. Immunol. 2002;32:2179–2187. doi: 10.1002/1521-4141(200208)32:8<2179::AID-IMMU2179>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallis RJ, Bai K, Arthanari H, Hussey RE, Handley M, Li Z, Chingozha L, Duke-Cohan JS, Lu H, Wang JH, Zhu C, Wagner G, Reinherz EL. Pre-TCR ligand binding impacts thymocyte development before αβTCR expression. Proc Natl Acad Sci U S A. 2015;112:8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yassai M, Gorski J. Thymocyte maturation: selection for in-frame TCR alpha-chain rearrangement is followed by selection for shorter TCR beta-chain complementarity-determining region 3. J Immunol. 2000;165:3706–3712. doi: 10.4049/jimmunol.165.7.3706. [DOI] [PubMed] [Google Scholar]
- 33.Stadinski BD, Trenh P, Duke B, Huseby PG, Li G, Stern LJ, Huseby ES. Effect of CDR3 sequences and distal V gene residues in regulating TCR-MHC contacts and ligand specificity. J Immunol. 2014;192:6071–6082. doi: 10.4049/jimmunol.1303209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.