Abstract
Retinol saturase (RetSat) catalyzes the saturation of double bonds of all-trans-retinol leading to the production of dihydroretinoid metabolites. Beside its role in retinoid metabolism, there is evidence that RetSat modulates the cellular response to oxidative stress and plays critical roles in adipogenesis and the accumulation of lipids. Here, we explore the relationship between RetSat, lipid metabolism and oxidative stress using in vitro and in vivo models with altered expression of RetSat. Our results reveal that RetSat is a potent modulator of the cellular response to oxidative stress and the generation of reactive oxygen species (ROS). The levels of reactive aldehydes products of lipid peroxidation, as measured based on thiobarbituric acid reactivity, are increased in RetSat overexpressing cells and, conversely, reduced in cells and tissues with reduced or absent expression of RetSat compared to controls. Despite increased weight gain, neutral lipid accumulation and alterations in hepatic lipid composition, RetSat−/− mice exhibit normal responses to insulin. In conclusion, our findings further expand upon the role of RetSat in oxidative stress and lipid metabolism and could provide insight in the significance of alterations of RetSat expression as observed in metabolic disorders.
Keywords: adiposity, lipid metabolism, oxidative stress, retinoid metabolism, vitamin A
INTRODUCTION
Vitamin A, obtained in the diet in the form of retinol, retinyl esters, or provitamin A carotenoids is required for essential life processes such as embryonic development, cell differentiation and vision. It has been a long-held belief that all the non-visual functions of vitamin A are mediated by all-trans-retinoic acid (ATRA). This assertion is supported by the broad transcriptional regulatory roles mediated by ATRA and by its ability to rescue growth defects associated with vitamin A deficiency [1]. However, an increasing number of studies report observations of regulatory activities which are carried out by metabolites of retinol and carotenoids other than ATRA. These include activities mediated by retinol, retinaldehyde, retro-retinoids as well as by longer β-apocarotenoids and uncleaved carotenoids (reviewed in [2–5].
Dihydroretinoids are a class of retinoid metabolites which are generated via the reduction of one of the C-C double bonds present in retinoids. We have shown that all-trans-dihydroretinoids can be generated in vivo via the enzyme activity of retinol saturase (RetSat) which catalyzes the reduction of either the 13–14 or the 7–8 double bonds in the polyene chain of all-trans-retinol [6, 7]. RetSat resembles the sequence and evokes the enzymatic activity of carotene isomerase (CRTISO), a plant enzyme which carries out double bond isomerization reactions necessary for the synthesis of carotenoids [8]. The product of the mammalian RetSat-mediated saturation of all-trans-retinol is 13(R)-all-trans-13,14-dihydroretinol [9], which is further oxidized to 13(R)-all-trans-13,14-dihydroretinoic acid; both metabolites being detectable in animal tissues [6, 10–12]. We previously showed that all-trans-13,14-dihydroretinoic acid can activate the retinoic acid receptors (RAR), albeit, less potently than the cognate ligand of these receptors, namely ATRA [13]. In addition to trans-configured dihydroretinoids, others have detected the presence in serum and tissues of cis-configured dihydroretinoids, which have also been suggested to act as ligands of nuclear receptors [11, 14, 15]. Altogether, these observations suggest that the activity of RetSat could be involved in the generation of novel retinoid metabolites, with interesting and potentially important biological functions. Nevertheless, to fully understand the physiological role of RetSat, studies employing genetic approaches are required.
Several observations suggest that the physiological role of RetSat is associated with the regulation of lipid metabolism and adipogenesis. The expression of RetSat is induced by PPARα and PPARγ, important transcriptional regulators of lipid metabolism [16, 17], and is often deregulated in diabetic patients and in various models of insulin resistance [18]. Moreover, in vitro studies have uncovered the requirement of RetSat in the adipogenic differentiation of 3T3-L1 cells [17]. However, it was suggested that the effect of RetSat on adipogenesis involves a different product than its known product, all-trans-13,14-dihydroretinol [17]. In vivo, the role of RetSat proved to be even more complex to elucidate. In this case, RetSat-deficient mice exhibit increased fat accumulation in comparison with control littermates; in contrast to what would be expected based on its requirement for adipogenesis in vitro [19]. Thus, there is discordance between the role of RetSat in the regulation of lipid accumulation and adipogenesis, in vivo versus in vitro, and in relation to its proposed enzymatic activity in saturating all-trans-retinol.
Here, we explore the potential role of RetSat in lipid metabolism as well as oxidative stress. A genome-wide shRNA screen had previously identified RetSat as one of several genes involved in the cellular response to tert-butylhydroperoxide (tert-BHP), and hydrogen peroxide [20]. This study raised important questions regarding the relationship between RetSat’s potential involvement in oxidative stress. Using both in vivo and cell culture models with altered expression of RetSat, we demonstrate that RetSat affects the formation of reactive oxygen species (ROS). At the same time, deficiency of RetSat is associated with changes in the adiposity and the composition of lipids in mice including decreases in the levels of many species of unsaturated fatty acids. As these roles appear to be independent of the formation of all-trans-13,14-dihydroretinol, this study provides a new direction in exploring the function of RetSat and its possible association with pathophysiological processes related to lipid deposition and oxidative stress.
MATERIAL AND METHODS
Animal husbandry and dietary treatment
RetSat wild-type (WT) and knockout (KO) mice were generated as previously described [19] on a mixed 129sv/C57BL6 background and were backcrossed to the C57BL/6N strain (Charles River) for six generations. All mice were housed under controlled conditions of 12-hour dark and light cycle and fed on chow or high fat diet (HFD) with free access to water before experiments. All animal experiments were performed under guidelines approved by the University of Kansas or Yale University Animal Care and Use Committees. Seven, six-week-old, male WT or KO mice were treated with HFD containing 45 kcal % fat (Research Diet Inc., NJ) or chow for ten weeks. Body weight and food intake was monitored and at the completion of treatment, liver and white fat were dissected for lipid analysis.
Cell culture and RNA interference experiments
The HEKK-RetSat cells are stably transfected with the tetR-expressing plasmid pcDNA6-TR and pcDNA4/TO expressing the mouse RetSat enzyme under the control of tetracycline-inducible promoter and have been previously described [6]. HEKK-RetSat, and commercially available NIH/3T3 (ATCC, VA) and 3T3-L1 cells (ZenBio, NC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (SAFC, KS) or fetal calf serum (SAFC, KS) at 37 °C under humidified atmosphere with 5% CO2. Cells were seeded into 24-well plates (3×104 cells/well) or 6-well plates (3×105 cells/well) for 24 hours before experiments were performed. To induce the expression of RetSat, HEKK-RetSat cells were treated with or without 2 µg/ml tetracycline for 30 hours. NIH/3T3 or 3T3-L1 cells were transfected 30 hours before being assayed with 3 fmol RetSat siRNAs targeting mouse RetSat exon 2 or 9, siRNA1 and siRNA2, respectively, or control siRNAs (Ambion - Thermo Fisher Scientific). The effect of suppression of the expression of RetSat via siRNA was validated (Supplemental Fig. 1). RNA was extracted from NIH/3T3 cells transfected with siRNAs targeting mouse RetSat or control siRNAs using Trizol (Invitrogen-Thermo Fisher Scientific). Complementary DNA (cDNA) was synthesized from 1 µg total RNA using SuperScript III reverse transcriptase kit (Invitrogen-Thermo Fisher Scientific) using oligo dT-primed reactions. Reactions were performed in a 10 µl volume of 25 ng/µl cDNA, 250 nM of each primer and 5 µl of QuantiFast SYBR Green PCR Master Mix (Qiagen, CA) on the Biosystems StepOnePlus™ Real-Time PCR System. qPCR condition was at 95 °C for 10 min, 95 °C for 10 sec. and 60 °C for 30 sec for 40 cycles. The sequences of primers for RetSat were forward: 5’- CCGAGGATGTCAAGCGAC-3’, reverse: 5’- CCAGCTTCTCTGGTACTCGG -3’; for Gapdh were forward: 5’- GTCAAGCTCATTTCCTGGTATG-3’, reverse: 5’-CTTGCTCAGTGTCCTTGCTG-3’. The levels of cDNA were quantified by the comparative threshold cycle method using Gapdh as an internal standard. Alternatively, changes in the expression levels of the RetSat protein in NIH/3T3 cells transfected with small interfering RNAs (siRNAs) targeting mouse RetSat or control siRNAs were assessed. Cells were cultured and transfected in 6-well plate harvest in 100 µl SDS loading buffer, and then denatured at 100 °C for 15 min. Protein concentrations were determined by EZQ Protein Quantitation Kit (Invitrogen-Thermo Fisher Scientific). Equal amounts of protein were subjected to electrophoresis on a 12% Tris-HEPES polyacrylamide gel (Thermo Fisher Scientific, MA) and transferred to a PVDF membrane. The target protein was detected using a previously described anti-RetSat monoclonal antibody [19] and anti-GADPH monoclonal antibody was used to assess the levels of GAPDH as the loading control (Ambion-Thermo Fisher Scientific). The signal was detected with chemiluminescent agent (Thermo Scientific) after incubating with anti-mouse IgG conjugated with horseradish peroxidase (Promega, CA).
Mouse embryonic fibroblasts (MEFs) were isolated from 13.5-day-old embryos of RetSat WT and KO mice. Briefly, embryos were finely minced into pieces after removal of head and visceral organs, and incubated in 0.2% trypsin-EDTA solution at 37 °C for 15 min. The resulting cell suspension was filtered and centrifuged at 400 rpm for 5 min. The cell pellets were re-suspended and maintained in DMEM with 10% fetal calf serum at 37 °C under humidified atmosphere with 5% CO2 in 10 cm dishes. After achieving 80% confluence, the MEF cells were passaged and plated in 6-well plate (3×105 cells/well) for further experiments.
Measurements of cell viability
NIH/3T3 or 3T3-L1 cells transfected with siRNAs targeting mouse RetSat or control siRNAs were treated with 100, 200 and 400 µM tert-butyl hydroperoxide (tert-BHP) or vehicle control for one hour. Similarly, HEKK-RetSat cells with or without tetracycline-induction of the expression of RetSat were treated with 0, 50, 200 and 600 µM tert-BHP in culture medium for one hour. The viability of treated cells was examined the next day using the PrestoBlue Cell Viability Reagent (Invitrogen-Thermo Fisher Scientific) as described by the manufacturer. In experiments examining the effect of repletion with all-trans-13,14-dihydroretinol on the response of 3T3-L1 cells to tert-BHP, the cells were handled in reduced light conditions to prevent isomerization of the retinoids. Specifically, 3T3-L1 cells with or without suppression of the expression of RetSat were treated with various doses of tert-BHP in the presence of all-trans-13,14-dihydroretinol. At the end of the incubation period, the medium was replaced with fresh growth medium and the cell viability was measured 24 h later.
Measurements of the levels of lipid peroxides and ROS
Tetracycline-induced or uninduced HEKK-RetSat cells were treated with 100, 200 and 400 µM tert-BHP or vehicle control in Hanks' Balanced Salt Solution (HBSS) for one hour. Similarly, NIH/3T3 cells with or without suppression of the expression of RetSat were treated with 0, 200, 400 and 600 µM tert-BHP in HBSS for one hour. MEFs derived from WT or KO mice were treated with 0, 50, 100 and 200 µM tert-BHP in HBSS for one hour. The cell supernatant was collected for an assay of thiobarbituric acid-reactive substances (TBARS), and the cell pellets were harvested in 400 µl RIPA buffer for the determination of protein concentration by EZQ Protein Quantitation Kit (Invitrogen-Thermo Fisher Scientific). The TBARS assay was performed according to previously described methods [21, 22]. Briefly, 400 µl of supernatant or homogenate was mixed with 375 µl of 20% acetic acid (pH3.5) and 225 µl of 1.33% thiobarbituric acid (TBA). The mixture was heated at 95 °C for 20 min followed by centrifugation at 14,000 rpm for 10 min. The levels of malondialdehyde (MDA) -TBA adduct were measured fluorometrically using excitation of 515 nm and emission of 535 nm, and normalized to the protein concentration present in cells.
To measure TBARS in mouse tissues, tissue corresponding to approximately 30 mg of liver or 100 mg of fat was isolated from RetSat KO or WT mice treated with HFD or chow. The tissues were homogenized in 1 ml of 2.5% SDS with 35 µM BHT and 6.25 µM deferoxamine and hydrolyzed by the addition of 120 µl of 1 M KOH at 60 °C for 45 min followed by centrifugation at 14,000 rpm for 15 min. 400 µl supernatant was mixed with 375 µl of 20% acetic acid (pH3.5) and 225 µl of 1.33% TBA. The mixture was heated in 95 °C for 20 min followed by centrifugation at 14,000 rpm for 10 min. The levels of MDA-TBA adducts were measured at 532 nm by HPLC-UV. 20 µl supernatant was injected into a C8 (4.6 mm × 150 mm, 2.7 µm) column (MAC-MOD Analytical, PA) with the flow rate of 0.6 ml/min by using 80% 20 mM ammonium acetate and 20% methanol at 40 °C as a isocratic mobile phase.
To measure reactive oxygen species (ROS), RetSat overexpressing cells or cells with suppressed expression of RetSat and matched controls were pre-incubated with 10 µM 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) in HBSS for 30 min, and then were treated with or without tert-BHP for one hour. Fluorescent microscopy was used to measure the fluorescence intensity of 2',7'-dichlorofluorescein, the oxidized de-esterified product of H2DCFDA, whose levels correlate with levels of ROS. The images were captured and processed using Metamorph software, and displayed with pseudo color assignment. Fluorescence intensities were quantified by ImageJ software. For an alternative means to measure the levels of H2O2, induced or uninduced HEKK-RetSat cells and media were assayed using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit following the manufacturer’s instruction (Invitrogen-Thermo Fisher Scientific), and normalized by the protein concentration present in cells.
Measurement of the lipid profile in the liver of RetSat WT and KO mice
Approximately 50 mg or 100 mg liver samples obtained from seven, six-week-old, male WT or KO mice treated with HFD or chow were homogenized in 1 ml of 5% IGEPAL® CA 630 (Sigma-Aldrich, MO). Triacylglycerides were dissolved by repeated heating to 85 °C for 2 min and cooling to room temperature. The homogenates were cleared by centrifugation at 14,000 rpm for 5 min and the cleared supernatant was used to analyze triglyceride content using the Triglyceride Assay Kit (Zenbio, NC) as described by the manufacturer.
Alternatively, 50 mg liver samples obtained from seven, six-week-old, male mice treated with HFD were homogenized in 1.6 ml H2O with 35 µM butylated hydroxytoluene (BHT) and 6.25 µM deferoxamine followed by mixing with 2 ml chloroform and 4ml methanol. The mixture was vortexed before extraction with 2 ml chloroform and 2 ml H2O. The mixture was separated by centrifugation at 1000 rpm for 5 min. The aqueous layer was re-extracted with 2 ml chloroform twice. All organic layers were combined and washed once with 0.5 ml of 1 M potassium chloride and once with 0.5 ml H2O and then dried down in a vacuum concentrator (Thermo Scientific, IL). The residue was dissolved in 1 ml chloroform for mass spectrometry analysis. The profiles of phospholipids were measured by an automated electrospray ionization–tandem mass spectrometry method as previously described [23, 24]. Data were processed as previously described [23]. Briefly, the lipids in each class were quantified by their peak height and normalized by two internal standards of that class using a correction curve determined between standards. The amount of individual phospholipid to total liver weight was chosen to estimate the lipid changes in RetSat KO compared with WT mice. Statistical analyses were performed using student’s t-test or ANOVA followed by Post-hoc test where appropriate.
RESULTS
The expression of RetSat is positively correlated to the sensitivity of cells to inducers of oxidative stress
We studied the effect of altered expression of RetSat on the oxidative stress induced by tert-BHP in NIH/3T3 cells. We altered the expression levels of RetSat using two different siRNAs targeting exons 2 or 9 of mouse RetSat, and we compared their effect to two different non-targeting control siRNAs, siRNA-Ctr1 and siRNA-Ctr2. We confirmed that transfection of siRNA targeting RetSat leads to a consistent and significant reduction in the levels of RetSat mRNA and RetSat protein compared to the negative control siRNAs. (Supplemental Figure 1A, bottom panels).
We next compared the effect of the downregulation of the expression of RetSat expression on the cytotoxicity of tert-BHP on NIH/3T3 cells. Our results are consistent with the results reported previously [20]. As shown in Figure 1A, the cytotoxic effects of tert-BHP became noticeable in the cells transfected with control siRNA starting with a concentration of 100 µM while cell survival was not affected in cells transfected with siRNA targeting RetSat exposed to the same treatment. The trend continued as suppression of RetSat expression was associated with increasingly higher cell viability compared to controls cells at tert-BHP concentrations of 200 and 400 µM.
Figure 1.
Modulation of RetSat expression affects tert-BHP-induced toxicity. (A) Dose-response of the cytotoxic effect of tert-BHP on NIH/3T3 cells transfected with siRNA directed to RetSat (black bars) or with non-targeting control siRNAs (white bars). (B) Dose-response of the cytotoxic effect of tert-BHP on HEKK-RetSat cells induced with tetracycline (labeled RetSat Ovexp, black bars,) or not (labeled Ctr, white bars). (C) Dose-response of the cytotoxic effect of tert-BHP on 3T3-L1 cells transfected with siRNA directed to RetSat (black bars) or with nontargeting control siRNAs (white bars). (D) Dose-response of the cytotoxic effect of tert-BHP on 3T3-L1 cells incubated with 10 µM all-trans-13,14-dihydroretinol and with indicated concentrations of tert-BHP and either transfected with siRNA directed to RetSat (black bars) or with non-targeting control siRNAs (white bars). Comparison of the viability of control cells and RetSat-suppressed or RetSat-overexpressing cells for a given dose of tert-BHP: * p<0.05, ** p<0.01, ***<0.001. Comparison of the effect of various doses of tert-BHP versus vehicle-treated cells: # p<0.05, ## p<0.01, ### p<0.001. Data are presented as mean ± sd (n=3).
We complemented our studies by testing if overexpression of RetSat would decrease the cell survival rate in response to tert-BHP. We tried to test this hypothesis in NIH/3T3 cells, but RetSat overexpressing NIH/3T3 cells were no more sensitive to tert-BHP than the control cells (not shown). This could be due to the existing high endogenous levels of RetSat in NIH/3T3 cells which precludes any observable impact of its overexpression. Therefore, we chose the previously established cell line, HEKK-RetSat, which expresses very low levels of RetSat initially but high levels upon induction with tetracycline [6, 7, 9]. As expected, the cells overexpressing RetSat were more susceptible to tert-BHP than the uninduced control cells. As shown in Figure 1B, uninduced and tetracycline-induced HEKK-RetSat cells both showed a dose dependent cytotoxic response to tert-BHP but the tetracycline-induced cells have evidently reduced cell viability when compared to the control cells exposed to concentrations of 200 and 600 µM tert-BHP.
The inverse correlation of RetSat expression and resistance to tert-BHP was also observed in the case of the 3T3-L1 cell line [25]. It was reported that 3T3-L1 preadipocytes endogenously express RetSat and dramatically upregulate its expression via PPARγ during adipogenesis [17]. Importantly, suppression of the expression of RetSat in 3T3-L1 cells blocks their adipogenic differentiation [17]. Thus, we examined whether suppression of RetSat expression in 3T3-L1 preadipocytes also modulates their cellular response to tert-BHP treatment. Even though 3T3-L1 cells are inherently more sensitive to the toxicity of tert-BHP than NIH/3T3 cells, their viability in response to tert-BHP is increased by suppressing the expression of RetSat. Thus, 3T3-L1 cells transfected with siRNA targeting RetSat displayed a robust increase in cell viability when exposed to concentrations of 200 and 400 µM tert-BHP compared to the control cells (Fig. 1C). As RetSat converts all-trans-retinol to all-trans-13,14-dihydroretinol,[6] we asked whether repleting RetSat’s known product in 3T3-L1 cells with suppressed RetSat expression restores their sensitivity to tert-BHP. Surprisingly, addition of 10 µM all-trans-13,14-dihydroretinol does not affect the sensitivity of 3T3-L1 cells to tert- BHP in cells transfected with siRNA targeting RetSat (Fig. 1D). We also examined if RetSat promotes cellular sensitivity to tert-BHP via depletion of its putative substrate, all-trans-retinol; however, depletion of retinol from cell media did not restore the sensitivity of 3T3-L1 cells to tert-BHP (Supplemental Figure 2). Therefore, our results argue that the conversion of all-trans-retinol to all-trans-13,14-dihydroretinol by RetSat most likely does not play a role in the way this enzyme influences the cellular response to oxidative stress.
The expression of RetSat is directly correlated with the production of ROS and lipid peroxides
Since the expression of RetSat is inversely correlated with the cytotoxic effects of tert-BHP, we asked if this is a result of an effect of RetSat on the production of ROS as opposed to a more general effect on cell death pathways. To assess ROS production by cells treated with tert-BHP we employed the redox-sensitive dye H2DCFDA and fluorescent microscopy. Exposing cells to tert-BHP resulted in dose-dependent production of ROS in both NIH/3T3 and HEKK-RetSat cells. Importantly, suppression of the expression of RetSat leads to a significant reduction of ROS production in the NIH/3T3 cells compared to the control siRNA transfected cell. NIH/3T3 cells transfected with siRNA targeting RetSat produced 3.6 and 8.3-fold less ROS in the response to tert-BHP (Fig. 2A) at doses of 200 and 400 µM tert-BHP, respectively. On the other hand, overexpression of RetSat results in 3.9 and 2.9-fold elevation of ROS levels in RetSat overexpressing cells compared to the control cells in response to 50 and 100 µM tert-BHP, respectively (Fig. 2B).
Figure 2.
Modulation of RetSat expression affects the formation of ROS (A) Imaging of ROS production in response to tert-BHP in NIH/3T3 cells transfected with siRNA directed to RetSat (bottom panel) or with non-targeting control siRNAs (top panel). (B) Imaging of ROS production in response to tert-BHP in HEKK-RetSat cells induced with tetracycline (bottom panel) or uninduced (top panel). Quantification of the fluorescence associated with ROS for each treatment condition. The staining was visualized by fluorescence microscopy and quantified using ImageJ software. Fluorescence quantification of the signal associated with ROS for each treatment condition is shown in bar graphs at right. The graphs indicate fold change of relative fluorescence intensity over untreated cells for either control cells (white bars) or RetSat-suppressed or RetSat-overexpressing cells at a given dose of tert-BHP (black bars): * p<0.05, ** p<0.01, *** p<0.001. Data are presented as mean ± sd (n=3).
We next examined whether RetSat affects the levels of reactive free radicals and lipid peroxide produced in response to tert-BHP. Lipid peroxidation leads to the generation of malondialdehyde which was monitored via the TBARS assay. Acknowledging that TBARS can also be produced via alternative metabolic pathways [26], we noted that all cell types studied produced TBARS in response to tert-BHP in a dose-dependent manner, with little to no TBARS being generated in the absence of tert-BHP-induced oxidative damage (Fig. 3).
Figure 3.
Modulation of RetSat expression affects the formation of TBARS in response to tert-BHP. (A) Dose-response of the TBARS levels in response to tert-BHP in NIH/3T3 cells transfected with siRNA directed to RetSat (black circles) or with non-targeting control siRNAs (white circles). (B) Dose-response of the TBARS levels in response to tert-BHP in HEKK-RetSat cells induced with tetracycline (labeled RetSat Ovexp, black circles,) or not (labeled Ctr, white circles). (C) Dose-response of the TBARS levels in response to tert-BHP in MEFs from RetSat WT (white circles) or KO (black circles). (D) Levels of H2O2 released in the media by HEKK-RetSat cells either induced (black bar) or not (white bar) with tetracycline and represented as nmol/ gram protein. Comparison of the TBARS or H2O2 levels measured in control cells versus RetSat-suppressed or RetSat-deficient, or RetSat -overexpressing cells for a given dose of tert-BHP: * p<0.05, ** p<0.01, ***p<0.001. Data are normalized to protein levels and presented as mean ± sd (n=3).
Upon suppression of the expression of RetSat in NIH/3T3 cells, the levels of TBARS significantly declined by 2.3, 2.4 and 2.9-fold in response to concentrations of 200, 400 and 600 µM tert-BHP, respectively, compared with the control cells (Fig. 3A). Conversely, the levels of TBARS were dramatically elevated by 4.2, 4.6 and 4.2-fold in RetSat overexpressing cells in response to 100, 200 and 400 µM tert-BHP compared to controls (Fig. 3B). Similar to the effects of RetSat suppression in NIH/3T3 cells, the levels of TBARS detected in MEFs derived from RetSat KO mice were significantly decreased compared with the levels seen in MEFs from WT mice treated with the same amounts of tert-BHP concentrations (Fig. 3C). Moreover, as measured by Amplex Red, induction of RetSat expression in HEKK-RetSat cells leads to a higher level of cellular H2O2 being generated in comparison to control cells (Fig. 3D). Altogether, these results suggest that the expression of RetSat directly correlates with the levels of TBARS and ROS in cells.
We decided to further examine the effect of RetSat on the production of lipid peroxides in vivo by assessing the levels of TBARS in the liver and white fat of RetSat KO and WT mice. We employed both mice fed ad libitum for ten weeks on normal chow diet as well as mice fed a HFD containing 45% of kcal derived from fat, since HFD is associated with a higher level of oxidative stress in mice [27]. Consistent with previous reports of a positive association between HFD and levels of oxidative stress, we observed increased TBARS levels in the liver of HFD-fed compared with chow-fed WT mice (Fig. 4A versus C). In agreement with our observations in cell culture, we observed that the levels of TBARS in the liver of RetSat KO mice were modestly but consistently reduced compared to WT mice in case of chow- or HFD-fed mice (Fig. 4A and C). There were no obvious differences in the levels of TBARS in the white fat of RetSat WT and KO mice fed chow or HFD (Fig. 4B and D). Therefore, RetSat-deficiency is associated with lower levels of lipid peroxidation in the liver.
Figure 4.
RetSat deficiency is associated with reduced levels of TBARS in the liver of mice. Comparison of the levels of lipid peroxides in the liver (left graphs) or white fat (right graphs) of RetSat WT (white bars) and RetSat KO mice (black bars) maintained on either an chow (A and B) or HFD containing 45 kcal % fat (C and D) for 10 weeks. Lipid peroxides were measured by HPLC-UV. Comparison between WT and KO mice: *p<0.05. Data are presented as mean / gram protein ± sd (n=7 animals/genotype).
RetSat deficiency leads to body weight gain, and increased triacylglycerides and altered lipid composition in the liver of mice
RetSat was shown to have an interesting relationship with several important metabolic pathways, such as the fasting response, insulin signaling, and lipid metabolism [16–19]. Increased lipid peroxidation and ROS production is an early pathological feature of HFD-induced insulin resistance [27, 28]. Our results have, so far, shown that RetSat’s expression correlates with the levels of lipid peroxidation in mouse liver, therefore, we proceeded to study if RetSat also affects other pathophysiological process related to lipid accumulation and insulin responses in mice.
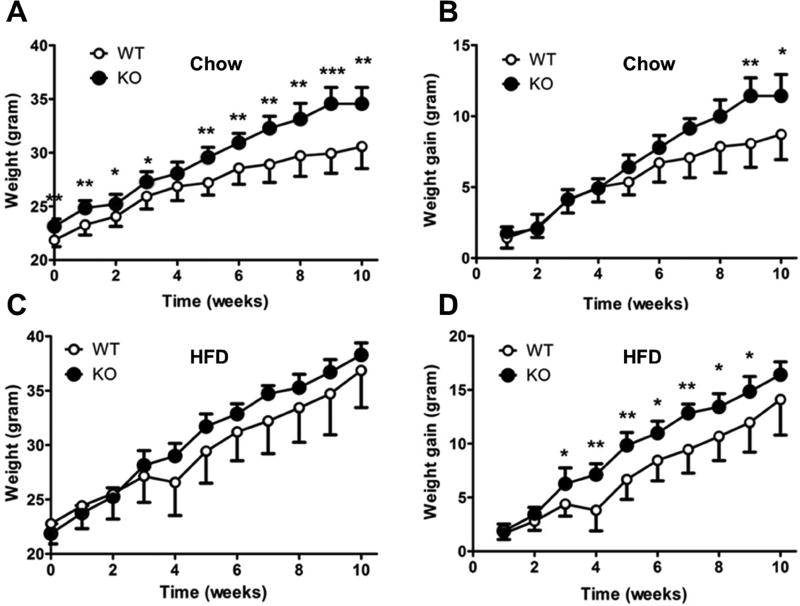
We first monitored the change in total body weight and the body weight gain of RetSat KO and WT mice fed chow for ten weeks. RetSat KO mice fed chow had a higher total body weight than chow-fed RetSat WT mice throughout the course of the experiment (Fig. 5A). Towards the end of the treatment, chow-fed RetSat KO mice also displayed a significantly higher weight gain than chow-fed RetSat WT mice (Fig. 5B). These results are consistent with our previous findings of increased adiposity in the RetSat KO mice [19], which have also been observed in a second RetSat-deficient mouse model [29]. Next, we examined RetSat WT and KO mice fed with HFD (45 kcal % fat). Though, RetSat KO mice on HFD displayed a consistently higher body weight gain from the third week to ninth week of treatment compared to HFD-fed RetSat WT mice (Fig. 5D) there was no significant difference in their total body weight (Fig. 5C). Metabolic cage studies of body weight matched HFD-fed RetSat WT and KO mice detected no differences in energy expenditure or feeding that could account for the differences in body weight gain (Supplemental Fig. 3). Interestingly, despite no detected differences in energy expenditure, RetSat KO mice were more active compared with WT mice, suggesting decreased energy efficiency may have contributed to their increased rate of weight gain during HFD and increased body weight and weight gain during chow studies, however, these possibilities will require future investigations (Supplemental Fig. 3).
Figure 5.
Comparison of body weight or body weight gain of RetSat WT and RetSat KO mice maintained on either chow (A and B) or HFD containing 45 kcal % fat (C and D) for 10 weeks. Body weight was measured once per week. Comparison between WT and KO mice: * p<0.05, ** p<0.01, *** p<0.0001. Data are presented as means ± sd (n=7 animals/genotype).
To corroborate our findings of decreased lipid peroxides in the livers of RetSat KO mice we measured triglyceride levels and the lipid composition in the livers of RetSat KO versus WT mice fed either chow or HFD. The level of triacylglycerides was elevated by 36% and 34% in the liver of RetSat KO mice fed chow or HFD, respectively, compared to RetSat WT mice (Fig. 6). In addition to neutral lipids, we also assessed the profile of 359 different species of polar lipids in the livers of RetSat KO and WT mice fed HFD by electrospray ionization (ESI) tandem mass spectrometry (ESI-MS/MS) including alkyl and acyl-phospholipids, lysophospholipids, and sphingomyelins. Figure 7 displays the changes associated with several species of polar lipids whose levels were significantly altered in the livers of HFD-fed RetSat WT versus KO mice. Many of the polar lipids species were decreased by 16% to 35% in the absence of RetSat. These include unsaturated phosphatidylcholines (PC), ether-linked PC, phosphatidylethanolamines (PE), and ether-linked PE, lysoPE and phosphatidylglycerols (PG). On the other hand, sphingomyelin (SM) 24:1, showed a 24% increase in levels in RetSat KO mouse versus WT. Altogether, our results indicate that RetSat plays a role in lipid metabolism and that its deficiency leads to increased weight gain and changes in the levels and composition of hepatic lipids. Because increased hepatic lipid levels are commonly associated with hepatic insulin resistance, we performed hyperinsulinemic euglycemic clamp studies in HFD-fed WT and RetSat KO mice [19] to evaluate whole-body and liver-specific insulin sensitivity directly. Plasma insulin levels were matched at 50.4 ± 8.2 and 64.7 ± 8.5 µU/ml for RetSat WT and RetSat KO mice, respectively, during the 140 min infusion. There was no difference in whole-body insulin sensitivity in RetSat WT compared with RetSat KO mice, as indicated by the similar rates of glucose infusion required to maintain euglycemia during the clamp. Hepatic insulin sensitivity, measured as the suppression of hepatic glucose production in response to insulin, was also not different between groups (Supplemental Fig. 4). These data are consistent with previous reports where no changes in insulin sensitivity or glucose homeostasis were observed in chow-fed RetSat-deficient mice subjected to glucose and insulin tolerance tests [19]. Therefore, in spite of results that suggest that RetSat deficiency is associated with increased adiposity and alterations in lipid metabolism we did not observe overt changes in insulin sensitivity.
Figure 6.
Comparison of hepatic level of triglycerides of RetSat WT and RetSat KO mice maintained on either chow (A) or HFD containing 45 kcal % fat for 10 weeks. Comparison between WT and KO: * p<0.05, ** p<0.01. Data are presented as means / milligram tissue ± sd (n=7 animals/genotype).
Figure 7.
Comparison of the level of different species of polar lipids isolated from the livers of RetSat WT and RetSat KO mice maintained on HFD containing 45 kcal % fat for 10 weeks. Phospholipid level were determined by shot-gun ESI-MS/MS. Comparison between WT and KO: * p<0.05, ** p<0.01, *** p<0.001. Data are presented as means ± sd (n=7 animals/genotype).
DISCUSSION
RetSat, a highly conserved chordate enzyme that carries out the reduction of double bonds of all-trans-retinol, has been implicated in the biology of adipocytes and the cellular response to oxidative stress [17, 20]. We present here results derived from both in vitro and in vivo models which indicate that the levels of RetSat are positively correlated with the sensitivity of cells to tert-BHP, and cellular production of ROS and lipid peroxides. In line with previous observations of increased lipid deposition in RetSat KO mice [19], RetSat KO mice display increased weight gain and increased levels of hepatic neutral lipids and a reduction in the levels of many species of unsaturated lipids. The reduction in the levels of unsaturated lipids might contribute to the observed decrease in the levels of lipid peroxides formed in the RetSat KO mouse. Overall, these result point towards a role of RetSat in lipid metabolism and in the formation of ROS in vitro and in vivo.
It is difficult to reconcile the influence of RetSat on the cytotoxic effects of tert-BHP and RetSat’s currently proposed enzymatic activity in saturating all-trans-retinol. Specifically, suppression of RetSat expression in NIH/3T3 cells leads to increased resistance to tert-BHP, however, the cellular sensitivity to tert-BHP cannot not be restored by either supplementation with its known product, all-trans-13,14-dihydroretinol, or by depletion of the substrate all-trans-retinol. This leads us to the hypothesis that an alternate, yet-to-be identified, enzymatic transformation carried out by RetSat is most likely responsible for its effects on ROS production. Though improbable, we cannot rule out the possibility that exogenous provision all-trans-13,14-dihydroretinol may not restore appropriate levels of this compound in the correct subcellular location. Despite our efforts we were not able to identify additional substrates of RetSat other than all-trans-retinol. However, results by Toomey et al. indicate that, in birds, RetSat can reduce the terminal double bond of the galloxanthin, a product of the asymmetric cleavage of the carotenoid zeaxanthin, to generate dihydrogalloxanthin [30]. Since carotenoids are known to influence the cellular response to oxidative stress [31] and mitochondrial function [32], it is possible that a better understanding of the enzymatic activity of RetSat in relation to other apocarotenoids could also shed light on its role in the formation of ROS.
Other possible explanations for the connection between RetSat and the formation of ROS could involve mechanisms independent of its known enzymatic product. It is formally possible that the effects of RetSat on the generation of ROS are mediated by a non-catalytic mechanism. In this case it is possible that RetSat, an endoplasmic reticulum-resident membrane protein [6], could interact with other cellular regulators of ROS within this compartment to promote the formation of ROS via protein-protein interactions. Another possibility is that the effect of RetSat on ROS is mediated by the saturation of the all-trans-retinol moiety associated with protein kinase Cδ (PCKδ). The π-conjugated system of all-trans-retinol mediates electron transfer from cytochrome c leading to the activation of PCKδ [33] signalosome with ensuing effects on glycolytic energy generation [34, 35]. It is, therefore, possible that RetSat may interfere with this pathway by breaking the π-conjugation system via the saturation of the C13-C14 double bond of all-trans-retinol. Though MEFs derived from RetSat KO mice exhibit a slightly higher mitochondrial O2 consumption rate, measurements of in vivo oxygen consumption and energy expenditure in RetSat KO mice did not reveal any significant difference from controls (not shown). The possibilities of the involvement of RetSat in ROS production via protein-protein interactions or via the regulation of PCKδ will require further studies.
Another unresolved issue is in terms of the role of RetSat in lipid metabolism or adipocyte differentiation. In data presented here and elsewhere, in a mouse model targeting exon 1 of mouse RetSat, we showed that RetSat deficiency in vivo is associated with increased adiposity and increased liver TGs [19]. Subsequently, our observations were confirmed by data derived from a different RetSat knockout mouse model produced through targeting exons 1–4, which was also reported to also display increased fat accumulation (Project ENZ775N1 reported by Lexicon Genetics-Genetech [29]. Importantly, studies by Schupp et al. have shown that RetSat-depletion via siRNA impairs the adipogenic differentiation of 3T3-L1 cells [17] and that siRNA-mediated ablation of RetSat expression in the liver leads to reduced hepatic TGs [36]. As RetSat is robustly expressed in both liver and fat [16, 17], it is not surprising that the phenotype expected based on the specific suppression of the expression of RetSat in liver or in adipocytes will differ from the one resulting from the global ablation of RetSat’s expression. Dissecting the physiological role of this important metabolic regulator would most likely require a tissue-specific conditional RetSat knockout. Nevertheless, we offer here support for a role of RetSat in regulating lipid metabolism by demonstrating that not only lipid levels but also their composition is altered in the liver of RetSat KO mice. Importantly, as seen here, in the case of RetSat’s role in ROS formation, the effect of RetSat on either adipogenic differentiation or liver lipogenesis was independent of its known enzymatic activity in saturating all-trans-retinol, which we also observed ([17, 36] and results not shown). All things considered, the available data from us and others suggests that alterations in the expression of RetSat affect lipid metabolism and that this effect appears to be independent of the saturation of all-trans-retinol.
The relationship of RetSat to lipid metabolism and ROS generation is intriguing. A comprehensive meta-analysis of multiple data sets obtained from human tissues and mouse models found RetSat to be the top ranked gene in association with insulin resistance [18]. Generally, RetSat is found to be upregulated in models of insulin resistance but often downregulated in obesity [17, 18]. We show here that while RetSat deficiency leads to increased lipid accumulation it also reduces the formation of ROS and lipid peroxides. Interestingly, despite the increased accumulation of neutral lipids, RetSat KO mice do not exhibit increased insulin resistance compared to controls (results presented here and [19]). It is interesting to speculate whether the surprisingly normal insulin responses despite the significantly increased adiposity seen in RetSat KO mice can be attributed to the reduced formation of ROS. Notably, we did not measure hepatic levels of bioactive lipids such as diacylglycerol [37] or ceramide [38] that are directly implicated in the pathogenesis of insulin resistance; thus, we cannot rule out that these species are not affected by RetSat deficiency. Since ROS are known to play an important role in adipogenic differentiation [39] and in the etiology of insulin resistance, metabolic syndrome and other pathophysiological processes [27, 28]. These interesting possibilities warrant further studies of the enzymatic activity of RetSat leading to the potential identification of novel biochemical pathways that affect lipid metabolism or the generation of ROS.
Supplementary Material
HIGHLIGHTS.
Retinol saturase is shown to play an important role in the generation of ROS
In vivo and in vitro ROS levels correlate with the expression of retinol saturase
Retinol saturase-deficient mice deposit more lipids but are sensitive to insulin
Acknowledgments
We are grateful to Dr. Kris Palczewski (Case Western Reserve University, Cleveland, OH) for the use of the RetSat KO mouse strain, the HEKK-RetSat cells and the monoclonal against RetSat, and to Drs. Qin Yang and Barbara Kahn (Beth Israel Deaconess Medical Center, Boston, MA) for their assistance in assessing glucose metabolism in the RetSat KO mouse. We thank Dr. Michael Schupp (Charité University, Berlin) for helpful discussions and Drs. Mary Roth and Ruth Welti (Kansas State University, Manhattan, KS) for their assistance in examining the lipid levels of RetSat KO and WT mice. Lipid analysis was performed at the Kansas Lipidomics Research Center (KLRC). Instrument acquisition and method development at KLRC was supported by NSF grants MCB 0455318 and 0920663 and DBI 0521587, and NSF EPSCoR grant EPS-0236913 with additional support from the State of Kansas through Kansas Technology Enterprise Corporation and Kansas State University and a K-INBRE grant, P20 RR16475 from the NIH. This work was supported in part by grants P20 GM103420, P20RR017708 and R01HD077260 (A.R.M.) from the National Institutes of Health.
ABBREVIATIONS
- ATRA
all-trans-retinoic acid
- BHT
butylated hydroxytoluene
- H2DCFDA
2’,7’-dichlorodihydrofluorescein diacetate
- HFD
high-fat diet
- HPLC
high performance liquid chromatography
- LC
liquid chromatography
- MEFs
mouse embryonic fibroblasts
- MDA
malondialdehyde
- MS
mass spectrometry
- PCKδ
protein kinase C δ
- PPAR
peroxisome proliferator-activated receptor
- qRT-PCR
quantitative real-time reverse transcription-polymerase chain reaction
- RAR
retinoic acid receptor
- RetSat
retinol saturase
- RNAi
RNA interference
- ROS
reactive oxygen species
- siRNAs
small interfering RNAs
- tert-BHP
tert-butylhydroperoxide
- TBA
thiobarbituric acid
- TBARS
thiobarbituric acid-reactive substances
- UV
ultraviolet
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: none.
References
- 1.Dowling JE, Wald G. The biological function of vitamin-A acid. Proc. Natl. Acad. Sci. U.S.A. 1960;46(5):587–608. doi: 10.1073/pnas.46.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziouzenkova O, Plutzky J. Retinoid metabolism and nuclear receptor responses: New insights into coordinated regulation of the PPAR-RXR complex. FEBS Lett. 2008;582(1):32–8. doi: 10.1016/j.febslet.2007.11.081. [DOI] [PubMed] [Google Scholar]
- 3.von Lintig J. Provitamin A metabolism and functions in mammalian biology. Am J Clin Nutr. 2012;96(5):1234S–44S. doi: 10.3945/ajcn.112.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eroglu A, Harrison EH. Carotenoid metabolism in mammals, including man: formation, occurrence, and function of apocarotenoids. Journal of Lipid Research. 2013;54(7):1719–1730. doi: 10.1194/jlr.R039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammerling U. Retinol as electron carrier in redox signaling, a new frontier in vitamin A research. Hepatobiliary Surg Nutr. 2016;5(1):15–28. doi: 10.3978/j.issn.2304-3881.2016.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moise AR, Kuksa V, Imanishi Y, Palczewski K. Identification of all-trans-retinol:all-trans-13,14-dihydroretinol saturase. J Biol Chem. 2004;279(48):50230–42. doi: 10.1074/jbc.M409130200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moise AR, Isken A, Dominguez M, de Lera AR, von Lintig J, Palczewski K. Specificity of zebrafish retinol saturase: formation of all-trans-13,14-dihydroretinol and all-trans-7,8-dihydroretinol. Biochemistry. 2007;46(7):1811–20. doi: 10.1021/bi062147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moise AR, von Lintig J, Palczewski K. Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 2005;10(4):178–86. doi: 10.1016/j.tplants.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Moise AR, Dominguez M, Alvarez S, Alvarez R, Schupp M, Cristancho AG, Kiser PD, de Lera AR, Lazar MA, Palczewski K. Stereospecificity of retinol saturase: absolute configuration, synthesis, and biological evaluation of dihydroretinoids. J Am Chem Soc. 2008;130(4):1154–5. doi: 10.1021/ja710487q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moise AR, Kuksa V, Blaner WS, Baehr W, Palczewski K. Metabolism and transactivation activity of 13,14-dihydroretinoic acid. J Biol Chem. 2005;280(30):27815–25. doi: 10.1074/jbc.M503520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruhl R, Krzyzosiak A, Niewiadomska-Cimicka A, Rochel N, Szeles L, Vaz B, Wietrzych-Schindler M, Alvarez S, Szklenar M, Nagy L, de Lera AR, Krezel W. 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice. PLoS Genet. 2015;11(6):e1005213. doi: 10.1371/journal.pgen.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazhin AV, Bleul T, de Lera AR, Werner J, Ruhl R. Relationship Between All-trans-13,14-Dihydro Retinoic Acid and Pancreatic Adenocarcinoma. Pancreas. 2016;45(6):e29–31. doi: 10.1097/MPA.0000000000000601. [DOI] [PubMed] [Google Scholar]
- 13.Moise AR, Alvarez S, Dominguez M, Alvarez R, Golczak M, Lobo GP, von Lintig J, de Lera AR, Palczewski K. Activation of retinoic acid receptors by dihydroretinoids. Mol Pharmacol. 2009;76(6):1228–37. doi: 10.1124/mol.109.060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirley MA, Bennani YL, Boehm MF, Breau AP, Pathirana C, Ulm EH. Oxidative and reductive metabolism of 9-cis-retinoic acid in the rat. Identification of 13,14-dihydro-9-cis-retinoic acid and its taurine conjugate. Drug Metab Dispos. 1996;24(3):293–302. [PubMed] [Google Scholar]
- 15.Schmidt CK, Volland J, Hamscher G, Nau H. Characterization of a new endogenous vitamin A metabolite. Biochim Biophys Acta. 2002;1583(2):237–51. doi: 10.1016/s1388-1981(02)00212-3. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Ng L, Lam W, Lo CK, Chan PT, Yuen YL, Wong PF, Tsang DS, Cheung WT, Lee SS. Identification and characterization of a novel mouse peroxisome proliferator-activated receptor alpha-regulated and starvation-induced gene, Ppsig. Int J Biochem Cell Biol. 2008;40(9):1775–91. doi: 10.1016/j.biocel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Schupp M, Lefterova MI, Janke J, Leitner K, Cristancho AG, Mullican SE, Qatanani M, Szwergold N, Steger DJ, Curtin JC, Kim RJ, Suh MJ, Albert MR, Engeli S, Gudas LJ, Lazar MA. Retinol saturase promotes adipogenesis and is downregulated in obesity. Proc Natl Acad Sci U S A. 2009;106(4):1105–10. doi: 10.1073/pnas.0812065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park PJ, Kong SW, Tebaldi T, Lai WR, Kasif S, Kohane IS. Integration of heterogeneous expression data sets extends the role of the retinol pathway in diabetes and insulin resistance. Bioinformatics. 2009;25(23):3121–7. doi: 10.1093/bioinformatics/btp559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moise AR, Lobo GP, Erokwu B, Wilson DL, Peck D, Alvarez S, Dominguez M, Alvarez R, Flask CA, de Lera AR, von Lintig J, Palczewski K. Increased adiposity in the retinol saturase-knockout mouse. FASEB J. 2010;24(4):1261–70. doi: 10.1096/fj.09-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaoka-Yasuda R, Matsuo N, Perkins B, Limbaeck-Stokin K, Mayford M. An RNAi-based genetic screen for oxidative stress resistance reveals retinol saturase as a mediator of stress resistance. Free Radic Biol Med. 2007;43(5):781–8. doi: 10.1016/j.freeradbiomed.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Liu R, Liu IY, Bi X, Thompson RF, Doctrow SR, Malfroy B, Baudry M. Reversal of age-related learning deficits and brain oxidative stress in mice with superoxide dismutase/catalase mimetics. Proc Natl Acad Sci U S A. 2003;100(14):8526–31. doi: 10.1073/pnas.1332809100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott SJ, Doan TN, Schilling WP. Role of lipid peroxidation in tert-butylhydroperoxide-induced inhibition of endothelial cell calcium signaling. J Pharmacol Exp Ther. 1993;264(3):1063–70. [PubMed] [Google Scholar]
- 23.Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277(35):31994–2002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- 24.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A. 1997;94(6):2339–44. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green H, Kehinde O. Sublines of mouse 3T3 cells that accumulate lipid. Cell. 1974;1(3):113–116. [Google Scholar]
- 26.Janero DR. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radic Biol Med. 1990;9(6):515–40. doi: 10.1016/0891-5849(90)90131-2. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzawa-Nagata N, Takamura T, Ando H, Nakamura S, Kurita S, Misu H, Ota T, Yokoyama M, Honda M, Miyamoto K, Kaneko S. Increased oxidative stress precedes the onset of high-fat diet-induced insulin resistance and obesity. Metabolism. 2008;57(8):1071–7. doi: 10.1016/j.metabol.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440(7086):944–8. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 29.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, Forrest W, Ghilardi N, Oravecz T, Platt KA, Rice DS, Hansen GM, Abuin A, Eberhart DE, Godowski P, Holt KH, Peterson A, Zambrowicz BP, de Sauvage FJ. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28(7):749–55. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 30.Toomey MB, Lind O, Frederiksen R, Curley RW, Riedle KM, Wilby D, Schwartz SJ, Witt CC, Harrison EH, Roberts NW, Vorobyev M, McGraw KJ, Cornwall MC, Kelber A, Corbo JC. Complementary shifts in photoreceptor spectral tuning unlock the full adaptive potential of ultraviolet vision in birds. Elife. 2016;5 doi: 10.7554/eLife.15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl W, Sies H. Antioxidant activity of carotenoids. Mol. Asp. Med. 2003;24(6):345–351. doi: 10.1016/s0098-2997(03)00030-x. [DOI] [PubMed] [Google Scholar]
- 32.Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25(3):948–59. doi: 10.1096/fj.10-173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acin-Perez R, Hoyos B, Gong J, Vinogradov V, Fischman DA, Leitges M, Borhan B, Starkov A, Manfredi G, Hammerling U. Regulation of intermediary metabolism by the PKCdelta signalosome in mitochondria. FASEB J. 2010;24(12):5033–42. doi: 10.1096/fj.10-166934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher-Wellman KH, Gilliam LA, Lin CT, Cathey BL, Lark DS, Neufer PD. Mitochondrial glutathione depletion reveals a novel role for the pyruvate dehydrogenase complex as a key H2O2-emitting source under conditions of nutrient overload. Free Radic Biol Med. 2013;65:1201–8. doi: 10.1016/j.freeradbiomed.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shabrova E, Hoyos B, Vinogradov V, Kim YK, Wassef L, Leitges M, Quadro L, Hammerling U. Retinol as a cofactor for PKC delta-mediated impairment of insulin sensitivity in a mouse model of diet-induced obesity. Faseb Journal. 2016;30(3):1339–1355. doi: 10.1096/fj.15-281543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heidenreich S, Witte N, Weber P, Goehring I, Tolkachov A, von Loeffelholz C, Docke S, Bauer M, Stockmann M, Pfeiffer AFH, Birkenfeld AL, Pietzke M, Kempa S, Muenzner M, Schupp M. Retinol saturase coordinates liver metabolism by regulating ChREBP activity. Nat Commun. 2017;8(1):384. doi: 10.1038/s41467-017-00430-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet (London, England) 2010;375(9733):2267–77. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Summers SA. Sphingolipids and insulin resistance: the five Ws. Current opinion in lipidology. 2010;21(2):128–35. doi: 10.1097/MOL.0b013e3283373b66. [DOI] [PubMed] [Google Scholar]
- 39.Lee H, Lee YJ, Choi H, Ko EH, Kim JW. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J Biol Chem. 2009;284(16):10601–9. doi: 10.1074/jbc.M808742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.