Abstract
Patient-derived xenografts (PDXs) have become a prominent cancer model system, as they are presumed to faithfully represent the genomic features of primary tumors. Here we monitored the dynamics of copy number alterations (CNAs) in 1,110 PDX samples across 24 cancer types. We observed rapid accumulation of CNAs during PDX passaging, often due to selection of pre-existing minor clones. CNA acquisition in PDXs was correlated with the tissue-specific levels of aneuploidy and genetic heterogeneity observed in primary tumors. However, the particular CNAs acquired during PDX passaging differed from those acquired during tumor evolution in patients. Several CNAs recurrently observed in primary tumors gradually disappeared in PDXs, indicating that events undergoing positive selection in humans can become dispensable during propagation in mice. Importantly, the genomic stability of PDXs was associated with their response to chemotherapy and targeted drugs. These findings have important implications for PDX-based modeling of human cancer.
Cancer research relies on interrogating model systems that mirror the biology of human tumors. Cell lines cultured from human tumors have been the workhorse of cancer research, but the marked differences between the in vitro cell culture environment and the in vivo tumor environment raise concerns that they may not be fully representative of human tumors. Recently, there have been increasing efforts to utilize patient-derived xenografts (PDXs) as models to study drug response1–4. These in vivo models are assumed to capture the cellular and molecular characteristics of human cancer better than simpler cell line-based models 1,2.
As the value of PDX models depends on their faithful representation of primary tumors, it is important to assess whether PDXs retain their genomic and phenotypic characteristics throughout propagation. To date, the genomic stability of PDX models has primarily been evaluated indirectly, leading to the notion that PDXs are highly stable3,5,6. Consistent with this perception, PDX-based studies often involve the analysis of tumors from multiple passages3. However, hints that PDXs may be more genomically unstable than assumed have recently begun to emerge7,8, emphasizing the need for a comprehensive analysis of PDX genomic evolution (Supplementary Note).
Here, we systematically analyzed landscapes of aneuploidy and large CNAs in PDX models across multiple human cancers. We generated a comprehensive CNA catalogue of 1,110 PDX samples from 24 cancer types, and used these data to characterize CNA dynamics during PDX derivation and propagation, to study the origin of passaging-acquired CNAs, and to compare PDX genomic stability across cancer types. We also compared the CNA dynamics observed in PDXs to those of newly-derived tumor cell lines and cell line-derived xenografts (CLDXs). Finally, we compared the CNA landscapes of PDXs to those of human primary and advanced tumors. We found that despite an overall similarity, the CNA landscapes of PDXs diverge substantially from their parental tumors during passaging. We discuss the potential implications of this divergence, including its effect on therapeutic response.
Results
Generating a catalogue of aneuploidy and CNAs in PDXs
To enable a comprehensive analysis of aneuploidy and CNAs in PDXs, we created an integrated CNA dataset representing 1,110 PDXs. We first assembled data from DNA-based copy number measurements across multiple PDX passages, using published SNP arrays, CGH arrays and DNA sequencing data. Unfortunately, such DNA copy number data were only available for 177 PDX samples from 5 studies – too few to support a comprehensive analysis of CNA stability (Supplementary Table 1)6,7,9–11. In contrast, gene expression profiles were available for 933 PDX samples collected from 511 PDX models across 17 studies (Supplementary Table 1)3,5,6,10–23. To reconstruct chromosomal aneuploidy and large (>5 Mb) CNAs from these expression profiles, we used previously described computational inference algorithms that accurately identify CNAs based on induced coordinated gene expression changes24–26. We validated the accuracy of this approach by analyzing PDXs from which both gene expression and SNP array data were available (Supplementary Fig. 1, Supplementary Table 2 and Supplementary Note). Our final dataset comprised CNA data of 1,110 PDX samples from 543 unique PDX models across 24 cancer types (Fig. 1a and Supplementary Data 1 and 2). For 342 of these PDX models, data were available from both the primary tumor and its derived PDX model(s), or from multiple PDX passages, thus enabling an analysis of tumor evolution (Fig.1a and Supplementary Table 1).
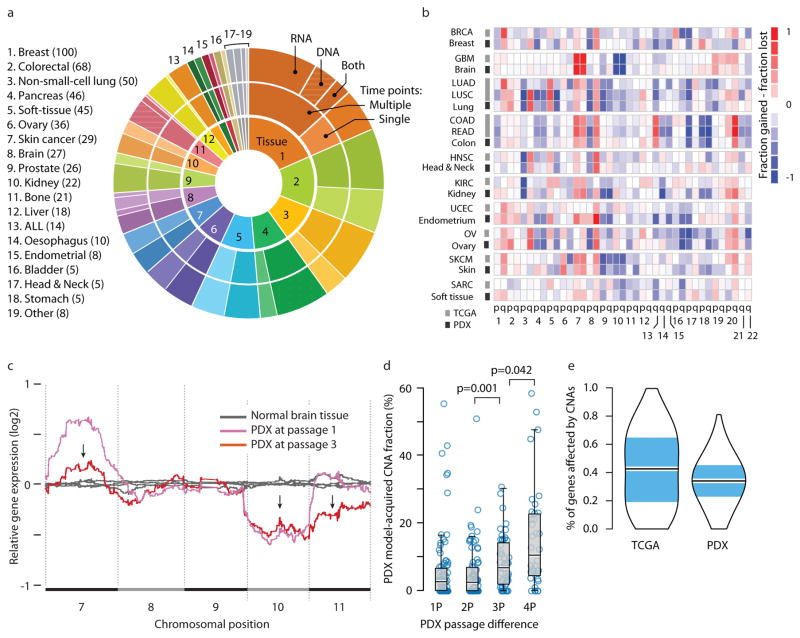
Figure 1. The landscape of aneuploidy and copy number alterations in PDXs.
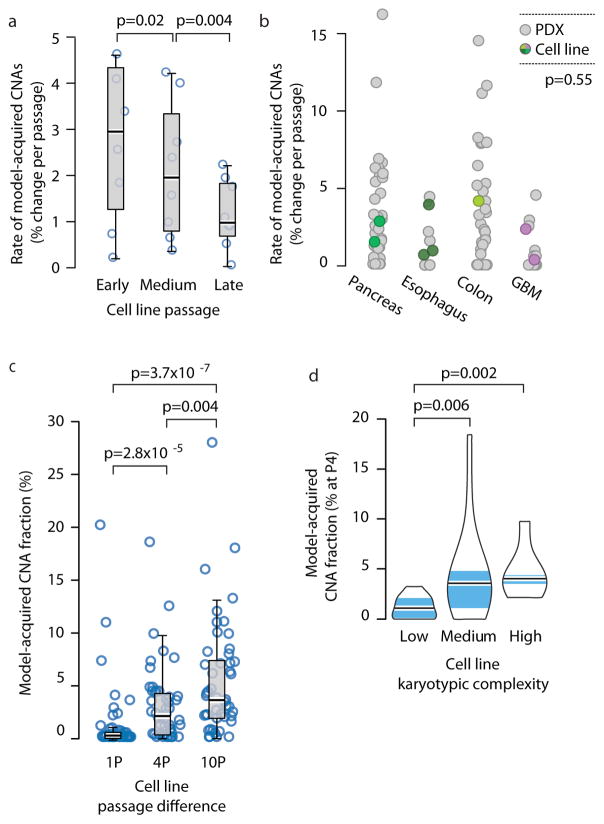
(a) Distribution of cancer types in our PDX dataset (n=543 unique models). In the inner circle models are divided by their lineage: each cancer type is denoted by a color and a number. In the middle circle models are divided by the number of time points analyzed: multiple time points are denoted by a darker color, and enable to follow PDX evolution throughout in vivo propagation. In the outer circle models are divided by the biological material from which CNAs were inferred: DNA (stripes), RNA (dots) or both (stripes and dots). (b) A heatmap comparing the landscapes of lineage-matched arm-level CNAs of PDXs and of primary TCGA tumors, showing an overall high degree of concordance (mean Pearson’s r = 0.79). The color of each chromosome arm represents the fraction difference between gains and losses of that arm. (c) A representative example of PDX model evolution. Shown are gene expression moving average plots of normal brain tissue (gray), GBM PDX model at p1 (pink) and GBM model at p3 (red), revealing the disappearance of trisomy 7, the retention of monosomy 10, and the emergence of monosomy 11, within two in vivo passages. (d) Gradual evolution of CNA landscapes throughout PDX passaging. Box plots present model-acquired CNA fraction as a function of the number of passages between measurements. Bar, median; box, 25th and 75th percentiles; whiskers, data within 1.5*IQR of lower or upper quartile; circles: all data points. P-values indicate significance from a Wilcoxon rank-sum test. (e) Violin plots present the proportion of genes affected by CNAs in TCGA and in PDX tumor samples (all tissue types combined), showing an overall similarity between both datasets. Bar, median; colored rectangle, 25th and 75th percentiles; width of the violin indicates frequency at that CNA fraction level.
We found the CNA landscapes of PDXs to be highly similar to those of their respective tumor types in The Cancer Genome Atlas (TCGA) (mean Pearson’s r=0.79; Fig. 1b and Supplementary Fig. 2), consistent with prior reports6,8,10,11. This confirms that, at the cohort level, PDX models are generally genomically representative of primary human tumors.
Tracking CNA dynamics during PDX derivation and propagation
We set out to follow CNA dynamics in individual PDX models, in order to assess their stability as well as their similarity to the tumors from which they were derived. For each model, the earliest passage (in most cases, P0 or P1) was compared to later passages (range, P1–P16; median: P3) in order to determine the changes that occurred throughout passaging. A representative example of PDX model evolution is shown in Fig. 1c.
We found that large (>5Mb) CNAs arose in PDXs rapidly: 60% of the PDX models acquired at least one large chromosomal aberration within a single in vivo passage, and 88% acquired at least one large aberration within four passages (Supplementary Fig. 3a). The CNA landscape of PDX models thus gradually shifted away from that of the original primary tumors, with a median of 12.3% of the genome (range, 0% to 58.8%) affected by model-acquired CNAs within four passages (Fig. 1d). Of note, similar results were obtained using three different definitions of CNA prevalence: the proportion of the genome affected by CNAs (CNA fraction), the number of discrete events, or the proportion of altered genes (Fig. 1d and Supplementary Fig. 3b–c), thereby highlighting the robustness of this finding.
There was no significant change in the overall number of CNAs throughout passaging (Supplementary Fig. 3d), indicating equal rates of emergence of new events and disappearance of existing ones. A median of 35.6% of the genome was affected by CNAs, consistent with prior estimates in primary tumors27 (Fig. 1e and Supplementary Fig. 3c). The disappearance of CNAs during passaging was not due to changes in tumor sample purity (for instance, contamination with mouse tissue might dilute the CNA signal), as other primary events were readily detected at similar signal strength. Importantly, approximately one out of six large CNAs identified in PDX models at P4 was not observed in the respective primary tumor. A similar proportion of primary clonal CNAs could no longer be detected in PDXs by P4. This observed tumor evolution was not limited to large CNAs, but also affected mutations in cancer-related genes (Supplementary Fig. 4 and Supplementary Note). We conclude that individual PDX models can quickly genomically diverge from their parental primary tumors.
Selection of pre-existing subclones underlies CNA dynamics
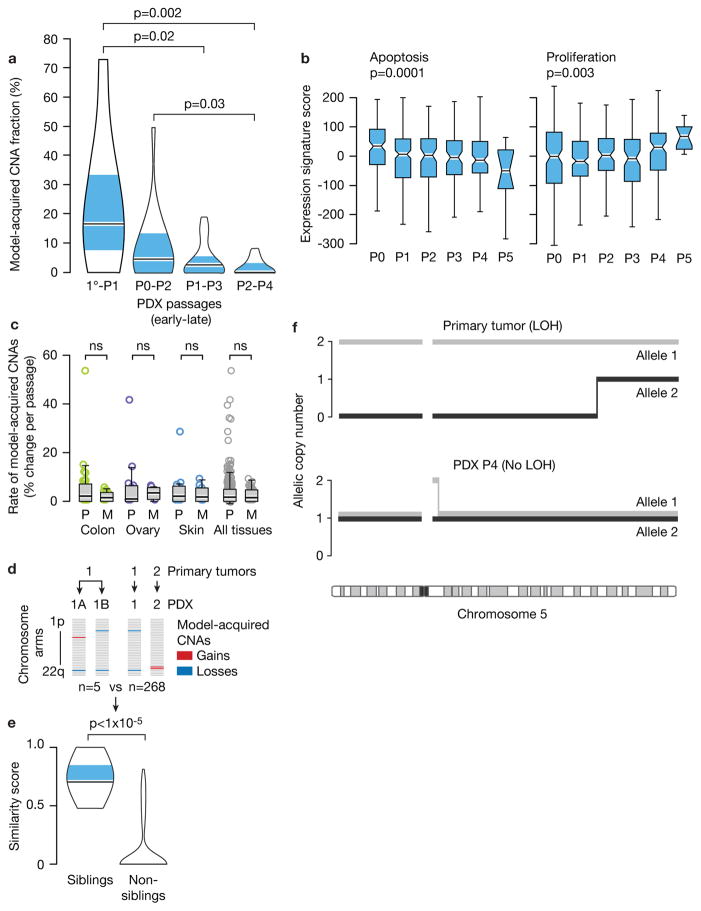
Our observation that CNAs were often gained or lost during PDX passage might be explained by expansion of pre-existing subclones, the acquisition and expansion of de novo events, or a combination of both. Several lines of evidence suggest that clonal selection of pre-existing subclones plays a major role in shaping the CNA landscape of PDXs. First, CNAs accumulated with each passage, but their acquisition rate decreased over time (Fig. 2a). Second, apoptosis expression signatures gradually decreased, while signatures of proliferation increased, with PDX passage number, in line with clonal selection of fitter clones (Fig. 2b). Third, the rates of model-acquired CNAs were similar in PDXs from primary tumors and from metastases (Fig. 2c), despite metastasis-derived PDXs being more aneuploid and exhibiting higher expression of genes associated with chromosomal instability (Supplementary Fig. 5a–b)28; this suggests that model-acquired CNAs predominantly result from clonal dynamics, rather than from genomic instability.
Figure 2. Selection of pre-existing subclones underlies CNA dynamics.
(a). The rate of model-acquired CNAs decreases with PDX passaging. Violin plots present the fraction of CNAs acquired within two in vivo passages as a function of passage number. P-value indicates significance from a Wilcoxon rank-sum test. 1°, primary tumor. (b) Apoptosis decreases and proliferation increases with PDX passaging. Box plots present the apoptosis (left panel) and proliferation (right panel) gene expression signature scores as a function of passage number. P-values indicate significance from a Kruskal-Wallis test. (c) Similar CNA acquisition rates in PDXs from primary tumors and from metastases. Box plots present the rate of model-acquired CNAs as a function of tumor source (P=primary, M=metastasis), across three available tissue types. n.s., non-significant (Wilcoxon rank-sum test). (d) Schematics showing the calculation of pair-wise similarity scores for PDX models coming from the same primary tumor but propagated independently in the mouse (“sibling” PDXs; n=5) and for PDX models coming from distinct primary tumors (“non-sibling” PDXs; n=268). (e) “Sibling” PDXs tend to acquire more similar aberrations than lineage-matched “non-sibling” PDXs. Violin plots present the similarity scores of “sibling” and “non-sibling” PDXs. P-value indicates significance from a lineage-controlled permutation test. (f) Alleles that seem to have been lost in primary tumors can “re-appear” in PDXs, demonstrating expansion of rare pre-existing subclones throughout PDX propagation. Plots present the copy number of both of chromosome 5 alleles in a primary tumor and its derived PDX. Loss of heterozygosity (LOH) is identified in the primary tumor along most of chromosome 5, but both alleles are detected in a 1:1 ratio in the PDX derived from that primary tumor.
If our hypothesis that acquired CNAs were the result of positive biological selection of existing minor subclones is correct, one would expect that the same minor clones would be enriched in multiple independent grafts of the same tumor (i.e., transplanted into “sibling” P0 mice). Five such PDX pairs (representing breast, lung, pancreas and skin cancer PDXs) were available for analysis (Fig. 2d). The similarity in model-acquired CNAs between “sibling” PDXs was indeed significantly higher than the similarity between lineage-controlled “non-sibling” PDXs (p<1E-5; Fig. 2e). This finding suggests that clonal evolution occurs through directional selection of pre-existing subclones, consistent with observations in breast and hematopoietic cancers7,29.
To further test this, we analyzed loss of heterozygosity (LOH) events. Because LOH is an irreversible event at the cellular level, an LOH “reversion” at the population level can only be explained by expansion of cells that did not undergo LOH in the first place. We queried previously published whole-genome sequencing data from 15 breast cancer pairs of primary tumors and PDXs7, and identified five cases of LOH “reversion” (Fig. 2f and Supplementary Fig. 6). These analyses thus confirm that rare pre-existing subclones not readily detected in the primary tumor, can expand and become the dominant clone in PDXs.
We conclude that CNA dynamics are strongest during engraftment and the first few in vivo passages, continue at a reduced rate throughout model propagation, and result primarily from selection of pre-existing subclones.
The degree of genomic instability in PDXs mirrors that of primary tumors
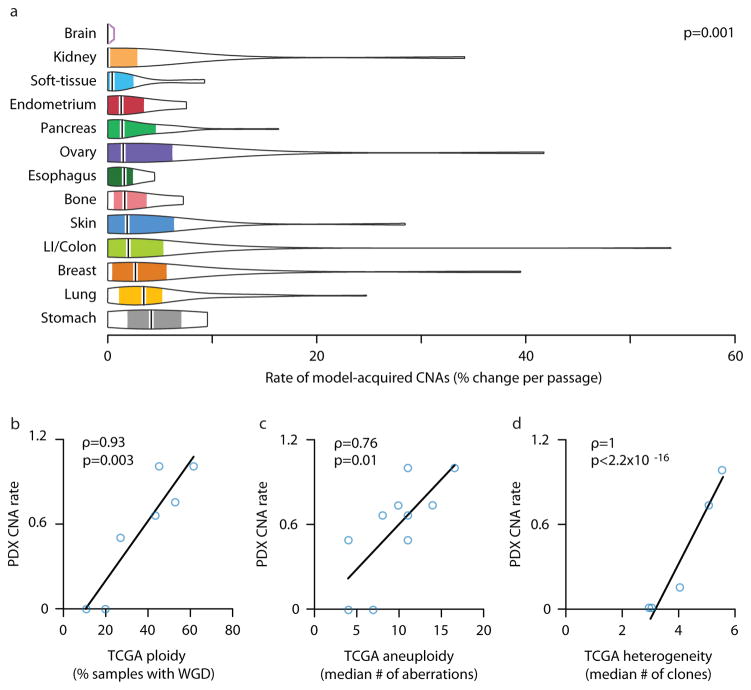
As human cancer types differ considerably in their CNA prevalence and acquisition rate (also referred to as degree of genomic instability, or DGI), we next compared CNA dynamics in PDXs across cancer types. We found that the rate of model-acquired CNAs varies significantly (p=0.001, comparing the most stable to the most unstable tumor types), with brain tumor PDXs being the most stable and gastric tumors being the most unstable (a median of 0% and 4.2% of the genome affected by CNAs per passage, respectively) (Fig. 3a).
Figure 3. Genomic instability of PDXs mirrors that of primary tumors.
(a) The degree of genomic instability (DGI) of PDXs is cancer type-specific. Violin plots present the rate of CNA acquisition throughout PDX propagation of 13 cancer types, for which data were available from at least three PDX models. P-value indicates significance from a Wilcoxon rank-sum test. (b) The DGI of PDXs and that of primary tumors correlate extremely well. In PDXs, tissue DGI was defined as the median number of CNAs per passage. In TCGA tumors, tissue DGI was defined as the fraction of samples with whole-genome duplication (WGD). (c) This correlation holds when the tissue DGI is defined, both for PDXs and for TCGA tumors, by the median number of arm-level CNAs. (d) The DGI of PDXs also correlates extremely well with intra-tumor heterogeneity (ITH) of primary tumors (excluding the skin tissue). The DGI of PDXs was defined as the median number of arm-level CNAs per passage. The heterogeneity of primary tumors was defined as the median number of clones per tumor. Spearman’s rho values and p-values indicate the strength and significance of the correlations, respectively.
We therefore asked whether this spectrum of PDX aneuploidy reflected the aneuploidy levels of human cancer types. We measured aneuploidy in TCGA data according to two metrics. First, we used the percentage of samples with whole-genome duplication (WGD)27. Across seven tissues for which data were available from both TCGA and PDX datasets, the CNA acquisition rate in PDXs correlated strongly with WGD prevalence in TCGA samples (Spearman’s rho=093, p=0.003; Fig. 3b and Supplementary Fig. 7a). Second, we found that the median number of PDX-acquired arm-level CNAs and the median number of arm-level events acquired during tumor development in TCGA samples were correlated across 10 different cancer types (Spearman’s rho=0.76, p=0.010; Fig. 3c). We thus conclude that DGI variation among PDX tumors types parallels that of primary tumors.
As we found that expansion of pre-existing subclones played a major role in shaping the CNA landscape of PDXs, we next examined whether the tissue-specific rate of CNA dynamics correlates with the degree of heterogeneity that characterizes each cancer type. Indeed, the CNA acquisition rate in PDXs correlated well with the median number of clones of the respective primary tumor type30, across six cancer types that could be matched (Spearman’s rho=0.82, p=0.044; Supplementary Fig. 7b). Interestingly, melanoma had the highest degree of intra-tumor heterogeneity, but only a moderate level of DGI in PDXs, and was therefore the only cancer type that significantly deviated from the observed correlation; the correlation became even stronger when melanoma was removed from the analysis (Spearman’s rho=1, p<2.2E-16; Fig. 3d and Supplementary Note).
The combined results of these analyses suggest that PDX models have characteristic tissue-specific levels of CNA dynamics that correspond both to the DGI and to the degree of heterogeneity of the respective primary tumor type. As genetic heterogeneity is closely associated with aneuploidy levels and DGI in primary tumors30–32, either of these factors – or both of them together – may explain the observed correlations.
CNA recurrence analysis reveals distinct selection pressures in PDXs vs. primary tumors
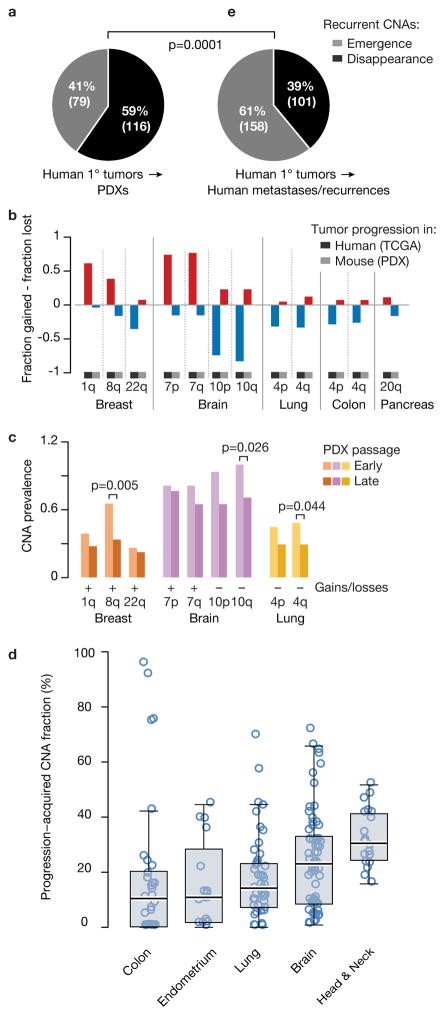
A key question is whether the clonal dynamics observed in PDXs mimic that observed in human patients. To address this, we asked whether recurrent arm-level genetic events that are observed in human tumors remain under selection pressure when transplanted into mice; loss of these signature events would signal significant differences in selection pressures between human and mouse hosts.
We identified 61 recurrent arm-level CNAs across TCGA tumor types, and followed them in PDXs. Surprisingly, events that were recurrent in the TCGA dataset (reflecting positive selection in humans) tended to disappear throughout PDX passaging. Specifically, among lineage-matched PDXs, we observed 116 model-acquired events that were in the opposite direction to the recurrent TCGA CNAs, and only 79 model-acquired events in the same direction (p=0.01; Fig. 4a). We identified twelve recurrent events in TCGA samples that were preferentially lost throughout PDX passaging, across five cancer types (GBM, breast, lung, colon and pancreatic cancer; Fig. 4b and Supplementary Fig. 8). Events that tend to disappear throughout PDX propagation should be less prevalent at high passage compared to low passage PDXs. Indeed, nine of the twelve events that PDXs tend to lose, including the hallmark gains of chr1q and 8q in breast cancer and chr7 in GBM, and the hallmark losses of chr10 in GBM and chr4q in non-small cell lung cancers, were less common at high passage PDXs (Fig. 4c and Supplementary Fig. 9).
Figure 4. Tumor evolution of PDXs diverges from that of primary tumors.
(a) Recurrent arm-level TCGA CNAs tend to disappear throughout PDX passaging. Pie chart presents the number of model-acquired events that were in the opposite direction to the recurrent TCGA CNAs vs. the number of events in the same direction. (b) Opposite propensities to gains and losses in human tumors and PDX models. Bar plots present the fraction difference between gains and losses of 12 recurrent TCGA arm-level CNAs. The PDX fractions represent the model-acquired CNAs, rather than the absolute prevalence of these events. (c) Recurrent TCGA arm-level CNAs are more common in early passage PDXs than in late passage PDXs. Bar plots present the absolute prevalence of each event in the relevant cancer type. P-values indicate significance from a Fisher’s exact test. (d) Evolution of CNA landscapes during tumor progression to advanced disease. Box plots present progression-acquired CNA fraction in the five tumor types analyzed. Bar, median; box, 25th and 75th percentiles; whiskers, data within 1.5*IQR of lower or upper quartile; circles: all data points. (e) Recurrent arm-level TCGA CNAs tend to emerge throughout tumor progression in patients. Pie chart presents the number of progression-acquired events that were in the opposite direction to the recurrent TCGA CNAs vs. the number of events in the same direction. P-value indicates significance from a Chi-squared test.
Together, these data demonstrate that PDXs can lose recurrent chromosomal aberrations that are believed to play causal roles in the development of human tumors. This suggests that selection pressures that led to the acquisition and retention of these hallmark CNAs in patients may no longer exist in the murine model environment.
Distinct CNA dynamics during tumor progression in PDXs and in human patients
In order to further assess whether the clonal dynamics observed in PDXs are indeed fundamentally different from those occurring during tumor evolution in patients, we next analyzed CNA dynamics during the progression of primary tumors into advanced disease (metastases and recurrences). We predicted that if recurrent CNAs tended to disappear in PDXs due to murine-specific selection pressures, this trend should not be found during tumor evolution in humans.
To address this, we compiled CNA data from 306 tumor samples of matched primary tumors (n=132) and their derived metastases or recurrences (n=174), across five cancer types represented in our PDX dataset (colon, lung, endometrial, brain and head and neck)33–40 (Supplementary Table 3). By comparing each metastasis/recurrence to its matched primary tumor, we found that tumor progression in patients is associated with a dramatic shift in CNA landscapes, with a median of 18.2% of the genome (range, 0% to 95.4%) affected by progression-acquired CNAs (Fig. 4d). This change is greater, on average, than the change observed in PDXs (p<1E-5; Fig. 1d), likely reflecting the much longer time periods (often years) between paired resections and the strong treatment-associated selection pressures.
However, in contrast to the disappearance of recurrent CNAs during PDX passaging, the opposite was observed during tumor evolution in patients: recurrent CNAs more often emerged than disappeared during tumor progression. We observed 158 progression-acquired events that were in the same direction to the recurrent TCGA CNAs, and only 101 model-acquired events in the opposite direction (p=0.0005; Fig. 4e). The relative proportion of recurrent CNAs that emerged, compared to those that disappeared, during tumor evolution was significantly different between PDXs and advanced human disease (p=0.0001; Fig. 4a and Fig. 4e). Therefore, these data further demonstrate that distinct selection pressures shape the CNA landscapes of tumors during their evolution in human and in murine hosts.
Genomic instability of PDXs is comparable to that of cell lines and CLDXs
PDXs are generally considered to reflect primary human tumors more faithfully than cell lines, due to the artificial cell culture environment1,41. However, the immunodeficient, subcutaneous murine microenvironment also differs considerably from the natural human host. To address the assumption that PDXs better preserve the fidelity of human tumors, we compared CNA dynamics of PDXs in vivo to those of cell lines in vitro.
We found that the prevalence of model-acquired CNAs in newly-derived cell lines is similar to that in PDXs. We analyzed the CNA landscapes of 38 samples of nine new cell lines derived in our lab from five cancer types (colon, GBM, pancreas, esophagus and thyroid; Supplementary Table 4). Similar to our observations with PDXs, newly-derived cell lines acquired CNAs with passaging, and their CNA landscape gradually shifted away from that of the earliest passage (Supplementary Fig. 10a). As seen in PDXs, changes occurred mostly during the first few passages, and the rate of model-acquired CNAs decreased throughout culture propagation (Fig. 5a). Notably, while CNA rates (defined as the fraction of the genome affected by model-acquired CNAs per passage) varied considerably among cell lines, they fell well within the range seen in PDXs, in a lineage-matched comparison (p=0.55; Fig. 5b). These results were recapitulated with newly-derived cell lines from three independent studies of GBM42,43, kidney44 and head and neck cancer22 (n=31; Supplementary Fig. 10b and Supplementary Table 4), suggesting that clonal selection is not unique to a particular cell line propagation method.
Figure 5. Genomic instability of PDXs is comparable to that of cell lines and CLDXs.
(a) The rate of CNA acquisition decreases with cell line passaging. Box plots present the rate of CNA acquisition as a function of in vitro passage number. P-values indicate significance from a Wilcoxon rank-sum test. (b) Similar rates of CNA acquisition in PDXs and in newly-derived cell lines. Dot plots present the distribution of model-acquired CNA rates across four available cancer types. P-value indicates lack of significance from a lineage-controlled permutation test. (c) Gradual evolution of CNA landscapes throughout CLDX passaging. Box plots present model-acquired CNA fraction as a function of the number of passages between measurements. P-values indicate significance from a Wilcoxon rank-sum test. (d) The CNA acquisition rate of CLDXs is associated with the numerical karyotypic complexity of the parental cell lines. Violin plots present the fraction of CNAs acquired by passage 4 as a function of numerical karyotypic complexity. P-values indicate significance from a Wilcoxon rank-sum test.
Next, we compared CNA dynamics between PDXs and cell line-derived xenografts (CLDXs). To assess CNA dynamics during the in vivo propagation of established cancer cell lines, we turned to the NCI MicroXeno project, which profiled gene expression of 49 human cancer cell lines across multiple in vivo passages45. We used the same computational algorithms24–26 that we applied to the PDX models to infer aneuploidy and CNAs from these gene expression profiles, resulting in 823 copy number profiles (Supplementary Data 3 and 4). We found that CNAs accumulate with in vivo passaging of CLDXs (Fig. 5c), and that the DGI of CLDXs correlates with the karyotypic complexity of their parental cell lines (Fig. 5d and Supplementary Fig. 10c), in line with what we observed in PDXs. However, the rate of CNA acquisition was lower in CLDXs: within four passages, the median model-acquired CNA fraction was 2.2% in CLDXs, compared to 12.3% in PDXs (p=1.6E-6), likely reflecting the reduced heterogeneity of established cell lines compared to primary tumors at the time of xenograft initiation46.
Taken together, our data from three types of cancer models (PDXs, cell lines, and CLDXs) demonstrate that switching the environment in which a model is propagated results in CNA dynamics that gradually alter its CNA landscape. All cancer models are subject to such clonal selection. PDXs do not appear to be spared.
CNA dynamics in PDXs and cell lines may affect drug response
It is conceivable that while PDXs undergo selection in the mouse, such selection is unimportant with respect to modeling therapeutic response. To address this, we turned to a dataset of PDXs with accompanying responses to both genotoxic chemotherapies and targeted therapeutics3.
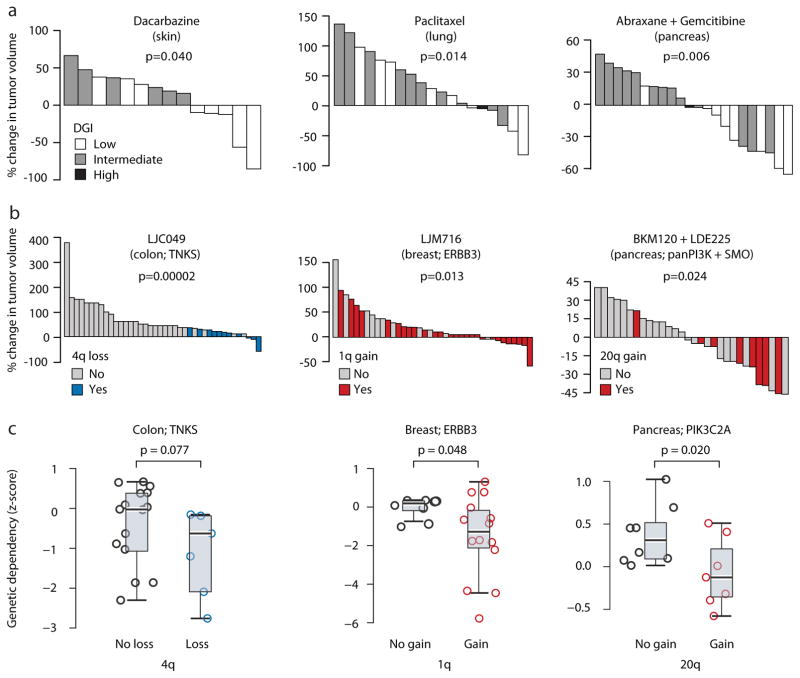
Both very low and very high levels of aneuploidy have been associated with response to genotoxic drugs and improved patient survival28,30,47,48. Importantly, CNA acquisition rate (DGI), rather than absolute levels of aneuploidy, determines sensitivity to further perturbation of chromosome segregation49. We therefore determined the DGI of PDX models, and asked whether it similarly predicts response to chemotherapies. For three of five chemotherapies tested, extreme (either very low or very high) levels of DGI – but not overall aneuploidy levels – were associated with favorable therapeutic response (Fig. 6a): dacarbazine in skin PDXs, paclitaxel in lung PDXs, and abraxane/gemcitabine in pancreas PDXs (p=0.04, 0.014 and 0.006, respectively). The biological activity and clinical efficacy of these drugs were previously linked to chromosomal instability50–54. PDXs thus recapitulate the correlation observed in patients between genomic instability and response to cytotoxic chemotherapies.
Figure 6. CNA dynamics affect PDX drug response.
(a) Extreme levels of genomic instability are associated with better therapeutic response to chemotherapies. Waterfall plots present the response to dacarbazine (n=14), paclitaxel (n=19), and the combination of abraxane and gemcitabine (n=22) in skin, lung and pancreas PDXs, respectively. DGI, degree of genomic instability. P-values indicate significance from a Wilcoxon rank-sum test. (b) The status of recurrent arm-level CNAs is associated with response to targeted therapies. Waterfall plots present the response to the TNKS inhibitor LCJ049 (n=40), the ERBB3 inhibitor LJM716 (n=38), and the combination of the PI3K inhibitor BKM120 and the SMO inhibitor LDE225 (n=31). P-values indicate significance from a Wilcoxon rank-sum test. (c). The status of recurrent arm-level CNAs is associated with the genetic depletion of the genes targeted by the identified drugs. Box plots present the dependency scores to RNAi-mediated knockdown of the indicated genes. Colon cancer cell lines with chromosome 4q loss are more sensitive to knockdown of TNKS, breast cancer cell lines with chromosome 1q gain are more sensitive to knockdown of ERBB3, and pancreatic cancer cell lines with chromosome 20q are more sensitive to knockdown of multiple PI3K genes, including PIK3C2A. P-values indicate significance from a Wilcoxon rank-sum test. -
We next asked whether particular model-acquired CNAs might affect PDX responses to targeted therapies, given that specific recurrent arm-level or whole-chromosome aberrations have been reported to alter the cellular response to certain drugs55–57. We evaluated the association between PDX response to targeted therapies and the presence or absence of individual arm-level CNAs, focusing on the twelve driver CNAs found to be selected against during PDX passaging. We identified three statistically significant drug response-CNA associations (Fig. 6b): chr4 loss was associated with increased response of colon PDXs to the TNKS inhibitor LCJ049 (p=0.005, q=0.04 for 4p loss, and p=0.00002, q=0.0003, for 4q loss); chr20q gain was associated with increased response of pancreatic PDXs to the combination of the PI3K inhibitor BKM120 and the SMO inhibitor LDE225 (p=0.024, q=0.19); and chr1q gain was associated with increased response of breast PDXs to the ERBB3 inhibitor LJM716 (p=0.013, q=0.23). These results indicate that it is not unusual for CNAs (and presumably for other genomic events) that undergo negative selection in the murine host to be associated with changes in sensitivity to specific targeted agents. Such associations may affect the stability of PDX drug response.
To further assess the potential functional relevance of model-acquired chromosomal changes we turned to the Cancer Cell Line Encyclopedia (CCLE) project and its associated datasets of genomic features, genetic dependencies and drug response58–60. For the twelve recurrent CNAs, we compared cell lines with and without the aberration with regard to their gene expression profiles, genetic dependencies and drug sensitivity(controlling for cell lineage). In line with the PDX drug response data, colon cancer cell lines with chr4q loss were more sensitive to knockdown of TNKS (p=0.077), breast cancer cell lines with chr1q gain were more sensitive to knockdown of ERBB3 (p=0.048), and pancreatic cancer cell lines with chr20q gain were more sensitive to knockdown of multiple PI3K genes (p=0.020, p=0.076, p=0.005 and p=0.014 for PIK3C2A, PIK3CD, PIK3CG and PIK3R2, respectively; Fig. 6c and Supplementary Fig. 11a). The analysis of cell lines also revealed that arm-level CNAs were commonly associated with a significant up- or down-regulation of genes that reside within the aberrant arm, and that these changes were significantly associated with cell line genetic and pharmacologic dependencies (Supplementary Figures 11 and 12, Supplementary Tables 5 and 6, and Supplementary Note). We conclude that recurrent arm-level CNAs are associated with drug response, and their gradual disappearance throughout PDX propagation may therefore be functionally important.
Discussion
The ability to directly transfer human tumors into mice, and propagate them for multiple passages in vivo, offers unique opportunities for cancer research and drug discovery, making PDXs a valuable cancer model. Like any other model system, however, understanding its limitations – and the ways in which it differs from human tumors in their natural environment – is required for optimal application. Our findings suggest that the genomic instability of PDXs has been underappreciated: the CNA landscapes of PDXs change continuously, and so their propagation distances them from the primary tumors from which they were derived (Supplementary note). Indeed, the comparison of PDXs to newly-derived cell lines revealed that PDXs do not necessarily capture the genomic landscape of primary tumors better than cell lines, in contrast to common belief1 (Supplementary Note). The similar CNA rate suggests that multiple cell line models from a single primary tumor may capture more of the original genomic landscape – and its heterogeneity – than a single PDX model. Moreover, the acquisition of genetic alterations throughout model propagation is unlikely to be restricted to CNAs (Supplementary Fig. 13 and Supplementary Note).
As our analysis was based on bulk-population measurements, the cellular origin of each model-acquired event could not be definitively determined. Our study strongly suggests that pre-existing alterations play a major role in model-acquired CNAs, especially at the early stages of PDX derivation and propagation, but that de novo events also contribute to genomic instability (Supplementary Fig. 14 and Supplementary Note). Regardless of their origin, we found that CNAs often became fixed in the population quickly; a single in vivo passage sometimes rendered an undetected chromosomal aberration readily detectable at the population level. Such strong clonal dynamics suggest that distinct selection pressures between patients and animal models result in divergent tumor evolution trajectories (Supplementary Note).
Recent genomic analyses revealed that metastases evolve independently from primary tumors, often representing common ancestral subclones that are not detected in individual biopsies of the primary tumor. In contrast to the considerable heterogeneity between primary tumors and metastases, distinct metastatic sites tend to be relatively homogeneous33,61,62. Our findings from PDXs echo those from metastasis. The dominant clones in PDXs often come from minor subclones of the primary tumor, and PDXs that originate from the same primary tumor (the equivalence of multiple metastatic sites) tend to evolve in similar trajectories. It has been suggested that caution is required when inferring the genetic composition of metastatic disease from a primary tumor biopsy, and vice versa33,61,62; similarly, we propose that the genetic composition of a PDX tumor may differ from its matched primary tumor, potentially in therapeutically meaningful ways (Supplementary Note). This should be considered when using PDXs as avatar models for personalized medicine, or to identify biomarkers of drug sensitivity.
Online Methods
PDX data assembly and processing
CGH array, SNP array and gene expression microarray data were obtained from the GEO (http://www.ncbi.nlm.nih.gov/geo) and EMBL-EBI (http://www.ebi.ac.uk) repositories. RNA sequencing data were obtained from the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/). Accession numbers are provided in Supplementary Table 1. Normalized matrix files were downloaded, and samples were curated manually to identify the cancer tissue type and the PDX passage number. Arrays were analyzed for quality control and outliers were removed. The final database consisted of 1,110 PDX tumor samples, from 543 unique PDX models across 24 cancer types. The analysis was performed in batches, and normal tissue samples included in each study served as internal controls, whenever available. Data were processed using the R statistical software (http://www.r-project.org/). For all platform types, probe sets were organized by their chromosomal location, and log2-transformed values were used. Probe sets without annotated chromosomal location were removed. For gene expression data, all the probe sets of each gene were averaged (as well as their chromosomal location), in order to obtain one intensity value per gene. A threshold expression value was set, and genes with lower expression values were collectively raised to that level: flooring values were 6–7 for the Affymetrix and Illumina platforms, and −0.5 for the Agilent platforms. Probe sets not expressed in >20% of the samples within a batch were removed. The 10% of the probe sets with the most variable expression levels were also excluded, to reduce expression noise.
Generation of CNA profiles
CNA profiles from SNP arrays were generated using the Copy Number Workflow of the Partek Genomics Suite software (http://www.partek.com/pgs), as reported by the original studies. CNA profiles from CGH arrays were generated using the CGH-Explorer software (http://heim.ifi.uio.no/bioinf/Projects/CGHExplorer/), using the program’s piecewise constant fit (PCF) algorithm, with the following set of parameters: Least allowed deviation = 0.3; Least allowed aberration size = 30; Winsorize at quantile = 0.001; Penalty = 12; Threshold = 0.01. CNA profiles from gene expression data were generated using the protocols developed by Ben-David et al 24 and by Fehrman et al26. For all gene expression platforms, the e-karyotyping method was applied24: whenever normal tissue samples were available, the median expression value of each gene across the normal samples was subtracted from the expression value of that gene in the tumor samples, in order to obtain comparative values. These relative gene expression data were then subjected to a CGH-PCF analysis, with the following set of parameters: Least allowed deviation = 0.25; Least allowed aberration size = 30; Winsorize at quantile = 0.001; Penalty = 12; Threshold = 0.01. For Affymetrix gene expression platforms, Human Genome U133A and U133Plus2.0, the functional genomic mRNA profiling (FGMP) method 26 was also applied: gene expression data were corrected for the first 25 previously-identified transcriptional components, and the corrected data were subjected to the same processing steps and CGH-PCF analysis described above, with the following set of parameters: Least allowed deviation = 0.15; Least allowed aberration size = 30; Winsorize at quantile = 0.001; Penalty = 12; Threshold = 0.01. CNA profiles from DNA sequencing were obtained in a processed table form from the publication by Eirew et al.7. CNA profiles from SNP arrays were obtained in a processed form from the publication by Gao et al. 3 and compared to CNA profiles from RNA sequencing of the same PDX models. For visualization purposes, moving average plots were generated using the CGH-Explorer moving average fit tool.
Identification of model-acquired CNAs
To identify CNAs emerging during the generation and propagation of PDXs, 342 PDX models in which data were available from multiple time points were analyzed. These PDX models were compared either to the primary tumors from which they were derived, or to their earliest available passage. For gene expression data, model-acquired CNAs were identified by e-karyotyping. For each probe set, a relative value was obtained by subtracting the early time point value from the late time point value. CGH-PCF analysis was then performed, with the same parameters described above. For visualization purposes, moving average plots were generated using the CGH-Explorer moving average fit tool.
CNA recurrence analysis
For each tissue type, the arm-level CNA recurrence was computed and compared between the PDX dataset and the human patient TCGA dataset (http://cancergenome.nih.gov). Chromosome arm-level events in TCGA samples were called using a novel approach to be described in Taylor, Shih, Ha et al. (manuscript in preparation). Briefly, segments of CNAs identified by ABSOLUTE63 were determined as loss, neutral, and gain relative to each sample’s predicted tumor ploidy. Consecutive segments were iteratively joined such that the combined segment is no less than 80% altered in a given direction (i.e. gain or loss, not both). For every combination of arm/chromosome and direction of alteration within each TCGA tumor type, the start coordinates, end coordinates, and proportion of chromosome arm altered (based on the longest joined segment) were clustered across samples using a 3-dimensional Gaussian Mixture Model. The optimal clustering solution was chosen based on the Bayesian Information Criterion. Clusters whose mean fraction altered in either specific direction was >=80% were considered “aneuploid”, those whose mean fraction altered (in both directions) was <=20% were considered “non-aneuploid”. Chromosome arm-level events in PDX samples were determined using the CNA status of the largest overlapping segment from the e-karyotyping analysis. The comparisons of absolute CNA landscapes were performed using the FGMP-derived CNA profiles, and the comparisons of model-acquired CNAs were performed using the e-karyotyping-derived CNA profiles. The comparisons between early and late passage PDXs were performed using FGMP-derived CNA profiles: samples from p<=1 were defined as early passage, and samples from p>=3 were defined as late passage. Heatmaps were generated using the ‘pheatmap’ R package, and clustering was performed using euclidean distance and complete linkage.
DGI comparison across passages and tissue types
The degree of genomic instability (DGI) of each sample was determined in three ways: 1) the fraction of the genome affected by model-acquired CNAs per passage, 2) the number of discrete events per passage, and 3) the fraction of altered genes per passage. For each cancer type, the median number of model-acquired arm-level CNAs across all PDX samples was determined, and compared to several TCGA statistics: the percentage of samples with whole-genome duplication taken from the publication by Zack et al27, the median number of arm-level CNAs per sample, and the median number of clones per sample taken from the publication by Andor et al30.
Similarity analysis
PDX samples derived from the same primary tumors, but propagated in different animals starting from their initial transplantation (i.e., transplanted into different P0 mice) were defined as “siblings”. PDX samples derived from the same primary tumors, and propagated in the same animal at some point during PDX propagation were excluded from the analysis. PDX samples from distinct primary tumors were defined as “non-siblings”. Similarity scores were calculated for each pair of samples, based on the arm-level events that occurred during their in vivo passaging (i.e., model-acquired CNAs), using a modified Jaccard similarity coefficient. This similarity coefficient was inversely weighted to account for the observed prevalence of each CNA in each PDX tissue type. Therefore, the similarity score was calculated using the following equation: , where , and freq(k) is the frequency of event k in that tumor type.
Loss of heterozygosity analysis
Allelic copy number data were obtained from the publication by Eirew et al7. Using 10Mb windows along the genome, we identified the following scenarios: (1) the minor allele was 0 (LOH) in a primary tumor but > 0 (presence of both alleles) in the tumor-derived PDX model, and (2) the minor allele was 0 (LOH) in an early passage PDX but > 0 in a later passage of the same PDX model. These instances of apparent “reversion” of LOH were visualized using the Integrative Genomics Viewer (IGV; https://www.broadinstitute.org/igv/) and re-plotted in Figure 2f.
Gene expression signature scores
The apoptosis and proliferation gene sets were derived from the Molecular Signature Databse (http://software.broadinstitute.org/gsea/msigdb), using the “Hallmark_Apoptosis”64 and the “Benporath_Proliferation”65 gene sets, respectively. The CIN70 gene set was derived from the publication by Carter et al13. Signature scores were generated for all PDXE models3. For each gene set, genes not expressed at all in the PDX dataset were removed, and the remaining gene expression values were log2-transformed and scaled by subtracting the gene expression means. The signature score was defined as the sum of these scale-normalized gene expression values.
Advanced disease data assembly, processing and CNA profiling
CGH array and SNP array data were obtained from the GEO (http://www.ncbi.nlm.nih.gov/geo) and EMBL-EBI (http://www.ebi.ac.uk) repositories. CNA profiles from DNA sequencing were obtained in a processed table form from the authors of the respective publications. Accession numbers are provided in Supplementary Table 4. The final database consisted of 306 tumor samples across five cancer types. Chromosome arm-level events in advanced disease samples were determined as described above. Progression-acquired CNAs were determined as events that were identified in the primary tumor but not in its advanced disease (metastasis, recurrence or progressed sample), or vice versa.
CLDX data assembly, processing and CNA profiling
Gene expression microarray data from the National Cancer Institute MicroXeno project were downloaded from the GEO repository (http://www.ncbi.nlm.nih.gov/geo), under accession number GSE48433 (ref 45). Data were processed as described above. CNA profiles were generated using the FGMP method, and model-acquired CNAs were identified by e-karyotyping, as described above. The in vitro cultured (P0) cell line gene expression values were used as reference in the e-karyotyping analysis. The numerical karyotypic complexity categorization of the cell lines was obtained from the publication by Roschke et al66.
Cell line data assembly, processing and CNA profiling
Whole-exome sequencing data from 9 newly-derived cell lines were obtained from Tseng et al. (manuscript in preparation). CNA profiles were generated from these data using the ReCapSeg program (http://gatkforums.broadinstitute.org/gatk/categories/recapseg-documentation), from the ratio of tumor read depth to the expected read depth (as determined from a panel of normal samples). Gene expression microarray data were obtained from the GEO repository (http://www.ncbi.nlm.nih.gov/geo). Accession numbers are provided in Supplementary Table 3. Data were processed as described above. CNA profiles were generated using the FGMP method, and model-acquired CNAs were identified by e-karyotyping, as described above. Renal cancer CNA data were obtained directly from the publication by Cifola et al44, and model-acquired CNAs were identified as described above. For the comparison of model-acquired CNA rates across passages, samples were compared to the earliest available passage (p=0 or p=1). Samples from p<=7 were defined as early passage, samples from p=10 were defined as medium passage, and samples from p>=19 were defined as late passage.
PDX drug response association analyses
PDX drug response data were obtained from the publication by Gao et al3. For the analysis of the association between chemotherapy response and absolute levels of aneuploidy, the CNA fraction was determined according to the FGMP-derived CNA profiles of the latest passage sample available from each model. Low CNA levels were determined as CNA fraction<0.3; intermediate CNA levels were determined as 0.3<CNA fraction<0.7; high CNA levels were determined as CNA fraction>0.7. For the analysis of the association between chemotherapy response and the degree of genomic instability (DGI), the DGI level of each model was determined as the number of discrete model-acquired CNAs per passage, using the latest passage sample available from each model: low DGI=0, 0<intermediate DGI<4, high DGI>4. The BestAvgResponse values were used to make response calls. Association tests were conducted in each available tissue type independently, yielding a total of six drug-tissue association tests (representing five chemotherapies in five tissue types). For the analysis of the association between targeted therapy response and the existence of specific arm-level events, the arm-level copy number status of each model was set according to the FGMP-derived CNA profiles of the latest passage sample available from that model. The BestAvgResponse values were used to make response calls. For each of the 12 recurrent TCGA events that tend to disappear throughout PDX passaging, its association with PDX drug response was evaluated in the relevant cancer type. All targeted drugs that were used as single agents, and that showed at least partial response in at least one animal, were evaluated. Drug combinations were also evaluated, if one (or both) of the drugs in the combination was not tested as a single agent. A total of X association tests were performed (representing 15 single agent drugs and five drug combinations in three tissue types).
Cell line gene expression, genetic dependency and drug response association analyses
Chromosome arm-level events in cell line samples were called using the same approach used to call arm-level events in TCGA samples (described above). Gene expression was measured for CCLE cell lines using RNAseq (n = 936 cell lines with arm-level CNV calls), and was normalized to RPKM values for each gene60. Gene essentiality profiles for each cell line were derived from genome-wide RNAi screens across 503 cell lines (n = 446 with arm-level CNA calls), using the DEMETER algorithm to isolate gene knockdown effects from off-target effects60. Drug sensitivity measurements were taken from the Cancer Therapeutics Response Portal (CTRP v2) dataset (downloaded from https://ctd2.nci.nih.gov/dataPortal/). These data represented dose-response curve AUC measures for 887 cell lines (n = 804 with arm-level CNV calls) across 545 compounds. Comparisons of gene expression, RNAi-based gene essentiality, and compound sensitivity, for cell lines with and without particular arm-level CNAs were made using standard linear modeling tools developed for differential expression analysis: the R package limma was used to derive p-values from empirical Bayes moderated t-statistics67. In all cases we tested the effect of arm-level CNVs while controlling for inter-lineage differences by including lineage as a covariate in the model67. Gene set testing was performed using a parametric approximation to permutation-based testing, implemented in the R package npGSEA68. Inter-lineage differences were controlled for by regressing lineage out of both the arm-level CNA calls and the variable of interest68.
Mutation allelic fraction analysis
The allelic fraction (AF) and predicted effects of point mutations were obtained from the publication by Eirew et al7. AF shifts were determined as |AF[Primary]−AF[PDX]|>0.2. Missense and nonsense coding mutations were considered separately from the rest of the mutations, and their AF shifts were plotted in Supplementary Fig. X.
Comparison of CNA-based and mutation-based phylogenetic trees
Copy number and mutation data were obtained from the publications by Gibson et al33 and Bi et al69. To construct SNV-based trees, SNVs present at low allelic fractions, and SNVs from regions of low sequencing coverage, were first excluded. To construct arm-level CNA-based trees, copy number data from the tumors were converted to arm-level calls. Phylogenetic trees were then generated separately. In each case, branch points in the tree were assigned based on the overall similarity in shared events, and branch lengths were set to be proportional to the number of shared events. Patients for which trees could not be generated due to insufficient information (e.g., no branch points identified) were excluded from the analysis. CNA-based trees were then compared to SNV-based trees to determine their sensitivity (the proportion of SNV-based branch points identified in CNA-based trees) and specificity (the proportion of CNA-based branch points identified in SNV-based trees).
Statistical analyses
The significance of the differences in prevalence and rate of absolute CNAs and of model-acquired CNAs between PDX passages, between primary and metastatic PDXs, between P53-WT and P53-mutated/deleted PDXs, between PDXs from the most stable (upper quartile) and least stable (lower quartile) tissue types, between cell line passages, between CLDX passages, and between CLDXs from cell lines of distinct numerical karyotypic complexities, was determined using the two-tailed Wilcoxon rank-sum test. The significance of the difference in similarity scores between “sibling” and “non-sibling”, the significance of the difference in CNA rates between PDXs and cell lines, and the significance of the differences between PDX-acquired and progression-acquired CNA fractions, were determined using a stratified bootstrap test, permuting the data 100,00 times within each tissue type. The significance of the gene expression signature trends observed throughout PDX passaging was determined by the Kruskal-Wallis rank-sum test. The significance of correlations between PDX and TCGA data was determined using a Spearman’s correlation test. To evaluate the tendency to acquire or to lose recurrent TCGA CNAs during PDX propagation and during disease progression in patients, recurrent CNAs were defined for each tissue type as those that recur in over 40% of the samples, and the number of events that involve these CNAs were computed in the lineage-matched PDX cohorts; the significance of the difference between the emergence frequency and the loss frequency in PDXs and in advanced disease was determined using the McNemar’s test, and the significance of the difference between the two groups was determined using a Chi-squared test for equality of proportions (using the proportion observed in humans as the expected proportion). The significance of the difference in CNA prevalence between early and late passage PDX samples was evaluated using the one-tailed Fisher’s exact test. The significance of the association between chromosome arms and drug response was determined using the Wilcoxon rank-sum test, with FDR multiple test correction performed for each tissue type independently. Box plots show the median, 25th and 75th percentiles, lower whiskers show data within 25th percentile −1.5 times the IQR, upper whiskers show data within 75th percentile +1.5 times the IQR, and circles show the actual data points. Violin plots show the combination of a box plot and a kernel density plot, in which the width is proportional to the relative frequency of the measurements. All of the statistical tests were performed, using the R statistical software (http://www.r-project.org/), and the box plots and violin plots were generated using the ‘boxplot’ and ‘vioplot’ R packages, respectively.
Code availability
The codes used to generate and/or analyze the data during the current study are publically available, or available from the authors upon request.
Data availability
The datasets generated during and/or analyzed during the current study are available within the article, its supplementary information files, or available from the authors upon request.
Supplementary Material
Acknowledgments
The authors thank Lude Franke for assistance with functional genomic mRNA profiling; Adam Bass, Keith Ligon, Andrew J. Aguirre and Jochen Lorch for providing the clinical samples for cell line derivation; Andrew Tubelli for assistance with figure preparation; Matthew Meyerson, Andrew J. Cherniack, Alison Taylor, Alexander Pearson and Zuzana Tothova for helpful discussions; William J. Gibson for copy number data. U.B.-D. is supported by a Human Frontiers Science Program postdoctoral fellowship, R.B received support from the NIH (R01 CA188228) and the Gray Matters Brain Cancer Foundation, and T.R.G. received support from the Howard Hughes Medical Institute.
Footnotes
Contributions
U.B.-D. conceived the project, collected the data and carried out the analyses; G.H., N.F.G. and J.M.M. assisted with computational analyses; Y.-Y.T. and J.S.B. provided cell line data. C.O. assisted with the copy number analysis of cell lines. B.W. assisted with figure design and preparation. J.S.B. assisted with the copy number analysis of TCGA samples. R.B. and T.R.G. directed the project. U.B.-D., R.B. and T.R.G. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Tentler JJ, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews. Clinical Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao H, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgson JG, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11:477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruna A, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016 doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kresse SH, Meza-Zepeda LA, Machado I, Llombart-Bosch A, Myklebost O. Preclinical xenograft models of human sarcoma show nonrandom loss of aberrations. Cancer. 2012;118:558–570. doi: 10.1002/cncr.26276. [DOI] [PubMed] [Google Scholar]
- 10.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin D, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–1283. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 12.Huynh H, et al. Bevacizumab and rapamycin induce growth suppression in mouse models of hepatocellular carcinoma. J Hepatol. 2008;49:52–60. doi: 10.1016/j.jhep.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, et al. SRRM4 Expression and the Loss of REST Activity May Promote the Emergence of the Neuroendocrine Phenotype in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2015;21:4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakes SR, et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci U S A. 2012;109:2766–2771. doi: 10.1073/pnas.1104778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager W, et al. Patient-derived bladder cancer xenografts in the preclinical development of novel targeted therapies. Oncotarget. 2015;6:21522–21532. doi: 10.18632/oncotarget.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monsma DJ, et al. Using a rhabdomyosarcoma patient-derived xenograft to examine precision medicine approaches and model acquired resistance. Pediatr Blood Cancer. 2014;61:1570–1577. doi: 10.1002/pbc.25039. [DOI] [PubMed] [Google Scholar]
- 17.Wong NC, et al. Stability of gene expression and epigenetic profiles highlights the utility of patient-derived paediatric acute lymphoblastic leukaemia xenografts for investigating molecular mechanisms of drug resistance. BMC Genomics. 2014;15:416. doi: 10.1186/1471-2164-15-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu Q, et al. Genomic characterization of a large panel of patient-derived hepatocellular carcinoma xenograft tumor models for preclinical development. Oncotarget. 2015;6:20160–20176. doi: 10.18632/oncotarget.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KB, et al. Novel dedifferentiated liposarcoma xenograft models reveal PTEN down-regulation as a malignant signature and response to PI3K pathway inhibition. Am J Pathol. 2013;182:1400–1411. doi: 10.1016/j.ajpath.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma CX, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–1552. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morisot S, et al. High frequencies of leukemia stem cells in poor-outcome childhood precursor-B acute lymphoblastic leukemias. Leukemia. 2010;24:1859–1866. doi: 10.1038/leu.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng S, et al. Tumor grafts derived from patients with head and neck squamous carcinoma authentically maintain the molecular and histologic characteristics of human cancers. J Transl Med. 2013;11:198. doi: 10.1186/1479-5876-11-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou J, et al. Phenotypic and transcriptional fidelity of patient-derived colon cancer xenografts in immune-deficient mice. PLoS One. 2013;8:e79874. doi: 10.1371/journal.pone.0079874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-David U, Mayshar Y, Benvenisty N. Virtual karyotyping of pluripotent stem cells on the basis of their global gene expression profiles. Nat Protoc. 2013;8:989–997. doi: 10.1038/nprot.2013.051. [DOI] [PubMed] [Google Scholar]
- 25.Ben-David U, et al. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat Commun. 2016;7:12160. doi: 10.1038/ncomms12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehrmann RS, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 27.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 29.Clappier E, et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med. 2011;208:653–661. doi: 10.1084/jem.20110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakker B, et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016;17:115. doi: 10.1186/s13059-016-0971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson WJ, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–855. doi: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TM, et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Resh. 2015;21:4461–4472. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 35.Bai H, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedberg ML, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126:1606. doi: 10.1172/JCI86862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Um SW, et al. Molecular Evolution Patterns in Metastatic Lymph Nodes Reflect the Differential Treatment Response of Advanced Primary Lung Cancer. Cancer Res. 2016;76:6568–6576. doi: 10.1158/0008-5472.CAN-16-0873. [DOI] [PubMed] [Google Scholar]
- 38.Sveen A, et al. Intra-patient Inter-metastatic Genetic Heterogeneity in Colorectal Cancer as a Key Determinant of Survival after Curative Liver Resection. PLoS Genet. 2016;12:e1006225. doi: 10.1371/journal.pgen.1006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SY, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS ONE. 2014;9:e90459. doi: 10.1371/journal.pone.0090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim B, et al. Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels. Oncotarget. 2015;6:22179–22190. doi: 10.18632/oncotarget.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel VC, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunther HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 43.Schulte A, et al. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia. 2011;59:590–602. doi: 10.1002/glia.21127. [DOI] [PubMed] [Google Scholar]
- 44.Cifola I, et al. Renal cell carcinoma primary cultures maintain genomic and phenotypic profile of parental tumor tissues. BMC Cancer. 2011;11:244. doi: 10.1186/1471-2407-11-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollingshead MG, et al. Gene expression profiling of 49 human tumor xenografts from in vitro culture through multiple in vivo passages--strategies for data mining in support of therapeutic studies. BMC Genomics. 2014;15:393. doi: 10.1186/1471-2164-15-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silk AD, et al. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci U S A. 2013;110:E4134–4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zasadil LM, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med. 2014;6:229ra243. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burrell RA, et al. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem. 2010;111:782–790. doi: 10.1002/jcb.22781. [DOI] [PubMed] [Google Scholar]
- 53.Tempest HG, et al. Sperm aneuploidy frequencies analysed before and after chemotherapy in testicular cancer and Hodgkin’s lymphoma patients. Hum Reprod. 2008;23:251–258. doi: 10.1093/humrep/dem389. [DOI] [PubMed] [Google Scholar]
- 54.Khan F, Sherwani AF, Afzal M. Analysis of genotoxic damage induced by dacarbazine: an in vitro study. Toxin Rev. 2010;29:130–136. doi: 10.3109/15569543.2010.516463. [DOI] [Google Scholar]
- 55.Ben-David U, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- 56.Cai Y, et al. Loss of Chromosome 8p Governs Tumor Progression and Drug Response by Altering Lipid Metabolism. Cancer Cell. 2016;29:751–766. doi: 10.1016/j.ccell.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Chen G, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–784. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rees MG, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsherniak A, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576 e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brastianos PK, et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roschke AV, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 67.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larson JL, Owen AB. Moment based gene set tests. BMC Bioinformatics. 2015;16:132. doi: 10.1186/s12859-015-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bi WL, et al. Genomic landscape of high-grade meningiomas. npj Genom Med. 2017;2 doi: 10.1038/s41525-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available within the article, its supplementary information files, or available from the authors upon request.