ABSTRACT
Elevated lipid metabolism promotes cancer cell proliferation. Clear cell renal cell carcinoma (ccRCC) is the most common subtype of kidney cancers, characterized by ectopic lipid accumulation. However, the relationship between aberrant lipid metabolism and tumorigenesis in ccRCC is not thoroughly understood. Here, we demonstrate that ring finger protein 20 (RNF20) acts as a tumor suppressor in ccRCC. RNF20 overexpression repressed lipogenesis and cell proliferation by inhibiting sterol regulatory element-binding protein 1c (SREBP1c), and SREBP1 suppression, either by knockdown or by the pharmacological inhibitor betulin, attenuated proliferation and cell cycle progression in ccRCC cells. Notably, SREBP1c regulates cell cycle progression by inducing the expression of pituitary tumor-transforming gene 1 (PTTG1), a novel target gene of SREBP1c. Furthermore, RNF20 overexpression reduced tumor growth and lipid storage in xenografts. In ccRCC patients, RNF20 downregulation and SREBP1 activation are markers of poor prognosis. Therefore, RNF20 suppresses tumorigenesis in ccRCC by inhibiting the SREBP1c-PTTG1 axis.
KEYWORDS: RNF20, SREBP1c, PTTG1, ccRCC, tumorigenesis
INTRODUCTION
During excessive proliferation of cancer cells, increased demands for building blocks and membrane biogenesis are satisfied by complex metabolic reprogramming (1, 2). In cancer cells, most lipid metabolites are derived from de novo lipogenesis rather than from extracellular lipid uptake (3, 4). It has been reported that lipogenesis and lipid accumulation are upregulated in various cancers (5, 6). Particularly, ectopic lipid accumulation is a hallmark of clear cell renal cell carcinoma (ccRCC), which is the most common subtype of kidney cancers (7, 8). According to current knowledge, the primary etiology of ccRCC involves the loss or inactivation of the von Hippel-Lindau tumor suppressor gene (VHL) and the consequent activation of hypoxia-inducible factor (HIF) (9, 10).
Ring finger protein 20 (RNF20) is an E3 ubiquitin ligase that plays various roles in transcription regulation, DNA damage responses, stem cell differentiation, and lipid metabolism (11–13). It promotes the monoubiquitination of histone H2B, which regulates the transcription of a subset of genes and contributes to chromatin remodeling (13, 14). RNF20 has been suggested to act as a tumor suppressor in inflammation-associated cancers (15).
Recently, we demonstrated that hepatic RNF20 is an E3 ubiquitin ligase for sterol regulatory element-binding protein 1c (SREBP1c), a key transcription factor in de novo lipogenesis (16, 17). RNF20 promotes the polyubiquitination and degradation of SREBP1c upon protein kinase A activation, thereby suppressing hepatic lipid metabolism (11). In mammals, SREBP1a and SREBP1c are encoded by the SREBF1 gene, whereas SREBP2 is encoded by SREBF2 (18, 19). SREBP1 primarily stimulates the expression of genes involved in fatty acid synthesis, including those encoding fatty acid synthase (FASN), stearoyl coenzyme A desaturase 1 (SCD1), and long-chain fatty acid elongase (ELOVL6), whereas SREBP2 promotes mainly cholesterol homeostasis (20, 21). It has been reported that activated SREBP1c upregulates lipogenic genes and enhances lipogenesis in certain cancers such as glioblastoma, prostate, and colon cancers (22, 23). In addition, SREBP1c-mediated lipogenic activation promotes tumor development, progression, and migration, leading to poor prognosis in several cancers (24, 25). Although it is well known that lipid metabolic pathways are dysregulated in ccRCC (26, 27), the underlying mechanisms that lead to aberrant lipogenesis and cell proliferation in ccRCC remain poorly understood.
In this study, we observed that RNF20 downregulation stimulates SREBP1c-mediated lipogenesis and cell proliferation in ccRCC. RNF20 suppressed ccRCC tumorigenesis by inhibiting SREBP1c, whereas genetic and pharmacological inhibition of SREBP1 decreased lipogenesis and proliferation in ccRCC cells. Furthermore, downregulation of RNF20 in ccRCC cells augmented cell cycle progression by activating SREBP1c-mediated pituitary tumor-transforming gene 1 (PTTG1). Intriguingly, in patients with ccRCC, RNF20 was downregulated, which was closely associated with SREBP1c activation and poor prognosis. Together, these findings suggest that RNF20 functions as a tumor suppressor in ccRCC by attenuating SREBP1c-dependent lipogenesis and PTTG1 signaling.
RESULTS
RNF20 is downregulated in ccRCC.
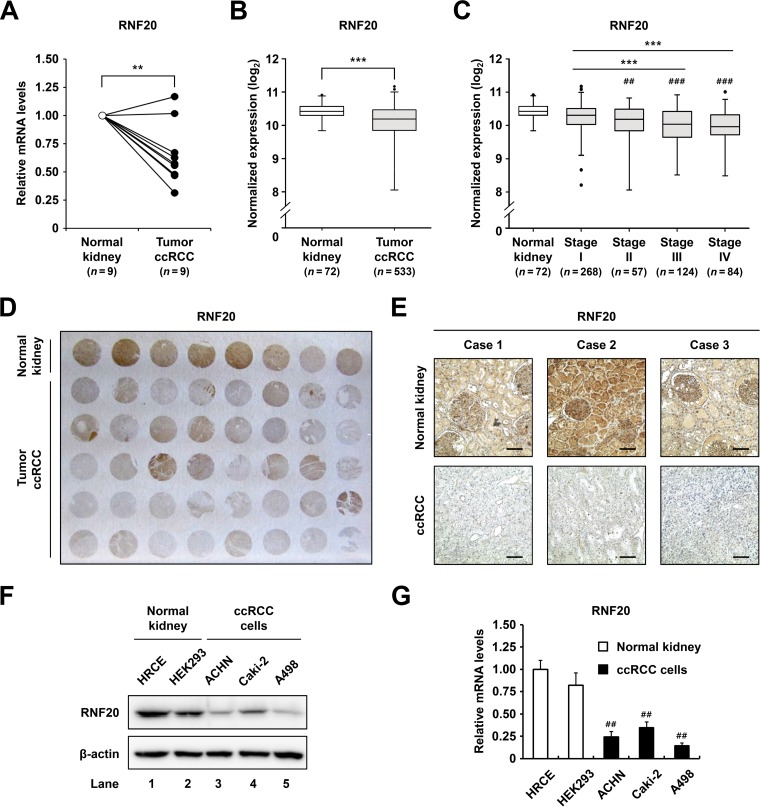
Ectopic lipid accumulation is profoundly upregulated in ccRCC (7, 8). Since RNF20 acts as a negative regulator of de novo lipogenesis by inhibiting SREBP1c (11), we investigated whether RNF20 might be dysregulated in ccRCC tumors. As shown in Fig. 1A, RNF20 mRNA expression was downregulated in ccRCC tumors compared to that in patient-matched normal kidney tissues. Similarly, RNA sequencing (RNA-seq) data from the Cancer Genome Atlas (TCGA) revealed significant reductions in RNF20 mRNA expression in ccRCC tumors (Fig. 1B) and indicated that low RNF20 expression is closely correlated with advanced tumor stages (Fig. 1C). Furthermore, immunohistochemistry (IHC) assays showed that RNF20 protein expression was lower in ccRCC tumors than in adjacent normal kidney tissues (Fig. 1D). In agreement, RNF20 staining data from patient-matched normal kidney and ccRCC tumor tissues revealed decreased RNF20 expression in the tumors (Fig. 1E). Moreover, protein and mRNA expression of RNF20 was lower in ccRCC cell lines A498, Caki-2, and ACHN than in human primary renal cortical epithelial (HRCE) and HEK293 normal kidney cell lines (Fig. 1F and G).
FIG 1.
RNF20 is downregulated in ccRCC. (A) qRT-PCR analysis of RNF20 mRNA expression in patient-matched normal kidney (n = 9) and ccRCC tumor (n = 9) samples. RNF20 mRNA levels were normalized to those in matched normal kidney samples. (B) Normalized RNA-seq reads of RNF20 in normal kidney (n = 72) and ccRCC tumor (n = 533) samples. RNA-seq data were obtained from TCGA. (C) RNF20 expression in ccRCC tumors was analyzed according to tumor stages. Significance versus normal kidney samples: ##, P < 0.01; ###, P < 0.001. (D) Representative ccRCC tissue microarray used for IHC with RNF20 antibody. (E) IHC staining of patient-matched adjacent normal kidney and ccRCC tumor tissues. Representative tissue sections stained for RNF20 are shown. Bar, 100 μm. (F) RNF20 protein expression levels in normal kidney cell lines, such as HRCE and HEK293, and ccRCC cell lines, including ACHN, A498, and Caki-2, were determined by Western blotting. (G) RNF20 mRNA expression in normal kidney and ccRCC cell lines was determined by qRT-PCR. Significance versus HRCE: ##, P < 0.01.
RNF20 suppresses ccRCC cell proliferation.
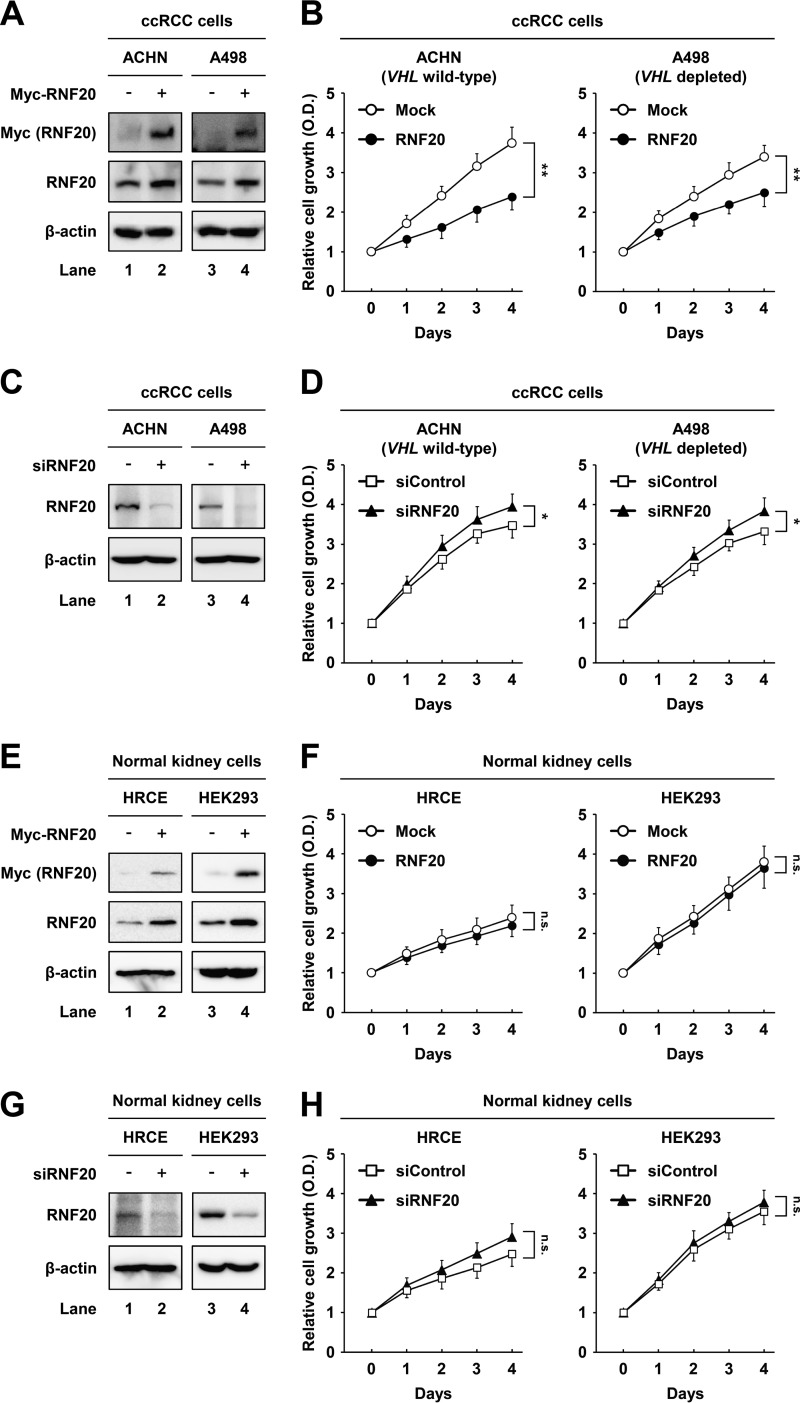
To evaluate the tumorigenic consequences of low RNF20 expression, we determined whether RNF20 affects proliferation in wild-type-VHL ACHN and VHL-depleted A498 ccRCC cells. In these experiments, RNF20 overexpression suppressed cell proliferation in both ACHN and A498 ccRCC cells (Fig. 2A and B). Conversely, small interfering RNA (siRNA)-mediated suppression of RNF20 increased cell growth in both ccRCC cell lines (Fig. 2C and D). In contrast, neither overexpression nor knockdown of RNF20 affected the growth of HRCE and HEK293 normal kidney cells, which express high levels of RNF20 (Fig. 2E to H). These data suggest that RNF20 suppresses ccRCC cell proliferation independently of VHL mutation status.
FIG 2.
RNF20 suppresses cell growth in ccRCC but not in normal kidney cell lines. (A) ACHN and A498 ccRCC cells were infected with adenovirus expressing GFP alone (−) or Myc-RNF20(+). After infection for 24 h, total cell lysates were subjected to Western blotting. (B) ACHN and A498 ccRCC cells were infected with adenovirus expressing GFP alone (Mock) or RNF20, and proliferation was monitored using the Cell Counting Kit-8 (CCK-8) assay. (C) ACHN and A498 ccRCC cells were transfected with siControl or siRNF20, and RNF20 expression was determined by Western blotting. (D) ACHN and A498 ccRCC cells were transfected with siControl or siRNF20, and relative growth rates were determined using the CCK-8 assay. (E) HRCE and HEK293 normal kidney cells were infected with adenovirus expressing GFP alone (−) or Myc-RNF20(+), and cell lysates were examined using Western blotting. (F) HRCE and HEK293 cells were infected with adenoviral RNF20, and cell proliferation was monitored using the CCK-8 assay. (G) HRCE and HEK293 cells were transfected with siControl or siRNF20, and cell lysates were determined by Western blotting. (H) HRCE and HEK293 cells were transfected with siRNF20, and cell proliferation rates were monitored using the CCK-8 assay. Cell proliferation data are presented as the means ± SD from five individual samples. *, P < 0.05; **, P < 0.01; n.s., not significant.
RNF20 decreases lipogenesis and cell proliferation by downregulating SREBP1c in ccRCC cells.
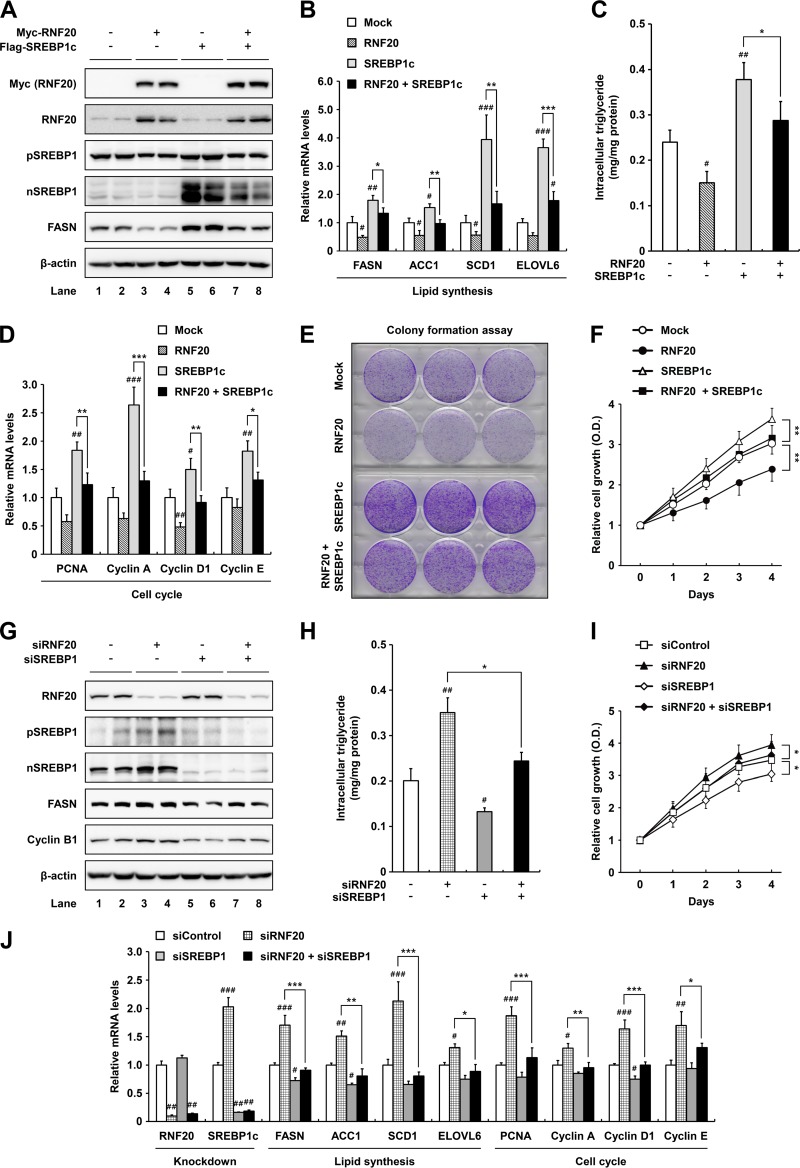
To examine the effect of RNF20 on lipid metabolism and proliferation in ccRCC cells, the level of RNF20 was modulated by either overexpression or suppression. In wild-type-VHL ACHN ccRCC cells, ectopic expression of RNF20 downregulated both precursor and nuclear SREBP1 proteins (Fig. 3A). In agreement, SREBP1c overexpression augmented mRNA levels of lipogenic genes, including FASN, ACC1, SCD1, and ELOVL6 in ACHN cells (Fig. 3B), in accordance with previous reports (20, 21). In contrast, RNF20 potently inhibited mRNA expression of lipogenic genes in both control and SREBP1c-overexpressing ACHN cells (Fig. 3B). Accordingly, intracellular triglyceride accumulation was greater in SREBP1c-overexpressing than in control ACHN cells, whereas it was lowered by RNF20 overexpression (Fig. 3C). It has been reported that loss of SREBP1 expression markedly decreases glioma cell proliferation through cell cycle regulation (25, 28). In accordance with these data, SREBP1c overexpression promoted the mRNA expression of cell cycle regulators, including PCNA, cyclin A, D1, and E in ccRCC cells (Fig. 3D), and concomitant RNF20 overexpression reduced the effects of overexpressed SREBP1c (Fig. 3D). Moreover, RNF20 overexpression reduced colony formation and cell growth whereas ectopic SREBP1c expression elevated cell proliferation in ACHN cells (Fig. 3E and F). Suppression of RNF20 via siRNA enhanced SREBP1 expression, intracellular triglyceride levels, and cell proliferation, accompanied with upregulated expression of lipogenic and cell cycle genes (Fig. 3G to J). However, double knockdowns of both RNF20 and SREBP1 abolished these effects (Fig. 3H to J). These data suggest that RNF20 inhibits ccRCC cell proliferation by suppressing SREBP1c-induced lipogenesis and cell cycle progression.
FIG 3.
RNF20 represses ccRCC cell proliferation by inhibiting SREBP1. (A) ACHN ccRCC cells were infected with adenovirus containing Myc-RNF20 and/or Flag-SREBP1c. After infection, total cell lysates were subjected to SDS-PAGE followed by Western blotting. pSREBP1, precursor SREBP1; nSREBP1, nuclear SREBP1. (B) ACHN ccRCC cells were transduced with lentivirus for stable overexpression of RNF20 and/or SREBP1c. Relative mRNA levels were determined by qRT-PCR. (C) Intracellular triglyceride contents were measured in lentiviral RNF20- and/or SREBP1c-overexpressing ACHN cells. (D) RNF20- and/or SREBP1c-overexpressing ACHN ccRCC cells were subjected to qRT-PCR. (E) ACHN ccRCC cells were transduced with RNF20 and/or SREBP1c lentivirus, and their ability to form colonies was determined by crystal violet staining. (F) ACHN ccRCC cells were transduced with RNF20 and/or SREBP1c lentivirus, and cell proliferation was monitored by the Cell Counting Kit-8 (CCK-8) assay. (G) RNF20 and/or SREBP1 was suppressed in ACHN ccRCC cells using siRNAs, and cell lysates were subjected to Western blotting. (H) Intracellular triglyceride contents were measured in RNF20- and/or SREBP1-suppressing ACHN cells. (I) ACHN ccRCC cells were transfected with siRNF20 and/or siSREBP1, and relative cell growth rates were monitored using the CCK-8 assay. (J) ACHN ccRCC cells were transfected with siRNAs for suppression of RNF20 and/or SREBP1, and relative mRNA levels were determined by qRT-PCR. The data shown are representative results for at least three independent experiments. Data presented are the means ± SD. Significance versus negative control: #, P < 0.05; ##, P < 0.01; ###, P < 0.001. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The SREBP inhibitor betulin represses lipid metabolism and proliferation of ccRCC cells.
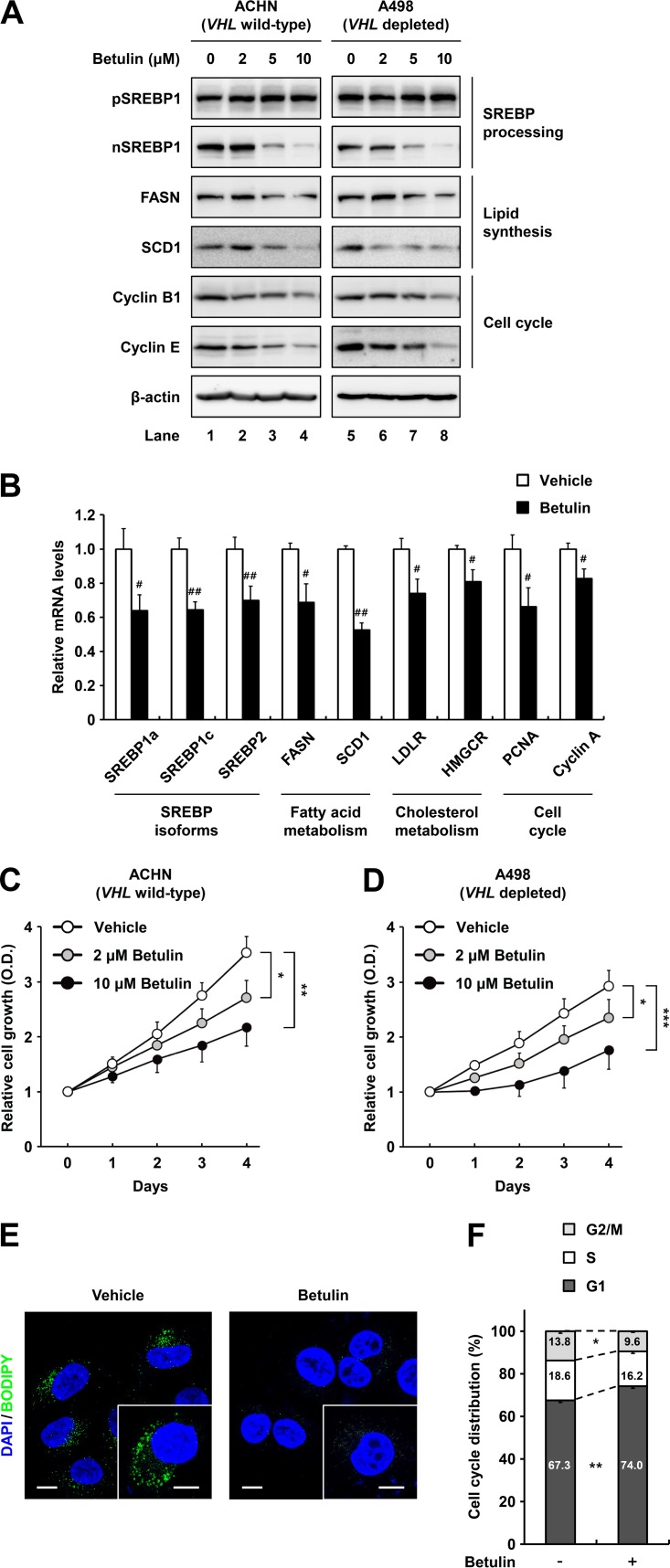
Betulin was developed as a pharmacological inhibitor that prevents proteolytic processing of both SREBP1 and SREBP2 to achieve lipid-lowering effects (29, 30). In addition, betulin attenuates the growth of various cancers by inhibiting multiple oncogenic factors, including cell cycle regulators (31, 32). Thus, we determined the effects of betulin on SREBP inhibition and assessed its antitumorigenic properties in ccRCC cells. Following treatment of wild-type-VHL ACHN and VHL-depleted A498 ccRCC cells with betulin, the nuclear SREBP1 protein level was decreased in a dose-dependent manner, whereas the precursor form of SREBP1 was unaffected (Fig. 4A). Simultaneously, betulin treatment led to decreased protein expression of the lipogenic enzymes such as FASN and SCD1 (Fig. 4A). Moreover, protein levels of cell cycle regulators, including cyclin B1 and E, were repressed in betulin-treated ccRCC cells (Fig. 4A). Betulin also decreased the expression of genes involved in fatty acid and cholesterol metabolism by inhibiting SREBP1 and -2 in ccRCC cells (Fig. 4B). To explore whether betulin suppresses ccRCC cell growth, we measured its effect on cell proliferation in ACHN and A498 cells. As shown in Fig. 4C and D, betulin showed dose-dependent antiproliferative effects in ccRCC cells. Lipogenic gene expression changes corresponded with decreased intracellular lipid accumulation in betulin-treated ccRCC cells (Fig. 4E). In addition, betulin substantially increased the population of ccRCC cells in G1 phase (Fig. 4F). These data indicate that betulin represses ccRCC cell proliferation by suppressing SREBP-dependent lipid metabolism and cell cycle progression.
FIG 4.
The SREBP inhibitor betulin inhibits ccRCC cell proliferation. (A) ACHN and A498 ccRCC cells were treated with increasing concentrations of betulin for 12 h, and total cell lysates were subjected to SDS-PAGE and Western blotting with the indicated antibodies. pSREBP1, precursor SREBP1; nSREBP1, nuclear SREBP1. (B) After treatment with betulin (10 μM) for 24 h in ACHN cells, relative mRNA levels were determined by qRT-PCR. (C and D) ACHN and A498 ccRCC cells were treated or not treated with betulin, and cell proliferation rates were determined using the Cell Counting Kit-8 (CCK-8) assay. Data are presented as the means ± SD from five individual samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) ACHN ccRCC cells were treated with betulin (10 μM). After incubation for 24 h, the cells were fixed and stained with BODIPY (green) and DAPI (blue). Images were acquired using a confocal microscope. Bars, 10 μm. (F) After treatment with betulin (10 μM) for 24 h, ACHN ccRCC cells were fixed and stained with propidium iodide, and DNA contents were analyzed by flow cytometry. Percentages of cells in each phase of the cell cycle are indicated. Similar results were obtained in at least three independent experiments.
In ccRCC cells, PTTG1 is a novel target gene of SREBP1c and is involved in cell cycle regulation.
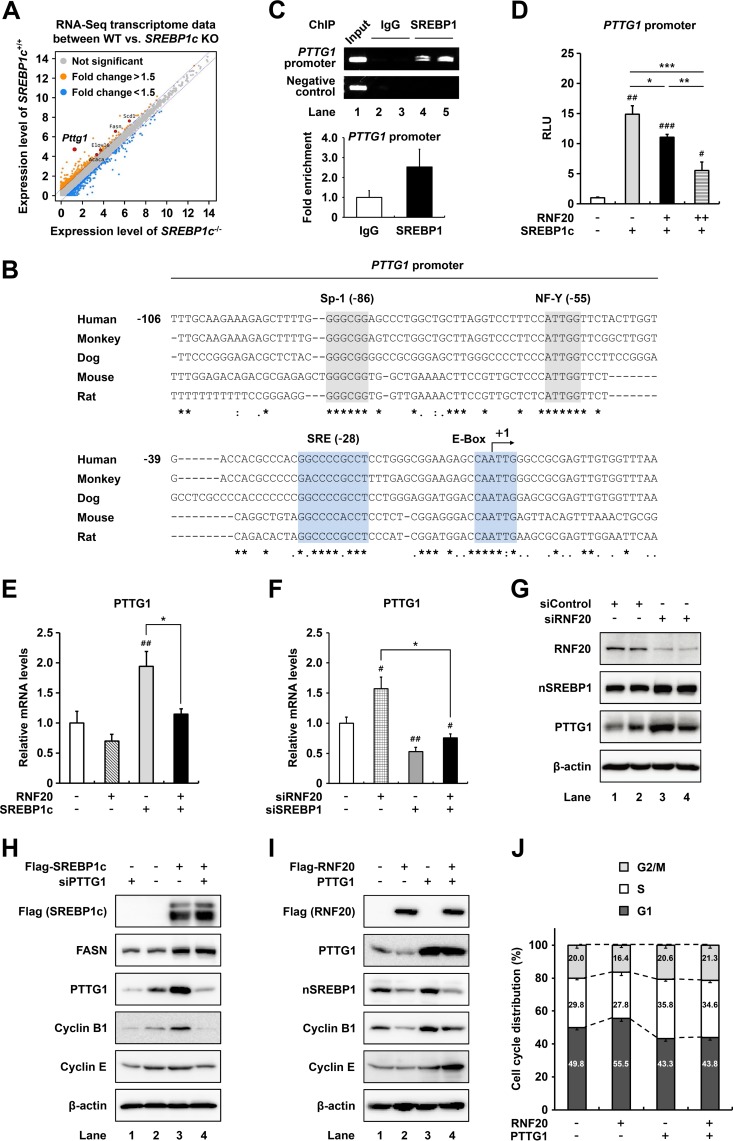
As a transcriptional activator, SREBP1c stimulates fatty acid metabolism and cell cycle progression (28, 33, 34). To elucidate the target gene(s) potentially involved in SREBP1c-induced cell cycle progression in ccRCC, we attempted to identify SREBP1c target genes that are associated with cell cycle regulation, by RNA-seq of liver tissues of wild-type and SREBP1c-deficient mice. The analysis revealed PTTG1 as a novel target gene of SREBP1c (Fig. 5A). Expectedly, PTTG1 expression was lower in kidney, liver, and adipose tissues of SREBP1c-deficient mice than in those of wild-type mice (data not shown). Analysis of the proximal promoter sequences of PTTG1 showed that putative binding sites for SREBP1c are well conserved in several species (Fig. 5B). As shown in Fig. 5C, chromatin immunoprecipitation (ChIP) assay revealed that SREBP1 indeed bound to the promoter of PTTG1. In luciferase reporter assays, SREBP1c stimulated PTTG1 promoter activity, whereas coexpression of RNF20 downregulated the SREBP1c-mediated promoter activation (Fig. 5D).
FIG 5.
PTTG1 is a novel target gene of SREBP1c in ccRCC cells. (A) RNA-seq data for transcriptome profiling of liver tissues from wild-type and SREBP1c-deficient mice are presented as a scatter plot. (B) SRE motifs and E-box sequences in the PTTG1 promoters from several species. (C) ChIP assay results showing the human PTTG1 promoter occupancy by SREBP1 in ACHN ccRCC cells. (D) HEK293 cells were cotransfected with luciferase reporter plasmid containing the PTTG1 promoter and expression vectors for β-galactosidase, RNF20, and/or SREBP1c. Total cell lysates were subjected to luciferase and β-galactosidase assays. RLU, relative luminescence units. (E) ACHN ccRCC cells were transduced with RNF20 and/or SREBP1c lentivirus, and mRNA levels were determined by qRT-PCR. (F) ACHN ccRCC cells were transfected with siRNF20 and/or siSREBP1, and relative mRNA levels were determined using qRT-PCR. #, P < 0.05; ##, P < 0.01 versus negative control; *, P < 0.05. (G) ACHN ccRCC cells were transfected with siControl or siRNF20. After incubation for 48 h, Western blotting was conducted with indicated antibodies. nSREBP1, nuclear SREBP1. (H) ACHN ccRCC cells were infected with SREBP1c adenovirus and transfected with PTTG1 siRNA. After incubation for 48 h, Western blotting was conducted with the indicated antibodies. (I) ACHN ccRCC cells were transfected with Flag-RNF20 and/or PTTG1. At 48 h after transfection, Western blotting was conducted with the indicated antibodies. (J) After transfection with Flag-RNF20 and/or PTTG1 for 48 h, ACHN ccRCC cells were stained with propidium iodide and analyzed by flow cytometry. The percentages of cells in each phase of the cell cycle are indicated.
Next, we investigated the expression profile of PTTG1 in SREBP1c-overexpressing ACHN cells and found that ectopic SREBP1c expression enhanced PTTG1 mRNA expression, whereas RNF20 coexpression attenuated this effect (Fig. 5E). On the contrary, knockdown of RNF20 in ccRCC cells increased the levels of PTTG1 mRNA (Fig. 5F) and protein (Fig. 5G). However, under suppression of SREBP1, RNF20 knockdown failed to increase the PTTG1 mRNA level (Fig. 5F). In accordance herewith, SREBP1c promoted PTTG1 protein expression in ccRCC cells (Fig. 5H). On the other hand, siRNA-mediated suppression of PTTG1 did not affect SREBP1c or FASN protein expression (Fig. 5H). While RNF20 overexpression reduced protein levels of PTTG1 and several cell cycle regulators in ccRCC cells, PTTG1 overexpression increased protein levels of these cell cycle regulators but not SREBP1c in both control and RNF20-overexpressing ccRCC cells (Fig. 5I). Consistent with these findings, RNF20 overexpression led to an increase in the G1 phase population, whereas PTTG1 coexpression attenuated it (Fig. 5J). Taken together, these data suggest that activated SREBP1c in ccRCC stimulates PTTG1 expression, promoting cell cycle progression.
SREBP1c independently controls PTTG1 and lipid metabolism, which leads to ccRCC proliferation.
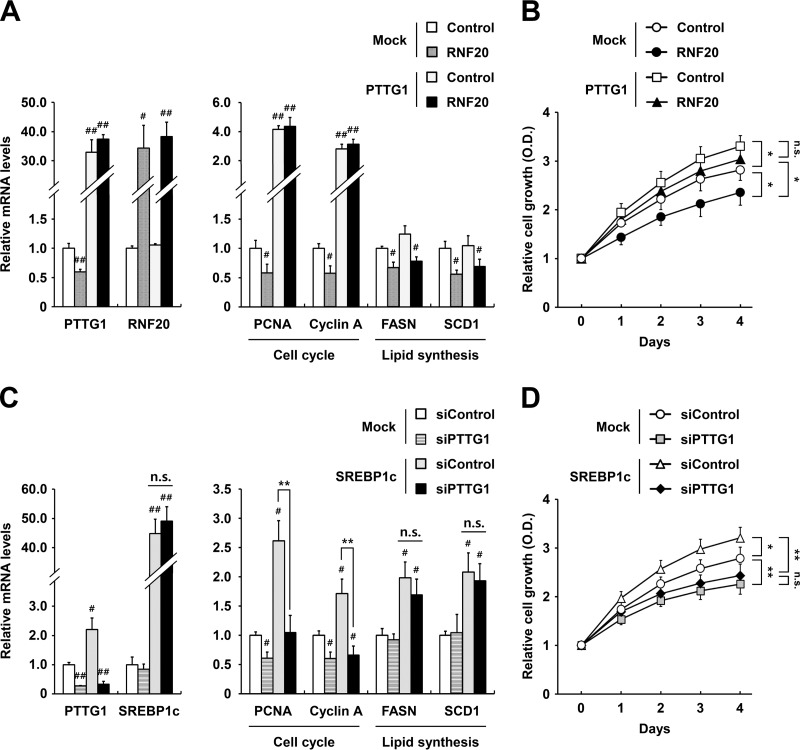
The above data suggested a potential relationship between PTTG1 and lipid metabolism during ccRCC cell proliferation. Thus, we next investigated the effects of PTTG1 on lipid metabolism and/or cell proliferation in RNF20-overexpressing ccRCC cells. RNF20 overexpression reduced mRNA expression levels of PTTG1 and cell cycle genes in ccRCC cells, whereas PTTG1 overexpression increased the mRNA expression levels of these cell cycle genes but not those of lipogenic genes, including FASN and SCD1, in both control and RNF20-overexpressing ccRCC cells (Fig. 6A). Ectopic PTTG1 expression markedly augmented cell growth in both control and RNF20-transduced ccRCC cells (Fig. 6B). Accordingly, ectopic SREBP1c expression promoted mRNA expression of PTTG1 and the cell cycle regulators, including PCNA, cyclin A, D1, and E, but PTTG1 suppression did not alter mRNA expression of SREBP1c or FASN (Fig. 6C). In contrast, suppression of PTTG1 expression in ccRCC cells downregulated the cell cycle regulatory genes (Fig. 6C). In addition, PTTG1 downregulation inhibited cell proliferation in both control and SREBP1c-overexpressing ACHN cells (Fig. 6D).
FIG 6.
In ccRCC cells, PTTG1 is involved in cell cycle regulation. (A) Lentivirus-mediated negative-control-infected (Mock) or RNF20-overexpressing ACHN cells were transfected with PTTG1 for 48 h. Relative mRNA levels were determined using qRT-PCR. Expression data are presented relative to the negative control. (B) ACHN ccRCC cells were transfected with RNF20 and/or PTTG1, and cell proliferation was monitored by the Cell Counting Kit-8 (CCK-8) assay. (C) Lentivirus-mediated negative-control-infected (Mock) or SREBP1c-overexpressing ACHN cells were transfected with PTTG1 siRNA for 48 h. Relative mRNA levels were determined using qRT-PCR. (D) ACHN ccRCC cells were transfected with SREBP1c and/or siPTTG1, and cell proliferation was monitored by the CCK-8 assay. Significance versus negative control: #, P < 0.05; ##, P < 0.01. *, P < 0.05; **, P < 0.01; n.s., not significant.
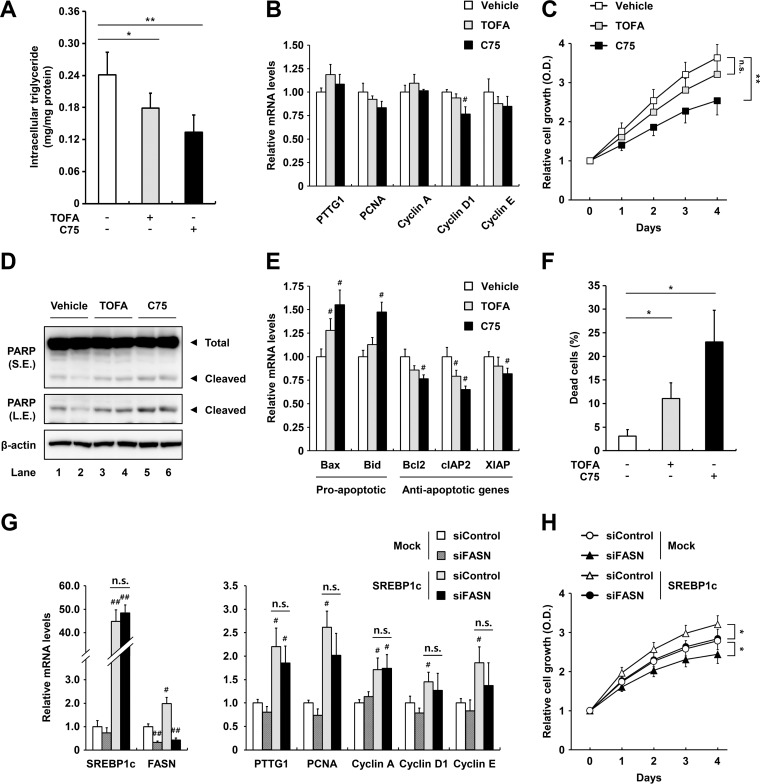
Next, to elucidate regulation of PTTG1 by lipogenic activities, we examined PTTG1 expression and cell proliferation in the presence or absence of the ACC inhibitor TOFA or the FASN inhibitor C75. As shown in Fig. 7A, TOFA and C75 treatments reduced intracellular triglyceride accumulation in ACHN cells. However, decreased lipogenic activity by TOFA or C75 did not greatly affect expression of PTTG1, PCNA, cyclin A, D1, and E (Fig. 7B). However, FASN inhibition with C75 decreased cell growth rates in ccRCC cells (Fig. 7C). Moreover, we measured the degrees of apoptosis of ccRCC cells in the presence of TOFA or C75 by evaluating poly(ADP-ribose) polymerase (PARP) cleavage, apoptotic gene expression, and annexin V staining. Lipogenic inhibition by TOFA or C75 elevated PARP cleavage in ACHN cells (Fig. 7D). In addition, mRNA levels of the proapoptotic genes, including Bax and Bid, were increased by C75, whereas those of the antiapoptotic genes, such as Bcl2, cIAP2, and XIAP, were reduced in ACHN cells (Fig. 7E). As shown in Fig. 7F, TOFA or C75 increased the percentage of dead cells among ccRCC cells. However, siRNA-mediated knockdown of FASN did not alter mRNA levels of PTTG1 or the cell cycle regulatory genes (Fig. 7G). Suppression of FASN reduced the effects of SREBP1c overexpression on proliferation in ACHN cells, implying that lipogenic activity is crucial for ccRCC cell proliferation in an SREBP1c-dependent manner (Fig. 7H). Taken together, these data indicate that PTTG1 and lipogenic pathways are independently regulated by SREBP1c, which promotes ccRCC cell proliferation.
FIG 7.
Inhibition of lipogenic activity does not affect PTTG1 expression but induces apoptosis in ccRCC cells. (A) ACHN ccRCC cells were treated with TOFA (10 μg/ml) or C75 (10 μg/ml) for 24 h, and intracellular triglyceride contents were measured. (B) ACHN ccRCC cells were treated with TOFA (10 μg/ml) or C75 (10 μg/ml) for 24 h. Relative mRNA levels are shown relative to those in the vehicle group. (C) ACHN ccRCC cells were treated with TOFA (10 μg/ml) or C75 (10 μg/ml), and cell proliferation was monitored by the Cell Counting Kit-8 (CCK-8) assay. (D) After treatment with TOFA (10 μg/ml) or C75 (10 μg/ml) for 24 h in ACHN ccRCC cells, PARP cleavage was detected by Western blotting. S.E., short exposure; L.E., long exposure. (E) ACHN ccRCC cells were treated with TOFA (10 μg/ml) or C75 (10 μg/ml) for 24 h, and relative mRNA levels were determined using qRT-PCR. (F) ACHN ccRCC cells were stained with annexin V after incubation with TOFA (10 μg/ml) or C75 (10 μg/ml) for 24 h, and apoptotic cells were measured by fluorescence-activated cell sorter (FACS) analysis. (G) Lentivirus-mediated negative-control-infected (Mock) or SREBP1c-overexpressing ACHN cells were transfected with FASN siRNA for 48 h, and relative mRNA levels were determined by qRT-PCR. mRNA levels are shown relative to those in the negative-control group. Significance versus negative control: #, P < 0.05; ##, P < 0.01. (H) ACHN ccRCC cells were transfected with SREBP1c and/or siFASN, and cell proliferation was monitored by CCK-8 assay. *, P < 0.05; **, P < 0.01; n.s., not significant.
RNF20 overexpression attenuates tumor growth in ccRCC xenografts.
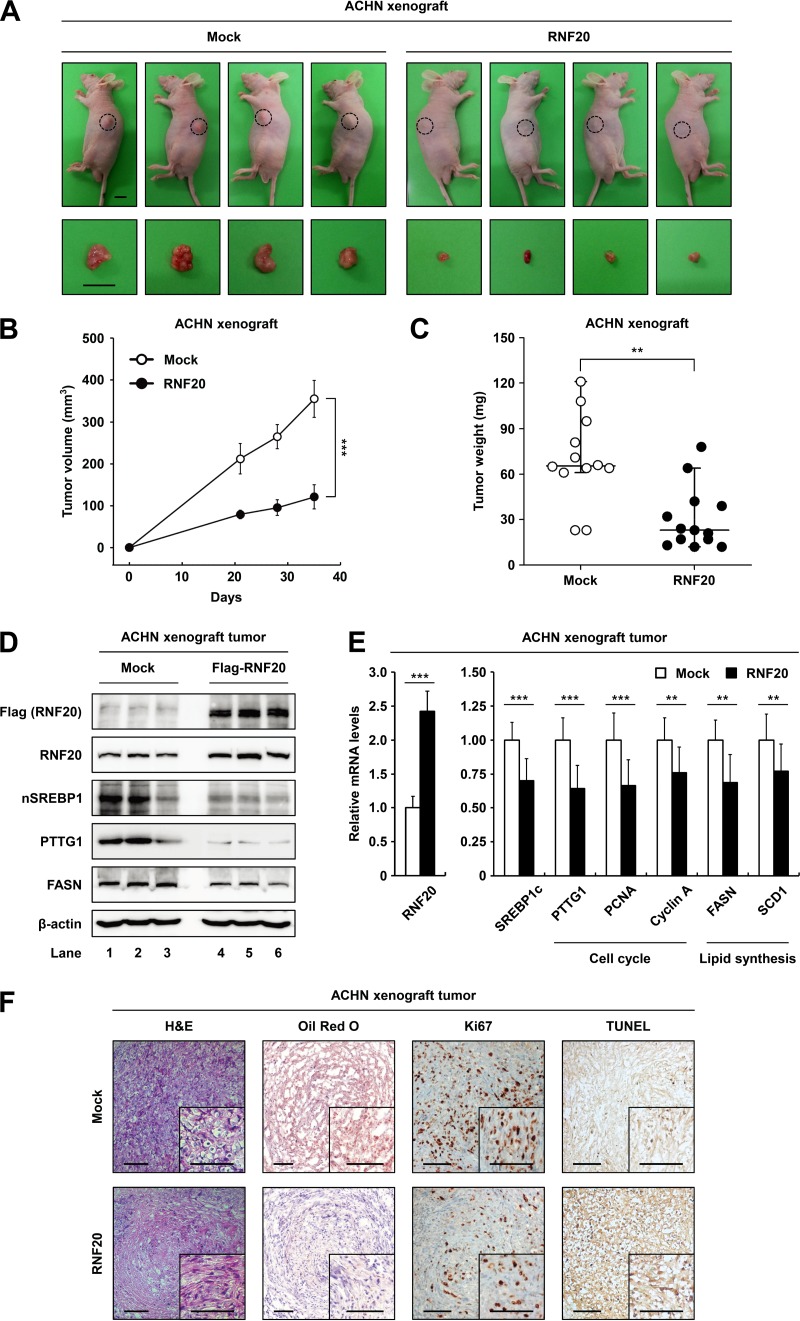
To validate the roles of RNF20 in ccRCC tumor growth in vivo, we performed xenograft experiments in nude mice. In ACHN xenograft tumors, ectopic RNF20 expression significantly inhibited the tumor growth rate (Fig. 8A and B) and decreased tumor mass (Fig. 8C), indicating that RNF20 can suppress ccRCC tumor growth. Western blot analyses showed that ectopic expression of RNF20 reduced the protein levels of SREBP1, PTTG1, and FASN (Fig. 8D) as well as the mRNA levels of SREBP1c, cell cycle regulators, and lipogenic genes in xenograft tumors (Fig. 8E). In hematoxylin and eosin (H&E) staining analyses, ACHN tumors with elevated RNF20 expression had fewer cells with clear cell morphology (Fig. 8F). Oil Red O staining indicated that ectopic RNF20 expression reduced lipid accumulation (Fig. 8F). Furthermore, Ki67 staining revealed that exogenous RNF20 decreased cell proliferation in xenograft tumors (Fig. 8F). Terminal deoxynucleotidyltransferase dUTP-biotin nick end labeling (TUNEL) analyses showed that RNF20 overexpression induced apoptosis in xenograft tumors (Fig. 8F). These data suggest that RNF20 acts as a tumor suppressor in vivo by inhibiting SREBP1c-mediated lipogenesis and cell cycle regulation in ccRCC.
FIG 8.
RNF20 overexpression attenuates tumor growth in xenograft mice. (A) Subcutaneous tumors of ACHN cells expressing either the negative control (Mock) or ectopic RNF20 were generated in female BALB/c nude mice. Representative images of tumors dissected at the end of the study showing the effect of RNF20 overexpression on the growth of xenograft tumors in vivo. Bar, 10 mm. (B) Xenograft tumor volumes (in cubic millimeters) of ACHN cells with or without ectopic RNF20 expression were determined over 35 days. The graph shows the means ± SEM; n = 10 for each group. (C) Endpoint xenograft tumor weights were determined and plotted. Data are represented as the means ± SEM; n = 10 for each group. (D) Expression levels of RNF20, nuclear SREBP1, PTTG1, and FASN protein in ACHN xenograft tumors were monitored using Western blotting. (E) The effects of ectopic RNF20 expression on cell cycle and lipogenic gene expression in ACHN xenograft tumors were determined using qRT-PCR. Relative mRNA levels are shown relative to the control group (Mock) levels. Data are presented as the means ± SD; n = 10 for each group; **, P < 0.01; ***, P < 0.001. (F) Histological analysis of xenograft tumors. Representative hematoxylin and eosin (H&E)- and Oil Red O-stained sections of negative-control (Mock)- or RNF20-transduced ACHN xenograft tumors. IHC of xenograft tumors stained with Ki67 and TUNEL. Bars, 100 μm.
In ccRCC patients, low RNF20 expression is accompanied by SREBP1 activation and poor prognosis.
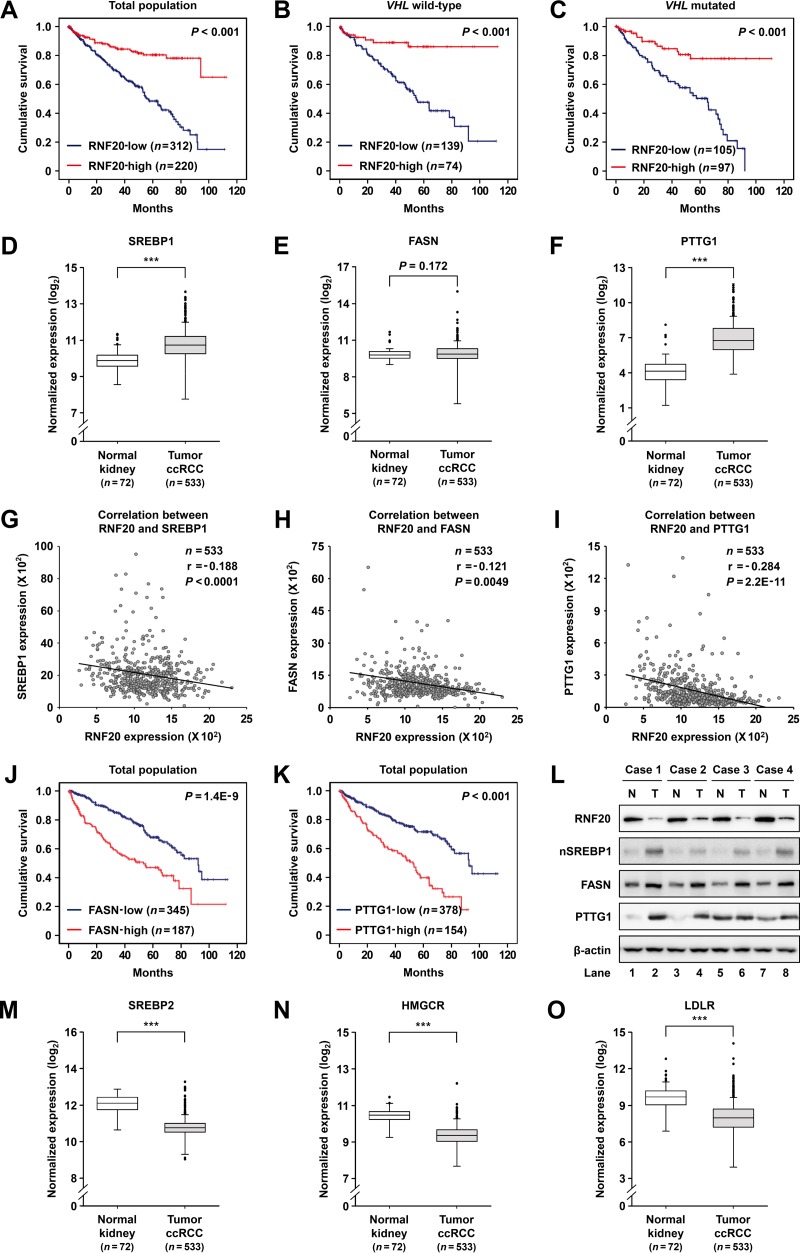
Given that RNF20 was downregulated in ccRCC tumors (Fig. 1), we explored the relationship between RNF20 expression and clinical outcomes. We found that low expression of RNF20 is significantly correlated with poor survival (Fig. 9A) in wild-type-VHL as well as VHL-mutated ccRCC patients (Fig. 9B and C). Several types of tumors, such as glioblastoma, hepatic, prostate, and pancreatic cancers, express high levels of SREBP1 and lipogenic genes, which are positively correlated with malignant progression and worse outcomes (22–25). However, it remains unclear whether SREBP1 and its target genes might have clinical significance in ccRCC patients. Thus, we analyzed the expression patterns of SREBP1, FASN, and PTTG1 in normal kidney and ccRCC tumor tissues. TCGA RNA-seq data revealed that SREBP1 expression is elevated in ccRCC tumors (Fig. 9D) and is associated with advanced tumor stages (data not shown). Accordingly, mRNA levels of SREBP1 target genes such as FASN and PTTG1 were enhanced in ccRCC tumors (Fig. 9E and F). Further TCGA analyses revealed an inverse correlation between RNF20 and SREBP1 mRNA expression in ccRCC tumors (Fig. 9G). Similarly, mRNA expression of RNF20 was inversely correlated with that of FASN or PTTG1 (Fig. 9H and I). Moreover, high expression of FASN or PTTG1 was correlated with poor survival in ccRCC patients (Fig. 9J and K). Consistent with these data, protein levels of SREBP1, FASN, and PTTG1 were increased in ccRCC tumors compared to those in patient-matched normal kidney tissues, whereas that of RNF20 was decreased (Fig. 9L). However, TCGA analyses indicated that the mRNA levels of SREBP2 and its target genes, such as those encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), low-density lipoprotein receptor (LDLR), 3-hydroxy-3-methylglutaryl coenzyme A synthase (HMGCS), and farnesyl-diphosphate farnesyltransferase 1 (FDFT1), were decreased in ccRCC tumors (Fig. 9M to O and data not shown). Together, these results imply that RNF20 downregulation enhances SREBP1 activation and determines poor clinical outcomes in ccRCC patients.
FIG 9.
SREBP1 activities are upregulated in ccRCC and are negatively correlated with RNF20 expression. (A) Kaplan-Meier survival curves of ccRCC patients enrolled in the TCGA database. Patients were divided into two groups according to median RNF20 mRNA levels, and differences were identified using the log rank test. (B and C) Survival stratified according to VHL mutation status. (D to F) Normalized RNA-seq reads of SREBP1 (D), FASN (E), and PTTG1 (F) in normal kidney and ccRCC tumor samples using the TCGA database. (G) Correlations between RNF20 and SREBP1 mRNA levels in ccRCC tumor samples identified from TCGA data sets using Pearson correlation tests. Numbers of cases (n), Pearson correlation coefficient (r), and P values are indicated. (H) Correlation of RNF20 and FASN mRNA expression in ccRCC samples identified by Pearson correlation tests. (I) Correlation of RNF20 and PTTG1 mRNA expression in ccRCC samples identified by Pearson correlation tests. (J) Kaplan-Meier survival analysis for ccRCC patients from low or high FASN expression groups. P values were calculated by the log rank test. (K) Kaplan-Meier survival analyses of ccRCC patients in low and high PTTG1 expression groups. (L) Protein levels in patient-matched normal kidney (n = 4) and ccRCC tumor (n = 4) samples were determined by Western blotting. N, normal kidney; T, ccRCC tumor; nSREBP1, nuclear SREBP1. (M to O) Normalized RNA-seq reads of SREBP2 (M), HMGCR (N), and LDLR (O) in normal kidney and ccRCC tumor samples using the TCGA database.
DISCUSSION
It is well established that constitutive activation of HIF due to the loss of VHL in ccRCC causes pathogenic metabolic alterations (9, 10). However, kidney-specific VHL-deficient mice did not exhibit ccRCC-like metabolic phenotypes (35), suggesting additional mechanisms of ccRCC tumor formation. The present data demonstrate that RNF20 downregulation promotes ccRCC tumorigenesis by activating SREBP1c. Several lines of evidence support the idea that RNF20 would suppress ccRCC tumor growth. First, RNF20 expression was decreased in ccRCC tumors compared to that in normal kidney tissues and was inversely correlated with the expression of SREBP1 and its target genes, including FASN and PTTG1. Second, low RNF20 expression was closely associated with poor survival in ccRCC patients, regardless of VHL mutation status. Third, ectopic expression of RNF20 repressed SREBP1c expression and cell proliferation in both wild-type-VHL and VHL-depleted ccRCC cell lines but did not affect proliferation of HRCE and HEK293 normal kidney cells, which have high basal RNF20 expression. Lastly, RNF20 overexpression decreased tumor growth in ccRCC xenografts and was accompanied by reduced expression of SREBP1c and its target genes.
Consistent with previous observations of abundant lipid storage and increased lipogenesis in ccRCC (36, 37), the present data show upregulated SREBP1 and lipogenic activities. However, the mRNA level of SREBP2 and its target genes, such as HMGCR, LDLR, HMGCS, and FDFT1, were decreased in ccRCC tumors, implying that SREBP1 and de novo lipogenesis would play crucial roles in ccRCC. These observations warrant the consideration of pharmacological inhibitors of lipogenesis as anticancer drugs for ccRCC. Accordingly, the FASN inhibitor C75 and the SCD1 inhibitor A939572 suppress tumor growth and invasiveness of ccRCC (38, 39), and in this study, the SREBP inhibitor betulin repressed ccRCC cell proliferation by downregulating SREBP1 and lipogenesis regardless of VHL gene mutations. Betulin potently abolished increased cell proliferation and lipogenic activity upon RNF20 suppression (data not shown). Given the hyperactivation of SREBP1 and lipogenesis along with RNF20 downregulation in ccRCC, it seems that unfettered SREBP1 may promote ccRCC tumor development, at least partly, via lipogenic activation.
SREBP1c has been associated with de novo lipogenesis and cell cycle progression (28, 33, 34). Moreover, cyclin-dependent kinase 1 (CDK1)/cyclin B phosphorylates and activates SREBP1c during mitosis (33). Similarly, the SREBP-responsive microRNA (miRNA) miR-33 reportedly inhibits CDK6 and cyclin D1 expression, thereby reducing proliferation and cell cycle progression in certain cancer cells (40). In glioma and cervical cancers, suppression of SREBP1 represses tumor growth by inducing G1 cell cycle arrest and apoptosis (28, 33). In addition, it has been shown that betulin inhibits lung cancer cell proliferation by downregulating cell cycle regulators such as cyclin B1, D, and E (32). This study demonstrated that PTTG1, a novel target gene of SREBP1c, is involved in cell cycle progression and tumorigenesis in ccRCC. PTTG1 (also known as securin, EAP1, and TUTR1) is an anaphase inhibitor that prevents premature chromosome separation by inhibiting separase activity (41, 42). Previous studies have suggested that PTTG1 participates in various pathways that modulate the cell cycle, proliferation, and survival. For example, PTTG1 has been shown to interact with p53 and prevent p53-dependent transcription and apoptosis (43, 44). Further, PTTG1 promotes the expression of several genes, including c-Myc, cyclin D3, FGF2, and MMP2, suggesting that it acts as a transcription factor (45–47). In addition, PTTG1 is overexpressed in certain tumors, including pituitary, thyroid, glioma, hepatic, colorectal, and renal cancers, and may drive tumorigenesis (48, 49). Here, we found that high PTTG1 expression was closely associated with advanced tumor stages and poor survival in ccRCC patients. Moreover, SREBP1c potently stimulated mRNA and protein expression of PTTG1 and several cell cycle regulators, potentiating cell proliferation in ccRCC. Furthermore, RNF20 overexpression downregulated PTTG1 in both ccRCC cells and xenograft tumors, whereas RNF20 suppression led to increased PTTG1 mRNA and protein levels. As PTTG1 expression was negatively associated with RNF20 expression, it is likely that RNF20 downregulation promotes ccRCC development and progression, in part, by upregulating PTTG1 via SREBP1c. In ccRCC cells, PTTG1 overexpression promoted mRNA levels of cell cycle regulatory genes without changing lipogenic gene expression, whereas PTTG1 suppression attenuated the effects of activated SREBP1c on the induction of cell cycle regulatory genes and cell proliferation. Collectively, these data imply that the RNF20-SREBP1c-PTTG1 axis is one of the key players in ccRCC cell proliferation and tumorigenesis.
It is well established that SREBP1 regulates lipid metabolism predominantly by increasing de novo lipogenesis. Thus, we tested whether lipogenesis is associated with PTTG1 expression by inhibiting de novo lipogenesis. In ccRCC cells, the mRNA level of PTTG1 was not affected by pharmacological inhibition of lipogenesis using TOFA or C75 or by siRNA-mediated knockdown of FASN, indicating that SREBP1c would stimulate PTTG1 in a lipogenesis-independent manner. Thus, SREBP1c may affect lipid metabolism and cell cycle progression by regulating different sets of target genes, which eventually coalesce to drive tumor development in ccRCC. In accordance with the present data, several studies have shown that lipogenic inactivation by TOFA or C75 induces cell cycle arrest and apoptosis in various cancers, including lung, colorectal, and renal cancers (38, 50). Further studies are required to clarify whether lipogenic pathways might affect PTTG1 and cell cycle progression during SREBP1 activation in other tissues and/or cancers. Recently, it has been reported that RNF20 would exhibit tumor-suppressive roles by regulating histone H2B monoubiquitination (13, 14). For example, RNF20+/− mice are prone to inflammation-associated colon cancer by augmenting NF-κB activity (15). In this regard, we cannot exclude the possibility that RNF20 suppresses ccRCC tumorigenesis with other substrates as well as SREBP1c.
Here, we propose a model in which RNF20 acts as a tumor suppressor by inhibiting SREBP1c-mediated lipogenesis and cell cycle progression. Conversely, RNF20 downregulation promotes tumorigenesis by activating SREBP1c in ccRCC tumors. Particularly, we identified a novel pathway by which SREBP1c stimulates cell cycle progression by inducing PTTG1 expression, and our data provide important clues to the molecular mechanisms that link SREBP1c and cell cycle regulation in ccRCC. Taken together, our findings suggest that RNF20 is a novel tumor suppressor that acts by modulating the SREBP1c-PTTG1 axis in ccRCC.
MATERIALS AND METHODS
Cell culture and chemicals.
ACHN, A498, HEK293, Caki-2, and human primary renal cortical epithelial (HRCE) cells were obtained from the American Type Culture Collection (ATCC). ACHN and A498 cells were grown in in Eagle's minimum essential medium (MEM; HyClone catalog number SH30024.01) supplemented with 10% fetal bovine serum (FBS; HyClone catalog number SH30919.03) and penicillin (100 U/ml)-streptomycin (100 μg/ml). HEK293 and Caki-2 cells were grown in Dulbecco's modified Eagle's medium (DMEM; HyClone catalog number SH30243.01) supplemented with 10% FBS and penicillin-streptomycin. HRCE cells were cultured in renal epithelial cell basal medium (catalog number PCS-400-030; ATCC) with the following supplements: 0.5% FBS, 10 nM triiodothyronine, 10 ng/ml epidermal growth factor, 100 ng/ml hydrocortisone, 5 μg/ml insulin, 1 μM epinephrine, 5 μg/ml transferrin, 2.4 mM l-alanyl-l-glutamine, and penicillin-streptomycin. All cells were cultured at 37°C in a 5% CO2 incubator. Betulin and BODIPY 493/503 were purchased from Sigma-Aldrich (catalog numbers B8936 and D3922, respectively). C75 and TOFA were obtained from Abcam (catalog numbers ab141397 and ab141578, respectively). Propidium iodide (PI) was provided by BD Biosciences (catalog number 51-66211E).
Human ccRCC samples.
Freshly frozen human ccRCC and matched normal kidney tissue samples were obtained from the Seoul National University Hospital (SNUH). The Institutional Review Board at SNUH approved this study (approval number H-1501-011-636). Informed consent documents from patients were not required because of the retrospective nature of the study.
TCGA RNA-seq analysis.
RNA-seq data, VHL mutation status information, and clinicopathological data for 533 ccRCC and 72 normal kidney samples were downloaded from the TCGA ccRCC project (http://cancergenome.nih.gov) in September 2015. Box and whisker plots are presented with 1st to 99th percentiles (bars), 25th to 75th percentiles (box), and median values (line in box). For survival analysis, the top half of ranked patients was defined as the “high” group, and the lower half was defined as the “low” group. Overall survival was estimated by Kaplan-Meier survival analyses, and survival outcomes were compared between the two groups by the log rank test.
Tissue array and immunohistochemistry.
Immunohistochemistry was conducted using tissue microarrays of ccRCC and normal kidney tissue sections (SuperBioChips Laboratories catalog number CL2) according to the supplier's protocol (Ventana Medical Systems catalog number 760-700). Briefly, the streptavidin-biotin complex method was followed to detect RNF20 and SREBP1 with corresponding primary antibodies from Abcam (1:30; catalog number ab32629) and BD Biosciences (1:25; catalog number 557036), respectively.
Preparation of recombinant adenovirus.
Adenovirus plasmids were constructed as previously described (11). Briefly, rat nuclear SREBP1c encoding amino acids 1 to 403 and full-length mouse RNF20 cDNAs were incorporated into the AdTrack-CMV shuttle vector, and recombinant vectors were generated using Ad-Easy adenoviral vector systems. An adenovirus encoding green fluorescent protein (GFP) alone was used as a negative control in all experiments. Adenoviruses were amplified in HEK293A cells and were purified by CsCl gradient centrifugation as described previously (51).
Lentivirus production and viral transduction.
Full-length RNF20 or nuclear SREBP1c cDNA with Flag tag was incorporated into the lentiviral vector pLVX-EF1α-AcGFP1-N1 (Clontech catalog number 631983). Lentiviruses were transfected into HEK293T cells with the indicated expression vectors, pAX2 (Addgene catalog number 35002) and pMD2.G (Addgene catalog number 12259), using Lipofectamine 2000 reagent (Invitrogen catalog number 11668-027). At 48 h after transfection, viruses were harvested and filtered through 0.45-μm filters. Subsequently, ACHN cells were incubated with medium containing virus and 8 μg/ml Polybrene (Sigma-Aldrich catalog number 107689) for 18 h. Infected cells were allowed to recover for 48 h before selection of puromycin-resistant colonies for experiments.
Western blot analysis.
Cells and tissues were lysed on ice in modified radioimmunoprecipitation assay (RIPA) buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 1% NP-40, 0.25% Na-deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (GeneDEPOT catalog number P3100). Equal amounts of protein were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Merck Millipore catalog number IPVH00010). After transfer, the membranes were blocked with 5% nonfat milk or 3% bovine serum albumin in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) and were probed with primary antibodies against RNF20 (1:1,000; Abcam catalog number ab32629), SREBP1 (1:1,000; BD Biosciences catalog number 557036), FASN (1:1,000; Cell Signaling catalog number 3180), SCD1 (1:500; Santa Cruz Biotechnology catalog number SC-58420), PTTG1 (1:1,000; Thermo Fisher Scientific catalog number MS-1511-P0), cyclin B1 (1:500; Santa Cruz Biotechnology catalog number SC-752), cyclin E (1:500; Santa Cruz Biotechnology catalog number SC-198), Myc-tag (1:1,000; Cell Signaling catalog number 2276), Flag-tag (1:1,000; Sigma-Aldrich catalog number F1804), or β-actin (1:2,000; Sigma-Aldrich catalog number A5316). Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse IgG antibody (Sigma-Aldrich catalog numbers A0545 and A9044, respectively), and protein bands were visualized using enhanced chemiluminescence with a LuminoImager (LAS-3000).
RNA isolation and qRT-PCR.
Total RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific catalog number 15596026). Equal amounts of RNA were subjected to cDNA synthesis using RevertAid reverse transcriptase (Thermo Fisher Scientific catalog number EP0441). Relative mRNA expression was evaluated using a CFX real-time system (Bio-Rad Laboratories) and was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA expression. The primer sequences used for real-time quantitative PCR (qRT-PCR) are listed in Table S1 in the supplemental material.
siRNA transfection.
siRNA duplexes for RNF20, SREBP1, PTTG1, and FASN were synthesized from the Bioneer, Inc. (Daejeon, South Korea). ACHN cells were transfected using Lipofectamine RNAiMAX Reagent (Invitrogen catalog number 13778-150) according to the manufacturer's protocol. Sequence information for the siRNAs is provided in Table S2 in the supplemental material.
CCK-8 cell proliferation assay.
Cell proliferation rates were determined using a Cell Counting Kit-8 (CCK-8) reagent as described previously (52). Briefly, cell growth curves were generated using the sensitive colorimetric assay for viable cells according to the manufacturer's protocol (Dojindo Molecular Technologies catalog number CK04-11).
Colony formation assay.
ACHN cells overexpressing RNF20 and/or SREBP1c were seeded in 6-well plates (5,000 cells/well), and cells were cultured at 37°C in 5% CO2. After 7 days of culture, colonies were fixed with formaldehyde and stained with crystal violet.
Cell cycle analysis.
Trypsinized cells were washed with phosphate-buffered saline (PBS) and fixed in 70% ethanol at 4°C for 30 min. Fixed cells were washed with PBS twice and incubated with propidium iodide solution containing 0.1% Nonidet P-40, 100 μg/ml RNase, and 2.5 μg/ml PI for 30 min. Stained cells were analyzed by flow cytometry using a FACSCanto II (BD Biosciences), and cells in each stage were quantified using the ModFit LT cell cycle analysis program (Verity Software House) according to the manufacturer's instructions.
Apoptosis assay.
ACHN cells were stained with fluorescein isothiocyanate (FITC)-conjugated annexin V (BD Biosciences) and propidium iodide according to the manufacturer's protocol, and cells were analyzed by the FACSCanto II instrument (BD Biosciences). The percentage of dead cells in the population represented the sum of annexin V-positive (early apoptosis) and annexin V/PI-double-positive (late apoptosis) cells.
Intracellular triglyceride measurement.
Intracellular triglycerides were determined in cell lysates by a colorimetric assay and were expressed as milligrams of lipid per milligram of cellular protein, as described previously (11). Briefly, total cell contents were extracted using 5% Triton X-100, incubated in a water bath at 80°C, and cooled to room temperature twice. After centrifugation at 12,000 rpm for 5 min at room temperature, supernatants were collected and intracellular triglycerides were assayed using the Infinity triglyceride reagent (Thermo Fisher Scientific catalog number TR22321). Values were normalized to total protein contents, which were estimated using a bicinchoninic acid (BCA) protein assay kit compatible with reducing agents (Thermo Fisher Scientific catalog number 23250).
BODIPY staining.
ACHN cells were treated or not with betulin (10 μM) for 24 h, rinsed twice with PBS, and fixed in 4% paraformaldehyde for 10 min. Fixed cells were washed twice with PBS containing Tween 20 and stained with FITC-conjugated BODIPY 493/503 (Thermo Fisher Scientific catalog number D3922) for 1 h in the dark at room temperature. Then, the samples were stained using a Vectashield solution (Vector Laboratories catalog number H-1200) containing 4′,6-diamidino-2-phenylindole (DAPI) and were observed using a Zeiss LSM 700 confocal microscope (Carl Zeiss).
ChIP assay.
ChIP assays using ACHN ccRCC cells were performed as described previously (53). Extracted proteins from total cell lysates were immunoprecipitated with anti-SREBP1 (BD Biosciences catalog number 557036) or IgG (Santa Cruz Biotechnology catalog number sc-2025) for 2 h. Precipitated DNA fragments were analyzed by qRT-PCR, using primer sets that encompassed the proximal region (nucleotides −116 to +84) of the human PTTG1 promoter and negative-control region (nucleotides −7432 to −7214). The sequences of ChIP assay primers were as follows: proximal PTTG1 promoter region, sense, 5′-TGCCACAAAGTTTGCAAGAA, and antisense, 5′-AATGCGGCTGTTAAGACCTG; negative-control primer, sense, 5′-TCTGGTTGTGGTCTCTGGTG, and antisense, 5′-TATTGCAGTGGTAGGGGGAA.
Luciferase assay.
The PTTG1 luciferase reporter containing nucleotides −908 to +25 from the transcription start site of the human PTTG1 promoter was cloned into a pGL3-basic vector (Promega catalog number E1751). HEK293 cells were transiently transfected with various DNA plasmids by the calcium-phosphate method, as described previously (54). After incubation for 36 h, transfected cells were harvested and extracted using lysis buffer containing 25 mM Tris-phosphate (pH 7.8), 10% glycerol, 2 mM EDTA, 2 mM dithiothreitol (DTT), and 1% Triton X-100, and luciferase and β-galactosidase activities were measured according to the manufacturer's protocol (Promega catalog number E1500). Relative luciferase activity was normalized to β-galactosidase activity in each sample.
Xenograft studies.
Subcutaneous xenograft experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of SNUH (IACUC number 13-0080). Five female BALB/c athymic nude mice (Central Lab Animal) were subcutaneously injected in both flanks with 1 × 107 vector control ACHN cells or ACHN cells stably expressing RNF20. Before injections, cells were resuspended in 200 μl of PBS and mixed with equal volumes of Matrigel (Corning catalog number 354234). Following establishment of palpable tumors, tumor sizes were measured once a week using calipers, and tumor volumes were calculated according to the following formula: volume = (length × width2) × π/6, where the volume is measured in cubic millimeters. At 5 weeks after transplantation, the mice were euthanized by CO2 inhalation, and xenograft tumors were dissected and weighed.
Xenograft tissue samples were fixed in 4% paraformaldehyde, equilibrated in 30% sucrose, and embedded in OCT (Scigen Scientific catalog number 4583). Sections (10 μm) were stained with H&E and Oil Red O as previously described (55). Immunohistochemical analysis of xenograft tumor sections was performed according to the supplier's protocol (SuperBioChips Laboratories). Briefly, Ki67 and TUNEL proteins were detected by the streptavidin-biotin complex method with primary antibodies against Ki67 (Abcam catalog number ab66155) and TUNEL (R&D Systems catalog number 4810-30-K). Images were obtained using an Evos Original microscope (Thermo Fisher Scientific; Advanced Microscopy group) and a Nikon TMS inverted microscope.
RNA-seq analysis of SREBP1c-deficient mice.
SREBP1c-deficient mice were kindly provided by Jay Horton at the University of Texas Southwestern Medical Center. Liver transcriptome profiles of 12-week-old wild-type littermates and SREBP1c-deficient mice were generated by mRNA-seq, in duplicate, using Illumina HiSeq4000 by Macrogen, Inc. (South Korea).
Statistical analysis.
All results illustrated in the figures are presented as the means ± standard deviations (SD) or the means ± standard errors of the means (SEM) (for Fig. 8B and C). Multiple comparisons were performed by one-way analysis of variance (ANOVA) or by two-way ANOVA when two conditions were involved. Statistical significance was assessed by the two-tailed Student t test. Statistical analyses were performed using Prism (GraphPad Software), and differences were considered significant at P values of <0.05.
Accession number(s).
The RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) public database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE100827.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the laboratory of adipocyte and metabolism research for helpful discussion. We also thank Jay Horton at the University of Texas Southwestern Medical Center for providing SREBP1c-deficient mice and Cho-Rok Jung of the Korea Research Institute of Bioscience and Biotechnology for sharing the pFlagCMV2-PTTG1 vector.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the South Korea government (Ministry of Science, ICT and Future Planning; 2011-0018312 and NRF-2012M3A9B6055344) to J.B.K. This research was also supported by the NRF grant funded by the South Korea government (MSIP; 2015R1A2A2A01006813) to C.K. Y.G.J. was supported by the BK21 program.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00265-17.
REFERENCES
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. 2008. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Schulze A, Harris AL. 2012. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 3.Medes G, Thomas A, Weinhouse S. 1953. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res 13:27–29. [PubMed] [Google Scholar]
- 4.Ookhtens M, Kannan R, Lyon I, Baker N. 1984. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol 247:R146–R153. [DOI] [PubMed] [Google Scholar]
- 5.Menendez JA, Lupu R. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 6.Kuhajda FP. 2000. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16:202–208. doi: 10.1016/S0899-9007(99)00266-X. [DOI] [PubMed] [Google Scholar]
- 7.Rezende RB, Drachenberg CB, Kumar D, Blanchaert R, Ord RA, Ioffe OB, Papadimitriou JC. 1999. Differential diagnosis between monomorphic clear cell adenocarcinoma of salivary glands and renal (clear) cell carcinoma. Am J Surg Pathol 23:1532–1538. doi: 10.1097/00000478-199912000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Valera VA, Merino MJ. 2011. Misdiagnosis of clear cell renal cell carcinoma. Nat Rev Urol 8:321–333. doi: 10.1038/nrurol.2011.64. [DOI] [PubMed] [Google Scholar]
- 9.Shen C, Kaelin WG Jr. 2013. The VHL/HIF axis in clear cell renal carcinoma. Semin Cancer Biol 23:18–25. doi: 10.1016/j.semcancer.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin WG., Jr 2008. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Lee GY, Jang H, Choe SS, Koo SH, Kim JB. 2014. Ring finger protein20 regulates hepatic lipid metabolism through protein kinase A-dependent sterol regulatory element binding protein1c degradation. Hepatology 60:844–857. doi: 10.1002/hep.27011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 15.Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H, Boocholez H, Hubert A, Rotkopf R, Baniyash M, Pikarsky E, Gorgoulis VG, Oren M. 2016. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep 14:1462–1476. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tontonoz P, Kim JB, Graves RA, Spiegelman BM. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol 13:4753–4759. doi: 10.1128/MCB.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MS, Goldstein JL. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. 1993. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75:187–197. doi: 10.1016/S0092-8674(05)80095-9. [DOI] [PubMed] [Google Scholar]
- 19.Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. 1993. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A 90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton JD, Goldstein JL, Brown MS. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131. doi: 10.1172/JCI0215593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein JL, DeBose-Boyd RA, Brown MS. 2006. Protein sensors for membrane sterols. Cell 124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, Wakelam M, Harris AL, Schulze A. 2013. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab 1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo D, Bell EH, Mischel P, Chakravarti A. 2014. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des 20:2619–2626. doi: 10.2174/13816128113199990486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang WC, Li X, Liu J, Lin J, Chung LW. 2012. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res 10:133–142. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo D, Prins RM, Dang J, Kuga D, Iwanami A, Soto H, Lin KY, Huang TT, Akhavan D, Hock MB, Zhu S, Kofman AA, Bensinger SJ, Yong WH, Vinters HV, Horvath S, Watson AD, Kuhn JG, Robins HI, Mehta MP, Wen PY, DeAngelis LM, Prados MD, Mellinghoff IK, Cloughesy TF, Mischel PS. 2009. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal 2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linehan WM, Ricketts CJ. 2013. The metabolic basis of kidney cancer. Semin Cancer Biol 23:46–55. doi: 10.1016/j.semcancer.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, Trott JF, Aboud OA, Stirdivant S, Neri B, Wolfert R, Stewart B, Perego R, Hsieh JJ, Weiss RH. 2015. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res 75:2541–2552. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG, Kidani Y, Pourzia AL, Akhavan D, Lisiero DN, Komisopoulou E, Henkin AH, Soto H, Chamberlain BT, Vergnes L, Jung ME, Torres JZ, Liau LM, Christofk HR, Prins RM, Mischel PS, Reue K, Graeber TG, Bensinger SJ. 2013. An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res 73:2850–2862. doi: 10.1158/0008-5472.CAN-13-0382-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, Song BL. 2011. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab 13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Soyal SM, Nofziger C, Dossena S, Paulmichl M, Patsch W. 2015. Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol Sci 36:406–416. doi: 10.1016/j.tips.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Chintharlapalli S, Papineni S, Ramaiah SK, Safe S. 2007. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res 67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- 32.Li XD, Zhang YJ, Han JC. 2014. Betulin inhibits lung carcinoma proliferation through activation of AMPK signaling. Tumour Biol 35:11153–11158. doi: 10.1007/s13277-014-2426-7. [DOI] [PubMed] [Google Scholar]
- 33.Bengoechea-Alonso MT, Ericsson J. 2006. Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell Cycle 5:1708–1718. doi: 10.4161/cc.5.15.3131. [DOI] [PubMed] [Google Scholar]
- 34.Jeon TI, Esquejo RM, Roqueta-Rivera M, Phelan PE, Moon YA, Govindarajan SS, Esau CC, Osborne TF. 2013. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab 18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rankin EB, Tomaszewski JE, Haase VH. 2006. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res 66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drabkin HA, Gemmill RM. 2010. Obesity, cholesterol, and clear-cell renal cell carcinoma (RCC). Adv Cancer Res 107:39–56. doi: 10.1016/S0065-230X(10)07002-8. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Kaelin WG Jr. 2011. New insights into the biology of renal cell carcinoma. Hematol Oncol Clin N Am 25:667–686. doi: 10.1016/j.hoc.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horiguchi A, Asano T, Asano T, Ito K, Sumitomo M, Hayakawa M. 2008. Pharmacological inhibitor of fatty acid synthase suppresses growth and invasiveness of renal cancer cells. J Urol 180:729–736. doi: 10.1016/j.juro.2008.03.186. [DOI] [PubMed] [Google Scholar]
- 39.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, Tan WW, Tun HW, Copland JA. 2013. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res 19:2368–2380. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Baldan A, Esplugues E, Fernandez-Hernando C. 2012. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 11:922–933. doi: 10.4161/cc.11.5.19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jallepalli PV, Lengauer C. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 42.Draviam VM, Xie S, Sorger PK. 2004. Chromosome segregation and genomic stability. Curr Opin Genet Dev 14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. 2002. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet 32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- 44.Yu R, Heaney AP, Lu W, Chen J, Melmed S. 2000. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J Biol Chem 275:36502–36505. doi: 10.1074/jbc.C000546200. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez A, Ramos-Morales F, Romero F, Rios RM, Dreyfus F, Tortolero M, Pintor-Toro JA. 1998. hpttg, a human homologue of rat pttg, is overexpressed in hematopoietic neoplasms. Evidence for a transcriptional activation function of hPTTG. Oncogene 17:2187–2193. [DOI] [PubMed] [Google Scholar]
- 46.Tong Y, Eigler T. 2009. Transcriptional targets for pituitary tumor-transforming gene-1. J Mol Endocrinol 43:179–185. doi: 10.1677/JME-08-0176. [DOI] [PubMed] [Google Scholar]
- 47.Tong Y, Tan Y, Zhou C, Melmed S. 2007. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene 26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vlotides G, Eigler T, Melmed S. 2007. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocr Rev 28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- 49.Wondergem B, Zhang Z, Huang D, Ong CK, Koeman J, Hof DV, Petillo D, Ooi A, Anema J, Lane B, Kahnoski RJ, Furge KA, Teh BT. 2012. Expression of the PTTG1 oncogene is associated with aggressive clear cell renal cell carcinoma. Cancer Res 72:4361–4371. doi: 10.1158/0008-5472.CAN-11-2330. [DOI] [PubMed] [Google Scholar]
- 50.Wang C, Xu C, Sun M, Luo D, Liao DF, Cao D. 2009. Acetyl-CoA carboxylase-alpha inhibitor TOFA induces human cancer cell apoptosis. Biochem Biophys Res Commun 385:302–306. doi: 10.1016/j.bbrc.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker TC, Noel RJ, Coats WS, Gomez-Foix AM, Alam T, Gerard RD, Newgard CB. 1994. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol 43(Part A):161–189. [DOI] [PubMed] [Google Scholar]
- 52.Kim KH, Lee GY, Kim JI, Ham M, Won Lee J, Kim JB. 2010. Inhibitory effect of LXR activation on cell proliferation and cell cycle progression through lipogenic activity. J Lipid Res 51:3425–3433. doi: 10.1194/jlr.M007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen K, Yu G, Gumireddy K, Li A, Yao W, Gao L, Chen S, Hao J, Wang J, Huang Q, Xu H, Ye Z. 2015. ZBRK1, a novel tumor suppressor, activates VHL gene transcription through formation of a complex with VHL and p300 in renal cancer. Oncotarget 6:6959–6976. doi: 10.18632/oncotarget.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jang H, Lee GY, Selby CP, Lee G, Jeon YG, Lee JH, Cheng KK, Titchenell P, Birnbaum MJ, Xu A, Sancar A, Kim JB. 2016. SREBP1c-CRY1 signalling represses hepatic glucose production by promoting FOXO1 degradation during refeeding. Nat Commun 7:12180. doi: 10.1038/ncomms12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ham M, Choe SS, Shin KC, Choi G, Kim JW, Noh JR, Kim YH, Ryu JW, Yoon KH, Lee CH, Kim JB. 2016. Glucose-6-phosphate dehydrogenase deficiency improves insulin resistance with reduced adipose tissue inflammation in obesity. Diabetes 65:2624–2638. doi: 10.2337/db16-0060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.