Abstract
DNA gyrase is a molecular motor that harnesses the free energy of ATP hydrolysis to introduce negative supercoils into DNA. A critical step in this reaction is the formation of a chiral DNA wrap on a similar scale to the nucleosome. Here we observe gyrase structural dynamics using a single-molecule assay in which gyrase drives the processive, stepwise rotation of a nanosphere attached to the side of a stretched DNA molecule. Analysis of rotational pauses and measurements of DNA contraction reveal multiple ATP-modulated structural transitions. DNA wrapping is coordinated with the ATPase cycle and proceeds via an unanticipated structural intermediate that dominates the kinetics of supercoiling. Our findings reveal a conformational landscape of loosely coupled transitions funneling the motor toward productive energy transduction, a feature that may be common to the reaction cycles of other DNA and protein remodeling machines.
INTRODUCTION
The motor function of DNA gyrase is essential for bacterial life1: DNA supercoiling facilitates compaction of the chromosome, relieves positive strain ahead of the advancing replication fork, favors the initiation of transcription and replication, and acts as a sophisticated global regulator of gene expression profiles2,3. Gyrase belongs to the GHL (Gyrase, Hsp90, MutL) family of ATPases4, distinct from the broad P-loop NTPase superfamily5 that includes well-characterized motors such as kinesin, myosin, and F1 ATPase. Gyrase and other GHL proteins are validated targets for antibiotic and anticancer drugs that inhibit distinct steps in their mechanochemical cycles6,7.
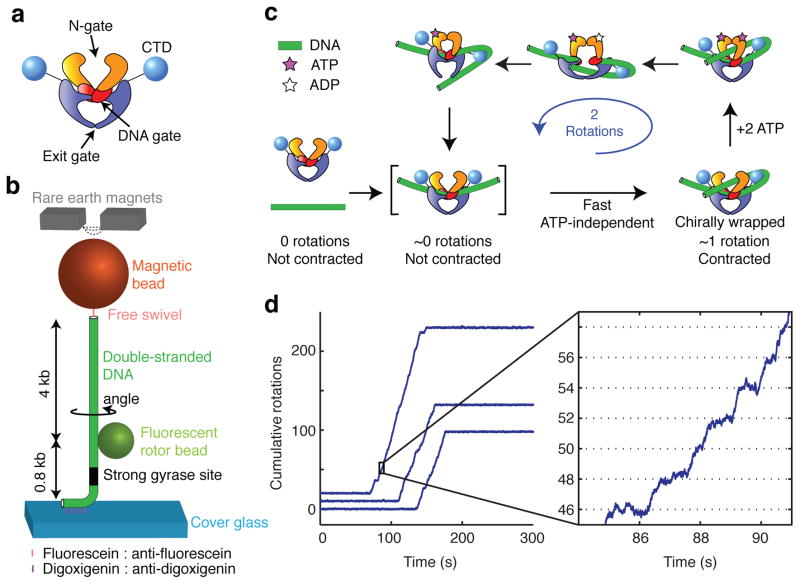
Gyrase alters DNA topology by passing one segment of DNA duplex (known as the transfer- or T-segment) through a transient gap in another duplex segment (known as the gate- or G-segment)8. Gyrase and other type IIA topoisomerases are thought to operate via a three-gate mechanism9 in which the enzyme initially binds to the G-segment, then captures a T-segment via ATP-dependent dimerization of the N-terminal domains (closing the “N-gate”), then cleaves and separates the G-segment (opening the “DNA gate”), and finally passes the T-segment through the G-segment before resealing the DNA gate and ejecting the T-segment through a third opening known as the “exit gate” (Fig. 1a).
Figure 1.
Single molecule assay for DNA gyrase activity. (a) Diagram of the gyrase tetramer showing approximate relationships between protein domains as described in the text. (b) Schematic of the rotor bead tracking (RBT) assay. The DNA template was stretched using magnetic tweezers, and the fluorescent rotor bead (diameter: ~300 nm) was imaged from below at 250 frames per second. (c) Mechanochemical model based on earlier work29,32. The expected positions of intermediates are indicated along the repeating two-rotation reaction coordinate probed by the RBT assay. (d) Plots of cumulative rotor angle acquired under 0.5 pN tension after the introduction of DNA gyrase and 1 mM ATP. Long periods of inactivity are interrupted by processive bursts of directional rotation, always in multiples of two rotations. The mean waiting time between presumptive single-enzyme bursts under these conditions was 825 ± 30 s (mean ± SEM). Detailed stepping behavior (seen in the expanded view) can be analyzed for each processive burst. At 1 mM ATP, pauses can be resolved at a spacing of two rotations (dotted lines) in phase with the 0 rotation mark established prior to enzyme binding.
Gyrase is unique among type IIA topoisomerases in its ability to unidirectionally introduce — rather than merely relax — negative supercoils10. Control of directionality was proposed to arise from a mechanism in which DNA forms a positive chiral wrap around the enzyme prior to strand passage11, yielding a positive node between the T- and G-segments. Strand passage inverts this node, resulting in the introduction of two negative supercoils12. Evidence for a chirally wrapped intermediate comes largely from analysis of nucleotide-free gyrase complexes, which trap ~ 0.8 positive supercoils per enzyme13,14 and protect ~140 bp of DNA from hydroxyl radicals and DNA digestion11,15,16. This footprint is much larger than other type II topoisomerases17 and similar in scale to the nucleosome18.
Unlike the nucleosome, DNA gyrase has not been cocrystallized with a full-length DNA binding site, and the detailed path of DNA around the holoenzyme remains unknown. The chirally wrapped conformation is thought to involve extensive DNA bending around the specialized C-terminal domain (CTD) (Figure 1A). The CTD is required for trapping of positive supercoils and for the directional supercoiling activity of DNA gyrase19, and is able to bend DNA and trap supercoils as an isolated fragment20–22. In addition to bending of the outer DNA arms around the CTDs, the full path of DNA wrapping is likely to include a sharp bend of the central G-segment, as observed in structures of the binding and cleavage core23–25.
A key step toward understanding the motor mechanism of gyrase is to define a cycle of global nucleoprotein conformations and characterize transitions between these conformations coupled to substeps in ATP consumption. Structural intermediates in the mechanochemical cycles of molecular motors can be dynamically observed using single-molecule tracking. For example, high-speed nanosphere tracking has been used to identify distinct substeps coupled to ATP binding26 and phosphate release27 within the 120° step of the F1 ATPase rotary motor. In principle, the rotor bead tracking (RBT) assay28,29 enables analogous observations of DNA gyrase activity.
In RBT, a DNA molecule is stretched between a surface and a magnetic bead (Fig. 1b). The angular position of a fluorescent ‘rotor’ bead attached to the side of the DNA reports on changes in twist of the lower, torsionally constrained DNA segment. A free swivel above the rotor prevents the permanent accumulation of torsional strain. Changes in linking number are thus immediately relaxed via rotation of the fluorescent bead, allowing visualization of enzymatic steps. Supercoils trapped by structural intermediates can also be visualized because the rotor spins to relax compensatory strain, in a single molecule analog of topology footprinting. Substeps in the cycle of DNA gyrase can thus be mapped along a repeating two-rotation reaction coordinate, analogous to the repeating 120° reaction coordinate defined by rotation of the gamma shaft in F1 ATPase.
In a previous RBT study of DNA gyrase29, processive supercoiling was analyzed at saturating [ATP] in order to map a major rate-limiting transition that may be coupled to release of hydrolysis products30. [ATP]-limited substeps were studied under distributive (but not processive) conditions, and were consistent with the formation of a chirally wrapped ATP-waiting state at ~1 rotation. DNA extension was not directly monitored, but the authors used the steep tension dependence of processivity to argue for a dynamic model in which rapid contraction occurs during DNA wrapping, and contour length is released later in the cycle. The proposed mechanochemical model maintained the core features of a mechanistic picture that has persisted for decades31,32 (Fig. 1c): ATP-independent formation of a wrapped intermediate that traps a positive supercoil, followed by ATP-dependent strand passage.
We set out to characterize gyrase mechanochemistry using new RBT assays that permit simultaneous measurements of DNA rotation and contraction, direct characterization of ATP-waiting states during processive supercoiling, and high spatiotemporal resolution. Unexpectedly, we distinguish [ATP]-dependent dwells at multiple locations along the reaction coordinate, and show that the dominant dwell corresponds to a novel structural intermediate that sequesters substantial DNA contour length but does not trap supercoils. Conversion of the novel intermediate to a chiral wrap depends on an ATP-accelerated conformational rearrangement, contradicting prevailing models in which chiral wrapping is fast and ATP-independent32.
RESULTS
A high resolution assay for DNA gyrase
To observe substeps in the enzymatic cycle of DNA gyrase, we made modifications to the RBT assay (Fig. 1b) including reducing the drag of the rotor bead and shortening the DNA tether, allowing high spatiotemporal resolution angular measurements at sufficiently low tensions to permit processive gyrase activity. In the presence of low concentrations of gyrase and 1 mM ATP, rotor angle traces contain long flat regions of constrained thermal fluctuations, interrupted by bursts of directional rotation as expected for processive introduction of negative supercoils by individual gyrase enzymes (Fig. 1d). As seen earlier, bursts occur in strict multiples of two rotations, allowing the straightforward definition of a repeating two-rotation reaction coordinate. Our high-resolution results (Fig. 1d) confirm a rate-limiting pause at ~0 rotations in saturating [ATP]29.
The dominant ATP waiting pause occurs at ~0 rotations
At limiting [ATP], previous models predict a dominant pause at ~1 rotation, since the enzyme is expected to dwell primarily in a chirally wrapped conformation awaiting nucleotide-gated strand passage14. Previous RBT measurements of individual isolated cycles at low [ATP] show that there is sometimes a pause at ~1 rotation, as expected29. However, any pauses near 0 rotations would be undetectable in these experiments. There have been no previous single molecule measurements of processive stepping at low [ATP], and it has remained unknown whether there are ATP-dependent pauses at additional positions in the cycle.
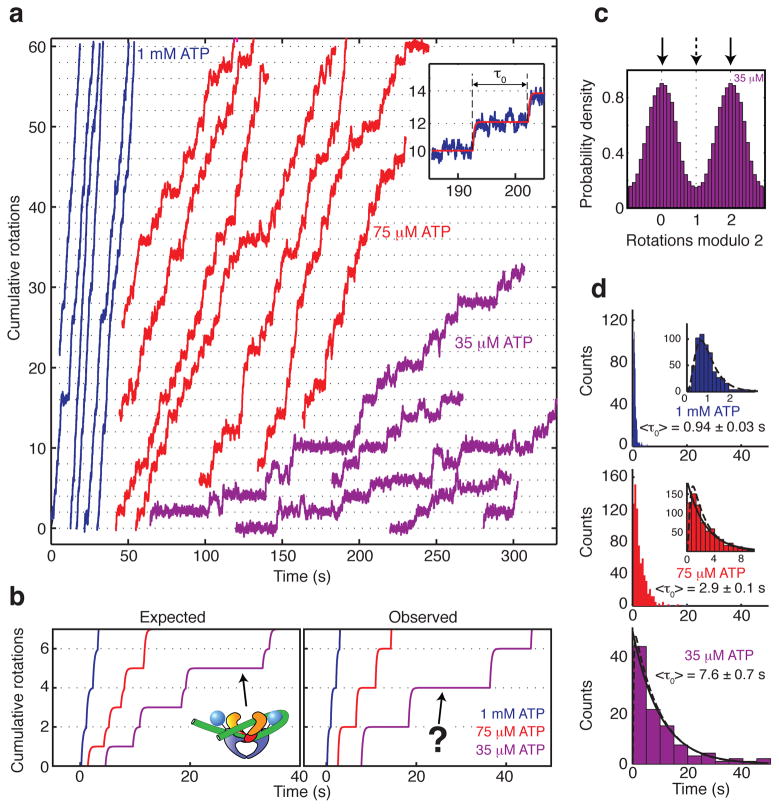
To directly investigate ATP waiting intermediates, we collected processive RBT traces under varying [ATP] (Fig. 2). Unexpectedly, lowering [ATP] primarily lengthens a pause at ~0 rotations. This result implies that the dominant ATP waiting intermediate does not trap DNA supercoils, and falsifies the previously proposed model29 in which the rate limiting pause at ~0 rotations is followed by a rapid transition to a chirally wrapped ATP-waiting intermediate at ~1 rotation (Fig. 2b).
Figure 2.
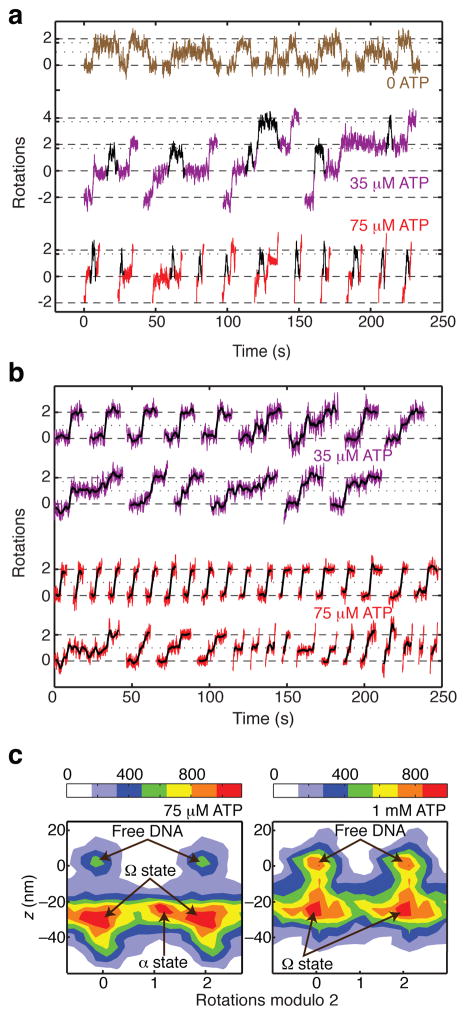
[ATP]-dependent pausing in gyrase stepping traces. All data are taken from processive bursts of gyrase activity at 0.5 pN tension. (a) Cumulative rotations as a function of time with different concentrations of ATP. Traces were fit to a stepwise model (inset; see Methods) to obtain the duration τ0 of each pause. (b) Stepwise fits to observed data are compared with a previous model30,39 which predicted that lowering [ATP] should specifically lengthen a midcycle pause at ~1 rotation. (c) Periodic histogram of rotor angles during processive activity in limiting (35 μM) ATP. Angles are plotted modulo 2 rotations, with 0 defined as the mean angle of the rotor prior to enzyme activity. Solid arrows mark the dominant pause at ~0 rotations; dashed arrow marks the expected intermediate location. (d) Waiting time histograms compiled from a total of 570 pauses at 1 mM ATP, 740 pauses at 75 μM ATP, and 97 pauses at 35 μM ATP. Solid lines are exponential functions with decay times equal to <τ0>. Dashed lines are fits to a multistep model (see Methods) in which [ATP]-independent processes occupy a substantial fraction of the waiting time at 1 mM ATP.
Distributions of waiting times in single molecule traces can provide information about hidden kinetic steps26,33. At low [ATP], waiting time distributions for DNA gyrase stepping are closely approximated by single exponentials, as expected if the kinetics are dominated by waiting for a single chemical event (Fig. 2d). Our observations are compatible with a two-ATP mechanism if the first ATP binds in rapid equilibrium, or if one ATP binds much faster than the other34. Mechanisms can be further distinguished by examining the dependence of mean waiting times on [ATP], as discussed later.
Dwell time distributions at saturating ATP are highly peaked, as expected for a multistep process (Fig. 2d). Distributions at all [ATP] are consistent with a model in which up to 0.5 s of the cycle are occupied by irreversible, [ATP]-independent processes (see Methods), explaining a rising phase that is prominent at 1 mM but nearly negligible at 75 μM ATP. Slow release of ATP hydrolysis products might thus account for a substantial fraction of the cycle time at saturating ATP, as suggested earlier for both gyrase and the related yeast topoisomerase II34–36.
Contour length remains trapped during processive bursts
Having identified a dominant ATP waiting intermediate at ~0 rotations, we used DNA extension measurements to further probe the structure of this intermediate and analyze the dynamics of DNA contraction throughout processive activity. In previous RBT experiments, DNA extension was not directly monitored — changes in DNA extension were only indirectly probed via analysis of force perturbations29.
Previous models make conflicting predictions about the dynamics of DNA contraction. Earlier single molecule studies29,37 suggested a model in which the enzyme releases its DNA wrap each cycle to dwell in a vulnerable state at ~ 0 rotations that does not trap contour length. AFM studies38 also suggested that the DNA wrap is lost and regained during each cycle, since trapped contour length was released upon nucleotide binding. In contrast, structural observations of multiple alternative CTD positions39–41 inspired “hand over hand” models in which mobile CTDs guide strand passage without fully releasing the wrapped DNA.
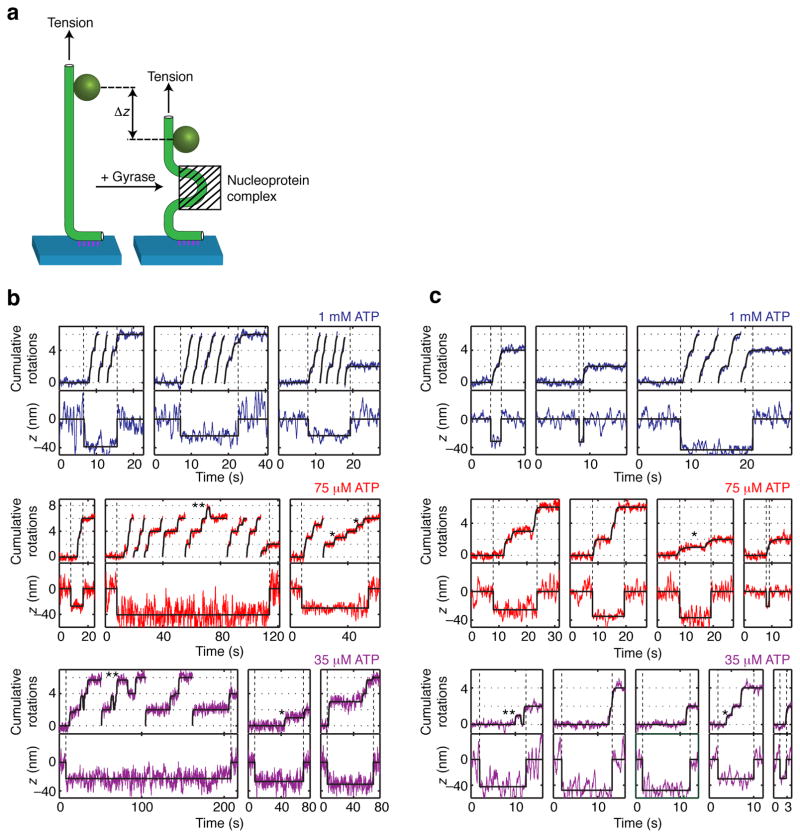
We used a defocus tracking method42 to measure the focal depth of the rotor bead simultaneously with its angular position (Fig. 3; see Methods). Changes in rotor depth reflect changes in overall DNA extension resulting from enzyme binding or structural rearrangements of the nucleoprotein complex. For all [ATP], the DNA extension drops immediately prior to each burst of rotation, and remains contracted throughout the processive burst. These measurements rule out models in which the enzyme dwells substantially in conformations where DNA is released from the enzyme.
Figure 3.
Simultaneous measurements of DNA rotation and contraction. (a) Changes in z reflect changes in overall DNA extension due to formation or rearrangement of a nucleoprotein complex. Simultaneous measurements of angle and z taken under (b) 0.5 pN or (c) 1 pN tension show that the DNA extension becomes contracted upon gyrase binding and remains contracted throughout processive bursts of activity. Traces that reach the top of the angle axis are continued from the bottom of the axis. Solid black lines show stepwise fits (see Methods). Contraction persists during the dominant [ATP]-dependent pause at 0 rotations as well as during secondary pauses at ~1 rotation (examples marked with *) and reversible excursions to ~1.7 rotations (examples marked with **). Contraction precedes angular rotation at the start of processive bursts, as indicated by the vertical dashed lines.
The dominant ATP waiting intermediate at ~0 rotations is thus revealed to be a highly contracted conformation of the nucleoprotein complex, which differs from the chirally wrapped conformation in that it does not trap supercoils. Contraction without supercoil trapping could be explained by G-segment bending, which has been observed in cocrystal structures of gyrase and other type IIA topoisomerases with DNA23–25. However, contraction due to DNA bending is suppressed by tension in the 0.2 pN – 1 pN range, and even a sharp bend is expected to generate only 10–15 nm of contraction under 1 pN of tension43,44. In contrast, we observe that the average contraction actually increases from 29 ± 1 nm at 0.5 pN (mean ± SEM, N=53) to 34 ± 2 nm at 1 pN (mean ± SEM, N=28), a larger contraction than expected from bending alone but consistent with sequestering >100 bp of contour length.
At the start of a processive burst, contraction always precedes rotation (Fig. 3b,c), indicating that the enzyme initially binds in a conformation that sequesters contour length without trapping supercoils. We propose that this initial conformation is then revisited every cycle during the dominant ATP waiting dwell.
Interconverting conformations in the absence of ATP
We have identified a structural intermediate that has two of the expected features of the chirally wrapped DNA gyrase complex: it extensively sequesters DNA contour length, and it accumulates at low [ATP]. However, this intermediate lacks a critical feature of the chirally wrapped complex, in that it does not trap any supercoils. In the absence of ATP, DNA gyrase traps ~ 0.8 positive supercoils per enzyme in topology footprinting experiments14, and generates reversible excursions to ~ +1.3 rotations in previous RBT assays29. To provide context for our measurements of structural intermediates during processive supercoiling, we conducted new measurements of contraction and rotation in the absence of ATP, matching the nucleotide condition that has previously been used to study chiral wrapping.
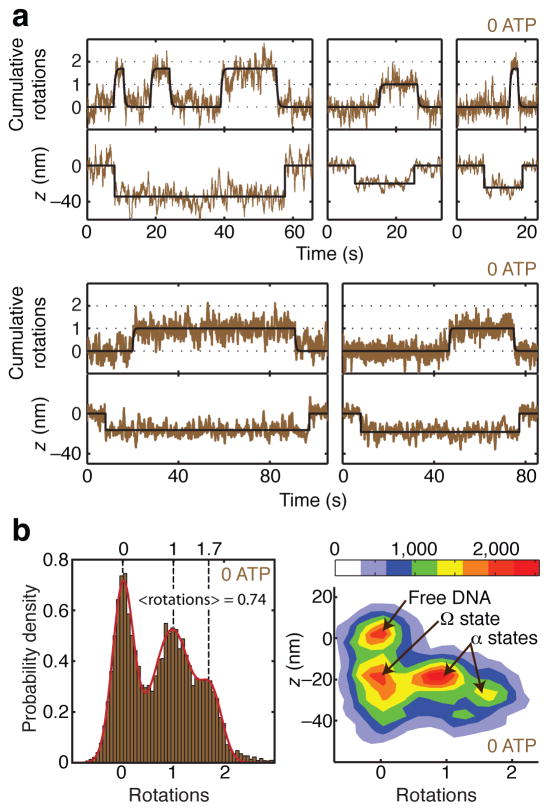
When gyrase is introduced in the absence of ATP (Fig. 4) we observe events in which DNA contraction initially occurs without rotation, and then the rotor makes reversible excursions to dwell at positive angles, as expected for reversible trapping of positive supercoils. During contraction events, the rotor appears to dwell in three distinct states centered at ~0 rotations, ~1 rotation, and ~1.7 rotations. Averaged over all three states, the mean angle visited during contraction events is 0.74 rotations (Fig. 4b).
Figure 4.
Distinct conformations of the nucleotide-free complex. (a) Measurements of angle and z during gyrase binding events in the absence of ATP, showing transitions between three distinct contracted states. Contraction initially occurs without rotation, and the rotor subsequently makes long-lived excursions to ~1 rotation and ~1.7 rotations. (b) Histograms of angle and z during gyrase binding events in the absence of ATP, comprising 41 reversible excursions. For the 1D histogram, only portions of the rotor trace showing z contraction were selected for analysis. Solid line shows a fit to the sum of three Gaussians centered at ~0 rotations, ~1 rotation, and ~1.7 rotations. The mean angle is 0.74 ± 0.07 rotations (error estimated as SEM of waiting angles based on N=87 dwells). For the 2D histogram of paired (angle, z) data, z contraction events were included along with 2000 frames (8 s) of flanking data on either side of each event, yielding a distinct population corresponding to enzyme-free DNA. The newly identified Ω state, which is contracted in z but does not trap supercoils, can be distinguished from states (designated α) that trap positive supercoils. The legend shows the correspondence between color coding and histogram counts, where each count represents a data point from a 4 ms frame.
These measurements reconcile previous studies of supercoil trapping in the absence of ATP. Previous RBT experiments did not measure DNA compaction and thus ignored the complex that traps ~0 supercoils, which was indistinguishable from free DNA. The earlier RBT study also did not distinguish between dwells at ~1 rotation and ~1.7 rotations, and reported an angle intermediate between these values. Our observed average number of supercoils trapped in contracted complexes (0.74 ± 0.07) closely matches results from bulk topology footprinting13,14. The bulk measurement can now be seen to reflect an equilibrium average over three interconverting complexes at 0 ATP that differ in the number of supercoils trapped.
To differentiate among substates of the “wrapped DNA gyrase complex”, we have introduced the term “α state” to designate complexes that trap supercoils, and “Ω state” to designate complexes that trap contour length without trapping supercoils (Fig. 4b). We can now use the term “Ω state” to describe the dominant ATP-waiting intermediate we observed during processive supercoiling.
[ATP] controls exit from the α state
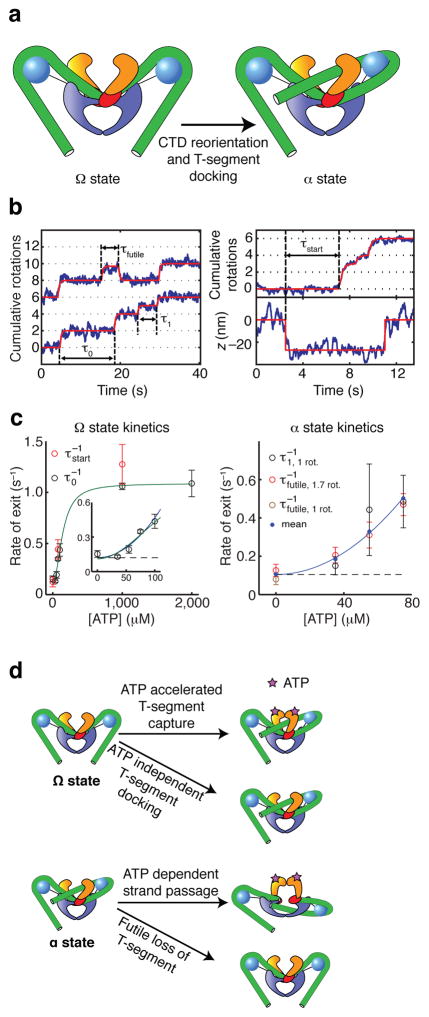
Trapping of positive supercoils at 0 ATP has long been proposed to reflect a critical on-pathway intermediate that confers directionality on the subsequent ATP-dependent strand passage reaction12,13. However, slow excursions to supercoil-trapping α states at 0 ATP could instead represent an off-pathway process unrelated to the supercoiling function of DNA gyrase. In order to test the on-pathway model in which ATP binding to the α state leads to directional strand passage, we examined whether [ATP] affects exit from α states.
Reversible excursions to α states are observed interspersed with complete forward cycles at 35 μM and 75 μM ATP (examples marked with ** in Figure 3, and compiled in Figure 5a). If these states are competent for ATP-dependent strand passage, then they should have reduced lifetimes at higher [ATP], due to the increased contribution of ATP binding to the rate of exit from the α state. Mean dwell times for these reversible excursions indeed decrease monotonically from 9 ± 3 s at 0 ATP (mean ± SEM, N=50) to 2.0 ± 0.3 s at 75 μM ATP (mean ± SEM, N=40), supporting a model in which α is an on-pathway state.
Figure 5.
[ATP]-dependent dwells in supercoil-trapping intermediates. (a) Excised regions of rotor traces showing reversible excursions to ~1.7 rotations at 35 and 75 μM ATP. Excursions are highlighted in black. The duration of excursions is reduced at higher [ATP]. (b) Excised steps from rotor traces, showing midcyle pauses at low [ATP]. Raw data are overlaid with averaged traces (2 s rolling window). Examples of steps are shown with either no detectable pause, a midcycle pause at ~1 rotation, or a midcycle pause at ~1.7 rotations. Pauses at ~1.7 rotations are rarely long enough to be discriminated from the subsequent plateau at our spatiotemporal resolution. (c) Two-dimensional histograms of paired (angle, z) data, taken from processive runs at 0.5 pN tension (comprising a total of 261 steps at 1 mM ATP and 118 steps at 75 uM ATP) together with flanking regions of inactivity (800 frames on either side of each event at 1 mM ATP, and 1500 frames on either side of each event at 75 μM ATP). At both ATP concentrations, populations can be distinguished corresponding to enzyme-free DNA and the contracted Ω state. At low [ATP], an additional population can be distinguished intermediate between 0 and 2 rotations, as expected for midcycle α states that trap positive supercoils. The mean contraction observed for a given event was 27 ± 2 nm (mean ± SEM) at 1 mM ATP, and 30 ± 2 nm (mean ± SEM) at 75 μM ATP. The legend shows the correspondence between color coding and histogram counts, where each count represents a data point from a 4 ms frame.
Direct visualization of on-pathway intermediates
On-pathway α states should also be detectable as pauses at ~1 rotation or ~1.7 rotations in the middle of forward steps. Midcycle pauses can be seen in some, but not all steps at low [ATP] (examples marked with * in Figure 3, and compiled in Figure 5b), agreeing with previous reports that a midcycle pause can be seen in some isolated cycles at 25 μM ATP29. Midcycle pauses at ~1.7 rotations are harder to detect than midcyle pauses at ~1 rotation, because of the proximity of the subsequent plateau at 2 rotations.
Distinct contracted intermediates of the DNA:gyrase complex can be distinguished as populations in 2D histograms of angle and z under varying [ATP] (Fig. 4b, 5c). Ω states are clearly separated from free DNA along the z coordinate and from α states along the angle coordinate. α states at ~1 rotation and ~1.7 rotations are sampled at 0 ATP, and an α state at ~1 rotation remains detectable at low [ATP]. Our measurements suggest that for all [ATP], the enzyme initially binds in an Ω conformation, and then undergoes a transition to an α conformation before either completing a forward cycle or returning unproductively from α to Ω.
The Ω to α transition may correspond to T-segment docking
The kinetics of supercoiling are dominated by dwells in the newly identified Ω state, which is distinguished by sequestering substantial contour length without trapping supercoils. The properties of this state can be explained (Fig. 6) if the DNA is bent around the CTDs but traces a nearly planar path, or a path that fluctuates to trap zero writhe on average. The slow transition to the α state may then correspond to docking of a T-segment between the ATPase domains, trapping positive writhe. The strong [ATP] dependence of the rate of exit from the Ω state (Fig. 6c) indicates that ATP favors the Ω to α transition. Rapid T-segment capture may be mediated by ATP-dependent closure of the N-gate.
Figure 6.
Structure and kinetics of intermediates. (a) Cartoons showing hypothetical conformations of the nucleoprotein complex (see also Supplementary Figure 1). In the Ω state, the DNA (green tube) makes extensive contacts with the CTDs but does not form a chiral loop, explaining the absence of supercoil trapping. Ω can be converted to the previously proposed chirally wrapped α states via reorientation of a CTD and docking of a T-segment between the ATPase domains, trapping positive writhe. (b) Stepwise fits (solid lines) to angle and z traces were used to extract four categories of dwell times. Durations were determined as illustrated for dominant pauses (τ0), midcycle pauses (τ1), reversible excursions (τfutile), and lag times between contraction and rotation at the start of processive bursts (τstart). (c) [ATP]-dependent kinetics of α and Ω states. Error bars indicate SEM. <τ0> and <τstart> are similar for all [ATP], as expected if both measurements reflect the lifetime of the Ω state. <τ1> and <τfutile> are also similar for all [ATP], as expected if both measurements reflect the lifetime of the α state. Ω state kinetics saturate at high [ATP] (solid green line; see Supplementary Notes). At low [ATP], the [ATP] dependence of exit rates from both the α state and the Ω state can be approximated by a parabola (solid blue line) with a non-zero intercept (dashed line), as expected for models (d) in which an ATP-independent process competes with a process requiring two ATP molecules. The kinetics of the Ω state are explained by a model in which T-segment docking can occur via either a slow ATP-independent process or an ATP-accelerated process requiring 2 ATP. Similarly, the kinetics of the α state can be explained by competition between forward progress (requiring 2 ATP) and slow reversion to Ω.
The extent of DNA contraction remains approximately constant during the Ω to α transition (Fig. 3–5). The conformational rearrangement may involve active or passive reorientation of the CTDs, while retaining extensive contact between the CTDs and the DNA. Detailed structural proposals for the conformations of these states must remain speculative at this stage, but some possible geometries under tension are illustrated in Supplementary Figure 1.
Kinetics reflect parallel pathways for the Ω to α transition
Our results build toward a model in which dominant pauses at ~0 rotations correspond to dwells in the Ω state awaiting supercoil trapping via T-segment docking, which can occur slowly in the absence of ATP or quickly when ATP is bound. We challenged this model by analyzing the lifetimes of rotational pauses (Fig. 6). In our model, the lag τstart between initial contraction and subsequent rotation at the beginning of a processive bursts corresponds to a dwell in Ω, and the enzyme dwells in the same Ω state during measured pauses τ0 that recur each cycle during processive supercoiling (Fig. 6b). As expected, the mean values of τstart and τ0 are similar as a function of [ATP] (Fig. 6c). The rate of exit from Ω has a quadratic dependence on [ATP], consistent with a model in which rapid T-segment capture requires binding of two ATP molecules. A non-zero intercept for the rate of exit as a function of [ATP] reflects an alternative slow process in which a T-segment can be reversibly docked in the absence of nucleotide (Fig. 6d). Increasing the tension from 0.5 pN to 1 pN does not substantially affect the rate of exit from Ω (Supplementary Fig. 2).
Strand passage competes with futile T-segment release
At low [ATP], we often observe pauses at ~1 rotation or ~1.7 rotations, which we have ascribed to dwells in a chirally wrapped α state awaiting ATP-dependent strand passage. Our measurements of midcycle pauses τ1 as well as reversible excursions τfutile are both expected to reflect the lifetime of the α state, with τfutile reflecting events in which futile T-segment release wins a kinetic competition with strand passage. As expected, τ1 and τfutile have similar mean values as a function of [ATP] (Fig. 6c). As with the Ω state, ATP-dependent kinetics of exit are described by a parabola with a non-zero intercept, as expected if a process requiring two ATP molecules (in this case, strand passage) competes with an ATP-independent process (in this case, futile T-segment release) (Fig. 6d).
Our results imply that the T-segment can remain docked for ~10 s in the absence of nucleotide. The docked state may be stabilized by partial N-gate closure: a recent FRET study reported narrowing of the N-gate in nucleotide-free DNA:gyrase complexes, as compared with gyrase alone45.
A branched kinetic model for gyrase mechanochemistry
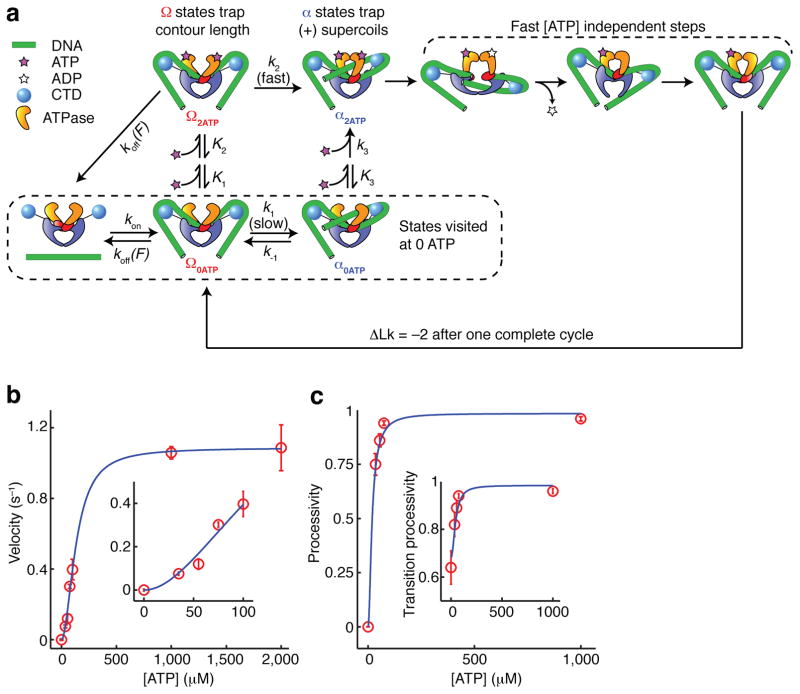
In the mechanochemical model that emerges from this study, the principal pathway involves binding of two ATP molecules in the Ω state, followed by an accelerated Ω→α transition in which a T-segment is docked in the upper cavity between the ATPase domains. Subsequent strand passage is rapid, explaining why many observed cycles contain no detectable dwell in the α state, even at low [ATP] (Fig. 2a, 5b). In a parallel pathway, T-segment docking can occur via a slow ATP-independent process. In this case, the enzyme reaches an ATP-free α state, and must wait for two ATP molecules to bind before proceeding to strand passage, yielding a substantial dwell in α. We have incorporated this branching model into an overall mechanochemical cycle (Fig. 7) that quantitatively explains many of our single molecule observations, including the dependence of α and Ω lifetimes on [ATP] (Fig. 6c), the distributions of Ω lifetimes at low [ATP] (Fig 2d), and the [ATP] dependence of supercoiling velocity (Fig. 7b) and processivity (Fig. 7c).
Figure 7.
Branched kinetic model for structural transitions and ATP coupling in DNA gyrase. (See also Supplementary Figure 3.) (a) Diagram of allowed transitions in the model. Several ATP binding transitions are approximated as rapid equilibria with state-dependent affinities K1, K2, and K3. Rate constants are labeled for all other transitions. Some reverse rates are assumed to be negligible and are not shown. The dissociation rate koff(F) varies with the tension F in the DNA molecule; all other rates are tension-independent. Unlabeled arrows represent transitions of unknown kinetics but are fast relative to k2. In aggregate, they may occupy as much as 0.5 s of the cycle (Fig. 2b). (b) Overall supercoiling velocity (expressed in strand passages per second) as a function of [ATP], as measured experimentally and predicted from the kinetic model. Inset expands the low [ATP] region. (c) Processivity of the enzyme, defined as the probability of completing a forward step before dissociating, as a function of [ATP]. The transition processivity (inset) is defined as the probability of docking a T-segment before dissociating, and remains measurable at 0 ATP. Error bars indicate SEM.
Parameters for our model are summarized in Figure 7a. Transitions are modeled as requiring two ATP binding events, with the first ATP binding weakly and reversibly34. We assume that the contracted Ω state is vulnerable to dissociation, which explains the high sensitivity of processivity to force, and further predicts that processivity should decrease at low [ATP], as we have observed (Fig. 7c). This trend is in the opposite direction from the [ATP] dependence of processivity in dimeric walking myosins46, and contradicts a previous model29 in which processivity should be insensitive to [ATP].
DISCUSSION
In a longstanding model for directional supercoiling by DNA gyrase14,29,31,37, formation of a chirally wrapped intermediate is an ATP-independent process that closely coincides with DNA binding, and ATP is only required to drive the subsequent strand transfer reaction. We have now shown that ATP is required to overcome a kinetic barrier to chiral wrap formation. By mapping out structural intermediates on a twist-extension plane, we have distinguished chirally wrapped “α states” from a newly defined “Ω state” that sequesters the DNA contour length involved in the wrap but does not trap a positive supercoil. The Ω→α remodeling transition dominates the kinetics of processive supercoiling over a wide range of [ATP], reflecting a high barrier to spontaneous rearrangement. ATP binding accelerates the Ω→α transition by at least an order of magnitude (Fig. 7a), via a mechanism that we propose is linked to T-segment capture (Fig. 6).
Multiple conformations of the DNA:gyrase complex
Our results prompt a re-examination of the “wrapped” DNA:gyrase complex. In the absence of ATP, the DNA:gyrase complex samples several functionally relevant conformations that all sequester contour length but differ in the number of supercoils trapped (Fig. 4). Previous bulk studies of the nucleotide-free wrapped complex (using topology footprinting14, hydroxyl radical footprinting15, and SAXS41) likely measured averaged properties of coexisting conformations, whereas previous RBT measurements in the absence of ATP29 selectively measured α states, overlooking Ω states because of the absence of extension measurements.
In the presence of AMPNP, gyrase retains a large DNA footprint15 but does not trap supercoils14, and shows an altered pattern of DNA protection relative to the nucleotide-free enzyme15. These properties are consistent with specific stabilization of the Ω state; after a single round of strand passage supported by AMPNP47,48, the enzyme may adopt an Ω-like configuration. The T-segment is not recaptured because the N-gate is clamped shut; the enzyme cannot reset without hydrolysis.
Subdivision of the “wrapped” conformation also enables reinterpretation of previous dynamic single molecule data29,37, whose interpretation relied on the erroneous assumption that sequestering contour length is always coincident with trapping supercoils. Since the rate-limiting dwell was observed in a state that did not trap supercoils, it was assumed that this state did not sequester contour length. Processivity was found to be a strong function of DNA tension, which was explained by a rapid tension-sensitive wrapping step that competes with dissociation. In our new model, the force sensitivity of processivity is explained by a force-sensitive dissociation rate for the highly contracted Ω state.
Structural rearrangements during the gyrase cycle have been proposed to involve changes in the positions of the CTDs, based on observations of complexes with CTDs in widely different positions: in plane with the DNA gate in a crystal structure of the related topoisomerase IV enzyme39, and positioned near the exit gate in SAXS studies of GyrA40 and holoenzyme41. FRET measurements further indicate that the CTD orientation in the DNA:gyrase complex differs from the orientation in gyrase alone49. Dynamic cycling of CTD positions has still not been directly observed, but our measurements of the Ω→α transition would be difficult to reconcile with a model in which the CTD positions remain static, and it is likely that this kinetically limiting transition involves dramatic CTD reorientation.
While our measurements reveal a central role for the newly defined Ω state, we also confirm the role of chirally wrapped intermediates (α states) as on-pathway intermediates for directional supercoiling. Additionally, we observe two conformational variants of the α state in which either ~1 or ~1.7 supercoils are trapped (Supplementary Fig. 1). These variants are structurally distinct but functionally and kinetically indistinguishable in our assays. We can only speculate on the structural details of the variants, but it is plausible that trapping ~1 supercoil requires rearrangement of only 1 CTD, while trapping ~1.7 supercoils requires a concerted rearrangement of both CTDs (Supplementary Figure 1).
Mechanochemical coupling in type II topoisomerases
Molecular motor mechanisms are often described using models with a strict correspondence between chemical states and structural intermediates. However, loose coupling between individual mechanical transitions and substeps of the ATP hydrolysis cycle has emerged as an important feature of type II topoisomerase mechanisms. Strand passage was initially thought to be independent of ATP hydrolysis, because one round of passage can be supported by non-hydrolysable ATP analogs47,48. Single turnover assays of yeast topoisomerase II ultimately showed that strand passage is slow unless it is accelerated by ATP hydrolysis50. For DNA gyrase, we have now similarly shown that trapping a positive supercoil is slow unless it is accelerated by ATP binding.
In addition to the possibility of strand passage without hydrolysis, it is also possible for hydrolysis to occur without strand passage, with detrimental effects on the fuel efficiency of the enzyme. In yeast topoisomerase II, ~8 ATP are hydrolyzed for each strand passage event at saturating [ATP]51. In DNA gyrase, more efficient coupling ratios have been reported close to 1 ATP per strand passage when the substrate is a relaxed DNA molecule52, although coupling becomes poor in the presence of high levels of negative supercoiling. A comparison of our measured overall reaction velocity at saturating [ATP] (1.1 s−1) with the previously measured ATPase rate (0.9 s−1) on nicked DNA53 is consistent with efficient coupling, and our kinetic model (Fig. 7) can be extended to suggest a mechanism in which futile hydrolysis is avoided by coordination between T-segment docking and N-gate closure (Supplementary Figure 4).
Chiral DNA wrapping is the critical step that confers directionality to the supercoiling function of DNA gyrase. We have shown that wrapping is a coordinated multistep process in which sequestering contour length is followed by an ATP-accelerated conformational rearrangement that traps positive supercoils. This study contributes to an emerging picture of loosely coupled mechanochemical transitions in GHL ATPases, in which nucleotide states cannot be uniquely identified with structural intermediates54. Our results illustrate the ability of dynamic measurements to complement structural investigations of trapped intermediates, by identifying functional states, directly characterizing transitions, and revealing mixtures of conformations. Our methods may further be used to identify substeps in the gyrase cycle affected by antibiotic drugs that target the enzyme6, and characterize the mechanisms of novel inhibitors developed during ongoing drug discovery efforts24.
METHODS
Preparation of DNA, proteins, and beads
DNA constructs for rotor bead tracking were prepared by ligation of three modified PCR fragments digested with BbsI (New England Biolabs, NEB) to generate non-palindromic overhangs. The lower segment (302 bp) was body-labeled with digoxigenin–dUTP (Roche) as described28; the middle segment (802 bp from pMP1000, containing a variant of the μ phage strong gyrase site56) was singly labeled with biotin using an internally modified PCR primer; and the top segment (3900 bp amplified from pBiex-1, Novagen) was labeled with fluorescein using a 5′ modified primer. 1 μm carboxy-modified superparamagnetic beads (MyOne, Invitrogen) were crosslinked with rabbit anti-fluorescein (Invitrogen)29. Rotor beads were ‘yellow-green’ neutravidin-coated Fluospheres (Invitrogen) with a nominal diameter of 200 nm; measured diameters were 300 ± 20 nm (mean ± s.d., N=20) based on rotor orbits. E. coli GyrA and GyrB subunits were individually expressed and purified as described57, mixed to reconstitute tetramers, and stored at −80°C in 50 mM Tris-HCl, pH 7.5, 100 mM potassium glutamate, 2 mM DTT, 1 mM EDTA, and 10% (v/v) glycerol.
Chamber preparation
Flow chambers were assembled using Nescofilm gaskets and hole-punched coverslips, with the objective-side coverslip spin-coated with 0.1 % (w/v) nitrocellulose (Ernest F. Fullam). To prepare rotor:DNA complexes, Fluospheres were centrifuged, resuspended in PB1 (400 μl of 5 mg ml−1 acetylated BSA (Bovine Serum Albumin, Invitrogen) in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5) to match their original suspended volume, sonicated (20 min), and incubated (1h) with DNA (~5 pM). Chambers were incubated with 0.79 μg ml−1 anti-digoxigenin (Roche) in PBS (30 min), washed with 400 μl PB1, and incubated in PB1 (1h) followed by rotor:DNA complexes (1h). Chambers were then washed with 400 μl PB1, incubated with magnetic beads in PB1 (30 min), and finally washed with 400 μl of 0.5 mg ml−1 BSA (NEB) in PBS.
Assays
Gyrase tetramer (0.6 nM – 6 nM) was added in GB (35 mM Tris-HCl pH 7.6, 24 mM potassium glutamate, 4 mM MgCl2, 2mM DTT, 0.25 mg ml−1 BSA (NEB), 0.2 mM spermidine) containing indicated [ATP], 10 mM phosphocreatine and 1.23 μM creatine phosphokinase (Calbiochem) for ATP regeneration, and an oxygen scavenging system consisting of 0.4% (w/v) glucose, 216 μg ml−1 glucose oxidase (Calbiochem), 36 μg ml−1 A. niger catalase (Calbiochem) and 26 μg ml−1 bovine liver catalase (Sigma). Assays were performed at ambient temperature (22 ± 1 °C).
Instrumentation
Magnetic tweezers were implemented on a modified Nikon TE2000 inverted microscope. The fluorescent rotor bead was excited with a 488 nm diode-pumped solid-state laser (Spectra Physics) using diagonal illumination58 through a 1.49 NA 60X objective (Nikon). Fluorescence and brightfield images were acquired simultaneously using dualview optics on an EMCCD camera (Andor iXon DV860) at a frame rate of 250 Hz. Forces were applied using permanent magnets (K & J magnetics) mounted on a motorized translation state (Physik Instrumente M-126). An XYZ piezo stage (Mad City Labs) was used to control the position and focus of the sample.
Angle and z tracking
Rotor angle was determined from the centroid position of the orbiting fluorescent rotor bead29. The centroid position of the magnetic bead was simultaneously recorded to allow corrections for stage drift and for lateral fluctuations of the DNA molecule. To measure the z height of the rotor, we used a calibrated defocus method42 in which a cylindrical lens is placed in the imaging path to introduce astigmatism in the rotor image. For each frame, we computed an empirical metric of point spread function ellipticity , where IX and IY are pixel intensities binned along each respective axis. ϕ(z) was calibrated for each rotor bead by stepping the objective focal plane using the piezo stage, accounting for the focal shift (0.78, measured as described59) arising from index mismatch between the immersion oil and the aqueous sample.
Spatiotemporal resolution
Thermal fluctuations of harmonically constrained rotors are described by an angular variance and a relaxation time , where κ is the torsional stiffness of the tether and γ is the rotational drag of the rotor. Under gyrase assay conditions, we measured 〈Δθ2〉 = 4.2 ± 0.4 rad2 and τ 0.29 ± 0.09 s (mean ± s.d., N=20), where τ was obtained by fitting the mean square deviation of the rotor angle as described60.
Substep analysis
The angular trace was modeled as a series of stepwise changes in the equilibrium rotor angle. The instantaneous rotor angle follows the equilibrium angle with a lag, modeled as an exponential relaxation with the characteristic decay time τ determined independently for each rotor as above. Step locations were obtained by least squares fitting, assuming that each forward cycle contains a dwell at 0 rotations, and may contain either a detectable (> 1 s) dwell at 1 rotation, a detectable (> 4 s) dwell at 1.7 rotations, or no detectable midcycle pause. 〈τ1〉 was shifted by the detection limit cutoff to correct for undetected short pauses46.
Waiting time distributions
Dashed lines in Figure 2d were calculated from a multistep model in which ATP-independent processes are approximated as three irreversible steps with equal rate constants kfast, leading to a distribution that is the convolution of four exponential functions representing one [ATP]-dependent and three [ATP]-independent steps. kfast = 6.5 s−1 was determined from a least squares fit to the 1 mM ATP histogram, and maintained for other [ATP]. The total time occupied by [ATP]-independent processes in this model is .
Software Used
Analysis was performed using custom scripts in MATLAB (Mathworks). Figures were prepared using MATLAB and Adobe Illustrator.
Supplementary Material
Table 1.
Parameters in the mechanochemical model for DNA gyrase
| Quantity | Value | Comments | |
|---|---|---|---|
|
|
114 ± 14 μM | Modeling assumes K1 ≫ K2. A previous model55 predicted K1 = 590 μM and K2 = 70 μM, giving | |
| k2 | 1.05 ± 0.09 s−1 | For simplicity, the value of k2 used here corresponds to the maximum supercoiling velocity, underestimating the raw transition rate. It is interesting to compare k2 with the ATPase rate of 0.9 s−1 measured on nicked DNA substrates53. | |
| k1 | 0.08 ± 0.03 s−1 | Rate of ATP-independent T-segment docking. | |
| k−1 | 0.10 ± 0.03 s−1 | Rate of futile T-segment release. | |
| koff(0.5 pN) | 0.038 ± 0.008 s−1 | All other rates are assumed to be tension independent. | |
|
|
7 ± 2 × 10−5 μM−2s−1 | Data are insufficient to independently constrain k3 and K3. |
For fitting procedures and error analysis, see Supplementary Notes.
Acknowledgments
We would like to thank J. Rubin and M. Yi for assistance with early assay development; and A. Spakowitz, L. Koslover, A. Vologodskii, A. Maxwell, A. Bates, and members of the Bryant and Berger labs for helpful discussions and critical comments on the manuscript. This work was supported by a Pew Scholars Award and NIH Grant DP2 OD004690 to Z.B. and by a Stanford Bio-X Graduate Fellowship to A.B.
Footnotes
AUTHOR CONTRIBUTIONS
A.B. and Z.B. designed the single molecule experiments and developed the mechanochemical model. A.B. established the high-resolution assay, performed the experiments, and analyzed the data. A.J.S. and J.M.B. provided purified DNA gyrase subunits and gyrase expertise. A.B. and Z.B. wrote the paper in consultation with J.M.B. Z.B. supervised the project. All authors discussed the results and commented on the manuscript.
MAIN REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nature Reviews Molecular Cell Biology. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Peter BJ, et al. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biology. 2004;5 doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Molecular microbiology. 2008;69:1071–90. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saraste M, Sibbald PR, Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends in biochemical sciences. 1990;15:430–4. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 6.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chemistry & biology. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YS, et al. Update on Hsp90 inhibitors in clinical trial. Current topics in medicinal chemistry. 2009;9:1479–92. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–92. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 9.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochemical Society transactions. 2005;33:1465–70. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 10.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–6. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LF, Wang JC. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–84. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 12.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–3. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 13.Liu LF, Wang JC. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc Natl Acad Sci U S A. 1978;75:2098–102. doi: 10.1073/pnas.75.5.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orphanides G, Maxwell A. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 1994;22:1567–75. doi: 10.1093/nar/22.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–9. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 17.Lee MP, Sander M, Hsieh T. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. The Journal of biological chemistry. 1989;264:21779–87. [PubMed] [Google Scholar]
- 18.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 19.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc Natl Acad Sci U S A. 2004;101:7293–8. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J Biol Chem. 2005;280:26177–84. doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 22.Reece RJ, Maxwell A. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 1991;19:1399–405. doi: 10.1093/nar/19.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–5. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 24.Bax BD, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–40. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 25.Laponogov I, et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nature structural & molecular biology. 2009;16:667–9. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 26.Yasuda R, Noji H, Yoshida M, Kinosita K, Jr, Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 27.Adachi K, et al. Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–21. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–41. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 29.Gore J, et al. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–4. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates AD. DNA topoisomerases: single gyrase caught in the act. Current biology: CB. 2006;16:R204–6. doi: 10.1016/j.cub.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 31.Cozzarelli NR. DNA gyrase and the supercoiling of DNA. Science. 1980;207:953–60. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- 32.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–9. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Rief M, et al. Myosin-V stepping kinetics: a molecular model for processivity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9482–6. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harkins TT, Lewis TJ, Lindsley JE. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998;37:7299–312. doi: 10.1021/bi9729108. [DOI] [PubMed] [Google Scholar]
- 35.Ali JA, Orphanides G, Maxwell A. Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry. 1995;34:9801–8. doi: 10.1021/bi00030a018. [DOI] [PubMed] [Google Scholar]
- 36.Ali JA, Jackson AP, Howells AJ, Maxwell A. The 43-Kilodalton N-Terminal Fragment of the DNA Gyrase-B Protein Hydrolyzes Atp and Binds Coumarin Drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 37.Nollmann M, et al. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat Struct Mol Biol. 2007;14:264–71. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 38.Heddle JG, Mitelheiser S, Maxwell A, Thomson NH. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J Mol Biol. 2004;337:597–610. doi: 10.1016/j.jmb.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 39.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–61. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Costenaro L, Grossmann JG, Ebel C, Maxwell A. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure. 2005;13:287–96. doi: 10.1016/j.str.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 41.Baker NM, Weigand S, Maar-Mathias S, Mondragon A. Solution structures of DNA-bound gyrase. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–3. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vologodskii A. Determining protein-induced DNA bending in force-extension experiments: theoretical analysis. Biophys J. 2009;96:3591–9. doi: 10.1016/j.bpj.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JY, Nelson PC, Betterton MD. Entropic elasticity of DNA with a permanent kink. Macromolecules. 2006;39:8816–8821. [Google Scholar]
- 45.Gubaev A, Klostermeier D. DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14085–90. doi: 10.1073/pnas.1102100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elting MW, Bryant Z, Liao JC, Spudich JA. Detailed tuning of structure and intramolecular communication are dispensable for processive motion of myosin VI. Biophysical journal. 2011;100:430–9. doi: 10.1016/j.bpj.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4838–42. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–40. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 49.Lanz MA, Klostermeier D. Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic acids research. 2011 doi: 10.1093/nar/gkr680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc Natl Acad Sci U S A. 1999;96:13685–90. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. The Journal of biological chemistry. 1993;268:8096–104. [PubMed] [Google Scholar]
- 52.Sugino A, Cozzarelli NR. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980;255:6299–306. [PubMed] [Google Scholar]
- 53.Bates AD, ODea MH, Gellert M. Energy coupling in Escherichia coli DNA gyrase: The relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry. 1996;35:1408–1416. doi: 10.1021/bi952433y. [DOI] [PubMed] [Google Scholar]
- 54.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nature structural & molecular biology. 2009;16:281–6. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 55.Kampranis SC, Maxwell A. The DNA gyrase-quinolone complex. ATP hydrolysis and the mechanism of DNA cleavage. The Journal of biological chemistry. 1998;273:22615–26. doi: 10.1074/jbc.273.35.22615. [DOI] [PubMed] [Google Scholar]
- 56.Pato ML, Howe MM, Higgins NP. A DNA Gyrase-Binding Site at the Center of the Bacteriophage Mu Genome Is Required for Efficient Replicative Transposition. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8716–8720. doi: 10.1073/pnas.87.22.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoeffler AJ, May AP, Berger JM. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic acids research. 2010;38:7830–44. doi: 10.1093/nar/gkq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature methods. 2008;5:159–61. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 59.Sun Y, McKenna JD, Murray JM, Ostap EM, Goldman YE. Parallax: high accuracy three-dimensional single molecule tracking using split images. Nano Lett. 2009;9:2676–82. doi: 10.1021/nl901129j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gore J. Thesis. Univ. California Berkeley; 2005. Single-Molecule Studies of DNA Twist Mechanics and Gyrase Mechanochemistry. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.