Summary
Srs2 is a Super-Family 1 helicase that promotes genome stability by dismantling toxic DNA recombination intermediates. However, the mechanisms by which Srs2 remodels or resolves recombination intermediates remain poorly understood. Here, single molecule imaging is used to visualize Srs2 in real time as it acts on single-stranded DNA (ssDNA) bound by protein factors that function in recombination. We demonstrate that Srs2 is highly processive and translocates rapidly (~170 nucleotides per second) in the 3′→5′ direction along ssDNA saturated with replication protein A (RPA). We show that RPA is evicted from DNA during the passage of Srs2. Remarkably, Srs2 also readily removes the recombination mediator Rad52 from RPA-ssDNA, and in doing so promotes rapid redistribution of both Rad52 and RPA. These findings have important mechanistic implications for understanding how Srs2 and related nucleic acid motor proteins resolve potentially pathogenic nucleoprotein intermediates.
eTOC
De Tullio et al., develop a single molecule assay for directly visualizing the behavior of the yeast helicase Srs2 as it acts upon single-stranded DNA (ssDNA). These experiments demonstrate that Srs2 is capable of rapid and processive translocation on ssDNA coated with the homologous recombination accessory proteins RPA and Rad52.
Introduction
Homologous recombination (HR) allows for repair of DNA double-strand breaks (DSBs), and that also functions in the recovery of stalled or collapsed replication forks (Paques and Haber, 1999; Symington et al., 2014). Several helicases help regulate recombination by dismantling aberrant or toxic HR intermediates (Bernstein et al., 2010; Brosh, 2013). Defects in the helicases that participate in HR are associated with genetic disorders, cancers and cancer-prone syndromes, and premature ageing (Bernstein et al., 2010; Brosh, 2013).
During HR, the DNA break ends are processed to yield long 3′ ssDNA overhangs (Symington et al., 2014). These overhangs are bound by the heterotrimeric single-strand binding protein Replication Protein A (RPA) (Chen and Wold, 2014; Wold, 1997). RPA protects the ssDNA against gratuitous nuclease action, helps eliminate secondary structure in the DNA, and the RPA-ssDNA complex serves an important signaling role in triggering the DNA-damage checkpoint (Zou and Elledge, 2003). For HR to occur, RPA must be replaced by the recombinase Rad51, a process that is facilitated by mediator proteins (Heyer et al., 2010; San Filippo et al., 2008). Rad52 is the key HR mediator in S. cerevisiae (Symington et al., 2014), and also mediates second DNA strand capture during a late stage of recombination (Lao et al., 2008; McIlwraith and West, 2008; Nimonkar et al., 2009; Sugiyama et al., 2006b). Moreover, Rad52 promotes Rad51-independent repair through its ability to anneal complementary DNA strands that are coated by RPA (Mortensen et al., 1996; Sugiyama et al., 2006a; Sugiyama et al., 1998). Rad52 co-localizes with both RPA and Rad51 at induced DSBs in vivo (Gasior et al., 1998; Lisby et al., 2004), and also forms spontaneous foci during S–phase, reflecting HR–dependent repair of stalled replication forks (Lisby et al., 2004). The importance of Rad52 is underscored by the susceptibility of yeast rad52 mutants to DNA damage, and depletion of human RAD52 engenders synthetic lethality in combination with a deficiency in BRCA1, BRCA2, PABL2, or RAD51 paralogs (Feng et al., 2011).
S. cerevisiae Srs2 participates in DNA replication and recombination, removal of mis-incorporated ribonucleoside monophosphates from newly replicated DNA, prevention of triplet repeat expansion, DNA-damage checkpoint responses, post-replication repair (PRR), and non-homologous end joining (Marini and Krejci, 2010; Niu and Klein, 2016). SRS2 mutations result in sensitivity to DNA damaging agents (Palladino and Klein, 1992). The near lethal phenotype of srs2Δ sgs1Δ and srs2Δ rad54Δ double mutants is restored by deletion of Rad51, suggesting that lethality stems from toxic recombination intermediates (Ira et al., 2003; Klein, 2001). Indeed, Srs2 is well established as a prototypical anti-recombinase due to its ability to remove Rad51 from ssDNA (Antony et al., 2009; Krejci et al., 2003; Marini and Krejci, 2010; Niu and Klein, 2016; Qiu et al., 2013; Sasanuma et al., 2013; Vasianovich et al., 2017; Veaute et al., 2003). Srs2 is orthologous to E. coli UvrD helicase, which can remove the bacterial recombinase RecA from ssDNA (Petrova et al., 2015; Veaute et al., 2005). Human FBH1, RECQ5, and other helicases have been shown to remove RAD51 from ssDNA as well (Bernstein et al., 2010; Branzei and Szakal, 2017; Brosh, 2013). These examples highlight helicase-mediated disruption of recombinase-ssDNA complexes as a mechanism of avoiding potentially pathogenic recombination intermediates (Branzei and Szakal, 2017; Brosh, 2013).
Little is known about how Srs2 is targeted to recombination intermediates, how Srs2 activities are regulated during different stages of recombination, or how Srs2 affects the stability of nucleoprotein complexes that harbor proteins other than Rad51. Here, we have examined the behavior of Srs2 on ssDNA bound by RPA and Rad52. These substrates mimic intermediates present during the earliest stages of HR prior to assembly of the Rad51 presynaptic complex. We demonstrate that Srs2 rapidly translocates in the 3′→5′ direction along both RPA-coated ssDNA and Rad52-RPA-ssDNA. We also show that Srs2 strips RPA and Rad52 from ssDNA to promote the recycling of both proteins. Together, our findings provide insights into how Srs2 mediates the clearance of potentially pathogenic recombination intermediates, and they also suggest that Srs2 helps redirect assembly of presynaptic complexes capable of the catalysis of recombination at proper locations.
Results
Visualizing the behaviors of Srs2 on RPA-coated ssDNA
We have established ssDNA curtains for examining proteins that interact with single-stranded DNA (Gibb et al., 2014b; Ma et al., 2017; Qi et al., 2015). In brief, long ssDNA substrates (≥50 kilo-nucleotides) are generated by rolling circle replication. The resulting ssDNA is tethered to a supported lipid bilayer on the surface of a microfluidic sample chamber through a biotin-streptavidin linkage. The ssDNA is then aligned at nanofabricated barriers to lipid diffusion, which are deposited onto the fused silica by electron beam lithography (Ma et al., 2017). Addition of GFP- or mCherry-RPA allows the ssDNA to be extended by hydrodynamic force, and also provides means of visualizing the ssDNA by total internal reflection fluorescence microscopy (TIRFM)(Figure S1a, b).
Srs2 has a robust ssDNA-dependent ATPase activity, and ATP hydrolysis is observed even in the presence of RPA, raising the possibility that Srs2 can translocate along ssDNA substrates bound by RPA (Marini and Krejci, 2012). Analysis of Srs2 bound to RPA-ssDNA by electron microscopy also suggests that Srs2 translocates along RPA-bound ssDNA (Dupaigne et al., 2008). However, RPA binds to ssDNA with affinities on the order of ~10−9 – 10−10M (Chen and Wold, 2014; Wold, 1997), and we detect no turnover of ssDNA-bound RPA for ≥2 hours in ssDNA curtain assays when free RPA is absent (Gibb et al., 2014a; Gibb et al., 2014b). These findings raise the question of how RPA might impact Srs2. To address this question, we visualized mCherry-tagged Srs2 molecules as they interacted with ssDNA bound by GFP-RPA, specifically aiming to characterize binding and translocation events (Figure 1a, b). GFP-tagged Srs2 supports DNA replication and recombination in vivo (Burgess et al., 2009) and mCherry-Srs2 retained near wild-type levels of ATP hydrolysis activity in vitro (Figure S1c). Full-length Srs2 has a tendency to aggregate, so unless stated otherwise, the single-molecule experiments were conducted with a C-terminally truncated version of Srs2 comprised of amino acids 1 to 898 (Srs2898). The truncated Srs2 retains wild-type levels of ATPase, DNA helicase, and Rad51 filament disruption activities (Antony et al., 2009; Colavito et al., 2009) and is referred herein as Srs2. Experiments were performed at 32°C and all buffers contained 100 pM RPA and 2 mM ATP, unless otherwise stated. Fluorescent Srs2 (150 μl at 100 pM) was injected into the sample chamber, and we then observed its interactions with the RPA-coated ssDNA under constant buffer flow (0.2 ml/min). These reaction conditions, allowed us to observe only those Srs2 molecules that associated with the RPA-ssDNA substrate during the ~45 second incubation time window. This strategy helps minimize overlapping Srs2 binding and translocation events (see below).
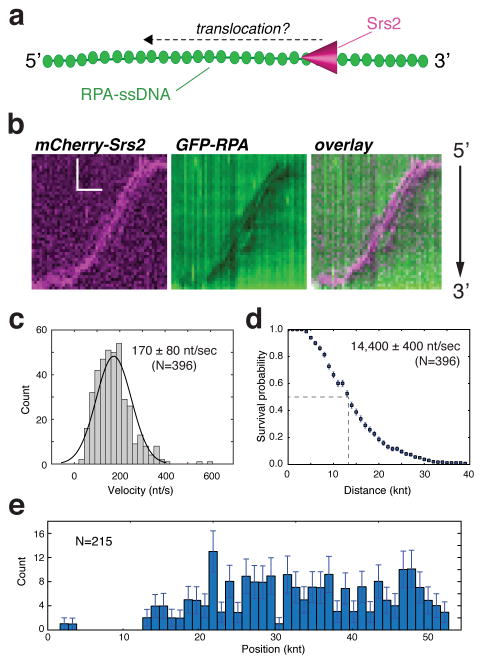
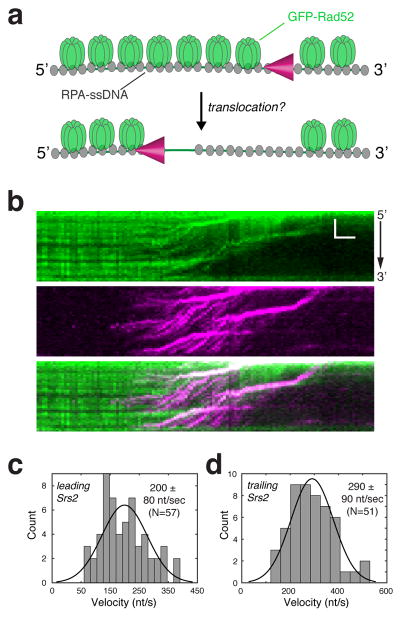
Figure 1. Srs2 translocation on ssDNA molecules bound by S. cerevisiae GFP-RPA.
(a) Schematic depiction of the assay used to visualize mCherry-Srs2 translocation on GFP-RPA-coated ssDNA. (b) Kymographs demonstrating the 3′→5′ movement of mCherry-Srs2 (magenta) along GFP-RPA bound ssDNA (green). The orientation of the ssDNA is indicated, and the images show the mCherry-Srs2 fluorescence signal, the GFP-RPA fluorescence signal, and the overlaid fluorescence signals, as indicated. (c) Velocity distribution and (d) survival probability measurement for mCherry-Srs2 translocating on RPA bound ssDNA; error bars represent s.d. calculated from bootstrapping analysis. The dashed line in (d) highlights the distance at which 50% of the Srs2 complex stop translocating, which we report as processivity values in the text. (e) Distribution of initial Srs2 binding sites; error bars represent s.d. calculated from bootstrapping analysis. The data in (c–e) represent the combined data sets for GFP-Srs2 and mCherry-Srs2 (see Figure S2).
The mCherry-Srs2 was able to interact with RPA-ssDNA and translocated rapidly over surprisingly long distances (Figure 1b & Figure S2a). Srs2 translocation occurred in the 3′→5′ direction, as expected (Rong and Klein, 1993). Similar findings were made for mCherry-Srs2 on ssDNA bound by either GFP-tagged or unlabeled RPA, confirming that the properties of RPA are not altered by either the GFP or mCherry tag (Figure S2a–c). The combined data sets for mCherry- and GFP-tagged Srs2 revealed that Srs2 translocated at an apparent velocity of 170 ± 80 nucleotides per second (nt/sec) (mean ± s.d.) (N=396) (Figure 1c), and exhibited an average processivity of 14,400 ± 370 nucleotides (Figure 1d). Srs2 translocation initiated at seemingly random positions along the RPA-ssDNA, indicating that there was no preferred site for initial Srs2 binding (Figure 1e). Experiments using the ATPase deficient Srs2-K41A mutant protein (100 pM), which bears a lysine to arginine mutation in the Walker A box (Krejci et al., 2004) showed that ATP hydrolysis is indispensable for translocation (Figure S3a–b).
Deletion of the Rad51-interaction domain reduces Srs2 association with RPA-ssDNA
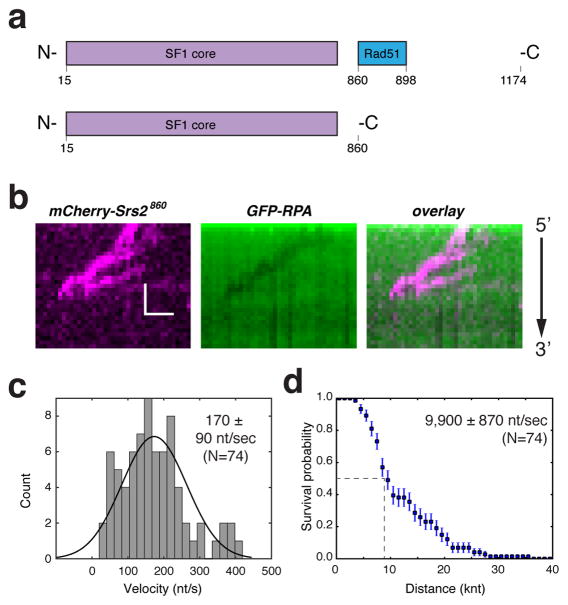
Srs2 possesses a domain that allows for interaction with Rad51 (Colavito et al., 2009)(Figure 2a) and the truncation mutant Srs2860 lacking this domain has ATPase and helicase activities that are comparable to wild-type Srs2. However, cells expressing Srs2860 exhibit a hyper-recombination phenotype (Colavito et al., 2009), and the truncated mutant is unable to efficiently disrupt Rad51 filaments (Antony et al., 2009; Colavito et al., 2009). It remains unclear whether the deficiencies ascribed this mutant stem from impaired Rad51 interaction, or that the mutant protein harbors other functional deficiencies. To our surprise, we were unable to detect significant association of mCherry-Srs2860 with RPA-ssDNA at the same concentration (100 pM) of Srs2898 described above. Specifically, we observed a total of only nine Srs2860 translocation events, revealing an average velocity of 160 ± 70 nt/sec (N=9) and a processivity of 12,700 ± 2,750 nucleotides. These results suggested that Srs2860 is compromised for initial association with the RPA-ssDNA complex, but once bound, the Srs2 mutant is nonetheless proficient in translocation through the RPA-coated DNA. In agreement with this conclusion, we detected many more translocation events when the concentration of mCherry-Srs2860 was increased to 1 nM (Figure 2b), revealing an average apparent velocity of 170 ± 90 nt/sec and an average processivity of 9,900 ± 870 nucleotides (Figure 2c, d). Together, these observations suggest that the C-terminal domain of Srs2 is necessary for efficient association with RPA-ssDNA.
Figure 2. Srs2860 is impaired in association with the RPA-ssDNA complex.
(a) Schematic depiction of full length Srs2 and Srs2860. (b) Kymograph showing mCherry-Srs2860 translocation on an GFP-RPA-ssDNA molecule for data collected at 1 nM mCherry-Srs2860. (c) Velocity distribution and (d) Survival probability plot for mCherry-Srs2860; error bars represent s.d. calculated from bootstrapping analysis.
Srs2 evicts RPA from ssDNA
Inspection of the GFP-RPA signal in the kymographs suggested that Srs2 might be stripping RPA from the ssDNA during translocation (e.g. Figure 1b). Specifically, short tracts of dark ssDNA that were transiently devoid of GFP-RPA were observed, and these tracts always coincided with the passage of mCherry-Srs2. However, in the experiments described above, free GFP-RPA (100 pM) was always present, and this available protein pool could easily replenish any GFP-RPA that might have been displaced by Srs2. As a consequence, the transient tracts of naked ssDNA did not persist for more than a few seconds after the passage of Srs2 (Figure 1b).
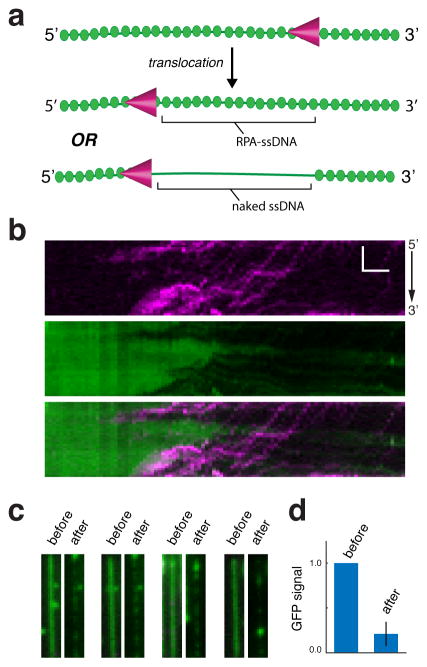
We sought to more definitely determine whether Srs2 removes RPA during translocation using mCherry-Srs2 and GFP-RPA (Figure 3a). Free GFP-RPA was omitted from the buffer to minimize the possibility that free RPA would occupy ssDNA after the passage of Srs2. These experiments demonstrated that mCherry-Srs2 clears GFP-RPA from ssDNA during translocation, such that ~80% of the GFP-RPA becomes dislodged (Figure 3b–d). Given the high binding affinity of RPA for ssDNA (Chen and Wold, 2014; Wold, 1997), the residual GFP-RPA that remained after passage of Srs2 may have been due to rebinding of free GFP-RPA that had been displaced by Srs2 elsewhere.
Figure 3. Srs2 strips RPA from ssDNA during translocation.
(a) Schematic showing Srs2 translocation without concomitant removal of RPA (top) and Srs2 translocation coinciding with the removal of Srs2 (bottom). (b) mCherry-Srs2 translocation on GFP-RPA bound ssDNA when free GFP-RPA is absent from solution. The three kymographs show the mCherry-Srs2 fluorescence signal (top), the GFP-RPA signal (middle), and the overlaid images (bottom), as indicated. (c) Images of GFP-RPA bound ssDNA before and after injection of mCherry-Srs2. (d) Fractional loss of normalized GFP-RPA signal (integrated over entire ssDNA molecules; N=20) due to the action of Srs2.
Srs2 translocation on near naked versus RPA-bound ssDNA
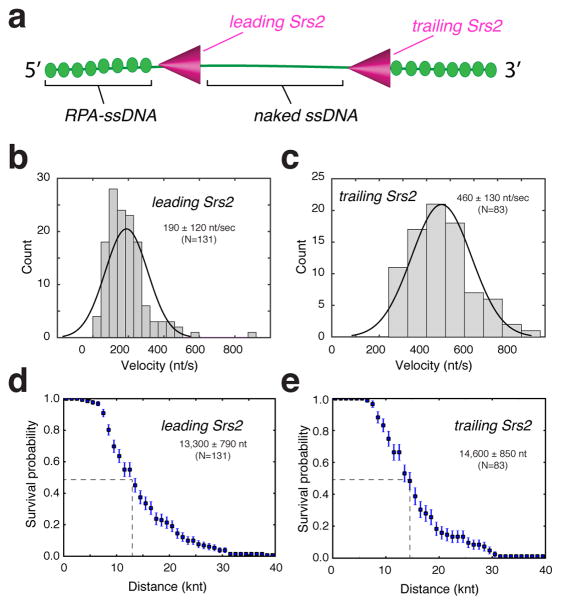
Inspection of the Srs2 kymographs revealed additional features reflecting the interactions between Srs2 and RPA-ssDNA. Under the conditions of our experiments, the initial Srs2 binding events were relatively rare; we typically observed no more than two to three initiation events per ssDNA molecule (e.g. Figure 1b & 3b). However, once Srs2 began translocating, many new Srs2 binding events occurred in its wake, and these latter events were especially prevalent in experiments in which free RPA was absent (i.e. Figure 3b). Indeed, upon stripping GFP-RPA from the ssDNA, subsequent Srs2 binding events appeared to occur more frequently on the newly created near naked ssDNA (Figure 3b). We will refer to Srs2 molecules that became associated with the RPA-ssDNA substrate initially as the “lead” Srs2 molecules, and the Srs2 molecules that bound the newly created near naked ssDNA as “trailing” Srs2 molecules (Figure 4a). We use to the term “near naked DNA” to indicate that there is a small amount of GFP-RPA still present (as indicated above; Figure 3d). At the concentration of mCherry-Srs2 required to efficiently nucleate the initial translocation events (100 pM), subsequent loading of Srs2 onto the near naked ssDNA became so prevalent that we were unable to accurately document its frequency. However, we were able to quantitate the translocation characteristics of the leading and trailing Srs2 molecules. This analysis revealed that the lead Srs2 had an apparent velocity of 190 ± 120 nts/sec and processivity of 13,300 ± 790 nts (N=131)(Figure 4b & 4d), slightly faster, but otherwise comparable to reactions in which free RPA was present (Figure 1c & 1d). Remarkably, the trailing Srs2 translocated ~2.5-fold faster, displaying an apparent velocity of 460 ± 130 nts/sec and processivity of 14,600 ± 850 nts (N=83)(Figure 4c & 4e).
Figure 4. Translocation properties of the leading and trailing Srs2.
(a) Schematic designating the identity of the leading and trailing Srs2. Velocity distributions for the leading (b) and trailing (c) mCherry-Srs2. Survival probability of the (d) leading and (e) trailing mCherry-Srs2; error bars represent s.d. calculated from bootstrapping analysis.
Srs2 evicts Rad52 from ssDNA
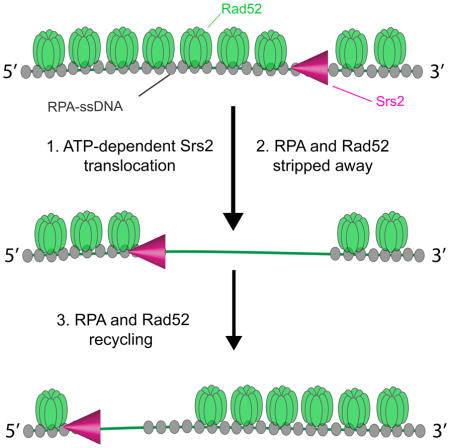
Rad52 is one of the first recombination proteins to arrive at RPA-coated ssDNA present at the ends of processed DSBs (Symington et al., 2014). Interestingly, Rad52 is thought to counteract the anti-recombinase activity of Srs2 (Burgess et al., 2009; Seong et al., 2009), although the molecular basis for these observations remains unknown. We have shown that Rad52 is readily recruited to RPA-ssDNA, and remains tightly bound to these complexes with lifetimes ≥2 hours (Gibb et al., 2014b). This remarkable stability raises the question of whether Rad52 might regulate Srs2 by restricting its translocation on ssDNA. To address this issue, we determined how Srs2 would behave in the presence of Rad52 (Figure 5a). For these experiments, RPA-ssDNA complex was prepared with unlabeled RPA. Then, GFP-tagged Rad52 (5 nM, 150 μl) was injected into the sample chamber at a constant flow rate of 0.2 ml/min, and any unbound GFP-Rad52 was flushed from the sample chamber. As reported before (Gibb et al., 2014b), Rad52 associated with the RPA-ssDNA complex avidly. Next, mCherry-Srs2 (100 pM) was injected in buffer containing 100 pM unlabeled RPA and 2 mM ATP. Remarkably, Srs2 was able to translocate on the ssDNA complexes, with Rad52 being rapidly stripped from the ssDNA during Srs2 translocation (Figure 5b). Inspection of the GFP-Rad52 kymographs revealed that Srs2 was able to push at least some of the Rad52 along the ssDNA, although there was not a pronounced accumulation of GFP-Rad52 towards the 5′ ends of the ssDNA, indicating that Rad52 was being evicted from the ssDNA (Figure 5b).
Figure 5. Srs2 can strip Rad52 from the RPA-ssDNA complex.
(a) Schematic for experiment to determine whether Srs2 can remove GFP-Rad52 from the RPA-ssDNA complex. (b) Kymographs depicting examples of what takes place when mCherry-Srs2 acts upon unlabeled RPA-ssDNA in the presence of bound GFP-Rad52. During data collection, the reaction buffer contained 100 pM free RPA (unlabeled), but did not contain free GFP-Rad52. Velocity distribution histograms for the (c) leading and (d) trailing mCherry-Srs2 complexes taken from data collected in the presence of bound GFP-Rad52.
Analysis of the translocation velocities revealed that the leading Srs2 molecules displayed an apparent translocation velocity of 200 ± 80 nt/sec (Figure 5c) and a processivity of 14,000 ± 1,500 nt on the Rad52-RPA-ssDNA complexes (Figure S4), indicating that the presence of Rad52 has no inhibitory effect on Srs2 translocation. Interestingly, the trailing Srs2 molecules translocated more rapidly than the leading ones, displaying an apparent translocation velocity of 290 ± 90 nt/sec and a processivity of 18,600 ± 1,100 nt (Figure 5d & Figure S4). Moreover, we found that the apparent velocity of Srs2 increases from 170 ± 80 nt/sec in the absence of Rad52 (Figure 1c) to 200 ± 80 nt/sec in the presence of Rad52 (Figure 5c). However, Rad52 has little or no effect on the rate of ATP hydrolysis by Srs2, which would seem to argue against a stimulatory effect of Rad52 on Srs2 translocation velocity (Figure S5). We speculate that the faster translocation rate of Srs2 stems from an alteration in the tension on the tethered ssDNA by Rad52.
Srs2 promotes redistribution of both RPA and Rad52
We next sought to determine whether Srs2 could promote Rad52 recycling. To address this issue, we conducted experiments where GFP-Rad52 (5 nM) and unlabeled RPA (100 pM) were present in the reaction buffer together with Srs2 (Figure S6a). The results demonstrated that GFP-Rad52 re-associates with ssDNA after the passage of Srs2 (Figure S6b). Under these conditions, Srs2 traveled at an apparent velocity of 215 ± 95 nt/sec (Figure S6c) and exhibited an average processivity of 13,000 ± 880 nts (Figure S6d). In addition, experiments using unlabeled Rad52 (5 nM) and GFP-RPA (100 pM) confirmed that GFP-RPA is also able to rebind ssDNA after passage of the translocating Srs2 when Rad52 was present (Figure S7). Note that these data were not ascribed to lead and trailing Srs2 molecules as they would experience a similar environment when free Rad52 and RPA were present and both proteins would rapidly re-associate with the ssDNA. This treatment of the Srs2 translocation data is supported by the observation of a single well-defined peak for the translocation velocity when free RPA and Rad52 were present (Figure S6c & FigureS7c). We conclude that Srs2 can remove both RPA and Rad52 from ssDNA, and as a consequence promotes the redistribution of both proteins.
Discussion
Srs2 plays crucial roles in DNA repair and DNA replication, and in fulfilling these roles it acts upon ssDNA intermediates bound by RPA as well as other DNA binding proteins. Here, we have demonstrated that Srs2 is capable of ATP hydrolysis-dependent translocation along crowded ssDNA molecules that are bound by either RPA alone, or both RPA and Rad52. The ability of Srs2 to remove and redistribute RPA and Rad52 is especially remarkable given the high affinity of these proteins for ssDNA. These findings suggest that Srs2 may play a role in channeling recombination factors away from aberrant intermediates and in the removal and recycling of these proteins after they have fulfilled their functions during recombination.
Srs2 translocation on RPA-coated ssDNA
We have shown that Srs2 translocates rapidly on ssDNA that is saturated with RPA and can travel over distances spanning thousands of nucleotides, giving no indication that that RPA offers a serious impediment despite it’s extremely high affinity for ssDNA (Chen and Wold, 2014; Wold, 1997). Assuming that RPA has a maximum binding site size of ~30 nucleotides under the buffer conditions used for our measurements (Kumaran et al., 2006), and that the ssDNA was saturated with RPA, then an apparent translocation rate of ~170 nt/sec and processivity of ~15,000 nt would correspond to the removal of ~6 molecules of RPA per second and ~500 molecules of RPA removed from the ssDNA per Srs2 translocation event.
Recent studies have shown that S. cerevisiae Pif1, and the E. coli UvrD and Rep helicases can push isolated E. coli SSB tetramers along ssDNA (Sokoloski et al., 2016). However, it remained unclear how helicases might behave when the ssDNA is fully coated by single strand binding proteins. Our results point to an alternative scenario for Srs2 acting on ssDNA substrates densely populated by RPA, namely, highly efficient eviction of RPA from the ssDNA. Interestingly, RPA-ssDNA complexes act as conserved signal to help trigger the DNA damage checkpoint by activating the ATR (ataxia telangiectasia mutated- and Rad3-related; Mec1 in S. cerevisiae)-ATRIP (ATR-interacting protein; Ddc2 in S. cerevisiae) protein kinase complex (Zou and Elledge, 2003). However, srs2Δ strains fail to recover from the DNA damage checkpoint-mediated growth arrest even after DNA repair has taken place, and these recovery defects are alleviated by mec1Δ (Vaze et al., 2002). Consonant with our findings, one possible explanation for these genetic results is that checkpoint recovery requires a clean-up process involving Srs2-mediated removal of persistent RPA from repaired intermediates.
Srs2 disruption of Rad52-containing recombination intermediates
Although the most well characterized role of Srs2 is to dismantle recombination intermediates containing Rad51, we have little understanding of how this activity is regulated. The clearest evidence for Srs2 regulatory control have come from experiments showing that the Rad55/57 complex, which is needed for Rad51 filament stability (Fortin and Symington, 2002), acts as a negative regulator of Srs2 (Liu et al., 2011). Recent studies have also implicated Rad52 and the SHU complex, which is comprised of Rad51 paralogs, as negative regulators of Srs2 (Bernstein et al., 2011; Burgess et al., 2009), although the mechanisms by which these HR factors antagonize the action of Srs2 remain unknown. Future studies will be necessary to explore the impact of Rad55/57 and the SHU complex on Srs2 translocation. However, our results argue against a direct action of Rad52 as a direct antagonist of Srs2 binding or translocation. Instead, our findings suggest that, by releasing Rad52 from ssDNA, Srs2 helps redirect Rad52-dependent Rad51 presynaptic filament assembly at alternative locations (Burgess et al., 2009). The ability of Srs2 to remove Rad52 from ssDNA may help avoid a futile cycle of Rad51 presynaptic filament assembly and disassembly at aberrant locations, and instead channel the recombination machinery to sites of DNA damage to mediate their repair. The finding that Srs2 removes Rad52 from RPA-ssDNA also suggests the possibility that Srs2 may play unforeseen roles in regulating the DNA repair processes that are directed by Rad52.
EXPERIMENTAL PROCEDURES
Protein Purification
S. cerevisiae RPA, GFP-RPA, mCherry-RPA, and GFP-Rad52 were purified as described (Gibb et al., 2014a; Gibb et al., 2014b). pET11c vector encoding 9xHis-tagged Srs2, and pET15b vectors encoding GFP-Srs2898, mCherry-Srs2898, mCherry-Srs2860, and GFP-Srs2K41A:898 were introduced into E. coli Rosetta2 (DE3) cells (Novagen). Cells were grown in 3 liters of LB at 37°C to an OD600 of 1–2. The temperature was reduced to 16°C before addition of 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After 20 hours of growth, cells were pelleted and frozen at −80°C. The pellet was then resuspended in Lysis Buffer (40 mM NaHPO4 [pH 7.5], 600 mM KCl, 5% glycerol, 10 mM imidazole [pH 7.8], 0.1mM TCEP, 0.05% Tween-20, 10 μM E-64, 1 pill per 100 ml of Protease inhibitor cocktail tablets, 1 mM Benzamidine, 1 mM PMSF, 0.125% myo-inositol) and lysed by sonication on ice. The lysate was clarified by ultracentrifugation and incubated for 30 min with a Ni-NTA resin (Clontech) that was equilibrated with Buffer Nickel A (40 mM NaHPO4 [pH 7.5], 300 mM KCl, 5% glycerol, 15 mM imidazole, 0.02% Tween-20, 1 mM Benzamidine, 1 mM PMSF, 0.125% myo-inositol). Before elution, the Ni-NTA column was washed with Buffer Nickel A. The proteins were eluted with a step of Buffer Nickel A containing 400 mM imidazole [pH 7.8]. The eluate was then dialyzed against Heparin Buffer (20 mM NaHPO4 [pH 7.5], 100 mM KCl, 5% glycerol, 0.01% Tween-20, 1 mM TCEP, 2 mM EDTA, 0.125% myo-inositol) during 2 hours. The eluate was then loaded onto a 5 ml HiTrap Heparin column (GE Lifesciences) equilibrated with Heparin Buffer, and proteins were eluted with a step of Heparin Buffer containing 500mM KCl. The purified fraction was applied to a Superdex 200 size exclusion column equilibrated with Storage Buffer (40 mM NaHPO4 [pH 7.5], 300 mM KCl, 10% glycerol, 0.01% Tween-20, 1 mM TCEP, 0.5 mM EDTA, 0.125% myo-inositol). Fractions corresponding to monomeric Srs2 were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Single Molecule Experiments
All experiments were conducted with a prism-type total internal reflection fluorescence (TIRF) microscope (Nikon) equipped with a 488-nm laser (Coherent Sapphire, 200 mW) and a 561-nm laser (Coherent Sapphire, 200 mW). Flowcells and ssDNA curtains were prepared as previously described (Ma et al., 2017). In brief, lipid bilayers were prepared with 91.5% DOPC, 0.5% biotinylated-PE, and 8% mPEG 2000-DOPE. The ssDNA substrate was generated using a biotinylated primer and a circular M13 ssDNA template, and rolling circle replication was accomplished with Phi29 DNA Polymerase. Biotinylated ssDNA was injected into the sample chamber and attached to the bilayer through a biotin-streptavidin linkage. Buffer was delivered to the sample chambers using a syringe pump (Kd Scientific), and sample delivery was controlled by two SCIVEX HPLC valves equipped with 150-μl loops. The ssDNA was aligned at the barriers by application of flow in HR buffer (30 mM Tris-Ac [pH 7.5], 50 mM KCl, 5 mM MgAc, 1 mM DTT, 0.3 mg/ml BSA, and 2 mM ATP) at a 0.8 ml/min. Secondary structure was reduced with a 150 μl injection of 8 M urea in 30 mM Tris-HCl [pH 7.4], immediately followed by 0.1 – 3.0 ml of HR buffer containing 100 pM unlabeled RPA, GFP-RPA, or mCherry-RPA, as indicated.
All Srs2 measurements were conducted in HR buffer at 32°C. Srs2 (100 pM, unless stated otherwise) was injected in a 150 μL aliquot while maintaining a constant buffer flow rate of 0.2 ml/min. Therefore, free Srs2 should be flushed from the flowcell after this 45 second injection interval. Videos were recorded beginning approximately 2 minutes before Srs2 entered the sample chamber and continued for a total of 20 minutes. For experiments where free RPA was absent in the buffer during Srs2 activity, the ssDNA-RPA filaments were generated as indicated above, and the buffer was then switched to HR buffer minus RPA. Sample chambers were flushed of free RPA for several minutes prior to the Srs2 injections. For experiments involving Rad52-RPA-ssDNA complexes in the absence of free Rad52 in solution, the RPA-ssDNA complexes were prepared as described above. A 150 μl sample of GFP-Rad52 (5 nM) in HR buffer was then injected into the sample chamber at a flow rate of 0.2 ml/min, and flow was continued for ~10 minutes to ensure removal of any unbound GFP-Rad52. Srs2 (150 μl, 100 pM) was then injected into the sample chamber at a constant flow rate of 0.2 ml/min while recording images, as described above. Experiments conducted in presence of free Rad52 were performed similarly, with the exception that all buffers were supplemented with 5 nM GFP-Rad52 or unlabeled Rad52, as indicated.
Single molecule data analysis
All data were collected at 1 frame per 4 seconds with 100 millisecond integration time, and the laser was shuttered between each acquired image to minimize photo-bleaching. Raw TIFF images were imported as image stacks into ImageJ, and kymograph were generated from the image stacks by defining a 1 pixel wide region of interest (ROI) along the long axis of the individual ssDNA molecules. Within the resulting kymographs, individual Srs2 trajectories were analyzed using NeuronJ (ImageJ). NeuronJ records the xy coordinates of each trace in pixel values. To transform pixel values into nucleotides, we assumed that 1 px corresponds to ~1087 nucleotides of ssDNA-RPA filament, as previously described (Qi et al., 2015). It must be emphasized we can only report apparent velocities, the values of which depend upon the assumed contour lengths of the extended RPA-ssDNA and RPA-Rad52-ssDNA complexes within the ssDNA curtains. Translocation velocity (nt/sec) was then calculated from the slope of each trajectory, and the velocity distribution histograms and Gaussian fitting were generated using Matlab. The distance of each translocation event was defined as the total length in nucleotides from each initial Srs2 binding position to the end of the translocation trajectory and defined by the location were Srs2 either dissociated from the ssDNA or stopped moving. The resulting values were used to generate the survival probability plots. The reported processivity values reflect the translocation distance at which one half of the total number of complexes dissociate from the ssDNA based upon the survival probability plots. The reported error bars for the survival probability correspond to standard deviation (s.d.) calculated by bootstrap analysis using a custom Python script. Srs2 binding distributions were determined by using the chromium barriers as a fiducial marker. The Srs2 starting positions relative to the barrier were then obtained with a custom Python script that located the Srs2 starting point to the nearest barrier. The error bars in the binding distribution plot represent s.d., and were obtained from 1000 bootstrap samples using a custom Python script.
Supplementary Material
Highlights.
Srs2 is visualized in real time as it translocates on ssDNA.
Srs2 is able to translocate rapidly on ssDNA coated with RPA.
Srs2 can also translocate on ssDNA bound by both RPA and Rad52.
RPA and Rad52 are removed from ssDNA by Srs2.
Acknowledgments
We thank members of the Greene laboratory for comments on the manuscript, and we thank Tsuyoshi Terakawa for assistance with data analysis. This research was funded by NIH grants to E.C.G. (R35GM118026) and P.S. (R01ES007061 and RO1ES015632). L.D.T. was funded by a PEW Latin American postdoctoral fellowship, the Williams Foundation, and by a program for Assistant Researchers, CONICET, Argentina. J.B.C. was funded by a Damon Runyon Postdoctoral Fellowship.
Footnotes
Author Contributions
L.D.T. designed and conducted the single molecule experiments and data analysis with assistance from K.K. L.D.T. and K.K. cloned, purified and characterized Srs2 constructs. Y.K. expressed and purified Rad51, and J.B.C. assisted with bulk biochemical analysis of Srs2. E.C.G. supervised the project and wrote the manuscript with input from L.D.T., K.K. Y.K., J.B.C., and P.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Molecular cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell. 2011;22:1599–1607. doi: 10.1091/mbc.E10-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Szakal B. Building up and breaking down: mechanisms controlling recombination during replication. Critical reviews in biochemistry and molecular biology. 2017:1–14. doi: 10.1080/10409238.2017.1304355. [DOI] [PubMed] [Google Scholar]
- Brosh RM., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Wold MS. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays: news and reviews in molecular, cellular and developmental biology. 2014;36:1156–1161. doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavito S, Macris-Kiss M, Seong C, Gleeson O, Greene EC, Klein HL, Krejci L, Sung P. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic acids research. 2009;37:6754–6764. doi: 10.1093/nar/gkp748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Molecular cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. The EMBO journal. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, Sung P, Greene EC. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014a;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B, Ye LF, Kwon Y, Niu H, Sung P, Greene EC. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nature structural & molecular biology. 2014b;21:893–900. doi: 10.1038/nsmb.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Macris M, Li Y, Van Komen S, Villemain J, Ellenberger T, Klein H, Sung P. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. The Journal of biological chemistry. 2004;279:23193–23199. doi: 10.1074/jbc.M402586200. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Molecular cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CJ, Steinfeld JB, Greene EC. Single-Stranded DNA Curtains for Studying Homologous Recombination. Methods in enzymology. 2017;582:193–219. doi: 10.1016/bs.mie.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V, Krejci L. Srs2: the “Odd-Job Man” in DNA repair. DNA repair. 2010;9:268–275. doi: 10.1016/j.dnarep.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V, Krejci L. Unwinding of synthetic replication and recombination substrates by Srs2. DNA repair. 2012;11:789–798. doi: 10.1016/j.dnarep.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Molecular cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Klein HL. Multifunctional Roles of Saccharomyces cerevisiae Srs2 protein in Replication, Recombination and Repair. FEMS Yeast Res. 2016 doi: 10.1093/femsyr/fow111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Klein HL. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V, Chen SH, Molzberger ET, Tomko E, Chitteni-Pattu S, Jia H, Ordabayev Y, Lohman TM, Cox MM. Active displacement of RecA filaments by UvrD translocase activity. Nucleic acids research. 2015;43:4133–4149. doi: 10.1093/nar/gkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Redding S, Lee JY, Gibb B, Kwon Y, Niu H, Gaines WA, Sung P, Greene EC. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. The Journal of biological chemistry. 1993;268:1252–1259. [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sasanuma H, Furihata Y, Shinohara M, Shinohara A. Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase. Genetics. 2013;194:859–872. doi: 10.1534/genetics.113.150615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong C, Colavito S, Kwon Y, Sung P, Krejci L. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. The Journal of biological chemistry. 2009;284:24363–24371. doi: 10.1074/jbc.M109.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski JE, Kozlov AG, Galletto R, Lohman TM. Chemo-mechanical pushing of proteins along single-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:6194–6199. doi: 10.1073/pnas.1602878113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. The EMBO journal. 2006a;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. The EMBO journal. 2006b;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasianovich Y, Altmannova V, Kotenko O, Newton MD, Krejci L, Makovets S. Unloading of homologous recombination factors is required for restoring double-stranded DNA at damage repair loci. The EMBO journal. 2017;36:213–231. doi: 10.15252/embj.201694628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Molecular cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. The EMBO journal. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annual review of biochemistry. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.