Abstract
Dysregulated adipocyte physiology leads to imbalanced energy storage, obesity, and associated diseases, imposing a costly burden on current health care. Cannabinoid receptor type-1 (CB1) plays a crucial role in controlling energy metabolism through central and peripheral mechanisms. In this work, adipocyte-specific inducible deletion of the CB1 gene (Ati-CB1–KO) was sufficient to protect adult mice from diet-induced obesity and associated metabolic alterations and to reverse the phenotype in already obese mice. Compared with controls, Ati-CB1–KO mice showed decreased body weight, reduced total adiposity, improved insulin sensitivity, enhanced energy expenditure, and fat depot–specific cellular remodeling toward lowered energy storage capacity and browning of white adipocytes. These changes were associated with an increase in alternatively activated macrophages concomitant with enhanced sympathetic tone in adipose tissue. Remarkably, these alterations preceded the appearance of differences in body weight, highlighting the causal relation between the loss of CB1 and the triggering of metabolic reprogramming in adipose tissues. Finally, the lean phenotype of Ati-CB1–KO mice and the increase in alternatively activated macrophages in adipose tissue were also present at thermoneutral conditions. Our data provide compelling evidence for a crosstalk among adipocytes, immune cells, and the sympathetic nervous system (SNS), wherein CB1 plays a key regulatory role.
Keywords: Genetics, Metabolism
Keywords: Adipose tissue, Mouse models, Obesity
Introduction
In mammals, 2 different types of adipose tissues coexist; these exert specific functions in the context of energy balance regulation. White adipose tissue (WAT) is the main site for the organism’s energy storage and is a highly active endocrine organ able to regulate energy homeostasis by affecting the activity of other organs, including the brain (1–4). Conversely, brown adipose tissue (BAT) is specialized in thermogenesis and energy dissipation (5, 6). The confirmation of the presence of BAT in human subjects has stimulated interest in the potential clinical relevance of BAT activation for the treatment of obesity (7, 8).
Adipose tissue can undergo dynamic remodeling, including quantitative and qualitative alterations of adipose tissue–resident cells. In obesity, changes in number and size of adipocytes have been extensively described (9). New insights have also revealed that a cell subpopulation of WAT, called beige or brite adipocytes, under certain stimuli are able to express uncoupling protein 1 (Ucp1) and are engaged in thermogenesis, a process known as adipose tissue browning (10). Human BAT shares more similarities with mouse beige cells than classical brown adipocytes (11, 12), which is therapeutically relevant, since human BAT thermogenesis can be activated by β3 adrenergic receptor agonists (13). Recent insights have suggested that alternatively activated macrophages in adipose tissue act as a source of catecholamines, contributing to adaptive thermogenesis and browning of WAT (14–17). However, the latest investigations have questioned this proposition (18, 19). Thus, understanding the mechanisms involved in the development of functional beige cells is a key element in finding novel therapeutic targets for tackling obesity.
The endocannabinoid system plays a critical role in energy balance via several mechanisms, including regulation of adipocyte physiology (20–22). Activation of the cannabinoid receptor type-1 (CB1) stimulates adipogenesis and lipogenesis, leading to impaired mitochondrial function in diet-induced obesity (DIO) (20, 23, 24). CB1 activation in mouse and human WAT reduces mitochondrial mass and function by downregulation of PPARγ coactivator 1α (Ppargc1a), while genetic and pharmacological blockade of CB1 function increases mitochondrial biogenesis by enhancing Ppargc1a expression (24, 25). Accordingly, body weight loss induced by chronic administration of CB1 antagonists is largely due to increased energy expenditure (EE) and consequent activation of lipolysis and fatty acid oxidation (21, 26–28). However, the therapeutic use of CB1 antagonists, such as rimonabant, is hampered by serious side effects, which include depression and anxiety, primarily caused by the action of this drug in the brain (29, 30). Nonetheless, peripherally restricted CB1 antagonists were developed as a safer alternative and have been shown to alleviate obesity (31–33). However, the exact sites and mechanisms of action leading to the observed effects are currently poorly defined. Thus, to clearly determine the role of CB1 in adipose tissue physiology and its relevance as a target of treatment, we generated an adipocyte-specific CB1-KO mouse line and demonstrated that adipocyte CB1 plays a crucial role in controlling adipocyte physiology and in regulating the energy balance of the entire organism.
Results
Generation of adipocyte-specific CB1-deficient mice.
We generated adipocyte-specific CB1-KO mice by crossing CB1-floxed mice (34) with transgenic AdipoqCreERT2 mice (35), which express Cre recombinase under the regulatory sequences of the adiponectin gene. Tamoxifen-induced recombination at 5 weeks of age led to adipocyte-specific CB1-deficient mice (named Ati-CB1–KO). Genomic DNA analysis revealed that CB1 deletion required tamoxifen treatment and occurred specifically in epididymal fat (EF), subcutaneous fat (SF), and BAT Ati-CB1–KO mice (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI83626DS1), while CB1 deletion was excluded from nonadipose tissues, such as hypothalamus, soleus muscle, and liver (Supplemental Figure 1B). In addition, lack of recombination in various brain areas relevant for CB1-mediated control of energy homeostasis (anterior olfactory nucleus, nucleus accumbens, hypothalamus) was shown by analyzing β-galactosidase staining in the double-transgenic mouse line AdipoqCreERT2;ROSA26LacZ (35) (Supplemental Figure 1D). As revealed by quantitative reverse-transcriptase PCR (RT-qPCR), standard diet–fed (SD-fed) (11% of calories from fat) Ati-CB1–KO mice displayed a strong reduction in CB1 mRNA levels in EF (70%–80%), SF (60%–70%), mesenteric fat (MF) (70%–80%), and BAT (40%) at the age of 20 weeks (Supplemental Figure 1E) as compared with their WT littermate controls (named Ati-CB1–WT). CB1 mRNA levels were also reduced when animals were exposed to a high-fat diet (HFD) (40% of calories from fat) for 12 weeks (Supplemental Figure 1F). Immunohistochemical analysis showed the absence of CB1 protein in adipocytes of Ati-CB1–KO mice (Supplemental Figure 1G), and no genotype differences in CB1 mRNA levels were found in nonadipose tissues, including brain, liver, and soleus muscle (Supplemental Figure 1H).
CB1 deletion in adipocytes prevents DIO.
Ati-CB1–KO and Ati-CB1–WT mice were monitored while being maintained on SD and HFD for 12 weeks, respectively. In order to exclude confounding factors of Cre recombinase expression per se on the metabolic phenotype, the body weight growth curve of heterozygous AdipoqCreERT2tg/+ mice was analyzed. As these mutant mice were previously shown to have no alteration in body weight under SD conditions as compared with WT (35), we determined whether HFD could differentially affect AdipoqCreERT2tg/+ mice and their WT littermates. No differences in body weight were observed between genotypes (Supplemental Figure 2A), further validating the suitability of this genetic tool.
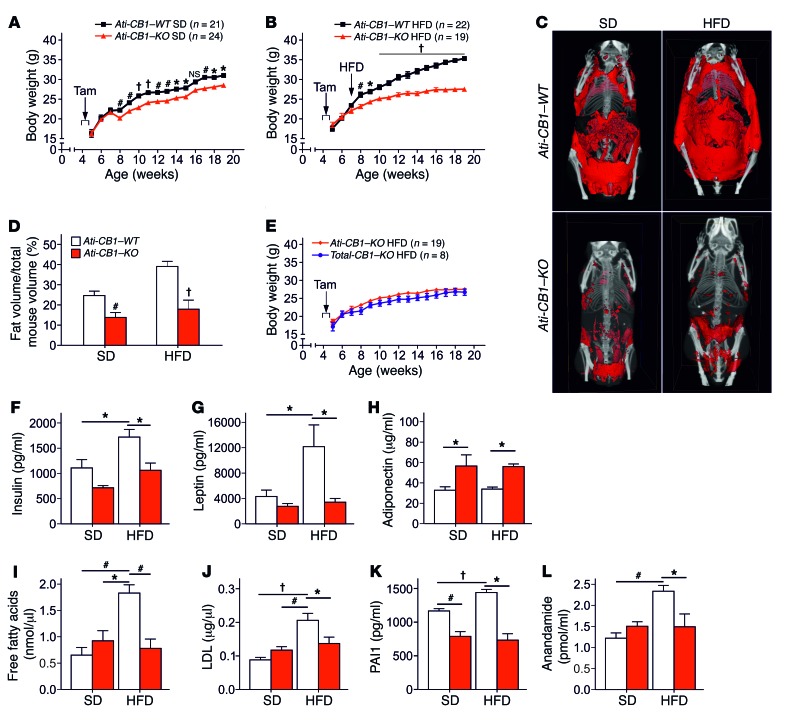
Under SD, Ati-CB1–KO mice displayed a significantly reduced body weight gain compared with WT (Figure 1A). Under HFD, WT mice became obese, while Ati-CB1–KO littermates were resistant to DIO (Figure 1B). Accordingly, Ati-CB1–KO displayed reduced total adiposity under both SD and HFD (Figure 1, C and D). No significant differences in body weight were observed between CB1–/– mice (herein referred to as Total-CB1–KO) and Ati-CB1–KO mice under HFD (Figure 1E), suggesting that CB1 deletion in adipocytes is sufficient to protect against DIO. Plasma insulin, leptin, and glucose levels were not significantly different between Ati-CB1–KO and WT littermates under SD (Figure 1, F and G, and Supplemental Figure 2B). HFD exposure increased these parameters in Ati-CB1–WT mice, but it failed to alter them in Ati-CB1–KO mice (Figure 1, F and G, and Supplemental Figure 2B). Plasma adiponectin levels were significantly increased in Ati-CB1–KO as compared with WT mice, independently of the diet (Figure 1H). In contrast, we did not observe a decrease in adiponectin levels in Ati-CB1–WT under this HFD as compared with SD, most likely due to the mild increase in caloric fat content of the HFD we used. Adiponectin is an adipokine with insulin-sensitizing functions (36, 37). Accordingly, Ati-CB1–KO displayed increased insulin sensitivity as compared with Ati-CB1–WT both in SD and HFD, as assessed by the insulin tolerance test (Supplemental Figure 2, C and D). HFD-fed Ati-CB1–WT mice were also characterized by increased plasma free-fatty acid, LDL, and triglyceride levels (Figure 1, I and J, and Supplemental Figure 2E). However, no signs of dyslipidemia were observed in Ati-CB1–KO exposed to HFD (Figure 1, I and J, and Supplemental Figure 2E). Similarly, no signs of hepatic steatosis were observed in HFD-fed Ati-CB1–KO mice (Supplemental Figure 2F). Obesity is a chronic low-inflammatory condition (38). Plasma levels of the inflammatory markers plasminogen-activator inhibitor 1 (PAI1) and IL-6 were increased in HFD-fed Ati-CB1–WT as compared with SD-fed WT. Ati-CB1–KO had reduced PAI1 levels as compared with Ati-CB1–WT on SD, and these levels were not altered by HFD (Figure 1K). Similarly, HFD did not affect plasma IL-6 levels in Ati-CB1–KO mice, but these values were instead increased in WT littermates (Supplemental Figure 2G). Obesity is also characterized by an overactive endocannabinoid system (39, 40). Indeed, plasma levels of the endocannabinoid anandamide (AEA) (Figure 1L), but not of 2-arachidonoyl glycerol (data not shown), were increased in Ati-CB1–WT on HFD as compared with SD; this increase was absent in Ati-CB1–KO.
Figure 1. Adipocyte CB1 deletion prevents DIO and metabolic dysregulation.
(A) Body weight growth curves of Ati-CB1–KO (n = 21) and Ati-CB1–WT (n = 24) littermates on SD. Tamoxifen-induced recombination occurred for 5 days at 5 weeks of age (arrow). Tam, tamoxifen. (B) Body weight growth curves of Ati-CB1–WT (n = 22) and Ati-CB1–KO (n = 19) mice on HFD. Tam-induced recombination occurred for 5 days at 5 weeks of age (arrow). The switch from SD to HFD was at the beginning of week 7. (C) 3D visualization (ventral view) of skeleton (white) and total adipose tissue (red) from in vivo micro-CT images of Ati-CB1–WT and Ati-CB1–KO mice (19 weeks of age) on SD and HFD, respectively. (D) Quantification of total fat content (as percentage of total mouse volume) by in vivo micro-CT analysis in Ati-CB1–WT and Ati-CB1–KO mice on SD and HFD, respectively. (E) Comparison of body weight growth curves of Ati-CB1–KO (n = 19) mice and Total-CB1–KO (n = 8) mice on HFD. (F–L) Plasma profiles of Ati-CB1–WT (n = 4–9) and Ati-CB1–KO (n = 4–8) on both diet treatments (at 19 weeks of age). Data are shown as mean ± SEM. *P < 0.05; #P < 0.01; †P < 0.001. Two-way ANOVA (A, B, E); Student’s t test (D); 1-way ANOVA (F–L).
CB1 deletion in adipocytes leads to fat depot–specific alterations and lowers energy storage capacity.
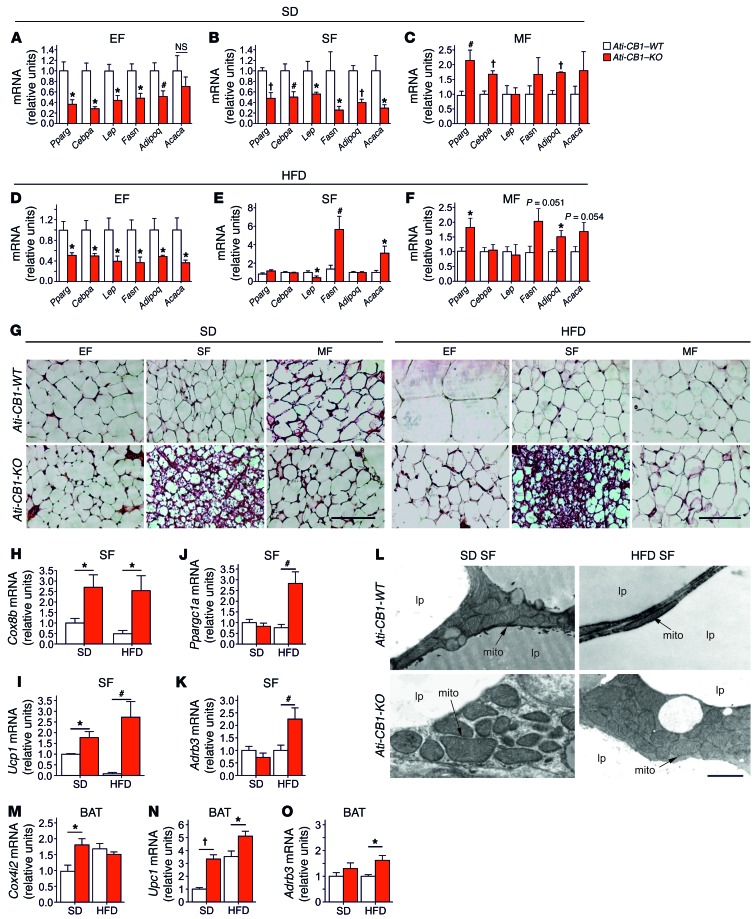
Lack of adipocyte CB1 led to profound modifications in adipocyte morphology and physiology. Interestingly, these changes were fat-depot specific. EF from SD- and HFD-fed Ati-CB1–KO displayed reduced mRNA levels of Pparg, CAAT/enhancer binding protein α (Cebpa), and adiponectin (Adipoq) (Figure 2, A and D), suggesting an impaired differentiation state of EF. Accordingly, EF from Ati-CB1–KO mice displayed decreased leptin (lep), fatty acid synthase (Fasn), and acetyl coenzyme A carboxylase α (Acaca) (latter significant only under HFD) mRNA levels. Similar changes were also observed in SF of SD-fed Ati-CB1–KO mice (Figure 2B). However, under HFD, SF adipocytes from Ati-CB1–KO did not display differences in mRNA levels of Pparg, Cebpa, and Adipoq as compared with WT (Figure 2E). In contrast, Lep mRNA levels were downregulated and, surprisingly, Fasn and Acaca mRNA levels were upregulated in SF of HFD-fed Ati-CB1–KO (Figure 2E). MF of SD- and HFD-fed Ati-CB1–KO displayed an upregulation of Pparg and Adipoq mRNAs as compared with Ati-CB1–WT littermates (Figure 2, C and F), while Cebpa was increased only in SD (Figure 2C). The upregulation of Adipoq was confirmed by immunofluorescence analysis (Supplemental Figure 3A). In MF, no significant differences were observed in Lep, Fasn, and Acaca mRNA levels, independently from the diet (Figure 2, C and F). Thus, adipocyte CB1 is an important regulator of WAT differentiation and adiponectin production. These features were paralleled by histological modifications. SD- and HFD-fed Ati-CB1–KO mice did not show cellular hypertrophy in EF and MF as compared with Ati-CB1–WT mice (Figure 2G and Supplemental Figure 3, B and C). Furthermore, histological analysis of SF from SD- and HFD-fed Ati-CB1–KO revealed the presence of adipocytes with reduced cell size and lipid content and increased multilocular aspect (Figure 2G) (41). Consistent with these histological features, mRNA levels of the mitochondrial markers cytochrome c oxidase VIIIb (Cox8b), cytochrome c oxidase IV isoform 2 (Cox4i2) (only on SD), and mitochondrial transcription factor A (Tfam) were significantly upregulated in SF of Ati-CB1–KO as compared with Ati-CB1–WT (Figure 2H and Supplemental Figure 3, D and E). Notably, mRNA levels of Ucp1, Ppargc1a, and β3 adrenergic receptor (Adrb3) were significantly increased in SF from Ati-CB1–KO on HFD as compared with Ati-CB1–WT (Figure 2, I–K). Also, mRNA levels of deiodinase iodothyronine type II (Dio2) were strongly upregulated in SF of Ati-CB1–KO as compared with Ati-CB1–WT mice, independently of the diet (Supplemental Figure 3F). mRNA levels of the fatty acid elongase 3 (Elovl3), an enzyme whose expression positively correlates with the amount of BAT (42), were decreased in SF of Ati-CB1–WT on HFD, but increased in Ati-CB1–KO mice as compared with Ati-CB1–WT mice on SD (Supplemental Figure 3G). Electron microscopic analysis showed that mitochondria from SF of Ati-CB1–KO were larger in size and had increased cristae content as compared with Ati-CB1–WT, independently of the diet (Figure 2L). This was associated with an increase in UCP1 protein levels in SF of Ati-CB1–KO (Supplemental Figure 3H). Similarly, BAT of Ati-CB1–KO also had increased mRNA levels of Cox4i2 (only on SD), Ucp1, and Adrb3 (Figure 2, M–O). Brown adipocytes are able to synthesize and release adiponectin (43). Accordingly, Adipoq mRNA was upregulated in BAT from Ati-CB1–KO on HFD (Supplemental Figure 3I).
Figure 2. CB1-dependent gene expression profile in adipocytes is depot specific.
(A–F) Gene expression analysis (relative units) of Pparg, Cebpa, Lep, Fasn, Adipoq, and Acaca in EF, SF, and MF from Ati-CB1–WT and Ati-CB1–KO on SD (WT, n = 5–9; KO, n = 5–8), and HFD (WT, n = 5–8; KO, n = 5–6). (G) Representative images of EF, SF, and MF from Ati-CB1–WT and Ati-CB1–KO on both SD and HFD. H&E staining. Scale bars: 100 μm. (H–K) Gene expression analysis (relative units) of Cox8b, Ucp1, Ppargc1a, and Adrb3 in SF from Ati-CB1–WT (n = 4–7) and Ati-CB1–KO (n = 4–7) on both SD and HFD. (L) Electron microscopy of SF from Ati-CB1–WT and Ati-CB1–KO on both SD and HFD. Scale bar: 1 μm. mito, mitochondrium; lp, lipid droplet. (M–O) Gene expression analysis (relative units) of Cox4i2, Ucp1, and Adrb3 in BAT from Ati-CB1–WT (n = 6) and Ati-CB1–KO (n = 6–8) on both SD and HFD. Data are shown as mean ± SEM. *P < 0.05; #P < 0.01; †P < 0.001 vs. WT, Student’s t test.
CB1 deletion in adipocytes affects energy homeostasis.
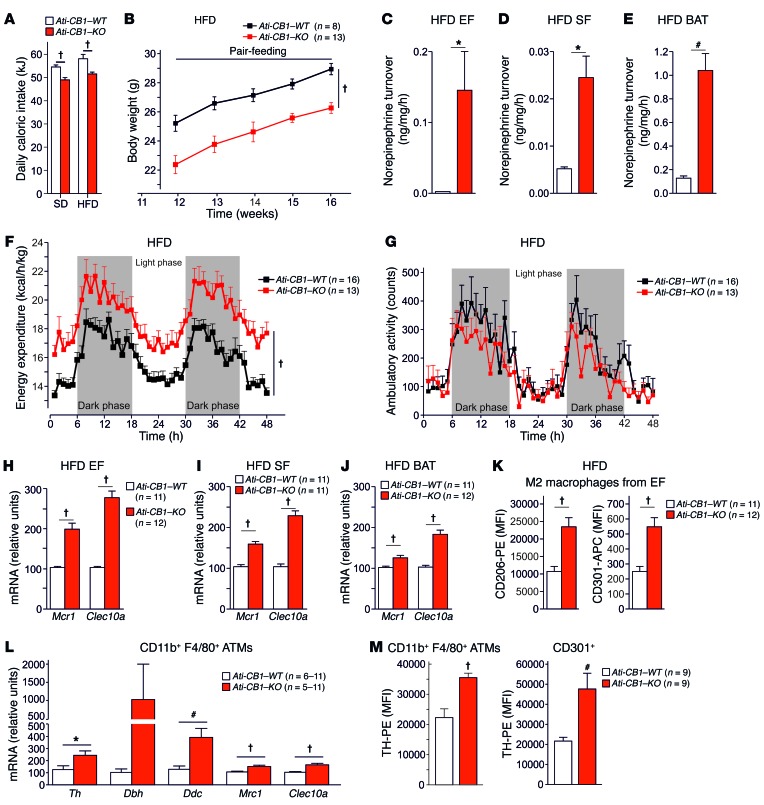
Daily caloric intake was significantly reduced in Ati-CB1–KO as compared with Ati-CB1–WT littermates under both SD and HFD (Figure 3A), and this decrease was associated with increased mRNA levels of the anorectic neuropeptide proopiomelanocortin (Pomc) in hypothalamus (Supplemental Figure 4A), while mRNA levels of other anorectic/orexigenic transcripts (neuropeptide Y [Npy]; cocaine- and amphetamine-regulated transcript [Cartpt]; pro-melanin-concentrating hormone [Pmch]) did not differ between the 2 genotypes (Supplemental Figure 4B). Pomc-expressing neurons in the arcuate nucleus of hypothalamus are one of the main targets of leptin action (44). Suppressor of cytokine signaling 3 (Socs3) plays an important role in the inhibition of leptin receptor signaling, and its expression leads to cellular leptin resistance (45). Accordingly, Socs3 mRNA levels were significantly increased in the hypothalamus of Ati-CB1–WT on HFD (Supplemental Figure 4C), while Ati-CB1–KO expressed Socs3 mRNA levels comparable to those observed under SD, suggesting preserved leptin sensitivity under HFD. In order to study the role of decreased food intake in the lean phenotype of Ati-CB1–KO on HFD, we performed a pair-feeding experiment (Figure 3B). The body weight increase in Ati-CB1–WT mice was unaffected by the reduced food intake, suggesting that the lean phenotype of the Ati-CB1–KO mice was primarily due to feeding-independent mechanisms.
Figure 3. CB1 deletion in adipocytes affects caloric intake and EE and promotes alternative macrophage activation.
(A) Daily caloric intake (in kJ) of Ati-CB1–WT (SD fed, n = 11; HFD fed, n = 21) and Ati-CB1–KO mice (SD fed, n = 17; HFD fed, n = 27) on SD and HFD. (B) Pair-feeding experiment. Body weight curves of Ati-CB1–WT (n = 8) and Ati-CB1–KO (n = 13) on HFD. Tissue NE turnover in EF (C), SF (D), and BAT (E) from Ati-CB1–WT (n = 3) and Ati-CB1–KO mice (n = 4) on HFD. (F) EE in Ati-CB1–WT (n = 16) and Ati-CB1–KO mice (n = 13) on HFD. (G) Ambulatory activity during indirect calorimetry recording in Ati-CB1–WT (n = 16) and Ati-CB1–KO (n = 13) on HFD. (H–J) Gene expression analysis (relative units) of markers for alternatively activated macrophages (Mrc1, Clec10a) in EF, SF, and BAT from Ati-CB1–WT and Ati-CB1–KO mice on HFD (n = 11–12). (K) Protein markers for alternatively activated macrophages (CD206 and CD301) were monitored by flow cytometry in EF from Ati-CB1–WT and Ati-CB1–KO mice on HFD (n = 11–12). (L) Gene expression (relative units) of the catecholamine-synthesizing enzymes (Th, Dbh, Ddc) and alternatively activated macrophage markers (Mrc1, Clec10a) in CD11b+F4/80+-sorted ATMs from WAT of Ati-CB1–WT (n = 6–11) and Ati-CB1–KO (n = 5–11) mice on HFD. Dbh mRNA levels were measured by ddPCR analysis. (M) TH protein levels were measured by flow cytometry in CD11b+F4/80+-sorted ATMs (left) and CD301+ cells (M2 macrophages, right) in WAT from Ati-CB1–WT (n = 9) and Ati-CB1–KO (n = 9) mice on HFD. Data are shown as mean ± SEM. *P < 0.05; #P < 0.01; †P < 0.001, Student’s t test (A, C–E, H–M); 2-way ANOVA (B, F, G).
Increased expression of Dio2 mRNA in SF (Supplemental Figure 3F) and of Adrb3 mRNA in SF and BAT of Ati-CB1–KO (Figure 2, K and O) implies possible changes in peripheral norepinephrine (NE) signaling. Indeed, NE turnover rate (Figure 3, C–E) and NE concentration (Supplemental Figure 4, D–F) in EF, SF, and BAT of Ati-CB1–KO on HFD were elevated as compared with WT. Furthermore, based on the observation of increased Adrb3 mRNA levels in SF (Figure 2K), we investigated whether β3 adrenergic receptor response is altered in WAT from Ati-CB1–KO. Indeed, the stimulation of in vivo lipolysis induced by the selective β3 adrenergic receptor agonist CL316243 was sustained in Ati-CB1–KO as compared with Ati-CB1–WT control mice under HFD (Supplemental Figure 4G).
The upregulation of thermogenic markers and the increased adipose NE turnover and NE concentration suggest that Ati-CB1–KO have increased energy dissipation. Accordingly, EE was higher in Ati-CB1–KO than in WT littermates on HFD (Figure 3F), whereas ambulatory activity was not different between genotypes (Figure 3G). The increased EE in Ati-CB1–KO mice was further confirmed by analysis of covariance (ANCOVA), in which the possible bias on EE analysis caused by differences in body weight is excluded (46). As shown, linear regression between mouse body weight and EE clearly demonstrates increased EE in Ati-CB1–KO mice as compared with their WT littermates under HFD (Supplemental Figure 4H). Similar results were found when we analyzed resting EE (REE) (Supplemental Figure 4I).
Increased abundance of alternatively activated macrophages in Ati-CB1–KO adipose tissue.
Next, we aimed at understanding the mechanistic underpinnings of the remodeling observed in adipose tissues of Ati-CB1–KO mice under HFD. Gene expression of the 3 catecholamine-synthesizing enzymes, tyrosine hydroxylase (Th), dopamine β hydroxylase (Dbh), and dopa decarboxylase (Ddc), were increased in whole EF, SF, and BAT in Ati-CB1–KO (Supplemental Figure 5A). Based on recent findings that alternative activation of macrophages (i.e., M2 polarization) contributes to adaptive thermogenesis and browning of WAT (14–17), whereby M2 macrophages act as a local source of catecholamines (14–17), we analyzed this process in Ati-CB1–WT and Ati-CB1–KO mice in HFD. We found significant increases in mRNA levels of 2 M2 markers analyzed (Mrc1, Clec10a) in all fat tissues investigated (EF, SF, BAT) in Ati-CB1–KO as compared with Ati-CB1–WT mice (Figure 3, H–J). These increases were further underlined at the protein level by flow cytometry analysis (Figure 3K, and Supplemental Figure 5B), wherein the median fluorescence intensities (MFI) for the M2 markers CD206 (mannose receptor C type 1, encoded by the Mrc1 gene) and CD301 (C-type lectin domain family 10 member A, encoded by the Clec10a gene) in EF were significantly increased in Ati-CB1–KO as compared with WT controls (Figure 3K).
In order to analyze whether these adipose tissue macrophages (ATMs) are able to synthesize NE, gene and protein expression analyses were performed in WAT-derived CD11b+F4/80+ stromal vascular cells, as defined by 2 well-described canonical markers of macrophages (Supplemental Figure 5C). First, we quantified gene expression of 3 enzymes (Th, Dbh, and Ddc) involved in catecholamine synthesis in CD11b+F4/80+ ATMs from Ati-CB1–KO and WT mice on HFD (Figure 3L). We found that Th and Ddc mRNA levels were significantly increased in FACS-sorted ATMs (CD11b+F4/80+ cells) from Ati-CB1–KO mice as compared with control WT littermates (Figure 3L), which is in congruence with gene expression data obtained from whole adipose tissue (Supplemental Figure 5A). We also observed that alternatively activated macrophage markers (Mrc1, Clec10a) were upregulated in sorted ATMs from mutant mice (Figure 3L). Importantly, Th gene expression levels in isolated ATMs were very low, but detectable by real-time PCR, while negative control samples never gave a signal (data not shown). In order to confirm this finding, we applied a second gene expression quantification method in an independent batch of sorted ATMs, using the more sensitive digital droplet PCR (ddPCR) system (Supplemental Figure 6A). In fact, we could only measure Dbh gene expression in sorted ATMs by ddPCR analysis (Supplemental Figure 6B). ddPCR analysis revealed almost 3-fold increased Th mRNA levels in ATMs in comparison with Dbh (Supplemental Figure 6, A and B). This marginal expression of Th and Dbh in ATMs was specific, since we could not detect any single copy in negative control samples among independent experiments using the ddPCR method (Supplemental Figure 6C). Next, using flow cytometry analysis, we measured TH protein levels in adipose tissue CD11b+F4/80+ cells and found enhanced TH protein levels in Ati-CB1–KO mice as compared with WT controls (Figure 3M and Supplemental Figure 5D). Furthermore, supporting the mRNA expression data, similarly increased TH protein levels were found in adipose tissue CD301+ M2 macrophages after deletion of CB1 in adipocytes (Figure 3M and Supplemental Figure 5D). We tested the selectivity of the anti-TH antibody used in Th-deficient mouse spleen samples (Supplemental Figure 5E).
Finally, we performed TH immunostaining anatomical studies in adipose tissue. First, immunoperoxidase staining of TH in adipose tissue showed an increased number of TH+ parenchymal fibers in BAT and SF from Ati-CB1–KO mice (Supplemental Figure 7), besides confirming an increased sympathetic tone in the mutant mice. Strikingly, double-fluorescence confocal analysis showed that CD206+ cells were in a tight anatomical proximity to TH+ sympathetic nerve fibers in both BAT and SF (Supplemental Figure 8). The close proximity between M2 macrophage cellular extensions and sympathetic nerve terminals was particularly evident in BAT, where we found a more pronounced increase in noradrenergic parenchymal nerve fibers than in SF.
In summary, the abundance of alternatively activated macrophages along with an enhanced sympathetic innervation was found to be increased in adipose tissues of Ati-CB1–KO mice. In addition, we clearly demonstrated that ATMs express the catecholamine-synthesizing machinery and that the absence of CB1 in adipocytes increased both mRNA and protein levels of TH in ATMs and in the alternatively activated M2 macrophages.
Lean phenotype of Ati-CB1–KO at thermoneutral conditions.
We next investigated whether a lean phenotype in Ati-CB1–KO could be observed at thermoneutrality (i.e., at 30°C) in the absence of chronic thermal stress. Animal housing conditions (22°C) demand extra energy, increasing total metabolism of the mice (50%–60% above basal) for the maintenance of body temperature (47). Interestingly, the same body weight difference between WT and KO groups was present at both 22°C and 30°C temperatures (Supplemental Figure 9A). Under thermoneutral conditions (30°C), both genotypes reduced food consumption as compared with at 22°C (Supplemental Figure 9B), in order to adjust to reduced adaptive thermogenesis. Accordingly, expression of thermogenic markers (Ucp1, Elovl3) in BAT was blunted in Ati-CB1–WT and Ati-CB1–KO mice at 30°C in comparison with Ati-CB1–WT at 22°C (Supplemental Figure 9C). Nevertheless, mRNA levels of markers for mitochondrial biogenesis (Ppargc1a, Tfam), M2 macrophage polarization (Clec10a, Mrc1), and NE production (Ddc) in SF were significantly increased in Ati-CB1–KO housed at thermoneutrality as compared with control WT littermates (Supplemental Figure 9D), although to a lower extent than at 22°C. Similarly, M2 macrophage polarization and NE synthesis machinery were enhanced in BAT of Ati-CB1–KO as compared with WT (Supplemental Figure 9E). Altogether, given that at thermoneutrality, SNS-induced BAT thermogenesis is most likely diminished, these experiments suggest that enhanced M2 macrophage polarization makes a major contribution to the lean phenotype of Ati-CB1–KO at thermoneutrality.
Adipocyte reprogramming precedes the appearance of body weight differences.
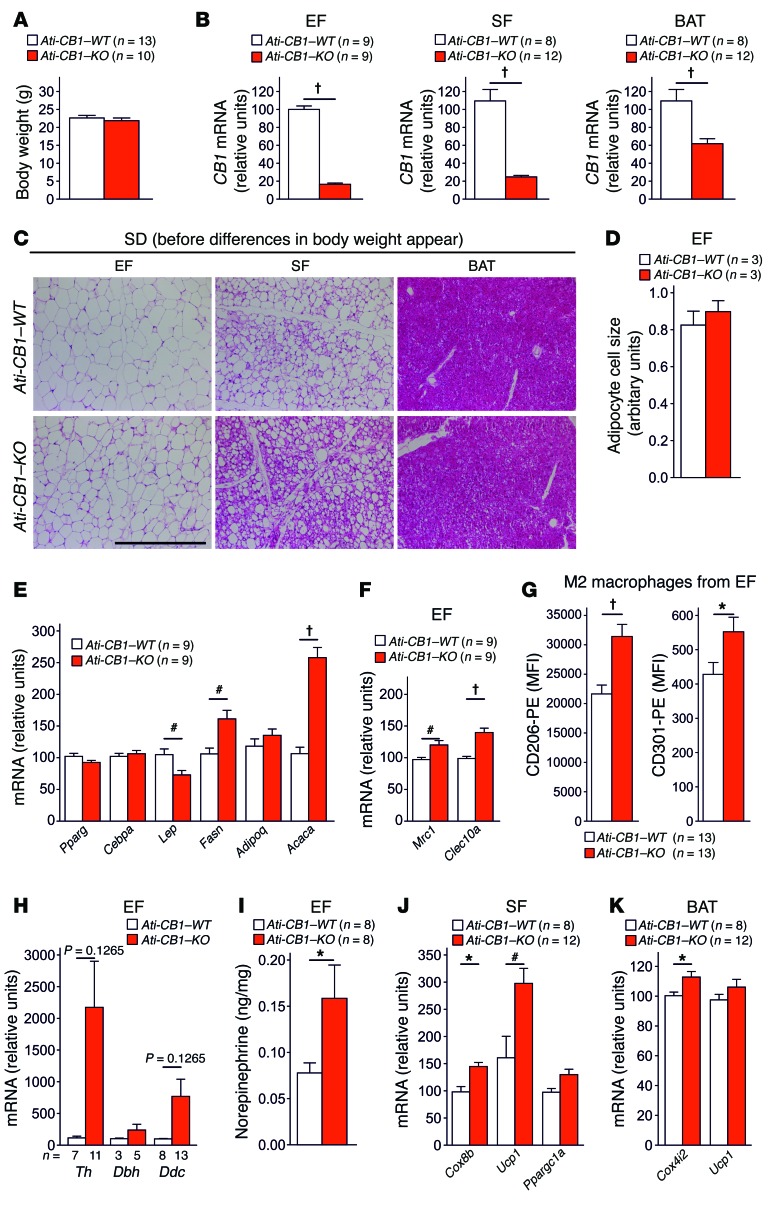
In order to determine whether adipocyte reprogramming is in causative relation with the loss of CB1 in adipocytes or whether it is merely an effect of the lack of obesity, we characterized SD-fed Ati-CB1–KO mice at 2 weeks after tamoxifen-induced genetic deletion of CB1 in adipocytes, when body weight differences had not yet occurred (Figure 4A). At this time, Ati-CB1–KO mice showed a strong reduction in CB1 mRNA levels in EF, SF, and BAT (Figure 4B). At the histological level, the deletion of CB1 promoted a profound remodeling in adipocytes in SF (Figure 4C) without alterations in adipocyte cell size in EF (Figure 4D). Interestingly, we observed strongly increased lipogenic (Fasn, Acaca) and lowered Lep gene expression in EF (Figure 4E), indicating a late differentiation status of adipocytes, similar to what was observed in adipocyte cell cultures (48, 49). This delayed differentiation in EF was associated with increased levels of alternatively activated macrophages and catecholamine production (Figure 4, F–I, and Supplemental Figure 10). Deletion of CB1 in SF led to WAT browning (Figure 4, C and J) and increase of mitochondrial marker (Cox4i2) in BAT (Figure 4K). Together, these experiments indicate that the loss of CB1 function in adipocytes leads to a strong remodeling of the different adipose tissue depots, preceding the emergence of body weight differences, implying that the effects observed after CB1 loss are of primary cause and not secondary to the lack of DIO.
Figure 4. CB1 deletion in adipocytes promotes alternatively activated macrophages and remodeling of adipose tissue preceding body weight changes.
(A) Body weight of 7-week-old SD-fed Ati-CB1–WT (n = 13) and Ati-CB1–KO mice (n = 10) 2 weeks after tamoxifen-induced CB1 deletion in adipocytes. (B) Relative CB1 mRNA levels are strongly decreased in EF, SF, and BAT from 7-week-old Ati-CB1–KO (n = 9–12) mice as compared with WT controls (n = 8–9) on SD. (C) Representative images of EF, SF, and BAT from Ati-CB1–WT and Ati-CB1–KO on SD. H&E staining. Scale bar: 100 μm. (D) Quantification of adipocyte cell size (arbitrary units) in EF from Ati-CB1–WT (n = 3) and Ati-CB1–KO (n = 3). (E) Gene expression analysis (relative units) in EF of markers of adipocyte differentiation (Pparg, Cebpa) and lipogenesis (Fasn, Acaca) and of adipokines (Lep, Adipoq) (n = 9). (F) Gene expression analysis (relative units) of alternatively activated macrophages (Mrc1, Clec10a) in EF (n = 9). (G) Protein analysis of markers for alternatively activated macrophages (CD206 and CD301) was monitored by flow cytometry in EF from Ati-CB1–WT (n = 13) and Ati-CB1–KO (n = 13) on SD. (H) Gene expression (relative units) of catecholamine-synthesizing enzymes (Th, Dbh, Ddc) in whole EF from Ati-CB1–WT (n = 3–8) and Ati-CB1–KO (n = 5–13) on SD. (I) Tissue NE levels in EF from Ati-CB1–WT (n = 8) and Ati-CB1–KO (n = 8) mice on SD. (J) Gene expression analysis (relative units) of thermogenic markers (Cox8b, Ucp1, Ppargc1a) in SF (n = 8–12). (K) Gene expression analysis (relative units) of thermogenic markers (Cox4i2, Ucp1) in BAT from Ati-CB1–WT (n = 8) and Ati-CB1–KO (n = 12) mice on SD. Data are shown as mean ± SEM. *P < 0.05; #P < 0.01; †P < 0.001 vs. WT, Student’s t test.
Reversal of obesity after inducible CB1 inactivation in adipocytes.
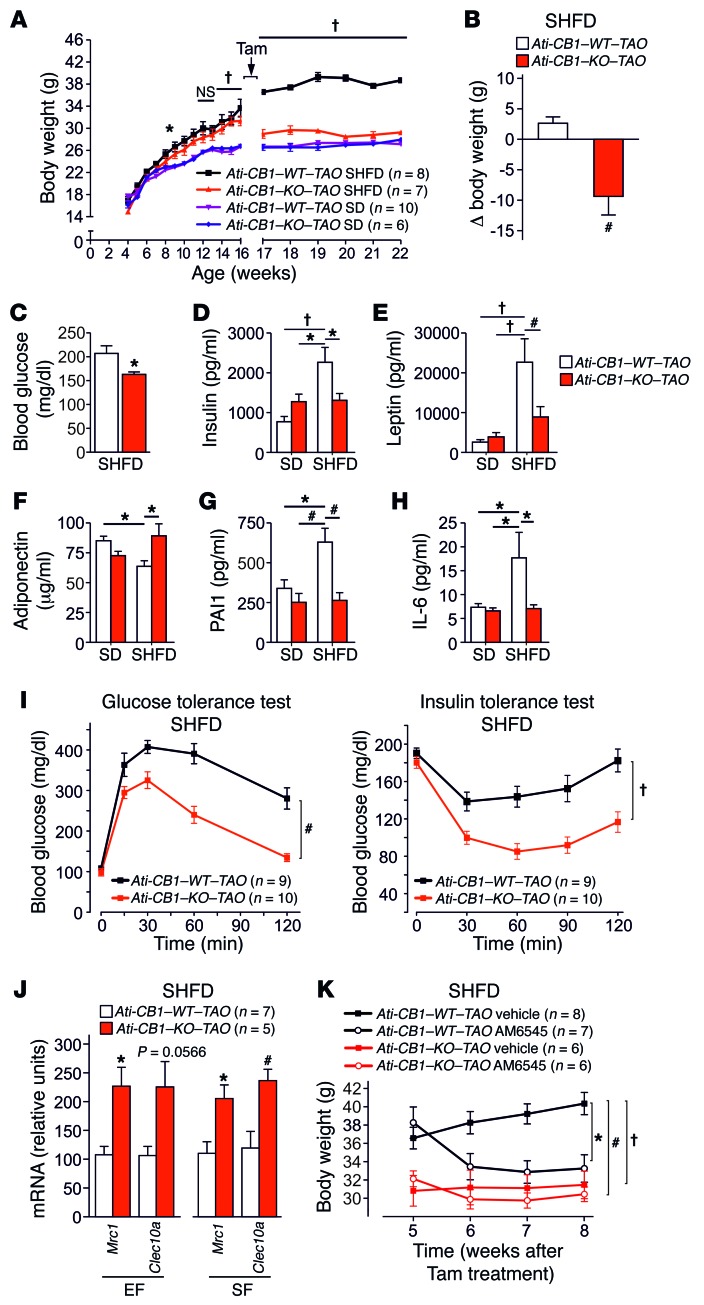
To evaluate whether adipocyte CB1 deletion can reverse the condition of preexisting obesity, we induced adipocyte CB1 inactivation in DIO mice. Mice were fed with a super-HFD (SHFD) (60% of calories from fat) for 12 weeks to reach severe obesity, and CB1 deficiency was induced in 16-week-old SD-fed and SHFD-fed animals (named Ati-CB1–KO–TAO, where TAO indicates tamoxifen after obesity). Without tamoxifen treatment, WT and KO mice developed DIO under SHFD (Figure 5A). The injection of tamoxifen induced a strong reduction of CB1 mRNA levels in EF (70%–80%), SF (70%–80%), MF (60%), and BAT (50%–70%) in both SD- and SHFD-fed Ati-CB1–KO–TAO as compared with their WT littermates, as analyzed at 22 weeks of age (Supplemental Figure 11, A and B). Adipocyte-specific CB1 deletion induced at 16 weeks of age in SD-fed Ati-CB1–KO–TAO did not cause significant changes in body weight and fat mass (Figure 5A and Supplemental Figure 11C), while adipocyte-specific CB1 deletion in SHFD-fed obese Ati-CB1–KO–TAO mice reduced body weight (Figure 5B) and the amount of EF and SF (Supplemental Figure 11D). The histology of adipocytes from Ati-CB1–KO–TAO showed the presence of multilocular adipocytes within SF, a visible decreased adipocyte size in EF, and a lack of lipid accumulation in BAT (Supplemental Figure 11E). Decreased body weight in Ati-CB1–KO–TAO was also associated with lowered fasting blood glucose levels as compared with Ati-CB1–WT–TAO, suggesting improved glucose homeostasis (Figure 5C). In addition, Ati-CB1–KO–TAO on SHFD had reduced plasma insulin and leptin levels as compared with Ati-CB1–WT–TAO (Figure 5, D and E), while plasma adiponectin levels were significantly increased in Ati-CB1–KO–TAO (Figure 5F). Furthermore, plasma levels of PAI1 and IL-6 were significantly decreased in Ati-CB1–KO–TAO as compared with Ati-CB1–WT–TAO on SHFD and were similar to levels observed in SD (Figure 5, G and H). The reversal of obesity in Ati-CB1–KO–TAO was also associated with significantly improved glucose homeostasis and insulin sensitivity (Figure 5I). Finally, also in this model of the reversal of obesity, CB1 loss promoted M2 polarization, as detected by increased expression of Mrc1 and Clec10a in EF and SF (Figure 5J).
Figure 5. Adipocyte-specific CB1 deletion in obese mice mediates weight loss and reverses obesity-related metabolic alterations.
(A) Body weight growth curves of Ati-CB1–WT–TAO (n = 8–10) and Ati-CB1–KO–TAO (n = 6–7) on both SD and SHFD before (weeks 4 to 16) and after (weeks 17 to 22) tamoxifen-induced CB1 deletion. (B) Body weight variation 6 weeks after tamoxifen-induced CB1 deletion in mice on SHFD. (C) Fasting blood glucose in Ati-CB1–WT–TAO (n = 8) and Ati-CB1–KO–TAO (n = 8) on SHFD. (D–H) Plasma levels of insulin, leptin, adiponectin, PAI1, and IL-6 in Ati-CB1–WT–TAO (n = 4–9) and Ati-CB1–KO–TAO (n = 4–8) on SD and SHFD. (I) Glucose and insulin tolerance tests in Ati-CB1–WT–TAO (n = 9) and Ati-CB1–KO–TAO (n = 10) on SHFD. (J) Gene expression for markers of alternatively activated macrophages (Mrc1, Clec10a) in EF and SF from Ati-CB1–WT–TAO (n = 7) and Ati-CB1–KO–TAO (n = 5) on SHFD. (K) Chronic treatment with the peripherally acting CB1 antagonist AM6545 (10 mg/kg, i.p.) or its vehicle and analysis of body weight in Ati-CB1–WT–TAO (n = 8–7) and Ati-CB1–KO–TAO (n = 6) on SHFD. Data are shown as mean ± SEM. *P < 0.05; #P < 0.01; †P < 0.001, 2-way ANOVA, Bonferroni’s post hoc test (A, K, I); Student’s t test (B, C, J); 1-way ANOVA (D–H).
We then tested the effect of AM6545 (10 mg/kg, i.p.), a peripheral CB1 antagonist (31, 32), on Ati-CB1–KO–TAO and Ati-CB1–WT–TAO mice (Figure 5K). As compared with the corresponding vehicle groups, chronic treatment with the peripherally restricted CB1 blocker caused a strong reduction in body weight (Figure 5K) and a slight nonsignificant decrease in food intake in Ati-CB1–WT–TAO mice (Supplemental Figure 11F), while having no effect in Ati-CB1–KO–TAO mice. Thus, the body weight of AM6545-treated Ati-CB1–WT–TAO became comparable to the body weight of Ati-CB1–KO–TAO mice (Figure 5K). Together, these data indicate that it is sufficient to abolish CB1 function in adipocytes in order to induce body weight loss and reach a body weight and a metabolic profile similar to those observed under SD.
Genetic inactivation of adipocyte CB1 in Ati-CB1–KO–TAO reduced obesity-related behavioral alterations.
It is known that obesity causes behavioral changes in humans (50) and in rodents (51). On the other hand, the anti-obesity drug Acomplia (rimonabant, CB1 inverse agonist) is associated with adverse effects such as anxiety and depression (52), similar to other weight loss therapies (53). Therefore, we analyzed obese-related anxiety-like phenotypes in Ati-CB1–KO–TAO using different behavioral paradigms. No significant differences were found in the total locomotion of Ati-CB1–KO–TAO and WT in the open field (OF) (Supplemental Figure 12A). However, SHFD-fed Ati-CB1–WT–TAO mice showed increased anxiety-like behavior in the elevated plus-maze (EPM), as indicated by the decreased percentage of entries into and time spent in the open arms of the maze. Conversely, in SHFD-fed Ati-CB1–KO–TAO mice, these parameters were comparable to those observed in SD (Supplemental Figure 12, B and C). Anxiety-like states have been described in combination with decreased motivation for exploratory activity, which can be analyzed by using the holeboard (HB) test (54). SHFD decreased head dippings in Ati-CB1–WT–TAO (Supplemental Figure 12D), whereas it showed no effect in Ati-CB1–KO–TAO. These data indicate that life-impairing behaviors, such as the increased anxiety-like behavior often observed in obesity, can be reversed in Ati-CB1–KO–TAO SHFD mice, which showed behavior similar to that of SD-fed mice.
Discussion
The ability of adipocytes to expand upon excessive energy intake determines the overall capacity of the body’s energy reserve (55). Our work demonstrates that the loss of adipocyte CB1 affects energy balance by promoting a profound remodeling in depot-specific adipocyte functions. These changes were associated with increased alternative macrophage activation and enhanced sympathetic outflow into adipose tissue.
These insights were obtained using a transgenic mouse line (AdipoqCreERT2) (35), allowing the specific and inducible inactivation of the CB1 gene in adiponectin-positive cells, thereby overcoming previously observed limitations using other transgenic mouse lines targeting adipocytes (56).
Adipose tissue plasticity has been studied extensively, and recent findings indicate a remarkable remodeling capacity of adipocytes (10, 41, 57, 58). Here, adipocyte-specific CB1 deletion led to smaller adipocytes and decreased lipid storage in EF, in both SD and HFD. Thus, in agreement with previously published evidence (40), CB1 is essential for the maintenance of the differentiation state and lipid storage capacity of white adipocytes. The reduced energy storage capacity was linked to decreased body weight in Ati-CB1–KO mice. Although gene deletion was induced in mature adipocytes at the age of 4 to 5 weeks, at this point, we cannot exclude the possibility that CB1 function is also required for proper developmental processes, such as the expansion of adipose tissue. In SF, we showed that CB1 deficiency in adipocytes led to an increase in the multilocular Ucp1-positive adipocytes. Gene expression analysis confirmed an upregulation of thermogenic genes and mitochondrial biogenesis in SF, suggesting the browning of this fat depot in Ati-CB1–KO mice. Furthermore, a similarly increased thermogenic program was found in BAT. Accordingly, energy dissipation was markedly enhanced in the Ati-CB1–KO mice under HFD. Altogether, CB1 inactivation in adipocytes was able to reduce lipid storage capacity, to increase the number of multilocular Ucp1-positive adipocytes in SF, and to induce BAT thermogenesis.
Ati-CB1–KO mice were leaner than Ati-CB1–WT controls independently of the diet, and their daily caloric intake was reduced. However, as suggested by pair-feeding experiments, the decreased caloric intake seems to play a minor role in the lean metabolic phenotype of Ati-CB1–KO mice.
Strikingly, the reprogramming induced by adipocyte CB1 deletion precedes the emergence of body weight changes. In particular, we observed a delayed differentiation of adipocytes in EF, adipose tissue browning in SF, and an increase in markers of mitochondrial activity in BAT. Independently of the emergence of body weight differences between genotypes, the thermogenic and metabolic remodeling in Ati-CB1–KO adipose tissue can be linked to the increased local abundance of alternatively activated macrophages (M2 polarization). Concomitantly, we found increased expression of biosynthetic enzymes of NE together with enhanced NE concentrations in adipose tissue. In summary, loss of adipose CB1 is a primary cause for early changes in metabolic, biochemical, and morphological features of adipocytes, preceding the diet-induced whole body changes, and thus, these alterations in adipocytes are not mere consequences of many weeks of HFD feeding.
The classical model proposes that BAT thermogenesis and browning of WAT are mediated by enhanced NE release from sympathetic nerve terminals and stimulation of β-adrenergic receptors in adipose tissue. BAT is densely innervated by sympathetic terminals. WAT is also innervated, although to a lesser degree, suggesting that alternative pathways participate in the regulation of Ucp1 expression in WAT. Recent investigations suggested that alternatively activated macrophages also contribute to BAT thermogenesis and browning of WAT by acting as a local source of catecholamines (14–17), but these are in contrast with other recent findings (18, 19). Here, we have demonstrated that ATMs express all enzymes necessary for catecholamine synthesis. In particular, we were able to detect expression of Th, the rate-limiting gene in NE production, in CD11b+F4/80+ adipose tissue cells using 2 independent experimental approaches, real-time PCR and the more sensitive ddPCR method. Both approaches showed very low gene expression levels, although TH protein levels were certainly measured in ATMs and M2 macrophages, as analyzed by flow cytometry. Confocal microscopy studies were not able to detect the presence of TH protein in CD206+ cells in adipose tissue, similar to recent studies (18, 59), this inability likely related to the detections limits of immunohistochemistry techniques. At this point, we do not know the functional relevance of these marginal expression levels; nonetheless, biochemical studies of NE synthesis in isolated ATMs should be able to shed light on this open issue.
Interestingly, double confocal microscope analysis showed that the CD206+ macrophage extensions are in tight anatomical proximity to TH+ sympathetic fibers, in agreement with recent findings (59). Due to this intimate proximity between M2 macrophage projections and sympathetic nerve terminals, we cannot exclude that macrophages may release other signaling molecules able to increase NE output from the sympathetic nerves or, alternatively, that the M2 macrophages may take up NE from the sympathetic cells and then release it. Wolf and colleagues (59) put forward the notion that certain macrophage subpopulations could modulate BAT sympathetic innervations via upregulation of Plexin4A. Indeed, there is rising and compelling evidence on the role of ATMs in adipose tissue remodeling and metabolic beneficial effects, via releasing NE and/or other factors or via other independent mechanisms, such as lysosome biogenesis (14–16, 19, 59–62).
Animal housing temperature (22°C) has been described as a chronic thermal stress for rodents (47). Under these housing conditions, mice have to increase whole energy metabolism and food intake for maintenance of a constant body temperature (42). Therefore, we analyzed HFD-fed Ati-CB1–KO mice at thermoneutrality (30°C). Strikingly, Ati-CB1–KO mice fed at thermoneutrality showed body weight reduction comparable to that of Ati-CB1–KO mice at standard animal housing temperature (22°C). Importantly, upregulation of mitochondrial biogenesis regulators (Ppargc1a, Tfam) as well as of Ucp1 and mitochondrial genes, M2 markers (Clec10a, Mrc1), and NE synthesizing genes (Ddc), were increased in the SF of Ati-CB1–KO mice compared with control littermates at thermoneutral conditions. Thereby, considering that mice housed at 30°C lack the thermal drive to activate BAT thermogenesis or to beige WAT (63), our findings may support the notion that the M2 macrophage polarization plays a prominent role in the lean phenotype observed in Ati-CB1–KO mice under HFD at thermoneutrality.
A pharmacological study has shown that the peripheral CB1 blockade using peripherally restricted CB1 inverse agonist JD5037 alleviates obesity by reversing leptin resistance in DIO mice (33). Genetic inactivation of adipocyte CB1 in Ati-CB1–KO–TAO mice reduced body weight and ameliorated obesity-related metabolic and behavioral alterations. Furthermore, treatment with AM6545, a restricted peripheral CB1 antagonist (31, 32), was unable to modify body weight and food intake in Ati-CB1–KO–TAO mice, suggesting that the adipocyte CB1 plays a major role in the actions of this compound.
The present findings reveal a critical role for the adipocyte CB1 in the regulation of energy balance. The selective deletion of CB1 in adipocytes promotes profound reprogramming, not only of adipocytes, but also of adipocyte-resident cells, including alternatively activated macrophages that work concomitantly with sympathetic neuronal efferents to play a pivotal role in reducing adipocyte differentiation and browning WAT as well as promoting a thermogenic program. Based on our current experimental approaches, we cannot estimate the contribution of alternatively activated macrophages in comparison with sympathetic innervation to the enhanced NE turnover observed in lean Ati-CB1–KO mice or determine whether the M2 macrophages are local sources of sufficient NE concentrations with physiological relevance. In summary, we conclude that deletion of CB1 in adipocytes leads to an increased SNS tone together with M2 macrophage abundance, both of which play critical roles in the emergence of the lean phenotype of Ati-CB1-KO. This compelling evidence illustrates the crosstalk between adipocytes and immune cells in concert with the SNS, in which CB1 occupies a key regulatory function.
Methods
Mice.
Male Total-CB1–KO (also named CB1–/–) mice were generated and genotyped as described (64). Ati-CB1–KO mice (CB1 deletion in adipocytes; CB1fl/fl;AdipoqCreERT2tg/+ with tamoxifen treatment) were generated by crossing the AdipoqCreERT2 line (35) with mice containing the 2 loxP sites flanking the open reading frame of the CB1 gene (64). Mice were genotyped by PCR as previously described (34, 35). For the AdipoqCreERT2 line, the primers 5′-TGGTGCATCTGAAGACACTACA and 5′-TGCTGTTGGATGGTCTTCACAG gave a band of 600 bp, when the transgene was present. For the genotyping of the CB1 gene locus, 3 primers were used: G50 (5′-GCTGTCTCTGGTCCTCTTAAA), G51 (5′-GGTGTCACCTCTGAAAACAGA), and G53 (5′-CTCCTGTATGCCATAGCTCTT). G50 plus G51 detected the floxed CB1 allele as a 500-bp band and the WT CB1 allele as a 400-bp band, G50 plus G53 detected the floxed CB1 allele after recombination (i.e., the inactivated CB1 gene) as a 600-bp band. Mutant animals were on a mixed genetic background with a predominant C57BL/6N contribution (at least 9 backcrosses). Mice were housed under conditions of controlled temperature (22°C) and illumination (12-hour light/12-hour dark cycle, light off at 7 pm). Generally, 4- to 5-week-old male Ati-CB1–KO mice and their littermate controls (CB1fl/fl, named Ati-CB1–WT) received tamoxifen (1 mg/mouse, stock solution in 33% ethanol, 33% dimethyl sulfoxide, 33% Tween-80, finally diluted 1:10 in 0.15 M NaCl; tamoxifen from Sigma-Aldrich, T5648) once a day i.p. for 5 consecutive days. For experiments shown in Figure 5, tamoxifen treatment was performed at 16 weeks of age. When treatment was at week 4, 2 weeks after the last tamoxifen injection, animals were single housed and sorted into their corresponding diet-specific group. Animals were fed with an SD containing 13.9 kJ/g (caloric contribution: 11% fat, 36% protein, 53% carbohydrate; Altromin, C1090/10), with an HFD containing 18.9 kJ/g (caloric contribution: 40% fat, 15% protein, 45% carbohydrate; Dottori Piccioni Laboratory), or with an SHFD containing 21.1 kJ/g (caloric contribution: 60% fat, 17% protein, 23% carbohydrate; Altromin, C1090/60).
Histology and immunofluorescence.
Paraffin-embedded tissue was sectioned on a microtome (8 μm) and mounted on microscope glass (Thermo Scientific, Superfrost Plus, 4951PLUS). Sections were deparaffinized. For histology, sections were stained with hematoxylin (2 g/l, Roth, CN04.1) for 15 minutes and with eosin (0.1%; in 0.0003% acetic acid) for 10 minutes. Cell size quantification was performed after H&E staining; sequential photographs from EF were taken and processed with ImageJ software (version 1.45, NIH). For immunofluorescence, epitopes were unmasked by the use of heat treatment in sodium citrate buffer (10 mM) at 95°C for 15 minutes. Sections were preincubated with 5% BSA and incubated overnight at 4°C in a 1:50 dilution of polyclonal anti-mouse CB1 antibody (rabbit L15 antiserum, directed against the last 15 amino acids of CB1; gift of Ken Mackie, Indiana University, Bloomington, Indiana, USA), in a 1:1,000 dilution of a monoclonal anti-mouse adiponectin antibody (Abcam, ab22554), or in a 1:1,000 dilution of a polyclonal anti-mouse UCP1 antibody (Santa Cruz Biotechnology Inc., sc-6529). Sections were incubated with the secondary antibody (1:1,000 coupled to Alexa Fluor 546 or Alexa Fluor 488, Invitrogen) for 2 hours at room temperature. For fluorescence microscopy, sections were analyzed using the Leica DMRA microscope (Leica Microsystems) equipped with appropriate excitation and emission filters.
β-Galactosidase staining.
Cre recombinase activity was evaluated by crossing the AdipoqCreERT2 mouse line with the Gt(ROSA)26Sor Cre reporter mouse line (35). β-Galactosidase staining was performed as described (65).
Electron microscopy.
Ati-CB1–WT and Ati-CB1–KO on SD and HFD (n = 3/group) were deeply anesthetized with pentobarbital and transcardially perfused with 2% glutaraldehyde in 0.1 M phosphate buffer pH 7.4. Fat depots were collected and cut in small pieces and post-fixed for 6 hours at 4°C in the same fixative. Small fragments of inguinal SF were fixed in 2% glutaraldehyde/2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for at least 4 hours, post-fixed in 1% osmium tetroxide and 1% potassium hexacyanoferrate (II), dehydrated in acetone, and epoxy resin embedded. Semi-thin sections (2 μm) were stained with toluidine blue, and thin sections obtained with an MT-X ultratome (RCM) were mounted on copper grids, stained with lead citrate, and examined with a CM10 transmission electron microscope (Philips).
Food intake, body weight monitoring, pair feeding.
Mice had ad libitum access to food and water. Food intake and body weight were measured at 9 am, twice a week from 8 to 22 weeks of age. For the pair-feeding experiment, from week 12 to week 16, Ati-CB1–WT mice received a daily amount of food that was calculated by determining the daily food intake of the Ati-CBO–KO mice based on the weekly average consumption.
Indirect calorimetry.
EE was assessed as described by measuring oxygen consumption (VO2) and carbon dioxide production (VCO2) by indirect calorimetry (46). Twelve-week-old male WT (n = 16) and Ati-CB1–KO (n = 12) mice maintained on an HFD for 5 to 6 weeks were individually housed in metabolic chambers (TSE Systems GmbH) in which water and food intake, locomotor activity, and gas exchanges were monitored. Following 48 hours of acclimation, O2 consumption, CO2 production, locomotor activity, and food and water intake were measured every 15 minutes for 48 hours. Mice had ad libitum access to HFD and water throughout the study. The values of EE corresponding to the lowest 10 values of ambulatory activity over the 48-hour measurement were averaged to provide a measure of REE (kcal/h). EE and REE were analyzed by ANCOVA using body weight as covariate.
RNA isolation and real-time PCR analysis.
RNA was isolated from Ati-CB1–WT and Ati-CB1–KO mice on SD and HFD and analyzed as described (24). Tissues were homogenized in TRIzol (Sigma-Aldrich, 12183-555), aqueous phase was collected, and RNA was isolated by using a QIAGEN RNA Kit (QIAGEN, 74106). For CD11b+F4/80+ FACS-sorted ATMs, RNA was isolated using a RNeasy Plus Micro Kit (QIAGEN, 74034). cDNA was synthesized by using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, 4390778). Primers for TaqMan Gene Expression Assay were purchased from Applied Biosystems. Real-time PCR analysis was carried out with the following primers (showing gene name followed by probe code): Pparg: Mm01184323_m1; Cebpa: Mm00514283_s1; Adipoq: Mm00456425_m1; Lep: Mm00434759_m1; Fasn: Mm00662291_g1; Acaca: Mm01304257_m1; Ucp1: Mm01244861_m1; Ppargc1a: Mm01208832_m1; CB1: Mm00432621_s1; Elovl3: Mm00468164_m1; Dio2:Mm00515664_m1; Cox8b: Mm00432648_m1; Cox4i2: Mm00446387_m1; Tfam: Mm00447485_m1; Adrb3: Mm02601819_g1; Pomc: Mm00435874_m1; Npy: Mm01410146_m1; Cartpt: Mm04210469_m1; Pmch: Mm01242886_g1; Socs3: Mm00545913_s1; Mrc1: Mm00485148_m1; Clec10a: Mm00546125_g1; Th: Mm00447557_m1; Dbh: Mm00460472_m1; Ddc: Mm00516688_m1; and Gusb: Mm01197698_m1.
Hormone and metabolite assays.
Plasma leptin, insulin, IL-6, and PAI1 were measured by Mouse Serum Adipokine Kit (Milliplex Map, Millipore, MADPK-71K), adiponectin by Mouse Adiponectin ELISA Assay Kit (Biovision, K4902-100), plasma free fatty acids by Fatty Acid Kit (Biovision, K612-100), plasma total cholesterol and LDL by HDL and LDL Cholesterol Quantification Kit (BioVision, K631-100), plasma and hepatic triglycerides by Triglyceride Quantification Kit (Biovision, K622-100), and blood glucose levels by an automated blood glucose reader (OneTouch Ultra2 Glucometer) (66).
NE turnover.
Ati-CB1–KO mice and their WT littermates on HFD were divided into 2 groups to study the NE turnover in EF, SF, and BAT at time point zero and 3 hours after i.p. injection of the TH inhibitor methyl ester of α-methyl-P-tyrosine (80 mg/kg, Sigma-Aldrich, M8131). EF, SF, and BAT were then rapidly removed and stored at –80°C for subsequent NE quantification. Tissue NE was measured by LDN ELISA Kit (Labor Diagnostika Nord, BA E-5200). NE turnover was calculated by plotting tissue NE content semilogarithmically, and the slope of the declining endogenous NE level versus time was used to calculate the turnover rate (67). Rates of NE turnover were then calculated by multiplying the turnover rate for each NE content of the organs of the individual mouse.
Glucose and insulin tolerance tests.
For glucose tolerance test, overnight-fasted mice were given i.p. glucose (2 mg/g). For insulin tolerance test, overnight-fasted mice were injected with insulin (0.75 IU/kg i.p.; Eli Lilly). Tail blood glucose was determined at defined time intervals (OneTouch Ultra, LifeScan). Concerning insulin tolerance tests, due to increased total adiponectin levels in Ati-CB1–KO mice and the insulin sensitization effect of this hormone on peripheral organs (36), we used a low insulin dose (0.5 IU/kg). Preliminary experiments with Ati-CB1–KO mice revealed that commonly used insulin doses induced severe hypoglycemia in KO mice.
In vivo quantification of adipose tissue.
Ati-CB1–WT (n = 6–12) and KO (n = 7–4) mice were placed under deep anesthesia with 5% sevoflurane and oxygen supplementation (1 l/min) and scanned using an in vivo micro-CT scanner (eXplore Locus) at an isometric resolution of 90 μm, as previously described (21).
Flow cytometric analysis and sorting of ATMs.
Adipose tissues were minced and digested with collagenase II (1 mg/ml, Sigma-Aldrich) for 30 minutes at 37°C on a shaker. The digested cell suspension was passed through a 70-μm strainer (MACS, Miltenyi Biotec) and centrifuged at 700 g for 10 minutes at 4°C to isolate the stromal-vascular fraction. Pelleted cells were resuspended in rbc lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 minutes and diluted with FACS buffer (PBS containing 1% FCS and 2 mM EDTA) to neutralize rbc lysis. Samples were centrifuged at 700 g for 10 minutes at 4°C and resuspended in FACS buffer. Antibodies against mouse CD206-PE (BioLegend, C068C2), CD301-APC (BioLegend, LOM-14), F4/80-APC (BioLegend, BM8), CD11b-FITC (Abcam, ab24874), and TH-PE (Abcam, ab209921) were used for flow cytometric analysis (acquired on LSRFortessa, BD, additionally equipped with a yellow-green laser for detecting PE and analyzed with FlowJo software, TreeStar). Sorting of ATMs (FACS Aria II SORP using DiVa software, BD) was based on anti-mouse CD11b-PE (BioLegend, M1/70) and anti-mouse F4/80-APC antibody (BioLegend, BM8). Samples were fixed in 4% PFA for 15 minutes at 4°C before staining of intracellular antigens. FACS-sorted CD11b+F4/80+ ATMs were directly resuspended in 750 μl of TRIzol Reagent (Thermo Fisher) for total RNA isolation.
ddPCR system.
Before droplet generation, ddPCR reactions were prepared in a manner similar to that of real-time reactions using TaqMan probes labeled with FAM (2× ddPCR Mastermix [Bio-Rad], 20× TaqMan probes and template in a final volume of 20 μl). As template, the maximum input (9 μl) from undiluted cDNA of CD11b+F4/80+-sorted ATMs were loaded per ddPCR reaction. Positive controls (1 μl from 1:1,000 dilution of cDNA from adrenal gland; and 1 μl from 1:100 dilution of cDNA from brain stem) and negative controls (1 μl from 1:1,000 dilution of no-RT adrenal gland; and 9 μl of H2O as no template control) were included. Each assembled ddPCR reaction mixture was applied into a sample droplet well of an 8-channel disposable droplet generator cartridge (Bio-Rad), and 70 μl of droplet generator oil (Bio-Rad) was loaded into an oil well for each channel. The cartridge was placed into the droplet generator (Bio-Rad) and 1 nl monodisperse droplets were formed in around 2 minutes, generating 15,000 droplets per sample. Droplets were transferred to a 96-well PCR plate, and droplet PCR amplification to end-point was performed in a conventional thermal cycler (95°C for 10 minutes; 40 cycles of 95°C for 30 seconds and 60°C for 1 minute; followed by 98°C for 10 minutes and hold at 4°C). After PCR, the 96-well plate was loaded and analyzed by a 2-color detector droplet reader (Bio-Rad), which automatically assigns droplets as positive or negative based on their fluorescence amplitude. Analysis of the ddPCR data was performed with QuantaSoft analysis software (Bio-Rad).
Peroxidase immunohistochemistry.
Immunohistochemistry was performed on 3-μm–thick paraffin-embedded sections of fat depots. In brief, paraffin sections were reacted with 0.3% H2O2 (in water; 10 minutes) to block endogenous peroxidase, rinsed with PBS, and incubated in a 3% normal-serum blocking solution for 60 minutes. Then they were incubated with the anti-TH primary antibody (1: 400, Millipore, AB1542) overnight at 4°C. After rinse in PBS, sections were incubated with a 1:200 biotinylated HRP-conjugated anti-sheep secondary antibody solution (Vector Laboratories) for 30 minutes. Histochemical reactions were performed using VECTASTAIN ABC Kit (Vector Laboratories) and SIGMAFAST 3,3′-diaminobenzidine (Sigma-Aldrich) as the substrate. Sections were finally counterstained with hematoxylin. Staining was not observed when the primary antibody was omitted. For morphometric analysis, adipocyte size was calculated as the mean adipocyte area of 200 randomly selected adipocytes, and TH-positive parenchymal fibers were calculated in 200 adipocytes. Tissue sections were analyzed using Nikon LUCIA IMAGE software (Laboratory Imaging).
Double-labeling and confocal microscopy.
For double-labeling experiments, frozen sections obtained with a cryostat (Leica CM1900) were incubated overnight in a mixture of the 2 primary antibodies (TH, Millipore AB1542; and CD206, Abcam ab64693). Sections were washed twice with PBS and incubated in a cocktail of fluorophore-linked secondary antibodies at a dilution of 1:400 in PBS for 1 hour at room temperature. The secondary antibodies were Alexa Fluor 488 donkey anti-sheep IgG and Alexa Fluor 555 donkey anti-rabbit IgG (Invitrogen). Sections were washed twice with PBS, counterstained with TO-PRO3 (1:4,000), and mounted using Vectashield mounting medium (Vector). Sections were viewed under a motorized Leica DM6000 microscope. Fluorescence was detected with a Leica TCS-SL spectral confocal microscope equipped with an Argon and He/Ne mixed gas laser. Fluorophores were excited with the 488 nm, 543 nm lines and imaged separately. Images (1024 × 1024 pixels) were obtained sequentially from 2 channels using a confocal pinhole of 1.1200. The brightness and contrast of the final images were adjusted using Photoshop 6 (Adobe Systems).
Western blot.
WT and Th–KO spleen sample preparations were obtained from Christoph Buettner (The Mount Sinai Hospital, New York, New York, USA)(18). Protein extracts were separated on 10% PAGE and blotted onto nitrocellulose membrane. Membranes were blocked at room temperature for 1 hour with blocking solution (5% powdered nonfat milk, 0.5% BSA, and 0.1% Tween-20) and incubated with primary antibody (1:5,000 in blocking solution) overnight at 4°C. Primary antibodies against mouse TH (Abcam, ab137869) and mouse actin (Millipore, 04-1040) were used. Membranes were washed 3 times for 5 minutes in PBS-T buffer, and blots were incubated with HRP-conjugated anti-rabbit secondary antibody (1:5,000 in blocking solution) for 1 hour at room temperature. After washing, signals were detected using a chemiluminescent enhancer solution (WESTAR NOVA 2.0, CYANAGEN). Images were captured using a Peqlab camera and analyzed with Fusion 1.0 software.
CL316243-induced in vivo lipolysis.
Lipolysis was carried out as described (68) with some modifications. After 4 hours starvation, HFD-fed Ati-CB1–KO mice and WT littermates were injected with the selective β3-adrenergic receptor agonist CL316243 (Sigma-Aldrich, 0.1 mg/kg, i.p.), and blood samples were collected from the submandibular vein at 0, 8, and 15 minutes. Plasma-free fatty acid concentration was measured using a fatty acid kit (Biovision, K612-100).
Thermoneutrality conditions.
Two weeks after tamoxifen injection, animals were single housed at standard housing temperature (22°C) and at thermoneutrality (30°C, ZOONLAB cabinets) on a 12-hour light/12-hour dark cycle for 4 weeks. Temperature in cabinets was continuously recorded. Mice had free access to HFD and water. Once per week, body weight and food weight were recorded.
Endocannabinoid measurements by LC–tandem mass spectrometry.
Plasma samples from Ati-CB1–WT mice on SD (n = 7), Ati-CB1–WT mice on HFD (n = 12), Ati-CB1–KO mice on SD (n = 6), and Ati-CB1–KO mice on HFD (n = 4) were stored at –80°C until extraction. Samples were allowed to thaw on ice water, and 50 μl aliquots were transferred to 1.5 ml centrifugation tubes. After adding 300 μl of ice-cold ethyl acetate/hexane (9:1, v/v) containing the deuterated endocannabinoids as internal standards, tubes were vortexed for 30 seconds with Vortex Disruptor (Scientific Industries). Tubes were immediately centrifuged for 15 minutes at 16,000 g at 4°C. The upper organic phase was removed, evaporated to dryness under a gentle stream of nitrogen at 37°C, and reconstituted in 50 μl acetonitrile. Concentrations of AEA and 2-arachidonoylglycerol (2-AG) were determined by liquid chromatography (LC) multiple reaction monitoring (MRM). LC and MRM conditions were as described (69). AEA and 2-AG levels were normalized to plasma volume.
Pharmacological treatment with peripheral CB1 antagonist.
Ati-CB1–KO–TAO and Ati-CB1–WT–TAO mice on HFD were injected i.p. with AM6545 (10 mg/kg), a neutral peripherally restricted CB1 antagonist (31, 32), or with vehicle for 3 weeks starting at 5 weeks after tamoxifen injection.
Behavioral experiments.
EPM and HB experiments were performed as previously described (54). Ati-CB1–WT–TAO (n = 8–10) and Ati-CB1–KO–TAO (n = 6–7) mice were analyzed.
Statistics.
Results are expressed as mean ± SEM. Data were analyzed by unpaired 2-tailed Student’s t test or by ANOVA followed by appropriate post hoc tests. Data with repeated measurements were analyzed by repeated-measures ANOVA followed by post-hoc test. Statistical comparison of NE turnover was made with Student’s t test. To determine whether differences in EE and REE were attributable simply to differences in body weight, ANCOVA was performed using as covariates appropriate measures of body weight. All the statistical analyses were performed by using GraphPad Prism 5.0 software. P < 0.05 was considered statistically significant.
Study approval.
Experimental protocols were carried out in accordance with the Council Directive 2010/63EU of the European Parliament and the Council of 22 September 2010 on the protection of animals used for scientific purposes and approved by the local ethical committees on animal care and use in Rhineland-Palatinate, Germany (reference number 23 177-07/G 10-1-038, and 23 177-07/G 16-1-005), by the French Ministry of Agriculture and Fisheries (animal experimentation authorization 3309004), by the ethical committee of the University of Bordeaux (N° 5012062-A), and by University of Bologna, Local Ethical Committee and Italian Ministry of Health (Project ID 09/59/09, and Project ID 54464-X/10).
Author contributions
IRDA, G Mancini, RKS, CQ, SC, KM, EN, UP, G Marsicano, DC, and BL designed the experiments. IRDA, G Mancini, RKS, AAR, CQ, PC, LT, CMZ, CS, AC, JH, MML, LB, and AG performed the experiments and analyzed the data. AS, NW, and SO provided the AdipoqCreERT2 line, VKV and AM provided AM6545. IRDA, G Mancini, RKS, SC, EN, UP, G Marsicano, DC, and BL wrote the paper.
Supplementary Material
Acknowledgments
This work was supported by EU-FP7 REPROBESITY (HEALTH-F2-2008-223713 to BL, G Marsicano, and UP); PAINCAGE (HEALTH-603191 to G Marsicano); the Cariplo Foundation (grants no. 2013-0786 and 2016-1006 to EN); DIABAT (HEALTH-F2-2011-278373 to SC), the German Research Council DFG (FOR926 and CRC1080 subproject B08 to BL), the European Foundation for the Study of Diabetes (to G Mancini, G Marsicano, and DC), INSERM (to G Marsicano and DC), Region Aquitaine (to G Marsicano and DC), the Agence Nationale de la Recherche (OPTOPATH ANR-10-EQPX-08, ANR-10-LABX-0043 and NeuroNutriSens ANR-13-BSV4-0006 to DC and G Marsicano), PhD Fellowship INSERM/Region Aquitaine (to PC), the European Research Council (ENDOFOOD, ERC-2010-StG-260515 and CannaPreg, ERC-2014-PoC-640923 to G Marsicano), the Fondation pour la Recherche Medicale (DRM20101220445 and DPP20151033974 to G Marsicano), the Human Frontiers Science Program (HFSP) (to G Marsicano), and the Ministero dell’Istruzione, dell’Università e della Ricerca (grant 2009E48P9M to EN and grant 2010329EKE004 to UP). We thank the animal facility and the genotyping platforms of the INSERM U1215 NeuroCentre Magendie, funded by INSERM and Labex Brain ANR-10-LABX-43, for animal care and mouse genotyping. We would like to thank Tina Zimmermann, Martin Purio, Anisa Kosan, and Samanha Clark for technical support. We are very grateful to the Core Unit Endocannabinoid Measurements (FOR926), and the Genomic, the Flow Cytometry and the Microscopy Core Facility of the Institute of Molecular Biology (IMB) Mainz. These IMB Core Facilities acknowledge the support by the German Research Foundation DFG (grants: INST 247/646-1 FUGG, and INST 247645-1 FUGG). We also would like to thank Ken Mackie for providing the anti-CB1 antibody, and Christoph Buettner for providing Th-deficient and WT control spleen samples.
Version 1. 10/16/2017
Electronic publication
Version 2. 11/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(11):4148–4162.https://doi.org/10.1172/JCI83626.
See the related Commentary at Cannabis in fat: high hopes to treat obesity.
Contributor Information
Inigo Ruiz de Azua, Email: inigo.azua@uni-mainz.de.
Raj Kamal Srivastava, Email: srivastr@uni-mainz.de.
Alejandro Aparisi Rey, Email: rey@uni-mainz.de.
Pierre Cardinal, Email: cardinalpierre_123@hotmail.com.
Laura Tedesco, Email: laura.tedesco@unimi.it.
Carmelo Quarta, Email: carmelo.quarta@helmholtz-muenchen.de.
Claudia Schwitter, Email: schwitte@uni-mainz.de.
Nina Wettschureck, Email: Nina.Wettschureck@mpi-bn.mpg.de.
V. Kiran Vemuri, Email: k.vemuri@neu.edu.
Alexandros Makriyannis, Email: a.makriyannis@neu.edu.
Jens Hartwig, Email: j_hart@gmx.de.
Maria Mendez-Lago, Email: M.Mendez-Lago@imb-mainz.de.
Laura Bindila, Email: bindila@uni-mainz.de.
Krisztina Monory, Email: monory@uni-mainz.de.
Antonio Giordano, Email: a.giordano@univpm.it.
Saverio Cinti, Email: s.cinti@univpm.it.
Giovanni Marsicano, Email: giovanni.marsicano@inserm.fr.
Stefan Offermanns, Email: stefan.offermanns@mpi-bn.mpg.de.
Enzo Nisoli, Email: enzo.nisoli@unimi.it.
Uberto Pagotto, Email: uberto.pagotto@unibo.it.
Daniela Cota, Email: daniela.cota@inserm.fr.
Beat Lutz, Email: beat.lutz@uni-mainz.de.
References
- 1.Henry SL, Bensley JG, Wood-Bradley RJ, Cullen-McEwen LA, Bertram JF, Armitage JA. White adipocytes: more than just fat depots. Int J Biochem Cell Biol. 2012;44(3):435–440. doi: 10.1016/j.biocel.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(5):367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enerbäck S. Human brown adipose tissue. Cell Metab. 2010;11(4):248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104(4):531–543. doi: 10.1016/S0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 10.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 11.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS ONE. 2012;7(11):e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinoda K, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21(4):389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cypess AM, et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab. 2015;21(1):33–38. doi: 10.1016/j.cmet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen KD, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480(7375):104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu Y, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157(6):1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suárez-Zamorano N, et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med. 2015;21(12):1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv Y, et al. Adrenomedullin 2 enhances beiging |in white adipose tissue directly in an adipocyte-autonomous manner and indirectly through activation of M2 macrophages. J Biol Chem. 2016;291(45):23390–23402. doi: 10.1074/jbc.M116.735563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer K, et al. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat Med. 2017;23(5):623–630. doi: 10.1038/nm.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spadaro O, et al. IGF1 shapes macrophage activation in response to immunometabolic challenge. Cell Rep. 2017;19(2):225–234. doi: 10.1016/j.celrep.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cota D, et al. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J Clin Invest. 2003;112(3):423–431. doi: 10.1172/JCI17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quarta C, et al. CB(1) signaling in forebrain and sympathetic neurons is a key determinant of endocannabinoid actions on energy balance. Cell Metab. 2010;11(4):273–285. doi: 10.1016/j.cmet.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 22.Silvestri C, Di Marzo V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metab. 2013;17(4):475–490. doi: 10.1016/j.cmet.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Bensaid M, et al. The cannabinoid CB1 receptor antagonist SR141716 increases Acrp30 mRNA expression in adipose tissue of obese fa/fa rats and in cultured adipocyte cells. Mol Pharmacol. 2003;63(4):908–914. doi: 10.1124/mol.63.4.908. [DOI] [PubMed] [Google Scholar]
- 24.Tedesco L, et al. Cannabinoid type 1 receptor blockade promotes mitochondrial biogenesis through endothelial nitric oxide synthase expression in white adipocytes. Diabetes. 2008;57(8):2028–2036. doi: 10.2337/db07-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tedesco L, et al. Cannabinoid receptor stimulation impairs mitochondrial biogenesis in mouse white adipose tissue, muscle, and liver: the role of eNOS, p38 MAPK, and AMPK pathways. Diabetes. 2010;59(11):2826–2836. doi: 10.2337/db09-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajzer M, et al. Cannabinoid receptor 1 (CB1) antagonism enhances glucose utilisation and activates brown adipose tissue in diet-induced obese mice. Diabetologia. 2011;54(12):3121–3131. doi: 10.1007/s00125-011-2302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herling AW, Kilp S, Juretschke HP, Neumann-Haefelin C, Gerl M, Kramer W. Reversal of visceral adiposity in candy-diet fed female Wistar rats by the CB1 receptor antagonist rimonabant. Int J Obes (Lond) 2008;32(9):1363–1372. doi: 10.1038/ijo.2008.105. [DOI] [PubMed] [Google Scholar]
- 28.Mølhøj S, Hansen HS, Schweiger M, Zimmermann R, Johansen T, Malmlöf K. Effect of the cannabinoid receptor-1 antagonist rimonabant on lipolysis in rats. Eur J Pharmacol. 2010;646(1–3):38–45. doi: 10.1016/j.ejphar.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Christensen R, Kristensen PK, Bartels EM, Bliddal H, Astrup A. Efficacy and safety of the weight-loss drug rimonabant: a meta-analysis of randomised trials. Lancet. 2007;370(9600):1706–1713. doi: 10.1016/S0140-6736(07)61721-8. [DOI] [PubMed] [Google Scholar]
- 30.Moreira FA, Grieb M, Lutz B. Central side-effects of therapies based on CB1 cannabinoid receptor agonists and antagonists: focus on anxiety and depression. Best Pract Res Clin Endocrinol Metab. 2009;23(1):133–144. doi: 10.1016/j.beem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Cluny NL, et al. A novel peripherally restricted cannabinoid receptor antagonist, AM6545, reduces food intake and body weight, but does not cause malaise, in rodents. Br J Pharmacol. 2010;161(3):629–642. doi: 10.1111/j.1476-5381.2010.00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tam J, et al. Peripheral CB1 cannabinoid receptor blockade improves cardiometabolic risk in mouse models of obesity. J Clin Invest. 2010;120(8):2953–2966. doi: 10.1172/JCI42551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam J, et al. Peripheral cannabinoid-1 receptor inverse agonism reduces obesity by reversing leptin resistance. Cell Metab. 2012;16(2):167–179. doi: 10.1016/j.cmet.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsicano G, et al. CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science. 2003;302(5642):84–88. doi: 10.1126/science.1088208. [DOI] [PubMed] [Google Scholar]
- 35.Sassmann A, Offermanns S, Wettschureck N. Tamoxifen-inducible Cre-mediated recombination in adipocytes. Genesis. 2010;48(10):618–625. doi: 10.1002/dvg.20665. [DOI] [PubMed] [Google Scholar]
- 36.Awazawa M, et al. Adiponectin enhances insulin sensitivity by increasing hepatic IRS-2 expression via a macrophage-derived IL-6-dependent pathway. Cell Metab. 2011;13(4):401–412. doi: 10.1016/j.cmet.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Yamauchi T, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7(8):941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 38.Iyer A, Fairlie DP, Prins JB, Hammock BD, Brown L. Inflammatory lipid mediators in adipocyte function and obesity. Nat Rev Endocrinol. 2010;6(2):71–82. doi: 10.1038/nrendo.2009.264. [DOI] [PubMed] [Google Scholar]
- 39.Blüher M, et al. Dysregulation of the peripheral and adipose tissue endocannabinoid system in human abdominal obesity. Diabetes. 2006;55(11):3053–3060. doi: 10.2337/db06-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matias I, et al. Regulation, function, and dysregulation of endocannabinoids in models of adipose and beta-pancreatic cells and in obesity and hyperglycemia. J Clin Endocrinol Metab. 2006;91(8):3171–3180. doi: 10.1210/jc.2005-2679. [DOI] [PubMed] [Google Scholar]
- 41.Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010;11(4):253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Westerberg R, et al. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J Biol Chem. 2006;281(8):4958–4968. doi: 10.1074/jbc.M511588200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Matheny M, Zolotukhin S, Tumer N, Scarpace PJ. Regulation of adiponectin and leptin gene expression in white and brown adipose tissues: influence of beta3-adrenergic agonists, retinoic acid, leptin and fasting. Biochim Biophys Acta. 2002;1584(2–3):115–122. doi: 10.1016/s1388-1981(02)00298-6. [DOI] [PubMed] [Google Scholar]
- 44.Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411(6836):480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 45.Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjørbaek C, Flier JS. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med. 2004;10(7):734–738. doi: 10.1038/nm1072. [DOI] [PubMed] [Google Scholar]
- 46.Tschöp MH, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2011;9(1):57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golozoubova V, Gullberg H, Matthias A, Cannon B, Vennström B, Nedergaard J. Depressed thermogenesis but competent brown adipose tissue recruitment in mice devoid of all hormone-binding thyroid hormone receptors. Mol Endocrinol. 2004;18(2):384–401. doi: 10.1210/me.2003-0267. [DOI] [PubMed] [Google Scholar]
- 48.MacDougald OA, Hwang CS, Fan H, Lane MD. Regulated expression of the obese gene product (leptin) in white adipose tissue and 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 1995;92(20):9034–9037. doi: 10.1073/pnas.92.20.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78(3):783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 50.Luppino FS, et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Arch Gen Psychiatry. 2010;67(3):220–229. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 51.Sharma S, Fulton S. Diet-induced obesity promotes depressive-like behaviour that is associated with neural adaptations in brain reward circuitry. Int J Obes (Lond) 2013;37(3):382–389. doi: 10.1038/ijo.2012.48. [DOI] [PubMed] [Google Scholar]
- 52.Teff KL, Kim SF. Atypical antipsychotics and the neural regulation of food intake and peripheral metabolism. Physiol Behav. 2011;104(4):590–598. doi: 10.1016/j.physbeh.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Lee S, Elmquist JK. Circuits controlling energy balance and mood: inherently intertwined or just complicated intersections? Cell Metab. 2014;19(6):902–909. doi: 10.1016/j.cmet.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rey AA, Purrio M, Viveros MP, Lutz B. Biphasic effects of cannabinoids in anxiety responses: CB1 and GABA(B) receptors in the balance of GABAergic and glutamatergic neurotransmission. Neuropsychopharmacology. 2012;37(12):2624–2634. doi: 10.1038/npp.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee KY, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spiegelman BM. Banting Lecture 2012: Regulation of adipogenesis: toward new therapeutics for metabolic disease. Diabetes. 2013;62(6):1774–1782. doi: 10.2337/db12-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf Y, et al. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat Immunol. 2017;18(6):665–674. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18(6):816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee YH, Kim SN, Kwon HJ, Maddipati KR, Granneman JG. Adipogenic role of alternatively activated macrophages in β-adrenergic remodeling of white adipose tissue. Am J Physiol Regul Integr Comp Physiol. 2016;310(1):R55–R65. doi: 10.1152/ajpregu.00355.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waqas SFH, et al. Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J Clin Invest. 2017;127(7):2842–2854. doi: 10.1172/JCI90152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J Exp Biol. 2011;214(Pt 2):242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- 64.Marsicano G, et al. The endogenous cannabinoid system controls extinction of aversive memories. Nature. 2002;418(6897):530–534. doi: 10.1038/nature00839. [DOI] [PubMed] [Google Scholar]
- 65.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 66.Ruiz de Azua I, et al. RGS4 is a negative regulator of insulin release from pancreatic beta-cells in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107(17):7999–8004. doi: 10.1073/pnas.1003655107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brodie BB, Costa E, Dlabac A, Neff NH, Smookler HH. Application of steady state kinetics to the estimation of synthesis rate and turnover time of tissue catecholamines. J Pharmacol Exp Ther. 1966;154(3):493–498. [PubMed] [Google Scholar]
- 68.Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS. Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology. 2000;141(9):3388–3396. doi: 10.1210/endo.141.9.7637. [DOI] [PubMed] [Google Scholar]
- 69.Schulte K, et al. Cannabinoid CB1 receptor activation, pharmacological blockade, or genetic ablation affects the function of the muscarinic auto- and heteroreceptor. Naunyn Schmiedebergs Arch Pharmacol. 2012;385(4):385–396. doi: 10.1007/s00210-011-0717-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.