Abstract
Few monogenic causes for severe manifestations of common allergic diseases have been identified. Via next generation sequencing on a cohort of patients with severe atopic dermatitis, some with comorbid infections, we found 8 individuals from 4 families with novel heterozygous mutations in CARD11, a scaffolding protein involved in lymphocyte receptor signaling. Disease improved over time in most patients. Transfection of mutant expression constructs into T cell lines demonstrated both loss of function and dominant interfering activity upon antigen receptor-induced NF-κB and mTORC1 activation. Patient T-cells had similar defects, as well as diminished IFN-γ cytokine production. The mTORC1 and IFN-γ production defects could be partially rescued by supplementing with glutamine, which requires CARD11 for import into T cells. Our findings indicate a single hypomorphic gene mutation in CARD11 can cause potentially correctable cellular defects that lead to atopic dermatitis.
Introduction
Monogenic causes for immune disorders have provided critical insights into the role of specific immune pathways in the pathogenesis of common diseases, including atopy and allergic diseases. In the case of some allergic disorders involving atopic dermatitis and elevated IgE coupled with infection, it may be difficult to separate impairment of host defense from the role of the defined genetic lesion in contributing to allergic disease. Patients with severe atopic disease and elevated serum IgE levels have now been linked to mutations in immune-mediated host defense pathways, including DOCK8, STAT3 or PGM3 1–3. In some patients with severe atopic dermatitis, elevated IgE and eosinophilia, there is little reason to suspect an immune deficiency, as infections beyond the skin are less common, and they lack comorbidities seen with the genetically defined disorders. Single gene mutations in common allergic disease without overt syndromic features have been less common4, but are being increasingly described. One example is found in a single gain of function (GOF) mutation in IL4RA identified initially in a few severely atopic patients5, which is a common risk allele in allergic disease6. Another example is in loss of function (LOF) IFNGR1 mutations leading to atopic dermatitis with eczema herpeticum, without additional immune phenotypes7. Other LOF mutations in cytokine signaling components such as STAT3 and STAT5b 8 can also contribute to atopic phenotypes, but with substantial multisystem comorbidity, seen also in putative antigen receptor signaling gene mutations such as WASP, DOCK89, and MALT1 10. Diminished antigen-driven TCR signaling can predispose to Th2 phenotypes in a variety of in vivo and in vitro settings9, and mouse models have shown that hypomorphic mutations in ZAP7011 and CARD1112 can also lead to severe atopy.
CARD11 (also known as CARMA1) encodes a membrane-associated guanylate kinase (MAGUK)-family protein which partners with BCL10 and MALT1 to form the “CBM complex”, a scaffold classically required for IKK and NF-κB activation in response to lymphocyte receptor ligation13. While homozygous null mutations in CARD11 lead to severe combined immune deficiency (SCID) in mice and in humans14–17, heterozygous GOF mutations give rise to a selective B cell lymphoproliferative disease known as BENTA 18,19. CARD11 has also been identified as a risk locus for atopic dermatitis20.
Here we describe rare hypomorphic dominant negative mutations in CARD11 in 4 unrelated families, which lead to dominantly inherited, severe atopy, with variable infection beyond the skin.
Results
Heterozygous CARD11 mutations found in 4 families with severe atopic dermatitis
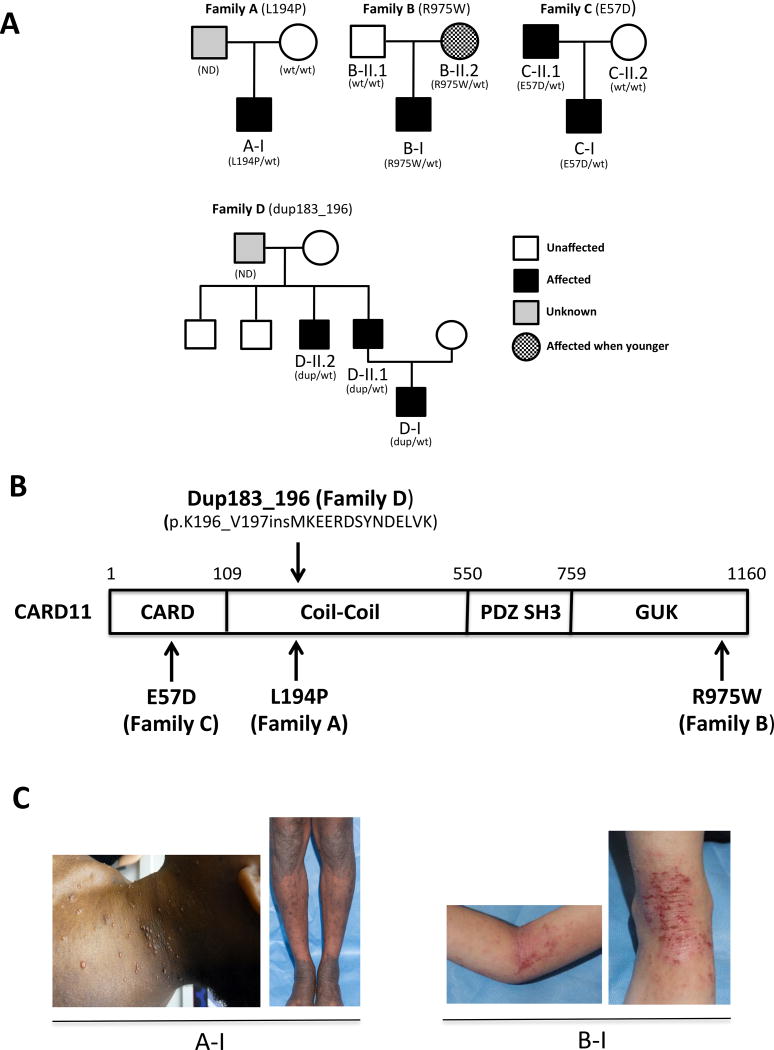
Next generation sequencing was performed on a series of patients with recalcitrant, severe atopic disease (Figure 1A–C). In four of these patients, novel, heterozygous CARD11 mutations were detected. These included 3 distinct missense mutations (p.Glu57Asp (E57D), p.Leu194Pro (L194)) p.Arg975Trp (R975W)) and one in-frame, 14aa insertion (p.Met183_Lys196; herein referred to as dup183_196) (Table 1). The PolyPhen values are 1 for all three missense mutations and the CADD scores are 21.2, 19.5, and 17.8, respectively. They are predicted to be deleterious. A parent and/or other relatives in 2 out of the 4 patients had a history of atopic dermatitis that waned in severity over time; these relatives were confirmed as carriers of the mutation (Figure 1A). Patient B-II.2, the mother of patient B-I, was unaffected but reported eczema when younger. Patients A-I and C-I had histories of pneumonia early in life, and C-II.1 had transient hypogammaglobulinemia of childhood. Patients A-I C-I, D-I, D-II.1 and D-II.2 had multiple episodes of respiratory distress in early childhood, which were treated as pneumonia. Patients A-I and C-I also were diagnosed with asthma. Oral steroids were required to resolve all asthmatic episodes for patient A-I, whose CT imaging showed normal lungs, raising the likelihood that his respiratory symptoms may not have been of infectious etiology. The infectious history of family D (dup183_196) includes abscesses, bacteremia and pulmonary infections. Affected members of family D had some other unique features which may or may not have been related to their primary genetic diagnosis, including a prominent forehead and broad nasal base from birth. Unlike STAT3LOF patients, there was no characteristic facial asymmetry or coarseness, and the birth onset differs as well (Table 1). Patient C-I has history of stroke and ulcerative colitis. C-II.1, his father, who also carries the E57D variant, has history of lymphoma. Cancer and stroke were not otherwise reported. Two of the seven patients had B cell lymphopenia with no decrease in other lymphocyte lineages; 3/7 patients had decreased IgM with no decrease in IgG and normal to elevated IgA in most recent testing. Elevated IgE (5/7) and eosinophilia (6/7) were prominent, and those without had more significant atopic phenotypes when younger without documentation of these laboratory values at that time (Supplementary Table 1).
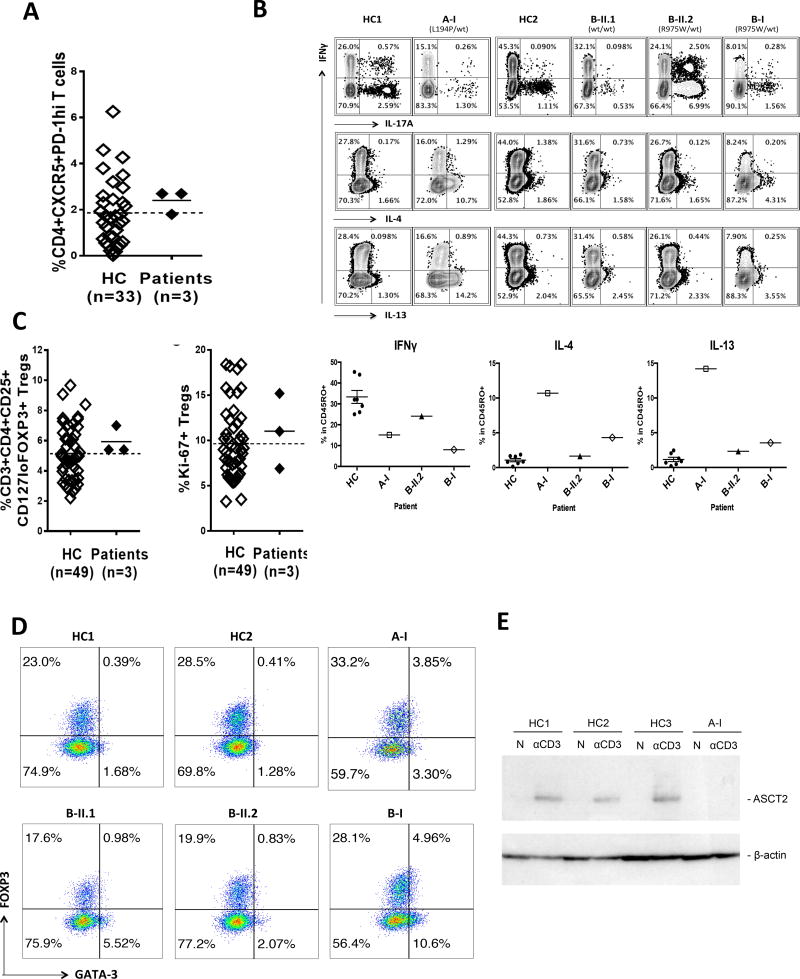
Figure 1. Novel heterozygous CARD11 mutations in the severe atopic dermatitis families.
(A) Pedigrees of the families who have the novel CARD11 mutations. (B) CARD11 protein domains and the position of CARD11 mutations found in the families. (C) Representative patient photographs depicting molluscum contagiosum in A-I (left) and atopic dermatitis in patients A-I (right) and B-I.
Table 1.
Summary of patient clinical data.
| Patient | Mutation | Age | Atopic Dermatitis (AD) |
Infections | Asthma | Other |
|---|---|---|---|---|---|---|
| A-I | L194P (C–C) | 15 | Severe SCORAD: 76 | molluscum | Yes | Food allergy, recurrent respiratory distress |
| B-I | R975W (GUK) | 5 | Severe SCORAD: 74 | Eczema herpeticum | Yes | Food allergy, treated with methotrexate for AD |
| B-II.2 | R975W (GUK) | 27 | Mild-moderate | none | No | AD improved over time |
| C-I | E57D (CARD) | 16 | Severe | Pneumonia, molluscum | Yes | Transient hypogammaglobinemia of childhood, ulcerative colitis, stroke, short stature (<5% for height), food allergy, AD improved over time |
| C-II.1 | E57D (CARD) | 51 | Severe | Pneumonia | Yes | AD improved over time, peripheral T cell lymphoma treated with autologous transplant |
| D-I | 196dup | 1.5 | Severe, erythrodermic desquamating | Lumbar abscess, catheter related infection, CMV pneumonitis | No | Eosinophilic coloproctitis, bilateral nystagmus, seizure, temporal sylvian opercular dysplasia ASMA & Anti-actin autoantibodies, prominent forehead, broad nose |
| D-II.1 | 196dup | 21 | Moderate | Pneumonia, pulmonary TB, bronchiectasis, boils, molluscum | No | Scoliosis, poor dentition, prominent forehead, broad nose, AD improved over time |
| D-II.2 | 196dup | 15 | Severe | Pneumonia, bacteremia, superinfected varicella, Kaposi varicelliform, boils, molluscum | Severe | Poor dentition, dental fractures, chronic diarrhea, prominent forehead, broad nose, AD improved over time |
Age in years. AD: atopic dermatitis. SCORAD: SCORing Atopic Dermatitis Score >50 indicates severe AD.
CARD11 mutations are hypomorphic and interfere with NF-κB and mTORC1 activation
The four heterozygous mutations detected in the patients are in markedly different domains of the CARD11 protein (accession code Q9BXL7) (Figure 1B). L194P and dup183_196 are found in the coiled coil (CC) domain. E57D is in the N-terminal CARD domain, while R975W is in the C-terminal GUK domain. None are found in public polymorphism databases (e.g. ExAC). Interestingly, Goodnow and colleagues noted elevated IgE levels and severe eczema in the unmodulated (UNM) mouse strain harboring an ENU mutagenesis-induced, homozygous Card11 mutation in the CC domain (p.Leu298Gln), raising the possibility that the patients’ CARD11 mutations might lead to hypomorphic activity. However, UNM +/− mice had no atopic phenotype, and the heterozygous parental carriers of patients with CARD11-associated SCID were reported as clinically asymptomatic14,15 and did not have a history of atopy (A. Kuzolik & P. Stepensky, personal communication), suggesting that haploinsufficiency was not a likely explanation for the patients’ presentation (Figure 1C).
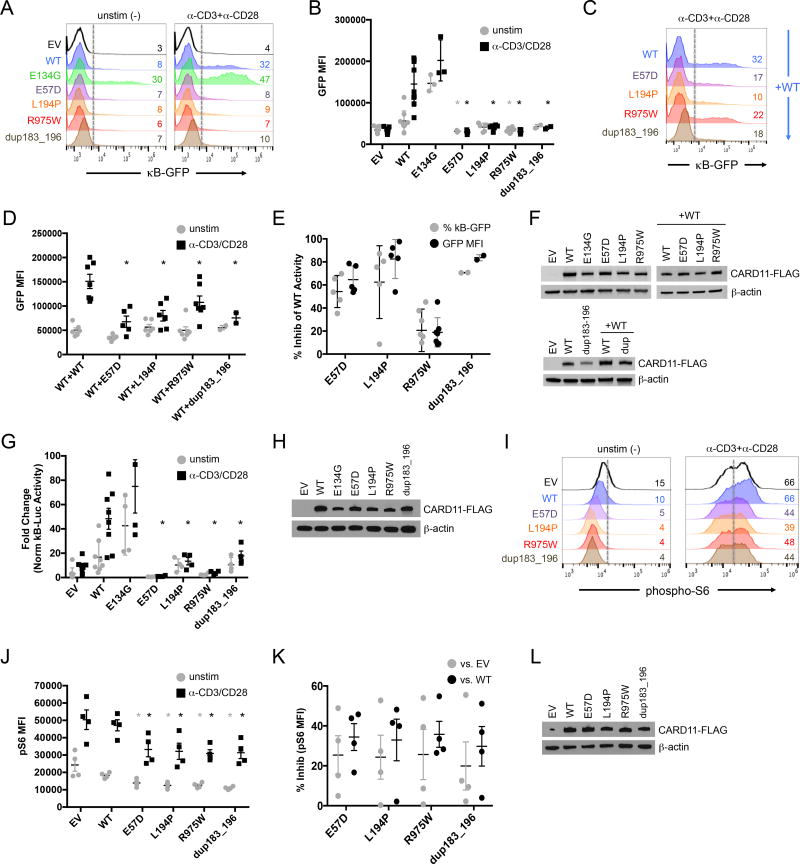
Two principal signaling pathways in which CARD11 participates are NF-κB 21–23 and, as identified more recently, mTORC1 24,29,25. We first assessed the effect of the four CARD11 mutations on NF-κB activation by transfecting WT and mutant CARD11 expression constructs into a CARD11-deficient Jurkat T cell line (JPM50.6) expressing a NF-κB-driven GFP reporter. As shown previously, the GOF mutant E134G, described in a family with BENTA disease, induced constitutive GFP expression in the absence of antigen receptor (AgR) ligation (Figure 2A) 18,19. When compared to WT, all four CARD11 mutants induced little NF-κB activation upon CD3/CD28 stimulation compared to WT, confirming loss of function (Figure 2A–B, Supplementary Fig. 1A). Furthermore, co-transfection of each mutant construct disrupted the ability of WT CARD11 to activate NF-κB (Figure 2C–E, Supplementary Fig. 1B), suggesting dominant interference; the R975W mutant was least disruptive. Importantly, disruption of WT CARD11-induced NF-κB activation was not observed in co-transfections with empty vector (EV), E134G, or Q945X, a homozygous, functionally “null” nonsense mutant described in an autosomal recessive SCID patient born to healthy, heterozygous parents 14 (Supplementary Fig 1C–H). Similar results were obtained upon transfection of normal Jurkat T cells expressing endogenous WT CARD11, as measured by an NF-κB-dependent luciferase assay (Figure 2G–H, Supplementary Fig. 1I). All mutants were comparably expressed in transfected cells (Figure 2F,H,L).
Figure 2. Atopy-associated CARD11 mutations are hypomorphic and dominantly interfere with WT CARD11 signaling to NF-κB and mTORC1.
(A) Flow cytometric histograms measuring NF-κB-driven GFP reporter expression in CARD11-deficient JPM50.6 cells transfected with empty vector (EV), WT, or mutant CARD11 constructs, ± anti-CD3/CD28 stimulation for 24 hr. (B) Quantification of NF-κB-driven GFP reporter expression in transfected JPM50.6 cells (mean fluorescence intensity (MFI) of GFP+ cells). Data are mean ± SEM over 8 separate experiments (EV/WT n=8; E134G n=3); E57D/L194P/R975W n=5; dup183_196=2). Asterisks denote statistically significant differences (Student’s t-test) for each LOF mutant versus WT ± stimulation (E57D p=0.008, 8.5×10−5 (±stim); L194P p=0.065, 1.7×10−4; R975W p=0.013, 8.8×10−5; dup183_196 p=0.08, 1.6×10−4). (C) Flow cytometric histograms for JPM50.6 cells transfected with WT CARD11 plus WT or mutant constructs and stimulated as in (A). (D) GFP MFI (mean ± SEM for 5 separate experiments) for JPM50.6 cells transfected in (C); asterisks denote significance versus stimulated WT+WT (E57D p=6.1×10−4; L194P p=1.4×10−3; R975W p=0.024; dup183_196 p=4.1×10−3). (E) Percent inhibition of WT CARD11 activity (mean ± SD) in (D) calculated for each mutant, based on the change in %GFP+ cells (black) or GFP MFI (gray) ± stimulation. (F) Cropped immunoblot for CARD11-FLAG expression in transfected JPM50.6 lysates (A,C); β-actin serves as a loading control. Data representative of 3 independent experiments. (G) NF-κB-driven luciferase activity in WT Jurkat cells transfected with CARD11 constructs plus luciferase reporter plasmids. Data represent fold change (mean ± SD) in B-driven luciferase activity ± 24 hr stimulation, normalized to Renilla luciferase activity over 8 separate experiments (EV/WT n=8; E134G/E57D/L194P/R975W/dup183_196 n=4). Asterisks denote significance for each LOF mutant versus WT in stimulated cells (E57D p=4.7×10−4; L194P p=2.3×10−3; R975W p=6.7×10−5; dup183_196 p=5.1×10−3). (H) Cropped immunoblot for CARD11-FLAG expression in transfected Jurkat lysates (G). Data representative of 3 independent experiments. (I) Flow cytometric histograms measuring phospho-S6 in Jurkat cells transfected as above ± anti-CD3/CD28 stimulation for 20 min. (J) Quantification of phospho-S6 (MFI) in transfected Jurkat cells. Data are mean ± SEM for 4 separate experiments. Asterisks denote significance for each LOF mutant versus EV or WT ± stimulation (see p values in Methods). (K) Percent inhibition of pS6 signal (mean ± SEM) for each mutant versus EV (gray) or WT (black) transfected cells in (B), based on the change in pS6 MFI ± stimulation. (L) Cropped immunoblot for CARD11-FLAG expression in transfected Jurkat lysates (B). Data representative of 2 independent experiments.
Next, we examined mTORC1 activity via measurement of the phosphorylation of ribosomal protein S6 29, and found that all 4 hypomorphic CARD11 mutants resulted in significantly decreased mTORC1 activity at baseline (unstimulated) and upon CD3/CD28 stimulation in Jurkat T cells (Figure 2I–J, Supplementary Fig. 1J). Similar attenuation of phospho-S6 signaling was noted for each mutant relative to both ectopic WT CARD11 and empty vector (EV) transfected cells, suggesting interference with endogenous WT CARD11 (Figure 2K, Supplementary Fig. 1J–K). Q945X expression had little effect (Supplementary Fig. 1L). Taken together, these findings show that each CARD11 mutant was hypomorphic and reduced WT CARD11 activity in a heterozygous context, as illustrated by decreased downstream activation of both NF-κB and mTORC1.
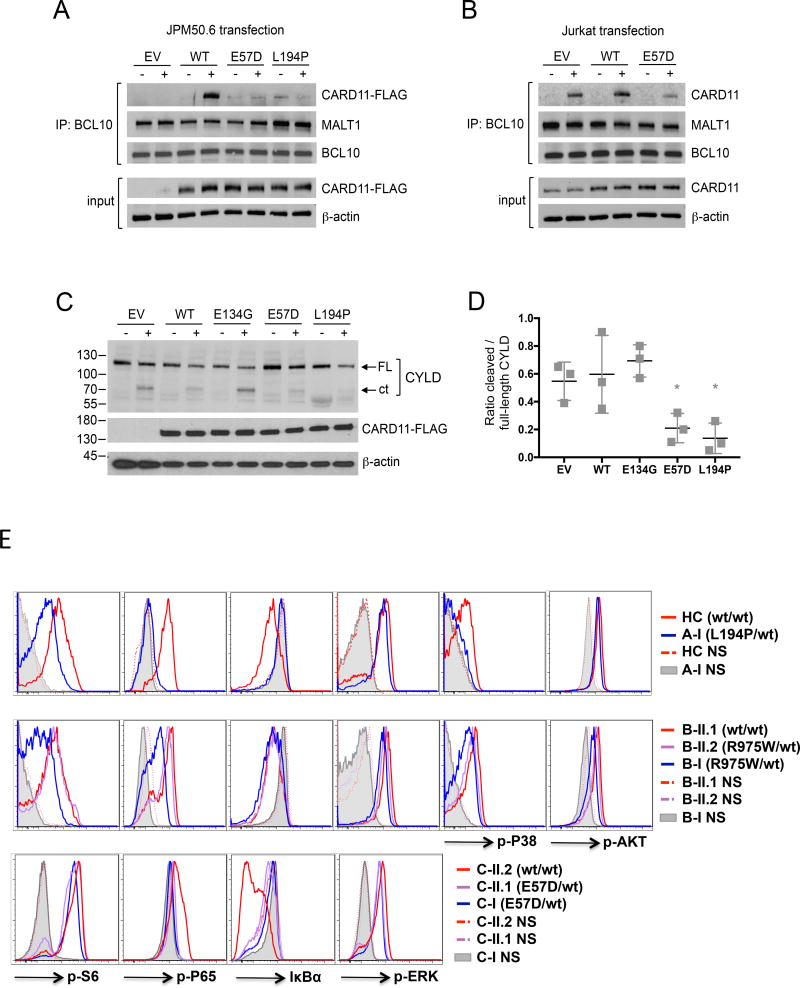
Defective NF-κB activation likely results from impaired CBM complex assembly, as we noted significantly less association of CARD11 LOF mutants with BCL10 and MALT1 in JPM50.6 co-immunoprecipitations (IPs) following PMA/ionomycin stimulation (Figure 3A). In fact, we recovered less CARD11 in BCL10 co-IPs from WT Jurkat cells transfected with CARD11 E57D, suggesting assembly of the endogenous CBM complex was disrupted (Figure 3B). Consistent with dominant interference, MALT1 protease activity was also diminished in Jurkat T cells in the presence of CARD11 LOF mutants, as measured by cleavage of the MALT1 substrate CYLD (Figure 3C–D).26
Figure 3. Impaired CBM complex formation leads to defective signaling in CARD11mut patient T cells.
(A) BCL10 immunoprecipitates from JPM50.6 cells transfected with EV, WT, E57D, and L194P CARD11 mutants ± 15 min PMA/ionomycin stimulation were immunoblotted to detect association of CARD11-FLAG, MALT1, and BCL10. Input lysates (bottom) were immunoblotted with FLAG and β-actin to confirm equivalent CARD11-FLAG expression. Data representative of 3 independent experiments. (B) BCL10 immunoprecipitates from WT Jurkat cells transfected with EV, WT, or E57D CARD11 mutants −/+ 15 min PMA/ionomycin stimulation were immunoblotted as above. Data representative of 2 independent experiments. (C) Lysates from transfected Jurkat cells −/+ 2 hr PMA/ionomycin stimulation were immunoblotted to detect MALT1-dependent cleavage of CYLD and CARD11-FLAG expression. FL=full-length, ct = C-terminal fragment. (D) Spot densitometric quantitation of the ratio of cleaved (ct) to full length CYLD in immunoblots represented in (C); data are mean ± SD of 3 independent experiments. Asterisks denote significance versus stimulated EV (E57D p=0.028; L194P p=0.016. (E) Flow cytometric assay of CARD11mut patient CD4+ T cells compared with unaffected family members (wt/wt) vs. healthy controls (HC) stimulated with PMA for 20 min (2 ng/mL for p-S6 and p-AKT, 5 ng/mL for p-P65, IκBα, p-P38 and p-ERK on Family A and B; 10 ng/mL on Family C, NS: no stimulation). Data representative of three independent experiments for A-I, two for Family B, and one for Family C.
Primary T-cells from CARD11mut patients display decreased activation responses, Th1/Th17 cytokine production and proliferation
We next investigated CARD11-dependent signaling in primary patient T cells. PMA stimulation of patient lymphocytes revealed marked impairment of NF-κB activation, based on reduced p65 phosphorylation and IκB degradation in T cells from families A, B and C (Figure 3E). Similarly, mTORC1 activity (indicated by p-S6) was also attenuated in stimulated patient cells (Figure 3E, Supplementary Fig. 2), while S473 AKT phosphorylation (a readout of mTORC2) was normal (Figure 3E). Reduced p38 activity was also noted in some patient cells (Figure 3E), whereas robust ERK activity was comparable in all patients and controls, consistent with normal ERK activity noted in the UNM mice12. Patient B-I, who harbors the R975W mutation which had the least NF-κB inhibitory activity, and who had the fewest comorbidities beyond atopy within this cohort, had recently been treated with corticosteroids and methotrexate when blood was drawn, potentially complicating the assessment of T cell phenotype and function. Of note, milder NF-κB and mTORC1 signaling defects could also be detected in CD19+ B cells from certain patients (Supplementary Fig. 3A). Overall B-cell development was generally unaffected in these patients (Supplementary Fig. 3B–C).
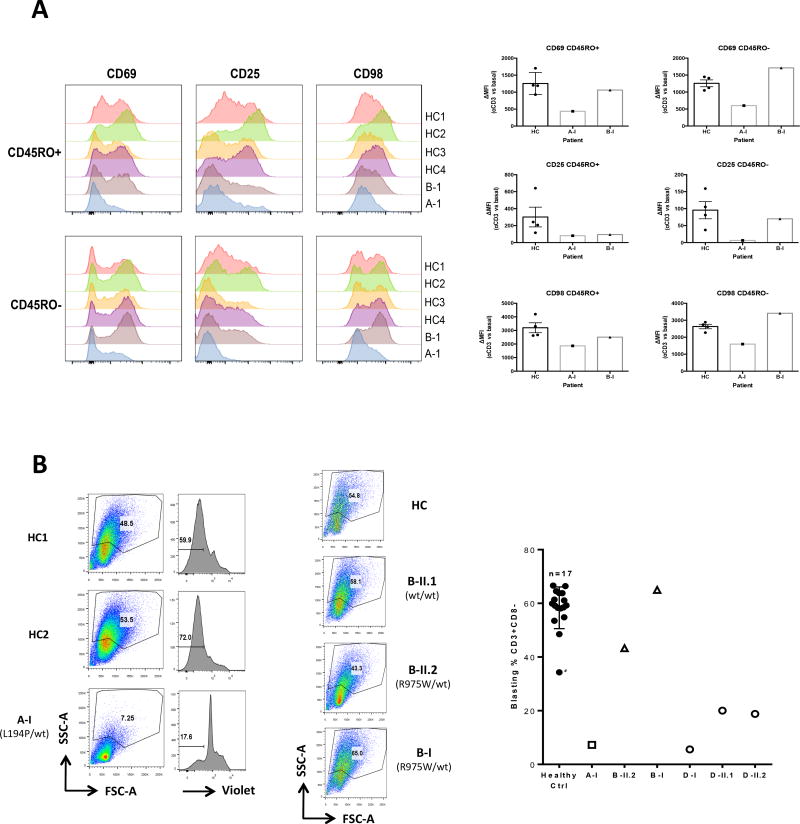
We also observed poor upregulation of key cell surface markers such as CD69, CD25 and CD98 after overnight anti-CD3/CD28 stimulation, particularly for Patient A-I (Figure 4A). Both PBMC (Figure 4B, left panel) and sorted naïve T-cells (Supplementary Fig. 4A) from patient A-I showed decreased short-term proliferation upon anti-CD3/CD28 stimulation. PBMC from patient B-II.2 also showed mildly decreased blastogenesis upon stimulation (Figure 4B, middle panel), and patients D-I, D-II.1 and D-II.2 showed a moderate defect (Figure 4B, right panel). These data indicated that the activation and proliferation of CARD11mut patient T cells were variably impaired, consistent with defective activation of NF-κB and mTORC1.
Figure 4. Decreased surface expression of activation markers and defective proliferation following TCR stimulation in CARD11mut patients.
(A) Flow cytometric measurement of expression of the surface activation markers CD69, CD25, and CD98 in healthy control (HC) vs. A-I and B-I patient naïve (CD45RO) and activated/memory (CD45RO+) CD4 cells after anti-CD3 stimulation for 16 hours. Graphs shown at right indicate ΔMFI for each marker between stimulated vs. untreated samples (HC, n=4; mean±SEM; patients, n=1). (B) Blastogenesis and/or proliferation of PBMCs from representative healthy control (HC) vs. CARD11 patients after anti-CD3/CD28 activation for 5 days. CellTrace Violet (Violet) staining was used for tracking the number of the cell divisions. Quantitation of blasting CD3+CD8− cells for HC (mean ± SEM, n=17), A-I (n=2), B-I, D-I, D-II.1, D-II.2 (n=1 for each) is graphed at right. Asterisk denotes travel control included with family D.
Further flow cytometric phenotyping of patient PBMC showed variability in CD45RA− memory T cells and circulating CXCR5+ PD-1hi T follicular helper (cTFH) cells in families A–C (Figure 5A, Supplementary Table 1). Direct ex vivo (Figure 5B, Supplementary Fig. 4B) and 5 day culture (Supplementary Fig. 4C) of patient T cells showed impaired Th1, normal Th17 and elevated Th2 cytokine production within the CD45RO+ compartment, mirroring the phenotype of both UNM mice and, with the exception of the IL-17 production, both mTORC1 and ASCT2 KO mice 27–30. Numbers of CD4+ FOXP3+ regulatory T cells, whose diminished frequency is thought to contribute to the UNM mouse phenotype 31, were normal in all patients (Figure 5C, Supplementary Table 1). The percentage of dividing Tregs (Ki-67+) was also comparable to controls (Figure 5C), as was their frequency and suppressive function (Supplementary Fig. 5). However, aberrant co-expression of GATA3 in patient A-I and B-I Tregs suggests impaired GATA3 regulation (Figure 5D)—a finding associated with atopy in a variety of settings32,33. Collectively these results indicate that patient T cells harboring CARD11 LOF/DN mutations are hyporesponsive and skew toward a Th2 phenotype, consistent with their atopic predisposition.
Figure 5. Impaired IFN-γ, augmented Th2 cytokine productions and Treg phenotype in CARD11mut patients.
(A) CXCR5+ PD-1hi peripheral cTFH cells in CARD11 mutated patients. (B) Top: Flow cytometry for intracellular cytokine staining of PBMCs after PMA/ionomycin stimulation. Bottom: Quantitation of IFN-γ+, IL-4+, and IL-13+ CD4+ CD45RO+ T cells. (HC, n=7, mean ± SEM; patients, n=1). (C) Quantitation of FOXP3+CD25+CD127lo Tregs (left), including Ki-67+ Tregs (right), in HC vs. patients. (D) Measurement of GATA3 expression in Tregs among CD45RO+ CD127low CD4 T cells for family members possessing CARD11 mutations vs. healthy controls (HC). (E) Immunoblot showing impaired expression of ASCT2 in purified naïve CD4 T-cells from patient A-I relative to HC (n=3) after 48 hours with plate-bound anti-CD3 (N: non-stimulated).
CARD11 mutations result in poor upregulation of glutamine transporter ASCT2; mTORC1 activation cellular proliferation, and interferon gamma production improves with glutamine supplementation
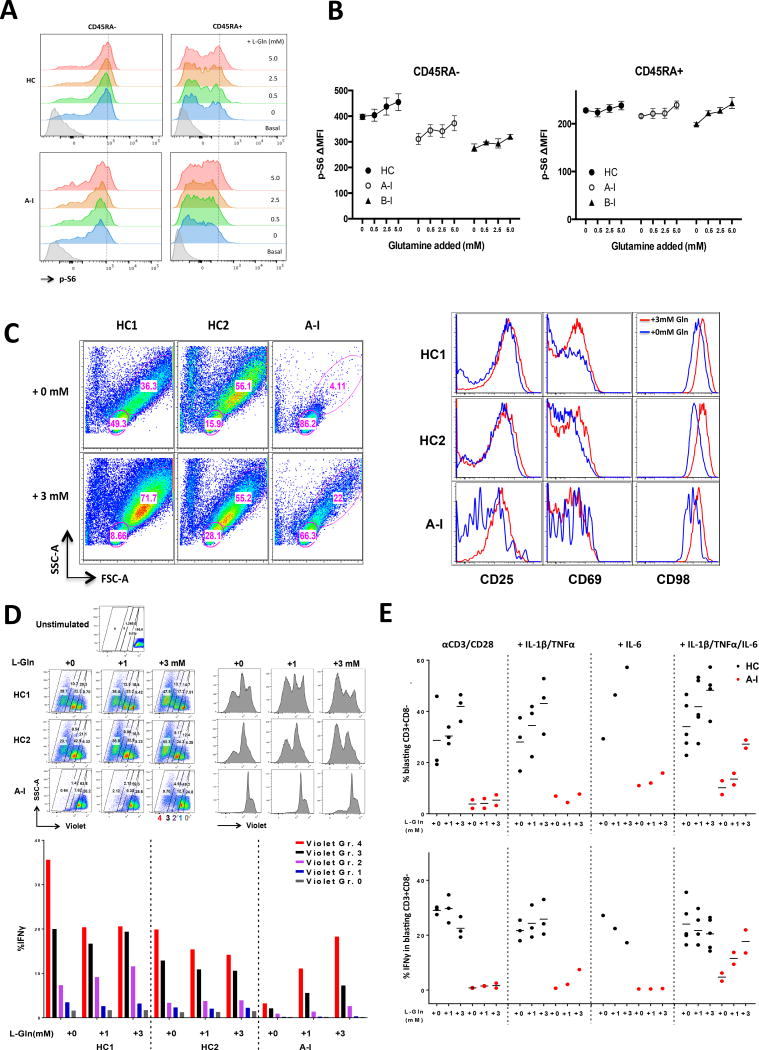
CARD11 was recently shown to be critical for mTORC1 activation25, at least in part by facilitating TCR-induced upregulation and/or activation of ASCT2, an essential glutamine transporter required for extracellular glutamine import during cellular activation24. Similar to CARD11-deficient murine T cells, we observed reduced ASCT2 upregulation following TCR activation of CARD11mut patient T cells (Figure 5E, Supplementary Data). Mice lacking ASCT2 also display mTORC1 activation defects, impaired Th1 differentiation and a Th2-skewed phenotype30. However, the addition of exogenous glutamine could rescue mTORC1 signaling and reverse Th1 defects24. Supplementing culture medium with exogenous glutamine boosted mTORC1 activation (phospho-S6) in Patient A-I T cells, particularly in CD45RA+ naïve T cells (Figure 6A–B), without affecting NF-κB or ERK phosphorylation (data not shown). We then cultured naïve patient A-I T-cells in the presence of supplemented glutamine (3mM) under a variety of stimulation conditions. We found that the combined addition of cytokines capable of activating NF-κB (IL-1β and TNF) and STAT3 (IL-6) partially rescued defects in proliferation and CD98 induction, but only with exogenous glutamine supplementation (Figure 6C–D, Supplementary Fig. 6). While these conditions are not ideal for measuring biases for cell intrinsic Th differentiation, we did find that divided cells were more likely to produce IFNγ upon glutamine supplementation (Figure 6E). Together these results suggest that the poor T cell responses in these patients might be restored in part through simple glutamine supplementation.
Figure 6. Glutamine supplementation in combination with cytokines can partially restore the TCR-induced proliferation and IFN-γ defects in a CARD11mut patient.
(A) Improved phospho-S6 activation by PMA after addition of glutamine. PBMC from the HC and CARD11 patient were rested in media then PBS supplemented with glutamine (L-Gln) and stimulated with PMA. Phospho-S6 was measured by intracellular flow cytometric staining of CD4 CD45RA- memory and CD45RA+ naïve T-cells. The dashed line indicates the maximum phospho-S6 activation peak (MFI) by PMA with 5mM glutamine addition in HC. (B) ΔMFI plot (PMA treated vs. untreated) for phospho-S6 in (A) (HC, n=5; three independent experiments for A-I and two for B-I; mean±SEM). (C) Left: Increased proliferation of CD4+ naive T cells isolated from patient A-I patient in serum-free medium stimulated with anti-CD3/CD28 and cytokines (IL-1β, TNFα, and IL-6) for 5 days with glutamine supplementation. Right: Increased surface activation marker expression with glutamine addition plus cytokines in patient A-I (gated on FSChiSSChi blasts from the left panel). Data are representative of three experiments. (D) Top: Impaired naive CD4+ T cell proliferation was partially restored by anti-CD3/CD28 stimulation with excess glutamine and cytokines IL-1β, TNFα, and IL-6 in patient A-I. Bottom: Rescue of IFN-γ expression was proliferation dependent. Dividing cells were gated into four groups based on CellTrace Violet intensity (top left), and IFN-γ was measured by intracellular staining in each group (bottom). Data are representative of two independent experiments. (E) Quantification of the percentage of proliferative CD3+CD8− blasts (top), and the percentage of IFNγ-producing CD3+CD8− blasts (bottom) with the treatments by increasing glutamine concentration. Data combined of the experiments: αCD3/CD28: HC (n=3), A-I (n=2); +IL1β/TNFα: HC (n=3), A-I (n=1); +IL-6: HC (n=1), A-I (n=1); + IL1β/TNFα/IL-6: HC (n=5), A-I (n=2).
Discussion
Here we show that different heterozygous hypomorphic mutations in CARD11 can lead to severe atopic disease in humans, associated with weaker, Th2-skewed T cell responses. These patients mostly lack the non-immunologic features seen in PGM3, STAT3 and DOCK8 deficiencies, but share features of severe atopic dermatitis often with elevated serum IgE levels and peripheral blood eosinophilia. A similar atopic disease phenotype in UNM mice homozygous for a hypomorphic mutation provides strong evidence for the association of hypomorphic CARD11 activity and atopy12. However, in contrast to the UNM mouse, our results demonstrate that the four unique CARD11 LOF mutations found in our patient cohort can dominantly interfere with WT CARD11 signaling, providing an autosomal dominant inheritance with variable penetrance. In addition to identifying a new cause of an inherited syndrome associated with atopic dermatitis, peripheral blood eosinophilia and elevated IgE, these findings raise the possibility that DN CARD11 mutations could potentially be a common cause of severe allergy in the absence of syndromic features. Heterozygous CARD11 mutations should therefore be considered in genetic testing of families with an autosomal dominant inheritance of a history of significant atopic dermatitis—current or remote-- elevated IgE and eosinophilia, independent of other comorbidities.
The scaffold function of CARD11 in lymphocytes is normally tightly regulated, ensuring CARD11-BCL10-MALT1 (CBM) complex formation and downstream signaling is only triggered after antigen receptor-dependent phosphorylation of the inhibitory linker domain 34–36. Two of the patient mutations were located in the CC domain, a demonstrated hotspot for both GOF and LOF mutations that likely perturb intramolecular regulation via the linker, thereby enhancing or debilitating self-oligomerization of CARD11 required for signalosome assembly, respectively37. Likewise, the E57D mutation found in the CARD domain likely directly disrupts interaction with BCL10, a key initiating event in CBM signaling. The importance of CARD and CC domain protein interactions for nucleating the larger CBM signalosome likely explains the substantial reduction in CBM complex formation and NF-κB activation observed in transfected cells. On the other hand, the unusual MAGUK domain mutation R975W may interfere with higher order, intermolecular “clustering” of activated CARD11 molecules via SH3-GUK interactions38, explaining a weaker dominant negative effect on NF-κB stimulation (Figure 2E). Indeed, a lesser NF-κB defect associated with this specific GUK-domain mutation may help explain the milder infectious disease history and cellular phenotypes observed in Family B. In contrast, the LOF/DN activity we observed in relation to mTORC1 signaling was comparable for all four CARD11 mutants in transfection studies, suggesting more discrete roles for the NF-κB and mTORC1 pathways in preventing infection and atopy, respectively.
It remains unclear which CARD11 domains might be required for direct interaction and/or transcriptional regulation of ASCT2, the glutamine transporter connecting AgR engagement with mTORC1 activation 24,25. Attenuated p38 signaling in some CARD11mut patients was also surprising, as CARD11 has not been specifically implicated in driving this MAPK pathway. Given that the MAGUK protein DLGH1 governs the TCR-driven “alternative p38 pathway”39, perhaps these CARD11 mutants interfere with related MAGUK family scaffold proteins in as yet undefined ways.
Problems with T cell proliferation and balanced Th differentiation in the patients likely result from defects in multiple TCR-driven signaling pathways that rely on CARD11. Disruption of TCR signal strength, which can occur an a variety of settings and TCR signaling pathways, has been associated with Th2 bias40 and this phenomenon likely contributes to the pathogenesis of these patients . Numerous studies have shown that disruption of either NF-κB or mTORC1 signaling blocks proper T cell expansion, likely by compromising both the IL-2 signaling loop and metabolic reprogramming required for T cell proliferation. For these reasons, it was difficult to definitively characterize the effect of these CARD11 LOF mutations on Th differentiation, since typical unbiased in vitro conditions did not support T cell proliferation. However, recent work indicates that mTOR activity profoundly affects Th differentiation41. The attenuation of CARD11-dependent mTORC1 activation ostensibly contributes to impaired Th1 differentiation in our patients, allowing an mTORC2-dependent Th2 response to dominate. NF-κB signal blockade in mouse T cells was also shown to preferentially disrupt the Th1 response and permit Th2-driven allergic inflammation in vivo42. Whether reduced (but not absent) IFN-γ production explains the atopic and skin infection phenotype entirely—as hinted to by patients with hypomorphic IFNGR mutations—is not clear. An intrinsic Th2 bias alone that could drive the atopic phenotype and predispose to skin infection43.
Recent work suggests CARD11-dependent NF-κB stimulation is particularly important for Th17 differentiation of murine T cells in vitro under specific polarizing conditions44. In vivo, CARD11-deficient mice fail to mount a Th2 response capable of driving allergic inflammation45, whereas UNM mice show decreased frequency of FOXP3+ Tregs and a gradual accumulation of Th2 cells, resulting in allergic disease31. In contrast, we found the patients possess normal numbers of Th17 cells and Tregs, although the patients Tregs appear “Th2-like” based on aberrant expression of GATA3, a recognized hallmark of atopic disease susceptibility32. Further study is required to confirm a causal role of this observation. These disparate findings underscore the importance of identifying and characterizing “CARD11-opathies” in humans versus mice. Indeed, BENTA patients harboring GOF CARD11 mutations show no signs of allergic disease, even though adoptively transferred T cells expressing constitutively active CARD11 can drive allergic disease in mice33,46.
Abnormal CARD11 signaling likely has intrinsic consequences for B cell fate as well. In contrast to the selective expansion of naïve B cells seen in BENTA, we noted reduced numbers of CD19+ B cells in some CARD11 LOF patients. However, both GOF and LOF CARD11 mutations can reduce circulating memory B cells and can affect immunoglobulin production in humans, suggesting CARD11 is a critical gatekeeper for tuning proper BCR signals required for normal B cell maturation and differentiation47. Of interest here, although IgE signaling normally forces murine B cells to terminally differentiate into short-lived plasmablasts prone to apoptosis, loss of key BCR signaling molecules facilitates the survival and subsequent development of IgE+ memory and long-lived plasma cells48. Our findings imply that attenuated CARD11 signaling may mirror this phenomenon in humans, permitting an atypical accumulation of longer-lived IgE-secreting plasma cells that promote allergic disease, particularly in childhood.
Additional work is required to identify the key pathogenic events related to the atopic phenotype and infection. Nevertheless, it may be that disease severity generally lessens over time, consistent with the phenotype in aged UNM mice (C. Goodnow, personal communication). Importantly, our findings suggest that exogenous glutamine could partially correct specific defects in CARD11mut T cell responses, including proliferation and IFN-γ secretion. Glutamine supplementation has been explored as a treatment for reducing allergic disease in low birth weight infants, with a promising decrease in atopic dermatitis reported49. This simple therapeutic intervention may ameliorate disease in atopic patients harboring functional mutations in CARD11 or related genes by restoring normal glutamine uptake and mTORC1 signaling in activated T cells.
Materials and Methods
Patients
Informed consent was obtained and the study was approved by the Institutional Review Boards of National Jewish Health and Yale University. Patients evaluated at NIH were enrolled on clinicaltrials.gov identifier NCT00557895. Informed consent was obtained from all participating patients.
Whole Exome Sequencing (WES)
WES was performed using the Ion Torrent AmpliSeq RDY Exome Kit (Life Technologies) and the Ion Chef and Proton instruments (Life Technologies). Briefly, 100 ng of gDNA was used as the starting material for the AmpliSeq RDY Exome amplification step following the manufacturer’s protocol. Library templates were clonally amplified and enriched using the Ion Chef and the Ion PI Hi-Q Chef Kit (Chef package version IC.4.4.2, Life Technologies), following the manufacturer’s protocol. Enriched, templated Ion Sphere Particles were sequenced on the Ion Proton sequencer using the Ion PI chip v3 (Life Technologies).
Bioinformatics Analysis
Read mapping and variant calling were performed using the Ion Torrent Suite software v4.4.2. In short, sequencing reads were mapped against the UCSC hg19 reference genome using the Torrent Mapping Alignment Program (TMAP) map4 algorithm. SNPs and INDELS were called by the Torrent Variant Caller plugin (v.4.414-1) using the ‘Generic-Proton-Germ Line – Low Stringency’ configuration. Only reads that were unambiguously mapped were used for variant calling. Variants were annotated using ANNOVAR (http://annovar.openbioinformatics.org/). Data mining, biological interpretation, and candidate gene discovery were performed using various online tools including The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov), and GeneCards (http://www.genecards.org). Target coverage was evaluated using the Torrent Coverage Analysis plugin (v.4.414-1), and the output was further evaluated using in-house, custom Perl scripts.
Sanger Sequencing
CARD11 Sanger sequencing was performed to confirm WES-detected variants and to screen family members. gDNA was PCR-amplified using GoTaq polymerase (Promega) and exon specific primers. Amplicons were bi-directly sequenced using the Big Dye Terminator version 1.1 cycle sequencing kit and an Applied Biosystems 3130xl Genetic Analyzer (Life Technologies).
Ex-vivo Treg and cytokine analysis
Patients’ and healthy control (HC) PBMCs were purified by Ficoll density gradient centrifugation, washed with RPMI 1640 plus penicillin, streptomycin, and L-glutamine along with 10% FBS (R10) and filtered through a 40 µM strainer. Cells were resuspended to a concentration of 1 × 106 cells/mL and aliquoted (1 mL/well) into 48-well plates. PMA (final concentration 20 ng/mL), ionomycin (1 µM) and brefeldin A (5 µg/mL, Sigma-Aldrich) were added prior to incubation at 37°C for 5–6 hours. After incubation, cells were washed with FACS buffer and stained with LIVE/DEAD Blue (Life Technologies). Intracellular staining was performed using BD CytoFix/CytoPerm (BD Biosciences) reagents according to the manufacturer’s instructions using the following antibodies: CD3 (SK7, BD Biosciences), CD4 (RPA-T4, BD Biosciences), CD8 (SK1, BD Biosciences), CD45RO (UCHL1, Beckman Coulter), CXCR5 (RF8B2, BD Biosciences), CD25 (M-A251, BD Biosciences), IL-2 (MQ1–17H12, BioLegend), IL-4 (8D4–8, BD Biosciences), IL-5 (JES1–39D10, BD Biosciences), IL-13 (JES10-5A2, BD Biosciences), IFNγ (B27, BD Biosciences), IL-17A (eBio64DEC17, eBioscience). Cells were gated on viable CD3+ CD4+ CD45RO+ cells. Ex-vivo Treg staining was performed using anti-CD3 (UCHT1), CD4 (OKT4), CD25 (M-A251),, CD45RO (UCHL1), CD127 (HIL-7R–M21),, GATA3 (L50–823) and FOXP3 (236A/E7) antibodies as described 33.
Intracellular phospho-flow cytometric analysis
PBMCs were isolated by density gradient centrifugation and resuspended in R10. PBMCs were rested in RPMI without glutamine and FBS (R0) for two hours. In some cases, PBMC are incubated in R10 for 1–2 days, passed through a 40µµ strainer and adjusted to 0.5 × 106/mL in PBS and rested prior to PMA stimulation. For glutamine supplementation experiments, PBMCs were rested in PBS containing 0.2mM glutamine and increasing concentrations of glutamine (0–5 mM) were added before PMA stimulation. PMA (1–20ng/mL) stimulation was performed in 1 mL PBS for 5–30 minutes in 37°C CO2 incubator. After stimulation, cells were fixed in 1.6% paraformaldehyde for 10 minutes at room temperature, spun down and resuspended in 100% cold methanol and incubated overnight at −20°C. Cells were then washed and stained with fluorochrome-conjugated phospho-antibodies: phospho-S6 (N7–548, BD Biosciences), phospho-ERK1/2 (20A, BD Biosciences), phospho-P65 (K10–895.12.50, BD Biosciences), phospho-P38 (36/p38, BD Biosciences), phospho-AKT (pS473, M89-61, BD Biosciences), IκBα (L35A5, Cell Signaling Technology), along with CD3 (HIT3a, BioLegend), CD4 (L200, BD Biosciences), CD45RO (UCHL1, BD Biosciences), CD45RA (HI100, BD Biosciences) antibodies.
Flow cytometric analysis of activation markers
Cells were stained after stimulation with LIVE/DEAD Blue (Life Technologies) and antibodies against CD69 (FN50, BD Biosciences), CD25 (M-A251, BD Biosciences), CD98 (UM7F8, BD Biosciences),, CD45RO (UCHL1, Beckman Coulter), CD3 (HIT3a, Biolegend) and CD4 (L200, BD Biosciences).
ProcartaPlex ELISA analysis
ELISA analysis for cytokine productions of IFNγ, IL-4, IL-13 and IL-5 was performed using the ProcartaPlex ELISA kits (eBioScience, Thermo-Fisher) and analyzed by Bio-Plex 200 systems (Bio-Rad). PBMCs were cultured in RPMI plus 10% FBS with 20 ng/mL PMA and 1mM ionomycin for 5–6 hours in 106 cells/mL. The media were harvested, diluted and analyzed.
Thelper activation and differentiation
Human PBMCs and naïve CD4+ T cells were isolated from PBMCs using negative selection MACS microbeads (Miltenyi Biotec) as per manufacturer’s protocol. Cells were cultured in X-VIVO-15 medium (Lonza), in the presence of plate-bound antibody to CD3 (1 µg/ml, OKT3, eBiosience) and soluble antibody to CD28 (0.5 µg/ml, L293, BD Biosciences) for 5 days, with recombinant human IL-6 (20 ng/ml, Peprotech), IL-1β (10 ng/ml, Peprotech) , Tumor necrosis factor α (TNF-α, 10ng/ml, R&D), and L-Glutamine (Gln) as indicated. On day 5, the cells were stimulated with 20 ng/mL PMA and 1 µM ionomycin for 5 hours. Brefeldin A (10 µg/mL) was added after 1 hour. Cells were stained with LIVE/DEAD Blue (Life Technologies), then fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) as per manufacturer’s protocol, and stained with antibodies against CD3 (UCHT1 or SP34-2, BD Biosciences), CD4 (RPA-T4, BD Biosciences), CD8 (SK1, BD Biosciences), IFNγ (B27, BD Biosciences), IL-4 (8D4–8, BD Biosciences), and CD45RO (UCHL1, Beckman Coulter) antibodies.
Treg suppression assay
CD4+ T cells were isolated by negative selection using magnetic beads (Miltenyi Biotec). For isolation of CD4+CD25− effector T (Teff) cells, CD25+ cells were depleted using anti-CD25 beads (Miltenyi Biotec). Treg cells were isolated by sorting for CD4+CD25+CD127− cells. Teff cells were labeled with CellTrace Violet or CFSE (Life Technologies) and stimulated with anti-CD3, CD28, and CD2 coated beads (Treg suppression inspector human, Miltenyi Biotec), and autologous or allogeneic Treg cells were added at given ratios; cell divisions were evaluated by flow cytometry.
Plasmid DNA cloning
The human pUNO-CARD11 plasmid (Invivogen) was modified to include a 3’ 3X FLAG tag using annealed, overlapping oligonucleotides encoding the tag and inserted via BamHI (5’) and NheI (3’) overhangs. Single point mutations were introduced into the WT CARD11 construct by site-directed mutagenesis, using specific primers for linear amplification using 2x Pwo DNA polymerase (Roche) and subsequent digestion with DpnI to destroy methylated template DNA (ThermoFisher Scientific). A 42 bp fragment encoding aa183–196 duplication was generated by overlap extension PCR and ligated into a BsrG1 site in pUNO-CARD11 using a rapid dephosphorylation/ligation kit (Roche). Mutations were confirmed by Sanger sequencing. All plasmids were purified using a GenElute HP Plasmid Maxi-Prep Kit (Sigma) from transformed competent DH5α E. coli (New England Biolabs) selected with blastidicin (InvivoGen).
Cell transfection assays
The wild-type Jurkat T cell line (clone E6.1) was obtained from ATCC (TIB-152). CARD11-deficient Jurkat cells (JPM50.6) were originally provided by Dr. Xin Lin (MD Anderson Cancer Center); for authentication, absence of CARD11 expression was periodically monitored by immunoblotting. Neither cell line tested positive for mycoplasma contamination. Cell lines were cultured in complete RPMI 1640 (Lonza) supplemented with 10% fetal calf serum (Sigma), 2 mM glutamine, and 100U/ml each of penicillin and streptomycin (Life Technologies). JPM50.6 cells (4–5 ×106/cuvette) were electroporated with 5 µg plasmid DNA in 0.4 mL RPMI plus 10% FBS (no antibiotics) using a BTX Electroporator (BTX Harvard Apparatus: 260 V, 950 µF). For luciferase assays, Jurkat cells were simultaneously transfected with 5 µg NF-κB-dependent firefly luciferase reporter plasmid (pNF-κB-Luc), and 0.25 µg of Renilla luciferase plasmid (pRL-TK) for transfection normalization. A portion of these cells were stimulated 24 hrs post-transfection with 1 µg/ml anti-CD3 + anti-CD28 (BD Biosciences). NF-κB activity was measured 24 hrs later via detection of κB-GFP reporter expression in JPM50.6 cells using an Accuri C6 flow cytometer (BD), or by dual luciferase assay in Jurkat T cells (Promega) using a Tecan Infinite M200 Mulimode Microplate Reader. Relative NF-κB activation in Jurkat transfectants was calculated by normalizing the relative ratio of firefly to Renilla luciferase signals. Phospho-S6 was measured 24 hrs post stimulation via intracellular flow cytometry as previously described (26), using an AlexaFluor488-conjugated anti-phospho-S6 Ab (2F9, Cell Signaling Technology) and an Accuri C6 cytometer.
Immunoprecipitations and immunoblotting
Transfected cell lysates were prepared in 1% NP-40 lysis buffer as previously described (17). To verify CARD11 expression, lysates (5–20 µg) were separated on 4–20% Tris-Glycine SDS gels (Bio-Rad), transferred to nitrocellulose (TransBlot Turbo, Bio-Rad). Blocked membranes were immunoblotted using the following Abs: anti-FLAG (M2) and anti-β-actin (AC-15, Sigma); anti-CARD11 (1D12, Cell Signaling Technology); anti-BCL10 (A-6), anti-MALT1, (H-300), anti-CYLD (E-10, Santa Cruz). Abs were detected using HRP-conjugated secondary Abs (Southern Biotech) and Pierce ECL (Thermo Scientific). To assay MALT1-dependent cleavage of CYLD, transfected cells were stimulated with 50 ng/mL PMA and 1 µM ionomycin for 2 hours prior to lysis. For CBM complex IPs, cells were stimulated for 15 min with PMA/ionomycin as above, lysed in 0.1% Triton X-100 buffer (20 mM Tris pH 8, 150 mM NaCl, 5 mM NaF, 5 mM sodium glycerophosphate, 1 mM DTT + Protease Complete/PhosStop inhibitors (Roche)), and incubated overnight with 2 µg anti-BCL10 (A-6) at 4°C. Protein G Sepharose beads (20 uL, Sigma) were then added and rocked for 2 hrs at 4°C. Beads were washed 3x in lysis buffer and boiled prior to SDS-PAGE and immunoblotting.
Primary human naïve CD4+ T cells were isolated from PBMCs using negative selection MACS microbeads (Miltenyi Biotec) as per manufacturer’s protocol. Naïve cells were stimulated in coated anti-CD3 (OKT3, eBioscience) plate for 48 hours in RPMI plus 10% FBS prior to lysis and SDS-PAGE. Anti-ASCT2 antibody (D7C12, Cell Signaling Technology) was used for detection.
Full-length immunoblots are shown in the Supplementary Data.
Statistics
Sample size was dictated by the small number of patients identified with CARD11 mutations (8 patients, 4 mutations). For Jurkat cell transfections, paired, one-way Student’s t-tests were utilized to test whether GFP or phospho-S6 signals (% positive and mean fluorescence intensity (MFI)) induced by each putative LOF CARD11 mutant were significantly less than WT. Variance (S.D. for % positive, S.E.M. for MFI) was comparable for each transfected construct. P values are included in each figure legend except Figure 2J (p values vs. EV (unstim): E57D=0.015, L194P=0.015, R975W=0.014, dup183_196=0.02; vs. EV (stim): E57D=0.004, L194P=0.0014, R975W=0.02, dup183_196=0.006; vs. WT (unstim): E57D=0.032, L194P=0.002, R975W=0.005, dup183_196=5.3×10−4; vs. WT (stim): E57D=0.001, L194P=0.013, R975W=0.002, dup183_196=0.009). There is no statistics p-value for figures 4, 5 and 6.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. Public deposition of sequence data is in progress.
Supplementary Material
Acknowledgments
We thank Wanxia Tsai, Massimo Gadina, and Claire Malinverni for technical assistance. We thank the patients and their families for participating in this research. The patients were enrolled on an IRB-approved protocol and were provided informed consent. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, the NIAID Clinical Genomics Program, and grants from the National Institutes of Health (1R21AI109187, 1R01GM105821 to A.L.S. and AI061093 to E. M.), the Henry M. Jackson Foundation (Val Hemming Fellowship to J.R.S.), Telethon (GGP13254 for E.R.), and the Joanne Siegel Fund (to E.W.G.).
Footnotes
Contributions
C.A.M, Y.Z., M.A.W., and S.G. performed experiments with primary patient cells. J.R.S. and B.D. produced all CARD11 mutant constructs; J.R.S., E.R., S.A., K.V. and B.D. conducted cell transfection experiments. J.J.L., C.G.N., T.D., K.D.S., H.M. and J.D.M. were involved in clinical workup of patient A-I. J.S., J.N., S.D.R. performed sequence analysis on patient A-I. J.K.A., P.J.H, and P.R.R., and E.W.G. were involved in clinical care and sequence analysis of family B. Y.Z., B.K., M.A.C., and N.R., S.G., E.M. were involved in clinical workup of family C. A.P., M.O., E.P., A.B., G.D., S.D. were involved in clinical care and workup of family D; J.Z. and M.M. performed sequence analysis of family D.C.M., M.A.W., J.R.S., A.L.S. and J.D.M. co-wrote the manuscript. E.W.G., A.L.S. and J.D.M. supervised the project. All authors discussed the results and contributed to the manuscript.
Conflict of interest disclosure
The authors declare no conflict of interests.
Accession codes
Exome and genome sequencing data have been deposited in the dbGaP under accession ---.
References
- 1.Mogensen TH. Primary Immunodeficiencies with Elevated IgE. Int Rev Immunol. 2016;35:39–56. doi: 10.3109/08830185.2015.1027820. [DOI] [PubMed] [Google Scholar]
- 2.Freeman AF, Olivier KN. Hyper-IgE Syndromes and the Lung. Clin Chest Med. 2016;37:557–67. doi: 10.1016/j.ccm.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan SK, Gelfand EW. Primary Immunodeficiency Masquerading as Allergic Disease. Immunol Allergy Clin North Am. 2015;35:767–78. doi: 10.1016/j.iac.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Bonnelykke K, Sparks R, Waage J, Milner JD. Genetics of allergy and allergic sensitization: common variants, rare mutations. Curr Opin Immunol. 2015;36:115–26. doi: 10.1016/j.coi.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 6.Chatila TA. Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–9. doi: 10.1016/j.molmed.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Gao L, et al. Targeted deep sequencing identifies rare loss-of-function variants in IFNGR1 for risk of atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2015;136:1591–600. doi: 10.1016/j.jaci.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernasconi A, et al. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 2006;118:e1584–92. doi: 10.1542/peds.2005-2882. [DOI] [PubMed] [Google Scholar]
- 9.Datta S, Milner JD. Altered T-cell receptor signaling in the pathogenesis of allergic disease. J Allergy Clin Immunol. 2011;127:351–4. doi: 10.1016/j.jaci.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinnon ML, et al. Combined immunodeficiency associated with homozygous MALT1 mutations. J Allergy Clin Immunol. 2014;133:1458–62. 1462e1–7. doi: 10.1016/j.jaci.2013.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Siggs OM, et al. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity. 2007;27:912–26. doi: 10.1016/j.immuni.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun JE, et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751–62. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 13.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greil J, et al. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J Allergy Clin Immunol. 2013;131:1376–83. e3. doi: 10.1016/j.jaci.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Stepensky P, et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol. 2013;131:477–85. e1. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs S, et al. Omenn syndrome associated with a functional reversion due to a somatic second-site mutation in CARD11 deficiency. Blood. 2015;126:1658–69. doi: 10.1182/blood-2015-03-631374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara H, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–75. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 18.Turvey SE, et al. The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency. J Allergy Clin Immunol. 2014;134:276–84. doi: 10.1016/j.jaci.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snow AL, et al. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J Exp Med. 2012;209:2247–61. doi: 10.1084/jem.20120831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–6. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 21.Gaide O, et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:836–43. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 22.Lin CY, Graca L, Cobbold SP, Waldmann H. Dominant transplantation tolerance impairs CD8+ T cell function but not expansion. Nat Immunol. 2002;3:1208–13. doi: 10.1038/ni853. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21:5184–94. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakaya M, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton KS, et al. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci Signal. 2014;7:ra55. doi: 10.1126/scisignal.2005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staal J, et al. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–52. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurebayashi Y, et al. PI3K–Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1:360–73. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–53. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng H, et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity. 2016;45:540–54. doi: 10.1016/j.immuni.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altin JA, et al. Decreased T-cell receptor signaling through CARD11 differentially compromises forkhead box protein 3-positive regulatory versus T(H)2 effector cells to cause allergy. J Allergy Clin Immunol. 2011;127:1277–85. e5. doi: 10.1016/j.jaci.2010.12.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noval Rivas M, et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42:512–23. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lexmond WS, et al. FOXP3+ Tregs require WASP to restrain Th2-mediated food allergy. J Clin Invest. 2016;126:4030–4044. doi: 10.1172/JCI85129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCully RR, Pomerantz JL. The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol Cell Biol. 2008;28:5668–86. doi: 10.1128/MCB.00418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sommer K, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561–74. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto R, et al. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–85. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Jattani RP, Tritapoe JM, Pomerantz JL. Intramolecular Interactions and Regulation of Cofactor Binding by the Four Repressive Elements in the Caspase Recruitment Domain-containing Protein 11 (CARD11) Inhibitory Domain. J Biol Chem. 2016;291:8338–48. doi: 10.1074/jbc.M116.717322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara H, et al. Clustering of CARMA1 through SH3-GUK domain interactions is required for its activation of NF-kappaB signalling. Nat Commun. 2015;6:5555. doi: 10.1038/ncomms6555. [DOI] [PubMed] [Google Scholar]
- 39.Round JL, et al. Scaffold protein Dlgh1 coordinates alternative p38 kinase activation, directing T cell receptor signals toward NFAT but not NF-kappaB transcription factors. Nat Immunol. 2007;8:154–61. doi: 10.1038/ni1422. [DOI] [PubMed] [Google Scholar]
- 40.Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev. 2013;252:12–23. doi: 10.1111/imr.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–46. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aronica MA, et al. Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J Immunol. 1999;163:5116–24. [PubMed] [Google Scholar]
- 43.Ong PY, Leung DY. Bacterial and Viral Infections in Atopic Dermatitis: a Comprehensive Review. Clin Rev Allergy Immunol. 2016;51:329–337. doi: 10.1007/s12016-016-8548-5. [DOI] [PubMed] [Google Scholar]
- 44.Molinero LL, Cubre A, Mora-Solano C, Wang Y, Alegre ML. T cell receptor/CARMA1/NF-kappaB signaling controls T-helper (Th) 17 differentiation. Proc Natl Acad Sci U S A. 2012;109:18529–34. doi: 10.1073/pnas.1204557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medoff BD, et al. CARMA1 is critical for the development of allergic airway inflammation in a murine model of asthma. J Immunol. 2006;176:7272–7. doi: 10.4049/jimmunol.176.12.7272. [DOI] [PubMed] [Google Scholar]
- 46.Blonska M, Joo D, Zweidler-McKay PA, Zhao Q, Lin X. CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J Immunol. 2012;188:3160–8. doi: 10.4049/jimmunol.1102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arjunaraja S, Snow AL. Gain-of-function mutations and immunodeficiency: at a loss for proper tuning of lymphocyte signaling. Curr Opin Allergy Clin Immunol. 2015;15:533–8. doi: 10.1097/ACI.0000000000000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haniuda K, Fukao S, Kodama T, Hasegawa H, Kitamura D. Autonomous membrane IgE signaling prevents IgE-memory formation. Nat Immunol. 2016;17:1109–17. doi: 10.1038/ni.3508. [DOI] [PubMed] [Google Scholar]
- 49.van Zwol A, Moll HA, Fetter WP, van Elburg RM. Glutamine-enriched enteral nutrition in very low birthweight infants and allergic and infectious diseases at 6 years of age. Paediatr Perinat Epidemiol. 2011;25:60–6. doi: 10.1111/j.1365-3016.2010.01173.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. Public deposition of sequence data is in progress.