Abstract
Genomic integrity is constantly threatened by sources of DNA damage, internal and external alike. Among the most cytotoxic lesions is the DNA double-strand break (DSB) which arises from the cleavage of both strands of the double helix. Cells boast a considerable set of defences to both prevent and repair these breaks and drugs which derail these processes represent an important category of anticancer therapeutics. And yet, bizarrely, cells deploy this very machinery for the intentional and calculated disruption of genomic integrity, harnessing potentially destructive DSBs in delicate genetic transactions. Under tight spatiotemporal regulation, DSBs serve as a tool for genetic modification, widely used across cellular biology to generate diverse functionalities, ranging from the fundamental upkeep of DNA replication, transcription, and the chromatin landscape to the diversification of immunity and the germline. Growing evidence points to a role of aberrant DSB physiology in human disease and an understanding of these processes may both inform the design of new therapeutic strategies and reduce off-target effects of existing drugs. Here, we review the wide-ranging roles of physiological DSBs and the emerging network of their multilateral regulation to consider how the cell is able to harness DNA breaks as a critical biochemical tool.
1. Introduction
Just as DNA breakage can devastate genomic integrity, it can also be deliberately and precisely exploited by cells in feats of genetic craftsmanship. Indeed, various integral cellular processes and genetic transactions are underpinned by the measured use of such potentially deleterious breaks.
The genome is continuously exposed to a panoply of DNA damaging agents, both endogenous and exogenous, which pose a considerable threat to genomic integrity. These genotoxic insults result in diverse DNA lesions including mismatches, base adducts, pyrimidine dimers, intra- and interstrand cross links, DNA-protein adducts, and strand breaks [1]. Among the most noxious lesions are DNA double-strand breaks (DSBs) wherein both strands of the double helix are broken through cleavage of phosphodiester linkages in the backbone of the duplex [2]. There is great variation among DSBs both structurally and in terms of the mechanisms of their generation. While simple DSBs such as those generated by restriction endonucleases may have either blunt or staggered ends, DSBs induced by physical or chemical agents, such as by ionising radiation and radiomimetic chemicals, both of which are used in cancer therapy, can display increased complexity. This includes chemical modifications of termini, differing latency of generation, indirect DSB formation from processing of other forms of DNA damage, and chromatin destabilisation from clustering of DSBs [3].
DSBs are highly mutagenic and can induce potentially tumorigenic chromosomal translocations. If unrepaired, DSBs may also lead to cell death [4]. It is thus of utmost primacy that such cytotoxic breaks are rapidly detected, signalled, and repaired. Indeed, DSBs elicit a potent DNA damage response (DDR) comprising DNA repair, cell cycle arrest, and/or apoptosis [2]. Two major pathways operate in eukaryotic cells in the repair of endogenously or exogenously induced DSBs: nonhomologous end-joining (NHEJ) and homologous recombination (HR). Unlike error-prone NHEJ which operates throughout the cell cycle, HR is essentially error-free and is limited to the S and G2 phases [5].
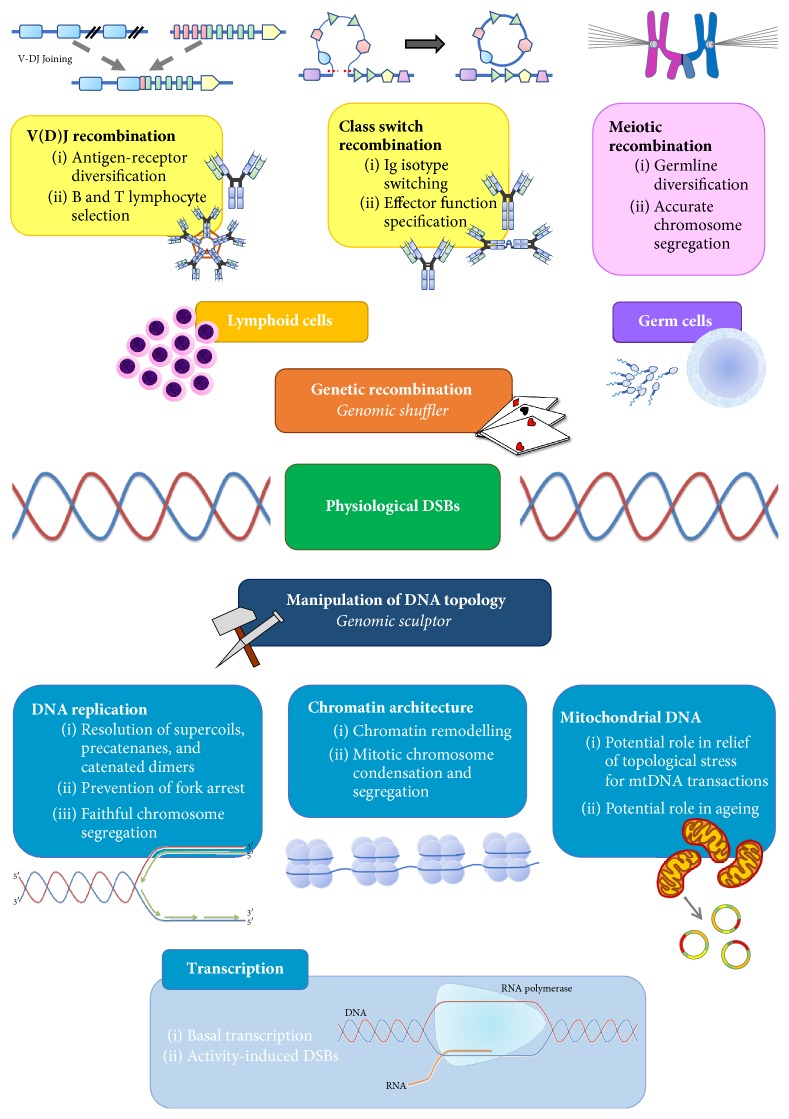
Despite the dangers inherent in such lesions, DSBs are precisely employed for the intentional but controlled disruption of genomic integrity in biological processes. The cellular roles of physiological DSBs can be broadly considered as one of two major functionalities: genetic recombination or manipulation of DNA topology. In the former role, DSBs can act as genomic “shufflers,” carefully but permanently recombining DNA segments for genomic diversification in lymphocytes [6, 7] and germ cells [8]. In contrast, topoisomerase-mediated DSBs serve as genomic “sculptors,” modulating higher-order DNA structure and serving to facilitate DNA replication and transcription [9–11], regulate gene expression [12–15], and alter chromatin state [16, 17]. While this latter form of DSB may, at first glance, appear to be little more than a transient intermediate, these “cleavage complexes” are structurally DSBs, also exist as longer-lived species, can be endogenously or exogenously converted into abortive breaks, and are highly spatiotemporally regulated to both produce functionally diverse genomic contortions and prevent genotoxicity or failure of genetic transactions [18, 19]. Hence, both sources of DSB will be considered in this discussion.
Here, we review the wide-ranging biological functions of these various physiological DSBs and the multilayered regulation thereof, together constituting cellular “DSB physiology” (Figure 1). By considering DSB physiology in its various forms, we explore how the cell is able to harness potentially destructive DSBs in delicate genetic operations.
Figure 1.
Summary of the diverse roles of physiological DSBs in biological processes.
2. Genetic Recombination: DSBs as Genomic Shufflers
2.1. Physiological DSBs in V(D)J Recombination
The role of physiological DSBs in the diversification of the adaptive immune response is well documented. V(D)J recombination describes the process whereby lymphoid cells recombine a repertoire of germline variable (V), diversity (D), and joining (J) exon gene segments in various permutations to generate enormous antigen-receptor diversity in immunoglobulins (Ig) and T cell receptors (TCRs). All three exon segments are used for assembling the variable region of the Ig heavy chain and the TCR β and δ chains, whereas only V and J segments are required for the Ig light chain and TCR α and γ chains [20]. The process involves large genomic rearrangements and DSBs act in these processes as “recombinogenic biomarkers,” specifying the recruitment of the recombinatorial machinery. It thus follows that the spatiotemporal regulation of DSB induction and repair is critical to the precise execution of this genetic shuffling, and perturbations thereof can produce disease in humans. The mechanism proceeds as two broad phases, cleavage and joining, corresponding to the processes of DSB formation and DSB resolution, respectively.
2.1.1. Cleavage: DSB Formation
Each V, D, and J segment is flanked by a recombination signal sequence (RSS), comprising conserved heptamer-spacer-nonamer sequence elements [21, 22]. Cleavage is initiated by the recognition of these RSS elements by the lymphoid-cell specific recombinase complex [23, 24]. Components of this complex, RAG-1 and RAG-2, bind one RSS from each of the paired gene segments, to form RAG-RSS complexes associated with each signal sequence [25, 26]. The two RSS groups are then approximated and synapsis proceeds to form a precleavage synaptic complex (PcSC). The PcSC is typically directed between gene segments with different RSS spacer lengths (either 12 or 23 base pairs) conforming to the 12/23 restriction rule [27]. PcSC formation is facilitated by the DNA-bending high-mobility group proteins HMGB1 and HMGB2 which are widely involved in chromatin architecture [28]. Here, HMGB1/2 act to enhance RAG1/2 binding to and cleavage of the 23 RSS, possibly via stabilisation of RAG-mediated bending of the 23 RSS spacer [29–33]. The RAG proteins proceed to introduce a nick at the junction between the RSS heptamer and coding sequence. While RAG-1 alone but not RAG-2 possesses DNA-binding activity and can mediate signal recognition, association of RAG-1 with RAG-2 increases sequence specificity and the stability of the enzyme-DNA complex [34, 35]. Nick formation is followed by a RAG1-mediated transesterification reaction forming a DSB comprising a blunt 5′-phosphorylated signal end and a hairpin coding end [23, 36]. The DSB is then enclosed in a postcleavage synaptic complex before NHEJ repair in the joining phase [37].
2.1.2. Joining: DSB Resolution
Prior to joining, hairpin coding ends are nicked open through the endonuclease activity of the nuclear enzyme Artemis, which complexes with the serine/threonine kinase DNA-PKcs, which, in turn, activates Artemis by phosphorylation [38, 39]. However, studies also indicate a postcleavage role for RAG proteins in opening hairpin coding ends through 3′-flap endonuclease activity [40–42] as well as serving as a scaffold to shield the DNA ends from aberrant nuclease digestion [43]. Modifications during recombination are introduced through Artemis-mediated asymmetric hairpin opening and/or terminal deoxynucleotidyl transferase- (TdT-) mediated nucleotide addition. The former mechanism enables palindromic repeat or “P” nucleotide addition as well as nucleotide loss from coding ends [44], while TdT-mediated addition inserts random “N” nucleotides at coding ends in a 5′-to-3′ direction [38, 45]. TdT, a template-independent polymerase, is recruited by a complex of NHEJ repair proteins comprising XRCC4, DNA ligase IV, and DNA-PK (itself a complex of Ku70, Ku86, and DNA-PKcs) [46–49]. The subsequent activity of DNA polymerase λ and μ combined with exonucleases generates compatible coding ends [50, 51] which are then ligated by the XRCC4-DNA ligase IV-XLF complex to produce the recombined V(D)J product [46, 47, 52].
2.1.3. Regulation of DSBs in V(D)J Recombination
An important point of control in the DSB formation step of V(D)J recombination is the cell cycle-dependent regulation of RAG expression. Cyclin A-CDK2 phosphorylation-mediated degradation of RAG-2 at the G1/S transition [53] exerts temporal control over RAG-induced DSBs by restricting RAG2 expression to the G1 phase of the cell cycle [54]. Furthermore, in p53-deficient mice a nonphosphorylatable T490A mutation of the phosphodegron in the C-terminus of RAG-2 results in increased lymphoma formation characterised by an increased frequency of clonal chromosomal translocations [55], illustrating the importance of this regulation for the integrity of recombination.
Additional temporal control is exerted over RSS selection for DSB formation and only certain RSS sites are available for recombination, exhibiting cell- and stage-specific biases [56, 57]. The great accuracy of the recombinatorial machinery, both in RSS specification and in avoiding inauthentic RSS-like (cryptic RSS; cRSS) sites, which occur around once per 600 bases in random DNA sequences [58], involves controlling the accessibility of the recombinase to its substrate for DSB generation. The discovery of the concurrence of IgH variable region (VH) germline transcription with its joining with DJH suggested an open chromatin state was required for recombination [57]. This “accessibility hypothesis” is now thought to involve chromatin architectural changes brought about by DNA and epigenetic modifications, chromatin-binding proteins, and cis-acting enhancer elements. These ultimately produce a transcriptionally active locus characterised by DNA hypomethylation, activating histone modifications, DNase I sensitivity, and increased RNA polymerase II occupancy [59–64]. RAG2 has been shown to specifically recognise the histone modification H3K4me3, a marker enriched at active promoters, via its PHD finger, an interaction which also stimulates RAG complex activity [65, 66]. This recognition likely serves to localise the recombinase to target RSSs by active promoters and upregulate recombination thereat [64]. Modulation of chromatin packing also determines the order of rearrangement observed in B cell Ig (heavy chain before light chain) and T cell TCR (β chain before α chain), as well as the process of allelic exclusion [56]. Aside from modulation of accessibility of RSS sites to binding and cleavage by the recombination machinery, target locus position within chromatin compartments of the nucleus and the structure of the locus itself are also directed to facilitate recombination [64].
2.1.4. Dysregulation of DSB Physiology in V(D)J Recombination
Broadly, perturbations in V(D)J recombination can be from either an inability to form DSBs, an inability to repair them, or inaccurate pairing of RSSs. Although viable, Rag-1- or Rag-2-deficient mice are unable to carry out V(D)J recombination and lack mature B and T lymphocytes, presenting with severe combined immune deficiency (SCID) [67, 68]. Missense mutations in either RAG-1 or RAG-2 genes are also observed in humans with an analogous form of autosomal recessive SCID known as Omenn syndrome [69]. Genetic studies of polymorphisms in wild-type and Omenn syndrome patients as well as biochemical data characterising recombinatorial competence of Omenn syndrome mutants suggest that RAG-1 and RAG-2 mutants exhibit lower efficiency of V(D)J recombination [69]. These mutations are associated with decreased RAG DNA-binding and cleavage activity and a lower efficiency of interaction between RAG proteins [69].
Impairment of the joining phase of V(D)J recombination can also cause immunodeficiency. While all mammalian NHEJ pathway mutants display sensitivity to ionising radiation and a lack of B and T lymphocytes, a spectrum of phenotypes is observed in mouse models. DNA-PKcs and Artemis mutants exhibit subtle defects in DSB repair [70] but display no growth retardation [71]. Hairpin opening is lost in Artemis-deficient SCID [72] and DCLRE1C hypomorphism in humans has been found to underlie an Omenn syndrome phenotype associated with defective endonuclease activity [73]. Ku70 and Ku80 mutants display similar phenotypes of radiosensitivity and growth retardation [74, 75] likely due to the interdependence conferred by their heterodimerisation [76]. Mutants of the critical end-joining proteins XRCC4 and DNA ligase IV have the most severe phenotype, both displaying V(D)J recombination defects, impaired lymphocyte development, p53-dependent neuronal apoptosis, and embryonic lethality [77–79]. These findings thus point towards a crucial role of DSB repair in B/T cell physiology and adaptive immune diversification.
Lymphomas have been found to exhibit a range of chromosomal aberrations as a result of erroneous DSB handling in V(D)J recombination. Oncogenic lesions have been found to include chromosomal translocations, insertions, deletions, and inversions and broadly result from errors in RSS pairing or joining [80–82]. Inaccurate RSS recognition can produce RSS/cRSS pairings which may exert tumorigenic effects via amplification of a protooncogene or juxtaposition of transcriptional regulatory elements from the antigen-receptor locus with a protooncogene, driving dysregulated expression [6]. cRSS/cRSS pairings may also arise which can produce translocations or deletions [6]. Aberrant DSB formation has also been found to underlie the reciprocal (t14; 18) translocation found in 90% of follicular lymphomas [83, 84]. In this case, it has been shown that cleavage of non-B-form DNA at a non-RSS “translocation fragile zone” called the major breakpoint region (MBR) at the Bcl-2 gene produces a translocation with the DSB at the Ig heavy chain locus on chromosome 14 [84]. Recent work on minichromosomal substrates has revealed the importance of CpG methylation in the localisation of aberrant DSB formation at the Bcl-2 MBR and that breakage is AID-dependent (activation-induced cytidine deaminase) [85]. Lymphomas have also been found to exhibit defects in DSB joining producing chromosome translocations by the “end donation” model. This involves the erroneous joining of a RAG-induced DSB at an authentic RSS with a non-RAG-mediated DSB [86].
2.2. Physiological DSBs in Class Switch Recombination
Class switch recombination (CSR) is another example of a DSB-dependent recombination event, occurring in mature B cells. In response to antigenic stimuli and costimulatory signals from other immune cells, B cells undergo CSR to alter their Ig constant heavy chain while retaining the same antigen specificity, in effect, generating an alternative Ig isotype with the same paratope but distinct effector functions [87]. This enables a permanent switch of expression from the default IgM and IgD to alternative Ig isotypes (IgA, IgG, or IgE) by recombination of the heavy chain (IgH) locus. Recombination is achieved through cleavage and excision of constant heavy chain (CH) genes via DSB generation, followed by ligation of the remaining segments through DSB resolution [87].
Conserved motifs called switch (S) regions upstream of each CH gene are the site of DSB generation [88, 89]. AID initiates recombination by conversion of multiple S region deoxycytidine residues into deoxyuracil [90–93]. The resulting UG mismatch can be converted into a DSB through base-excision repair (BER) and/or mismatch repair (MMR) DNA repair pathways [87]. BER proceeds through excision of deoxyuracil by uracil DNA glycosylase (UNG) [93, 94] followed by nicking of the resulting abasic site by apurinic/apyrimidinic endonucleases (APE) [87, 95]. AID-mediated deamination on the opposite strand introduces a second nick in close proximity, thereby generating a staggered DSB [89, 96]. Processing of these overhangs forms blunt free DNA ends which are permissive for shuffling [87, 97].
As aforementioned, MMR is another pathway employed in CSR DSB generation and is thought to provide auxiliary support to BER-mediated mismatch excision [98]. Indeed, multiple studies of MMR-deficient murine B cells have shown impaired and aberrant CSR implicating MMR proteins Msh2, Msh6, Mlh1, Pms2, and Exo1 in the recombination process [99–104]. One model proposes that while the aforementioned BER-induced SSBs can automatically form DSBs if near one another, distant SSBs within the large S regions can be converted into DSBs by assistance from MMR [105]. This first involves recognition of an AID-induced UG mismatch by a heterodimer of Msh2-Msh6, followed by recruitment of the endonuclease heterodimer Mlh1-Pms2. These proteins, in turn, recruit the 5′-3′ exonuclease Exo1 to a nearby BER-induced 5′ SSB and Exo1 proceeds to excise the strand towards the mismatch and beyond until it reaches another SSB, thus producing a DSB [105]. Alternatively it has also been proposed that if Exo1-generated single-stranded DNA gaps are formed on opposite strands, this too may form a DSB [106].
After intrachromosomal deletion of the intervening unwanted CH genes, the free end of the variable domain region is ligated to that of the new constant domain exon by the NHEJ machinery [107–110].
2.2.1. Regulation of DSBs in CSR
Recombination underlying isotype switching of constant heavy chain genes is regulated by germline transcripts which modify switch region accessibility to the DNA handling machinery [105]. Transcription beginning at germline promoters, upstream of a target S region, generates a sterile (noncoding) transcript known as a germline transcript (GT) [111, 112]. Activation of specific cytokine-inducible transcription factors and their action at germline promoters determines which GT is generated [113]. These germline transcripts direct AID to specific S regions thus spatially regulating DSB generation and, in turn, CSR [105, 114].
AID deamination can only occur on single-stranded DNA (ssDNA) [115] and one proposed mechanism of GT-mediated regulation is optimisation of S region structure for AID recognition [115]. This could be achieved through the creation of short ssDNA tracts in the S regions, paralleling somatic hypermutation [116], or through RNA-DNA hybrid R-loop formation [117, 118]. AID can also be directly recruited to RNA Pol II at the initiation or elongation phase of GT formation, which may facilitate AID targeting [105, 119]. Further regulation may also occur at the level of higher-order chromatin structure whereby histone modifications in the transcribed region may modulate AID accessibility [120]. While the exact mechanism of this architectural control is unclear, it appears to be associated with B cell activation, AID expression, and RNA Pol II enrichment [120, 121]. It is thus clear that various levels of regulation act to spatiotemporally restrain DSB formation and repair in CSR.
2.3. Physiological DSBs in Meiotic Recombination
Beyond its roles in lymphoid cells, DSB-induced recombination also acts in meiosis where it is essential for both the fidelity of chromosome segregation and diversification of the germline. Meiosis is a form of reductive cell division exclusive to gametogenesis and involves two successive divisions following replication, known as MI and MII [122]. A hallmark of meiosis is meiotic recombination between homologous chromosome pairs or dyads, which is reliant on DSB induction and HR repair, resulting in exchange of genetic information between nonsister chromatids [122]. These “crossover” events essentially produce shuffling of alleles in each chromatid, giving rise to a unique haploid complement in the resultant gametes, ultimately creating genetic diversity within the population. Furthermore, meiotic recombination establishes physical linkages between homologs allowing accurate segregation by the spindle apparatus [123]. Just as in V(D)J recombination and CSR, careful spatiotemporal regulation of DNA break induction and resolution is critical to the integrity of the process.
2.3.1. DSB Formation
Meiotic recombination is initiated by DSB induction by a dimer of the topoisomerase-like transesterase Spo11 which acts via a covalently linked protein tyrosyl-DNA intermediate complex [124–127]. Interestingly, the human SPO11 homolog maps to position 20q13.2-q13.3, a region known to be amplified in some breast and ovarian tumours, possibly reflecting genomic instability due to dysregulation of DSB formation [128]. Spo11 action is critical for gametogenesis and loss in mice produces infertility and results in defective meiotic recombination, DSB formation, and synapsis in Spo11−/− mouse spermatocytes [129]. DSB formation is also dependent on an extensive protein-protein interaction network, most studied in S. cerevisiae, between Spo11 and other meiotic proteins. These proteins include components of the Mre11-Rad50-Xrs2 (MRX) complex and multiple Spo11-accessory proteins including Mei4, Mer2, Rec102, Rec104, Rec114, and Ski8 which facilitate DSB formation [126, 130]. The functions of these various proteins are incompletely understood; however, the WD repeat protein Ski8 has been found to stabilise association of Spo11 on meiotic chromatin and promotes recruitment of other accessory proteins [130] and Rec102 and Rec104 are known to form a multiprotein complex with Spo11 and act with Rec114 to regulate Spo11 self-association [131–133]. Rec 114, Mer2, and Mei4 (RMM) constitute a separate subcomplex [134] which plays a role in the spatiotemporal regulation of DSB formation as will be discussed below.
2.3.2. DSB Resolution
The resulting DSBs then undergo endonucleolytic processing which releases Spo11 to allow repair [135]. This initial DSB resection is coordinated with cell cycle progression and is dependent on the activity of the multisubunit nuclease MRX complex and Sae2 protein in budding yeast [126, 136–138]. The former complex also acts to facilitate HR independently of its nucleolytic activity [139]. Analogous to the MRX complex, the MRE11-RAD50-NBS1 (MRN) complex possesses a similar role in SPO11 removal in other eukaryotes [140]. Spo11 removal is followed by extension of the end resection, involving the helicase Sgs1 and nucleases Exo1 and Dna2, forming 3′ single-stranded overhangs for HR [138, 141]. The overhangs then undergo homology search and strand invasion catalysed by the strand exchange proteins Rad51 and meiosis-specific Dmc1 alongside various other cofactors [142, 143]. The resulting interactions are processed to form recombination products with either noncrossovers or crossovers, defined, respectively, as gene conversion or reciprocal genetic exchange between homologous chromosomes [142]. During this pairing or synapsis of dyads, a proteinaceous structure called the synaptonemal complex establishes a zipper-like connection between homologous chromosomes [144]. Reciprocal interhomolog recombination results in the formation of physical linkages between homologous chromosomes known as chiasmata, which aid coalignment of the resultant bivalents along the spindle axis for accurate segregation [145].
2.4. Regulation of DSBs in Meiotic Recombination
A multitude of interacting mechanisms operate in meiotic recombination to control DSB formation and resolution. This tight spatiotemporal regulation of DSB handling allows the intentional disruption of gamete DNA integrity for diversification, while ensuring coordinated cell cycling and, ultimately, generation of an intact haploid germline.
2.4.1. Temporal Control of Meiotic DSBs
DSB induction is mechanistically coupled to DNA replication [146] and is subject to strict temporal regulation [147–149], occurring only between replication and chromosome segregation [150]. This ensures both coordinated chiasma formation and effective homology search for synapsis [150]. In addition, a multitude of checkpoints operate to couple DSB induction and resolution to the cell cycle to protect against the deleterious effects of unrepaired DSBs and also act to arrest or eliminate aberrant cells [151–154].
Posttranslational modifications on the Spo11-accessory protein Mer2 have been shown to contribute to the timely execution of DSB formation in meiotic recombination. Phosphorylation of Mer2 by the budding yeast cyclin-dependent kinase Cdc28 stimulates DSB formation via promotion of interactions both with itself and with other proteins involved in DSB generation including Mei4, Rec114, and Xrs2 [155]. It is known that Cdc28-Clb5 (CDK-S) activity increases during premeiotic replication and peaks before MI [156, 157] and thus may help orchestrate a concerted progression of meiotic recombination and prophase I [155]. Mer2 is also phosphorylated by Cdc7 kinase in S. cerevisiae to control Spo11 loading on DSB target sites [158]. It has been found that CDK-S phosphorylation on Mer2 S30 promotes DDK (Cdc7-Dbf4)-dependent phosphorylation at the adjacent S29 [157]. As well as being involved in meiotic recombination, CDK-S and DDK are both required for premeiotic S phase [156, 159] which may contribute to coordination therewith. Furthermore, it has been found that the level of DDK activity required for premeiotic S phase is lower than that supporting its postreplicative role in meiosis [159]. It has been proposed that different thresholds for both CDK-S and DDK in these processes may ensure replication precedes DSB formation [160].
Additional temporal control is afforded by meiosis-specific expression of Spo11 and accessory proteins Rec102, Rec104, Rec114, and Mei4, largely achieved via transcriptional regulation [150]. In contrast, Mer2 has been found to undergo meiosis-specific posttranscriptional regulation by a splicing factor Mer1, which itself is only expressed in meiosis [161, 162]. Although Mer2 transcription does occur in mitosis, efficient splicing by Mer1 to generate functional Mer2 is exclusive to meiosis [161].
2.4.2. Spatial Control of Meiotic DSBs
Meiotic recombination and DSB formation exhibit nonrandom patterning across the genome, being more frequently localised to particular sites on chromosomes called “hotspots” [163, 164]. While exchange usually takes place between alleles, homologous recombination between nonallelic sequences with high sequence identity may occur when DSBs form in repetitive DNA sequences such as transposable elements and low-copy repeats/segmental duplications, leading to genomic rearrangements [165]. In addition, it has been shown that DSBs formed in pericentromeric regions are associated with disruption of sister chromatid cohesion leading to missegregation and aneuploidy [166]. Therefore, the location of DSBs and subsequent recombination is controlled for the maintenance of genomic integrity.
In mice, recombination hotspots have been found to be enriched for H3K4me3 histone modifications [167, 168]. These hotspots are determined by the DNA-binding histone H3K4 trimethyltransferase PRDM9, which has been found to regulate activation of recombination loci and is a key factor in specification of meiotic DSB distribution [169, 170]. Indeed, Prdm9-deficiency results in infertility and disruption of early meiotic progression in mouse models [171]. PRDM9 has been shown to directly and specifically bind a 13-mer DNA motif enriched at hotspots via its C2H2-type zinc finger array [169, 172, 173] and changes in the array affect H3K4me3 enrichment, hotspot recombination, and crossover patterns [173]. Furthermore, sequence variation in PRDM9 may underlie the intra- and interspecies differences observed in recombination hotspot distribution [169, 174]. While the exact mechanism linking hotspot specification with targeting of the DSB machinery is unclear, it has been proposed that PRDM9 may recruit Spo11, itself or via a partner protein, or may promote chromatin remodelling permissive for Spo11 binding [173].
Negative feedback systems acting both in cis and in trans also exert an inhibitory effect from one DSB on further break formation, thus limiting DSB frequency [8]. In addition, DSB formation occurs at unsynapsed chromosome segments, implicating interhomolog interactions in the control of DSB site and numbers [8, 175]. Recombination exhibits an interhomolog bias as opposed to occurring between sister chromatids [176] and it has recently been shown that total DSB number itself is a determinant of DSB repair pathway selection and development of biased recombination [177].
DSB formation is also subject to spatial regulation via control of chromosome ultrastructure. During the leptotene stage of prophase I, DSB formation occurs alongside the development of the axial element, a proteinaceous axis comprising various proteins including cohesin [145]. The sister chromatids form linear arrays of chromatin loops which are connected at their bases by the axial element, which is the precursor of the lateral element of the synaptonemal complex at pachytene [145]. Sites of DSB formation are found to map to loop sequences which are “tethered” to the axis during recombination in “tethered loop-axis complexes” [178]. ChIP-chip studies in S. cerevisiae have shown that Rec114, Mer2, and Mei4 stably associate with chromosome axis association sites between loops, in a manner dependent on components of the axial element [179]. Meiotic cohesin was found to control DSB formation and axial element protein and RMM recruitment at a subset of chromatin domains, while cohesin-independent DSB formation was observed in other domains [179]. Such differences in DSB machinery deposition between chromatin domains may contribute to the establishment of different recombination proficiencies, potentially providing a form of spatial regulation of DSB induction [179]. Furthermore, this interaction of RMM with axis association sites is dependent on Mer2 phosphorylation by Cdc28 which likely affords additional temporal coordination of DSB formation with the end of premeiotic replication [179].
3. Manipulation of DNA Topology: DSBs as Genomic Sculptors
3.1. Physiological DSBs in the Relief of Topological Stress
To access the information stored in compacted nuclear DNA in such processes as transcription, replication, and repair requires the repeated tangling and untangling of the DNA double helix. Such molecular contortions introduce topological entanglements into the duplex which if left unchecked could compromise genomic stability [180, 181]. Relief of such topological strain is achieved by a family of essential enzymes, the type I and type II topoisomerases. The former generate transient single-strand breaks (SSBs) while the latter family hydrolyse ATP to cut the DNA double helix and generate a transient DSB. The type II enzyme then passes one DNA duplex through the gap, and the gap is subsequently resealed [182]. Type II topoisomerase (Top II) activity in mammals is mediated by two isoenzymes, Top IIa and Top IIb, which exhibit considerable homology except in their less conserved C-termini [183, 184] and display different subcellular distributions during the cell cycle [185, 186]. Studies on Top IIa/b chimeric “tail swap” proteins and C-terminal truncations have shown that the C-terminal regions contribute to isoform-specific functionality and may modulate decatenation activity [187, 188]. Immunohistochemical studies of Top II isoform expression patterns in normal and neoplastic human tissues show that Top IIa expression is largely restricted to normal proliferative tissue and is highly expressed in certain tumour types. In contrast, Top IIb is widely expressed across both proliferative and quiescent cell populations, although at a higher level in normal proliferative tissue and tumours [189, 190].
While, in general, a Top II-induced DSB is resolved in the enzyme's catalytic cycle, it does possess a capacity for formation of abortive breaks [19] and can form longer-lived complexes in certain functions [12, 191] and both inappropriately low or excessive formation of DSB intermediates can compromise cellular viability, thus necessitating commensurate regulation [18]. The critical involvement of DNA topoisomerases in various essential cellular processes has led to the development of topoisomerase poisons for use as antibacterial [192, 193] or anticancer therapeutics [194, 195]. While catalytic inhibitors block enzymatic activity without trapping of the covalently linked DNA-enzyme intermediate cleavage complex, the majority of clinically used Top II-targeting drugs act as Top II poisons, accumulating complexes by either blocking religation or increasing complex formation. The resultant DSBs drive mitotic arrest and apoptotic cell death, illustrating the toxicity of uncontrolled DSB physiology [196]. The tight spatiotemporal regulation of Top II-mediated DSB formation enables the measured usage of these potentially deleterious DNA breaks as a physiological tool for the relief of topological stress.
3.1.1. Type II Topoisomerase DSB Handling
Supercoiling, entanglement, and knotting are frequent topological obstacles encountered in DNA physiology. Type II DNA topoisomerases, through DSB induction and repair, act to resolve such obstacles using a repertoire of supercoil introduction/relaxation, catenation/decatenation, and knotting/unknotting processes [197, 198]. Eukaryotic Top II enzymes are homodimeric proteins with symmetrically paired domains, whereof each interface serves as a “gate” to control the passage of DNA strands [180]. While catalytic efficiency is a function of the structural interplay between the DNA substrate and the type II topoisomerase isoenzyme, the general mechanism is conserved across the enzyme family. The overall cycle acts to relax DNA, involving DSB induction, strand passage, and DSB repair and proceeds via a phosphotyrosine-linked DNA-enzyme intermediate [199]. The aforementioned DSB-dependent relief of DNA topological tension is of particular importance in the processes of DNA replication, transcription, and compaction into higher-order structures.
3.2. Physiological DSBs in DNA Replication
Replication fork arrest, such as due to impassable DNA damage, can give rise to DSBs [200, 201] which are highly mutagenic and cytotoxic and elicit a potent DDR comprising HR-mediated repair, cell cycle arrest, and possibly apoptosis, to ensure the faithful inheritance of genetic information [202–204]. Indeed, drugs which promote DSB formation or target DSB repair represent an important category of chemotherapeutics for cancer [205, 206]. In spite of all this, Top II-induced DSBs play an integral role in replication, illustrating how the careful regulation of a potentially destructive lesion can be exploited for cellular physiology.
Top II enzymes are essential in the elongation of replicating DNA chains and during segregation of newly replicated chromosomes. During semiconservative replication, the replisomal machinery at the advancing replication fork forces the intertwined DNA strands ahead of it to become overwound or positively supercoiled. These positive supercoils are redistributed behind the fork by the rotation of the replicative machinery around the helical axis of the parental duplex. This causes intertwinement of the newly replicated DNA double helices, forming precatenanes [197]. Studies in yeast reveal a necessity of either Top I or Top II in elongation, acting as replication “swivels” to relax positive supercoiling and resolve precatenanes, respectively [9, 207–209]. As replication nears completion, the unreplicated double-stranded DNA (dsDNA) segment in between the converging forks becomes too short for the action of a type I topoisomerase and replication must first be completed resulting in intertwinement of the replicated duplexes into catenated dimers [208, 210]. These intertwines are decatenated by the action of Top II to allow for chromosome separation in mitosis [211, 212]. In yeast, which only possesses a single type II topoisomerase, loss of the enzyme results in chromosome nondisjunction, accumulation of catenated dimers, and inviability at mitosis [211, 213, 214]. In contrast, it has been shown in human cells that Top IIa but not Top IIb is essential for chromosome segregation [215].
3.3. Physiological DSBs in Transcription
Transcription of nascent mRNA presents topological obstacles akin to those seen during replication [216]. Transcription elongation is accompanied by changes in the local supercoiled state of DNA, forming positive supercoils ahead and negative supercoils behind the transcriptional machinery [197, 198, 216, 217]. While loss of Top I or Top II alone in yeast generally does not compromise transcription, top1 top2 temperature-sensitive double mutants exhibit significant inhibition of rRNA synthesis and, to a lesser extent, mRNA synthesis at nonpermissive temperatures [207]. In addition, double mutants accumulate negative supercoils in extrachromosomal plasmids which appear to promote transcription initiation of ribosomal minigenes on such plasmids [218, 219]. More recent studies of yeast rRNA synthesis suggest distinct roles of Top II and Top I, acting, respectively, to relieve positive supercoiling ahead of and negative supercoiling behind the advancing polymerase [11]. Pharmacological inhibition of Top II in human cells is found to decrease Pol I transcription and Top IIa depletion both impairs Pol I preinitiation complex (PIC) formation and produces a loss of DSBs at these sites. It has thus been proposed that Top IIa-induced DSBs are involved in topological changes at rDNA promoters to facilitate PIC formation [220].
As aforementioned, top2 yeast exhibits normal levels of transcription; however it has been shown in budding yeast that loss of Top2 produces a specific reduction in Pol II-dependent transcription of genes longer than around 3 kb, attributed to defective elongation due to Pol II stalling [221]. More recently, Top II-induced DSBs have been proposed to act in a mechanism involving the transcription factor TRIM28 and DNA damage signalling to promote Pol II pause release and transcription elongation [222].
3.3.1. Activity-Induced DSBs in Transcription Regulation
Transcription has long been known to stimulate HR in a locus-specific manner [223–225] and it is emerging that physiological DSBs may be involved in regulated transcription [191, 222]. Furthermore, it has recently been found that DSBs induced at transcriptionally active sites in human cells preferentially promote HR over NHEJ and this repair is dependent on the transcription elongation-associated epigenetic marker H3K36me3 [226]. It is thus becoming increasingly clear that transcription and HR-dependent DSB repair may be reciprocally regulated. Of particular interest is the emerging role of so-called “activity-induced DSBs” in transcriptional control, adding additional complexity to the integration of environmental stimuli and cell state to the modulation of transcriptional programmes. Indeed, Top IIb has previously been shown to be required for 17β-estradiol-dependent transactivation, via the regulated formation of site-specific longer-lived DSBs in target gene promoters [191]. Top IIb-mediated DSB formation in a target promoter has also been shown to be required for glucocorticoid receptor-dependent transactivation [227]. More recently, the spotlight has shifted to the broadening role of activity-induced DSBs in neuronal physiology and neuropathology.
3.3.2. Role of Activity-Induced DSBs in Neuronal Function
Neuronal early response genes or “immediate early genes” (nIEGs) are a subset of activity-regulated genes induced with shortest latency following neuronal stimulation and typically encode transcription factors involved in transcriptional modulation for synaptic plasticity [228, 229]. Treatment of neuronal cells with etoposide, a Top II inhibitor which induces Top II-mediated DSBs [230], has been found to promote upregulation of a subset of nIEGs including Fos, FosB, and Npas4 [12]. This increase was not observed with other DSB-inducing agents including neocarzinostatin, bleomycin, and olaparib, suggesting a Top II-mediated mechanism [12]. Furthermore, etoposide-induced upregulation of Fos and Npas4 is not affected by inhibition of DSB signalling following ATM blockade, which also suggests that nIEG induction is independent of the effects of a generic DDR to nonspecific DSBs [12]. Similar to etoposide treatment, CRISPR-Cas9-induced DSBs targeted to the Fos and Npas4 promoters also produced upregulation thereof in the absence of neuronal activity. Together these indicate a role of DSBs acting within target gene promoters in the regulation of nIEG transcription [12].
Furthermore, electrical or pharmacological stimulation of cultured primary neurons revealed upregulation of Fos and Npas4 mRNA accompanied by elevated γH2AX levels [12], a known biomarker of DSB formation [231], suggesting the possible involvement of “activity-induced DSBs” in nIEG expression. Genome-wide ChIP-seq of γH2AX following stimulation with NMDA (N-methyl-D-aspartate) demonstrated preferential clustering of yH2AX elevations at actively transcribed genes and downstream sequences. Differential peak calling on NMDA-treated samples showed that yH2AX enrichment was confined to only 21 loci including the nIEGs Fos, FosB, Npas4, Egr1, Nr4a1, and Nr4a3 among other transcription factors and noncoding RNAs, pointing to the specific localisation of DSB induction following neuronal stimulation [12]. Interestingly, γH2AX was also observed to increase following pharmacological or electrical stimulation of mouse acute hippocampal slices as well as in hippocampal lysates from mice exposed to a fear-conditioning paradigm [12]. This corroborates recent evidence supporting DSB induction in mouse neurons in response to environmental stimuli in the form of spatial exploration [232]. These DSBs were found to predominate in the dentate gyrus suggesting a physiological role in spatial learning and memory [232]. Overall, these results suggest that neuronal activity specifically induces targeted DSBs involved in transcriptional regulation of nIEGs. Interestingly, studies of repair kinetics of DSBs generated in the Fos promoter following NMDA treatment demonstrated long-lived breaks, being repaired within 2 hours of stimulation. However, the significance of this extended lifespan is as yet unclear [12].
Other experiments showed that short hairpin RNA-mediated knockdown of Top2b in cultured primary neurons reduces yH2AX enrichment at nIEG exons following NMDA stimulation, pointing towards a role of Top IIb in activity-induced DSB formation at nIEGs [12]. In ChIP-seq experiments, genome-wide binding of Top IIb was found to almost quintuple following NMDA treatment and it was observed that Topo IIb enrichment clustered alongside yH2AX signals in both unstimulated and NMDA-treated cells [12]. In addition, yH2AX-enrichment following NMDA stimulation and etoposide treatment were strongly correlated, lending further support to a model of Top IIb-generated activity-induced DSBs [12]. Linking Top IIb-mediated DSB formation with activity-dependent nIEG transcription, it was found that Top2b knockdown in cultured primary neurons inhibits induction of nIEGs following NMDA stimulation but could be rescued by Cas9-generated DSBs targeted to the nIEG promoter [12].
While the mechanism whereby Top IIb-mediated DSBs could induce nIEG transcription is as yet unclear, one model proposed involves a functional interaction with the architectural protein CTCF [12]. CTCF organises genomic superstructure, defining chromatin architecture to control long-range interactions between different genomic loci. Major effects of CTCF action include the establishment of chromatin domains and blockade of promoter-enhancer interactions [233]. Searches for motif enrichment at Top IIb binding sites have revealed greatest clustering of CTCF under both basal conditions and NMDA treatment [12] and Top IIb itself is found to concentrate within CTCF motifs. Furthermore, Top IIb is found to coimmunoprecipitate with CTCF both under basal conditions and to a greater extent with NMDA stimulation. Genome-wide CTCF binding distribution also corresponds well with yH2AX signals at NMDA-stimulated activity-induced DSBs and following etoposide [12]. Combining these results Madabhushi et al. [12] propose a model whereby neuronal activity drives Top IIb-induced DSB formation which relieves CTCF-based topological impediments to nIEG transcription. While a recent proteomic analysis of the Top IIb proximal protein interaction network revealed interaction of Top IIb with CTCF at topologically associating domain (TAD) boundaries [234], the precise mechanism whereby activity-induced DSBs may overcome CTCF-generated obstacles is unknown. It was recently postulated that activity-induced DSBs in nIEGs lead to genomic rearrangements involving transposable elements producing a distinct form of somatic mosaicism in neurons, with implications on plasticity, network formation, and potential interplay with age-related epigenetic changes and disease [235].
Additional mechanistic complexity derives from the need for concerted repair of activity-induced DSBs. Top IIb exhibits preferential binding and cleavage at the promoters of target nIEGs, cooccupying the site with the transcriptional activator ELK1 and histone deacetylase HDAC2, the latter repressing nIEG expression [236]. Also found enriched at these sites is the tyrosyl-DNA phosphodiesterase TDP2 which acts via the NHEJ pathway to repair abortive Top IIb-induced DSBs [19, 237, 238], suggesting coordinated resolution of such physiological breaks [12]. Furthermore, TDP2-deficiency in human cells produces hypersensitivity to Top II-induced DSBs and reduces Top II-dependent transcription, illustrating the importance of regulated break repair in DSB physiology [19].
3.3.3. Dysregulation of Physiological DSBs in Neuropathology
Defective DSB repair and/or loss of DSB repair components is a feature of various neuropathologies and age-associated neurodegenerative disorders, including ataxia-telangiectasia (ATM), ataxia-telangiectasia-like disorder (MRE11), Nijmegen breakage syndrome (NBS1), and Alzheimer's disease [239–245]. In addition, Ercc1Δ/- mice, which have compromised nucleotide excision repair, interstrand crosslink repair, and DSB repair pathways, display age-associated neurodegeneration and progressive loss of motor function [246, 247].
Increased incidence of DNA strand breakage has been identified in the brains of Alzheimer's disease (AD) patients [248, 249], and AD mouse models exhibit an elevated baseline frequency of neuronal DSBs [232]. Furthermore, amyloid-β (Aβ), aggregates of which are widely recognised as a key factor in AD pathogenesis, is found to increase DSB formation in primary neuronal cultures via dysregulation of synaptic transmission [232, 250]. Expression of BRCA1, an important HR DSB repair factor [251, 252], is found to be reduced in AD brains and exhibits neuronal activity-dependent regulation as well as downregulation by Aβ oligomers [253]. Moreover, short hairpin RNA knockdown of BRCA1 in wild-type mice impairs repair of activity-induced DSBs [253].
There is growing evidence suggesting that aberrant Top II activity may play a role in the aetiology of neurodevelopmental defects. Recent work involving clinical whole-exome sequencing identified a novel TOP2B mutation associated with intellectual disability, autistic traits, microcephaly, and developmental retardation [254] and homozygous mutations in TDP2 have been linked with cases of intellectual disability, epilepsy, and ataxia [19]. Top IIb has been shown to be important in neuronal differentiation and survival and is upregulated during the transition from mouse embryonic stem cells to postmitotic neurons with reciprocal downregulation of Top IIa. Furthermore, loss of Top IIb in mice results in premature degeneration and apoptosis of neurons at later stages of differentiation concomitant with transcriptional deregulation of genes involved in neurogenesis and cell division [13, 14]. Pharmacological inhibition of Top I in mouse cortical neurons results in preferential downregulation of long genes, many of which are associated with autism spectrum disorder. As Top IIa/b inhibition by ICRF-193 produces a similar length-dependent transcriptional reduction, it is possible that Top II is also responsible for the regulation of certain autism-associated candidate genes [255]. Furthermore, it has been proposed that environmental and dietary sources of topoisomerase poisons and inhibitors in early neurodevelopment could be a potential cause of autism and there have been calls to factor this into food safety and risk assessments [256, 257].
Together, these findings point towards the importance of DSB repair regulation in neuronal physiology and activity-induced DSB handling, but further studies will be required to elucidate more specific roles of activity-induced DSBs in neuropathology.
3.4. Physiological DSBs in Chromatin Architecture
Top II is a highly abundant chromatin protein, constituting 1-2% of total chromosomal protein content in mitosis [258]. Studies of isolated chromosomes reveal association of one Top II molecule per 23,000 base pairs of DNA, largely at and around loop bases, suggesting a structural role in higher-order DNA architecture [258, 259]. Indeed, Top II binding and DSB formation sites have been identified in matrix-attachment regions, cis-elements which are involved in long-range chromatin organisation [260, 261]. However, photobleaching experiments have revealed both Top II isoforms to be highly mobile in the chromosomal scaffold, arguing for a more complex dynamic interaction with chromatin structure [262].
DNA topology and chromatin structural changes are known to be intricately related [263]. Early work with temperature-sensitive top2 yeast [212] and complementation experiments with Top II-immunodepleted Xenopus egg extracts [264] revealed Top II to be important in chromosome condensation and segregation in mitosis, presaging the discovery of a more active role of the enzyme in chromatin remodelling. Indeed, Top II has been identified as a component of the Drosophila ATP-dependent chromatin remodelling complex CHRAC (chromatin-accessibility complex) [265], and mammalian BAF chromatin remodelling complexes are found to promote Top IIa activity [266]. Furthermore, Top II-mediated DNA supercoil relaxation is stimulated by the Drosophila protein Barren [267], which is involved in chromosome condensation and disjunction [268]. ChIP experiments in S. pombe demonstrate an intergenic localisation of both Top I and Top II which strongly correlates with distributions of the chromodomain ATPase Hrp1 and the histone chaperone Nap1, both involved in nucleosome disassembly [269–271]. Top II-dependent chromatin remodelling is also important in apoptotic chromosome condensation, and caspase-activated DNase (CAD) association is found to promote Top IIa activity [272]. While the exact mechanistic role of DSB handling in chromatin packing is unclear, AFM and fluorescence imaging have revealed an H1-dependent ATP-independent mechanism of Top II-mediated chromatin compaction involving clamping of DNA strands [273]. Thus, it appears that Top II DSB handling is important in highly diverse manipulations of chromatin packing and further studies are needed to elucidate the precise regulation thereof.
3.5. Mitochondrial DNA Maintenance and Ageing
Mitochondrial DNA (mtDNA) in humans is typically a closed circle of dsDNA [274]. As in genomic DNA, replication and transcription processes exert topological stresses on mtDNA molecules which are relieved by native topoisomerases [197, 275]. Mammalian mitochondria contain 3 topoisomerases encoded in the nucleus: Top1mt (TOP1MT) and Top IIIα (TOP3A) of the type I family and Top IIb (TOP2B) from the type II family of enzymes [275]. Although mitochondrial Top IIb is C-terminally truncated compared to its nuclear counterpart, it still retains basic type II topoisomerase DSB turnover activity, albeit with reduced processivity, and exhibits a similar pharmacological profile [276]. Top1mt deletion in mice results in elevated mtDNA transcription and induction of a stress response accompanied by considerable upregulation of Top IIb, suggesting a potential compensatory role of the type II enzyme [277]. However, both TOP2B−/− MEFs and TOP1MT−/−TOP2B−/− display wild-type levels of transcription and only the double-knockout elicits a stress response, suggesting nonoverlapping functions of the two molecules [277]. This picture is further complicated by the recent discovery of active full-length Top IIa and Top IIb in both murine and human mitochondria, which, although as yet uncharacterised, are likely involved in classical functions in replication and transcription like their nuclear counterparts [278].
Top IIIa is a type IA topoisomerase which is found in both the nucleus and mitochondria and has been shown to play a crucial role in the maintenance of mtDNA genomic integrity in Drosophila. Indeed, loss of mitochondrial import of Top IIIa in Drosophila is associated with a reduction in mtDNA copy number, mitochondrial dysfunction, impaired fertility, and accelerated ageing [279, 280]. The interplay between these type I and II topoisomerases in the maintenance of mtDNA and the precise roles of DSB handling therein will require further investigation.
A recently proposed but unexplored theory suggests that mitochondrial Top II may be involved in ageing [275]. With age, mtDNA undergoes attrition through entanglement and/or recombination such that mtDNA deletion is increasingly observed over time. These deletion events are found to contribute to dysfunction of the respiratory chain [281] and other tissue impairments associated with ageing [282–287]. One hypothesis postulates that age-related attrition occurs as a result of mitochondrial “poisoning” by exogenous agents, resulting in dysregulation of topoisomerase II-mediated DSB handling [275]. Many such “poisons” disrupt the ligation step of Top II, resulting in accumulation of DSB-enzyme intermediates [275]. Mammalian mitochondrial Top IIb has been found to be inhibited by the anticancer drugs amsacrine and teniposide [288] and it is possible that various known nuclear Top II poisons, ranging from the fungal toxin alternariol [289] and chemotherapeutics [290] to dietary components such as bioflavonoids from fruit and vegetables [291], may also inhibit the truncated or full-length type II topoisomerases in mitochondria. Furthermore, base oxidations, abasic sites, and other exogenously induced base modifications can act, via an unknown mechanism, to abnormally enhance Top II DSB formation [292] and promote accumulation of DNA-enzyme intermediates at sites of damage [293]. Combined with the finding that DSBs in the mitochondrial genome can induce considerable mtDNA deletion [294], Sobek and Boege speculate that exposure to mitochondrial topoisomerase “poisons” over time with age could lead to potential DSB-dependent mtDNA attrition [275]. While it is likely that mitochondrial Top II performs similar DSB-related functions to its nuclear counterpart, further studies will be needed to elucidate the precise nature of its role in mitochondria and its contribution if any to the ageing process.
Further insight into the role of Top II-induced DSBs in mitochondria may be drawn from parallels in Kinetoplastids, a group of flagellated protozoa which includes the parasitic Trypanosoma and Leishmania species, which possess a characteristic dense granule of DNA called a kinetoplast within their single mitochondrion. The kinetoplast consists of an enormous network of catenated circular mtDNA (kDNA) comprising short minicircles and much larger maxicircles and thus represents a considerable topological challenge in replication and segregation [295]. The mitochondrion contains a type II topoisomerase which localises to the kDNA [296, 297] and has been shown to be important for the maintenance of structural integrity in the kinetoplast [298], mending of holes in the kDNA following individual minicircle release for replication [299], segregation of daughter minicircles [300], and their reattachment to the kDNA network [301].
4. Concluding Remarks and Future Prospects
DSB physiology is thus, at its very core, a molecular surgery, reliant on both calculated incisions and timely suturing thereof, all the while, under the intense scrutiny of numerous regulatory mechanisms. While the basic biochemical mechanisms of the processes discussed above are well established, their multilateral spatiotemporal regulation remains a field of active research.
The roles of recombination defects in human disease and tumorigenesis are being increasingly appreciated and further studies will be needed to examine the nature of the dysregulation of DSB physiology in pathogenesis. New work may reveal novel pharmacological targets for human disease such as the possibility of targeting DSB signalling and repair proteins in neurodegenerative conditions such as Alzheimer's disease, to lessen aberrant DSB formation.
Novel tissue-specific roles of topoisomerases in neurophysiology are only just being uncovered with future studies required to elucidate the function of DSBs therein to understand mechanisms of learning, memory, and cognition. In addition, future work is necessary to characterise the nature of mitochondrial DSB physiology and the as yet unexplored implications on mitochondrial metabolism, disease, and ageing.
Furthermore, both topoisomerases and cancer DSB repair deficiencies are important areas of investigation in the development of anticancer therapeutics, and an understanding of the physiological roles of these targets in multiple disparate processes will inform the design of specific inhibitors with fewer off-target effects.
This account illustrates the breadth of DSB physiology across cellular biology and adds a new dimension of complexity to the regulation of genomic transactions on a basic biochemical level. The emerging role of DSB-dependent changes in topology, not only in replication and basal transcription, but also in activity-induced transcriptional changes, and potential roles in the chromatin landscape and mitochondria paint a picture of a far more active role of DSBs in fundamental DNA metabolism than previously thought.
Acknowledgments
The authors are very grateful to Dr. Joseph Maman for his guidance and support during the production of this work and for his ever helpful advice whenever needed.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.O'Connor M. J. Targeting the DNA Damage Response in Cancer. Molecular Cell. 2015;60(4):547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 2.Jackson S. P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schipler A., Iliakis G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Research. 2013;41(16):7589–7605. doi: 10.1093/nar/gkt556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeggo P. A., Löbrich M. DNA double-strand breaks: their cellular and clinical impact? Oncogene. 2007;26(56):7717–7719. doi: 10.1038/sj.onc.1210868. [DOI] [PubMed] [Google Scholar]
- 5.Brandsma I., Gent D. C. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integrity. 2012;3, article 9 doi: 10.1186/2041-9414-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth D. B. V(D)J Recombination: Mechanism, Errors, and Fidelity. Microbiology Spectrum. 2014;2(6) doi: 10.1128/microbiolspec.MDNA3-0041-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z., Zan H., Pone E. J., Mai T., Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nature Reviews Immunology. 2012;12(7):517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam I., Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harbor Perspectives in Biology. 2015;7(1) doi: 10.1101/cshperspect.a016634.a016634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas I., Germe T., Chevrier-Miller M., Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO Journal. 2001;20(22):6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxter J., Diffley J. F. X. Topoisomerase II Inactivation Prevents the Completion of DNA Replication in Budding Yeast. Molecular Cell. 2008;30(6):790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 11.French S. L., Sikes M. L., Hontz R. D., et al. Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Molecular and Cellular Biology. 2011;31(3):482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madabhushi R., Gao F., Pfenning A. R. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161(7):1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari V. K., Burger L., Nikoletopoulou V., et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proceedings of the National Acadamy of Sciences of the United States of America. 2012;109(16):E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyu Y. L., Lin C.-P., Azarova A. M., Cai L., Wang J. C., Liu L. F. Role of topoisomerase IIβ in the expression of developmentally regulated genes. Molecular and Cellular Biology. 2006;26(21):7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakurela S., Garding A., Jung J., Schübeler D., Burger L., Tiwari V. K. Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nature Communications. 2013;4, article 2478 doi: 10.1038/ncomms3478. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez R. E., Lim C.-U., Cole K., et al. Effects of conditional depletion of topoisomerase II on cell cycle progression in mammalian cells. Cell Cycle. 2011;10(20):3505–3514. doi: 10.4161/cc.10.20.17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vas A. C. J., Andrews C. A., Matesky K. K., Clarke D. J. In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Molecular Biology of the Cell (MBoC) 2007;18(2):557–568. doi: 10.1091/mbc.E06-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deweese J. E., Osheroff N. The DNA cleavage reaction of topoisomerase II: Wolf in sheep's clothing. Nucleic Acids Research. 2009;37(3):738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-Herreros F., Schuurs-Hoeijmakers J. H. M., McCormack M., et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nature Genetics. 2014;46(5):516–521. doi: 10.1038/ng.2929. [DOI] [PubMed] [Google Scholar]
- 20.Jung D., Alt F. W. Unraveling V(D)J recombination: insights into gene regulation. Cell. 2004;116(2):299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- 21.Akira S., Okazaki K., Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(d)-J joining. Science. 1987;238(4830):1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- 22.Ramsden D. A., Baetz K., Wu G. E. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Research. 1994;22(10):1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBlane J. F., van Gent D. C., Ramsden D. A., et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83(3):387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 24.Swanson P. C., Desiderio S. V(D)J recombination signal recognition: Distinct, overlapping DNA- protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9(1):115–125. doi: 10.1016/S1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 25.Hiom K., Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88(1):65–72. doi: 10.1016/S0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 26.Mo X., Bailin T., Sadofsky M. J. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. The Journal of Biological Chemistry. 1999;274(11):7025–7031. doi: 10.1074/jbc.274.11.7025. [DOI] [PubMed] [Google Scholar]
- 27.Eastman Q. M., Leu T. M. J., Schatz D. G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380(6569):85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 28.Štros M. HMGB proteins: Interactions with DNA and chromatin. Biochimica et Biophysica Acta. 2010;1799(1-2):101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Swanson P. C. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Molecular and Cellular Biology. 2002;22(5):1340–1351. doi: 10.1128/MCB.22.5.1340-1351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawchuk D. J., Weis-Garcia F., Malik S., et al. V(D)J recombination: Modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. The Journal of Experimental Medicine. 1997;185(11):2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aidinis V., Bonaldi T., Beltrame M., Santagata S., Bianchi M. E., Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Molecular and Cellular Biology. 1999;19(10):6532–6542. doi: 10.1128/MCB.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodgers K. K., Villey I. J., Ptaszek L., Corbett E., Schatz D. G., Coleman J. E. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Research. 1999;27(14):2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Gent D. C., Hiom K., Paull T. T., Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO Journal. 1997;16(10):2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Difilippantonio M. J., McMahan C. J., Eastman Q. M., Spanopoulou E., Schatz D. G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87(2):253–262. doi: 10.1016/S0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 35.Akamatsu Y., Oettinger M. A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Molecular and Cellular Biology. 1998;18(8):4670–4678. doi: 10.1128/MCB.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Gent D. C., Mizuuchi K., Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271(5255):1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 37.Agrawal A., Schatz D. G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89(1):43–53. doi: 10.1016/S0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y., Pannicke U., Schwarz K., Lieber M. R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108(6):781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 39.Schlissel M. S. Does Artemis end the hunt for the hairpin-opening activity in V(D)J recombination? Cell. 2002;109(1):1–4. doi: 10.1016/S0092-8674(02)00694-3. [DOI] [PubMed] [Google Scholar]
- 40.Besmer E., Mansilla-Soto J., Cassard S., et al. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Molecular Cell. 1998;2(6):817–828. doi: 10.1016/S1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 41.Shockett P. E., Schatz D. G. DNA hairpin opening mediated by the RAG21 and RAG2 proteins. Molecular and Cellular Biology. 1999;19(6):4159–4166. doi: 10.1128/MCB.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santagata S., Besmer E., Villa A., et al. The RAG1/RAG2 complex constitutes a 3' flap endonuclease: Implications for junctional diversity in V(D)J and transpositional recombination. Molecular Cell. 1999;4(6):935–947. doi: 10.1016/S1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- 43.Tsai C.-L., Drejer A. H., Schatz D. G. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes & Development. 2002;16(15):1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis S. M. The Mechanism of V(D)J Joining: Lessons from Molecular, Immunological, and Comparative Analyses. Advances in Immunology. 1994;56(C):27–150. doi: 10.1016/S0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 45.Gauss G. H., Lieber M. R. Mechanistic constraints on diversity in human V(D)J recombination. Molecular and Cellular Biology. 1996;16(1):258–269. doi: 10.1128/MCB.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Critchlow S. E., Bowater R. P., Jackson S. P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Current Biology. 1997;7(8):588–598. doi: 10.1016/S0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 47.Grawunder U., Wilm M., Wu X., et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388(6641):492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 48.Costantini S., Woodbine L., Andreoli L., Jeggo P. A., Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair. 2007;6(6):712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Mickelsen S., Snyder C., Trujillo K., Bogue M., Roth D. B., Meek K. Modulation of terminal deoxynucleotidyltransferase activity by the DNA- dependent protein kinase. The Journal of Immunology. 1999;163(2):834–843. [PubMed] [Google Scholar]
- 50.McElhinny S. A. N., Havener J. M., Garcia-Diaz M., et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Molecular Cell. 2005;19(3):357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Bertocci B., De Smet A., Weill J.-C., Reynaud C.-A. Nonoverlapping Functions of DNA Polymerases Mu, Lambda, and Terminal Deoxynucleotidyltransferase during Immunoglobulin V(D)J Recombination In Vivo. Immunity. 2006;25(1):31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 52.Ahnesorg P., Smith P., Jackson S. P. XLF interacts with the XRCC4-DNA Ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124(2):301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 53.Li Z., Dordai D. I., Lee J., Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5(6):575–589. doi: 10.1016/S1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 54.Lin W. C., Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proceedings of the National Acadamy of Sciences of the United States of America. 1994;91(7):2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L., Reynolds T. L., Shan X., Desiderio S. Coupling of V(D)J Recombination to the Cell Cycle Suppresses Genomic Instability and Lymphoid Tumorigenesis. Immunity. 2011;34(2):163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanhope-Baker P., Hudson K. M., Shaffer A. L., Constantinescu A., Schlissel M. S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85(6):887–897. doi: 10.1016/S0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 57.Yancopoulos G. D., Alt F. W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 58.Lewis S. M., Agard E., Suh S., Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Molecular and Cellular Biology. 1997;17(6):3125–3136. doi: 10.1128/MCB.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sleckman B. P., Gorman J. R., Alt F. W. Accessibility control of antigen-receptor variable-region gene assembly: Role of cis-acting elements. Annual Review of Immunology. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 60.McBlane F., Boyes J. Stimulation of V(D)J recombination by histone acetylation. Current Biology. 2000;10(8):483–486. doi: 10.1016/S0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 61.Kwon J., Imbalzano A. N., Matthews A., Oettinger M. A. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Molecular Cell. 1998;2(6):829–839. doi: 10.1016/S1097-2765(00)80297-X. [DOI] [PubMed] [Google Scholar]
- 62.Golding A., Chandler S., Ballestar E., Wolffe A. P., Schlissel M. S. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO Journal. 1999;18(13):3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roth D. B., Roth S. Y. Unequal access: Regulating V(D)J recombination through chromatin remodeling. Cell. 2000;103(5):699–702. doi: 10.1016/S0092-8674(00)00173-2. [DOI] [PubMed] [Google Scholar]
- 64.Schatz D. G., Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nature Reviews Immunology. 2011;11(4):251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 65.Matthews A. G. W., Kuo A. J., Ramón-Maiques S., et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450(7172):1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimazaki N., Tsai A. G., Lieber M. R. H3K4me3 Stimulates the V(D)J RAG Complex for Both Nicking and Hairpinning in trans in Addition to Tethering in cis: Implications for Translocations. Molecular Cell. 2009;34(5):535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mombaerts P., Iacomini J., Johnson R. S., Herrup K., Tonegawa S., Papaioannou V. E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-G. [DOI] [PubMed] [Google Scholar]
- 68.Shinkai Y., Lam K.-P., Oltz E. M., Stewart V., Mendelsohn M., Charron J., et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-C. [DOI] [PubMed] [Google Scholar]
- 69.Villa A., Santagata S., Bozzi F., et al. Partial V(D)J recombination activity leads to omenn syndrome. Cell. 1998;93(5):885–896. doi: 10.1016/S0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 70.Wang J., Pluth J. M., Cooper P. K., Cowan M. J., Chen D. J., Yannone S. M. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair. 2005;4(5):556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 71.Gao Y., Chaudhuri J., Zhu C., Davidson L., Weaver D. T., Alt F. W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9(3):367–376. doi: 10.1016/S1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 72.Rooney S., Sekiguchi J., Zhu C., et al. Leaky Scid Phenotype Associated with Defective V(D)J Coding End Processing in Artemis-Deficient Mice. Molecular Cell. 2002;10(6):1379–1390. doi: 10.1016/S1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 73.Ege M., Ma Y., Manfras B., et al. Omenn syndrome due to ARTEMIS mutations. Blood. 2005;105(11):4179–4186. doi: 10.1182/blood-2004-12-4861. [DOI] [PubMed] [Google Scholar]
- 74.Nussenzweig A., Sokol K., Burgman P., Li L., Li G. C. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: The effects of ionizing radiation on growth, survival, and development. Proceedings of the National Acadamy of Sciences of the United States of America. 1997;94(25):13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu Y., Seidl K. J., Rathbun G. A., et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7(5):653–665. doi: 10.1016/S1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 76.Jin S., Weaver D. T. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO Journal. 1997;16(22):6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao Y., Ferguson D. O., Xie W., et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404(6780):897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 78.Frank K. M., Sharpless N. E., Gao Y., et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Molecular Cell. 2000;5(6):993–1002. doi: 10.1016/S1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 79.Gao Y., Sun Y., Frank K. M., et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95(7):891–902. doi: 10.1016/S0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 80.Taub R., Kirsch I., Morton C., et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proceedings of the National Acadamy of Sciences of the United States of America. 1982;79(24 I):7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monni O., Oinonen R., Elonen E., et al. Gain of 3q and deletion of 11q22 are frequent aberrations in mantle cell lymphoma. Genes, Chromosomes and Cancer. 1998;21(4):298–307. doi: 10.1002/(SICI)1098-2264(199804)21:4<298::AID-GCC3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 82.Baer R., Chen K.-C., Smith S. D., Rabbitts T. H. Fusion of an immunoglobulin variable gene and a T cell receptor constant gene in the chromosome 14 inversion associated with T cell tumors. Cell. 1985;43(3):705–713. doi: 10.1016/0092-8674(85)90243-0. [DOI] [PubMed] [Google Scholar]
- 83.Bende R. J., Smit L. A., van Noesel C. J. M. Molecular pathways in follicular lymphoma. Leukemia. 2007;21(1):18–29. doi: 10.1038/sj.leu.2404426. [DOI] [PubMed] [Google Scholar]
- 84.Raghavan S. C., Swanson P. C., Wu X., Hsieh C.-L., Lieber M. R. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428(6978):88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 85.Cui X., Lu Z., Kurosawa A., et al. Both CpG methylation and activation-induced deaminase are required for the fragility of the human bcl-2 major breakpoint region: Implications for the timing of the breaks in the t(14;18) translocation. Molecular and Cellular Biology. 2013;33(5):947–957. doi: 10.1128/MCB.01436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roth D. B. Restraining the V(D)J recombinase. Nature Reviews Immunology. 2003;3(8):656–666. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 87.Chaudhuri J., Alt F. W. Class-switch recombination: Interplay of transcription, DNA deamination and DNA repair. Nature Reviews Immunology. 2004;4(7):541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 88.Dunnick W., Hertz G. Z., Scappino L., Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Research. 1993;21(3):365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rush J. S., Fugmann S. D., Schatz D. G. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to Sμ in Ig class switch recombination. International Immunology. 2004;16(4):549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- 90.Catalan N., Selz F., Imai K., Revy P., Fischer A., Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch μ region. The Journal of Immunology. 2003;171(5):2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 91.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 92.Petersen-Mahrt S. K., Harris R. S., Neuberger M. S. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 93.Schrader C. E., Linehan E. K., Mochegova S. N., Woodland R. T., Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. The Journal of Experimental Medicine. 2005;202(4):561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rada C., Williams G. T., Nilsen H., Barnes D. E., Lindahl T., Neuberger M. S. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Current Biology. 2002;12(20):1748–1755. doi: 10.1016/S0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 95.Guikema J. E. J., Linehan E. K., Tsuchimoto D., et al. APE1- And APE2-dependent DNA breaks in immunoglobulin class switch recombination. The Journal of Experimental Medicine. 2007;204(12):3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stavnezer J., Schrader C. E. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends in Genetics. 2006;22(1):23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 97.Bastianello G., Arakawa H. A double-strand break can trigger immunoglobulin gene conversion. Nucleic Acids Research. 2017;45(1):231–243. doi: 10.1093/nar/gkw887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rada C., Di Noia J. M., Neuberger M. S. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Molecular Cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 99.Ehrenstein M. R., Neuberger M. S. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: Parallels with somatic hypermutation. EMBO Journal. 1999;18(12):3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martin A., Li Z., Lin D. P., et al. Msh2 ATPase Activity Is Essential for Somatic Hypermutation at A-T Basepairs and for Efficient Class Switch Recombination. The Journal of Experimental Medicine. 2003;198(8):1171–1178. doi: 10.1084/jem.20030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martomo S. A., Yang W. W., Gearhart P. J. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. The Journal of Experimental Medicine. 2004;200(1):61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schrader C. E., Vardo J., Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. The Journal of Experimental Medicine. 2002;195(3):367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen H. M., Tanaka A., Bozek G., Nicolae D., Storb U. Somatic hypermutation and class switch recombination in Msh6 -/-Ung-/- double-knockout mice. The Journal of Immunology. 2006;177(8):5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- 104.Bardwell P. D., Woo C. J., Wei K., et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nature Immunology. 2004;5(2):224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 105.Stavnezer J., Guikema J. E. J., Schrader C. E. Mechanism and regulation of class switch recombination. Annual Review of Immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matthews A. J., Zheng S., DiMenna L. J., Chaudhuri J. Regulation of immunoglobulin class-switch recombination: Choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Advances in Immunology. 2014;122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Casellas R., Nussenzweig A., Wuerffel R., et al. Ku80 is required for immunoglobulin isotype switching. EMBO Journal. 1998;17(8):2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Manis J. P., Gu Y., Lansford R., et al. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. The Journal of Experimental Medicine. 1998;187(12):2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. The Journal of Experimental Medicine. 2007;204(7):1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han L., Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. The Journal of Experimental Medicine. 2008;205(12):2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bottaro A., Lansford R., Xu L., Zhang J., Rothman P., Alt F. W. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO Journal. 1994;13(3):665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jung S., Rajewsky K., Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259(5097):984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 113.Kinoshita K., Tashiro J., Tomita S., Lee C.-G., Honjo T. Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequences of S regions. Immunity. 1998;9(6):849–858. doi: 10.1016/S1074-7613(00)80650-0. [DOI] [PubMed] [Google Scholar]
- 114.Nambu Y., Sugai M., Gonda H., et al. Transcription-Coupled Events Associating with Immunoglobulin Switch Region Chromatin. Science. 2003;302(5653):2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 115.Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422(6933):726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 116.Ronai D., Iglesias-Ussel M. D., Fan M., Li Z., Martin A., Scharff M. D. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. The Journal of Experimental Medicine. 2007;204(1):181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang F.-T., Yu K., Balter B. B., et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Sμ class switch region. Molecular and Cellular Biology. 2007;27(16):5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu K., Chedin F., Hsieh C.-L., Wilson T. E., Lieber M. R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunology. 2003;4(5):442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 119.Pavri R., Gazumyan A., Jankovic M., et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143(1):122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang L., Wuerffel R., Feldman S., Khamlichi A. A., Kenter A. L. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. The Journal of Experimental Medicine. 2009;206(8):1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang L., Whang N., Wuerffel R., Kenter A. L. AID-dependent histone acetylation is detected in immunoglobulin S regions. The Journal of Experimental Medicine. 2006;203(1):215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ohkura H. Meiosis: An overview of key differences from mitosis. Cold Spring Harbor Perspectives in Biology. 2015;7(5):1–15. doi: 10.1101/cshperspect.a015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bishop D. K., Zickler D. Early decision: Meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117(1):9–15. doi: 10.1016/S0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 124.Keeney S., Giroux C. N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–384. doi: 10.1016/S0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 125.Liu J., Wu T.-C., Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: Evidence for a covalently linked DNA-protein intermediate. EMBO Journal. 1995;14(18):4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Keeney S., Neale M. J. Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochemical Society Transactions. 2006;34(4):523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 127.Marston A. L., Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nature Reviews Molecular Cell Biology. 2004;5(12):983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 128.Romanienko P. J., Camerini-Otero R. D. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61(2):156–169. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 129.Romanienko P. J., Camerini-Otero R. D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular Cell. 2000;6(5):975–987. doi: 10.1016/S1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 130.Arora C., Kee K., Maleki S., Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Molecular Cell. 2004;13(4):549–559. doi: 10.1016/S1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 131.Kee K., Protacio R. U., Arora C., Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO Journal. 2004;23(8):1815–1824. doi: 10.1038/sj.emboj.7600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kee K., Keeney S. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics. 2002;160(1):111–122. doi: 10.1093/genetics/160.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sasanuma H., Murakami H., Fukuda T., Shibata T., Nicolas A., Ohta K. Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic Acids Research. 2007;35(4):1119–1133. doi: 10.1093/nar/gkl1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li J., Hooker G. W., Roeder G. S. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173(4):1969–1981. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Neale M. J., Pan J., Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436(7053):1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Usui T., Ohta T., Oshiumi H., Tomizawa J.-I., Ogawa H., Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95(5):705–716. doi: 10.1016/S0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 137.Haber J. E. The many interfaces of Mre11. Cell. 1998;95(5):583–586. doi: 10.1016/S0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 138.Manfrini N., Guerini I., Citterio A., Lucchini G., Longhese M. P. Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. The Journal of Biological Chemistry. 2010;285(15):11628–11637. doi: 10.1074/jbc.M110.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bressan D. A., Baxter B. K., Petrini J. H. J. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1999;19(11):7681–7687. doi: 10.1128/MCB.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Andersen S. L., Sekelsky J. Meiotic versus mitotic recombination: Two different routes for double-strand break repair: The different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays. 2010;32(12):1058–1066. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun H., Treco D., Szostak J. W. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 142.Baudat F., Imai Y., De Massy B. Meiotic recombination in mammals: Localization and regulation. Nature Reviews Genetics. 2013;14(11):794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 143.Neale M. J., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442(7099):153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fraune J., Schramm S., Alsheimer M., Benavente R. The mammalian synaptonemal complex: Protein components, assembly and role in meiotic recombination. Experimental Cell Research. 2012;318(12):1340–1346. doi: 10.1016/j.yexcr.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 145.Hunter N. Meiotic recombination: The essence of heredity. Cold Spring Harbor Perspectives in Biology. 2015;7(12) doi: 10.1101/cshperspect.a016618.a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Borde V., Goldman A. S. H., Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290(5492):806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 147.Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 148.Mahadevaiah S. K., Turner J. M. A., Baudat F., et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genetics. 2001;27(3):271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 149.Jang J. K., Sherizen D. E., Bhagat R., Manheim E. A., McKim K. S. Relationship of DNA double-strand breaks to synapsis in Drosophila. Journal of Cell Science. 2003;116(15):3069–3077. doi: 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- 150.Murakami H., Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes & Development. 2008;22(3):286–292. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Odorisio T., Rodriguez T. A., Evans E. P., Clarke A. R., Burgoyne P. S. The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nature Genetics. 1998;18(3):257–261. doi: 10.1038/ng0398-257. [DOI] [PubMed] [Google Scholar]
- 152.Lydall D., Nikolsky Y., Bishop D. K., Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383(6603):841–844. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 153.Roeder G. S., Bailis J. M. The pachytene checkpoint. Trends in Genetics. 2000;16(9):395–403. doi: 10.1016/S0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 154.Hochwagen A., Amon A. Checking your breaks: Surveillance mechanisms of meiotic recombination. Current Biology. 2006;16(6):R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 155.Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S. Cyclin-Dependent Kinase Directly Regulates Initiation of Meiotic Recombination. Cell. 2006;125(7):1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stuart D., Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes & Development. 1998;12(17):2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wan L., Niu H., Futcher B., et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes & Development. 2008;22(3):386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sasanuma H., Hirota K., Fukuda T., et al. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes & Development. 2008;22(3):398–410. doi: 10.1101/gad.1626608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Matos J., Lipp J. J., Bogdanova A., et al. Dbf4-Dependent Cdc7 Kinase Links DNA Replication to the Segregation of Homologous Chromosomes in Meiosis I. Cell. 2008;135(4):662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 160.Murakami H., Keeney S. Temporospatial coordination of meiotic dna replication and recombination via DDK recruitment to replisomes. Cell. 2014;158(4):861–873. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Engebrecht J., Voelkel-Meiman K., Roeder G. S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66(6):1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 162.Engebrecht J., Roeder G. S. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Molecular and Cellular Biology. 1990;10(5):2379–2389. doi: 10.1128/MCB.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lichten M., Goldman A. S. H. Meiotic recombination hotspots. Annual Review of Genetics. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 164.Gerton J. L., DeRisi J., Shroff R., Lichten M., Brown P. O., Petes T. D. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proceedings of the National Acadamy of Sciences of the United States of America. 2000;97(21):11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sasaki M., Lange J., Keeney S. Genome destabilization by homologous recombination in the germ line. Nature Reviews Molecular Cell Biology. 2010;11(3):182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rockmill B., Voelkel-Meiman K., Roeder G. S. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174(4):1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Buard J., Barthès P., Grey C., De Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO Journal. 2009;28(17):2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Smagulova F., Gregoretti I. V., Brick K., Khil P., Camerini-Otero R. D., Petukhova G. V. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472(7343):375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baudat F., Buard J., Grey C., et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Parvanov E. D., Petkov P. M., Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327(5967):p. 835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hayashi K., Yoshida K., Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438(7066):374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 172.Billings T., Parvanov E. D., Baker C. L., Walker M., Paigen K., Petkov P. M. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biology. 2013;14(4, article R35) doi: 10.1186/gb-2013-14-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Grey C., Barthès P., Friec G., Langa F., Baudat F., de Massy B. Mouse Prdm9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biology. 2011;9(10) doi: 10.1371/journal.pbio.1001176.e1001176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Myers S., Bowden R., Tumian A., et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327(5967):876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., Keeney S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes & Development. 2013;27(8):873–886. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schwacha A., Kleckner N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90(6):1123–1135. doi: 10.1016/S0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 177.Joshi N., Brown M. S., Bishop D. K., Börner G. V. Gradual Implementation of the Meiotic Recombination Program via Checkpoint Pathways Controlled by Global DSB Levels. Molecular Cell. 2015;57(5):797–811. doi: 10.1016/j.molcel.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kleckner N. Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115(3):175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 179.Panizza S., Mendoza M. A., Berlinger M., et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146(3):372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 180.Berger J. M., Gamblin S. J., Harrison S. C., Wang J. C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379(6562):225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 181.Schoeffler A. J., Berger J. M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Quarterly Reviews of Biophysics. 2008;41(1):41–101. doi: 10.1017/s003358350800468x. [DOI] [PubMed] [Google Scholar]
- 182.Roca J. The mechanisms of DNA topoisomerases. Trends in Biochemical Sciences. 1995;20(4):156–160. doi: 10.1016/S0968-0004(00)88993-8. [DOI] [PubMed] [Google Scholar]
- 183.Lang A. J., Mirski S. E. L., Cummings H. J., Yu Q., Gerlach J. H., Cole S. P. C. Structural organization of the human TOP2A and TOP2B genes. Gene. 1998;221(2):255–266. doi: 10.1016/S0378-1119(98)00468-5. [DOI] [PubMed] [Google Scholar]
- 184.Austin C. A., Marsh K. L. Eukaryotic DNA topoisomerase IIβ. BioEssays. 1998;20(3):215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 185.Null A. P., Hudson J., Gorbsky G. J. Both alpha and beta isoforms of mammalian DNA topoisomerase II associate with chromosomes in mitosis. Cell Growth & Differentiation: The Molecular Biology Journal of the American Association for Cancer Research American Association for Cancer Research. 2002;13(7):325–333. [PubMed] [Google Scholar]
- 186.Chaly N., Chen X., Dentry J., Brown D. L. Organization of DNA topoisomerase II isotypes during the cell cycle of human lymphocytes and HeLa cells. Chromosome Research. 1996;4(6):457–466. doi: 10.1007/BF02265053. [DOI] [PubMed] [Google Scholar]
- 187.Meczes E. L., Gilroy K. L., West K. L., Austin C. A. The impact of the human DNA topoisomerse II C-terminal domain on activity. PLoS ONE. 2008;3(3) doi: 10.1371/journal.pone.0001754.e1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Linka R. M., Porter A. C. G., Volkov A., Mielke C., Boege F., Christensen M. O. C-Terminal regions of topoisomerase IIα and IIβ determine isoform-specific functioning of the enzymes in vivo. Nucleic Acids Research. 2007;35(11):3810–3822. doi: 10.1093/nar/gkm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Turley H., Comley M., Houlbrook S., et al. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. British Journal of Cancer. 1997;75(9):1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bauman M. E., Holden J. A., Brown K. A., Harker W. G., Perkins S. L. Differential immunohistochemical staining for DNA topoisomerase II alpha and beta in human tissues and for DNA topoisomerase II beta in non-Hodgkin’s lymphomas. Modern Pathology: An Official Journal of the United States and Canadian Academy of Pathology. 1997;10(3):168–175. [PubMed] [Google Scholar]
- 191.Ju B.-G., Lunyak V. V., Perissi V., et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312(5781):1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 192.Lewis R. J., Tsai F. T. F., Wigley D. B. Molecular mechanisms of drug inhibition of DNA gyrase. BioEssays. 1996;18(8):661–671. doi: 10.1002/bies.950180810. [DOI] [PubMed] [Google Scholar]
- 193.Tse-Dinh Y.-C. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infectious Disorders - Drug Targets. 2007;7(1):3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 194.Salerno S., Da Settimo F., Taliani S., et al. Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs. Current Medicinal Chemistry. 2010;17(35):4270–4290. doi: 10.2174/092986710793361252. [DOI] [PubMed] [Google Scholar]
- 195.Li T.-K., Liu L. F. Tumor cell death induced by topoisomerase-targeting drugs. Annual Review of Pharmacology and Toxicology. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 196.Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chemistry & Biology. 2010;17(5):421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang J. C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Reviews Molecular Cell Biology. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 198.Champoux J. J. DNA topoisomerases: structure, function, and mechanism. Annual Review of Biochemistry. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 199.Chen S. H., Chan N.-L., Hsieh T.-S. New mechanistic and functional insights into DNA topoisomerases. Annual Review of Biochemistry. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 200.Limoli C. L., Giedzinski E., Bonner W. M., Cleaver J. E. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA doublestrand breaks, γ-H2AX formation, and Mre11 relocalization. Proceedings of the National Acadamy of Sciences of the United States of America. 2002;99(1):233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Michel B., Ehrlich S. D., Uzest M. DNA double-strand breaks caused by replication arrest. EMBO Journal. 1997;16(2):430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. Journal of Molecular Biology. 2001;307(5):1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 203.Lee J.-H., Paull T. T. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 204.Khanna K. K., Jackson S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genetics. 2001;27(3):247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 205.Montecucco A., Zanetta F., Biamonti G. Molecular mechanisms of etoposide. EXCLI Journal. 2015;14:95–108. doi: 10.17179/excli2014-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jekimovs C., Bolderson E., Suraweera A., Adams M., O'Byrne K. J., Richard D. J. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: The good, the bad, and the promising. Frontiers in Oncology. 2014;4 doi: 10.3389/fonc.2014.00086.Article 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Brill S. J., Dinardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication and for transcription of ribosomal RNA. Nature. 1987;326(6115) doi: 10.1038/326812a0. [DOI] [PubMed] [Google Scholar]
- 208.Nitiss J. L. DNA topoisomerase II and its growing repertoire of biological functions. Nature Reviews Cancer. 2009;9(5):327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kim R. A., Wang J. C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. Journal of Molecular Biology. 1989;208(2):257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- 210.Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: Dissection of the final stages of SV40 DNA replication. Cell. 1981;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 211.Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41(2):553–563. doi: 10.1016/S0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 212.Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 213.DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: Topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proceedings of the National Acadamy of Sciences of the United States of America. 1984;81(9):2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Holm C., Stearns T., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Molecular and Cellular Biology. 1989;9(1):159–168. doi: 10.1128/MCB.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Carpenter A. J., Porter A. C. G. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIα mutant human cell line. Molecular Biology of the Cell (MBoC) 2004;15(12):5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proceedings of the National Acadamy of Sciences of the United States of America. 1987;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wu H.-Y., Shyy S., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 218.Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 219.Schultz M. C., Brill S. J., Ju Q., Sternglanz R., Reeder R. H. Topoisomerases and yeast rRNA transcription: Negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes & Development. 1992;6(7):1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 220.Ray S., Panova T., Miller G., et al. Topoisomerase IIα promotes activation of RNA polymerase i transcription by facilitating pre-initiation complex formation. Nature Communications. 2013;4, article 1598 doi: 10.1038/ncomms2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Joshi R. S., Piña B., Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Research. 2012;40(16):7907–7915. doi: 10.1093/nar/gks626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bunch H., Lawney B. P., Lin Y.-F., et al. Transcriptional elongation requires DNA break-induced signalling. Nature Communications. 2015;6 doi: 10.1038/ncomms10191.10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nickoloff J. A., Reynolds R. J. Transcription stimulates homologous recombination in mammalian cells. Molecular and Cellular Biology. 1990;10(9):4837–4845. doi: 10.1128/MCB.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.González-Barrera S., García-Rubio M., Aguilera A. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics. 2002;162(2):603–614. doi: 10.1093/genetics/162.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gottipati P., Cassel T. N., Savolainen L., Helleday T. Transcription-associated recombination is dependent on replication in mammalian cells. Molecular and Cellular Biology. 2008;28(1):154–164. doi: 10.1128/MCB.00816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Aymard F., Bugler B., Schmidt C. K., et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nature Structural & Molecular Biology. 2014;21(4):366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Trotter K. W., King H. A., Archer T. K. Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2β and Ku70/86. Molecular and Cellular Biology. 2015;35(16):2799–2817. doi: 10.1128/MCB.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.West A. E., Greenberg M. E. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor Perspectives in Biology. 2011;3(6):1–21. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sheng M., Greenberg M. E. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4(4):477–485. doi: 10.1016/0896-6273(90)90106-P. [DOI] [PubMed] [Google Scholar]
- 230.Yang J., Bogni A., Schuetz E. G., et al. Etoposide pathway. Pharmacogenetics and Genomics. 2009;19(7):552–553. doi: 10.1097/FPC.0b013e32832e0e7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kuo L. J., Yang L.-X. γ-H2AX-a novel biomarker for DNA double-strand breaks. In Vivo (Brooklyn) 2008;22(3):305–309. [PubMed] [Google Scholar]
- 232.Suberbielle E., Sanchez P. E., Kravitz A. V., et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nature Neuroscience. 2013;16(5):613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ghirlando R., Felsenfeld G. CTCF: Making the right connections. Genes & Development. 2016;30(8):881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Uusküla-Reimand L., Hou H., Samavarchi-Tehrani P., et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biology. 2016;17(1, article 182) doi: 10.1186/s13059-016-1043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Newman A. G., Bessa P., Tarabykin V., Singh P. B. Activity-DEPendent Transposition. EMBO Reports. 2017;18(3):346–348. doi: 10.15252/embr.201643797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hendrickx A., Pierrot N., Tasiaux B., et al. Epigenetic regulations of immediate early genes expression involved in memory formation by the amyloid precursor protein of alzheimer disease. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0099467.e99467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ledesma F. C., El Khamisy S. F., Zuma M. C., Osborn K., Caldecott K. W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461(7264):674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 238.Gómez-Herreros F., Romero-Granados R., Zeng Z., et al. TDP2-Dependent Non-Homologous End-Joining Protects against Topoisomerase II-Induced DNA Breaks and Genome Instability in Cells and In Vivo. PLoS Genetics. 2013;9(3) doi: 10.1371/journal.pgen.1003226.e1003226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Madabhushi R., Pan L., Tsai L.-H. DNA damage and its links to neurodegeneration. Neuron. 2014;83(2):266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rao K. S. Mechanisms of disease: DNA repair defects and neurological disease. Nature Clinical Practice Neurology. 2007;3(3):162–172. doi: 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- 241.Taylor A. M. R., Byrd P. J. Molecular pathology of ataxia telangiectasia. Journal of Clinical Pathology. 2005;58(10):1009–1015. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Taylor A. M. R., Groom A., Byrd P. J. Ataxia-telangiectasia-like disorder (ATLD) - Its clinical presentation and molecular basis. DNA Repair. 2004;3(8-9):1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 243.Fernet M., Gribaa M., Salih M. A. M., Seidahmed M. Z., Hall J., Koenig M. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Human Molecular Genetics. 2005;14(2):307–318. doi: 10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 244.Stewart G. S., Maser R. S., Stankovic T., et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99(6):577–587. doi: 10.1016/S0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 245.The International Nijmegen Breakage Syndrome Study Group. Nijmegen breakage syndrome. Archives of Disease in Childhood. 2000;82(5):400–406. doi: 10.1136/adc.82.5.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Borgesius N. Z., de Waard M. C., van der Pluijm I., et al. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. The Journal of Neuroscience. 2011;31(35):12543–12553. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.De Waard M. C., Van Der Pluijm I., Zuiderveen Borgesius N., et al. Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathologica. 2010;120(4):461–475. doi: 10.1007/s00401-010-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Adamec E., Vonsattel J. P., Nixon R. A. DNA strand breaks in Alzheimer's disease. Brain Research. 1999;849(1-2):67–77. doi: 10.1016/s0006-8993(99)02004-1. [DOI] [PubMed] [Google Scholar]
- 249.Mullaart E., Boerrigter M. E. T. I., Ravid R., Swaab D. F., Vijg J. Increased levels of DNA breaks in cerebral cortex of alzheimer's disease patients. Neurobiology of Aging. 1990;11(3):169–173. doi: 10.1016/0197-4580(90)90542-8. [DOI] [PubMed] [Google Scholar]
- 250.Murphy M. P., LeVine H. Alzheimer’s disease and the amyloid-β peptide. Journal of Alzheimer's Disease. 2010;19(1):311–323. [Google Scholar]
- 251.Liu Y., West S. C. Distinct functions of BRCA1 and BRCA2 in double-strand break repair. Breast Cancer Research. 2002;4(1):9–13. doi: 10.1186/bcr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang J., Powell S. N. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Molecular Cancer Research. 2005;3(10):531–539. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 253.Suberbielle E., Djukic B., Evans M., et al. DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nature Communications. 2015;6, article 8897 doi: 10.1038/ncomms9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lam C.-W., Yeung W.-L., Law C.-Y. Global developmental delay and intellectual disability associated with a de novo TOP2B mutation. Clinica Chimica Acta. 2017;469:63–68. doi: 10.1016/j.cca.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 255.King I. F., Yandava C. N., Mabb A. M., et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501(7465):58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Marko D., Boege F. A possible link between nutritional uptake of ubiquitous topoisomerase inhibitors and autism? International Journal of Developmental Neuroscience. 2016;53:8–9. doi: 10.1016/j.ijdevneu.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 257.Vokálová L., Durdiaková J., Ostatníková D. Topoisomerases interlink genetic network underlying autism. International Journal of Developmental Neuroscience. 2015;47:361–368. doi: 10.1016/j.ijdevneu.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 258.Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. Journal of Molecular Biology. 1986;188(4):613–629. doi: 10.1016/S0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 259.Earnshaw W. C., Halligan B., Cooke C. A., Heck M. M. S., Liu L. F. Topoisomerase II is a structural component of mitotic chromosome scaffolds. The Journal of Cell Biology. 1985;100(5):1706–1715. doi: 10.1083/jcb.100.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 261.Razin S. V., Vassetzky Y. S., Hancock R. Nuclear matrix attachment regions and topoisomerase II binding and reaction sites in the vicinity of a chicken DNA replication origin. Biochemical and Biophysical Research Communications. 1991;177(1):265–270. doi: 10.1016/0006-291X(91)91977-K. [DOI] [PubMed] [Google Scholar]
- 262.Christensen M. O., Larsen M. K., Barthelmes H. U., et al. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. The Journal of Cell Biology. 2002;157(1):31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Peterson C. L. Chromatin remodeling enzymes: Taming the machines. Third in review series on chromatin dynamics. EMBO Reports. 2002;3(4):319–322. doi: 10.1093/embo-reports/kvf075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Adachi Y., Luke M., Laemmli U. K. Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell. 1991;64(1):137–148. doi: 10.1016/0092-8674(91)90215-K. [DOI] [PubMed] [Google Scholar]
- 265.Varga-Weisz P. D., Wilm M., Bonte E., Dumas K., Mann M., Becker P. B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388(6642):598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 266.Dykhuizen E. C., Hargreaves D. C., Miller E. L., et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497(7451):624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Bhat M. A., Philp A. V., Glover D. M., Bellen H. J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell. 1996;87(6):1103–1114. doi: 10.1016/S0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 268.Somma M. P., Fasulo B., Siriaco G., Cenci G. Chromosome Condensation Defects in barren RNA-Interfered Drosophila Cells. Genetics. 2003;165(3):1607–1611. doi: 10.1093/genetics/165.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Durand-Dubief M., Svensson J. P., Persson J., Ekwall K. Topoisomerases, chromatin and transcription termination. Transcription. 2011;2(2):66–70. doi: 10.4161/trns.2.2.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Durand-Dubief M., Persson J., Norman U., Hartsuiker E., Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO Journal. 2010;29(13):2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Walfridsson J., Khorosjutina O., Matikainen P., Gustafsson C. M., Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO Journal. 2007;26(12):2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Durrieu F., Samejima K., Fortune J. M., Kandels-Lewis S., Osheroff N., Earnshaw W. C. DNA topoisomerase IIα interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution. Current Biology. 2000;10(15):923–926. doi: 10.1016/S0960-9822(00)00620-5. [DOI] [PubMed] [Google Scholar]
- 273.Hizume K., Araki S., Yoshikawa K., Takeyasu K. Topoisomerase II, scaffold component, promotes chromatin compaction in a linker-histone H1-dependent manner. Nucleic Acids Research. 2007;35(8):2787–2799. doi: 10.1093/nar/gkm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Iborra F. J., Kimura H., Cook P. R. The functional organization of mitochondrial genomes in human cells. BMC Biology. 2004;2, article 9 doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sobek S., Boege F. DNA topoisomerases in mtDNA maintenance and ageing. Experimental Gerontology. 2014;56:135–141. doi: 10.1016/j.exger.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 276.Low R. L., Orton S., Friedman D. B. A truncated form of DNA topoisomerase IIβ associates with the mtDNA genome in mammalian mitochondria. European Journal of Biochemistry. 2003;270(20):4173–4186. doi: 10.1046/j.1432-1033.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 277.Sobek S., Rosa I. D., Pommier Y., et al. Negative regulation of mitochondrial transcription by mitochondrial topoisomerase i. Nucleic Acids Research. 2013;41(21):9848–9857. doi: 10.1093/nar/gkt768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zhang H., Zhang Y.-W., Yasukawa T., Dalla Rosa I., Khiati S., Pommier Y. Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Research. 2014;42(11):7259–7267. doi: 10.1093/nar/gku384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tsai H.-Z., Lin R.-K., Hsieh T.-S. Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. Journal of Biomedical Science. 2016;23(1, article 38) doi: 10.1186/s12929-016-0255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wu J., Feng L., Hsieh T.-S. Drosophila topo IIIα is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proceedings of the National Acadamy of Sciences of the United States of America. 2010;107(14):6228–6233. doi: 10.1073/pnas.1001855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Trounce I., Byrne E., Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. The Lancet. 1989;333(8639):637–639. doi: 10.1016/S0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- 282.Corral-Debrinski M., Horton T., Lott M. T., Shoffner J. M., Beal M. F., Wallace D. C. Mitochondrial DNA deletions in human brain: Regional variability and increase with advanced age. Nature Genetics. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 283.Bender A., Krishnan K. J., Morris C. M., et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genetics. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 284.Kraytsberg Y., Kudryavtseva E., McKee A. C., Geula C., Kowall N. W., Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nature Genetics. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 285.Hattori K., Tanaka M., Sugiyama S., et al. Age-dependent increase in deleted mitochondrial DNA in the human heart: Possible contributory factor to presbycardia. American Heart Journal. 1991;121(6):1735–1742. doi: 10.1016/0002-8703(91)90020-I. [DOI] [PubMed] [Google Scholar]
- 286.Cortopassi G. A., Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Research. 1990;18(23):6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yen T.-C., King K.-L., Lee H.-C., Yeh S.-H., Wei Y.-H. Age-dependent increase of mitochondrial DNA deletions together with lipid peroxides and superoxide dismutase in human liver mitochondria. Free Radical Biology & Medicine. 1994;16(2):207–214. doi: 10.1016/0891-5849(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 288.Lin J.-H., Castora F. J. DNA topoisomerase II from mammalian mitochondria is inhibited by the antitumor drugs, m-AMSA and VM-26. Biochemical and Biophysical Research Communications. 1991;176(2):690–697. doi: 10.1016/S0006-291X(05)80239-6. [DOI] [PubMed] [Google Scholar]
- 289.Fehr M., Pahlke G., Fritz J., et al. Alternariol acts as a topoisomerase poison, preferentially affecting the IIα isoform. Molecular Nutrition & Food Research. 2009;53(4):441–451. doi: 10.1002/mnfr.200700379. [DOI] [PubMed] [Google Scholar]
- 290.Soubeyrand S., Pope L., Haché R. J. G. Topoisomerase IIα-dependent induction of a persistent DNA damage response in response to transient etoposide exposure. Molecular Oncology. 2010;4(1):38–51. doi: 10.1016/j.molonc.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Bandele O. J., Osheroff N. Bioflavonoids as poisons of human topoisomerase IIα and IIβ. Biochemistry. 2007;46(20):6097–6108. doi: 10.1021/bi7000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Sabourin M., Osheroff N. Sensitivity of human type II topoisomerases to DNA damage: Stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Research. 2000;28(9):1947–1954. doi: 10.1093/nar/28.9.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mielke C., Christensen M. O., Barthelmes H. U., Boege F. Enhanced processing of UVA-irradiated DNA by human topoisomerase II in living cells. The Journal of Biological Chemistry. 2004;279(20):20559–20562. doi: 10.1074/jbc.C400032200. [DOI] [PubMed] [Google Scholar]
- 294.Srivastava S., Moraes C. T. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Human Molecular Genetics. 2005;14(7):893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Jensen R. E., Englund P. T. Network news: The replication of kinetoplast DNA. Annual Review of Microbiology. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 296.Melendy T., Sheline C., Ray D. S. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell. 1988;55(6):1083–1088. doi: 10.1016/0092-8674(88)90252-8. [DOI] [PubMed] [Google Scholar]
- 297.Shapiro T. A., Klein V. A., Englund P. T. Drug-promoted cleavage of kinetoplast DNA minicircles. Evidence for type II topoisomerase activity in trypanosome mitochondria. The Journal of Biological Chemistry. 1989;264(7):4173–4178. [PubMed] [Google Scholar]
- 298.Shapiro T. A., Showalter A. F. In vivo inhibition of trypanosome mitochondrial topoisomerase II: Effects on kinetoplast DNA maxicircles. Molecular and Cellular Biology. 1994;14(9):5891–5897. doi: 10.1128/MCB.14.9.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lindsay M. E., Gluenz E., Gull K., Englund P. T. A new function of Trypanosoma brucei mitochondrial topoisomerase II is to maintain kinetoplast DNA network topology. Molecular Microbiology. 2008;70(6):1465–1476. doi: 10.1111/j.1365-2958.2008.06493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Shapiro T. A. Mitochondrial topoisomerase II activity is essential for kinetoplast DNA minicircle segregation. Molecular and Cellular Biology. 1994;14(6):3660–3667. doi: 10.1128/MCB.14.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wang Z., Englund P. T. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO Journal. 2001;20(17):4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]