Abstract
Transcriptional deregulation caused by epigenetic or genetic alterations is a major cause of leukemic transformation. The Spi1/PU.1 transcription factor is a key regulator of many steps of hematopoiesis, and limits self-renewal of hematopoietic stem cells. The deregulation of its expression or activity contributes to leukemia, in which Spi1 can be either an oncogene or a tumor suppressor. Herein we explored whether cellular senescence, an anti-tumoral pathway that restrains cell proliferation, is a mechanism by which Spi1 limits hematopoietic cell expansion, and thus prevents the development of leukemia. We show that Spi1 overexpression triggers cellular senescence both in primary fibroblasts and hematopoietic cells. Erythroid and myeloid lineages are both prone to Spi1-induced senescence. In hematopoietic cells, Spi1-induced senescence requires its DNA-binding activity and a functional p38MAPK14 pathway but is independent of a DNA-damage response. In contrast, in fibroblasts, Spi1-induced senescence is triggered by a DNA-damage response. Importantly, using our well-established Spi1 transgenic leukemia mouse model, we demonstrate that Spi1 overexpression also induces senescence in erythroid progenitors of the bone marrow in vivo before the onset of the pre-leukemic phase of erythroleukemia. Remarkably, the senescence response is lost during the progression of the disease and erythroid blasts do not display a higher expression of Dec1 and CDKN1A, two of the induced senescence markers in young animals. These results bring indirect evidence that leukemia develops from cells which have bypassed Spi1-induced senescence. Overall, our results reveal senescence as a Spi1-induced anti-proliferative mechanism that may be a safeguard against the development of acute myeloid leukemia.
Introduction
Transcription factors (TFs) are major regulators of hematopoietic cell differentiation and are often deregulated in acute myeloid leukemia (AML). Spi1/PU.1 is a member of the ETS family, and accurate expression levels are critical for specifying cell fate and for proper hematopoietic differentiation.1 Spi1 plays a pivotal role in hematopoietic stem cell (HSC) self-renewal and in myeloid and B lymphoid differentiation.2–5 It acts by controlling the expression of a subset of lineage-specific genes involved in hematopoiesis6 and the expression of ubiquitous cell cycle regulators.5,7,8 Although the involvement of Spi1 alterations in tumor formation is well-established, the mechanisms by which Spi1 drives the development of AML are still not clear and seem to be complex. A reduction in Spi1 levels or an indirect inhibition of its activity by cooperating factors involved in leukemic transformation causes AML in humans.9–12 Rare cases of heterozygous inactivating mutations have also been described in human AML.13,14 Studies using several mouse models of Spi1 reduction have corroborated the involvement of Spi1 in the development of AML.15–19 Consistent with the role of Spi1 in controlling growth arrest and promoting myeloid differentiation, its re-expression in knocked down or mutated Spi1 cells or in leukemic progenitors in which Spi1 expression is suppressed induces growth arrest and monocytic differentiation.10,15,20 Despite this tumor-suppressor function, Spi1 is required for the maintenance of leukemic cells in AMLs with specific fusion genes.21–23 Spi1 also displays oncogenic activity, promoting the proliferation of erythroid progenitors in mice.24,25 High Spi1 expression levels in mice cause a pre-leukemic syndrome characterized by an increase in the number of hyper-proliferative erythroid progenitors in which differentiation and apoptosis are blocked.25–27 In these cells, Spi1 induces replication stress and accelerates genetic mutability.28
Increasing evidence points to a critical role for cellular senescence as a barrier to malignant transformation. This tumor suppressive mechanism is activated when cells are exposed to exogenous or endogenous stresses such as supraphysiological oncogenic signaling. Oncogene-induced senescence (OIS) is a mechanism that limits cell hyper-proliferation through a stable cell cycle arrest process,29 thus blocking the expansion of cells at the pre-cancerous stage in solid tumors.30,31 The expression of the hematopoietic oncogenes HRASV12, BCR-ABL, CBFB-MYH11 or RUNX1-ETO in primary HSCs and committed progenitors (HSCPs) elicits a senescence response,32 and OIS acts as an antitumoral barrier in NRASV12-induced lymphomas and MLL-ENL-induced AML.33,34 Senescence can be triggered, at least in part, by DNA replication stress, mainly due to the over-activation of replication origin firing, and an associated DNA-damage response (DDR)33,35–37 or independently of DNA replication stress.32 Although the role of OIS in limiting the proliferation of primary fibroblasts and epithelial cells and in protecting against the progression of solid tumorigenesis is now well characterized, the extent of the role of OIS in primary HSCPs and its protective effect against leukemic processes have yet to be fully explained.
Because Spi1 is required to maintain murine HSCs in a quiescent state and to restrict HSC division,5 we examined whether cellular senescence is a mechanism by which Spi1 restricts cell proliferation and if it protects against the development of AML. Our results reveal that Spi1 restrains cell expansion by inducing senescence in primary HSCPs as well as in primary fibroblasts in vitro. The mechanistic underpinnings of this response are distinct for the two cell types. We took advantage of the well-characterized Spi1 transgenic (TgSpi1) mouse model, in which Spi1 overexpression results in oncogenic behavior,25 to analyze senescence in vivo. We established that Spi1 overexpression induces senescence of bone marrow cells in mice, and that this anti-proliferative process is lost during the leukemic progression observed in TgSpi1 mice. Overall, the study herein identifies a new pathway by which Spi1 controls hematopoietic cell growth.
Methods
Mice and cell culture
TgSpi1 mice have been described previously.25 Wild-type (WT) mice were obtained from crossing heterozygous TgSpi1 mice. Early passage foreskin human fibroblasts BJ cells and fetal lung fibroblasts WI-38 cells obtained from American Type Cell Culture collection (BJ batch number: 59899913; WI-38 batch number: 58483158) were cultured using Dulbecco’s modified Eagle’s medium (DMEM) with high glucose supplemented with 10% (v/v) fetal bovine serum (FBS) at 37°C in a humidified atmosphere containing 5% CO2 and 3% O2. HEK293EBNA and HeLa cells were maintained in DMEM medium with high glucose, 10% FBS at 37°C and 5% CO2.
A detailed description of Methods is included in the Online Supplementary Appendix.
Results
Ectopic expression of Spi1 induces senescence in primary fibroblasts and primary hematopoietic cells
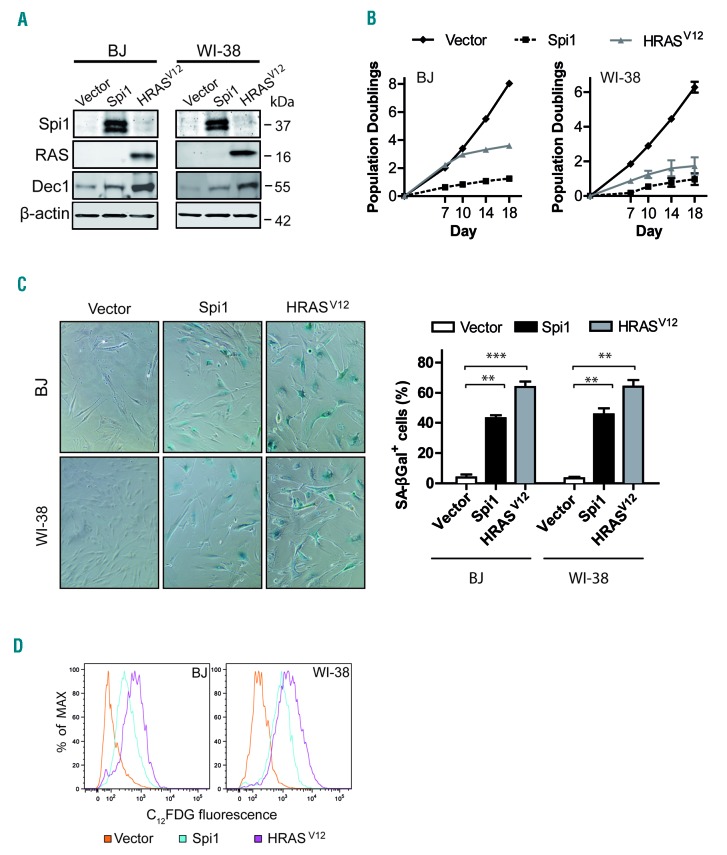
To understand the role of Spi1 in senescence, we first studied the consequences of its ectopic expression in the well-established senescence cell model based on normal human primary fibroblasts (strains BJ and WI-38).38 We transduced cells with a retroviral construct expressing Spi1 or an activated RAS mutant (HRASV12) as a positive control for OIS (Figure 1A and Online Supplementary Figure S1). The ectopic expression of Spi1 or HRASV12 both led to senescence that was characterized by stable cell cycle arrest (Figure 1B), increased senescence-associated beta-galactosidase (SA-βgal) activity, as measured by cytochemical staining and cytometric analyses (Figure 1C,D), and increased protein levels of the senescence biomarker Dec1 (Figure 1A).29,39
Figure 1.
Ectopic expression of Spi1 and HRASV12 induces growth arrest and senescence in BJ and WI-38 fibroblast cells. (A) Western blot analysis of Spi1, HRASV12, and the senescence marker Dec1 in BJ and WI-38 cells subjected to the retroviral-mediated expression of Spi1, HRASV12 or an empty vector as a control 10 days after puromycin selection. β-actin served as the loading control. (B) Population doublings (PDs) of BJ (left panel) and WI-38 (right panel) cells transduced as described in (A) over the indicated periods of time. Day 0 was the first day after puromycin selection. PDs for each time point are the mean of triplicate experiments. (C) Representative SA-βgal staining and percent of SA-βgal positive cells (histograms) in samples of BJ and WI-38 cells subjected to the retroviral-mediated expression of Spi1, HRASV12 or an empty vector as a control 10 days after puromycin selection. Magnification of images, 200X. The means ± SD of at least 3 independent experiments are shown. **P<0.005; ***P<0.0005 from two-tailed Student’s t-tests. (D) Flow cytometric detection of SA-βgal activity using C12FDG as a fluorogenic substrate in cells as described in (C).
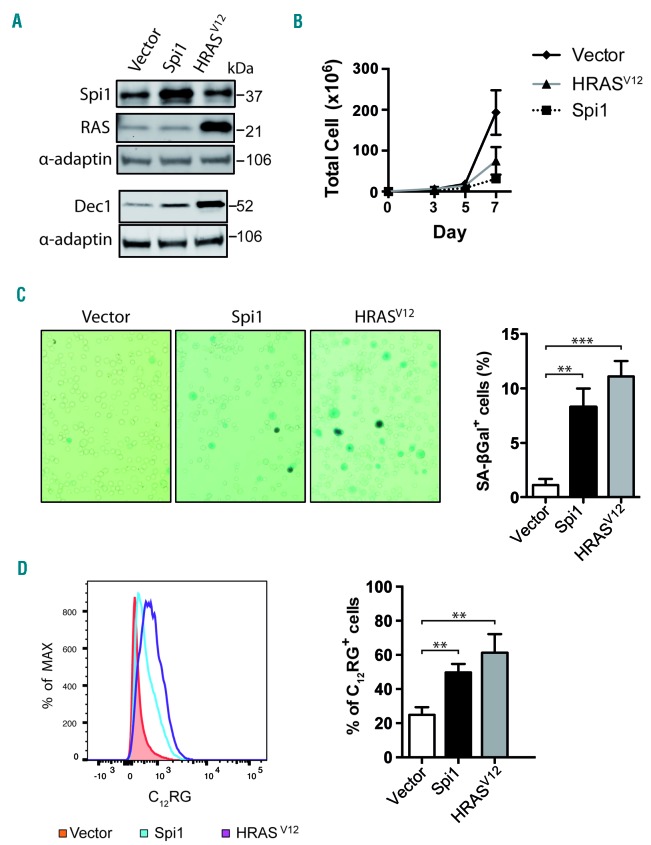
We next investigated the induction of senescence in primary hematopoietic stem/progenitor cells, defined as Lin−Kit+Sca1+ (LSK) cells (Online Supplementary Figure S2). LSK cells were transduced with retroviral vectors expressing green fluorescent protein (GFP) alone (vector) or co-expressing GFP with Spi1 or HRASV12 (Figure 2A). Total cell number was reduced over time by Spi1- and HRASV12 -overexpression compared with the numbers of cells infected with the control vector (Figure 2B). Moreover, Spi1 and HRASV12 expression caused an increase in the proportion of cells that stained positive for SA-βgal activity among GFP-positive cells compared with the control cells (8.3% and 11.1% for Spi1- and HRASV12 -expressing cells, respectively, and 1.1% for control cells; Figure 2C). We also observed an increase in SA-βgal activity using C12RG as a fluorogenic substrate in Spi1- or H-RASV12-expressing GFP-positive cells compared with activity in control cells (51% and 61 % for Spi1- and HRASV12-expressing cells, respectively, and 24% for control cells; Figure 2D) and Spi1- or HRASV12-expressing GFP-negative cells (Online Supplementary Figure S3A). The proportion of GFP-positive cells was positively correlated with the proportion of senescent cells (Online Supplementary Figure S3B). In agreement with the increase of SA-βgal activity, the expression level of the senescence marker Dec1 was increased in cells transduced with Spi1 and HRASV12 (Figure 2A).
Figure 2.
Overexpression of Spi1 and HRASV12 in Lin−Kit+Sca1+ (LSK) cells leads to senescence. (A) Western blot analysis of Spi1, HRASV12 and the senescence marker Dec1 in hematopoietic cells subjected to the retroviral-mediated expression of Spi1, HRASV12 or an empty vector. Protein extracts of GFP-positive sorted cells were analyzed 7 days post-infection as described in Online Supplementary Figure S2. α-adaptin served as the loading control. (B) Number of total living cells retrovirally transduced with Spi1 and HRASV12 or an empty vector (control), at the indicated periods of time. The means ± SEM of at least 3 independent experiments are shown. (C) Representative SA-βgal staining and mean percentages of SA-βgal positive cells (histograms) in samples of hematopoietic cells subjected to the retroviral-mediated expression of Spi1, HRASV12 or an empty vector as a control. SA-βgal assays for sorted GFP-positive cells were performed 7 days post-infection as described in Online Supplementary Figure S2. The counting of GFP-positive SA-βgal cells was performed in 9 randomly selected fields with a total of more than 2000 cells from each group. Magnification of images, 200X. The means ± SD of at least 3 independent experiments are shown. **P<0.005; ***P<0.0005 from two-tailed Student’s t-test. (D) Flow cytometric detection of SA-βgal activity using C12RG as a fluorogenic substrate in cells retrovirally transduced with Spi1, HRASV12 or an empty vector. The histograms represents the % of C12RG positive cells among GFP-positive cells. The means ± SD of at least 3 independent experiments are shown. **P<0.005 from two-tailed Student’s t-test.
Collectively, these results indicate that the ectopic expression of Spi1 in primary fibroblasts and hematopoietic cells induces cellular senescence.
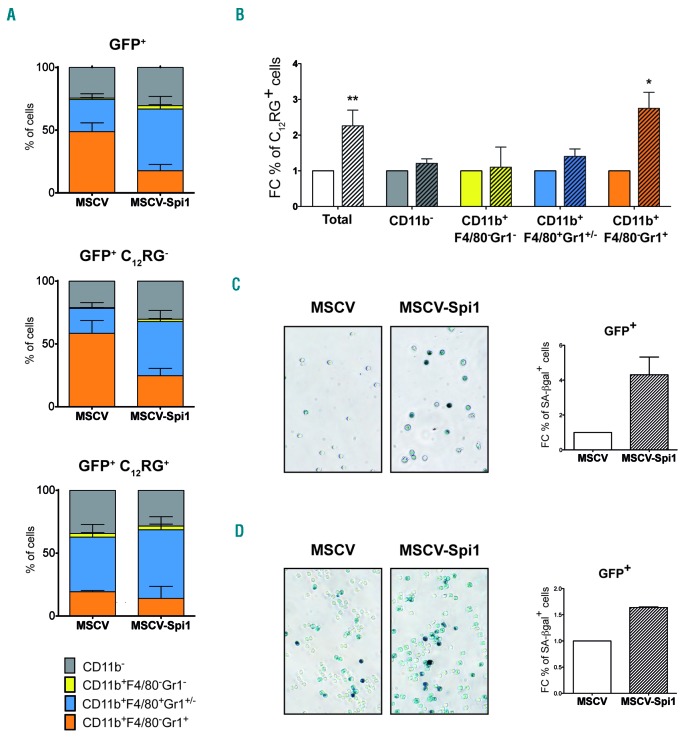
To characterize, in depth, which hematopoietic cells are prone to Spi1-induced senescence, we first combined measures of SA-βgal activity using the C12RG fluorogenic substrate and specific immunophenotypic markers for hematopoietic progenitors using flow cytometry. Mainly Kit−Sca1− cells were found at day 7, a time when senescence was measured (Online Supplementary Figure S4A), indicating that cells were engaged in differentiation. The distribution of cells in the different categories of Kit/Sca1 markers was similar in senescent (C12RG+) and in non-senescent (C12RG−) cells (Online Supplementary Figure S4A). As the cytokines used for LSK culture are prone to push cells towards myeloid differentiation, and overexpression of Spi1 in primary hematopoietic cells causes differentiation primarily in macrophages,10,15,20 we looked at the immunophenotypic myeloid markers CD11b, Gr1 and F4/80 (Online Supplementary Figure S4B). Spi1 overexpression (GFP+ cells) reduced the proportion of granulocytes (CD11b+F4/80−Gr1+) and increased the proportion of monocytes/macrophages (CD11b+F4/80+Gr1+/−) (Figure 3A, upper histogram). All types of cells contributes to the Spi1-induced senescent cells as observed from the distribution of the CD11b, Gr1 and F4/80 population among the GFP+C12RG+ cells (Figure 3A, lower histogram), which are not different to the non-senescent GFP+ C12RG− cells (Figure 3A, middle histogram). Moreover, Spi1 induced 2.75-fold more senescent cells among the granulocytes compared to murine stem cell virus (MSCV)-transduced control cells (27% to 10%, respectively), and 1.5-fold more among the monocytes/macrophages (55% to 40%, respectively) (Figure 3B). These results indicate that granulocytes and monocytes/macrophages are prone to Spi1-induced senescence even if Spi1 favors only differentiation of monocytes/macrophages.
Figure 3.
Spi1 triggers senescence in granulocytes, monocytes/macrophages and myeloid and erythroid progenitor cells. (A) Distribution of the cells according to CD11b, Gr1 and F4/80 myeloid markers and SA-βgal activity using C12RG as fluorogenic substrate by flow cytometry among total GFP+, GFP+C12RG− or GFP+C12RG+, 7 days after transduction of LSK cells with MSCV-Spi1 or MSCV control vectors. The means ± SEM of 3 independent experiments are shown. (B) Fold change (FC) of the % of C12RG positive cells between Spi1-overexpressing cells (hatched histograms) and control cells among GFP+ cells inside each indicated cells compartment, granulocytes (CD11b+F4/80−Gr1+), monocytes/macrophages (CD11b+F4/80+Gr1+), immature myeloid progenitors (CD11b+F4/80−Gr1−) or CD11b− cells. *P<0.05 from two-tailed Student’s t-test. (C and D) Representative SA-βgal staining using cytochemical staining and FC of the % of SA-βgal positive cells in MEP (C) and in GMP (D) progenitor cells subjected to the retroviral-mediated expression of Spi1 (MSCV-Spi1) relative to empty vector as a control (MSCV). SA-βgal assays for sorted GFP-positive cells were performed 4 or 6 days post-infection for MEP and GMP, respectively. The counting of GFP-positive SA-βgal cells was performed in 3 randomly selected fields with a total of more than 100 cells from each group. Magnification of images, 200X. Results are from 3 independent experiments. GFP: green fluorescent protein; MSCV: murine stem cell virus.
Spi1 serves as oncogene in erythroid but not in myeloid progenitors, wherein it is a tumor suppressor. Therefore, to investigate whether Spi1 overexpression is associated with senescence in both types of cells, we studied the senescence in Spi1-overexpressing megakaryocyte-erythroid (MEP) and granulocyte-monocyte (GMP) progenitor cells. Sorted MEP (Lin−Sca−Kit+CD34−CD16/32−) and GMP (Lin−Sca−Kit+CD34+CD16/32+) were transduced with retroviral vectors expressing GFP alone (vector) or co-expressing GFP with Spi1, and senescence was evaluated by measuring SA-βgal activity using cytochemical staining (Figure 3C). Spi1 overexpression caused an increase in the proportion of SA-βgal+ cells among GFP-positive GMP progenitor cells compared with the control cells (1.7-fold more for Spi1-overexpressing cells compared to the MSCV control cells; 80.7% and 48.8%, respectively), confirming that myeloid cells are prone to Spi1-induced senescence. As expected,26 we found that Spi1 blocked erythroid differentiation (Online Supplementary Figure S4C). Strikingly, Spi1 also triggered senescence when overexpressed in MEP progenitors (3.5-fold more SA-βgal+ cells for Spi1-overexpressing cells compared to the MSCV control cells; 61.6% and 17.4%, respectively; Figure 3D).
In conclusion, our results demonstrate that Spi1 is able to trigger senescence in myeloid and erythroid progenitors.
Spi1 triggers senescence through its DNA-binding activity
To determine whether the DNA binding and transcriptional activities of Spi1 were required for inducing senescence, we used the Δβ4-Spi1 mutant with a deletion in the β4 region of the ETS domain, which we previously showed as being unable to bind DNA and transactivate40 (Online Supplementary Figure S5). The expression of Δβ4-Spi1 or an empty vector (Online Supplementary Figure 6A) did not impact the proliferation of BJ or WI-38 fibroblasts when compared with the proliferation of Spi1-WT-expressing cells (Online Supplementary Figure 6B). Moreover, cells overexpressing Δβ4-Spi1 were not found to be SA-βgal-positive (Online Supplementary Figure 6C,D) and did not increase Dec1 expression when compared with cells expressing the vector control (Online Supplementary Figure S6A). Similar to the results for fibroblasts, the expression of Δβ4-Spi1 in primary sorted LSK cells neither altered cell proliferation nor the percentage of cells staining positive for SA-βgal or Dec1 expression levels in hematopoietic cells (Online Supplementary Figure S6E–G). Together, these results demonstrate that the DNA binding activity of Spi1 is required to induce senescence.
Spi1 induces senescence through distinct pathways in primary fibroblasts and hematopoietic cells
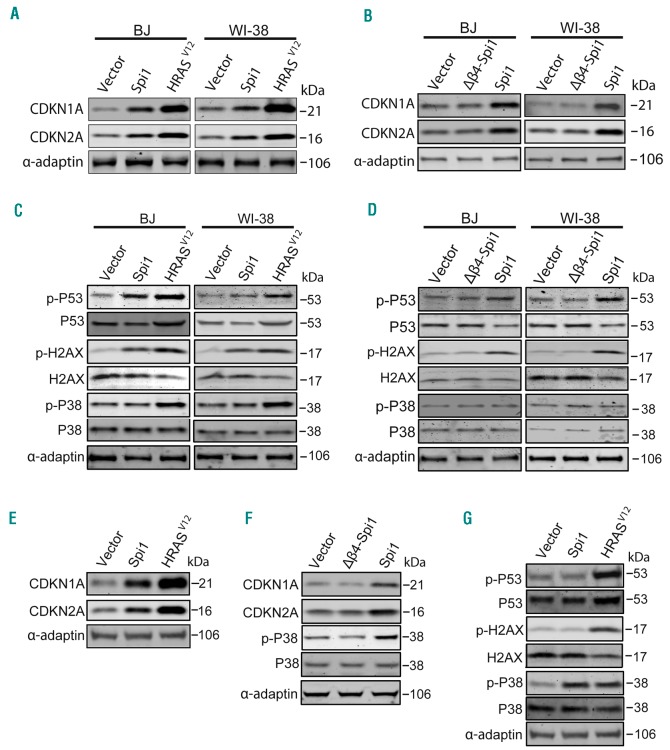
To further understand the mechanisms underlying Spi1-mediated senescence, we examined the expression of two cell cycle inhibitors, CDKN1A (alias p21/CIP) and CDKN2A (alias p16/INK4A), which are instrumental for inducing senescence. We detected a robust increase in CDKN1A and CDKN2A in Spi1- or HRASV12-expressing primary fibroblasts when compared with levels in cells transduced with the vector alone (Figure 4A). CDKN1A and CDKN2A expression levels were not increased in cells expressing the Δβ4-Spi1 protein (Figure 4B), which is consistent with the absence of senescence in those cells. It has been shown that OIS in primary fibroblasts is, at least in part, induced by DNA replication stress evoking a DDR, which is characterized by increased H2AX phosphorylation on Serine 139 (Ser139) and p53 phosphorylation on Serine 15 (Ser15). We found that Spi1, similar to the effects of HRASV12, induced the phosphorylation of p53 on Ser15 and of H2AX on Ser139 (Figure 4C), whereas this response was not detected in cells expressing the Δβ4-Spi1 mutant, as expected (Figure 4D). Next, we performed this analysis in HSCPs. Similar to the results observed in fibroblasts, the expression levels of CDKN1A and CDKN2A were increased in GFP-positive primary HSCPs 7 days post-infection with Spi1 or HRASV12 vectors when compared with levels in cells expressing control vectors (Figure 4E), whereas the overexpression of the Δβ4-Spi1 mutant had no effect (Figure 4F). Remarkably, in contrast to the results observed in fibroblasts and the effects of HRASV12, we did not detect any changes in the phosphorylation of p53 or H2AX in HSCPs overexpressing Spi1, indicating that Spi1 does not elicit a DDR in primary hematopoietic cells. These findings prompted us to test the role of the stress p38 mitogen-activated protein kinase (p38MAPK14) in Spi1-induced senescence, as OIS can be mediated by p38-dependent, p53-independent signaling pathways.41 We observed that HRASV12 induced the activation of p38MAPK14 based on an observed increase in the phosphorylated form (Thr180/182) of p38MAPK14 in both primary HSCPs and fibroblasts (Figure 4C,G). In contrast, Spi1 overexpression did not induce any increase in p38MAPK14 phosphorylation in BJ or WI-38 fibroblasts (Figure 4C,D), whereas in primary HSCPs, the overexpression of Spi1 produced an increase in the phosphorylation of p38MAPK14. Again, as shown by previous assays, Δβ4-Spi1 overexpression did not induce this response (Figure 4F).
Figure 4.
Spi1 induces senescence through distinct mechanisms in fibroblasts and in hematopoietic cells. (A-B) Western blot analysis of CDKN1A and CDKN2A in BJ and WI-38 cells subjected to the retroviral-mediated expression of Spi1, Δβ4-Spi1, HRASV12 or an empty vector (vector) 10 days after puromycin selection. α-adaptin served as the loading control. (C-D) Western blot analysis of S15-phosphorylated (p-p53) and total p53, S139-phosphorylated (p-H2AX) and total H2AX, and Thr180/182-phosphorylated and total p38MAPK14 (p-P38 and P38) in the same cells described in (A). (E, G) Western blot analysis of CDKN1A and CDKN2A (E) and S15-phosphorylated and total p53, S139-phosphorylated and total H2AX, and Thr180/182-phosphorylated and total p38MAPK14 (p-P38 and P38) (G) in hematopoietic cells subjected to the retroviral-mediated expression of Spi1, HRASV12 or an empty vector. Protein extracts of GFP-positive sorted cells were analyzed 7 days post-infection as described in Online Supplementary Figure S2. α-adaptin served as the loading control. (F). Thr180/182-phosphorylated and total p38MAPK14 (p-P38 and P38), and CDKN1A and CDKN2A expression were analyzed via Western blotting in hematopoietic cells subjected to the retroviral-mediated expression of Spi1, Δβ4-Spi1 or an empty vector. Protein extracts of GFP-positive sorted cells were analyzed 7 days post-infection as described in Online Supplementary Figure S2. α-adaptin served as the loading control.
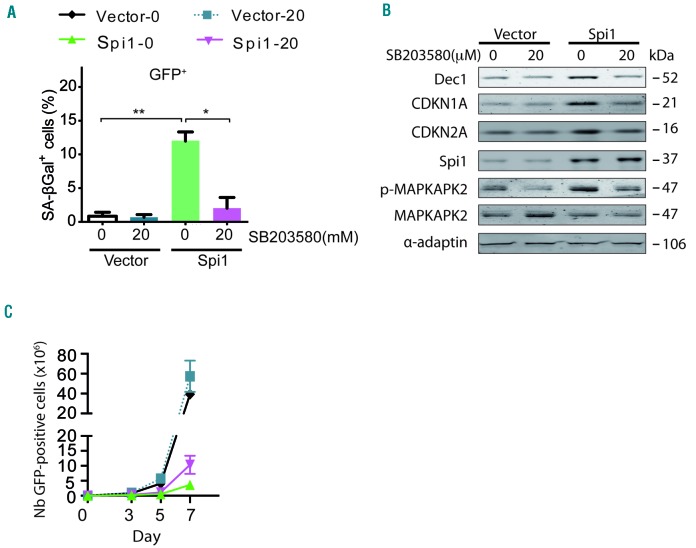
To corroborate the role of p38MAPK14 activity in Spi1-induced senescence in hematopoietic cells, we treated cells undergoing Spi1-mediated senescence with the selective p38MAPK14 pharmacological inhibitor, SB203580. We found that the fraction of cells stained positive for SA-βgal was strongly reduced in the presence of the inhibitor compared with the fraction in untreated cells (Figure 5A, compare Spi1-0 μM versus Spi1-20 μM). Spi1 did not increase Dec1, CDKN1A or CDKN2A levels of expression in the presence of the p38MAPK14 inhibitor (Figure 5B), further confirming the absence of senescence in SB203580-treated cells. The treatment of Spi1 overexpressing cells with SB203580 also increased, although partially, the number of GFP-positive cells (Figure 5C), indicating that p38MAPK14 controls senescence and additional mechanisms modulating cell number. The functional inhibition of p38MAPK14 activity by SB203580 was confirmed based on a decreased phosphorylation level of its downstream target mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2) (Figure 5B).
Figure 5.
Spi1-induced senescence requires P38MAPK14 signaling in hematopoietic cells. (A) Mean percentage of SA-βgal positive cells (histograms) subjected to the retroviral-mediated expression of Spi1 or an empty vector, and maintained in cultures with or without 20 μM of SB203580 in samples of GFP-positive sorted hematopoietic cells 7 days post-infection. The means ± SD of at least 3 independent experiments are shown. *P<0.05; **P<0.005 from two-tailed Student’s t-tests. (B) Hematopoietic cells transduced with empty vectors (vector) or Spi1 expression vectors and maintained in cultures with or without 20 μM of SB203580 were sorted for GFP-positive cells 7 days post-infection and subjected to Western blot analyses of CDKN1A, CDKN2A, p-MAPKAPK2, MAPKAPK2, Dec1 and Spi1. α-adaptin served as the loading control. (C) Number of GFP-positive cells retrovirally transduced with empty vectors (vector) or Spi1 expression vectors and maintained in cultures with or without 20 μM of SB203580 from day 1 to day 6 at the indicated periods of time. The means ± SD of at least 3 independent experiments are shown.
Together, these data support a model in which Spi1 elicits senescence in primary fibroblasts and hematopoietic cells through distinct molecular pathways. In fibroblasts, the induction of Spi1-mediated senescence involves a DDR, whereas in hematopoietic cells, it requires p38MAPK14 activation but is independent of a DDR.
Spi1-induced senescence in vivo is disrupted during leukemic progression
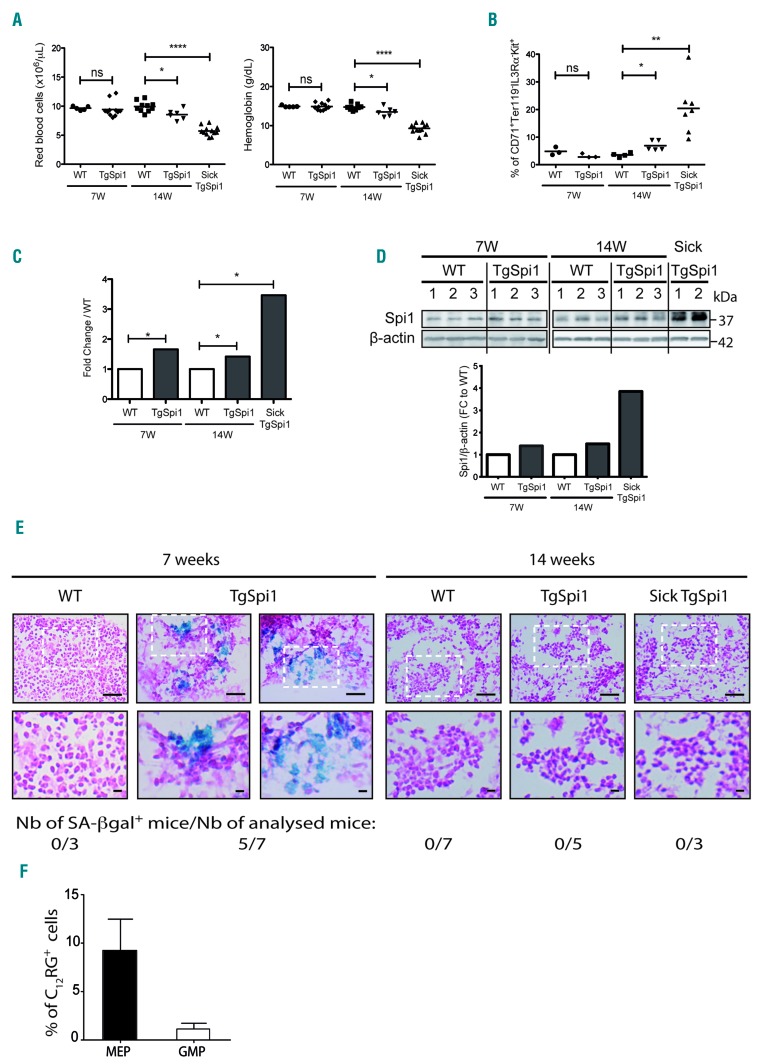
To investigate the impact of dysregulated Spi1 expression on senescence in vivo, we utilized the TgSpi1 mouse model, which overexpresses Spi1 and develops AML from the erythroid lineage.25,42 Within 3.6 ± 1.5 months (14 weeks) after birth, TgSpi1 mice developed anemia with infiltration of the bone marrow and spleen associated with erythroid cells blocked at the colony forming unit-erythroid (CFU-E) stage (CD71+Ter119−IL3Rα−Kit+). These mice are referred to as sick TgSpi1 mice (Figure 6A,B and Online Supplementary Figure S7A). These pre-leukemic cells, derived from sick TgSpi1 mice, are not tumorigenic when engrafted into nude mice.25 This is in contrast to the cells, referred to as leukemic cells, detected at a later stage that emerge due to the acquisition of mutations in the SCF receptor gene (Kit), and are characterized by fully malignant features.42 Here, we explored the effect of Spi1 up to the pre-leukemic stage.25,26 At this stage, the expansion of blast cells in the bone marrow and spleen of the sick TgSpi1 mice was mainly at the expense of the CD11bhigh Gr1high myeloid cells (Online Supplementary Figure S7B, compare 14-week-old WT and sick TgSpi1 mice). As expected, Spi1 was highly expressed at both the RNA and protein levels in the bone marrow cells of the pre-leukemic sick TgSpi1 mice (3.5- and 4-fold upregulation, respectively) compared with levels in age-matched WT mice (Figure 6C,D and Online Supplementary Figure S8). To explore the progression of the Spi1-mediated disease, we examined healthy TgSpi1 mice at 7 and 14 weeks of age. Mice were assumed to be healthy if their spleen weighed less than 0.2g and if there were no blast cells in the blood or bone marrow (Online Supplementary Figure 7C,D). The increase of Spi1 gene and protein expression levels was detectable in the whole bone marrow cells of TgSpi1 mice as early as 7 weeks after birth (Figure 6C,D). Moreover, using immunohistochemistry, we observed an increase in the number of cells that expressed Spi1 in the bone marrow of 7- and 14-week-old healthy TgSpi1 mice (Online Supplementary Figure S8). Seven-week-old TgSpi1 mice did not display any abnormalities compared with age-matched WT mice for any analyzed parameters, i.e., red blood cell and blast cell numbers, hemoglobin concentration, the fraction of CFU-E and the percentage of myeloid cells (Figure 6A,B, Online Supplementary Figure S7A,B and D). Early signs of pre-leukemic syndrome were detectable in 14-week-old TgSpi1 mice that presented a moderate but significant decrease in red cell number and hemoglobin concentration compared with WT mice concomitant with a moderate increase of CFU-E in the bone marrow (7% for 14-week-old healthy TgSpi1 mice compared with 3.5% for 14-week-old WT mice; Figure 6A,B). We then characterized the senescence program during the pre-leukemic progression phase using this well-characterized model. We detected an accumulation of SA-βgal in bone marrow sections from five out of seven TgSpi1 mice at 7 weeks of age, whereas this was not observed in age-matched WT mice (Figure 6E). A slightly increased expression of the senescence marker Dec1 was also observed in the total bone marrow of TgSpi1 mice compared to WT, consistent with the induction of senescence in the TgSpi1 mice (Online Supplementary Figure S9). Interestingly, more senescent cells were observed among the MEP cells of 7-week-old TgSpi1 mice compared with age-matched WT mice, while this was not the case for the GMP cells (Figure 6F and Online Supplementary Figure S10A). Consistently, Spi1 was significantly overexpressed in the MEP cells and weakly expressed in the GMP cells in TgSpi1 mice compared with age-matched WT mice (Online Supplementary Figure S10B). Overexpression of Spi1 in MEP cells was also found to be associated with an induction of Dec1 and IL1α gene expression, a cytokine of the senescence-associated secretory phenotype (SASP) (Online Supplementary Figure S10C), supporting senescence activation in those types of cells. These data indicate that only erythroid progenitors are prone to Spi1-induced senescence in the TgSpi1 mice in vivo. Importantly, even though Spi1 expression was increased (Online Supplementary Figure S10B and S11), we did not detect any SA-βgal activity in the bone marrow sections from TgSpi1 mice at 14 weeks of age or from sick TgSpi1 mice (Figure 6E), indicating that the senescence program was not effective in either the 14-week-old TgSpi1 mice that displayed only early and minimal signs of a pre-leukemic syndrome or in the TgSpi1 mice with a pre-leukemic syndrome. Remarkably, while sorted erythroid blastic cells from sick TgSpi1 mice (CFU-E like, CD71+Ter119−IL3Rα−Kit+) still displayed a higher expression of CDKN2A compared with equivalent WT CFU-E cells, Dec1 and CDKN1A RNA expression was decreased (Online Supplementary Figure S11).
Figure 6.
Spi1 induces senescence in vivo in the bone marrow of young TgSpi1 mice and is lost before the onset of the pre-leukemic syndrome. (A) Red blood cell numbers and hemoglobin concentrations of wild-type (WT) and TgSpi1 mice at the indicated ages. Bars indicate the mean values. *P<0.05; ****P<0.0001 from two-tailed Student’s t-tests. (B) Scatter plots represent the results of flow cytometry analyses of whole bone marrow cells for CFU-E markers (CD71+Ter119−Kit+IL3Rα−) in WT and TgSpi1 mice at the indicated ages. Bars indicate the mean values. *P<0.05; **P<0.001 from two-tailed Student’s t-tests. (C) Spi1 messenger ribonucleic acid (mRNA) levels in bone marrow cells from 7- and 14-week-old WT, healthy TgSpi1 and sick TgSpi1 mice were quantified via real-time quantitative polymerase chain reaction (qPCR) and normalized to the Polr2α mRNA level (ΔCt, Ctgene-CtPolr2α). Between 4 and 6 animals were analyzed for each category of mice. Bars represent the fold change relative to values for age-matched WT mice, as calculated from the 2−ΔΔCt values. Statistical analysis of the 2−ΔCt values was carried out using Student’s t-test; *P<0.05. (D) Spi1 protein levels in bone marrow cells from 7- and 14-week-old WT, healthy TgSpi1 and sick TgSpi1 mice were analyzed by Western blotting. The histograms represent the quantified results, using ImageJ, relative to β-actin and to values for age-matched WT mice. (E) SA-βgal activity was examined in fresh bone marrow sections. Staining was performed on bone marrow from 7- and 14-week-old WT, healthy TgSpi1 and sick TgSpi1 mice. The number of mice displaying SA-βgal positive cells in their bone marrow is indicated for each category of mice. Bars represent 50μm (top) and 10μm (bottom) pictures. (F) MEP (Lin−Sca−Kit+CD34−CD16/32−) and GMP (Lin−Sca−Kit+CD34+CD16/32+) from bone marrow cells of 7-week-old WT and healthy TgSpi1 mice were analyzed for SA-βgal activity using C12RG as a fluorogenic substrate. The histograms represent the means of percentage of C12RG+ cells in TgSpi1 mice considering WT mice as negative control, as presented in Online Supplementary Figure S10A. N= 4 animals for WT and 5 animals for TgSpi1 mice. Ns: non-significant.
These results are consistent with the lack of senescent phenotype in the pre-leukemic cells and bring molecular supports of the bypass of senescence in vivo. In conclusion, Spi1 overexpression is associated with senescence of erythroid progenitors in the bone marrow of asymptomatic young TgSpi1 mice. Erythroid cells give rise to blastic cells in sick TgSpi1 animals. The absence of a senescence program in older and sick TgSpi1 mice suggests that this process is lost during the completion of the pre-leukemic syndrome phase at a step that precedes the emergence of the pre-leukemic stage.
Discussion
In the study herein, we demonstrated that Spi1 TF is a driver of senescence in several types of cells, ie., in primary fibroblasts, hematopoietic stem cells, MEP and GMP progenitors.
It is well established that the OIS is a barrier that stands at pre-invasive stages of solid tumorigenesis. As Spi1 is not expressed in normal fibroblasts, and as its over-expression induces senescence through the activation of a DDR in primary fibroblasts, as shown for the HRASV12, our results suggest that Spi1 behaves as a “classical” oncogene for OIS which is associated with DNA replication stress.33,35–37 Of note, while HRASV12 -induced senescence is mediated by both DDR-dependent and DDR-independent processes, i.e., the activation of p38MAPK14, Spi1-induced senescence is only associated with DDR-dependent signaling in primary fibroblasts.
In contrast with the response observed in primary fibroblasts, DDR signaling is not involved in the Spi1-induced senescence of primary hematopoietic cells, consistent with data from our previous study showing that Spi1 overexpression is not associated with the presence of DNA strand breaks in the K562 cell line and in the pre-leukemic cells of TgSpi1 mice.28 We found that p38MAPK14 signaling is required to elicit Spi1-induced senescence in hematopoietic cells. Contributions from either DDR or p38MAPK14 signaling in OIS have also been reported for leukemogenic fusion proteins in hematopoietic cells.32 The P38MAPK14 activation mediated by reactive oxygen species (ROS) plays a role in the exhaustion of hematopoietic stem cells.43 Interestingly, a p38MAPK14/p16 axis independent of the serine/threonine-protein kinase ataxia telangiectasia (ATR)- or Rad3-related protein (ATM)-DDR has also been found to mediate senescence in epithelial cells.44 Therefore, p38MAPK14 seems to be an alternative and major signaling pathway by which cells others than fibroblasts can undergo senescence. In conclusion, we identified diverse molecular pathways for Spi1-induced senescence, either through DDR activation in primary fibroblasts or DDR-independent P38MAPK14 activation in primary hematopoietic cells.
Interestingly, even if Spi1 triggers senescence at a slightly lower degree than H-RASV12 in the two cell types, the effect of Spi1 overexpression on growth arrest was stronger than that due to overexpression of H-RASV12. These results suggest that Spi1 also limits cell proliferation by additional mechanisms, other than that of senescence. As we have shown that Spi1 overexpression favors monocytes/macrophage differentiation and that growth arrest is concomitant to terminal differentiation, the reduced number of proliferating cells in hematopoietic cells may be the combined consequence of at least two mechanisms on a heterogeneous population, senescence and differentiation-associated growth arrest. Consistently, not all cells overexpressing Spi1 underwent senescence (up to 50% of LSK cells). Additionally, SA-βgal positive cells may not accumulate in culture and be eliminated as reported for apoptotic hematopoietic cells.
We have shown that, in contrast to the opposite effects of Spi1 on erythroid and monocytic differentiation, monocytes/macrophages, granulocytes and erythroid cells undergo senescence when Spi1 was overexpressed in their respective immature progenitors, demonstrating that a wide range of cells are prone to Spi1-induced senescence. One query raised by the study herein is to determine the relationship between senescence and differentiation and, in particular, which program (senescence or differentiation) was the first to be initiated. However, the only parameter applicable to identify senescent cells and characterize their differentiation markers is the SA-βGal assay, which is a final parameter of senescence. Thus, the initial cell wherein senescence was instructed remains unknown.
Spi1 has been shown to play an important role in normal HSC homeostasis.2–5 In particular, Spi1 prevents the self-renewal and exhaustion of HSCs by restraining the cell cycle through multiple downstream targets.45 Thus, our findings that Spi1 triggers senescence in vitro in hematopoietic cells and in vivo in the bone marrow of TgSpi1 mice are consistent with senescence as a new growth arrest mechanism by which Spi1 limits expansion. This result raises the question of whether the ability of Spi1 to induce senescence is only of pathological relevance, or if it also plays a role in normal hematopoiesis. As a gradient of Spi1 expression is observed during normal lineage commitment and in divergent hematopoietic lineages,1 we speculate that the senescence-inducing activity of Spi1 may participate in the regulation of cell number, depending on its level of expression and the hematopoietic context.
While Spi1 is oncogenic in the erythroid lineage, considerable evidence points to a role for this TF as a tumor suppressor in myeloid malignancies.46 Thus, we propose that in addition to blocking myeloid differentiation, decreased Spi1 expression/activity may promote leukemogenesis through the loss of anti-proliferative activity. This may be particularly deleterious in the case of a cooperative proliferative signaling, such as replicative or oncogenic stress. In human acute promyelocytic leukemias (APLs) expressing the oncoprotein PML-RARα, a down-regulation of Spi1 occurs that is overridden by all-trans retinoic acid (ATRA) treatment.10 The restoration of Spi1 in APL cells triggers granulocytic differentiation.10 ATRA also rescues a senescence-like program, demonstrating that this treatment constitutes a senescence-driven cure for leukemia.47 As Spi1 expression and senescence are both increased by ATRA in APL cells, Spi1-induced senescence may contribute to the cure of APL.
We used the TgSpi1 model to examine the process of senescence up to the appearance of the pre-leukemic syndrome. Notably, even though Spi1 induces senescence in erythroid progenitors of young asymptomatic TgSpi1 mice, senescence was not detectable in the bone marrow of sick TgSpi1 mice. Because senescence was likewise not detectable in the bone marrow of 14-week-old TgSpi1 mice that were asymptomatic, the senescence program is turned off prior to the emergence of pre-leukemic blast cells. Even if not proved, several results argue that senescent cells give rise to blastic cells. Senescent cells were detected among the MEP cells in the young TgSpi1 mice; this was not found for the GMP. Consistently, Spi1 was significantly overexpressed in the MEP cells and weakly in the GMP cells in those TgSpi1 mice. Remarkably, while the erythroid blastic cells of the sick pre-leukemic mice still displayed a higher expression of Spi1 and CDKN2A compared with equivalent WT CFU-E cells, Dec1 and CDKN1A RNA expression was decreased, indicating that the pre-leukemic cells lost some marks of senescence which they displayed in young animals. Knocking out p53 in the TgSpi1 mice accelerates the speed of the disease,48 in agreement with the fact that growth arrest pathway delays the leukemic progression. Altogether, our data suggest that, as is the case for solid tumors, senescence represents a mechanism for constraining cell number and is disrupted during the development of AML. High MCL1 expression found in the TgSpi1 erythroid blasts27 may be involved in the erasure of the Spi1-induced senescence program, as MCL1 is a repressor of senescence.49
The anti-apoptotic function of Spi1 participates in the continuous expansion of pre-leukemic cells.26,27 Thus, the two main protective barriers to tumor development, apoptosis and senescence, are disrupted during erythroleukemic transformation in TgSpi1 mice. Further work will be needed to identify the pathways that are responsible for the bypass of senescence during leukemic progression and how targeting these pathways may rescue the senescence program in the pre-leukemic stage.
Supplementary Material
Acknowledgments
The authors would like to thank Y. Lecluse, P. Rameau and Z. Maciorowski from the cytometry platform and A. Nicolas, P. Opolon, O. Bawa, S. Arrufat, R. Corre and E. Louvet from the IHC platform. We thank the animal facility housing at Curie and Gustave Roussy Institutes. We thank O. Bernard, M. David for comments on the manuscript and F. Rosselli and F. Moreau-Gachelin for helpful scientific discussions and critical reading of the manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/11/1850
Funding
This work was supported by the Fondation de France, Inserm, the Institut National du Cancer (INCa-DGOS-INSERM 6043 and PL-BIO-06), ITMO Cancer de l’alliance nationale pour les sciences de la vie et de la santé (AVIESAN), Section régionale de la Ligue Nationale contre le Cancer. L. Delestré was supported by AVIESAN and Fondation de France; H. Cui by Cancéropole Ile-de-France; M. Esposito by the Institut National du Cancer (PL-BIO-06). Quiveron and E. Mylonas by a CDI-Mission (Institut Gustave Roussy).
References
- 1.DeKoter RP, Kamath MB, Houston IB. Analysis of concentration-dependent functions of PU.1 in hematopoiesis using mouse models. Blood Cells, Mol Dis. 2007;39(3): 316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106(5): 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HG, de Guzman CG, Swindle CS, et al. The ETS family transcription factor PU.1 is necessary for the maintenance of fetal liver hematopoietic stem cells. Blood. 2004;104(13):3894–3900. [DOI] [PubMed] [Google Scholar]
- 4.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201(2):221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staber PB, Zhang P, Ye M, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49(5):934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turkistany SA, DeKoter RP. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch Immunol Ther Exp (Warsz). 2011;59(6):431–440. [DOI] [PubMed] [Google Scholar]
- 7.Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341(6146):670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wontakal SN, Guo X, Will B, et al. A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet. 2011;7(6):e1001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steidl U, Steidl C, Ebralidze A, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117(9):2611–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller BU, Pabst T, Fos J, et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107(8):3330–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vangala RK, Heiss-Neumann MS, Rangatia JS, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101(1):270–277. [DOI] [PubMed] [Google Scholar]
- 12.Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173. [DOI] [PubMed] [Google Scholar]
- 13.Mueller BU, Pabst T, Osato M, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100(3):998–1007. [DOI] [PubMed] [Google Scholar]
- 14.Lavallee VP, Baccelli I, Krosl J, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47(9): 1030–1037. [DOI] [PubMed] [Google Scholar]
- 15.Cook WD, McCaw BJ, Herring C, et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104(12):3437–3444. [DOI] [PubMed] [Google Scholar]
- 16.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624–630. [DOI] [PubMed] [Google Scholar]
- 17.Metcalf D, Dakic A, Mifsud S, Di Rago L, Wu L, Nutt S. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci USA. 2006;103(5):1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Will B, Vogler TO, Narayanagari S, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 2015;21(10):1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, Park JS, Ries RE, et al. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARalpha. Proc Natl Acad Sci USA. 2005;102(35): 12513–12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado MD, Gutierrez P, Richard C, Cuadrado MA, MoreauGachelin F, Leon J. Spi-1/PU.1 proto-oncogene induces opposite effects on monocytic and erythroid differentiation of K562 cells. Biochem Biophys Res Commun. 1998;252(2):383–391. [DOI] [PubMed] [Google Scholar]
- 21.Staber PB, Zhang P, Ye M, et al. The Runx-PU.1 pathway preserves normal and AML/ETO9a leukemic stem cells. Blood. 2014;124(15):2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aikawa Y, Katsumoto T, Zhang P, et al. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16(5):580–585, 581,p following 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Wu J, Li B, et al. PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia. 2014;28(7):1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukemia. Nature (London). 1988;331(6153):277–280. [DOI] [PubMed] [Google Scholar]
- 25.Moreau-Gachelin F, Wendling F, Molina T, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16(5):2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimmele P, Kosmider O, Mayeux P, Moreau-Gachelin F, Guillouf C. Spi-1/PU.1 participates in erythroleukemogenesis by inhibiting apoptosis in cooperation with Epo signaling and by blocking erythroid differentiation. Blood. 2007;109(7):3007–3014. [DOI] [PubMed] [Google Scholar]
- 27.Ridinger-Saison M, Evanno E, Gallais I, et al. Epigenetic silencing of Bim transcription by Spi-1/PU.1 promotes apoptosis resistance in leukaemia. Cell Death Differ. 2013;20(9): 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimmele P, Komatsu J, Hupe P, et al. Spi-1/PU.1 oncogene accelerates DNA replication fork elongation and promotes genetic instability in the absence of DNA breakage. Cancer Res. 2010;70(17):6757–6766. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–740. [DOI] [PubMed] [Google Scholar]
- 30.Collado M, Gil J, Efeyan A, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. [DOI] [PubMed] [Google Scholar]
- 31.Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. [DOI] [PubMed] [Google Scholar]
- 32.Wajapeyee N, Wang SZ, Serra RW, et al. Senescence induction in human fibroblasts and hematopoietic progenitors by leukemogenic fusion proteins. Blood. 2010;115(24):5057–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takacova S, Slany R, Bartkova J, et al. DNA damage response and inflammatory signaling limit the MLL-ENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21(4):517–531. [DOI] [PubMed] [Google Scholar]
- 34.Braig M, Lee S, Loddenkemper C, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–665. [DOI] [PubMed] [Google Scholar]
- 35.Di Micco R, Fumagalli M, Cicalese A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. [DOI] [PubMed] [Google Scholar]
- 36.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035): 907–913. [DOI] [PubMed] [Google Scholar]
- 37.Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. [DOI] [PubMed] [Google Scholar]
- 38.Cristofalo VJ, Volker C, Allen RG. Use of the fibroblast model in the study of cellular senescence. Methods Mol Med. 2000;38(23–52. [DOI] [PubMed] [Google Scholar]
- 39.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Na Rev Cancer. 2006;6(6):472–476. [DOI] [PubMed] [Google Scholar]
- 40.Guillouf C, Gallais I, Moreau-Gachelin F. Spi-1/PU.1 oncoprotein affects splicing decisions in a promoter binding-dependent manner. J Biol Chem. 2006;281(28):19145–19155. [DOI] [PubMed] [Google Scholar]
- 41.Han J, Sun P. The pathways to tumor suppression via route p38. Trends Biochem Sci. 2007;32(8):364–371. [DOI] [PubMed] [Google Scholar]
- 42.Kosmider O, Denis N, Lacout C, Vainchenker W, Dubreuil P, Moreau-Gachelin F. Kit-activating mutations cooperate with Spi-1/PU.1 overexpression to promote tumorigenic progression during erythroleukemia in mice. Cancer Cell. 2005;8(6):467–478. [DOI] [PubMed] [Google Scholar]
- 43.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–451. [DOI] [PubMed] [Google Scholar]
- 44.Nassour J, Martien S, Martin N, et al. Defective DNA single-strand break repair is responsible for senescence and neoplastic escape of epithelial cells. Nat Commun. 2016;7:10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuchi Y, Ito M, Shibata F, Kitamura T, Nakajima H. Activation of CCAAT/enhancer-binding protein alpha or PU.1 in hematopoietic stem cells leads to their reduced self-renewal and proliferation. Stem Cells. 2008;26(12):3172–3181. [DOI] [PubMed] [Google Scholar]
- 46.Dakic A, Wu L, Nutt SL. Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis? Trends Immunol. 2007;28(3):108–114. [DOI] [PubMed] [Google Scholar]
- 47.Ablain J, Rice K, Soilihi H, de Reynies A, Minucci S, de The H. Activation of a promyelocytic leukemia-tumor protein 53 axis underlies acute promyelocytic leukemia cure. Nat Med. 2014;20(2):167–174. [DOI] [PubMed] [Google Scholar]
- 48.Scolan EL, Wendling F, Barnache S, et al. Germ-line deletion of p53 reveals a multistage tumor progression in spi-1/PU.1 transgenic proerythroblasts. Oncogene. 2001;20(39):5484–5492. [DOI] [PubMed] [Google Scholar]
- 49.Bolesta E, Pfannenstiel LW, Demelash A, et al. Inhibition of Mcl-1 promotes senescence in cancer cells: implications for preventing tumor growth and chemotherapy resistance. Mol cell Biol. 2012;32(10):1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.