Significance
Abscisic acid (ABA) accumulates to high levels during drought stress, and ABA levels, in turn, control many downstream responses that determine plant growth, productivity, and survival during drought. Despite the central importance of ABA, we know little of how drought stress elicits ABA accumulation, how different aspects of ABA metabolism are coordinated to match the level of ABA to stress severity, or how variation in ABA accumulation may contribute to environmental adaptation. When exposed to an equal severity of water limitation, Arabidopsis thaliana accessions exhibited wide variation in their ABA content. These results allowed us to identify effectors of ABA accumulation and also indicated that variation in ABA accumulation may contribute to variation in other drought-related traits.
Keywords: drought stress, abscisic acid, genome-wide association mapping, natural variation, START domain protein
Abstract
Accumulation of the stress hormone abscisic acid (ABA) in response to drought and low water-potential controls many downstream acclimation mechanisms. However, mechanisms controlling ABA accumulation itself are less known. There was a 10-fold range of variation in ABA levels among nearly 300 Arabidopsis thaliana accessions exposed to the same low water-potential severity. Genome-wide association analysis (GWAS) identified genomic regions containing clusters of ABA-associated SNPs. Candidate genes within these regions included few genes with known stress or ABA-related function. The GWAS data were used to guide reverse genetic analysis, which found effectors of ABA accumulation. These included plasma-membrane–localized signaling proteins such as receptor-like kinases, aspartic protease, a putative lipid-binding START domain protein, and other membrane proteins of unknown function as well as a RING U-box protein and possible effect of tonoplast transport on ABA accumulation. Putative loss-of-function polymorphisms within the START domain protein were associated with climate factors at accession sites of origin, indicating its potential involvement in drought adaptation. Overall, using ABA accumulation as a basis for a combined GWAS–reverse genetic strategy revealed the broad natural variation in low-water-potential–induced ABA accumulation and was successful in identifying genes that affect ABA levels and may act in upstream drought-related sensing and signaling mechanisms. ABA effector loci were identified even when each one was of incremental effect, consistent with control of ABA accumulation being distributed among the many branches of ABA metabolism or mediated by genes with partially redundant function.
Abscisic acid (ABA) is one of the classical plant hormones and regulates processes including seed dormancy and germination, stomatal regulation, and gene expression (1–3). ABA content increases rapidly when plants are exposed to low water potential (ψw) during drought. This ABA accumulation induces downstream drought responses via a core ABA-signaling pathway (1, 3). However, it is less clear how drought and low ψw cause ABA accumulation. Several experiments have indicated that dehydration of the plant tissue and resulting change in cell volume or loss of turgor pressure are key triggers of ABA accumulation (4–6). Beyond this, the sensing and signaling processes that influence ABA accumulation remain little known.
The level of ABA that accumulates in response to low ψw is determined by coordination of ABA synthesis and catabolism, ABA conjugation and deconjugation (particularly of ABA-glucose ester), and ABA transport between different tissues and cellular compartments (7, 8). ABA has a short half-life, even during drought stress (9), consistent with dynamic modulation of ABA level response to environmental signals. Conversely, there is also a surprisingly stable aspect to ABA accumulation. In plants allowed to acclimate to reduced ψw, ABA content is linearly related to the severity of low ψw stress (10–12). Efforts to perturb the relationship between ABA content and stress severity by manipulating only one aspect of ABA metabolism have met with limited success. For example, overexpression of the key ABA synthesis enzyme 9-cis-epoxycarotenoid dioxygenase (NCED) led to a relatively small increase in ABA but larger increases in the ABA catabolite phaseic acid (13), indicating that much of the extra ABA produced was catabolized. Likewise, manipulation of ABA conjugation could dramatically increase the level of ABA-glucose ester but had little effect on ABA levels (14). These observations suggest the existence of unknown regulatory mechanisms that integrate the activities of different ABA-metabolism pathways to match ABA levels to stress severity. Mechanisms that influence ABA accumulation are thus of great interest and are tied to the even broader question of understanding plant osmosensing systems that detect changes in water status (15, 16).
Genetic screens have identified factors acting downstream of ABA but in general have revealed less about the mechanisms controlling ABA levels. One screen of particular interest identified a mutant of Vacuolar Sorting Receptor 1 (VSR1), which had decreased ABA accumulation and NCED3 expression, possibly because of altered intracellular pH and tonoplast transport (17). While such forward genetic screens are promising, they are limited by a lack of knowledge of which promoters respond most directly to upstream stress signals and by functional redundancy within gene families. The NCED3 promoter, for example, is regulated by ABA accumulation, likely as part of a feed-forward mechanism to stimulate ABA production (7, 17, 18). Also, extensive genetic screens failed to find ABA receptors, and we now know that was because of genetic redundancy among the PYL/RCAR family (1). Similarly, persuasive arguments have been made that receptor-like kinases (RLKs) may have roles in stress sensing and signaling (19); yet, there is little data to show which of the more than 600 RLKs are involved in drought response. Conversely, reverse genetic approaches in which selected genes are analyzed in enough detail to find subtle effects on drought response require a priori assumptions about which genes are worthy to test, thus limiting the ability to find new and unexpected loci.
Natural variation and use of genome-wide association analysis (GWAS) followed by reverse genetic tests of candidate genes offer another path that combines an open-ended query of the genome with a more detailed analysis of selected candidate genes. In Arabidopsis, there is extensive natural variation in many drought-related traits (20–24). GWAS has been applied to analyze several of these traits (20, 22, 25) but has yet to be applied to ABA accumulation. Validation of candidate genes continues to be a rate-limiting step in utilizing GWAS results. However, when such validation is performed, it can identify genes of relatively small phenotypic effect (25, 26). We discovered a wide range of natural variation in low ψw-induced ABA accumulation among Arabidopsis thaliana accessions and used GWAS-guided reverse genetics to identify multiple loci affecting ABA accumulation.
Results
Arabidopsis Has Extensive Natural Variation in ABA Content at Low ψw.
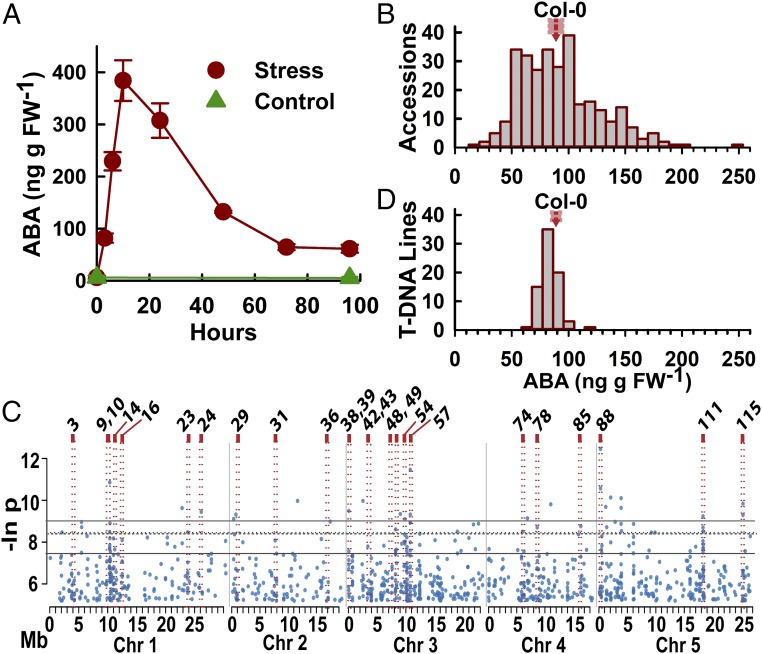
After transfer of 7-d-old seedlings from unstressed condition (ψw = −0.25 MPa) to low ψw (−1.2 MPa) there was a several hundred-fold increase in ABA content in the first 10 h (Fig. 1A). As the plants acclimated to the stress, ABA content stabilized at an intermediate level that was well below the peak ABA accumulation but still more than 50-fold above the unstressed control (Fig. 1A). Further ABA measurements were conducted at the 96-h time point as we were most interested to discover factors involved in ABA homeostasis during longer-term acclimation to low ψw. Measurements of ABA content at low ψw for 298 accessions found a 10-fold range of natural variation with the Col-0 reference accession near the median (Fig. 1B and Dataset S1).
Fig. 1.
Natural variation and GWAS analysis of low-ψw–induced ABA accumulation. (A) Time course of ABA accumulation after transfer of 7-d-old Col-0 seedlings to control (−0.25 MPa) or low-ψw stress (−1.2 MPa). Data are means ± SE (n = 6) combined from two independent experiments. (B) Distribution of ABA contents at 96 h after transfer to −1.2 MPa for 298 A. thaliana accessions. The mean and SE of the Col-0 reference accession is indicated by the red arrow and red box, respectively. ABA data for each accession are given in Dataset S1. (C) Manhattan plot of SNP P values (expressed as the natural log, base e) versus genomic position for the top 1,000 (T1000) SNPs from GWAS analysis using the ABA data shown in B. Gray lines indicate the cutoffs for the top 100, and top 20 low P value SNPs. For reference, the dashed line indicates an FDR of 0.75. Red boxes indicate the positions of regions of interest for which we analyzed ABA accumulation for T-DNA mutants of one or more candidate genes. A total of 116 regions of interest were identified based on the GWAS analysis, and these were numbered consecutively through the genome from chromosomes 1–5 (SI Appendix, Fig. S2 and Dataset S3). (D) Distribution of ABA contents after low-ψw treatment for 75 T-DNA mutants. ABA data for all T-DNA mutants are given in Dataset S5. The mean and SE of Col-0 wild type are indicated.
Interestingly, we found that ABA accumulation had limited association with accession climate-of-origin. ABA was not significantly associated with any of the first five principal components of climate (α = 0.05). We did find a significant association of higher ABA accumulation with wetter locations (total growing season precipitation, ρ = 0.16, P = 0.0202) although this relationship was nonsignificant after accounting for population structure. Further analysis found only limited association (not significant after accounting for population structure or driven by a few outlying accessions) of ABA with other climate factors (see SI Appendix, SI Materials and Methods for further details).
GWAS Analysis Identifies Genomic Regions Associated with Variation in Low-ψw-Induced ABA Accumulation.
We calculated genetic values of log ABA accumulation using linear mixed models and found that random effects of genotype explained 38% of the variation in ABA (broadsense heritability [H2] = 0.38). GWAS analysis was conducted using a linear mixed model approach (27) and published SNP data (28). After discarding low-frequency SNPs [minor allele frequency < 0.1 (29)], association with ABA content was tested for 171,619 SNPs. A substantial portion (75% averaged across SNP-specific models) of total variation in genotype ABA levels was explained by random effects correlated with the kinship matrix. The distribution of SNP P values closely followed the theoretical expectation, suggesting good control of false positives by the mixed model (SI Appendix, Fig. S1).
While GWAS studies sometimes focus only on the candidate genes around one or a few SNPs of lowest P value, our goal was to fully use the GWAS data to generate hypotheses about candidate genes affecting ABA accumulation. Thus, we followed a previously described heuristic approach (25) to identify and rank genomic regions associated with clusters of low P value SNPs while also giving greater weight to the lowest P value SNPs (further details of the analysis are given in SI Appendix, SI Materials and Methods). This approach identified 2,293 genes (Dataset S2) within 5 kb (an estimate of linkage disequilibrium) of one or more of the top 1,000 low-P-value SNPs (Fig. 1C; these are the top 0.58% of all SNPs analyzed and are hereafter referred to as the T1000 SNPs). Although it was not used in our analysis, Fig. 1C also shows a false-discovery-rate threshold that falls within the top 1,000 SNPs for reference. We then gave the genes associated with one or more of the top 100 SNPs a score based on the number and P value of T1000 SNPs within 5 kb (25). From this we identified 116 regions of interest containing one or more genes with a score above 3 (SI Appendix, Fig. S2 and Dataset S3; note that the regions of interest are numbered 1–116 sequentially through the genome from chromosome 1 to chromosome 5) and also ranked the candidate genes (Dataset S4). Candidate genes to analyze by reverse genetics were selected by considering both the top scoring regions as well as individual genes that had high score and were associated with tight clusters of the T1000 SNPs. In total, 75 T-DNA mutants of 44 genes from 24 regions of interest were analyzed for ABA content at low ψw (Dataset S5; the regions of interest analyzed are marked in Fig. 1C). Many of the T-DNA mutants were also tested by RT-PCR to verify knockout or knockdown of the targeted gene (SI Appendix, Fig. S3). In this analysis, it was clear that the effect of any single T-DNA mutant on ABA accumulation was relatively small compared with the wide range of natural variation (Fig. 1D). This suggested the existence of many ABA effector genes, each having incremental effect.
Plasma Membrane Proteins and a RING-U Box Protein Identified as Effectors of ABA Accumulation.
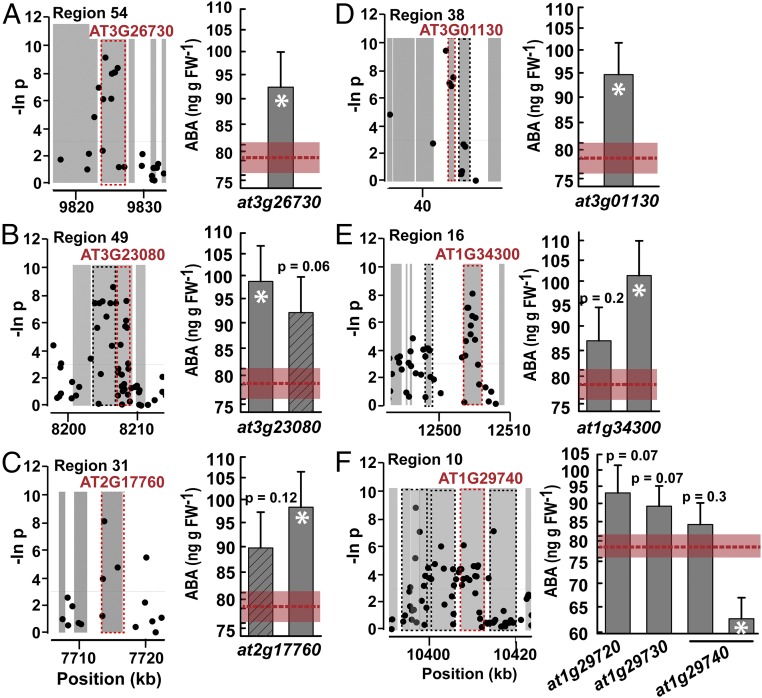
We found seven cases where T-DNA mutants had strongly significant (P < Bonferroni adjusted α) differences in ABA accumulation compared with wild type. Six of these cases are presented here (the remaining one is discussed below), and each one had roughly similar effects in that they altered ABA accumulation by 20–30%. None of these six genes were previously reported to affect ABA or abiotic stress response. In region 54, which was highly ranked in our scoring system (Dataset S4), there was a cluster of ABA-associated SNPs in the gene body of AT3G26730, and an at3g26730 mutant had significantly increased ABA accumulation at low ψw (Fig. 2A). This indicated that AT3G26730 is a negative effector (repressor) of ABA accumulation. AT3G26730 encodes a RING U-box protein that was previously reported to interact with C-terminal phosphatase domain phosphatase-like1 (CPL1) (30). CPL1 is a negative regulator of stress and ABA-responsive gene expression via effects on transcription and RNA processing (31, 32). Possibly, AT3G26730 affects ABA accumulation via control of CPL protein abundance.
Fig. 2.
Six regions of interest with T-DNA mutants found to have significant differences in low-ψw–induced ABA accumulation compared with Col-0 wild type. (A–F) Each panel shows data for a genomic region of interest for which the mutant analysis uncovered ABA effector genes. For each panel, the Left graph is a plot of SNP P values (−log base e) for a 30- to 50-kb window around the main candidate gene(s). Gene bodies (UTRs, introns, and exons) are shown in gray. A black dashed outline around a gene indicates that it was analyzed by ABA phenotyping of T-DNA mutants but no significant differences were found (Dataset S5). A red dashed outline and red gene label indicate that a significant difference in ABA content compared with wild type was found for at least one T-DNA mutant of that gene. The Right graph shows ABA contents for T-DNA mutants of the indicated genes. The red dashed line indicates the mean ABA content of the Col-0 wild type, and the red shaded area marks the SE. A white asterisk indicates ABA difference that was strongly significant (P < Bonferroni adjusted α). For other lines, the nominal P values are listed. Diagonal lines within a bar mark mutants that had reduced expression (knockdown) rather than knockout of the target gene. T-DNA mutant ABA data are listed in Dataset S5.
In other regions, we discovered plasma membrane proteins with significant effects on ABA accumulation. Region 49 had a cluster of SNPs with relatively low P values in the gene bodies of AT3G23070 and AT3G23080. An at3g23080 T-DNA mutant had significantly elevated ABA accumulation, and another knockdown mutant had a marginally nonsignificant effect (P = 0.06; Fig. 2B) while an at3g23070 mutant had no effect (Dataset S5). AT3G23080 encodes a polyketide cyclase-type protein containing a putative transmembrane domain and a START domain possibly involved in lipid binding (33). In region 31, an AT2G17760 mutant had elevated ABA accumulation while another T-DNA line of this gene had a marginally nonsignificant effect (Fig. 2C). This latter T-DNA line (SALK_011498) has an intron insertion and may still have a low level of AT2G17760 expression. AT2G17760, an aspartic peptidase, was identified in proteomic analyses of plasma-membrane–enriched proteins (34, 35) and increased in abundance by more than threefold during cold acclimation or in response to ABA (34). In region 38, significantly elevated ABA content was found for a mutant of at3g01130 (Fig. 2D). AT3G01130 encodes the small hydrophobic protein ATAucsisa1, which affects polar auxin transport via an unknown mechanism (36). ATAucsisa1 interacts with PROTEOLYSIS6 (PRT6), a Ubiquitin E3 ligase that affects ABA sensitivity via its function in N-end rule protein degradation (37, 38). Interestingly, we found that two prt6 T-DNA mutants also had elevated ABA accumulation (SI Appendix, Fig. S4). Whether AT3G01130 affects ABA accumulation via its interaction with PRT6 and N-end rule protein degradation or via its proposed cytoskeleton-related function (36) is of interest for further investigation.
RLKs, including lectin domain RLKs, are of particular interest based on their presumed roles in sensing stress-related stimuli at the cell wall–plasma membrane interface (19). Region 16 contained a cluster of ABA-associated SNPs in AT1G34300, which encodes a β-lectin domain RLK (39). An at1g34300 T-DNA mutant had significantly elevated ABA accumulation while another mutant tended to have high ABA but was more variable between experiments (Fig. 2E and Dataset S5). Mutants of an adjacent gene, AT1G34290, did not significantly differ from wild type (Dataset S5). Region 10 contained a broad band of ABA-associated SNPs covering several genes (AT1G29720, AT1G29730, AT1G29740, and AT1G29750) that encode closely related group VIII RLKs with leucine-rich repeat (LRR) extracellular domains (39, 40) and putative malectin domain. Both mutants of at1g29720 and at1g29730 had marginally nonsignificant increases in ABA accumulation (Fig. 2F and Dataset S5), raising the possibility that these genes act redundantly. For AT1G29740, one T-DNA line (SALK_027357) had a significant reduction in ABA accumulation (Fig. 2F and Dataset S5). This was the opposite effect as that of at1g29720 and at1g29730 mutants; however, we did not find any evidence that SALK_027357 had increased expression of other RLKs in region 10 (SI Appendix, Fig. S5). It remains possible that this mutant alters the activity of AT1G29750 or produces a truncated protein with an aberrant signaling function. Despite the uncertainty for individual mutants, the combined data for region 10 suggest that these RLKs affect ABA accumulation in a redundant manner. There is no previously reported physiological function of these RLKs (nor for AT1G34300).
The effect of AT3G01130 and AT1G34300 on ABA accumulation was further verified by generating transgenic lines ectopically expressing these genes (SI Appendix, Fig. S6). Ectopic expression of AT3G01130 strongly suppressed ABA accumulation while AT1G34300 decreased ABA to lesser extent, possibly because AT1G34300 has other developmental roles (see below) that precluded us from identifying transgenic lines with a high level of AT1G34300 expression.
Physiological and Climate Analysis of Candidate Genes.
Further physiological analyses were conducted for five genes with the most prominent effect on ABA accumulation (i.e., those that were significant after Bonferroni correction but excluding AT1G29740 and bZIP17). AT3G26730 and AT3G23080 mutants had strongly reduced water loss in detached leaf assays (SI Appendix, Fig. S7), consistent with higher ABA levels, and perhaps different ABA sensitivity, in the mutants causing more extensive stomatal closure. Mutants of AT2G17760 and AT1G34300 also had reduced leaf water loss. In other experiments, mutants of AT1G34300 and AT3G23080 had substantially increased seedling dry weight (SI Appendix, Fig. S8A). Mutants of AT1G34300 had the largest effect, suggesting that it may have additional functions in growth regulation. None of the mutants had altered proline accumulation (SI Appendix, Fig. S8B), consistent with stress and metabolic mechanisms other than ABA having more prominent roles in controlling proline (41) and with the fact that this ABA GWAS identified a different set of candidate genes than the GWAS of proline accumulation (25).
We also studied whether natural loss-of-function variants in these genes were associated with particular climates, suggesting a role in local adaptation. AT3G23080 has 25 native range accessions in the 1001 genomes panel (42) with putative loss-of-function variants (Dataset S6). These 25 accessions occupy locations of low temporal variability in growing-season precipitation compared with the rest of the native range accessions in the panel (t test, t = 2.49, P = 0.0192) and warm growing seasons (t test, t = −3.88, P = 0.0007) but not significantly so after accounting for population structure (linear mixed models, P = 0.1392 and P = 0.2486, respectively). However, only five of these accessions are in our ABA dataset; thus we cannot draw conclusions about the impact of these natural variants on ABA accumulation. In addition, we found that AT3G23080 was near six SNPs with strong climate associations (linear mixed model controlling for kinship, P < 0.01). The strongest of these (8,204,936 bp) was ranked 107 among ∼170,000 SNPs tested (P = 0.0005) while 4 other SNPs were in the top 200 (43).
Reverse Genetic Analysis of Top Genomic Regions and Stress-Related Candidate Genes Identified by ABA GWAS.
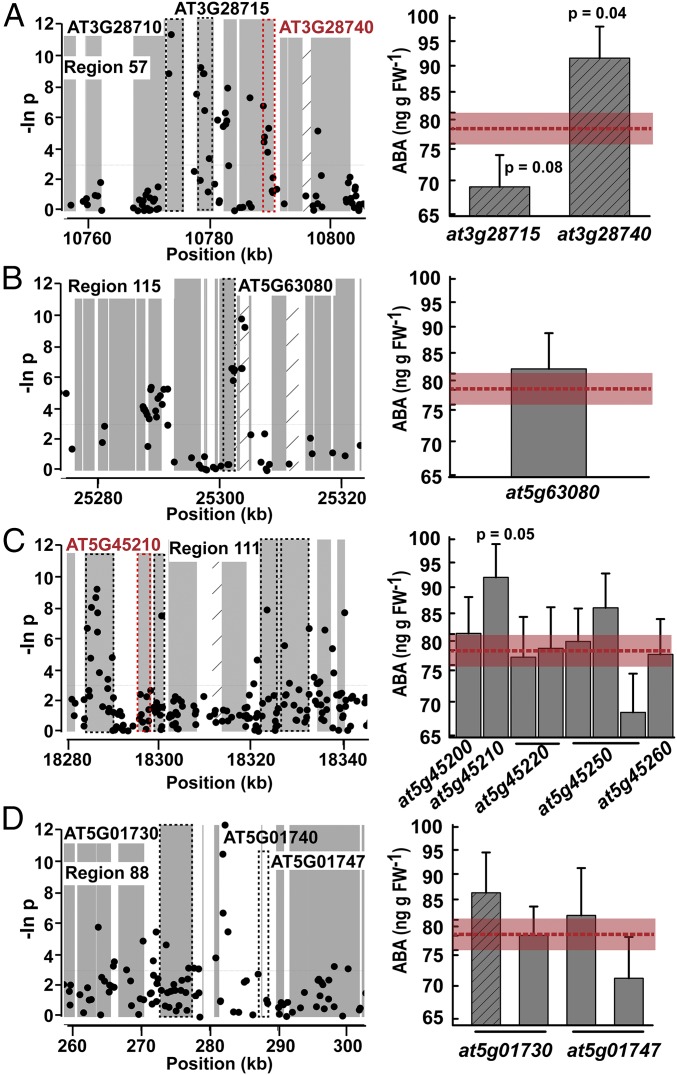
We also tested candidate genes in the five top scoring regions of interest (Dataset S4; region 54 is discussed above). The top scoring region (region 57) contained the second lowest-P-value SNP and had a band of low-P-value SNPs covering several genes (Fig. 3A). Of these, a mutant of AT3G28740, which encodes the cytochrome P450 family protein CYP81D11, the expression of which is regulated by jasmonic acid signaling and xenobiotic stress (44), had a marginally significant increase in ABA (significant based on nominal P value but not by Bonferroni-adjusted threshold). Another gene in this region, AT3G28715, had a marginally nonsignificant (P = 0.079) decrease in ABA accumulation (Fig. 3A). However, this line (SALK_039088) was a knockdown rather than knockout mutant (SI Appendix, Fig. S3). AT3G28715 encodes the ATPase, V0/A0 complex, subunit C/D VHA-d2, which is involved in tonoplast proton transport. The adjacent gene AT3G28710 encodes the nearly identical VHA-d1 (45). It is possible that AT3G28710 and AT3G28715 act redundantly to affect ABA accumulation. This hypothesis is consistent with previous results where a mutant of the vacuolar sorting receptor VSR1 altered ABA accumulation by affecting cytosolic pH (17). ABA-related function of AT3G28740, and putatively of AT3G28715 and AT3G28710, likely caused the broad band of strong ABA–SNP associations in this region. Interestingly, VSR6 (AT1G30900) is a high-scoring candidate gene in region 14 (Dataset S3). An effect of tonoplast transport on ABA accumulation also seems consistent with the finding that a vacuolar type H+ pyrophosphatase (identified by GWAS in maize) affected survival under severe water stress (46).
Fig. 3.
Analysis of top-scoring regions identified by ABA GWAS. Data are shown for four regions of interest (A–D) that had the highest scores in our heuristic ranking system (Dataset S4). The format of data presentation is the same as that described for Fig. 2 except that data of all T-DNA lines analyzed for a given region are shown (SNP P values are shown as natural log).
Region 115 was ranked second in our scoring system and had a cluster of low-P-value SNPs around AT5G63080, AT5G63085, and AT5G63087 (Fig. 3B). A T-DNA mutant of AT5G63080 did not differ from wild type in ABA accumulation. While these data do not rule out variation in AT5G63080 affecting ABA, they do suggest AT5G63085 and AT5G63087 as candidates. Both these genes encode cell wall thionins that may bind lectins. No publically available T-DNA lines exist for these genes. Region 111 had a band of SNPs covering several genes encoding closely related Toll Interluken Repeat-Nucleotide Binding Site-Leucine Rich Repeat (TIR-NBS-LRR) proteins. T-DNA lines for five genes in region 111 were evaluated but no strong effects on ABA found (Fig. 3C). As for region 10 described above (Fig. 2F), redundant function of the TIR-NBS-LRR genes in region 111 may obscure their effect on ABA accumulation. Region 88 contained the SNPs of lowest and fourth lowest P value in our GWAS analysis (Fig. 3D). Mutants of two genes, At5g01730 and At5g01747, in this region had no effect on ABA accumulation (Fig. 3D). While not ruling out these two genes, the data suggest AT5G01740 as a promising candidate. AT5g01740 encodes an uncharacterized Nuclear Transport Factor 2 family protein, the expression of which was up-regulated in plants overexpressing the stress-associated protein XERICO, which influences ABA accumulation and sensitivity (47). Lack of T-DNA mutants prevented us from testing the effect of AT5G01740.
The GWAS analysis identified few candidate genes with previously established stress or ABA-related function, and those that were identified had relatively weak association. Nonetheless, some of these were tested, including AT4G09570 (calcium-dependent protein kinase 4, region 74) and Phytocystatin 6 (CYS6, region 43), which affect ABA signaling and abiotic stress resistance, the senescence- and dehydration-related gene AT3G21600, and the Unfolded Protein Response genes bZIP28 and bZIP17. None of these mutants affected ABA accumulation with the exception of bZIP17 where three mutants gave conflicting results (SI Appendix, Fig. S9 and Dataset S5). For these cases, as well as other regions of interest, a further test of candidate genes as well as analysis of specific natural alleles of candidate genes is promising for finding additional factors affecting ABA accumulation and drought response.
Discussion
ABA phenotyping of nearly 300 Arabidopsis accessions dramatically illustrated the range of natural variation in low-ψw–induced ABA accumulation. This raises the possibility that variation in ABA content may be a factor in the natural variation of various other drought-related traits that has been observed (20, 21, 24). Interestingly, we did not detect any tendency of accessions originating from dry environments to accumulate more ABA. Although this may at first seem surprising, it is consistent with the idea that ABA accumulation is an emergency signal (7). Thus, accessions that habitually face drought may have other adaptations or have an altered response to ABA that make it unnecessary to accumulate more ABA than other accessions. The accession ABA dataset will be a resource for further ecophysiology and genomic analyses as well as for identification of new ABA effector genes.
In agreement with other studies (25, 26), we found that GWAS results contain a wealth of information beyond just a few lowest-P-value SNPs. As the SNPs analyzed by GWAS are in varying degrees of linkage disequilibrium with ABA accumulation and with each other, the association signal (SNP P values) can be influenced by effect size and age of the polymorphism as well as local patterns of recombination and linkage disequilibrium. Thus, there is a compelling logic to also consider clusters of SNPs with more moderate P values, and this proved to be an effective guide to identify ABA effector genes. Interestingly, when we compared the genes with significant or marginally significant effects on ABA (Dataset S5) to gene expression profiling conducted under the same stress condition (48), only AT3G28740 (Fig. 3A) was differentially expressed (3.9-fold increase in Col-0 at low ψw compared with the unstressed control). Thus, the GWAS–reverse genetics approach can identify effector genes that are not transcriptionally regulated by stress and is complementary to transcriptome analyses commonly used to identify drought-related genes. That said, alternative strategies that do incorporate gene expression to identify candidate genes or that consider different linkage intervals to associate candidate genes with low-P-value SNPs could also be effective. The optimal strategy also depends on the importance of reducing the false positive rate to minimize downstream validation versus generating a somewhat broader set of candidates for validation (as we did here). It should also be noted that lack of phenotype in T-DNA mutant cannot conclusively rule out a candidate gene as specific natural alleles may lead to altered function that cannot be recapitulated by knockout mutants or may be effective only in specific genetic backgrounds because of epistatic interactions with other loci.
The GWAS-guided reverse genetic approach could identify genes of moderate effect unlikely to be found in forward genetic screens where stronger phenotypes are typically required to selected a mutant from a large population. The mutants that we tested generally had incremental effects on ABA accumulation (20–30%) consistent with distributed control of ABA accumulation among the many branches of ABA metabolism and, possibly, among multiple signaling mechanisms affecting ABA levels. Gene redundancy and genetic interactions are still factors, and we anticipate that construction of multigene knockouts with close homologs of our identified ABA regulators, generation of additional overexpression lines, or study of natural alleles and use of genetic backgrounds other than the Col-0 reference accessions could reveal even stronger effects on ABA accumulation. Also, we identified more mutants with increased ABA accumulation than with decreased ABA, suggesting that during drought acclimation ABA accumulation is under negative regulation as part of homeostasis mechanisms that match ABA level to stress severity.
For specific ABA effector genes, a main theme that emerges from our data is the identification of multiple plasma membrane proteins affecting ABA accumulation. These include RLKs, START domain protein, aspartic peptidase, and ATAucsisa1 (Fig. 2). A number of additional regions of interest include candidate genes that encode proteins with known or putative plasma membrane localization (regions 15, 23, 99, and 114, Dataset S3). Also, a number of other regions (2, 3, 4, 7, 16, 24, 66, 76, 81, 94, and 102) contain candidate genes encoding RLKs (including genes annotated as encoding transmembrane TIR-NBS-LRR proteins or lectin domain protein kinases). Most of these are of unknown phenotypic function. Thus, even within the RLKs, our approach turned up unexpected candidates. As the plasma membrane–cell wall interface is a likely location for sensing and signaling events that directly respond to loss of water or turgor change (15, 16), these findings are consistent with the hypothesis that using ABA accumulation as a marker could find upstream drought sensing and signaling factors. Although the RLKs are of interest, perhaps the strongest candidate that emerged from our analyses was the START domain/polyketide cyclase AT3G23080, based on its effect on ABA accumulation and leaf water loss as well as the association of AT3G23080-adjacent SNPs and loss-of-function alleles with climate variation. As there is no known function for AT3G23080, we now have the opportunity (and challenge) to analyze its cellular function, and that of other ABA effector genes, in detail.
Materials and Methods
Low-ψw stress was imposed by transferring 7-d-old seedlings to polyethylene glycol-infused agar plates. ABA was quantified by gas chromatography–tandem mass spectrometry using deuterated ABA as an internal standard. GWAS was conducted using the efficient mixed-model association (EMMA) software package (27) followed by scoring and ranking of candidate genes (25). Further details can be found in SI Appendix, SI Materials and Methods and Dataset S7.
Supplementary Material
Acknowledgments
We thank Min May Wong for help with the transgenic plant analysis. This work was supported by Academia Sinica and Taiwan Ministry of Science and Technology Grant 102-2628-B-001-003 and Grant 105-2628-B-001-010 (to P.E.V.) and National Science Foundation Grant IOS-0618347 (to T.E.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705884114/-/DCSupplemental.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl 1):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Pierce M, Raschke K. Correlation between loss of turgor and accumulation of abscisic acid in detached leaves. Planta. 1980;148:174–182. doi: 10.1007/BF00386419. [DOI] [PubMed] [Google Scholar]
- 5.Creelman RA, Zeevaart JAD. Abscisic acid accumulation in spinach leaf slices in the presence of penetrating and nonpenetrating solutes. Plant Physiol. 1985;77:25–28. doi: 10.1104/pp.77.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sussmilch FC, Brodribb TJ, McAdam SAM. Up-regulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. J Exp Bot. 2017;68:2913–2918. doi: 10.1093/jxb/erx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verslues PE. ABA and cytokinins: Challenge and opportunity for plant stress research. Plant Mol Biol. 2016;91:629–640. doi: 10.1007/s11103-016-0458-7. [DOI] [PubMed] [Google Scholar]
- 8.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 9.Ren H, et al. Dynamic analysis of ABA accumulation in relation to the rate of ABA catabolism in maize tissues under water deficit. J Exp Bot. 2007;58:211–219. doi: 10.1093/jxb/erl117. [DOI] [PubMed] [Google Scholar]
- 10.Verslues PE, Bray EA. LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiol. 2004;136:2831–2842. doi: 10.1104/pp.104.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quarrie SA. Genotypic differences in leaf water potential, abscisic acid and proline concentrations in spring wheat during drought stress. Ann Bot. 1980;46:383–394. [Google Scholar]
- 12.Wang ZL, Huang BR, Bonos SA, Meyer WA. Abscisic acid accumulation in relation to drought tolerance in Kentucky bluegrass. HortScience. 2004;39:1133–1137. [Google Scholar]
- 13.Qin X, Zeevaart JAD. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priest DM, et al. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006;46:492–502. doi: 10.1111/j.1365-313X.2006.02701.x. [DOI] [PubMed] [Google Scholar]
- 15.Haswell ES, Verslues PE. The ongoing search for the molecular basis of plant osmosensing. J Gen Physiol. 2015;145:389–394. doi: 10.1085/jgp.201411295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins NE, 2nd, Dinneny JR. The divining root: Moisture-driven responses of roots at the micro- and macro-scale. J Exp Bot. 2015;66:2145–2154. doi: 10.1093/jxb/eru496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ZY, et al. The Arabidopsis vacuolar sorting receptor1 is required for osmotic stress-induced abscisic acid biosynthesis. Plant Physiol. 2015;167:137–152. doi: 10.1104/pp.114.249268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrero JM, et al. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006;29:2000–2008. doi: 10.1111/j.1365-3040.2006.01576.x. [DOI] [PubMed] [Google Scholar]
- 19.Marshall A, et al. Tackling drought stress: Receptor-like kinases present new approaches. Plant Cell. 2012;24:2262–2278. doi: 10.1105/tpc.112.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clauw P, et al. Leaf growth response to mild drought: Natural variation in Arabidopsis sheds light on trait architecture. Plant Cell. 2016;28:2417–2434. doi: 10.1105/tpc.16.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Des Marais DL, et al. Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell. 2012;24:893–914. doi: 10.1105/tpc.112.096180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bac-Molenaar JA, Granier C, Keurentjes JJB, Vreugdenhil D. Genome-wide association mapping of time-dependent growth responses to moderate drought stress in Arabidopsis. Plant Cell Environ. 2016;39:88–102. doi: 10.1111/pce.12595. [DOI] [PubMed] [Google Scholar]
- 23.Lovell JT, et al. Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell. 2015;27:969–983. doi: 10.1105/tpc.15.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney AM, McKay JK, Richards JH, Juenger TE. Direct and indirect selection on flowering time, water-use efficiency (WUE, δ (13)C), and WUE plasticity to drought in Arabidopsis thaliana. Ecol Evol. 2014;4:4505–4521. doi: 10.1002/ece3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN. Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol. 2014;164:144–159. doi: 10.1104/pp.113.224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtin SJ, et al. Validating genome-wide association candidates through quantitative variation in nodulation. Plant Physiol. 2017;173:921–931. doi: 10.1104/pp.16.01923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bang WY, Kim SW, Jeong IS, Koiwa H, Bahk JD. The C-terminal region (640-967) of Arabidopsis CPL1 interacts with the abiotic stress- and ABA-responsive transcription factors. Biochem Biophys Res Commun. 2008;372:907–912. doi: 10.1016/j.bbrc.2008.05.161. [DOI] [PubMed] [Google Scholar]
- 31.Koiwa H, et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA. 2002;99:10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong L, et al. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Takahashi D, Kawamura Y, Uemura M. Comparison of plasma membrane proteomic changes of Arabidopsis suspension-cultured cells (T87 line) after cold and ABA treatment in association with freezing tolerance development. Plant Cell Physiol. 2012;53:543–554. doi: 10.1093/pcp/pcs010. [DOI] [PubMed] [Google Scholar]
- 35.Zhang ZJ, Peck SC. Simplified enrichment of plasma membrane proteins for proteomic analyses in Arabidopsis thaliana. Proteomics. 2011;11:1780–1788. doi: 10.1002/pmic.201000648. [DOI] [PubMed] [Google Scholar]
- 36.Molesini B, Pandolfini T, Pii Y, Korte A, Spena A. Arabidopsis thaliana AUCSIA-1 regulates auxin biology and physically interacts with a kinesin-related protein. PLoS One. 2012;7:e41327. doi: 10.1371/journal.pone.0041327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holman TJ, et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4549–4554. doi: 10.1073/pnas.0810280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graciet E, et al. The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA. 2009;106:13618–13623. doi: 10.1073/pnas.0906404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiu S-H, Bleecker AB. Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verslues PE, Sharma S. Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book. 2010;8:e0140. doi: 10.1199/tab.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. 1001 Genomes Consortium. Electronic address: magnus.nordborg@gmi.oeaw.ac.at; 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166:481–491. [DOI] [PMC free article] [PubMed]
- 43.Lasky JR, et al. Natural variation in abiotic stress responsive gene expression and local adaptation to climate in Arabidopsis thaliana. Mol Biol Evol. 2014;31:2283–2296. doi: 10.1093/molbev/msu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Köster J, et al. Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 2012;159:391–402. doi: 10.1104/pp.112.194274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sze H, Schumacher K, Müller ML, Padmanaban S, Taiz L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H(+)-ATPase. Trends Plant Sci. 2002;7:157–161. doi: 10.1016/s1360-1385(02)02240-9. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet. 2016;48:1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- 47.Ko J-H, Yang SH, Han K-H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 48.Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160:379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.