Significance
Using a combined approach relying on mass spectrometric analysis and molecular phylogeny, a fungus was identified that produced the gichigamins, which are peptaibols that contain a remarkable combination of structural features. The gichigamins possess a repeating α-residue/α-residue/β-residue motif creating a 311-P-helix secondary structure. These structural elements confer upon the gichigamins the unique ability among peptaibols to enter into cells whereupon they disrupt mitochondrial function. Semisynthetic modifications further enhanced gichigamin mitochondrial depolarization and cytotoxicity, while removing virtually all plasma-membrane pore-forming capabilities. These discoveries open vistas for engineering peptaibols into potent cytotoxins and intracellular delivery tools that are devoid of ion leakage effects.
Keywords: peptaibol, fungi, natural products, gichigamin, mitochondria
Abstract
Mass-spectrometry-based metabolomics and molecular phylogeny data were used to identify a metabolically prolific strain of Tolypocladium that was obtained from a deep-water Great Lakes sediment sample. An investigation of the isolate’s secondary metabolome resulted in the purification of a 22-mer peptaibol, gichigamin A (1). This peptidic natural product exhibited an amino acid sequence including several β-alanines that occurred in a repeating ααβ motif, causing the compound to adopt a unique right-handed 311 helical structure. The unusual secondary structure of 1 was confirmed by spectroscopic approaches including solution NMR, electronic circular dichroism (ECD), and single-crystal X-ray diffraction analyses. Artificial and cell-based membrane permeability assays provided evidence that the unusual combination of structural features in gichigamins conferred on them an ability to penetrate the outer membranes of mammalian cells. Compound 1 exhibited potent in vitro cytotoxicity (GI50 0.55 ± 0.04 µM) and in vivo antitumor effects in a MIA PaCa-2 xenograft mouse model. While the primary mechanism of cytotoxicity for 1 was consistent with ion leakage, we found that it was also able to directly depolarize mitochondria. Semisynthetic modification of 1 provided several analogs, including a C-terminus-linked coumarin derivative (22) that exhibited appreciably increased potency (GI50 5.4 ± 0.1 nM), but lacked ion leakage capabilities associated with a majority of naturally occurring peptaibols such as alamethicin. Compound 22 was found to enter intact cells and induced cell death in a process that was preceded by mitochondrial depolarization.
Natural product chemical research has evolved in many extraordinary ways over the last decade largely due to the integration of powerful and accessible analytical chemistry (1, 2) and molecular biology (3, 4) tools. Some of these techniques have been applied toward the creation of natural product big-data sets in which genomic, metagenomic, metabolomic, and proteomic approaches are combined to obtain new insights into nature’s remarkable chemical bounty (4–8). The predictive capabilities of these methods have helped guide efforts in several fertile areas of natural products research including the DNA cross-linking enediynes (9), bioactive cyanobacterial metabolites (10, 11), cytotoxic pentangular polyphenols (12), and thiopeptide antibiotics (13).
The integration of analytical and molecular approaches is an effective way to find new natural product analogs that exhibit enhanced biological activities and improved pharmacological attributes (e.g., greater potency, better solubility, enhanced therapeutic index, etc.), but it also has the potential to uncover compounds with activities that are markedly dissimilar to other members of the queried chemical class. Nature is well known for its chemical repurposing capabilities, which allow populations of organisms to evolve new biological functions from existing molecular scaffolds. The gonanes (perhydrocyclopenta[a]phenanthrenes) are a classic example of chemical repurposing with members of this diverse natural-product family acting as hormones (e.g., progesterone), bile acids (e.g., cholic acid), membrane stabilizers (e.g., cholesterol), toxins (e.g., digitoxin), as well as serving other roles (14). However, identifying functional outliers from among a group of chemically related natural products is not trivial; it requires adding a bioassay search component to the gene- or chemistry-based search process to detect a metabolite’s biological attributes.
The peptaibols are a class of nonribosomal-peptide-synthetase (NRPS)-derived metabolites produced by several types of fungi (15, 16). These compounds have attracted considerable interest due in large part to their antimicrobial properties and potential for biocontrol applications. Peptaibols are typically defined as peptidic natural products that contain 5–20 amino acid residues, are rich in nonproteinogenic amino acids including α-aminoisobutyric acid (Aib) and isovaleric acid (Iva), exhibit a C-terminal alcohol, and contain an N-terminal acetyl group. The peptaibols have been intensively studied because of their abilities to form pores in the outer membranes of cells; these pore-forming capabilities result in most compounds in this class exhibiting nonspecific cytotoxic properties toward bacterial, fungal, and mammalian cells (15, 17). The effects of peptaibols on mammalian cells tend to be relatively modest, causing the loss of cell viability at concentrations in the mid-to-high micromolar range (18–25). With many hundreds of peptaibols reported, finding new peptaibol analogs that possess an alternative mode of biological activity would be an impracticable prospect if one relied solely on either chemically driven or genome-based approaches to identify candidate molecules. Instead, an exploration that coupled these techniques with a bioassay-guided search tool could conceivably provide a reasonable opportunity to capture natural products that are both chemically unique and functionally divergent from the pool of known structurally analogous metabolites.
In this article, we describe the gichigamins, which are members of the peptaibol class of natural products. While their initial detection arose from mass spectrometric and genomic-based screening, parallel bioassay testing revealed a marked change in the biological properties of these metabolites. A subtle shift in the combination of structural features present in the gichigamins has endowed these peptaibols with both a unique 3D structure and distinct cell-penetration properties that foster interaction with intracellular targets. Semisynthetic modifications were identified that have opened a path to transforming the gichigamins into peptides that are devoid of outer membrane ion channel-forming properties and set a benchmark within this metabolite class for mammalian-cell cytotoxicity.
Results and Discussion
Identification of Tolypocladium sp. Sup5-1 (T2) by Laser Ablation Electrospray Ionization Mass Spectrometry–Principal Component Analysis.
Our prior discovery of the unusual metabolite maximiscin from the Alaskan-soil-derived Tolypocladium sp. T1 isolate (26) prompted us to search our laboratory’s collection of >25,000 fungal isolates for additional Tolypocladium sp. specimens that produced rich assortments of natural products. Principal component analysis (PCA) of laser ablation electrospray ionization mass spectrometry (LAESIMS) data (Fig. 1) obtained for 61 putative Tolypocladium sp. isolates identified a single isolate that clustered with the Alaskan Tolypocladium sp. T1. The isolate, which was derived from a deep-water sediment sample collected from Lake Superior, Michigan (depth ∼152 m), showed high sequence similarity spanning the rRNA ITS1-5.8S-ITS2 region (i.e., fungal “barcode” region) (26) to the Alaskan isolate. The fungal isolate has been designated here as Tolypocladium sp. Sup5-1 (T2). Our interest in pursuing further studies of this fungal isolate were reinforced when bioassay data obtained using the crude organic fungal extract revealed that the sample exhibited remarkably potent cytotoxicity against a pancreatic cancer cell line (MIA PaCa-2).
Fig. 1.
Identification of Tolypocladium sp. T2 using the LAESIMS-PCA. (A and B) LAESIMS profiles of Tolypocladium sp. T1 (A) and T2 (B). (C) PCA analysis of the LAESIMS profiles for the pool of 61 selected fungal isolates (red and blue diamonds) investigated in this study.
Metabolomics Analysis of Tolypocladium sp. Sup5-1 (T2).
To explore the secondary metabolite capabilities of Tolypocladium sp. T2, the fungus was grown in several types of broth media. Growth of the fungus in a modified potato dextrose broth (PDB) resulted in the generation of numerous new and known members of the malettinin family of polyketides [malettinins A (27), B (28), and F–H (16‒20), respectively] (Fig. 2A). However, the addition of NaNO3 to the PDB resulted in the suppression of polyketide biosynthesis and instead led to the production of at least 15 peptide natural products (1‒15), including the 22-mer peptaibol, gichigamin A (1), and six analogs [gichigamins B‒G (2‒7, respectively)] (Fig. 2B). The remaining peptides consisted of a series of 11-mer lipopeptaibols (8‒13), which were identified as dakwaabakains A-E (8‒12) and LP237-F7 (13) (25).
Fig. 2.
Secondary metabolite profiles of Tolypocladium sp. T2. HPLC analysis (PDA scan at 200‒600 nm) of the secondary metabolite profiles of T2 grown in PDB medium (A) and PDB medium with 2 g/L NaNO3 (B). Isolated compounds are categorized in the dashed boxes according to their regions on the chromatograms. Structures of the secondary metabolites from T2 are shown in C and D. Additional structural information and detailed renderings of the compounds discussed in this report are provided in SI Appendix. Amino acid codes for the rare residues: BALA, beta-alanine; J, isovaline; PA, pipecolic acid; U, 2-aminoisobutyric acid.
Primary Structures of the Tolypocladium sp. T2 Metabolites.
The structures of the peptides (1–12) and polyketides (18–20) (Fig. 2 C and D) were determined by high-resolution multidimensional NMR [1H and 13C NMR, 1H-1H COSY (1H-1H correlation spectroscopy), HSQC (heteronuclear single quantum correlation), HMBC (heteronuclear multiple bond correlation), TOCSY (total correlation spectroscopy), and ROESY (rotating frame overhauser effect spectroscopy)] (SI Appendix, Fig. S3B), electronic circular dichroism (ECD) (SI Appendix, Fig. S6), high-resolution (accurate mass) and collision-induced dissociation mass spectrometry (SI Appendix, Fig. S4), as well as analysis of Marfey’s reaction products (29) (SI Appendix, Fig. S5). Gichigamin A (1) and its structural analogs (2 and 5–7) are particularly long (22-mer) among peptaibols that have been purified from a natural source (16) (SI Appendix, Fig. S8B). The gichigamins are rich in the uncommon residue, β-alanine (BALA), which is otherwise rare among the peptaibols (16, 18, 30, 31) (SI Appendix, Fig. S8A). It has been proposed that the introduction of β-amino acids into the peptides provides increased resistance to hydrolysis and contributes to their stability in biological systems (32, 33). Additionally, the gichigamins lack both glutamine (Gln) and glutamic acid (Glu) residues, which are reported to be important for peptaibols to form ion channels (15, 34) (SI Appendix, Fig. S8A).
Single-Crystal X-Ray Diffraction Analysis of 1.
After many attempts, crystals of 1 were obtained from a concentrated MeOH solution, which provided an opportunity for single-crystal X-ray diffraction analysis (see details in SI Appendix). The X-ray crystal diffraction studies were carried out on an in-house macromolecule diffractometer (Rigaku MicroMac 008HF and Dectris Pilatus 200K detector at 100 K, Cu Kα) due to the weak diffraction of the small crystals of 1 obtained on a small-molecule diffractometer (Bruker-AXS APEX CCD on a D8 platform goniometer, Mo Kα). As a consequence of the combined data collection process, structure determination methods, and structure refinement procedures, it was realized that the crystal data for compound 1 exhibited many macromolecule-like features, including a precipitous drop of R and Rfree upon anisotropic refinement that had stemmed from the relatively large number of atoms in the unit cell and C-terminal disorder. The 1.1-Å-resolution crystal structure of 1 (Fig. 3A), which was refined to a final R and Rfree of 10.98 and 12.06%, respectively, contained two molecules in the unit cell organized as compacted antiparallel lamellar sheets (SI Appendix, Table S17). Although a reliable Flack factor was not available for determining absolute configuration directly, the unambiguous relative configuration assigned based on the crystal structure of 1 was consistent with the NMR assignments and the Marfey analysis. Upon further examination of the secondary structure of 1, the N-terminal residues Aib2-Phe6 formed a type IV β-turn in an α-β-motif (35) with the Pro2 carbonyl hydrogen bonded to the i + 3 and i + 4 amines (Phe5 and Iva6) (Fig. 3B). Residues 7–20 were determined to form a unique 311-P helix with the final BALA and Gly-ol arranged in a coil (Fig. 3 A and C). The 311-P helix was designated as a right-handed helix with a 3-residue repeat and 11 backbone atoms per helix according to the peptide nomenclature proposed by Bragg et al. (36). Both the ECD and VCD (vibrational circular dichroism) spectra (SI Appendix, Figs. S6 and S7) revealed that in solution, the 311-P helix exhibited spectroscopic features that are similar to a 310 helix (37, 38) (Fig. 3D). The BALA-rich 311-P helix results from a repeating α-residue/α-residue/β-residue (ααβ) pattern with the β-residue carbon causing distortion to the helical backbone. Consequently, the 311-P helix in the gichigamins is longer by ∼0.5 Å per turn compared with a canonical 310 helix (39). As a result, the overall length of 1 was determined to be ∼41.6 Å (40). The gichigamins’ repeating ααβ motif and 311-P helix constitutes an unusual secondary structural element among naturally occurring peptide.
Fig. 3.
Single-crystal X-ray diffraction of gichigamin A (1). (A) The complete structure of the gichigamin A monomer generated using PyMol. The C-terminal Gly-ol hydroxyl is displayed with the highest occupancy model (73%). (B) N-terminal α-β-motif of 1. Pro2 is shown coordinating through two hydrogen bonds to the i + 3 and i + 4 residues. (C) A perpendicular view of the unique 311-P-helical structure resulted from the inclusion of BALA residues in 1. The N- and C-termini were removed for clarity. (D) For the purpose of comparison, a canonical 310 helix is shown that was generated with Φ- and Ψ-angles of −49° and −29° for poly-Ala dodecamer using the PepMake 1.2 server at pepmake.wishartlab.com/.
Genome Analysis of Tolypocladium sp. Sup5-1 (T2).
Genomic analysis of Tolypocladium sp. T2 (see details of genome sequencing and assembly in ref. 41) using the AntiSMASH secondary-metabolite gene-cluster search tool (version 3.0.2) (42) (see details of genome analysis in SI Appendix) revealed an NRPS-PKS (NPT2) cluster with a 21-module NRPS on contig 75, open-reading frames 0–6 (Fig. 4 and SI Appendix, Fig. S1 and Table S18). The 21-module NRPS was unequally split into an NRPS-PKS gene (ctg75_orf0, 30 kb, encoding one AT module and eight NRPS modules) and an NRPS gene (ctg75_orf3, 42 kb, encoding 13 NRPS modules), which were separated by an ABC transporter and a small hypothetical gene. Substrate prediction, utilizing the Stachelhaus et al. (43) and NRPSPredictor 2.0 (44) methods, revealed high similarities between the genomically derived peptaibol structure prediction (based on bioinformatics analysis; see SI Appendix, Table S16) and the experimentally determined structures of gichigamins A–G (1–7). Specifically, positions 2, 3, 4, 7, 9, 12, 18, and 19 were accurately predicted between both Stachelhaus and NRPSPredictor-2.0 algorithms with 100% agreement on the proline positions and volumetric agreement with d-isovaline and 2-aminoisobutyric acid positions. Substrate prediction indicated that this cluster was the only reasonable candidate gene cluster for the production of gichigamins A–G (1–7) in the Tolypocladium sp. T2 genome.
Fig. 4.
Comparison of the biosynthetic clusters NPT1, NPT2, and NPTi following analysis by AntiSMASH v3.0.2 (fungismash.secondarymetabolites.org/#!/start). The numbers and codes appearing in this figure identify the isolates, contigs, and genes discussed in this article. The three putative NRPS-PKS clusters each contained 21 NRPS modules, as well as the signature of a PKS module [an acyltransferase (AT) and a ketosynthase (KS)].
A plausible biosynthetic pathway was subsequently proposed for gichigamin A (1, SI Appendix, Fig. S1). The initiating AT module of the NRPS-PKS gene (ctg75_orf0) incorporated an acetate group followed by chain elongation with each NRPS module from ctg75_orf0 and ctg75_orf3 resulting in the 21-residue peptaibol product 3. Gichigamin A (1) formation was terminated by linkage of 2-aminoethanol to the carboxyl terminus of the peptide. We speculate that the 2-aminoethanol is a reaction product of a separate enzyme system (45). Analogous 21-module NRPS clusters have been reported from the genomes of Tolypocladium sp. T1 (NPT1) (41) and a recently sequenced Tolypocladium inflatum (NPTi) (46) (Fig. 4 and SI Appendix, Tables S16 and S18). However, despite efforts to manipulate the growth media for these fungi, no metabolite in the expected mass range for the products of NPT1 and NPTi has been reported from these isolates.
In Vitro Antiproliferative and Cytotoxic Activities of Gichigamins.
The antiproliferative and cytotoxic effects of the purified gichigamins A–G (1–7), dakwaabakains A-E (8–12), and LP237-F7 (13) were evaluated against a pancreatic cancer cell line (MIA PaCa-2). Compounds 1‒7 exhibited a wide range of antiproliferative and cytotoxic potencies (SI Appendix, Table S19). The concentrations at which gichigamins inhibited cell proliferation by 50% (GI50) ranged from 0.23 µM for 2 to 1.58 µM for 4, whereas the concentration that induced 50% cytotoxicity (LC50) ranged from 0.81 µM for 1 to >10 µM for 6 and 7. It was observed that replacement of an Iva residue with an Aib residue at positions 6, 12, and/or 19 led to reduced potencies among the gichigamins (1–7). This structure-activity pattern was repeated in a second pancreatic cancer cell line (PANC-1; refer to SI Appendix, Table S19).
The 11-mer lipopeptaibols (8‒13) exhibited less activity (GI50 >1 µM) relative to the gichigamins, which was anticipated based on published data for related metabolites (15). Alamethicin, an archetypical peptaibol, was eightfold less potent than 1 against PANC-1 cells (SI Appendix, Table S19). This agrees with widely reported cytotoxic properties described for most known peptaibols (18–25). To provide an overall better understanding of the antiproliferative and cytotoxic properties of 1, the compound was tested in the NCI-60 panel, which revealed that all cell lines were relatively sensitive to the peptaibol with average GI50 and LC50 values of 0.15 and 0.88 μM, respectively (SI Appendix, Table S20). Examination of these results by COMPARE analysis (47, 48) did not provide any specific indicators for a potential mode of action since there were no notable differences in potency or efficacy among the queried cell lines. Combined, these data revealed that the gichigamins were more potent cytotoxic agents than most peptaibols, which provided a strong indicator that the remarkable structural features of the gichigamins might have yielded peptaibol analogs with a distinct mode of action.
In Vivo Antitumor Activity of 1.
Based on the potent in vitro cytotoxicity of 1, the in vivo antitumor effects of 1 were evaluated in a MIA PaCa-2 xenograft mouse model (Fig. 5). Female athymic nude (nu/nu) mice were implanted bilaterally with MIA PaCa-2 tumor fragments that had been propagated in nu/nu mice. Stocks of 1 were made up at 2 mg/mL in ethanol (EtOH) and diluted in PBS to a final concentration of 2.6% EtOH immediately before i.p. injection. Statistically significant antitumor effects were observed 2–6 d after an initial dose of 0.25 mg/kg of 1. However, an average 10% weight loss was observed in animals in this treatment group on days 2 and 3 that precluded additional dosing. Lower doses of 0.1 mg/kg were also administered to a second treatment group on days 0 and 3, which also showed statistically significant antitumor effects on days 3 and 6 compared with the control group (Fig. 5). This dosing regimen was associated with minor, <3%, body weight loss throughout the trial. These results indicate that 1 has significant and potent antitumor activity in the MIA PaCa-2 xenograft model within a relatively narrow therapeutic window.
Fig. 5.
Antitumor effects of 1 in a pancreatic tumor xenograft model. Athymic mice were bilaterally implanted with MIA PaCa-2 tumors and treated with 1 at a dose of 0.1 mg/kg on days 0 and 3 (open triangles) or 0.25 mg/kg on day 0 (open circles). The results represent the average change in tumor volume ±SE. *P < 0.05; **P < 0.01; ****P < 0.0001 compared with untreated control (filled squares) by a two-way ANOVA with a Dunnett’s posthoc test.
Gichigamin A Penetrates into Cells.
The mechanism of cell cytotoxicity induced by the peptaibols has been extensively studied and it has been consistently demonstrated that their abilities to induce cell death are related to their pore-forming capabilities (18–25). Considering the increased potencies and unique structural features of the gichigamins, we first investigated whether these peptaibols incorporate into membranes to form pores. For these tests, alamethicin was selected as the positive control due to its well-established in vitro pore-forming capabilities. Atomic force microscopy (AFM) (49) revealed that alamethicin induced deformation of membranes at 10 µM, similar to what has been described (50). In contrast, membranes treated with 10 µM of 1 were indistinguishable from untreated control membranes (Fig. 6 A–C). To determine if the gichigamins were able to cross lipid bilayers, an artificial membrane permeability assay (PAMPA) was performed (51) (Fig. 6D and SI Appendix, Table S21). In contrast to most peptaibols, 1 was found to readily penetrate the lipid membrane at concentrations as low as 0.5 µM. The passage of 1 through the membrane was not accompanied by leakage of calcein dye, even at concentrations as high as 5 μM. In comparison, application of 1–5 μM alamethicin to the membrane led to the rapid leakage of calcein, although the compound itself could not be detected as having crossed the membrane. At 10 μM, alamethicin and calcein both crossed the membrane (Fig. 6D), which is not surprising since this concentration also formed large holes in the membrane (Fig. 6B). These data demonstrated that 1, which appeared to have successfully passed through an intact lipid bilayer, acted in a way that was distinct from alamethicin. These observations were extended to whole cells using a calcein release assay (Fig. 6F). PANC-1 cells were incubated (30 min) with nonfluorescent acetoxymethyl calcein, which was taken up by the live cells and enzymatically hydrolyzed to form fluorescent calcein before peptaibol treatment. In corroboration with published results (52) and our PAMPA assay (Fig. 6D), 5 μM alamethicin caused the near-immediate release of calcein from the cells, whereas the same concentration of 1 resulted in no detectable release of calcein from the cells 1 h after treatment (Fig. 6F). However, as the concentration of 1 was increased to 10 μM, calcein leakage was detected, indicating that 1 can compromise the plasma membrane, albeit well beyond the concentrations that initiate cytotoxicity.
Fig. 6.
Membrane permeability assays. (A–C) AFM of supported lipid bilayers treated with (A) vehicle, (B) 10 µM alamethicin, or (C) 10 µM gichigamin A (1). (Scale bar: 2 μm.) (D) PAMPA assay using Corning Gentest Pre-Coated Plate System. Permeability of gichigamin A (1) and alamethicin was determined using FPLC-ESIMS and calcein leakage was monitored to determine the integrity of the artificial membrane. (E) Single-cell MS analysis of PANC-1 cells treated with 0.5 µM of 1. (F) Cell-based calcein release assay. PANC-1 cells were preincubated with calcein AM and Hoechst 33342 before treatment of gichigamin A (1) and alamethicin. Real-time fluorescence was monitored using the Operetta system.
To test if 1 was capable of entering intact PANC-1 cells, single-cell mass-spectrometric analysis (53) was used to monitor cells before and after treatment with 1 or alamethicin. At a concentration of 0.5 µM, 1 was readily detected in PANC-1 cells based on the presence of an ion ([M+2Na]2+ m/z 1,030.6031) (Fig. 6E), which served as an unequivocal signal for the presence of the gichigamin. The DNA-staining membrane-penetrating dye, Hoechst 33342 (1 µM), which was coadministered as a positive control, was readily detected within the intracellular environment along with 1. In contrast, alamethicin (0.5−10 µM) could not be detected within PANC-1 cells, although an intense signal was observed in the extracellular medium ([M+2Na]2+ m/z 1,004.5470). These results provided compelling evidence that the gichigamins entered cells at concentrations well below those required by archetypal peptaibols to compromise cell outer membranes and indicated that at cytotoxic concentrations, the gichigamins are able to enter cells whereupon they have the potential to interact with an intracellular target(s).
Partial Hydrolysis of Gichigamins Reveals Structural Requirements for Cell Penetration.
We hypothesized that the distinct cell penetration and membrane permeabilization properties of the naturally occurring gichigamins could result from their unique conformations and that modest changes to one or more amino acid residues could have profound effects on the biological activities of these compounds. To probe the combination of structural features required for potent cytotoxicity, we subjected 1 to partial acid hydrolysis. The resulting products were purified, structurally characterized, and tested for cytotoxicity to gain further insights into the relationships between the primary and secondary structural features of the gichigamins and their biological properties. Nine major products were obtained (24–32) from the hydrolysis of 1 (SI Appendix, Fig. S3A) and ECD spectra were collected and analyzed for each compound (SI Appendix, Fig. S6). These data revealed that only the three longest hydrolysis products, 24–26, exhibited Cotton effects (Δε210:Δε235 = 3:1) comparable to the 311-P-helix pattern (MeOH solvent) observed for 1. Subsequent testing revealed that compounds 24 and 25 retained cytotoxic properties similar to 1 against MIA PaCa-2 and PANC-1 cell lines (SI Appendix, Table S19). Further truncation of amino acid residues from either end of the gichigamins only served to abolish their cytotoxic effects at concentrations of up to 10 µM. Single-cell mass spectrometry comparing compounds 24 (1 µM) and 26 (1 µM) showed that only 24 was able to enter PANC-1 cells (SI Appendix, Fig. S10A), demonstrating a correlation between cell penetration and cytotoxicity for the gichigamins. The results of these experiments revealed that an amalgamation of structural features (i.e., peptaibol length, sequence/type of amino acid residues, and ability to adopt an ααβ 311-P-helical secondary structure) were critical for endowing the gichigamins with their unique cell penetration capabilities and cytotoxic properties.
Gichigamin A Disrupts Mitochondrial Function.
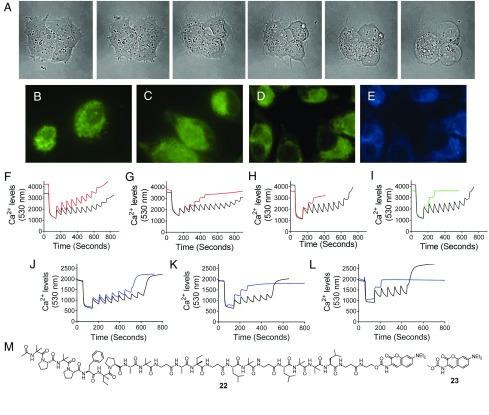
To garner insight into the potential intracellular target(s) of the gichigamins, we considered the sequence of distinctive morphological changes that occurred when cells were treated with these peptaibols. When PANC-1 cells were treated with 1 at twofold the GI50 value (1.6 μM), long, thin cellular protrusions began to form ∼5 h posttreatment (Fig. 7A). This phenomenon continued until ∼8 h posttreatment, whereupon the cells ballooned and the protrusions dissipated. This sequence of morphological changes preceding cell death was decidedly reminiscent of the cellular transformations induced by propofol, which causes mitochondrial depolarization, leading to increased intracellular calcium levels and restructuring of the actin skeleton (54). Indeed, TRITC-phalloidin staining of actin showed that 1 (0.5–2 μM) caused a concentration-dependent reorganization of the actin cytoskeleton along the cellular periphery (SI Appendix, Fig. S13). Follow-on microscopy experiments lent support to the supposition that gichigamins were disrupting mitochondrial function. Specifically, visualization of mitochondria using MitoTracker Green FM in cells treated with 1 (0.5 µM) revealed that mitochondrial morphology was markedly altered 4 h after treatment compared with vehicle treated cells (Fig. 7 B and C).
Fig. 7.
Effects of 1 and 22 on mitochondrial morphology, function, and colocalization. (A) PANC-1 cells were visualized by bright-field live microscopy 1, 3, 5, 6, 7, and 8 h after the addition of 1.6 µM 1. (B–E) The mitochondria of PANC-1 cells treated with vehicle (B), 0.5 μM 1 (C), or 0.05 μM 22 (D) were visualized using mitotracker Green FM (green) 4 h after compound addition. (E) Localization of 22 (blue) in the same field as mitochondrial staining in D. (F–L) Ca2+ uptake by purified mitochondria treated with vehicle (black traces), 1 (red traces) at 0.05 μM (F), 0.075 μM (G) or 0.1 μM (H); 0.075 μM alamethicin (I, green trace); or 22 (blue traces) at 0.005 μM (J), 0.01 μM (K), or 0.02 μM (L). (M) Structures of 22 and 23.
We next measured the effects of 1 on calcium uptake using purified mitochondria to determine whether the gichigamins could directly disrupt mitochondrial function. Control experiments showed that vehicle-treated mitochondria were able to readily uptake eight to nine doses of 5 μM calcium before becoming depolarized (Fig. 7 F–I, black traces). In comparison, acute administration of 1 blocked calcium uptake by the mitochondria in a concentration-dependent manner (0.05–0.1 μM). Although the indiscriminate membrane-disrupting agent alamethicin also had this effect on purified mitochondria (55) (Fig. 7I), our previous observations that the compound does not enter cells at up to 10 μM meant that this is not a relevant cytotoxic mechanism for this classical peptaibol at these concentrations. In comparison, our data demonstrated that 0.5–1 μM of 1 is sufficient to enter cells and cause disruption of the mitochondria, leading to calcium release and actin reorganization. These results suggested that disruption of mitochondrial structure and function occurred at concentrations of 1 that elicit cytotoxicity.
Despite the combination of remarkable chemical features and cell penetrating capabilities of 1, the gichigamins retained several aspects of chemical commonality with classic peptaibols. Since the calcein permeability assays could not rule out that the gichigamins were completely devoid of pore-forming properties, we turned to a series of patch-clamp electrophysiology experiments to investigate the effects of gichigamins on ion transport. Treatment of PANC-1 cells with the total growth inhibitory (TGI) concentration of 1 (1 µM) resulted in a modest increase to the inward and outward currents caused by both step and ramp voltage changes within 8 min of compound addition (Fig. 8A). Further increasing the amount of 1 applied to cells (10 µM) led to the rapid onset of linear leak currents, which agreed with results for the calcein leakage assay. Similar results were also observed in the MIA PaCa-2 cell line and are consistent with the known voltage dependence and pore-forming properties of the peptaibol alamethicin, which has long been used as a model compound for studying conductance through voltage-gated ion channels (56). Therefore, while 1 was clearly able to enter cells at cytotoxic concentrations without causing visible disruption to membranes in AFM and calcein release studies, the acute ion leakage initiated in patch-clamp studies, at its minimum cytotoxic concentration, suggested that 1 still retained a certain degree of plasma membrane-disrupting capacity, which likely contributed in part to its cytotoxic capabilities.
Fig. 8.
Effects of 1 and 22 on whole-cell patch-clamp electrophysiology and mitochondrial depolarization in PANC-1 cells. Shown are the effects of (A) 1 µM of 1 (n = 8) and (B) 0.025 μM of 22 (n = 8) on macroscopic whole-cell ionic currents elicited in individual representative PANC-1 cells. Control families of step-derived current recordings for A and B are in SI Appendix, Fig. S163 A and B, respectively. (A) Ramps recorded without compound (a), upon application (b), and after 5-min (c) and 8-min (d) exposure to 1. Acute and 5-min ramps exhibited small additional increases in inward currents. The 5-min ramp showed larger outward currents compared with the acute ramp. Subsequent rapidly developing linear leak currents (8 min) resulted in cell loss (all studied cells were lost between 10 and 15 min). (B) Shown are the first 12 ramps recorded over 15-min encompassing control, acute, and 5-min exposure to 22. Not shown for clarity are stable ramps recorded over the subsequent 30 min before cell loss due to breakdown of recording configuration. Ramps exhibited no changes in magnitude and voltage-dependent properties, nor did leak currents develop over the 35-min recording. Membrane potential (U) is shown in millivolts on the left vertical axis, current magnitude (I) is shown in picoampere on the right vertical axis, and time duration (time) of voltage steps and elicited currents is shown in milliseconds on the horizontal axis. (C and D) PANC-1 cells were treated for 4 h with 1, 22, or alamethicin at the indicated concentrations. The extent of cell death, as determined by 7-AAD staining (C), and mitochondrial depolarization, as measured by incorporation of mitopotential dye (D), were quantified by flow cytometry. n = 3–5 independent experiments ±SEM ****p < 0.0001 by two-way ANOVA with Dunnett’s posthoc test compared with vehicle control.
Semisynthetic Modification of Gichigamins Increases Potency.
While investigating the potential intracellular target(s) of the gichigamins, we prepared a semisynthetic coumarin-conjugated probe (22) of 1 for fluorescent microscopy studies (Fig. 7M). Although the chemical transformation was successful, we were taken by surprise that the semisynthetic derivative 22 was roughly 60-fold more potent than unconjugated gichigamin A (1) and over 700-fold more potent than a methylated coumarin control 23 (Fig. 7M) in both pancreatic cancer cell lines (SI Appendix, Table S19). To evaluate whether the change in the potency of 1 was specifically due to the addition of the coumarin group (22), 10 semisynthetic gichigamin analogs were prepared by adding different ester and carbamate groups to the C-terminus hydroxy (33–42, SI Appendix, Fig. S3C). Each of the new products showed increased potency in PANC-1 cells relative to 1 (SI Appendix, Table S19). The effect was most highly pronounced when aromatic moieties (e.g., phenyl, biphenyl, naphthyl, coumarin, anthracene, and anthraquinone) (33–39) were introduced. In contrast, the coumarin-linked analog of alamethicin (43) exhibited only a minor increase in potency in PANC-1 cells (GI50 6.75 μM for alamethicin and GI50 1.33 μM for 43), which buttressed the finding that the primary and secondary structural features of the gichigamins were essential to the overall unique biological activity of this group of peptaibols.
Intrigued by the dramatic increase in potency conferred by C-terminal modification of the gichigamins, we probed the biological effects of 22 versus 1. Costaining of PANC-1 cells with 0.05 μM of 22 and MitoTracker Green FM provided a visual indication that 22 entered cells and colocalized with mitochondria (Fig. 7 D and E and SI Appendix, Fig. S12). Similar to the results observed for 1 (Fig. 7C), 22 caused the mitochondria to appear morphologically disorganized and diffuse within 4 h of treatment (Fig. 7D), which is consistent with the ability of the gichigamins to alter mitochondrial structure and function. Additionally, 22 was found to be 10-fold more potent than 1 in its ability to directly depolarize purified mitochondria. We observed that concentrations of 22 as low as 5 nM greatly diminished mitochondrial capacity (Fig. 7J), whereas 0.01–0.02 μM was sufficient to completely inhibit mitochondrial function (Fig. 7 K and L). Entry of 22 into PANC-1 cells was confirmed by single-cell mass spectrometry; administration of 0.05 µM of 22 to PANC-1 cells produced a strong and unambiguous signal (m/z 1,159.6520 [M+2Na]2+ (SI Appendix, Fig. S10B), confirming the compound was present in the intracellular compartment at cytotoxic concentrations.
Semisynthetic Gichigamin Modification Alters the Mechanism of Cytotoxicity.
While 22 behaved similarly to 1 in its ability to penetrate cells and disrupt mitochondrial morphology, it was unique in that this intracellular mechanism of action was not associated with ion leakage. Whole-cell patch-clamp experiments revealed that treatment of both PANC-1 and MIA PaCa-2 cells with the TGI concentration of 22 (25 nM) did not produce any detectable increase in current magnitude when monitored for up to 35 min (Fig. 8B). Additionally, no ion leakage was observed with 22 at concentrations up to 1 μM, indicating that the structural modification used to generate compound 22 shifted its cytotoxic mechanism of action from acute ion permeabilization, which is a hallmark feature of the peptaibols, to one that could be related to its intracellular effect on mitochondrial depolarization.
To evaluate the relationship between mitochondrial disruption and cytotoxicity, a whole-cell mitochondrial depolarization assay (Muse Mitopotential kit; EMD Millipore) was performed. In this assay, mitochondrial integrity was monitored by tracking the accumulation of a cationic, lipophilic dye within inner mitochondrial membranes. PANC-1 cells were treated with 0.025–0.1 µM of 22 for 4 h and the number of live cells containing depolarized mitochondria was evaluated by flow cytometry. Concentrations of 0.025–0.1 μM of 22, which elicited cytotoxicity at 48 h (SI Appendix, Table S19), did not cause a significant increase in cell death within 4 h (Fig. 8C) and instead were associated with depolarization of the mitochondrial membrane at this early time point (Fig. 8D). The rapid accumulation of live cells with depolarized mitochondria at concentrations of 22 that eventually cause cytotoxicity suggested that the compound’s primary mechanism of cytotoxic action is due to its direct effects on mitochondrial membrane depolarization (MMP). In comparison, the TGI of 1 (1 μM) caused significant cytotoxicity within 4 h. This was identical to the effect of alamethicin at its cytotoxic concentration of 10 μM and was consistent with ion leakage being the primary cytotoxic mechanism of 1. Together, these results demonstrate that 1 and 22 have distinct early effects on cells at concentrations that eventually lead to cell death, with 22 causing a distinctive accumulation of cells with depolarized mitochondria before cell death.
The shift in the mechanism of action of 22 compared with 1 can also be inferred from the different concentration-response NCI-60 cytotoxicity profiles for the two compounds (SI Appendix, Fig. S11). As mentioned above, 1 exhibited similar potencies and efficacies against all cell lines with no clear indicators for a potential mode of action emerging from COMPARE analysis, which is consistent with a primary cytotoxic mechanism of action due to ion leakage. However, there was comparatively substantial variability in the potency of 22 against the NCI-60 cell lines. The data indicated a distinct mechanism of action for 22. COMPARE analyses with 22 showed highest homology to diverse compounds, including actinomycin D, chromomycin A3, and paclitaxel, which are all substrates for the P-glycoprotein (Pgp) drug efflux pump (57, 58). Additionally, the most resistant cell line to 22 in the NCI-60 panel was the Pgp-expressing NCI/ADR-RES OVCAR-8 derivative. In comparing the cytotoxicity profile of both 1 and 22, we found that 1 was twofold less potent in the Pgp-expressing NCI/ADR-RES cell line compared with parental OVCAR-8 cells, whereas 22 was 36-fold less potent in the Pgp-expressing line (SI Appendix, Table S20). These data further support that 22 has an internal mechanism of action, which can be attenuated by Pgp-efflux while internalization is not as critical for the cytotoxicity of 1. These results revealed that the unusual structural features of the gichigamins (e.g., 1 and its natural analogs) introduced a cell-penetration quality among these natural products and that a simple structural modification to the natural product (e.g., 22 and other semisynthetic analogs) was sufficient to shift this scaffold’s mechanism of cytotoxicity to an exclusively internal mechanism of action involving mitochondrial disruption.
Conclusions
The use of LAESIMS and PCA enabled us to identify and focus our chemical studies on a particularly exceptional secondary-metabolite producer, Tolypocladium sp. Sup5-1 (T2), which yielded the 22-mer peptaibol, gichigamin A (1) and several related gichigamin analogs. Although peptaibols had been extensively studied for their membrane-modifying and antimicrobial activities, comparatively little has been reported concerning their activities against mammalian cells. Chemical and biological investigations revealed several unique structural features in 1 (i.e., high content of β-amino acid residues and unique ααβ-repeat 311-P-helical structure), as well as potent cytotoxic activity against pancreatic tumor cell lines. Compared with other peptaibols, compound 1 readily entered cells and disrupted mitochondrial structure and function, although it also retained plasma membrane permeabilizing activities at cytotoxic concentrations. Semisynthetic modifications of the gichigamins dramatically improved potency and shifted the bioactivity profile among this family of natural products by diminishing ion leakage, which led to an internal mechanism of action associated with mitochondrial disruption. These results are important because this article documents the cell-penetrating capabilities for a member of peptaibol class of fungal metabolites (59, 60). Cell-penetrating peptides (CPPs) (61–63) are an important class of drug transporters that can deliver large-cargo molecules through cell membranes and thus may be useful in clinical applications. The unique structural features of gichigamins, which include the absence of positively charged amino acid residues (e.g., Arg and Lys) in contrast with common CPPs, provide a template for the design of cell-penetrating molecules that can carry a payload across the outer cell membranes of mammalian cells.
Materials and Methods
Details of all experimental methods, reagents, equipment, and protocols are provided in SI Appendix. This includes descriptions of the approaches used to solve the structures of the gichigamins and their semisynthetic analogs, in vitro and in vivo biological assays, single-cell mass spectrometry, LAESIMS-PCA experiments, and synthetic methods. Tables of NMR data along with MS, NMR, ECD, and VCD data, as well as accession codes for X-ray and genomic data, are also provided. Research pertaining to the use of mice was approved by the University of Texas Health Science Center at San Antonio (UTHSCSA) Institutional Review Board.
Supplementary Material
Acknowledgments
The authors are grateful for the help provided by Dr. Jiangnan Peng from UTHSCSA for collecting some of the NMR spectra. The authors also appreciate the help of Dr. Douglas R. Powell and Dr. Leonard M. Thomas from University of Oklahoma for collecting the X-ray diffraction data and providing suggestions to C.A.M. of elucidating the crystallographic structure of 1. The CytoPatch-4 automated patch-clamp instrument was gifted to the Center for Innovative Drug Discovery by CytoBioscience Incorporated (formerly Cytocentrics, Inc.). Research reported in this publication was supported in part by the National Institute of General Medical Sciences of the NIH, R01GM107490 (to R.H.C.) and San Antonio Area Foundation Grant (to A.L.R.). We also acknowledge Grant UL1 TR001120 from the National Center for Advancing Translational Sciences, NIH for its support of the Center for Innovative Drug Discovery and High Throughput Screening Facility at UTHSCSA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomes of Tolypocladium sp. MEA-2 (T1) have been deposited in the GenBank database (accession no. JPIJ00000000.2) and Tolypocladium sp. Sup5-1 (T2) (accession no. JPHH00000000.3). The X-ray data of compound 1 were deposited in the Worldwide Protein Data Bank, www.wwpdb.org (PDB ID code 4Z0W).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707565114/-/DCSupplemental.
References
- 1.Jarmusch AK, Cooks RG. Emerging capabilities of mass spectrometry for natural products. Nat Prod Rep. 2014;31:730–738. doi: 10.1039/c3np70121b. [DOI] [PubMed] [Google Scholar]
- 2.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: Current, emerging and future technologies. Nat Prod Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ongley SE, Bian X, Neilan BA, Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep. 2013;30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- 4.Adnani N, et al. Emerging trends for stimulating the discovery of natural products. In: Havlicek V, Spizek J, editors. Natural Products Analysis: Instrumentation, Methods, and Applications. Wiley; New York: 2014. pp. 115–161. [Google Scholar]
- 5.Letzel AC, Pidot SJ, Hertweck C. A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat Prod Rep. 2013;30:392–428. doi: 10.1039/c2np20103h. [DOI] [PubMed] [Google Scholar]
- 6.Milshteyn A, Schneider JS, Brady SF. Mining the metabiome: Identifying novel natural products from microbial communities. Chem Biol. 2014;21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlop-Powers Z, Milshteyn A, Brady SF. Metagenomic small molecule discovery methods. Curr Opin Microbiol. 2014;19:70–75. doi: 10.1016/j.mib.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cacho RA, Tang Y, Chooi YH. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front Microbiol. 2015;5:774. doi: 10.3389/fmicb.2014.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, et al. Strain prioritization and genome mining for enediyne natural products. MBio. 2016;7:e02104–e02116. doi: 10.1128/mBio.02104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss NA, et al. Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery. J Ind Microbiol Biotechnol. 2016;43:313–324. doi: 10.1007/s10295-015-1705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winnikoff JR, Glukhov E, Watrous J, Dorrestein PC, Gerwick WH. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J Antibiot (Tokyo) 2014;67:105–112. doi: 10.1038/ja.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HS, Brady SF. Mining soil metagenomes to better understand the evolution of natural product structural diversity: Pentangular polyphenols as a case study. J Am Chem Soc. 2014;136:18111–18119. doi: 10.1021/ja510606j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lednicer D. Steroid Chemistry at a Glance. Wiley; Hoboken, NJ: 2010. pp. 1–152. [Google Scholar]
- 15.de la Fuente-Nunez C, Whitmore L, Wallace BA. Peptaibols. In: Kastin A, editor. Handbook of Biologically Active Peptides. 2nd Ed. Academic; Cambridge, MA: 2013. pp. 150–156. [Google Scholar]
- 16.Stoppacher N, et al. The comprehensive peptaibiotics database. Chem Biodivers. 2013;10:734–743. doi: 10.1002/cbdv.201200427. [DOI] [PubMed] [Google Scholar]
- 17.Leitgeb B, Szekeres A, Manczinger L, Vágvölgyi C, Kredics L. The history of alamethicin: A review of the most extensively studied peptaibol. Chem Biodivers. 2007;4:1027–1051. doi: 10.1002/cbdv.200790095. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, et al. Microbacterins A and B, new peptaibols from the deep sea actinomycete Microbacterium sediminis sp. nov. YLB-01(T) Org Lett. 2015;17:1220–1223. doi: 10.1021/acs.orglett.5b00172. [DOI] [PubMed] [Google Scholar]
- 19.Oh SU, Yun BS, Lee SJ, Kim JH, Yoo ID. Atroviridins A-C and neoatroviridins A-D, novel peptaibol antibiotics produced by Trichoderma atroviride F80317. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot (Tokyo) 2002;55:557–564. doi: 10.7164/antibiotics.55.557. [DOI] [PubMed] [Google Scholar]
- 20.Shi M, et al. Antimicrobial peptaibols, novel suppressors of tumor cells, targeted calcium-mediated apoptosis and autophagy in human hepatocellular carcinoma cells. Mol Cancer. 2010;9:26. doi: 10.1186/1476-4598-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroux A, et al. Unprecedented 17-residue peptaibiotics produced by marine-derived Trichoderma atroviride. Chem Biodivers. 2013;10:772–786. doi: 10.1002/cbdv.201200398. [DOI] [PubMed] [Google Scholar]
- 22.Wilhelm C, Anke H, Flores Y, Sterner O. New peptaibols from Mycogone cervina. J Nat Prod. 2004;67:466–468. doi: 10.1021/np0302899. [DOI] [PubMed] [Google Scholar]
- 23.Mitova MI, et al. Evolving trends in the dereplication of natural product extracts. 2. The isolation of chrysaibol, an antibiotic peptaibol from a New Zealand sample of the mycoparasitic fungus Sepedonium chrysospermum. J Nat Prod. 2008;71:1600–1603. doi: 10.1021/np800221b. [DOI] [PubMed] [Google Scholar]
- 24.Ayers S, et al. Peptaibols from two unidentified fungi of the order Hypocreales with cytotoxic, antibiotic, and anthelmintic activities. J Pept Sci. 2012;18:500–510. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsantrizos YS, Pischos S, Sauriol F, Widden P. Peptaibol metabolites of Tolypocladium geodes. Can J Chem. 1996;74:165–172. [Google Scholar]
- 26.Du L, Risinger AL, King JB, Powell DR, Cichewicz RH. A potent HDAC inhibitor, 1-alaninechlamydocin, from a Tolypocladium sp. induces G2/M cell cycle arrest and apoptosis in MIA PaCa-2 cells. J Nat Prod. 2014;77:1753–1757. doi: 10.1021/np500387h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angawi RF, Swenson DC, Gloer JB, Wicklow DT. Malettinin A: A new antifungal tropolone from an unidentified fungal colonist of Hypoxylon stromata (NRRL 29110) Tetrahedron Lett. 2003;44:7593–7596. [Google Scholar]
- 28.Angawi RF, Swenson DC, Gloer JB, Wicklow DT. Malettinins B-D: New polyketide metabolites from an unidentified fungal colonist of Hypoxylon Stromata (NRRL 29110) J Nat Prod. 2005;68:212–216. doi: 10.1021/np049625r. [DOI] [PubMed] [Google Scholar]
- 29.Bhushan R, Brückner H. Marfey’s reagent for chiral amino acid analysis: A review. Amino Acids. 2004;27:231–247. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- 30.Grigoriev PA, Berg A, Schlegel B, Heinze S, Gräfe U. Formation of anion-selective membrane pores by texenomycin A, a basic lipopeptaibol antibiotic. J Antibiot (Tokyo) 2002;55:826–828. doi: 10.7164/antibiotics.55.826. [DOI] [PubMed] [Google Scholar]
- 31.He H, et al. Culicinin D, an antitumor peptaibol produced by the fungus Culicinomyces clavisporus, strain LL-12I252. J Nat Prod. 2006;69:736–741. doi: 10.1021/np058133r. [DOI] [PubMed] [Google Scholar]
- 32.Hook DF, et al. The proteolytic stability of “designed” beta-peptides containing alpha-peptide-bond mimics and of mixed alpha,beta-peptides: Application to the construction of MHC-binding peptides. Chem Biodivers. 2005;2:591–632. doi: 10.1002/cbdv.200590039. [DOI] [PubMed] [Google Scholar]
- 33.Steer DL, Lew RA, Perlmutter P, Smith AI, Aguilar MI. Beta-amino acids: Versatile peptidomimetics. Curr Med Chem. 2002;9:811–822. doi: 10.2174/0929867024606759. [DOI] [PubMed] [Google Scholar]
- 34.Sansom MS. The biophysics of peptide models of ion channels. Prog Biophys Mol Biol. 1991;55:139–235. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- 35.Golovin A, Henrick K. MSDmotif: Exploring protein sites and motifs. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bragg L, Kendrew JC, Perutz MF. Polypeptide chain configurations in crystalline proteins. Proc R Soc Lond A Math Phys Sci. 1950;203:321–357. [Google Scholar]
- 37.Toniolo C, Polese A, Formaggio F, Crisma M, Kamphuis J. Circular dichroism spectrum of a peptide 3(10)-helix. J Am Chem Soc. 1996;118:2744–2745. [Google Scholar]
- 38.Silva RAGD, et al. Discriminating 3(10)- from alpha-helices: Vibrational and electronic CD and IR absorption study of related Aib-containing oligopeptides. Biopolymers. 2002;65:229–243. doi: 10.1002/bip.10241. [DOI] [PubMed] [Google Scholar]
- 39.Vieira-Pires RS, Morais-Cabral JH. 3(10) helices in channels and other membrane proteins. J Gen Physiol. 2010;136:585–592. doi: 10.1085/jgp.201010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitmore L, Wallace BA. The peptaibol database: A database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamps BW, Du L, Mitchell CA, Cichecwicz RH, Stevenson BS. Draft genomes of two sordariomycete fungi that produce novel secondary metabolites. Genome Announc. 2015;3:e00291-15. doi: 10.1128/genomeA.00291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K, et al. antiSMASH 2.0–A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 44.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohr H, Kleinkauf H. Alamethicin biosynthesis: Acetylation of the amino terminus and attachment of phenylalaninol. Biochim Biophys Acta. 1978;526:375–386. doi: 10.1016/0005-2744(78)90129-8. [DOI] [PubMed] [Google Scholar]
- 46.Bushley KE, et al. The genome of tolypocladium inflatum: Evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013;9:e1003496. doi: 10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaharevitz DW, Holbeck SL, Bowerman C, Svetlik PA. COMPARE: A web accessible tool for investigating mechanisms of cell growth inhibition. J Mol Graph Model. 2002;20:297–303. doi: 10.1016/s1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- 48.Holbeck SL, Collins JM, Doroshow JH. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther. 2010;9:1451–1460. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mingeot-Leclercq MP, Deleu M, Brasseur R, Dufrêne YF. Atomic force microscopy of supported lipid bilayers. Nat Protoc. 2008;3:1654–1659. doi: 10.1038/nprot.2008.149. [DOI] [PubMed] [Google Scholar]
- 50.Oliynyk V, Kaatze U, Heimburg T. Defect formation of lytic peptides in lipid membranes and their influence on the thermodynamic properties of the pore environment. Biochim Biophys Acta. 2007;1768:236–245. doi: 10.1016/j.bbamem.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Reis JM, Sinkó B, Serra CHR. Parallel artificial membrane permeability assay (PAMPA)–Is it better than Caco-2 for human passive permeability prediction? Mini Rev Med Chem. 2010;10:1071–1076. doi: 10.2174/1389557511009011071. [DOI] [PubMed] [Google Scholar]
- 52.Hjørringgaard CU, et al. Cyclodextrin-scaffolded alamethicin with remarkably efficient membrane permeabilizing properties and membrane current conductance. J Phys Chem B. 2012;116:7652–7659. doi: 10.1021/jp2098679. [DOI] [PubMed] [Google Scholar]
- 53.Pan N, et al. The single-probe: A miniaturized multifunctional device for single cell mass spectrometry analysis. Anal Chem. 2014;86:9376–9380. doi: 10.1021/ac5029038. [DOI] [PubMed] [Google Scholar]
- 54.Garib V, et al. Propofol-induced calcium signalling and actin reorganization within breast carcinoma cells. Eur J Anaesthesiol. 2005;22:609–615. doi: 10.1017/s026502150500102x. [DOI] [PubMed] [Google Scholar]
- 55.Kredics L, Szekeres A, Czifra D, Vágvölgyi C, Leitgeb B. Recent results in alamethicin research. Chem Biodivers. 2013;10:744–771. doi: 10.1002/cbdv.201200390. [DOI] [PubMed] [Google Scholar]
- 56.Weidema AF, Kropacheva TN, Raap J, Ypey DL. Membrane permeabilization of a mammalian neuroendocrine cell type (PC12) by the channel-forming peptides zervamicin, alamethicin, and gramicidin. Chem Biodivers. 2007;4:1347–1359. doi: 10.1002/cbdv.200790115. [DOI] [PubMed] [Google Scholar]
- 57.Wu L, et al. Multidrug-resistant phenotype of disease-oriented panels of human tumor cell lines used for anticancer drug screening. Cancer Res. 1992;52:3029–3034. [PubMed] [Google Scholar]
- 58.Jang SH, Wientjes MG, Au JL. Kinetics of P-glycoprotein-mediated efflux of paclitaxel. J Pharmacol Exp Ther. 2001;298:1236–1242. [PubMed] [Google Scholar]
- 59.Wada S, Tanaka R. A functionalized 20-residue peptaibol derivative for nucleic acid delivery. Chem Biodivers. 2007;4:991–997. doi: 10.1002/cbdv.200790090. [DOI] [PubMed] [Google Scholar]
- 60.Wada S, Hitora Y, Tanaka R, Urata H. Translocation of an Aib-containing peptide through cell membranes. Bioorg Med Chem Lett. 2008;18:3999–4001. doi: 10.1016/j.bmcl.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 61.Copolovici DM, Langel K, Eriste E, Langel Ü. Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 62.Svensen N, Walton JG, Bradley M. Peptides for cell-selective drug delivery. Trends Pharmacol Sci. 2012;33:186–192. doi: 10.1016/j.tips.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Madani F, Lindberg S, Langel U, Futaki S, Gräslund A. Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys. 2011;2011:414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.