Significance
Poultry feed is usually prepared as a corn–soybean mixture. Because the only essential sulfur amino acid missing in this mixture is methionine, it is chemically synthesized and added separately, increasing the cost of major food supply. It appears to be difficult to circumvent the regulatory aspects of sulfur metabolism, which is controlled at many levels, without damage to plant growth. By using tissue-specific promoters to express a bacterial enzyme that increases the efficiency of assimilative sulfate reduction, seed methionine accumulation can be increased without the concomitant accumulation of toxic metabolites. We show that even in maize inbred lines with repressed seed methionine levels, sink strength can be increased to the benefit of feed consumption efficiency in chicks.
Keywords: APS reductase, sulfur assimilation, sulfur-rich zeins, 10-kDa δ-zein
Abstract
Sulfur assimilation may limit the pool of methionine and cysteine available for incorporation into zeins, the major seed storage proteins in maize. This hypothesis was tested by producing transgenic maize with deregulated sulfate reduction capacity achieved through leaf-specific expression of the Escherichia coli enzyme 3′-phosphoadenosine-5′-phosphosulfate reductase (EcPAPR) that resulted in higher methionine accumulation in seeds. The transgenic kernels have higher expression of the methionine-rich 10-kDa δ-zein and total protein sulfur without reduction of other zeins. This overall increase in the expression of the S-rich zeins describes a facet of regulation of these proteins under enhanced sulfur assimilation. Transgenic line PE5 accumulates 57.6% more kernel methionine than the high-methionine inbred line B101. In feeding trials with chicks, PE5 maize promotes significant weight gain compared with nontransgenic kernels. Therefore, increased source strength can improve the nutritional value of maize without apparent yield loss and may significantly reduce the cost of feed supplementation.
Maize is one of the most important agricultural commodities, with its production amounting to 1,065.1 million metric tons in the trade year 2016/2017, far exceeding that of wheat and rice. About 60% of this global production was used for animal feed (https://apps.fas.usda.gov/psdonline/circulars/grain-corn-coarsegrains.pdf). To provide for amino acid balance in a corn-based diet, the addition of soybean corrects the deficiency of corn in certain essential amino acids such as lysine and tryptophan. This corn–soybean formulation, however, is still deficient in the sulfur (S)-containing methionine (Met). Therefore, feeds are supplemented with synthetic Met. Inclusion of unnatural amino acids, such as the racemic Met used in feed formulation to replace protein-bound amino acids, provides suboptimal protein use and, in some cases, reduces growth rate (1). Maize with elevated Met content could obviate the need for supplementation of animal feed with synthetic Met.
The major seed storage proteins (SSPs) in maize, called zeins, are synthesized in the endosperm and serve as a reservoir of amino acids for the germinating seedling. The proline- and glutamine-rich zeins make up about 60% of the total seed proteins and are mostly devoid of essential amino acids such as lysine, threonine, tryptophan, methionine, and tyrosine (2, 3). Zeins such as the 10-kDa δ-zein have a higher proportion of Met, but they normally make up only a small proportion of the total zeins. Therefore, most maize inbred lines have low Met content. The variability of Met levels (4) and the complex regulation of kernel Met accumulation in different maize inbred lines (5–8) complicate traditional breeding approaches for high-Met maize. As a consequence, Met accumulation in maize seeds has been tested using transgenic means.
Two direct transgenic approaches that have met with less-than-optimal results involved seed-specific expression of a Met-rich protein (9–13) or targeted reduction of S-poor SSPs by gene silencing (14). Introduction of S-rich proteins in developing seeds resulted in simultaneous reduction in levels of endogenous S-rich proteins, suggesting a reallocation of protein S in seeds brought about by limitations of S availability (9–11). Knockdown of expression of S-poor zeins by RNA interference increased lysine but did not increase Met (15).
Another transgenic approach might be to increase the supply of sulfur amino acids (SAAs) by deregulating assimilative sulfate reduction. In this pathway, plants take up inorganic sulfate, reduce it to sulfide, and then assimilate it into cysteine (Cys) (Fig. S1A). A major metabolic control point in Cys synthesis is the enzyme adenosine 5′-phosphosulfate reductase (APS reductase or APR), which has been shown to increase flux through the pathway when constitutively overexpressed in maize. However, the plants were stunted because of the accumulation of toxic intermediates (16). If this problem could be solved, then S assimilation could potentially increase the source of Met for accumulation in the seed (17).
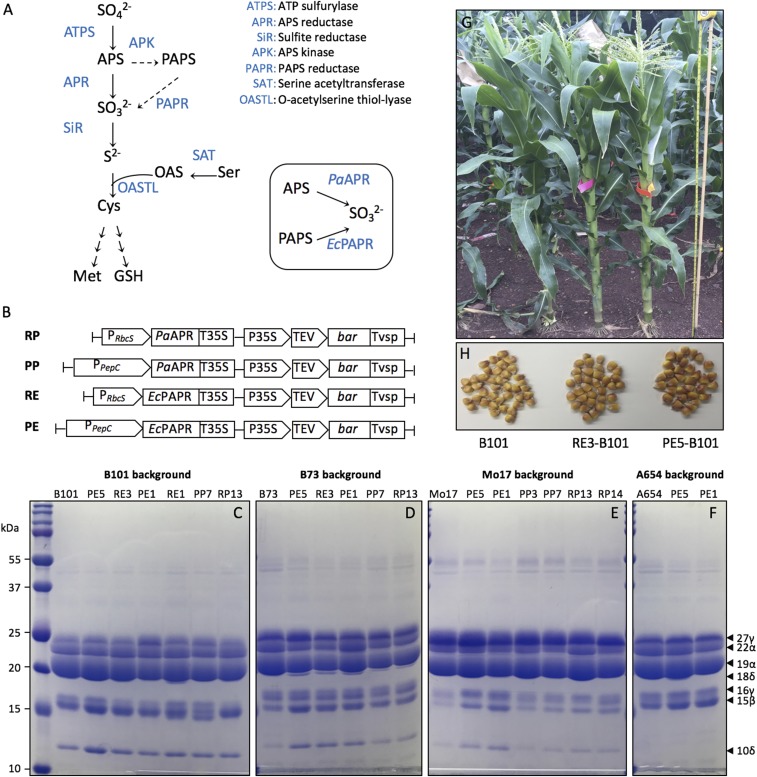
Fig. S1.
Accumulation patterns of the S-rich zeins in transgenic maize expressing the assimilatory reductases EcPAPR or PaAPR. (A) Assimilative sulfate reduction in plants. Sulfate is reduced to sulfite either by APS reductase (APR) via the primary (solid arrows) or an alternative secondary sulfate assimilation pathway (dashed arrows) with PAPS reductase (PAPR). Boxed are the P. aeruginosa APR, PaAPR, and E. coli PAPR, EcPAPR, that had been demonstrated to deregulate sulfate assimilation in Arabidopsis and maize (16, 20). Multiple arrows between steps indicate intervening steps in the pathway. (B) Transgenic expression cassettes of EcPAPR and PaAPR under PepC or RbcS promoters used for Agrobacterium transformation of the Hi-II A × B maize immature embryos. SDS/PAGE zein profiles of the transgenic events backcrossed to the (C) B101, (D) B73, (E) Mo17, and (F) A654 genetic backgrounds for two to five generations. The transgenic events PE, RE, PP, and RP contain the expression cassettes PepC-EcPAPR, RbcS-EcPAPR, PepC-PaAPR, and RbcS-PaAPR, respectively. (G and H) Events PE5 and RE3 introgressed for four and five generations, respectively, into the B101 background (PE5-B101 and RE3-B101) exhibit normal plant development and kernel phenotypes.
Plant APR exerts a strict control over the metabolic flux through S assimilation, but it is also more susceptible to regulatory control than other enzymes in the assimilative reduction pathway (18, 19). To circumvent the many control points, bacterial APR homologs may be better targets for transgenic plant studies. Ectopically expressed Escherichia coli and Pseudomonas aeruginosa enzymes EcPAPR and PaAPR, respectively, have both been shown to function in plants (16, 20). Although having differing substrate specificities (APS for PaAPR and the phosphorylated APS derivative 3′-phosphoadenosine-5′-phosphosulfate, PAPS, for EcPAPR; Fig. S1A), both enzymes are able to drive sulfate reduction (16). Because sulfate assimilation in maize is known to be compartmentalized in specific cell types (21), we achieved expression of the bacterial genes with two tissue-specific promoters: the mesophyll-specific PepC and the bundle sheath cell-specific RbcS promoters (22). The results showed a marked increase in seed Met sequestered in S-rich zeins regardless of APR or promoter used. Recurrent backcrosses of the transgenic plants to the high-Met maize inbred B101 exhibited a stable, high-Met seed phenotype. The transgenic maize kernels used in feed formulation enhanced the growth of chicks. These results represent a breakthrough in the nutritional quality of maize.
Results
Tissue-Specific Expression of EcPAPR.
Transgenic plants that harbor one of four different chimeric constructs were obtained via Agrobacterium infection of immature maize embryos. The constructs included PaAPR or EcPAPR, each under transcriptional control of the leaf- and cell-specific RbcS or PepC promoter (Fig. S1B). Transgenic events generated from each of the constructs showed no phenotypic abnormalities, and some exhibited high accumulation of S-rich storage proteins, SSPs (Fig. S1 C–F). Because the objective of the research was to examine the effect of increased S assimilation on SSP expression, further analysis focused only on two EcPAPR transgenic lines, PE5 and RE3, showing the highest accumulation of the Met-rich 10-kDa δ-zein. Both events, having single copies of the transgene (Table S1), were backcrossed to four different inbred lines (A654, B101, B73, or Mo17) for two to five generations. Subsequent analysis of SSP accumulation after every generation of backcrossing showed that transgenic events PE5 and RE3 introgressed into the inbred B101 (PE5-B101 and RE3-B101) had consistently high accumulation of the 10-kDa δ-zein, and PE5 had high accumulation of the 10-kDa δ-zein in different genetic backgrounds compared with the other transgenic events (Fig. S1 C–F). PE5-B101 and RE3-B101 were backcrossed to the inbred B101 for four and five generations, respectively, and resembled B101 with respect to plant height, tassel morphology, and anthesis-silking interval (Fig. S1G). Therefore, EcPAPR expression in these lines did not negatively affect plant growth or development. In addition to elevated 10-kDa δ-zein expression, the seeds had higher total protein content and no increase in total nitrogen (Table 1). The seeds also showed elevated fat and reduced fiber contents (Table S2). Transgenic plants displayed no apparent yield loss, as kernels had increased weight and kernel number per ear was not significantly different from that of the nontransgenic control (Table 1 and Fig. S1H). Actual yield would have to be determined with genotype x environment (GXE) performance by introgressing the transgene into elite lines and growth in different geographic field locations.
Table S1.
Genetic characterization of selected transgenic maize lines overexpressing EcPAPR
| Event | Characteristics of the standard curves* | Average 2ΔCt ± SD† | Estimated copy number‡ | Transgene segregation (+/−)§ | ||||
| Genes | Efficiency | R† | Slope | Intercept | ||||
| PE5-B101 | GAPDH | 1.0024 ± 0.0309 | 0.9953 ± 0.0064 | −3.3175 ± 0.0738 | 40.7405 ± 0.5947 | 51/48 | ||
| Bar | 1.0710 ± 0.0318 | 0.9901 ± 0.0091 | −3.1638 ± 0.0668 | 41.0330 ± 1.6136 | 1.1020 ± 0.0410 | 1 | ||
| RE3-B101 | GAPDH | 1.0026 ± 0.0039 | 0.9968 ± 0.0035 | −3.3158 ± 0.0093 | 40.5855 ± 0.1068 | 45/51 | ||
| Bar | 0.9805 ± 0.0390 | 0.9950 ± 0.0061 | −3.3715 ± 0.0972 | 42.9820 ± 0.863 | 0.6090 ± 0.0086 | 1 | ||
| PE5-B73 | ND¶ | 29/25 | ||||||
| RE3-B73 | ND | 10/8 | ||||||
| PE5-Mo17 | ND | 9/6 | ||||||
| PE5-A654 | ND | 11/11 | ||||||
The standard curve was generated with the SYBR Green qRT-PCR and used for determination of the transgene copy number.
Ratio of the copy number of the transgene (Bar, selectable marker gene for bialaphos resistance) to the reference gene (GAPDH, glyceraldehyde-3-phosphate dehydrogenase) was calculated using the equation: ratio = 2(Ctreference − Cttransgene).
DNA from leaf tissues of PE5-B101 and RE3-B101 were from germinated S1BC4 and BC5 seeds, respectively.
+ and − denote the number of plants that tested positive and negative for the EcPAPR transgene. These transgenic events were characterized at the BC3 generation with the exception of RE3-B101, which was analyzed at the BC4 generation.
ND, not determined.
Table 1.
Kernel composition analysis
| Mean (SD) | B101 | PE5-B101 | RE3-B101 |
| Protein, % | 10.58 (0.28) | 12.54 (0.20)** | 12.86 (0.21)*** |
| Nitrogen, % | 1.937 (0.029) | 1.947 (0.025) | 1.990 (0.030) |
| Sulfur, % | 0.140 (0.002) | 0.194 (0.002)**** | 0.191 (0.001)**** |
| 100-kernel wt., g | 19.65 (1.28) | 24.75 (1.26)**** | 22.02 (1.61)** |
| Kernel number per ear | 459.83 (50.82) | 494.50 (49.01) | 483.67 (46.81) |
Mature kernels were pooled and measured, and values (SD) represent the average of three measurements for protein, nitrogen, and sulfur contents; average kernel weight and number per ear were determined from 10 replicates of a 100-kernel sample and six ears, respectively. Statistical analysis was performed with the Student’s t test: significantly different at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Table S2.
Kernel composition analysis
| Mean (SD) | B101 | PE5-B101 | RE3-B101 |
| Fat | 3.80 (0.10) | 4.72 (0.04)*** | 4.25 (0.12)** |
| Fiber | 2.67 (0.14) | 2.24 (0.14)* | 2.25 (0.05)** |
Mature kernels were pooled and measured, and values (SD) represent the average of three measurements. Statistical analysis was performed with the Student’s t test: significantly different at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
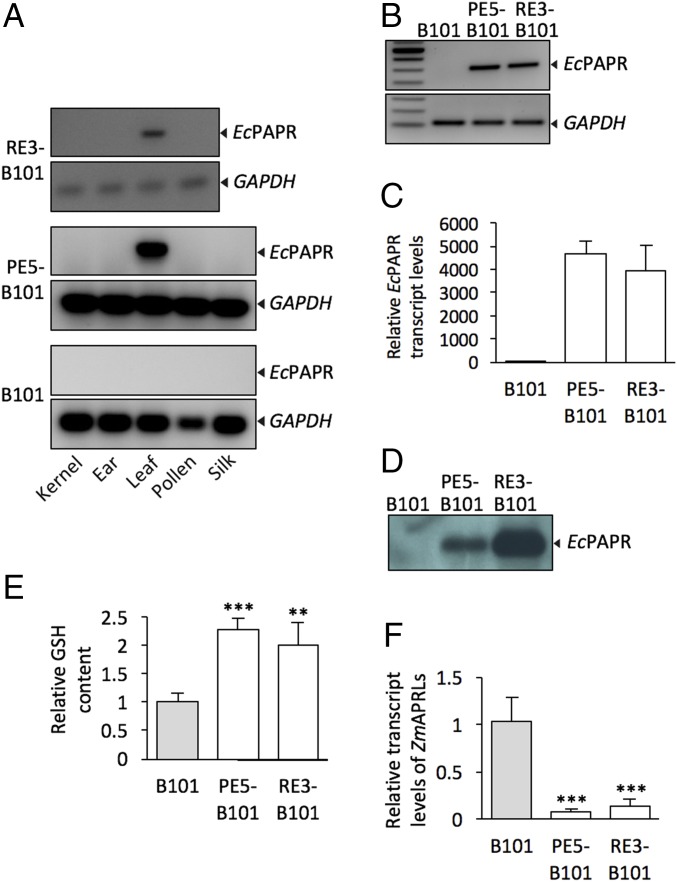
EcPAPR mRNA was detected in leaves but not in the silks, pollen, ears, and immature kernels of PE5-B101 and RE3-B101, demonstrating leaf-specific expression (Fig. 1A). EcPAPR protein localization showed that the PepC promoter directed specific expression in mesophyll cells, but the RbcS promoter resulted in EcPAPR expression in bundle sheath cells as well as leaky expression in the mesophylls (Fig. S2). EcPAPR transcripts in PE5-B101 and RE3-B101 accumulated at similar levels in mature leaves (Fig. 1 B and C), however, the protein was much more abundant for RE3-B101 (Fig. 1D).
Fig. 1.
Maize transformed with the bacterial assimilatory reductase EcPAPR. (A) Tissue-specific expression of EcPAPR under PepC and RbcS promoter control in transgenic maize. RT-PCR analysis was performed to detect EcPAPR expression in different maize tissues. First-strand cDNA from RE3-B101 was amplified for 30 cycles, whereas that from B101 and PE5-B101 were amplified for 40 cycles. (B) RT-PCR and (C) qRT-PCR analysis of EcPAPR transcript levels in mature leaves of PE5-B101 and RE3-B101. Data shown are means ± SD of four determinations each from two biological replicates. (D) Western blot analysis of protein extracts from leaves, using an antibody against EcPAPR. (E) Relative glutathione (GSH) content and (F) transcript levels of ZmAPRLs in transgenic mature leaves. Primers targeting two putative APR-like genes in maize, ZmAPRL1 and ZmAPRL2 (GenBank accession nos. AY739296 and AY739297), were used for qRT-PCR analysis. Glyceraldehyde-3-phosphate dehydrogenase, GAPDH, was used as the reference gene. Data shown are means ± SD of three measurements per three biological replicates. Statistical analysis was performed with the Student’s t test: significantly different from the B101 control at **P < 0.01 and ***P < 0.001.
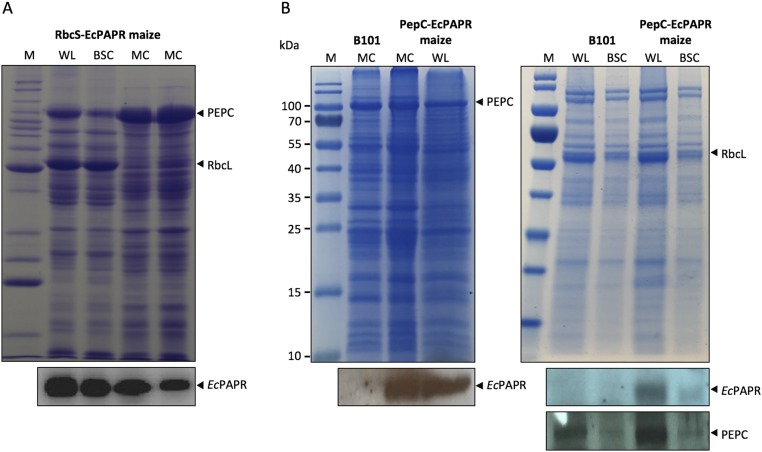
Fig. S2.
Nonspecificity and specificity of the RbcS and PepC promoters, respectively, used for tissue-specific protein localization of EcPAPR in transgenic maize leaves. Total proteins isolated from whole leaf (WL), bundle sheath cells (BSC), and mesophyll cells (MC) from RbcS-EcPAPR (A) and PepC-EcPAPR (B) maize. (Upper) SDS/PAGE analysis of protein extracts from the indicated cellular preparation. (Lower) Immunoblotting of these protein extracts, with antibodies indicated on the right.
Glutathione (GSH) acts as transport and storage form of reduced S, and its biosynthesis is limited by Cys concentration (23). GSH accumulated by more than twofold in the leaves of both transgenic events (Fig. 1E). Because plant APR transcription is known to be particularly sensitive to down-regulation by an end-product of S assimilation, expression of endogenous maize APR would be expected to decrease if S assimilation had been deregulated by EcPAPR expression. The maize genome contains two putative APR-like proteins ZmAPRL1 and ZmAPRL2 (GenBank accession nos. AY739296 and AY739296) (24). Both PE5-B101 and RE3-B101 show decreased abundance of ZmAPRL1 and ZmAPRL2 transcripts (Fig. 1F). These results indicate that EcPAPR expression has resulted in deregulation of sulfate reduction, but without a negative effect on plant growth and yield (Table 1 and Fig. S1 G and H), likely because of the leaf-specific promoters that were used compared with the constitutive promoter employed in prior studies (16).
Variation in Zein Expression Is a Function of Genetic Background.
The EcPAPR transgene is stable and heritable. PCR analysis of segregating plants of PE5 and RE3 introgressed into different backgrounds (Table S1) indicates the segregation ratios of transgenic versus the null segregants were about 1:1, suggesting the presence of a single copy of the EcPAPR transgene in these transgenic events. Backcrosses of transgenic events to maize inbreds that differ in their accumulation of the Met-rich 10-kDa δ-zein revealed that EcPAPR also induced expression of the S-rich δ-, β-, and γ-zeins dependent on the genetic background (Fig. S1 C–F).
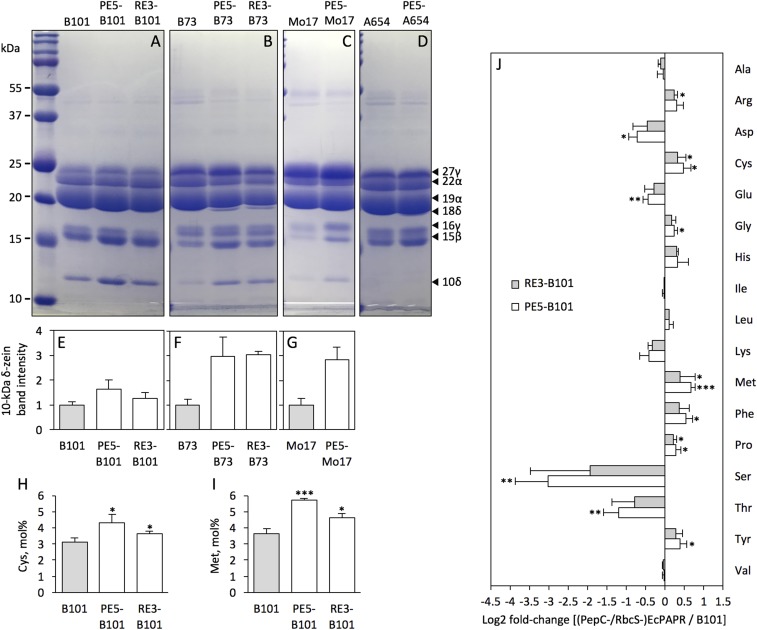
Varying expression levels of the Met-rich zeins were observed in events PE5 and RE3 (Fig. 2 A–G). Both PE5 and RE3 in the B73 and Mo17 backgrounds show global increases in the levels of the S-rich zeins (Fig. 2 B and C), whereas in the B101 background, only the 10-kDa δ- and 15-kDa β-zeins were increased (Fig. 2A). PE5-B101 has 14.4% more kernel Met (Fig. 2I) than those from an F3 ear of the PE5 event, illustrating that specific maize inbreds can be exploited to enhance Met level. The nonfunctional 10-kDa δ-zein gene in A654 (Fig. 2D) resulted in an increase of only the 15-kDa β-zein in PE5. Of the S-containing zeins, the 10-kDa δ-zein appears to be the most responsive to enhanced sulfate assimilation. The relative accumulation of the 10-kDa δ-zein in both transgenic events backcrossed to different inbreds is shown in Fig. 2 E–G. Elevated levels of the 15-kDa β- and 16-kDa γ-zeins were differentially regulated (Fig. 2A and Fig. S3) and observed only when the 10-kDa δ-zein was increased (Fig. S1 C–F). In the absence of the 10-kDa δ-zein, β-zein acted as the primary sink of Met among the SSPs (Fig. 2D).
Fig. 2.
Accumulation patterns of seed storage proteins in transgenic EcPAPR maize introgressed into different genetic backgrounds. (A–D) SDS/PAGE zein profiles of the transgenic events PE5 and RE3 in different genetic backgrounds and (E–G) relative band intensity of the 10-kDa δ-zein in these events. Events were backcrossed to the (A) B101, (B) B73, (C) Mo17, and (D) A654 genetic backgrounds for at least four generations of backcrossing. Quantification of the relative band intensity of the 10-kDa δ-zein in six to eight kernels (SI Materials and Methods), using the Image Studio Lite software. Relative accumulation levels of the 10-kDa δ-zein in transgenic events in the B101, B73, and Mo17 backgrounds are shown in E, F, and G, respectively. Sulfur amino acid contents (H and I) and changes in the composition of protein-bound amino acids (J) in mature dry seeds of PE5-B101 and RE3-B101. (H) Cys and (I) Met contents in mol% determined after protein hydrolysis and separation in a UPLC column. (J) Fold-changes in amino acid levels in transgenic seeds compared with B101 were log2-transformed and plotted in the bar graph. Bars to the left and right indicate a reduction and increase, respectively, in the amino acid content of the EcPAPR plants relative to B101. Student t test at *P < 0.05, **P < 0.01, and ***P < 0.001 were used to determine the statistical difference between the transgenic PE5-B101 and RE3-B101 and nontransgenic B101 kernels. Data shown are means ± SD of three replicates.
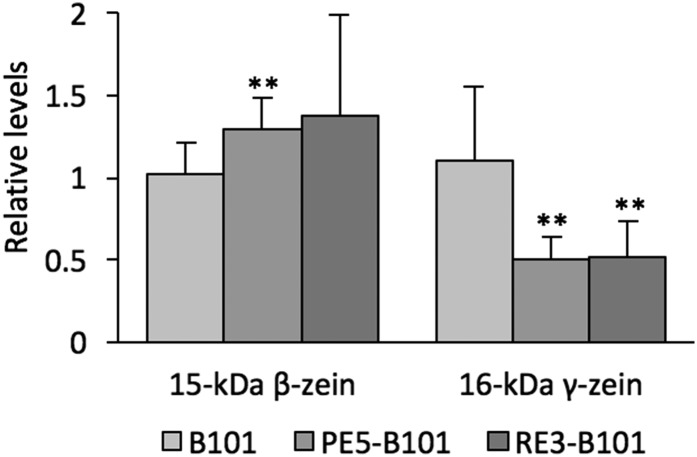
Fig. S3.
Relative expression levels of the 15-kDa β- and 16-kDa γ-zein transcripts in the endosperm of 16-d postpollination PE5-B101 and RE3-B101 kernels. Statistical analysis was performed with the Student’s t test: significantly different from the B101 control at **P < 0.01. Data shown are means ± SD of three determinations each from two biological replicates.
Amino Acid Analysis of Transgenic Maize Kernels.
Cys (Fig. 2H) and Met (Fig. 2I) were both increased in mature dry seeds of PE5-B101 and RE3-B101. Met was increased 57.6% in PE5-B101 and 27.8% in RE3-B101 compared with the B101 control. Cys was increased 39.4% and 17.7% in PE5-B101 and RE3-B101; expression of Cys-containing nonzein proteins were also increased in the PE5 event (Fig. S4). Total S content of transgenic seeds from PE5-B101 and RE3-B101 was increased by 38.6% and 36.4%, respectively (Table 1). In contrast, transgenic seeds expressing the S-rich sunflower seed albumin either had unchanged or slightly lower Cys content than nontransgenic controls. In addition, total seed S content did not change, presumably because of reallocation of S reserves from endogenous proteins to the transgenic products (9–13) (Table S3).
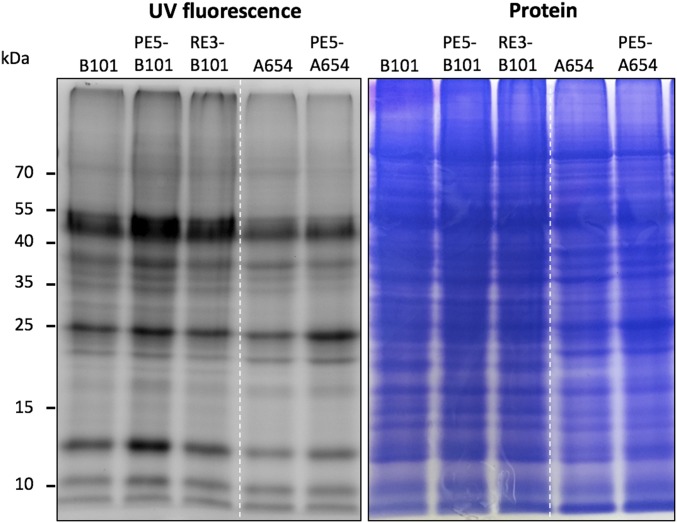
Fig. S4.
Protein S distribution in the kernels of transgenic high-Met maize. Accumulation patterns of cysteine-containing nonzein proteins in transgenic kernels. Nonzein proteins were derivatized with monobromobimane and separated in a 12% SDS/PAGE gel. Band intensity under UV fluorescence is proportional to the cysteine content and relative abundance of the protein.
Table S3.
Reports on transgenic seeds with elevated Met content expressing the Met-rich sunflower seed albumin (SSA) or the 10-kDa δ-zein
| Plant | Tissue-specific expression of a Met-rich protein | Sulfur content of transgenic seeds | Refs. |
| Lupins | Seed-specific expression of an SSA gene | No difference in total S content between control and transgenic seeds Marked differences in the distribution of S between the oxidized (sulfate) and the reduced C-bonded S fraction (cysteine and Met) | 12, 13 |
| Chickpea | Seed-specific expression of an SSA gene | Total seed S concentrations were not consistently different between transgenic and control genotypes Transgenic seeds have significantly higher reduced S than the parental control | 10 |
| Rice | Grain-specific expression of SSA | S content does not change in the transgenic grain Level of reduced S was not significantly different between control and transgenic lines | 11 |
| Maize* | Seed-specific overexpression of its own 10-kDa δ-zein | The null segregant kernels have 0.132% (0.002)†, whereas the transgenic kernels have 0.138% (0.002)† sulfur contents | 9 |
This transgenic maize is from our own laboratory stock.
The sulfur contents of the maize kernels are shown as means (SD) of three replicates. Statistical analysis performed with the Student’s t test showed that the value of the transgenic kernels is significantly different from the null segregant kernels at P < 0.01.
Concomitant with increased Met and Cys, total aspartic acid, lysine, threonine, and serine decreased in the transgenic seeds (Fig. 2J). Serine was reduced the most, amounting to only 14.1% of the B101 control in PE5-B101 seeds (Fig. 2J). It is not clear why these amino acids are reduced, but it is interesting to note that lysine, threonine, and isoleucine use aspartic acid as a precursor for their synthesis, and serine is used for Cys synthesis (Fig. S1A). Therefore, one might expect the free level of these amino acids to decline in plants engineered to increase Cys and Met biosynthesis. Other noteworthy changes included an increase in phenylalanine, tyrosine, and proline. Elevated phenylalanine and tyrosine were previously reported to correlate with higher Met levels in transgenic seeds (25, 26). The increase in proline content may be attributed to increased expression of the 10-kDa δ-zein, of which 15.5% of its residues are proline (27).
Chick Feeding Trials with the High-Met PE5.
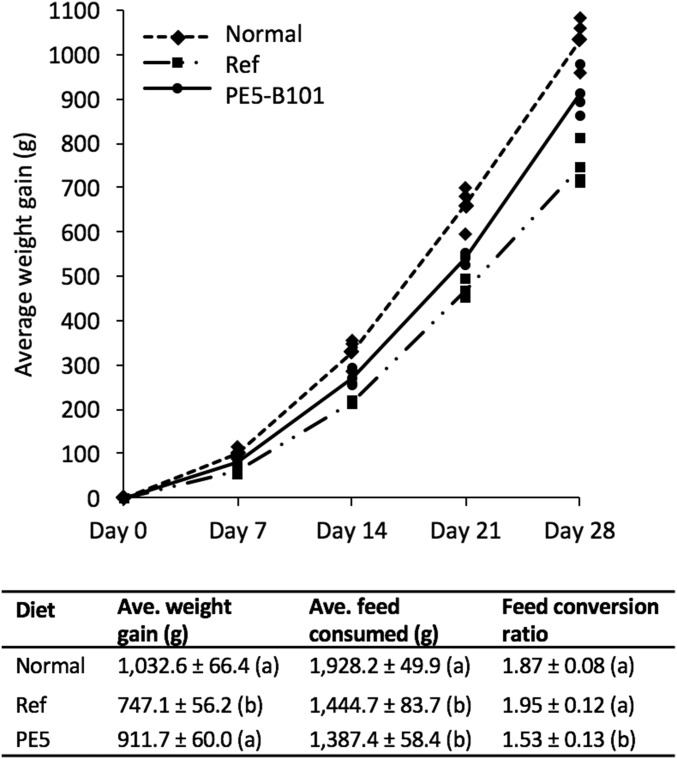
Ultimately, the usefulness of increased seed methionine must be judged on whether it improves nutritional value. PE5-B101 kernels were used in a 4-wk feeding trial of chicks with a corn–soybean meal formulation that is deficient in Met (28) (Table S4). Three diet rations, consisting of different corn meals, were tested with 5-d-old chicks: a complete diet consisting of a yellow dent corn supplemented with synthetic Met, corn meal from PE5-B101 without Met supplementation, and a reference diet composed of corn meal from null segregants derived from PE5-B101 without Met supplementation. Chicks receiving the normal diet had the biggest weight gain, although this is not significantly different from those fed with PE5-B101, whereas those fed the reference diet had the lowest weight gain (Fig. 3).
Table S4.
Composition of the experimental diets used in the chick feeding trials
| Ingredient | Amount | ||
| Normal | Reference | PE5 | |
| Corn meal, g* | 717.5 | 720.0 | 720.0 |
| Soy protein, g† | 150.0 | 150.0 | 150.0 |
| DL-Met, g‡ | 2.5 | — | — |
| Cellulose, BW200, g | 30.0 | 30.0 | 30.0 |
| Corn oil, g | 30.0 | 30.0 | 30.0 |
| Vitamin mixture, g§ | 10.0 | 10.0 | 10.0 |
| Salt mixture, g¶ | 60.0 | 60.0 | 60.0 |
| Total, g | 1,000 | 1,000 | 1,000 |
Cornish male chicks were used in a 4-wk feeding trial starting with 5-d-old chicks in triplicate groups of five per diet formulation.
Three diet rations with different corn meals were used in the feeding experiment: yellow dent corn supplemented with Met (normal group), null transgenic segregant from event PE5 without synthetic Met supplementation (reference group), and high-Met transgenic event PE5 without synthetic Met supplementation (PE5).
Supro isolated soy protein (DuPont) contains 1.1% Met and 1.1% cysteine.
DL-Met was added to the diet at a concentration of 2.5 g/kg (52).
Supplied per kilogram of diet: Vitamin A palmitate, 500,000 IU/g, 8 mg; Vitamin D3, 100,000 IU/g, 10 mg; Vitamin E acetate, 500 IU/g, 50 mg; menadione sodium bisulfite, 62.5% menadione, 3 mg; biotin, 1%, 20 mg; cyanocobalamin, 0.1%, 10 mg; folic acid, 2 mg; nicotinic acid, 70 mg; calcium panthothenate, 15 mg; pyridoxine-HCl, 10 mg; riboflavin, 5 mg; thiamine-HCl, 5 mg; sucrose, 9,792 mg.
Supplied per kilogram of diet: CaCO3, 900 mg; Ca3(PO4)2, 30 g; MgO, 1,002 mg; C6H5K3O7, 17.28 g; K2SO4, 1.65 g; NaCl, 4.5 g; Cu2(OH)2CO3, 13.80 mg; KIO3, 660 μg; C6H5FeO7, 376.80 mg; MnCO3, 126 mg; Na2SeO3, 420 μg; ZnCO3, 144 mg; sucrose, 4,006.32 mg.
Fig. 3.
Feeding trial with the transgenic high-Met PE5 maize. A 4-wk feeding trial with 5-d-old chicks was carried out with three types of diets consisting of yellow dent corn supplemented with synthetic methionine (normal group), PE5-B101 without synthetic methionine, and the null transgenic segregant from PE5-B101 without methionine supplementation (reference group). Shown in the graph is the average weight gain, denoted by the lines, during the course of the experiment, and the table shows the weight gain and feed intake per chick at the conclusion of the feeding trial. Weight gain is calculated as the difference between the finishing and starting weights, and the feed conversion ratio is the amount of food consumed per gained weight. Statistical analysis was performed with two-way ANOVA at P < 0.05, and significant differences between samples are indicated by different letters. Data shown are means ± SD of three replicates with five animals per replicate.
Discussion
Deregulation of the sulfate assimilation pathway in the source tissues led to increased accumulation of protein-bound S in seeds, resulting from the accumulation of specific S-rich zeins. Although previous work showed that deregulation of the reductive sulfate assimilation pathway by overexpression of the assimilatory reductases could be used to increase S flow from uptake to storage in seeds, it had no practical application because of detrimental plant phenotypes resulting from the accumulation of toxic intermediates, which could not be efficiently metabolized during plant development (16). In addition, different maize inbred lines exhibit variability in the amount of Met stored in the seed (4), and this variability appears to be mainly a result of the differential expression of the Met-rich 10-kDa δ-zein gene, Dzs10, with 22.5% Met codons (27), the same gene whose expression is up-regulated by increased S assimilation.
Remarkably, transgenic kernels from S-deregulated plants showed no apparent rebalancing of protein S that was previously observed for overexpression of the 10-kDa δ-zein gene and the overexpression of S-rich proteins in other species (9–13), or by reducing expression of an S-poor SSP through antisense RNA expression (14). The overall increase we observed in the accumulation of the S-rich zeins indicates another facet of the regulation of zeins achieved by altering the supply of SAA. The present work illustrates that by genetically engineering increased biosynthesis of SAA, seed development can be altered to increase the ability to accumulate and fix the SAA into SSP to produce seeds with improved nutritional quality.
The introgression events PE5-B101 and RE3-B101 accumulated more Met and Cys than B101, which already has the highest kernel Met content among common maize inbreds (4). In prior work, it was shown that when B101 was crossed with other inbreds or used as a high-Met male donor parent, the high expression level of the Dzs10-B101 allele was lost, suggesting the presence of more than one genetic factor that affects expression of the gene in trans (29). This regulation was eliminated with a chimeric storage protein gene that contained only the coding region of the Dzs10 gene (9). Thus, it appears that B101 exemplifies the maximum natural threshold of maize grain Met accumulation under limiting SAA availability. This threshold apparently can be overcome with increased S reduction and assimilation during photosynthesis. Even in the B73 and Mo17 backgrounds, overexpression of EcPAPR can induce an increase in the accumulation of the S-rich zeins, indicating SAA supply or availability is a critical limiting factor in maize seed Met accumulation.
Under increased S supply, the 10-kDa δ-zein has higher accumulation compared with the other S-containing zeins. This preferential accumulation of the 10-kDa δ-zein is probably a function of its content of SAAs (22.5% Met and 3.9% Cys). Based on the zein profiles of the different transgenic events, the 10-kDa δ-zein seems to be the most responsive to enhanced assimilative sulfate reduction followed by the 15-kDa β-, 16-kDa γ-, and 27-kDa γ-zein, respectively. This order also follows the number of SAA residues in these zeins. Therefore, it would seem that the higher the SAA residues of the zeins, the more responsive it would be to increased S supply. We do not consider here the 18-kDa δ-zein, although it is exceptionally rich in the SAAs, as its expression is highly variable across inbred lines and most inbred lines have very low levels of expression of this protein (8). As suggested by our data on transgenic events introgressed into different genetic backgrounds, the 10-kDa δ-zein seems to be the primary, and foremost, sink for Met in the seeds.
In maize, a C4 plant, Cys synthesis is localized to the bundle sheath cells and exhibits spatial separation from synthesis of glutathione, a downstream metabolite produced from Cys in mesophyll cells (21). For this work, we focused on obtaining transgenic plants with increased S assimilation that did not show negative effects on plant growth, to assess the effect of increased S assimilation on SSP. Therefore, we did not perform a rigorous comparison of EcPAPR and PaAPR under either the PepC or RbcS promoters. However, of all the transgenic events we have generated, EcPAPR plants appeared to accumulate more S-rich zeins than PaAPR plants. EcPAPR plants also showed the hallmarks of deregulated S assimilation, including the accumulation of glutathione and down-regulation of expression of endogenous APR. This result was unexpected, as maize has an APS reductase-type (PaAPR) sulfate assimilatory pathway, not the PAPS reductase type (EcPAPR) (30–32). Although maize, similar to other flowering plants, can produce PAPS, used as a sulfate donor in sulfation reactions of some secondary metabolites (33, 34), it was until now unclear whether PAPS could be directed toward sulfate assimilation. Our results show that endogenous PAPS in higher plants such as maize can be co-opted, with the use of an ectopic PAPS reductase, for reductive sulfate assimilation. Moreover, in prior studies it was shown that APS reductase overexpression in maize and Arabidopsis produces growth defects (16, 20), yet in the present study, EcPAPR expression was not associated with any apparent growth abnormalities. Thus, our results point to another aspect of metabolic engineering for enhanced crop value: using redundancies and alternative circuits for endogenous biosynthetic pathways to improve the nutritional value of crops. That Met accumulates in transgenic EcPAPR plants suggests Met synthesis in maize is not strictly controlled by the enzyme cystathionine γ-synthase responsible for synthesis of Met from Cys. Cystathionine γ-synthase is also not a limitation for Met synthesis in potato (35), whereas it is a bottleneck enzyme in Arabidopsis (36–38).
It is known that APR is expressed in leaf mesophyll or bundle sheath cells, not in developing kernels, indicating that changes in S metabolism in the leaf parenchyma are sufficient to drive increased Met accumulation in the kernel. Therefore, the evidence is consistent with the hypothesis that S assimilated in the leaf is transported to the kernel. Our evidence does not rule out the possibility that S can be assimilated in the kernel (39) or vascular cells resulting from low-level expression of APR. Still, the prevailing hypothesis is that S is transported in a S transport form from the leaf to the ear via the phloem sap. In wheat, S-methylmethionine is the major form in which reduced S moves in the phloem (40). However, insertional mutants of Arabidopsis and maize in Met S-methyltransferase, the enzyme that catalyzes synthesis of S-methylmethionine, produced plants that grew and reproduced normally, and the mutant seeds from Arabidopsis had normal S contents. These results rule out an indispensable role for S-methylmethionine in S transport in Arabidopsis and maize, and the S transport form is probably fulfilled by other reduced S form in these species (41). Whether there is an in situ biosynthesis of SAAs in the maize phloem sap is currently unknown.
Depending on source availability of S, two distinct features of the regulation of SAA levels in the seed emerge from our study. Enhanced S assimilation in maize, in which SAA is not limiting, leads to an overall increase in the expression of the β-, γ-, and δ-zeins. In the default state, in which SAAs are limiting, increased expression of the 10-kDa δ-zein decreases expression of the β- and γ-zeins, leading to rebalancing of protein S in the seeds (9). There seems to be two major limiting factors in the accumulation of Met and Cys in maize seeds: demand for S imposed by the S-rich zeins, and SAA availability or supply from the source tissues, which determines the uptake of SAA into the seeds (13). The demand, or S sink strength, is itself responsive to the SAA supply. These limitations constitute a conservative mechanism in the seeds that senses SAA availability from the source tissues and accordingly adjust the sink strength for SAA.
We have shown that by enhancing sulfate assimilation in the leaf by transgenic means coupled with traditional backcross breeding into desirable genetic backgrounds, maize kernels with high Met content were produced that was of significant increased nutritional value to livestock. Increased Met sequestered in the S-rich zeins was bioavailable in the diet fed to chicks and can supplant synthetic Met supplementation needed for optimal growth. From a nutritional point of view, increasing Met rather than Cys is beneficial because, although animals are not able to synthesize Met from Cys, they are able to convert Met to Cys (42).
Materials and Methods
Maize genetic stocks and methods to characterize the transgenic plants and kernels are described in SI Materials and Methods. Primers used for vector construction, genotyping, and qRT-PCR analysis are listed in Table S5.
Table S5.
List of primers used
| Primer name | Sequence (5′ to 3′) | Usage | Available restriction sites | Ref. |
| For expression vector construction* | ||||
| EcpFOR† | GGGgaattcccgggatccAAGCTTCATGGCGCCCACC | Amplification of EcPAPR fragment from #788 | EcoRI, SmaI, BamHIHI | |
| EcpREV† | GGGgagctcTTACCCTTCGTGTAACCCACAT | SacI | ||
| RbcsFOR‡ | CCCCcaattgGAGCTCGGTACCCGGGGATCC | Amplification of RbcS fragment from pPTN533 | MfeI | |
| RbcsREV‡ | CCCtgatcaGCCTGGCTGCCTAGTATGTATGTACTC | BclII | ||
| PepcFOR‡ | ATGATTACgaattcGAGCTCGGTACCC | Amplification of PepC fragment from pPTN512 | EcoRIRI | |
| PepcREV‡ | GCCATagatctATCATAGAAGCCATAGATCC | BglII | ||
| For genotyping | ||||
| ExpV_Ec_R | CTTCCGCCGTTGCTGACGTTGCCGAG | Verification of construct integrity after ligation | ||
| ExpVec_Pa_R | GAAGAGAGCATAGAGGAAGCCATTGT | |||
| ExpVec_F | AGGCTTTACACTTTATGCTTCCGGCTCGTATG | |||
| ExpVec_R | CTGGGAACTACTCACACATTATTCTGGA | |||
| rPepC_For | CTCCCCATCCCTATTTGAACCC | Genotyping of transgenic (P)APR events | ||
| rRbcS_For | CCGCTTCCTCCTATCTACAAGT | |||
| rEcPAPR_Rev | GGTAGGTTTCCGGGAACAAGTA | |||
| rPaAPR_Rev | GAAGGAGATCCACAGCTCGTC | |||
| For RT-PCR and RT-qPCR | ||||
| bar_F_2 | TGCACCATCGTCAACCACTACATCGAG | Determination of transgene copy number | 53 | |
| bar_R_2 | CAGGCTGAAGTCCAGCTGCCAGAAAC | |||
| ZmGAPDH_f | GACAGCAGGTCGAGCATCTTCGA | Reference gene for RT-PCR r qRT-PCR | 46 | |
| ZmGAPDH_r | GTCGACGACGCGGTTGCTGTA | |||
| EcPAPR_for | TTACTTGTTCCCGGAAACCTACC | qRT-PCR analysis of EcPAPR transcript levels | ||
| EcPAPR_rev | AATCGGCAGCACTTTAAATACGC | |||
| q15-kDa_for | CATGGGTGGACTCTACCAGTACC | |||
| q15-kDa_rev | CATGATAATGTGTGTCGTCTTACTGC | qRT-PCR analysis of zein transcript levels | ||
| q16-kDa_for | GCGGTGTCTACTACTGAGGAAACT | |||
| q16-kDa_rev | CATTCAGGTCATTGCTCACACT | |||
| JP-ZmAPRL_for | CAGGGCTACGTGTCCATCGGGTG | qRT-PCR analysis of maize APR-like transcript levels | ||
| JP-ZmAPRL_rev | TTGTGGAGGCCGCACTCCTTGG |
The cloning scheme for generation of the expression vectors involved two consecutive steps: first, generation of the pTF102-(P)APR vectors by ligation of EcoRI- and SacI-digested fragments of EcPAPR or PaAPR into pTF102 and second, ligation of the PepC or RbcS promoter fragments into EcoRI- and BamHIHI-digested pTF102-(P)APRs.
EcPAPR fragment was PCR-amplified from plasmid 788, whereas the PaAPR fragment was digested out of plasmid 793 with EcoRI and SacI.
Promoter fragments were digested with available restriction sites on their flanking primers and ligated into EcoRI- and BamHI-digested pTF102-(P)APRs.
SI Materials and Methods
Construction of Gene Cassettes, Maize Transformation, and Crosses.
Plasmids from our own collection (16, 20) designated as strain files 788 and 793 were used for cloning the EcPAPR and PaAPR fragments, respectively. The 1.6-kb maize phosphoenolpyruvate carboxylase (PepC) and 1.0-kb Rubisco small subunit 1 (RbcS) promoters were cloned from the binary vectors pPTN512 and pPTN533, respectively, and were kindly provided by Dr. David Stern (22). Four expression cassettes that harbored either EcPAPR or PaAPR, collectively called (P)APRs (16), were assembled under the control of either the maize PepC or RbcS promoters. These four expression cassettes were subcloned into the binary plasmid pTF102, which had an excised P35S-GUS INT gene cassette. Resulting vectors were referred to as PE, RE, PP, and RP for the PepC-EcPAPR, RbcS-EcPAPR, PepC-PaAPR, and RbcS-PaAPR expression cassettes, respectively.
Electrocompetent Agrobacterium tumefaciens EHA101 was prepared (43) and transformed with the binary (P)APR vectors. Maize transformations using immature embryos were carried out using a known Agrobacterium-mediated transformation protocol (44). Transformation efficiency was 1–2%.
Transgenic (P)APR events were backcrossed to the inbred lines A654, B101, B73, or Mo17 for two to five generations, with some events having additional one to two generations of self-pollination.
Biochemical and Molecular Biology Analysis of Transgenic Events.
Genomic DNA was extracted from maize leaf tissues at the 4- or 5-leaf stage, using a modified CTAB extraction method (45), and transgenic events were genotyped by PCR, using primer pairs listed in Table S5.
Maize tissues were processed for RNA extraction with NucleoSpin RNA Plant kit (Takara Bio USA). cDNA was synthesized from 1 μg RNA with the Prime Script RT Reagent kit (Takara Bio USA), using the poly-dT primer in a 20-µL reaction. To determine the localization of expression of EcPAPR in different tissues, semiquantitative RT-PCR was performed using 2 µL of the first-strand cDNA. Forty cycles were used for B101 and PE5-B101, whereas RE3-B101 cDNA was amplified for 30 cycles.
For RT-qPCR (StepOnePlus Real-Time PCR system; Applied Biosystems), 2 µL cDNA and primers at 400 nM each were used in 10-µL reactions with PowerUp SYBR Green Master mixture (Applied Biosystems). Four technical replicates from pooled tissues were run per sample, and each sample was analyzed using two or three biological replicates. No template control reactions were included and returned no CT values. GAPDH was used as the reference gene (46). For relative quantitation, the ΔΔCT or the comparative CT method was used, which gave values in fold-change of expression.
Ten micrograms total protein (47) was separated in a 12% Tris⋅glycine SDS/PAGE gel, and immunoblotting was performed using an antibody against EcPAPR (48). Western blotting was performed after each backcross generation to test for stability and heritability of the transgene. Three biological replicates were analyzed for Western blotting, and datum was presented from a representative blot.
Total glutathione was assayed using the Glutathione assay kit (Cayman Chemical Company). This assay reflects both GSH and its oxidized form, the disulfide dimer GSSG.
Bundle sheath and mesophyll cell preparations were obtained from seedling leaves at the third or fourth leaf stage, following the procedure of Sheen (49).
Characterization of Transgenic Kernels.
Zein and nonzein proteins were extracted from pooled endosperm samples of mature maize kernels, following the procedures of Wu et al. (17) and Wallace et al. (50), respectively. Proteins were separated in a 15% SDS/PAGE gel as previously described (17).
Densitometry analysis of the 10-kDa and 19-kDa zeins were determined from SDS/PAGE gels, using Image Studio Lite (LI-COR Biotechnology). To normalize for protein loading, the 10-kDa δ-zein intensity was divided by that of the 19-kDa α-zein. The relative intensity of the 10-kDa δ-zein of the transgenic kernels compared with the nontransgenic controls was calculated as the ratio of the normalized values of the transgenic and nontransgenic kernels.
Forty micrograms of the nonzein protein fraction from pooled samples of mature maize endosperm were labeled with monobromobimane (Sigma-Aldrich), as described (51). Protein separation was performed in a 12% SDS/PAGE gel for 50 min at 200 V, and the fluorescent conjugates were viewed under UV light.
Pooled mature dry seeds were ground to fine powder, and about 10 mg were used for amino acid composition analysis conducted by the Proteomics and Mass Spectrometry Facility, Donald Danforth Plant Science Center. For protein, fat, fiber, nitrogen, and sulfur content determinations, pooled mature dried kernels were sent to the NJ Feed Lab Inc. for analysis. Three independent measurements were done per sample.
The average kernel weight and number per ear were determined from 10 replicates of a 100-kernel sample and six ears, respectively. Ears that were well-filled and of similar size were chosen for determining the kernel number.
Statistical Analysis.
GraphPad Prism version 7.00 for Mac (GraphPad Software) was used to determine the statistical significance of the differences observed between the control nontransgenic and transgenic plants.
Chick Feeding Trials.
The research protocol (Protocol No. 16–049) for chick feeding trial was approved by Rutgers University’s Institutional Animal Care and Use Committee and followed the guidelines contained in the Guide for the Care and Use of Laboratory Animals (National Research Council) and applicable provisions of the Animal Welfare Act.
Day-old male broiler chicks (Cornish Cross) were housed in the brooder barn in pens provided with overhanging heaters maintained at 90 °F for the first 2 wk and reduced to 85 °F on the third week. A 14-h fluorescent illumination was provided per day, and feed and water were supplied ad libitum. The birds were acclimatized to the barn conditions until day 4. On day 5, 45 chicks within a weight range of 77–87 g were randomly assigned to nine pens of five chicks each, covering three dietary treatments of a corn–soybean meal preparation (28) (Table S4). Each experimental diet was fed to three replicate pens over the course of 4 wk. Group body weights and feed intake per pen were recorded at weekly and daily intervals, respectively. A notable difference was observed in the efficiency of feed use, which is the feed conversion ratio (Fig. 3), between the three experimental diets. Chicks fed the normal and reference diets consumed the same amount of feed that were converted into body weight, whereas those subsisting in PE5-B101 consumed less amount of feed for the same amount of weight gain.
Corn meals from PE5-B101 and its null segregant were obtained from ears collected from about 1,000 plants for each genotype. Ears were obtained from plants that were either selfed or backcrossed to the inbred B101 and were grown in the greenhouse during the winter of 2015–2016 and the summer of 2016.
Acknowledgments
We thank Dr. Yongrui Wu of the Chinese Academy of Sciences for advice and comments, Marc Probasco of the Waksman Institute for field and greenhouse management and plant care, and Dr. Renate Scheibe for providing antibodies against EcPAPR. This research was supported by the Selman A. Waksman Chair in Molecular Genetics of Rutgers University (J.M.) and the U.S. Department of Agriculture National Institute of Food and Agriculture Hatch Project 202535 through the New Jersey Agricultural Experiment Station Hatch Project NJ12136 and the Professor Charles Gilvarg Memorial Fund for Plant Metabolic Engineering (T.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714805114/-/DCSupplemental.
References
- 1.Sveier H, Nordås H, Berge GE, Lied E. Dietary inclusion of crystalline D- and L-methionine: Effects on growth, feed and protein utilization, and digestibility in small and large Atlantic salmon (Salmon salar L.) Aquac Nutr. 2001;7:169–181. [Google Scholar]
- 2.Messing J. The manipulation of zein genes to improve the nutritional value of corn. Trends Biotechnol. 1983;1:54–59. [Google Scholar]
- 3.Shewry PR, Halford NG. Cereal seed storage proteins: Structures, properties and role in grain utilization. J Exp Bot. 2002;53:947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- 4.Phillips RL, McClure BA. Elevated protein-bound methionine in seeds of a maize line resistant to lysine plus threonine. Cereal Chem. 1985;62:213–218. [Google Scholar]
- 5.Chaudhuri S, Messing J. Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc Natl Acad Sci USA. 1994;91:4867–4871. doi: 10.1073/pnas.91.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz-Alvarez M, Kirihara JA, Messing J. Post-transcriptional regulation of methionine content in maize kernels. Mol Gen Genet. 1991;225:331–339. doi: 10.1007/BF00269866. [DOI] [PubMed] [Google Scholar]
- 7.Schickler H, Benner MS, Messing J. Repression of the high-methionine zein gene in the maize inbred line Mo17. Plant J. 1993;3:221–229. [Google Scholar]
- 8.Swarup S, Timmermans MC, Chaudhuri S, Messing J. Determinants of the high-methionine trait in wild and exotic germplasm may have escaped selection during early cultivation of maize. Plant J. 1995;8:359–368. doi: 10.1046/j.1365-313x.1995.08030359.x. [DOI] [PubMed] [Google Scholar]
- 9.Lai J, Messing J. Increasing maize seed methionine by mRNA stability. Plant J. 2002;30:395–402. doi: 10.1046/j.1365-313x.2001.01285.x. [DOI] [PubMed] [Google Scholar]
- 10.Chiaiese P, et al. Sulphur and nitrogen nutrition influence the response of chickpea seeds to an added, transgenic sink for organic sulphur. J Exp Bot. 2004;55:1889–1901. doi: 10.1093/jxb/erh198. [DOI] [PubMed] [Google Scholar]
- 11.Hagan ND, Upadhyaya N, Tabe LM, Higgins TJV. The redistribution of protein sulfur in transgenic rice expressing a gene for a foreign, sulfur-rich protein. Plant J. 2003;34:1–11. doi: 10.1046/j.1365-313x.2003.01699.x. [DOI] [PubMed] [Google Scholar]
- 12.Molvig L, et al. Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proc Natl Acad Sci USA. 1997;94:8393–8398. doi: 10.1073/pnas.94.16.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabe LM, Droux M. Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol. 2002;128:1137–1148. doi: 10.1104/pp.010935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohno-Murase J, Murase M, Ichikawa H, Imamura J. Improvement in the quality of seed storage protein by transformation of Brassica napus with an antisense gene for cruciferin. Theor Appl Genet. 1995;91:627–631. doi: 10.1007/BF00223289. [DOI] [PubMed] [Google Scholar]
- 15.Segal G, Song R, Messing J. A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics. 2003;165:387–397. doi: 10.1093/genetics/165.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin MN, Tarczynski MC, Shen B, Leustek T. The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth Res. 2005;86:309–323. doi: 10.1007/s11120-005-9006-z. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Wang W, Messing J. Balancing of sulfur storage in maize seed. BMC Plant Biol. 2012;12:77. doi: 10.1186/1471-2229-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopriva S, et al. Influence of chilling stress on the intercellular distribution of assimilatory sulfate reduction and thiols in Zea mays. Plant Biol. 2001;3:24–31. [Google Scholar]
- 19.Vauclare P, et al. Flux control of sulphate assimilation in Arabidopsis thaliana: Adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 2002;31:729–740. doi: 10.1046/j.1365-313x.2002.01391.x. [DOI] [PubMed] [Google Scholar]
- 20.Tsakraklides G, et al. Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J. 2002;32:879–889. doi: 10.1046/j.1365-313x.2002.01477.x. [DOI] [PubMed] [Google Scholar]
- 21.Burgener M, Suter M, Jones S, Brunold C. Cyst(e)ine is the transport metabolite of assimilated sulfur from bundle-sheath to mesophyll cells in maize leaves. Plant Physiol. 1998;116:1315–1322. doi: 10.1104/pp.116.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sattarzadeh A, et al. Transgenic maize lines with cell-type specific expression of fluorescent proteins in plastids. Plant Biotechnol J. 2010;8:112–125. doi: 10.1111/j.1467-7652.2009.00463.x. [DOI] [PubMed] [Google Scholar]
- 23.Noctor G, et al. Glutathione: Biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot. 1998;49:623–647. [Google Scholar]
- 24.Houston NL, et al. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen H, Israeli H, Matityahu I, Amir R. Seed-specific expression of a feedback-insensitive form of cystathionine-γ-synthase in Arabidopsis stimulates metabolic and transcriptomic responses associated with desiccation stress. Plant Physiol. 2014;166:1575–1592. doi: 10.1104/pp.114.246058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song S, et al. Soybean seeds expressing feedback-insensitive cystathionine γ-synthase exhibit a higher content of methionine. J Exp Bot. 2013;64:1917–1926. doi: 10.1093/jxb/ert053. [DOI] [PubMed] [Google Scholar]
- 27.Kirihara JA, Hunsperger JP, Mahoney WC, Messing JW. Differential expression of a gene for a methionine-rich storage protein in maize. Mol Gen Genet. 1988;211:477–484. doi: 10.1007/BF00425704. [DOI] [PubMed] [Google Scholar]
- 28.Messing J, Fisher H. Maternal effect on high methionine levels in hybrid corn. J Biotechnol. 1991;21:229–237. [Google Scholar]
- 29.Olsen MS, Krone TL, Phillips RL. BSSS53 as a donor source for increased whole-kernel Methionine in maize: Selection and evaluation of high-methionine inbreds and hybrids. Crop Sci. 2003;43:1634–1642. [Google Scholar]
- 30.Hopkins L, Parmar S, Bouranis DL, Howarth JR, Hawkesford MJ. Coordinated expression of sulfate uptake and components of the sulfate assimilatory pathway in maize. Plant Biol (Stuttg) 2004;6:408–414. doi: 10.1055/s-2004-820872. [DOI] [PubMed] [Google Scholar]
- 31.Setya A, Murillo M, Leustek T. Sulfate reduction in higher plants: Molecular evidence for a novel 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bick JA, Leustek T. Plant sulfur metabolism–The reduction of sulfate to sulfite. Curr Opin Plant Biol. 1998;1:240–244. doi: 10.1016/s1369-5266(98)80111-8. [DOI] [PubMed] [Google Scholar]
- 33.Koprivova A, Kopriva S. Sulfation pathways in plants. Chem Biol Interact. 2016;259:23–30. doi: 10.1016/j.cbi.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Mugford SG, et al. Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell. 2009;21:910–927. doi: 10.1105/tpc.109.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreft O, Hoefgen R, Hesse H. Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol. 2003;131:1843–1854. doi: 10.1104/pp.102.015933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, et al. Constitutive overexpression of cystathionine gamma-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol. 2002;128:95–107. [PMC free article] [PubMed] [Google Scholar]
- 37.Chiba Y, et al. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- 38.Lee M, et al. Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. Plant J. 2005;41:685–696. doi: 10.1111/j.1365-313X.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- 39.Tabe LM, Droux M. Sulfur assimilation in developing lupin cotyledons could contribute significantly to the accumulation of organic sulfur reserves in the seed. Plant Physiol. 2001;126:176–187. doi: 10.1104/pp.126.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourgis F, et al. S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kocsis MG, et al. Insertional inactivation of the methionine s-methyltransferase gene eliminates the s-methylmethionine cycle and increases the methylation ratio. Plant Physiol. 2003;131:1808–1815. doi: 10.1104/pp.102.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkelstein JD, Mudd SH. Trans-sulfuration in mammals. The methionine-sparing effect of cystine. J Biol Chem. 1967;242:873–880. [PubMed] [Google Scholar]
- 43.Wise AA, Liu Z, Binns AN. Three methods for the introduction of foreign DNA into agrobacterium. In: Wang K, editor. Agrobacterium Protocols. Humana Press; Totowa, NJ: 2006. pp. 43–54. [DOI] [PubMed] [Google Scholar]
- 44.Frame BR, et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002;129:13–22. doi: 10.1104/pp.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawa S, Ito T, Okada K. A rapid method for detection of single base changes in Arabidopsis thaliana using the polymerase chain reaction. Plant Mol Biol Report. 1997;15:179–185. [Google Scholar]
- 46.Fletcher SJ. qPCR for quantification of transgene expression and determination of transgene copy number. Methods Mol Biol. 2014;1145:213–237. doi: 10.1007/978-1-4939-0446-4_17. [DOI] [PubMed] [Google Scholar]
- 47.Conlon HE, Salter MG. Plant protein extraction. Methods Mol Biol. 2007;362:379–383. doi: 10.1007/978-1-59745-257-1_28. [DOI] [PubMed] [Google Scholar]
- 48.Krone FA, Westphal G, Schwenn JD. Characterisation of the gene cysH and of its product phospho-adenylylsulphate reductase from Escherichia coli. Mol Gen Genet. 1991;225:314–319. doi: 10.1007/BF00269864. [DOI] [PubMed] [Google Scholar]
- 49.Sheen J. Methods for mesophyll and bundle sheath cell separation. Methods Cell Biol. 1995;49:305–314. doi: 10.1016/s0091-679x(08)61462-4. [DOI] [PubMed] [Google Scholar]
- 50.Wallace JC, Lopes MA, Paiva E, Larkins BA. New methods for extraction and quantitation of zeins reveal a high content of γ-zein in modified opaque-2 maize. Plant Physiol. 1990;92:191–196. doi: 10.1104/pp.92.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchanan BB, et al. Thioredoxin-linked mitigation of allergic responses to wheat. Proc Natl Acad Sci USA. 1997;94:5372–5377. doi: 10.1073/pnas.94.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.NRC . Nutrient Requirements of Poultry. National Academy Press; Washington, DC: 1994. [Google Scholar]
- 53.Grohmann L, Brünen-Nieweler C, Nemeth A, Waiblinger H-U. Collaborative trial validation studies of real-time PCR-based GMO screening methods for detection of the bar gene and the ctp2-cp4epsps construct. J Agric Food Chem. 2009;57:8913–8920. doi: 10.1021/jf901598r. [DOI] [PubMed] [Google Scholar]