We visualized the activation process of the MotPS complex of the Bacillus flagellar motor by high-speed atomic force microscopy.
Abstract
The bacterial flagellar motor consists of a rotor and a dozen stator units and regulates the number of active stator units around the rotor in response to changes in the environment. The MotPS complex is a Na+-type stator unit in the Bacillus subtilis flagellar motor and binds to the peptidoglycan layer through the peptidoglycan-binding (PGB) domain of MotS to act as the stator. The MotPS complex is activated in response to an increase in the Na+ concentration in the environment, but the mechanism of this activation has remained unknown. We report that activation occurs by a Na+-induced folding and dimer formation of the PGB domain of MotS, as revealed in real-time imaging by high-speed atomic force microscopy. The MotPS complex showed two distinct ellipsoid domains connected by a flexible linker. A smaller domain, corresponding to the PGB domain, became structured and unstructured in the presence and absence of 150 mM NaCl, respectively. When the amino-terminal portion of the PGB domain adopted a partially stretched conformation in the presence of NaCl, the center-to-center distance between these two domains increased by up to 5 nm, allowing the PGB domain to reach and bind to the peptidoglycan layer. We propose that assembly of the MotPS complex into a motor proceeds by means of Na+-induced structural transitions of its PGB domain.
INTRODUCTION
Intrinsically disordered (ID) proteins adopt an ensemble of conformations to permit rapid but specific interactions of the proteins with their binding partners to modulate their biological activities. The conformational flexibilities of ID motifs in proteins regulate a variety of biological activities by folding and/or binding to numerous cellular partners in a highly cooperative manner (1, 2).
The bacterial flagellum is a supramolecular motility machinery consisting of at least three parts: the basal body as a rotary motor, the hook as a universal joint, and the filament as a helical screw. The flagellar motor consists of a rotor and a dozen stator units (Fig. 1A) and is driven by ion-motive force across the cytoplasmic membrane. The flagellar motor also acts as a biosensor to respond to changes in the environment, inducing developmental changes such as cell differentiation and biofilm formation in planktonic cells for their survival. The stator unit is a transmembrane (TM) ion channel and couples ion flow through the channel with torque generation. It has been shown that the ion channel activity of the stator unit is linked with cell differentiation (3–5). The stator units show rapid exchange between the motor and the cytoplasmic pool while the motor is spinning. Furthermore, each stator unit associates with and dissociates from the motor in response to changes in the environment as well as load. These observations suggest that the stator unit acts as a molecular sensor to detect changes in the environment to regulate the number of active stator units in the motor (6–10).
Fig. 1. Schematic drawing of the bacterial flagellar motor.
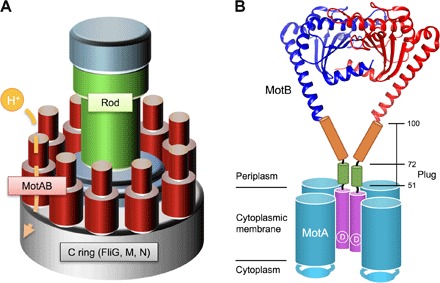
(A) Schematic diagram of the flagellar motor. The flagellar motor consists of a rotor ring complex and a dozen stator units made of two TM proteins, MotA and MotB. FliG, FliM, and FliN assemble into the C ring on the cytoplasmic face of the MS ring in this order and act as a rotor. (B) Schematic diagram of the MotAB complex. MotA and MotB together form a proton channel with four copies of MotA and two copies of MotB. Cα ribbon representation of the Salmonella MotBC structure (PDB code: 2ZVY) with the TM of MotAB and a flexible stalk of MotB (residues 51 to 100) connecting these two domains are shown. A highly conserved Asp33 residue (indicated as D) is directly involved in proton translocation through the MotAB proton channel complex. The stalk contains the plug segment (residues 53 to 66), which regulates the proton channel activity before stator assembly around the rotor.
The stator unit of the H+-driven motor consists of four copies of MotA and two copies of MotB (11). A specific interaction between MotA and the rotor protein FliG is required not only for motor rotation but also for efficient stator assembly around the rotor (12–14). MotA has four TM helices (TM1 to TM4), and MotB has a single TM helix. The TM3 and TM4 helices of MotA form a proton channel with MotB-TM (15). Asp33 of Salmonella MotB, which is a highly conserved aspartic residue among MotB orthologs, is located at the cytoplasmic end of MotB-TM (Fig. 1B) and is responsible for proton translocation through the channel in a load-dependent manner (16). MotB also has a large C-terminal periplasmic domain (MotBC), which consists of a flexible stalk (residues 51 to 100) and a compactly folded peptidoglycan-binding (PGB) domain (residues 101 to 309) responsible for the association with the peptidoglycan layer. The stalk, which is predicted to be ID (fig. S1A), not only suppresses the proton channel activity before stator assembly around the rotor (17, 18) but also regulates the assembly and disassembly dynamics of the stator unit in response to the environmental changes (9), although not essential for the torque generation mechanism in Salmonella enterica (19).
The Bacillus subtilis flagellar motor has two distinct types of stator units: H+-type MotAB and Na+-type MotPS (20, 21). MotP and MotS together form a Na+ channel in a manner similar to the MotAB proton channel, and the MotPS complex acts as a Na+ sensor to regulate its assembly into and disassembly from the motor (10, 20, 21). However, the mechanism remains unknown. Here, we used high-speed atomic force microscopy (HS-AFM) combined with mutational analysis and obtained structural insights into the assembly and activation mechanism of the Na+-type MotPS stator unit.
RESULTS
Effect of Na+ ions on the torque-speed relationship of the B. subtilis wild-type flagellar motor
Precise analyses of the torque versus speed relationship of the flagellar motor provide insights into the torque generation mechanism of the flagellar motor (3). It has been shown that the torque-speed curve of the MotPS motor is distinct from that of the MotAB motor (fig. S2A, blue and magenta lines) (10). A single MotPS stator unit produces the same torque as a single MotAB stator unit when operating at high load (10). However, the maximum rotational speed of the MotPS motor is about four times slower than that of the MotAB motor when operating at low load (10). To understand the Na+-sensing mechanism of the MotPS complex, we first characterized the torque-speed relationship of the B. subtilis wild-type motor over a wide range of Na+ concentrations by bead assays (fig. S2A and table S1). The maximum speed of the wild-type motor was decreased from ca. 200 to 80 Hz when the external Na+ concentration was increased from 0 to 200 mM, although the stall torque was not changed at all. It has been shown that only one MotPS stator unit can be installed into the motor under these experimental conditions (10). Because the stall torque produced by the motor is dependent on the number of active stator units around a rotor (3, 4), this result suggests that the wild-type motor contains both H+-type MotAB and Na+-type MotPS stator units in the presence of NaCl.
To clarify whether the wild-type motor shows rapid stator exchange in a Na+-dependent manner, we carried out buffer exchange experiments (fig. S2B and table S2). To quantitatively evaluate the speed stability of flagellar motor rotation, we calculated the average speed (ωav) and the SD of ω (σω). The wild-type motor with a 0.8-μm bead attached rotated at an average speed of 83.8 Hz with an SD of 7.5 Hz in a buffer containing 200 mM KCl. The speed fluctuation, evaluated as the value of σω/ωav, was 0.09. Upon exchanging the buffer to that containing 200 mM NaCl, the rotation rate of the wild-type motor with the 0.8-μm bead attached was decreased to an average speed of 65.8 Hz with an SD of 19.5 (σω/ωav = 0.3). When the buffer was exchanged back to the one containing 200 mM KCl, the motor rotated stably at the original level again (σω/ωav = 0.09). The rotation rate of the MotAB motor was stable in the presence and absence of 200 mM NaCl, whereas the MotPS motor rotated at an average speed of 9.8 Hz with an SD of 2.3 Hz only in the presence of 200 mM NaCl. These results indicate that the assembly and disassembly of the MotPS complex rapidly occur in a Na+-dependent manner.
Molecular shape of the MotPS complex in the presence and absence of Na+ ions
MotP and MotS together form a Na+ channel complex with four copies of MotP and two copies of MotS in a manner similar to the MotA4B2 proton channel complex (Fig. 2A). The C-terminal periplasmic domain of MotS (MotSC), which has a putative plug segment and a PGB motif (Fig. 2B), is postulated to act as a Na+ sensor (10). To clarify how it senses Na+, we purified the MotP4S2 complex, of which TM helices were covered by amphipol to make the TM domain stably solubilized in solution (fig. S3), and analyzed its structural dynamics by HS-AFM in a buffer solution with or without Na+ ions. HS-AFM is capable of not only visualizing conformational dynamics of protein complexes at a high spatiotemporal resolution (22, 23) but also observing the status of ID regions of proteins (24). Purified MotPS complex was visualized as two ellipsoid domains connected by a flexible linker (Fig. 2C and movies S1 and S2). The molecular shape of the small domain in the experimental AFM images was almost the same as that of a simulated image constructed from the crystal structure of the MotBC dimer [Protein Data Bank (PDB) code: 2ZVY] (fig. S4) (19). To confirm this, we constructed the MotPSΔperi complex generated by a deletion of MotSC (Fig. 2B) and found that it did not have the small domain (Fig. 2C and movie S3), leading to a conclusion that the small domain corresponds to the MotSC dimer. Because the molecular shape of the large domain looked similar to that of a simulated model image constructed from the three-dimensional (3D) density map of the MotA tetramer reconstructed from negatively stained electron microscopy images (fig. S4) (25), we suggest that the large domain corresponds to the MotP tetramer in complex with the MotS-TM dimer.
Fig. 2. HS-AFM imaging of wild-type MotPS and its mutant derivatives.
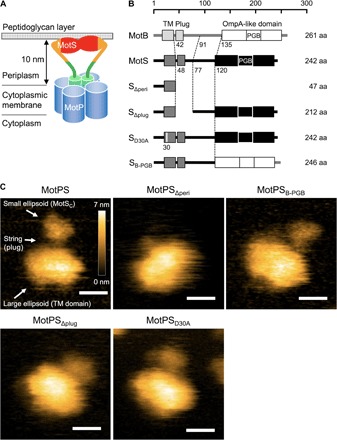
(A) Schematic diagram of the Na+-type MotPS complex. The MotPS complex is composed of four copies of MotP and two copies of MotS. The distance between the periplasmic surface of the cytoplasmic membrane and the inner surface of the peptidoglycan layer is about 10 nm. (B) Primary structures of Bacillus MotB, MotS, and various mutant variants of MotS. B. subtilis MotB and MotS have a single TM helix and a C-terminal large periplasmic domain containing a PGB motif termed an OmpA-like domain. MotPSΔperi and MotPSΔplug lack the indicated region of MotS. MotPSD30A has the D30A substitution at a putative Na+-binding site of MotS-TM. MotPSB-PGB has an OmpA-like domain of MotB instead of its original OmpA-like domain. aa, amino acid. (C) Typical HS-AFM images of wild-type MotPS, MotPSΔperi, MotPSB-PGB, MotPSΔplug, and MotPSD30A, placed on mica in a buffer containing 150 mM NaCl. All images were recorded at 200 ms per frame in a scanning area of 50 nm × 50 nm with 150 pixels × 150 pixels. Color bar shows a range of particle height (nanometers). Scale bars, 10 nm.
About 86% of the particles adopted the two-domain structure in the presence of 150 mM NaCl (n = 102). In the remaining 14% of the particles, two filamentous structures were seen to be attached to the large domain, indicating that MotSC was disordered. In the absence of NaCl, only 25% of the particles took the two-domain feature, and MotSC adopted unfolded conformations in 75% of the particles (n = 104). The buffer containing 150 mM KCl did not affect the folding of MotSC at all (n = 103) (fig. S5, A and B). These observations raise the possibility that Na+ ions directly stabilize the folded MotSC structure. To clarify this, we replaced the PGB domain of MotS by that of MotB to generate MotPSB-PGB (Fig. 2B) and carried out HS-AFM imaging (Fig. 2C and movie S4). The structural stability of the PGB domain of MotPSB-PGB was not affected by depletion of NaCl (fig. S5B). These results suggest that Na+ acts as a structural stabilizer to maintain the folded conformation of the PGB domain of MotSC.
Real-time imaging of Na+-induced structural transitions of MotSC
We next investigated whether the structural transitions of the MotSC dimer between the unfolded and folded states are reversible (movies S5 and S6). When MotPS was exposed to a buffer with 150 mM KCl, MotSC was unfolded in solution. Upon gradually exchanging the buffer to the one containing 150 mM NaCl, thereby increasing the Na+ concentration up to 150 mM (fig. S5C), MotSC became suddenly structured with a transition time within 1 s (Fig. 3A, second and third panels). When the Na+ concentration was reduced back to zero again (fig. S5D), the folded MotSC dimer also became suddenly unstructured with a transition time within 1 s (Fig. 3B, second and third panels). Because the temporal resolution of the present HS-AFM imaging was 250 ms per frame, it was not possible to estimate the rate constant for the folding and unfolding transitions. However, we obtained the Michaelis constant (Km) value of MotSC for Na+ and the Hill’s cooperativity coefficient by varying the Na+ concentration and estimated them to be about 130 mM and 15.0 ± 2.0, respectively (Fig. 3C). These results indicate that Na+ ions directly induce the disorder-to-order transition of MotSC, facilitating its dimerization to form the PGB domain in a highly cooperative manner. Therefore, we propose that Na+ ions induce the structural switching of the MotSC dimer from the unfolded to the folded states, thereby facilitating MotPS assembly into a motor to act as an active Na+-type stator unit.
Fig. 3. Na+-induced structural transitions of the MotSC dimer.
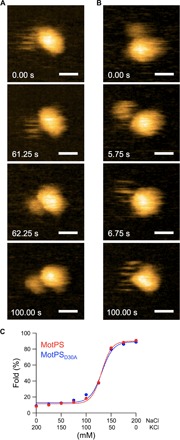
(A) Real-time imaging of MotPS with exchanging the salt in the buffer from 150 mM KCl to 150 mM NaCl. MotSC became suddenly folded after approximately 60 s (second and third panels) from the solvent exchange indicated as 0.00 s (0 mM NaCl in fig. S5C). (B) Real-time imaging of MotPS with exchanging the salt from 150 mM NaCl to 150 mM KCl. MotSC became suddenly unfolded after approximately 5 s (second and third panels) from the solvent exchange indicated as 0.00 s (150 mM NaCl in fig. S5D). All images in (A) and (B) were recorded at 250 ms per frame in a scanning area of 50 nm × 50 nm with 150 pixels × 150 pixels. Scale bars, 10 nm. (C) Effect of NaCl concentrations on the structural transitions of the PGB domain of MotSC. At least 100 individual particles were analyzed under each condition. The data points were fitted by the Hill’s equation.
Effect of an in-frame deletion of residues 48 to 77 of MotS on Na+-induced structural transitions of MotSC
ID regions in proteins are known to modulate their biological activities along with their binding partners (1, 2). Residues 48 to 77 of MotS, which form a putative motif termed a plug segment (17), are predicted to be ID, as judged by the predictor of naturally disordered regions (PONDR) program (fig. S1B). To examine whether the plug segment contributes to the Na+-induced structural transitions of MotSC, we constructed MotPSΔplug lacking residues 48 to 77 of MotS (Fig. 2B) and carried out its HS-AFM imaging (movie S7). The deletion of residues 48 to 77 in MotS caused a loss-of-function phenotype (fig. S6), indicating that the plug segment of MotSC is essential for motor function. The MotPSΔplug complex observed by HS-AFM showed the two ellipsoid domains, but the flexible linker connecting these two domains was missing (Fig. 2C). The population of particles with the unfolded PGB domain was increased to about 80% when the salt in the buffer was exchanged from 150 mM NaCl to 150 mM KCl (fig. S5B). This indicates that the PGB domain of MotSC itself contains a Na+-induced structural switch.
Effect of the MotSD30A mutation on Na+-induced structural transitions of MotSC
A highly conserved Asp24 residue of Vibrio alginolyticus PomB, which corresponds to Asp30 of B. subtilis MotS-TM, is a Na+-binding site (26) and is responsible for the Na+-dependent assembly and disassembly of the PomAB complex (27), which acts as a Na+-type stator unit of marine Vibrio (fig. S1C). Therefore, we tested whether the replacement of Asp30 of MotS by alanine affects the structural transitions of MotSC. The D30A substitution inhibited motility in soft agar (fig. S6), in agreement with a previous report (28). MotPSD30A observed by HS-AFM looked very similar in its entire architecture to wild-type MotPS (Fig. 2C and movie S8). Replacement of NaCl by KCl in the buffer still significantly increased the population of an unfolded conformation of MotSC from ca. 18 to 89% (fig. S5B), indicating that it is not the binding of Na+ to Asp30 of MotS that induces the disorder-to-order transition of MotSC. Consistently, the D30A mutation affected neither the Km value of MotSC for Na+ nor the Hill’s cooperativity coefficient (Fig. 3C). Therefore, we conclude that Asp30 of MotS is not directly involved in the Na+-induced structural transition of MotSC.
Structural stability of folded MotSC in the presence of Na+ ions
Structure-based functional analyses of MotBC and PomBC led to a proposal that drastic conformational changes of the N-terminal portion of the PGB domains of MotBC and PomBC are required not only for the binding of their PGB domains to the peptidoglycan layer but also for the ion channel formation upon MotAB and PomAB stator assembly into the flagellar motor, because the distance between the periplasmic surface of the cytoplasmic membrane and the inner surface of the peptidoglycan layer is about 10 nm, which is too far to reach for their PGB domains with only 5 nm in height above the TM domain (19, 29). To see whether the 5-nm extension of the PGB domain required for its binding to the peptidoglycan layer occurs in action, we analyzed the correlation between the center-to-center distance and the heights of these two ellipsoid domains of wild-type MotPS in HS-AFM images (Fig. 4A). The center-to-center distance between these two domains varied within a short time scale. The histogram of the center-to-center distance showed a broad distribution that can be fitted with three Gaussian peaks at 10.1, 12.5, and 15.4 nm, although these peaks do not seem to be meaningful. The image height of the small domain corresponding to MotSC decreased as the distance between these two domains increased, whereas the image height of the large domain did not change at all (Fig. 4B). The histogram of the height of MotSC showed two distinct peaks at 4.1 and 3.2 nm (Fig. 4B). Because the accuracy of our HS-AFM measurements is estimated to be about 1 nm for the distance between the two objects and 0.15 nm for the height (30), we suggest that the N-terminal portion of MotSC is less stable and easy to unfold, allowing the PGB domain of MotSC to extend from the large domain by about 5 nm.
Fig. 4. Correlation between the center-to-center distance between the small and large domains and the domain heights.
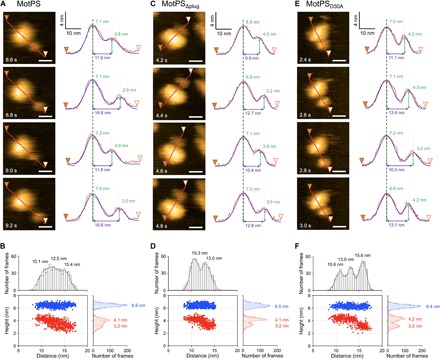
Sequential HS-AFM images and their cross sections are shown for MotPS (A), MotPSΔplug (C), and MotPSD30A (E). All images were recorded at 200 ms per frame in a scanning area of 50 nm × 50 nm with 150 pixels × 150 pixels. Scale bars, 10 nm. Surface profiles along the line (from filled to open arrowheads) were fitted by two Gaussian functions, and the peak-to-peak distance and peak heights were measured. Distance versus height plots were generated for MotPS (B), MotPSΔplug (D), and MotPSD30A (F) from data points collected from more than 600 frames of 20 individual molecules. Blue and red dots are height data for the large and small domains, respectively. Histograms were fitted by multiple Gaussian functions.
We found that residues 48 to 77 of MotSC correspond to the flexible linker connecting the two distinct domains of MotPS (Fig. 2). Therefore, we next tested whether the plug segment contributes to the 5-nm extension of its PGB domain from the large domain. The center-to-center distance between these two domains of MotPSΔplug also changed within a short time scale (Fig. 4C). In contrast to the wild type, the histogram of the center-to-center distance of MotPSΔplug showed two distinct peaks at 10.3 and 13.0 nm. The height of MotSC decreased as the distance between these two domains increased, whereas the height of the large domain did not change at all (Fig. 4D) in a manner similar to that of the wild type (Fig. 4B). This indicates that the N-terminal portion of the PGB domain of MotSC is less stable and becomes easily unstructured. Consistently, the N-terminal potion of the PGB domain is predicted to have a flexible conformation (fig. S1B). Therefore, we suggest that residues 48 to 77 of MotSC form a flexible string responsible for an approximately 2.5-nm distance of the PGB domain from the large domain and that the N-terminal portion of the PGB domain can adopt a stretched conformation, causing a further roughly 2.5-nm extension from 13.0 to 15.4 nm. Because the deletion of residues 48 to 77 caused loss of function (fig. S6), we propose that conformational fluctuations of the plug segment in MotSC allow the PGB domain to reach and bind to the peptidoglycan layer.
A highly conserved Asp30 residue of MotS-TM is postulated to be involved in the Na+ flow through the Na+ channel formed by MotP-TM3, MotP-TM4, and MotS-TM helices in a manner similar to Asp33 residue of Salmonella MotB-TM (4). The plug segment coordinates the ion channel activity with stator assembly into the motor (17), raising the possibility that the D30A mutation may also affect the extension and contraction processes of MotSC. To test this, we analyzed the correlation between the center-to-center distance between the small and large domains of MotPSD30A and their heights (Fig. 4E). Similar to the wild type, the distance distribution of MotPSD30A clearly showed three distinct peaks at 10.6, 13.0, and 15.6 nm (Fig. 4F). However, the height of the MotSC domain did not change when the distance was increased from 10.6 to 13.0 nm, indicating that the 2.4-nm extension between the two domains is a consequence of the dissociation of the MotSC plug segment from the TM domain (Fig. 4F). When the center-to-center distance was increased from 13.0 to 15.6 nm, the height of the MotSC domain decreased from 4.2 to 3.2 nm (Fig. 4F). Wild-type MotPS did not show this two-step extension so clearly, raising the possibility that the D30A mutation reduces the rate of extension and contraction processes of MotSC. Therefore, we propose that the binding of Na+ to Asp30 in MotS-TM may regulate the rates of detachment, following conformational dynamics of the plug segment of MotSC.
DISCUSSION
The assembly of Na+-type stator units into the flagellar motor is affected by changes in the external Na+ concentration. This suggests that the Na+-type stator unit acts as a Na+ sensor that regulates the number of active stator units in the motor in response to the external Na+ concentration (27, 31–33). Wild-type B. subtilis has two distinct types of stators: H+-type MotAB and Na+-type MotPS complexes (20, 21). When the PGB domain of MotB of B. subtilis is replaced by that of MotS, the chimeric motor shows the same torque-speed curve as the MotAB motor in the presence of Na+ ions. In contrast, the stall torque produced by this chimeric motor decreases significantly in the absence of NaCl (10). Because the stall torque is proportional to the number of active stator units in the motor (3, 4), it has been proposed that the PGB domain of MotS has the Na+-binding site to stabilize its association with the peptidoglycan layer (10). Here, we showed real-time observations of the Na+-induced folding and dimerization of the PGB domain of MotSC (Fig. 3) and dynamic changes in the distance between this PGB domain and the TM domain of MotPS connected by a flexible linker (Fig. 4). Because an about 5-nm extension of the PGB domain of MotSC from the TM domain allows the MotPS complex to reach and be anchored to the peptidoglycan layer to act as an active Na+-type stator unit, we suggest that the PGB domain of MotSC acts as a Na+ sensor to regulate the activation and assembly of the MotPS complex into the motor through its Na+-induced structural transition between the unfolded and folded states.
On the basis of the available information, we propose the following mechanism for assembly and activation of the Na+-type MotPS stator unit (Fig. 5). In the absence of Na+, MotPS exists as an inactive form in the cytoplasmic membrane because MotSC adopts an unfolded conformation. When the concentration of Na+ is increased, the binding of Na+ to MotSC induces the folding of the PGB domain and facilitates its dimerization in a highly cooperative manner. Then, an interaction between MotP and FliG stabilizes the MotPS complex assembled into the motor, which allows the opening of the Na+ channel by releasing the plug and the 5-nm extension of the PGB domain from the TM domain to bind to the peptidoglycan layer. As a result, MotPS becomes an active Na+-type stator unit to couple the Na+ flow with torque generation. When Na+ ions are depleted, the PGB domain becomes unstructured, inducing its rapid dissociation from the peptidoglycan layer and dissociation of the MotPS complex from the motor.
Fig. 5. Model for the Na+-induced assembly and disassembly of the MotPS stator.
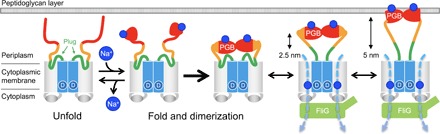
Structural transitions of MotSC between the unfolded and folded states occur in a Na+-dependent manner. The plug region of MotSC is detached from the TM Na+ channel formed by the TM helices of MotP and MotS, followed by partial unfolding of the N-terminal portion of the PGB domain of MotSC, causing a 5-nm extension of the PGB domain from the TM domain of MotPS to reach the peptidoglycan layer, allowing MotPS to become an active Na+-type stator unit.
MATERIALS AND METHODS
Bead assays
The wild-type Bacillus strain producing sticky filaments was grown in L-broth composed of 1% (w/v) Bacto tryptone, 0.5% (w/v) yeast extract, and 0.5% (w/v) NaCl with shaking at 37°C until the cell density had reached an optical density at 600 nm of 1.0. Polystyrene beads with diameters of 2.0, 1.5, 1.0, 0.8, 0.6, or 0.5 μm (Invitrogen) were attached to the sticky filament, as described previously (10). Bead assays were carried out at room temperature in motility buffer [10 mM potassium phosphate (pH 7.0), 0.1 mM EDTA, and 10 mM l-lactic acid] containing 200 mM K+, 150 mM K+ plus 50 mM Na+, 100 mM K+ plus 100 mM Na+, or 200 mM Na+, as described (10). The rotation speed analysis and torque calculation were carried out as described previously (34, 35). The average rotation rate, <ω>, and SD, σω, of each rotation data were calculated as described previously (36).
Expression and purification of His-tagged MotPS complex
DNA manipulations and site-directed mutagenesis were carried out as described previously (37). DNA fragments encoding MotPS with a His6-tag attached to the N terminus of MotP or the C terminus of MotS were amplified by polymerase chain reaction (PCR) and were cloned into the Nhe I and Xho I sites of the pET21b plasmid vector (Novagen), yielding pET-HPS or pET-PSH plasmid, respectively. A DNA fragment encoding MotPSB-PGB was amplified by PCR using a plasmid named pDR-PSp3sticky (10) as a template. To express MotPSΔperi and MotPSΔplug, the corresponding regions were deleted by inverse PCR using the pET-PSH plasmid as a template. The MotSD30A mutation was introduced into the pET-PSH plasmid by site-directed mutagenesis. All plasmids were confirmed by DNA sequencing with a 3130 Genetic Analyzer (Applied Biosystems). The obtained recombinant plasmids were introduced into the Escherichia coli BL21 (DE3) strain (Stratagene). BL21 (DE3) cells harboring a pET21b-based plasmid were grown in L-broth for 4 hours at 30°C. The expression of His-tagged MotPS was induced by adding isopropyl-β-d-1-thiogalactopyranoside at a final concentration of 0.6 mM, and then the incubation was continued for another 4 hours at 30°C. After washing with 50 mM tris-HCl (pH 8.0), cells were resuspended in 50 mM tris-HCl (pH 8.0) containing complete protease inhibitor cocktail (Roche) and deoxyribonuclease I (New England Biolabs) and were disrupted by passage through a French pressure cell (FA-032, Central Scientific Commerce). Undisrupted cells were removed by low-speed centrifugation (20,000g, 15 min, 4°C). Membranes were collected by ultracentrifugation (110,000g, 1 hour, 4°C) and suspended in 50 mM tris-HCl (pH 8.0) and 10% glycerol. The membranes were solubilized in 50 mM tris-HCl (pH 8.0), 10% glycerol, 500 mM NaCl, and 1% decyl maltose neopentyl glycol (DMNG) (Anatrace) for 30 min at 4°C, followed by ultracentrifugation. The solubilized membranes were mixed well with Ni–nitrilotriacetic acid (NTA) agarose (Qiagen) using a rotator for 30 min at 4°C. The Ni-NTA agarose resin, to which His-tagged MotPS bound, was packed into an open column (Bio-Rad), and then unbound proteins were removed by washing with 50 mM tris-HCl (pH 8.0), 5% glycerol, 500 mM NaCl, 50 mM imidazole, and 0.1% DMNG. His-tagged MotPS was eluted with 50 mM tris-HCl (pH 8.0), 5% glycerol, 500 mM NaCl, 400 mM imidazole, and 0.05% DMNG. Pooled fractions containing His-tagged MotPS were loaded onto a Superose 6 10/300 GL column (GE Healthcare) equilibrated with 20 mM tris-HCl (pH 8.0), 5% glycerol, 500 mM NaCl, and 0.05% DMNG (fig. S3A). To replace DMNG by amphipol, pooled fractions containing MotPS (fig. S3B) were mixed with amphipols A8-35 (Anatrace) at a 1-to-3 weight ratio with end-over-end rotation for 12 hours, and then DMNG was removed by the addition of Bio-Beads (Bio-Rad) at a final concentration of 15 mg ml−1 for 24 hours. After centrifugation to remove the Bio-Beads, supernatants were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with 20 mM tris-HCl (pH 8.0) and 150 mM NaCl (fig. S3, C and D).
HS-AFM imaging
HS-AFM imaging was performed in solution at room temperature using a laboratory-built HS-AFM setup (38, 39), as described previously (40). In brief, a glass sample stage (diameter, 2 mm; height, 2 mm) with a thin mica disc (1 mm in diameter and ~0.05 mm in thickness) glued to the top by epoxy was attached onto the top of a Z-scanner by a drop of nail polish. A freshly cleaved mica surface was prepared by removing the top layers of mica using a Scotch tape. Then, a drop (2 μl) of diluted protein sample (ca. 10 nM) with an observation buffer [20 mM tris-HCl (pH 8.0) with or without various concentrations of NaCl or KCl] was deposited onto the mica surface. After incubation for 3 min at room temperature, the mica surface was rinsed with 20 μl of the observation buffer to remove floating samples. The sample stage was then immersed in a liquid cell containing ~60 μl of the observation buffer. AFM imaging was carried out in a tapping mode using small cantilevers (BLAC10DS-A2, Olympus) (resonant frequency, ~0.5 MHz in water; quality factor, ~2 in water; spring constant, ~0.1 N m−1). The cantilever’s free oscillation amplitude A0 and set-point amplitude As were set at 1 to 2 nm and ~0.9 × A0, respectively. The imaging rate, scan size, and pixel size for each AFM image are described in the figure legends.
Buffer exchange system
To carry out buffer exchange during HS-AFM imaging, we installed a high-precision syringe pump system (11 Pico Plus Elite, Harvard Apparatus) to the liquid cell of HS-AFM, as previously described (41) with minor modifications. By setting 10-ml Terumo syringes (SS-10SZP, Terumo), the constant-pressure and constant-flow pump system concomitantly infuses buffer solutions on one side and withdraws them from the other side of the liquid cell at a rate ranging from ~0.2 nl s−1 to ~0.2 ml s−1, allowing the buffer composition to be gradually exchanged. Here, we connected two infusion syringes and one withdraw syringe to the pump system and operated the pump at the flow rate of 2.5 μl s−1. Each of the syringe was connected with a silicon tube of about 60 cm in length, and the other end of the silicon tube was connected with a gel-loading tip (1034-R-204, BIO-BIK). A buffer solution from one of the infusion syringes was infused into the liquid cell, and a buffer solution from the other infusion syringe was wasted on the sponge placed beside the liquid cell, while the withdraw syringe kept withdrawing the buffer solution from the liquid cell to keep the buffer volume (ca. 60 μl) constant. Here, NaCl buffer [20 mM tris-HCl (pH 8.0) and 150 mM NaCl] and KCl buffer [20 mM tris-HCl (pH 8.0) and 150 mM KCl] were degassed using an aspirator for 30 min to remove bubbles, each of which was loaded into each infusion syringe. Replacements of the tip ends connected to the infusion syringes were done using XYZ axis manual stages (M-152, Narishige). To measure precise Na+ concentrations of the buffer, we used a Na+-sensitive fluorescent dye, CoroNa Green (Molecular Probes), as a Na+ indicator. The fluorescence emission spectra of CoroNa Green were recorded on a fluorescence spectrophotometer (RF-5300PC, Shimadzu). The Na+ concentration in the buffer was determined by the fluorescence intensity of the CoroNa Green. To generate a calibration curve, the fluorescence intensities of CoroNa Green were determined over a wide range of 0 to 150 mM.
Analysis of AFM images
For AFM image analysis, AFM images were pretreated by a low-pass filter to remove spike noise and a flatten filter to make the overall xy plane flat, using a laboratory-built software as described previously (42). This software is available at https://elifesciences.org/content/4/e04806/article-data-fig-data-supplementary-material. The distances between two ellipsoid domains and the heights of these two domains were analyzed by ImageJ (https://imagej.nih.gov/ij/), as described previously (43). The surface profiles to go along two ellipsoidal centers of gravity were fitted by multiple Gaussian functions, and each height and center-to-center distance were measured. Data points were collected from more than 600 frames of 20 individual molecules. Because structural transitions of the MotSC domain rapidly occurred in solution, we decided that the MotSC domain took a steady-state conformation if its structure did not change at all in about a minute. At least 100 individual molecules under each condition were analyzed.
Simulation of AFM image
To simulate AFM images of the MotSC dimer and the MotA4 tetramer attached to a mica surface, we used a software (SPM simulator, Advanced Algorithm Systems), as described previously (23). The simulation was carried out with a simple hard sphere model. The cantilever tip was modeled as a circular cone (apex angle, 10°) with a small sphere (radius, 0.5 to 2 nm) at the apex. The crystal structure of the periplasmic domain of MotB (MotBC) (PDB code: 2ZVY) and the 3D density map of negatively stained MotA4 tetramer (25) were used as templates to generate simulated AFM images. Each atom in the MotBC protein was modeled as a hard sphere with a corresponding van der Waals radius. We simulated AFM images using various radii for the tip apex sphere and found that a radius of 1.0 nm produced almost the same images as experimental HS-AFM images. The simulated images were processed by a low-pass filter with a cutoff wavelength of 2 nm because a spatial resolution of our HS-AFM images was approximately 2 nm, as judged by the 2D Fourier transformation of actual images.
Supplementary Material
Acknowledgments
Funding: This work was supported, in part, by Japan Society for the Promotion of Science KAKENHI grant nos. JP15H04360 (to N.K.), JP15H03540 (to T.U.), JP24227005 (to T.A.), and JP25000013 (to K.N.); MEXT (Ministry of Education, Culture, Sports, Science and Technology) KAKENHI grant nos. JP16H00830 and JP16H00758 (to T.U.), JP26119003 (to T.A.), and JP24117004 and JP15H01640 (to T.M.); and Japan Science and Technology Agency grant nos. JPMJPR13L4 (to N.K.) and JPMJCR13M1 (to T.A.). Author contributions: N.T., N.K., K.N., and T.M. conceived and designed the research. N.T. and N.K. performed the research. N.K., T.U., and T.A. set up the HS-AFM imaging system including both hardware and software. N.T., N.K., and T.U. analyzed the data. N.T., N.K., K.N., and T.M. wrote the paper based on discussion with other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao4119/DC1
fig. S1. Primary structures of MotB and its homologs, MotS and PomB.
fig. S2. Effect of Na+ concentrations on motor rotation of the flagellar motor in wild-type Bacillus cells expressing both MotAB and MotPS.
fig. S3. Purification of His6-tagged MotPS by size exclusion chromatography.
fig. S4. Comparison of simulated AFM images of the MotBC and the MotA tetramer with experimental image of the MotPS complex.
fig. S5. Two distinct conformations of the PGB domain of MotS.
fig. S6. Motility of motS mutants.
table S1. Rotational speed and torque of the wild-type motor.
table S2. Speed fluctuations of the wild-type, MotAB, and MotPS motor.
movie S1. Real-time imaging of wild-type MotPS by HS-AFM.
movie S2. Typical HS-AFM imaging of wild-type MotPS in buffer containing 150 mM NaCl.
movie S3. Typical HS-AFM imaging of MotPSΔperi in buffer containing 150 mM NaCl.
movie S4. Typical HS-AFM imaging of MotPSB-PGB in buffer containing 150 mM NaCl.
movie S5. Real-time imaging of a disorder-to-order transition of MotPS with an increase in the concentration of NaCl.
movie S6. Real-time imaging of a order-to-disorder transition of MotPS with a decrease in the concentration of NaCl.
movie S7. Typical HS-AFM imaging of MotPSΔplug in buffer containing 150 mM NaCl.
movie S8. Typical HS-AFM imaging of MotPSD30A in buffer containing 150 mM NaCl.
REFERENCES AND NOTES
- 1.Tompa P., Multisteric regulation by structural disorder in modular signaling proteins: An extension of the concept of allostery. Chem. Rev. 114, 6715–6732 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Berlow R. B., Dyson H. J., Wright P. E., Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature 543, 447–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg H. C., The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Morimoto Y. V., Minamino T., Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 4, 217–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamino T., Imada K., The bacterial flagellar motor and its structural diversity. Trends Microbiol. 23, 267–274 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Lele P. P., Hosu B. G., Berg H. C., Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 110, 11839–11844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tipping M. J., Steel B. C., Delalez N. J., Berry R. M., Armitage J. P., Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol. Microbiol. 87, 338–347 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Tipping M. J., Delalez N. J., Lim R., Berry R. M., Armitage J. P., Load-dependent assembly of the bacterial flagellar motor. MBio 4, e00551–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo D. J., Nakamura S., Morimoto Y. V., Che Y.-S., Kami-ike N., Kudo S., Minamino T., Namba K., The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics 9, 173–181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terahara N., Noguchi Y., Nakamura S., Kami-ike N., Ito M., Namba K., Minamino T., Load- and polysaccharide-dependent activation of the Na+-type MotPS stator in the Bacillus subtilis flagellar motor. Sci. Rep. 7, 46081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima S., Blair D. F., Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43, 26–34 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Zhou J., Blair D. F., Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273, 428–439 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Morimoto Y. V., Nakamura S., Kami-ike N., Namba K., Minamino T., Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 78, 1117–1129 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Morimoto Y. V., Nakamura S., Hiraoka K. D., Namba K., Minamino T., Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 195, 474–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihara Y., Kitao A., Gate-controlled proton diffusion and protonation-induced ratchet motion in the stator of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 112, 7737–7742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Che Y.-S., Nakamura S., Morimoto Y. V., Kami-ike N., Namba K., Minamino T., Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol. Microbiol. 91, 175–184 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Hosking E. R., Vogt C., Bakker E. P., Manson M. D., The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364, 921–937 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Morimoto Y. V., Che Y.-S., Minamino T., Namba K., Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 584, 1268–1272 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Kojima S., Imada K., Sakuma M., Sudo Y., Kojima C., Minamino T., Homma M., Namba K., Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73, 710–718 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Ito M., Hicks D. B., Henkin T. M., Guffanti A. A., Powers B. D., Zvi L., Uematsu K., Krulwich T. A., MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 53, 1035–1049 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Ito M., Terahara N., Fujinami S., Krulwich T. A., Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J. Mol. Biol. 352, 396–408 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodera N., Yamamoto D., Ishikawa R., Ando T., Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Uchihashi T., Iino R., Ando T., Noji H., High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 333, 755–758 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Miyagi A., Tsunaka Y., Uchihashi T., Mayanagi K., Hirose S., Morikawa K., Ando T., Visualization of intrinsically disordered regions of proteins by high-speed atomic force microscopy. Chemphyschem 9, 1859–1866 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Takekawa N., Terahara N., Kato T., Gohara M., Mayanagi K., Hijikata A., Onoue Y., Kojima S., Shirai T., Namba K., Homma M., The tetrameric MotA complex as the core of the flagellar motor stator from hyperthermophilic bacterium. Sci. Rep. 6, 31526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudo Y., Kitade Y., Furutani Y., Kojima M., Kojima S., Homma M., Kandori H., Interaction between Na+ ion and carboxylates of the PomA–PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry 48, 11699–11705 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Fukuoka H., Wada T., Kojima S., Ishijima A., Homma M., Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71, 825–835 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y., Koyama K., Ito M., Suppressor mutants from MotB-D24E and MotS-D30E in the flagellar stator complex of Bacillus subtilis. J. Gen. Appl. Microbiol. 60, 131–139 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Zhu S., Takao M., Li N., Sakuma M., Nishino Y., Homma M., Kojima S., Imada K., Conformational change in the periplamic region of the flagellar stator coupled with the assembly around the rotor. Proc. Natl. Acad. Sci. U.S.A. 111, 13523–13528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto M., Kodera N., Tsunaka Y., Oda M., Tanimoto M., Ando T., Morikawa K., Tate S.-i., Phosphorylation-coupled intramolecular dynamics of unstructured regions in chromatin remodeler FACT. Biophys. J. 104, 2222–2234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulick A., Koerdt A., Lassak J., Huntley S., Wilms I., Narberhaus F., Thormann K. M., Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 71, 836–850 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Paulick A., Delalez N. J., Brenzinger S., Steel B. C., Berry R. M., Armitage J. P., Thormann K. M., Dual stator dynamics in the Shewanella oneidensis MR-1 flagellar motor. Mol. Microbiol. 96, 993–1001 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Sowa Y., Homma M., Ishijima A., Berry R. M., Hybrid-fuel bacterial flagellar motors in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 111, 3436–3441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Che Y.-S., Nakamura S., Kojima S., Kami-ike N., Namba K., Minamino T., Suppressor analysis of the MotB(D33E) mutation to probe the bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 190, 6660–6667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura S., Kami-ike N., Yokota J.-i. P., Kudo S., Minamino T., Namba K., Effect of intracellular pH on the torque–speed relationship of bacterial proton-driven flagellar motor. J. Mol. Biol. 386, 332–338 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Muramoto K., Magariyama Y., Homma M., Kawagishi I., Sugiyama S., Imae Y., Kudo S., Rotational fluctuation of the sodium-driven flagellar motor of Vibrio alginolyticus induced by binding of inhibitors. J. Mol. Biol. 259, 687–695 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Hara N., Namba K., Minamino T., Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLOS ONE 6, e22417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ando T., Kodera N., Takai E., Maruyama D., Saito K., Toda A., A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ando T., Uchihashi T., Fukuma T., High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 83, 337–437 (2008). [Google Scholar]
- 40.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Miyagi A., Chipot C., Rangl M., Scheuring S., High-speed atomic force microscopy shows that annexin V stabilizes membranes on the second timescale. Nat. Nanotechnol. 11, 783–790 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Ngo K. X., Kodera N., Katayama E., Ando T., Uyeda T. Q. P., Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4, e04806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodera N., Uchida K., Ando T., Aizawa S.-I, Two-ball structure of the flagellar hook-length control protein FliK as revealed by high-speed atomic force microscopy. J. Mol. Biol. 427, 406–414 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao4119/DC1
fig. S1. Primary structures of MotB and its homologs, MotS and PomB.
fig. S2. Effect of Na+ concentrations on motor rotation of the flagellar motor in wild-type Bacillus cells expressing both MotAB and MotPS.
fig. S3. Purification of His6-tagged MotPS by size exclusion chromatography.
fig. S4. Comparison of simulated AFM images of the MotBC and the MotA tetramer with experimental image of the MotPS complex.
fig. S5. Two distinct conformations of the PGB domain of MotS.
fig. S6. Motility of motS mutants.
table S1. Rotational speed and torque of the wild-type motor.
table S2. Speed fluctuations of the wild-type, MotAB, and MotPS motor.
movie S1. Real-time imaging of wild-type MotPS by HS-AFM.
movie S2. Typical HS-AFM imaging of wild-type MotPS in buffer containing 150 mM NaCl.
movie S3. Typical HS-AFM imaging of MotPSΔperi in buffer containing 150 mM NaCl.
movie S4. Typical HS-AFM imaging of MotPSB-PGB in buffer containing 150 mM NaCl.
movie S5. Real-time imaging of a disorder-to-order transition of MotPS with an increase in the concentration of NaCl.
movie S6. Real-time imaging of a order-to-disorder transition of MotPS with a decrease in the concentration of NaCl.
movie S7. Typical HS-AFM imaging of MotPSΔplug in buffer containing 150 mM NaCl.
movie S8. Typical HS-AFM imaging of MotPSD30A in buffer containing 150 mM NaCl.


