Abstract
A microcosm experiment was conducted for 112 d by spiking petroleum hydrocarbons into soils from four regions of China. Molecular analyses of soils from microcosms revealed changes in taxonomic diversity and oil catabolic genes of microbial communities. Degradation of total petroleum hydrocarbons (TPHs) in Sand from the Bohai Sea (SS) and Northeast China (NE) exhibited greater microbial mineralization than those of the Dagang Oilfield (DG) and Xiamen (XM). High-throughput sequencing and denaturing gradient gel electrophoresis (DGGE) profiles demonstrated an obvious reconstruction of the bacterial community in all soils. The dominant phylum of the XM with clay soil texture was Firmicutes instead of Proteobacteria in others (DG, SS, and NE) with silty or sandy soil texture. Abundances of alkane monooxygenase gene AlkB increased by 10- to 1000-fold, relative to initial values, and were positively correlated with rates of degradation of TPHs and n-alkanes C13-C30. Abundances of naphthalene dioxygenase gene Nah were positively correlated with degradation of naphthalene and total tricyclic PAHs. Redundancy analysis (RDA) showed that abiotic process derived from geographical heterogeneity was the primary effect on bioremediation of soils contaminated with oil. The optimization of abiotic and biotic factors should be the focus of future bioremediation of oil contaminated soil.
Introduction
Bioremediation is the most efficient, economical way to deal with contamination of soils by petroleum hydrocarbons (PHs). It also does not generate toxic metabolites. Thus, it has been widely accepted and used for remediation of areas that have been contaminated long-term with petroleum1. Besides the presence of PHs, the structure of communities of microbes capable of degrading PHs also depends on properties of soils in different geographic regions (i.e., physical-chemical as well as biological characteristics). Examples of this variability are Actinobacter sp., and Bacteroidete sp., which were the dominant alkane-degrading bacteria in soils of the Huabei Oilfield2, while the community on King George Island was composed mainly of Rhodococcus sp., Acinetobacter sp., Mycobacterium sp., Gordonia sp., and Aeromicrobium sp.3. Pedobacter sp. and Mycobacterium sp. were the major bacteria capable of degrading alkanes in areas where PHs are produced in the Karamay Oilfield4, while dominant alkane-degrading bacteria in soils from the Shengli Oilfield were Alcanivorax sp. and Acinetobacter sp.5. Based on profiles of phospholipid fatty acid, populations of gram-positive bacteria were significantly larger in petroleum contaminated upland soils than paddy fields6.
Previously, in contaminated ecosystems, relationships between dominant populations of microbes and intensity of contamination with PHs have been studied. Microorganisms inhabiting locations contaminated with PHs often contain enzymes that can degrade oil and thus might be of interest for scientific and industrial applications7,8. Recently developed molecular biotechnology, involving direct isolation of nucleic acids from environmental samples followed by denaturing gradient gel electrophoresis (DGGE), real-time quantitative polymerase chain reaction (qPCR) and high-throughput DNA sequencing (HTS) have been used to determine diversity of endogenous microorganisms9. Due to their highly conserved genetic topologies, genes capable of degrading oil have been used as biomarkers for determination of abundances and diversities of oil-degrading microorganisms from various environments10,11. Recently, real-time PCR initially developed for quantifying expressions of genes, has also been applied in microbial ecology and has become the most popular alternative for measurement of 16S rRNA for genes to describe specific microbial populations12. Alternatively, HTS using Illumina HiSeq technology provides a robust method to detect genome sequence information of all autochthonal organisms and describe their communities more comprehensively13.
Microbes capable of degrading PHs can operate on a wide range of metabolic substrates, and can even be adapted to polar regions14. Diverse microbial flora can degrade PHs anaerobically or aerobically by use of a variety of enzymes encoded by key functional genes, such as alkane hydroxylase genes, ring-hydroxylating dioxygenase α-subunit (RHDα) genes15, alkylsuccinate synthase gene (assA) and benzylsuccinate synthase gene (bssA)16. The main components of PHs are saturated aliphatic and aromatic hydrocarbons. During the initial activation step of aerobic aliphatic hydrocarbon metabolism, a non-heme integral membrane alkane monooxygenase encoded by AlkB gene is expressed. The AlkB can transfer electrons from nicotinamide adenine dinucleotide phosphate (NADPH) to PH substrates in the presence of the cofactors, rubredoxin (AlkG) and rubredoxin reductase (AlkT)17. Alternatively, during aerobic degradation of PAHs of smaller molecular masses, naphthalene dioxygenase systems encoded by Nah gene and phenol monooxygenase encoded by the Phe gene during the initial phase add single or two atoms of molecular oxygen to aromatic rings, respectively. Thus, quantification of these hydroxylase genes is interesting and important for assessment of potential for bioremediation by microbial communities of soils contaminated by PHs18.
In this study, laboratory-scale microcosms were used to study process and microbial responses to PHs spiked into soils from different geographic regions of China. Dynamic changes in genera within microbial communities and functional genes responsible for degradation of PHs were monitored using advanced techniques including DGGE, real-time quantitative PCR and high-throughput DNA sequencing. The studies were focused on enumeration of AlkB, Nah and Phe, which are thought to be responsible for hydroxylation of saturated aliphatic and aromatic hydrocarbons.
Results
Soil physical-chemical properties
The major physical-chemical characteristics of soils collected from different geographic sites are shown (Table 1). There was a great divergence in pH and salinity between soils from the north and south of China. In the north of China, DG and SS soil had pH values greater than 8 and salinities greater than 17 g kg−1. However, pH was 5.27 in XM soil which was from in the south of China. NE exhibited a pH of less than 7. DG and NE contained greater amounts of TOC exceeding 17 mg g−1, and a similar particle size distribution of about 5.6% clay, 77% silt and 17.5% sand as average value. However, SS and XM contained lesser amounts of TOC of less than 6 mg g−1. Sand content of soils from SS was greatest with a value of 96%, while contents of clay and silt were least at 0.56% and 3.37%, respectively. XM soil had the greatest clay and silt contents, in contrast to the least sand.
Table 1.
Physical-chemical properties of soils.
| Location | pH | Salinity (g/kg) | TOC (mg/g) | Particle size (%) | ||
|---|---|---|---|---|---|---|
| Clay (<2 μm) | Silt (2–50 μm) | Sand (>50 μm) | ||||
| DG | 9.23 | 17.657 | 20.82 | 5.8 | 78.4 | 15.8 |
| SS | 8.73 | 36.783 | 2.25 | 0.56 | 3.37 | 96.07 |
| NE | 4.78 | 3.725 | 17.62 | 5.47 | 75.41 | 19.12 |
| XM | 5.27 | 0.75 | 5.28 | 10.27 | 81.08 | 8.65 |
DG: Dagang Oilfield; SS: Sea Sand; NE: Northeast China; XM: Xiamen.
Degradation of petroleum hydrocarbons in different soil microcosms
Gravimetric estimation of total concentrations of PHs in soils initial and after various durations of incubation of microcosms was done to determine the degradation of PHs (Fig. 1a). All microcosms containing various soils resulted in approximately 50% loss of TPHs via degradation compared to sterile soils. Microbial flora in SS and NE soils exhibited greater potential to degrade TPHs, with maximum reduction of TPHs to 1.87 × 104 (SS) and 2.04 × 104 mg kg−1 (NE) after 112 d, from initial concentrations of 4.92 × 104 (SS) and 4.85 × 104 mg kg−1 (NE) (t = 0 d), which represented 62% and 58% of TPHs degradation, respectively. Residual concentrations of n-alkanes (C8-C40) and PAHs were also determined by use of GC-MS (Fig. 1b,c and Supplementary Figure S1). SS soils exhibited the greatest ability to degrade C8-C40 compounds and concentrations of C8-C40 alkanes were reduced from 1.29 × 104 (t = 0 d) to 0.27 × 104 mg kg−1 (t = 112 d), with degradation as great as 79%. Alkanes (C13-C30) which were the predominant alkanes were mineralized from 1.07 × 104 to 1.76 × 103 mg kg−1 by microbes in SS showing the highest mineralization of 84%. XM soil exhibited the least degradation of alkanes C8-C40 and C13-C30 with 58% and 59%, respectively. Similarly, degradation of PAHs in the four soils showed similar trends with SS showing the highest PAHs degradation up to 78%. Degradation of tricyclic and tetracyclic Ahs, which were the main components of 16 ΣPAHs, were as much as 78% and 79% in SS, respectively. SS showed the greatest biodegradability of PAHs compared to other soils especially soil DG, of which the PAHs degradation was only 65%. The degradation of tricyclic and tetracyclic AHs in DG were also least with 62% and 68%, respectively.
Figure 1.
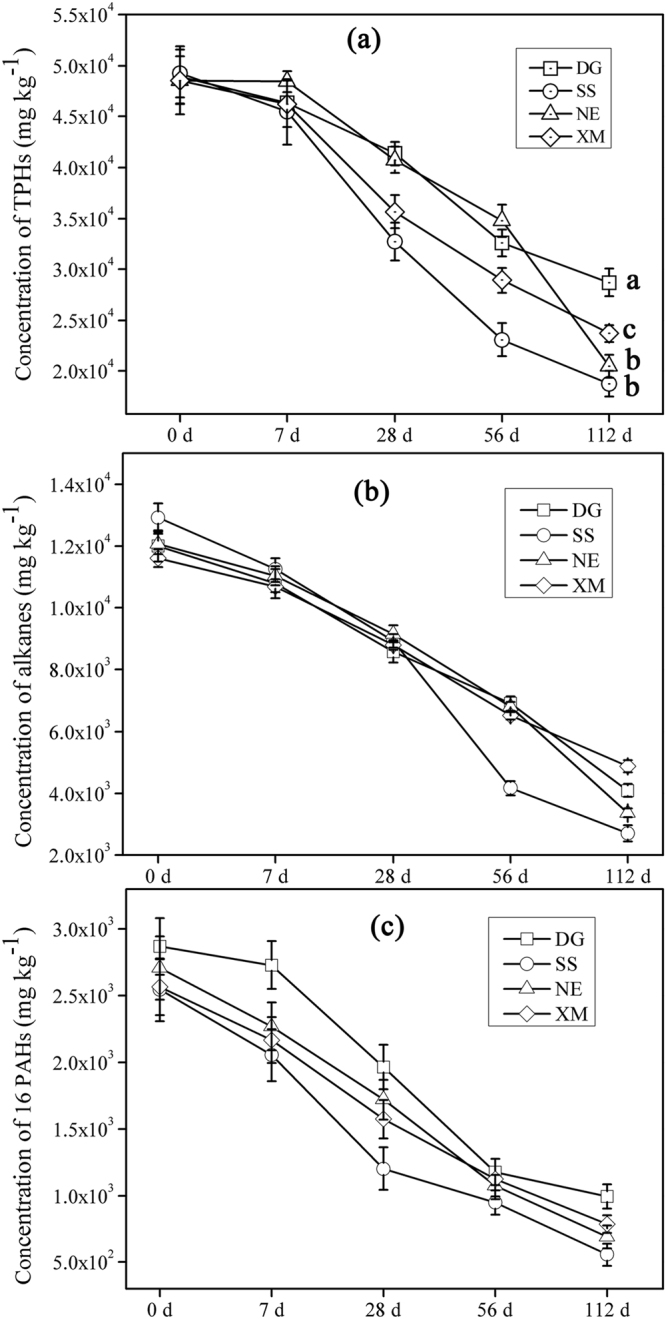
Concentrations of petroleum hydrocarbons (PHs) in soils from four geographic regions. (a) Concentrations of total petroleum hydrocarbons (TPHs) in microcosm soils; (b) Concentration of n-alkanes (C8-C40) in microcosm soils; (c) Concentration of 16 ΣPAHs in microcosm soils. Statistically significant differences between the sampling sites (p < 0.05) are indicated by the different letters in each column. Error bar represent standard deviation (n = 3).
Correlations between biodegradation of PHs and soil properties
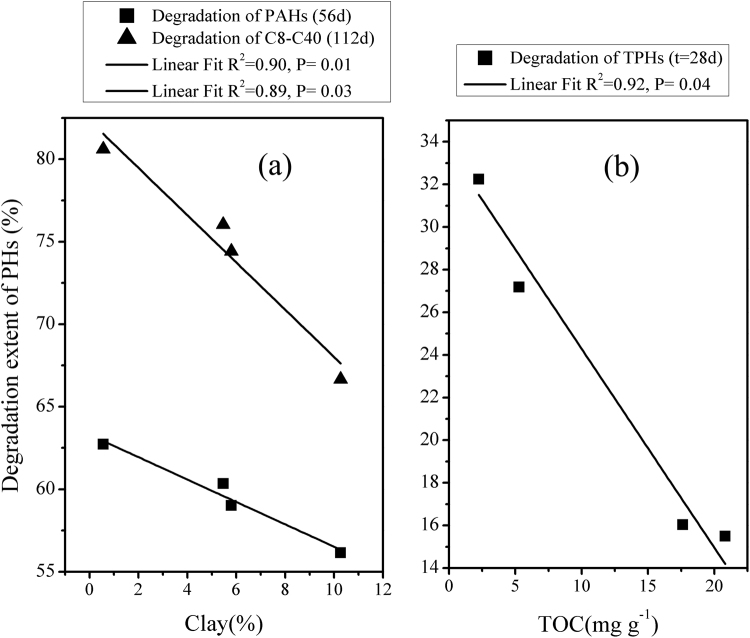
Biodegradation of PHs was significantly and negatively correlated with clay contents of soils. Degradation of total alkanes over 112 d was negatively correlated with clay content of the soils (R2 = 0.90, p = 0.01). Rates of degradation of PAHs over 56 d was also significantly and negatively correlated (R2 = 0.89, p = 0.03) (Fig. 2a) with clay content of soils. During the initial phase (t = 28 d) of the study, there was also a significant negative correlation (R2 = 0.92, p = 0.04) between TOC content and degradation of TPHs (Fig. 2b).
Figure 2.
Pairwise correlations between soil properties and degradation of PHs. (a) Pairwise correlations between clay content and PHs degradation; (b) Pairwise correlations between TOC contents and degradation of PHs.
High throughput sequencing
Based on partial 16 S rRNA sequences, numbers of bacteria in soils from four geographic regions ranged from 5777 sequences to 13854 sequences and contained 94–247 operational taxonomic units (OTUs) based on 97% nucleotide sequence identity (Supplementary Table S1). Results of a cluster analysis and relative abundance of taxonomic assignments for all samples at initial (t = 0 d) and last (t = 112 d) are shown (Supplementary Figure S2). Initially, Proteobacteria was the dominant phylum in DG and SS soils with relative abundances of 34.31% and 86.56%, respectively. However, the dominant microbial phylum in NE and XM were both Firmicutes of which the relative abundances were 36.90% and 78.67%, respectively. Over the duration of cultivation in the microcosms, the abundance of Proteobacteria increased in all the soils. Increases were from 34.31% to 73.56% in DG; from 86.56% to 97.65% in SS; from 21.48% to 74.62% in NE; from 12.65% to 59.53% in XM). In contrast, abundances of Actinobacteria, Bacteroidetes, and Firmicutes decreased in all soils.
Detailed changes in proportions of microbial genera detected by HTS are given in a heat map (Figure S3). In DG soil, the relative abundance of Marinobacter increased from 11.60% (t = 0 d) to 22.65% (t = 112d). In SS soil, the genus Pseudomonas was the primary alkane-degrading bacterium of which relative abundance decreased from 68.73% to 39.30%, while the proportion of Acinetobacter increased from 10.80% to 21.64% after the 112 d of cultivation. In NE soil, the abundance of Rhodanobacter increased from 1.26% to 36.24% to be the dominant genus. In XM soil, the relative abundances of dominant genera such as Lactococcus (belonging to Firmicutes) decreased from 39.78% to 16.96%. Sphingobium (α-Proteobacteria) and Burkholderia (β-Proteobacteria) were significant populations of microbes with the ability to degrade PHs of which the relative abundances were 14.03% and 13.51%, respectively.
Characterization of microbial community by DGGE fingerprints
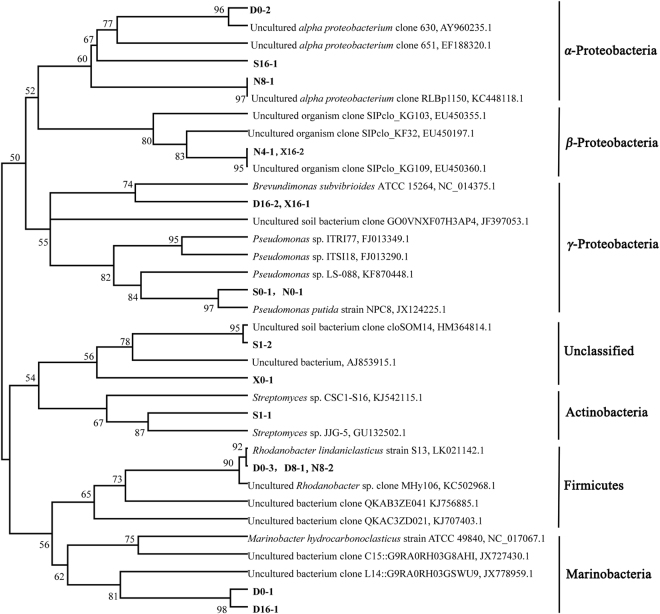
PCR-DGGE analysis was performed to determine shifts in patterns of relative abundances of bacterial populations in soils from different soil environments which had been amended with crude oil (Supplementary Figure S4). The sequences of V3 regions of 16S rRNA gene from gel were compared to the GenBank and homologous strains are shown (Supplementary Table S2). Partial sequences of 16S rRNA from genes 338 f to 518r were compared to the GenBank database by use of the Basic Local Alignment Search Tool (BLAST). Alignments were performed by Clustal X (version 1.81) with default values. A phylogenetic tree was constructed using the neighbor-joining method in the Mega 5.0 program (Fig. 3). A sequence having similarity less than 95% found in the database suggested a novel new genus of bacteria.
Figure 3.
Phylogenetic tree constructed based on 16S rRNA sequences using neighbor-joining methods. Sequences were referred from NCBI database, and the corresponding GenBank accession numbers were labeled after the name of the strains. Associated taxa were clustered in the bootstrap test (1000 replicates), and the bootstrap values were greater than 50%.
Sequencing bands in DGGE profiles and the phylogenetic tree demonstrate the shift in relative numbers of OTUs in microbial communities. In DG soils, the uncultured Rhodanobacter sp. sequenced from D0-3 band was the native bacterium, which gradually disappeared after treatment for 28 d, however, appeared again after 56 d (D8-1). Brevundimonas sp. (band D16-2) was the dominant species among microbial communities under this study. DNA sequences from bands D0-1 and D16-1 were identified as Marinobacter sp. During initial and final phases of incubation, Marinobacter sp. showed a great abundance of community population. Pseudomonas sp. sequenced from the S0-1 band from SS soil exhibited an intense band pattern on the DGGE gel throughout incubation. A strain of uncultured bacterium (X0-1) was initially the dominant autochthonal microorganism. However, culturable Brevundimonas sp. (X16-1) exhibited a greater abundance after 28 d. The Shannon-Weiner index was used to investigate changes in diversity of microbial communities among the four soils (Supplementary Table S3). The Shannon-Weiner index decreased after 7 days of incubation in all soils and then increased again after 28 days and decreased again at the late period of the process. While the Shannon-Weiner index values were different with the following order at initial: NE > DG > XM > SS, which was changed to the following order in the end: DG > SS > XM > NE.
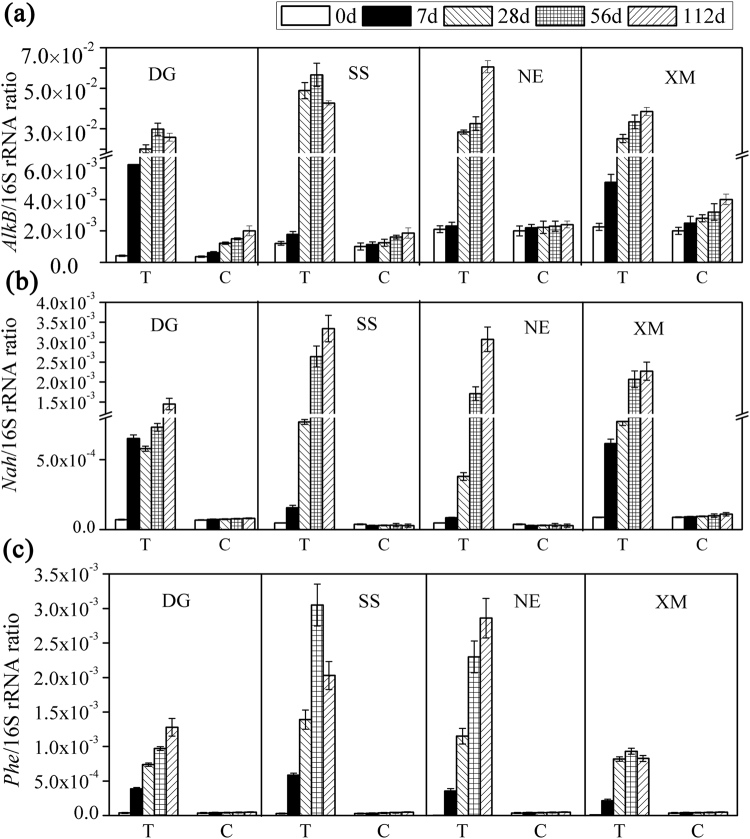
Abundances of typical oil catabolic genes
Relative abundances of metabolic genes normalized to total number of copies of 16S rRNA genes are shown (Fig. 4). Absolute copies of oil-degrading genes are listed in Supplementary Table S4. Relative abundances of genes capable of catabolism of PHs in soils changed significantly during incubation of soils in microcosms. Initially (t = 0 d), relative abundances of the AlkB gene in soils from the four geographic regions ranged from (4.22 ± 0.33) × 10−4 to (1.21 ± 0.12) × 10−3, while the AlkB gene accumulated during incubation. The maximum relative abundance of the AlkB gene in soil NE (6.06 ± 0.38) × 10−2 was observed on 112 d, followed by soil SS (5.67 ± 0.26) × 10−2 after 56 d. These values were 10 to 100-fold greater than those observed during initial stages of incubations of soils and consistent with greater degradation of alkanes in soils SS and NE.
Figure 4.
Relative abundances of oil-degrading genes in four soils spiked with crude oil. Abundances of genes were normalized to the total 16S rRNA genes. Note: C represents CK without addition of petroleum hydrocarbons; T represents test groups with addition of petroleum hydrocarbons.
Similarly, during incubation, the relative abundance of the Nah gene changed substantially in all four soils. An almost identical trend was observed for relative abundance of the Phe gene in soils NE and SS, which also increased 1000-fold from 10−6 to 10−3, whereas, the Phe gene increased from 10−5 to 10−3 in DG and XM. Results of statistical analysis (ANOVA) showed that relative abundances of degradation genes AlkB, Nah, and Phe were significantly different (p < 0.01) among the four soils and among sampling times, except for the AlkB gene in soils NE and XM after 56 d, and initial abundances of the Nah gene in soils SS and NE.
Correlations between mineralization of petroleum hydrocarbons and indices of microbial communities among soils
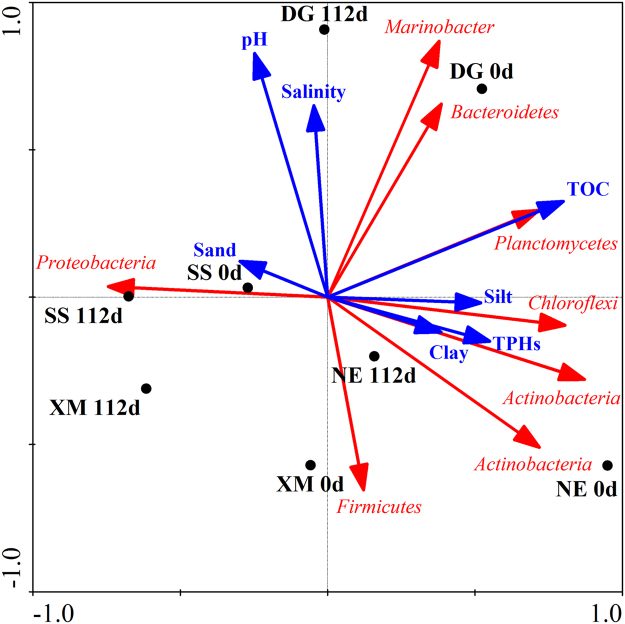
The results of which are presented here, significant positive correlations were observed between relative abundance of the AlkB gene and degradation of TPHs (R2 = 0.94, p < 0.05) as well as medium-short chain alkanes (C13-C30) (R2 = 0.85, p < 0.05) in all soils after 112 d (Supplementary Figure S5a). Positive correlation was also found between relative abundances of the Nah gene and mineralization of naphthalene (R2 = 0.88, p < 0.05) and total tricyclic PAHs (R2 = 0.81, p < 0.05) (Supplementary Figure S5b). Redundancy analysis (RDA) in Fig. 5 showed that soils DG, SS, NE and XM, which were collected from four geographic regions of China, varied greatly in physical properties and presence of microbial communities capable of degrading PHs.
Figure 5.
Redundancy analysis (RDA) biplot depicting the relationship between the bacterial communities and main physicochemical parameters of the four different geographic soil samples. Note: Solid circles represent the four different soil samples collected at initial and last experimental time. Environmental variables that significantly explained variability in composition of microbial communities were fitted to the ordination. Arrows indicate the direction and magnitude of environmental variables associated with the different bacterial phylum.
Discussion
Compared to the shorter microcosm cultivation used in previous studies, this study reached a stable stage of availabilities of alkanes and PAHs to microbes19,20. Physical properties and presence of microbial communities capable of degrading PHs varied among soils DG, SS, NE and XM, which were collected from four geographic regions of China, varied. Microbial flora in SS and NE soils exhibited greater potential to degrade TPHs as well as the PH components (16 ΣPAHs, alkanes C8-C40) than that in DG and XM soils. Bioavailability of petroleum hydrocarbons among soils, in part due to enrichments of various microbial communities which have been adapted to the specific environmental factors21,22.
Obvious changes in bacterial communities were observed, which demonstrated that there was no tendency for the bacterial community of the four different soil types to converge as a result of contamination with crude oil. The dominant genera of bacteria in DG, SS, NE, and XM were Marinobacter, Pseudomonas, Rhodanobacter, and Lactococcus, respectively after 112 d of cultivation. Dominant phylum of the XM with clay soil texture was Firmicutes instead of Proteobacteria in other sites (DG, SS, and NE) with silty or sandy soil texture. The dominant phylum in clay soils has been shown to be Actinobacteria, while that in sandy or sandy loam soil texture was Proteobacteria 23. A similar conclusion, based on divergence of microbial communities among six types of soil, for which textures ranged from clay to loam, and pH values ranged from 5.4 to 8.8 was reported24. DG soil, which had the greatest salinity, contained a great abundance of saline and alkaline-tolerant bacteria. Marinobacter has previously shown to be efficient at metabolism of alkanes25. Uncultured Rhodanobacter sp. was the native bacterium which gradually disappeared after treatment for 28 d, however, appeared again after 56 d. Previously, it has been shown that Rhodanobacter sp. was easily inhibited by PAHs, whereas it could degrade intermediate metabolites which enabled further degradation of hydrocarbons26. Brevundimonas sp., which was previously isolated and identified from bottom sludge in a crude oil tank and hydrocarbon-contaminated soil of Azzawiya oil refinery plant27 had a great accumulation in DG. Pseudomonas sp. was present in SS soil, which had the greatest capacity to degrade both PHs and alkanes. Previously, it had been reported that Pseudomonas putida GPo1 degraded alkanes by a monooxygenase enzyme encoded by the AlkB gene28. In NE soil, Rhodanobacter (γ-Proteobacteria) was the dominant genus which has been shown to have a large capacity to degrade various PAHs and play an important role in bioremediation of petroleum hydrocarbons26. Sphingobium (α-Proteobacteria) and Burkholderia (β-Proteobacteria) were significant populations of microbes with the ability to degrade PHs in XM soil. Previous studies have found that both Sphingobium and Burkholderia can mineralize the PAHs, especially for BTEX29,30. A new strain of uncultured bacterium was initially the dominant autochthonal microorganism in XM soil. Results of previous studies using clone libraries, hybridization probes and other molecular biological techniques have shown that new genotypes PHs-degrading genes are widely contained in native microorganisms in sea water, and marine sediments and soils of oilfield that are contaminated with PHs31,32.
Abundances of catabolic genes represent the degradation potential of microflora, and can be used as an indicator of the community’s response to bioavailable hydrocarbons. Therefore, differences in abundances of catabolic genes are assumed to result from divergent accumulation of bacterial populations within biogeographic sampling sites. These values of relative abundance of AlkB gene were 10 to 100-fold greater than those observed during initial stages of incubations of soils and consistent with greater rates of degradation of alkanes in soils SS and NE. Since each bacterial cell contained 3-4 copies of AlkB gene based on the previously described analytical approach33, the total population of alkane-degrading bacteria was calculated to be between 4.27% and 5.67% in SS, and between 2.85% and 6.06% in NE during the period from 56 d to 112 d. These values are similar to the proportion of bacteria reported to be capable of degrading alkanes reported by Xu et al.5 who determined the alkane-degrading bacteria to be between 4.7% and 7.4% in soils of the Shaozhuang oil and gas field soil, and Shengli Area, China, respectively. The proportion of alkane-degrading bacteria in soils from DG and XM were 2.98% and 3.86% in their respective maxima which were less than those in SS and NE. Absolute abundances of AlkB in four soils were similar to those in soils from the Daqing and Karamay Oil Fields, China, which had ranges of 106–107 and 105–107 copies/g, dm soil, respectively4, but greater than that in sediments from the Timor Sea Australia that had been contaminated with oil, where the values ranged from 1.1 × 105 to 2.9 × 105 copies/g, dm soil34. Aromatic hydrocarbon-degrading bacteria expressing the Nah gene in all soils showed same range of relative abundance as that of bacteria that are capable of hydroxylating PAHs quantified previously18. An almost identical trend was observed for relative abundance of the Phe gene in soils NE and SS. Phe encodes for the enzyme phenol monoxygenase, which can further mineralize intermediates in degradation of monoaromatic hydrocarbons35. Abundances of the Nah and Phe genes were used as indicators for estimating the PAH-degradation potential of aromatic oxygenase present in autochthonic bacterial communities36,37.
Bioremediation of soils contaminated with PHs depends on abundances of metabolic genes, which can encode for hydroxylase enzymes. In the study, except the higher degradation extent of PHs found in SS, due to the same order of relative abundance of three degradation genes, which had a range of 10-1000-fold increase, similar degradation was observed in other soils. While the microbial responses to oil contamination were different at the same site under the different oil exposure time, RDA analysis demonstrated that the soil properties were the dominant effect factors on the microbial communities and TPHs degradation. In detail, the pH and salinity were restrictive factors on the accumulation of degradation microbes in DG soil, and only the saline and alkaline-tolerant bacteria such like Marinobacter account for the great abundance. pH has been proved the important factor in determining the composition of microbial community especially in saline lake and tailing dam soils38,39. Particle size was the dominant determining factor of the microbial degradation potential in NE, XM, and SS soil. NE and XM, which contained greater fractions of clay and silt, exhibited lesser PHs degradation. Results of a previous study demonstrated that PHs absorbed on clay minerals by hydrogen bonding and electric dipole van der Waals forces were recalcitrant to degradation by soil microorganisms40. Soil SS, which contained a greater proportion (96.07%) of sand exhibited the maximum potential to degrade PHs. Soils containing greater contents of sand can provide the adequate oxygen to aerobic microbes which so that exhibited greater PHs degradation41. During the initial stage, TPHs were more likely to be degraded by indigenous microbes in soils with lesser concentrations of TOC. Meanwhile, TOC content was the key factor affecting the magnitude and relative compositions of microbial communities, especially Planctomycetes and Proteobacteria. Results of previous studies have demonstrated that some carbon sources derived from non-petroleum hydrocarbons (biogenic and anthropogenic carbonaceous organic compounds) are more readily metabolized by microorganisms which results in competitive metabolism with PHs42. Nevertheless, due to the accumulation of specialized PHs degrading microorganisms, as well as formation of co-metabolic pathways on PHs and non-petroleum hydrocarbon degradation by microbes43, the negative correlation between TOC content and degradation extent of PHs was not observed after initial stages of incubations of soils.
Relative abundances of metabolic genes are good biomarkers to evaluate potential of PHs to be degraded in soils. In addition, heterogeneity of physical-chemical properties of soils contaminated with PHs complicates the prediction of PHs mineralization. In silty soils (silt content: 75.41~78.4% wt), halophilic Marinobacter were the dominant bacteria capable of degrading PHs at greater pH (>9.0) and salinity (>17.65 g kg−1). However, indigenous bacteria capable of degrading PHs, such as Rhodanobacter prefer to accumulate in soils of lesser pH (<4.8) and salinity (>3.8 g kg−1). Pseudomonas played an important role in PHs metabolism in sea sand with higher pH (>8.7) and salinity (>36.78 g kg−1) and 96.07% wt of sand content, while Firmicutes had the greatest abundance at lower pH (<5.3) and salinity (<0.8 g kg−1) soil with 10.27% wt of clay content. Therefore, research on structures of microbial communities and abundances of genes is critical to predict favorable conditions for bioremediation of soils contaminated with crude oil.
Materials and Methods
Sampling and microcosm simulation
Soils which had not been contaminated with PHs (less than limits of detection) were collected from four geographic locations in China, Dagang Oilfield, Sand of Bohai Sea, Xiamen, and Northeast China (Fig. 6). The Dagang Oilfield is located in a coastal saline area, near Tianjin, China where soils contain greater salinity and alkalinity. The Bohai Bay Rim is one of the most important offshore bases for exploration and transportation of oil. Northeast China is famous for its fertile black soil, and is one of the three black earth terrains in the world. Red earth in Xiamen is typically acid and orthic ferralsols with unsaturated basicity. Soils were collected from a depth of 0–20 cm using sterile shovel. Samples of soil were air-dried prior to quantification of PHs and other physical and chemical properties. Samples of soil for analysis of diversity and abundance of microbial catabolic genes were thoroughly homogenized to obtain a uniform sample and stored at −20 °C.
Figure 6.
Locations from which soils were collected from four regions of China. DG: Dagang Oilfield; SS: Sand of Bohai Sea; NE: Northeast of China; XM: XiaMen. The figure map was generated by using software ArcGIS 10 (Environmental Systems Research Institute, Inc. Redlands, US). http://www.esri.com.
To study effects of types of soils and microbial communities on degradation of crude oil, soil was incubated in 850 mL volume polytetrafluoroethylene equipment (Inner diameter 8.5 cm; Height 15 cm) containing 500 g soil to which 5% (mass) crude oil (provided by No. 1 Oil Production Plant, PetroChina Dagang Oilfield Company, see Table 2) by use of previously described methods44. Original composition of the crude oil is shown (Table S5 of Supporting Information). Soils for the study of abiotic factors influence were sterilized by autoclaved sterilization. The blank, “control” was maintained without addition of any petroleum contaminants. The total volume of sterile, deionized water was determined and added to DG, SS, NE, and XM soils as per their water characteristic to achieve an equivalent matric potential of 40, 22, 34 and 35 kPa, respectively (32.2%, 39.6%, 30.5%, and 31.8% water content, respectively). Microcosms were established in triplicate and incubated for 112 days in dark condition at the room temperature. Soils were allowed to equilibrate after oil spiking, and then experiments were initiated. Approximately 10 g of soil were collected by use of sterilized shovel at the sampling time of 0, 7, 28, 56, and 112 d during microcosm process. Each treatment (soil type) was carried out in triplicate.
Table 2.
Original composition and element contents of the crude oil (provided by PetroChina Dagang Oilfield Company).
| Components of crude oil (%) | Saturate hydrocarbons | Aromatic hydrocarbons | Resin | Asphaltene | |
|---|---|---|---|---|---|
| 57.2 | 28.6 | 10.7 | 3.5 | ||
| Element contents (%) | C | H | S | N | O |
| 85.67 | 13.40 | 0.12 | 0.23 | — |
Determination of physical-chemical properties of soil
The pH was determined by pH meter (Sartorius, Germany) in a suspension of 1:5 (soil:deionized water). Salinity was determined gravimetrically45. In brief, a suspension of soil was heated in a water bath at 100 °C to dryness, and 10% H2O2 was added as oxidant subsequently for three times. Residual salt residual was then weighted and salinity of the soil calculated. Analyses of particle sizes of soils were determined via BT-9300S laser particle size analyzer (Dandong Bettersize Instruments Ltd. Liaoning province, China). Total organic carbon was measured through Multi N/C 3000 (Analytik Jena AG, Germany).
Determination of petroleum hydrocarbons
Concentrations of total petroleum hydrocarbons (TPHs) in soils were determined gravimetrically. A five-gram aliquant of moist soil (after deduction of moisture content) was Soxhlet-extracted for 12 h with 120 mL dichloromethane at 54 °C46. Extracts were concentrated to dryness using rotary evaporator, and masses of TPHs were measured by gravimetric method47. All extractions and quantifications were performed in triplicate.
PAHs and saturated hydrocarbons (SHs) were separated by silica gel-alumina column and quantified by use of a model Agilent 7890 gas chromatograph connected to a 5975 Agilent HP mass spectrometer (Agilent, CA, USA) as per previous study48. Mixtures of 16 target PAHs on the EPA priority pollutant list (see Supplementary Figure S6) and 33 target n-alkanes (C8-C40) were used as standards for external determination of components of extracts49. Extraction efficiencies of PAHs and SHs were ranged from 82% to 105%.
Molecular analysis
High throughput sequencing of partial 16S rRNA genes
Genomic DNA extraction was performed by a ZR microbe DNA MiniPrep™ kit (Zymo Research, CA, USA) according to the manufacturer’s protocol. Extracted DNA was checked by 1.5% agarose gel electrophoresis. To determinate changes in microbial communities during bioremediation of soils contaminated with PHs, from 0 d to 112 d, the V4-V5 fragment of the 16S rRNA gene was amplified with a set of primers 515f (GTG CCA GCM GCC GCG GTAA) and 907r (CCGTCAATTCMTTTRAGTTT) following a previously described protocol50. Amplicons were sequenced by using the Illumina HiSeq. 2000 platform (Majorbio co., Shanghai, China). Sequences were excluded from the analysis if the read length was less than 150 bp. Corrected sequences were then processed using the UPARSE pipeline51 within QIIME (Quantitative Insights Into Microbial Ecology), and operational taxonomic units (OTUs) were defined at 97% nucleotide similarity. Sequences for each OTU and the Ribosomal Database Project (RDP) classifier were used to assign taxonomic data50. Sequences of amplicons from Illumina HiSeq have been deposited at DDBJ (DNA Data Bank of Japan) database with accession numbers of SAMD00042819-SAMD00042826. The hierarchical clustering method using average linkage was used to analyze the relationships between samples. Specific differences in community composition of soil microbes were visualized by use of heat maps using the R package52. Chao1 and Shannon index were calculated to estimate the change of microbial diversity.
Community and phylogenic analysis using PCR-DGGE
PCR- DGGE analysis was carried out to assess the microbial community of soils by targeting the V3 region of the bacterial 16S rRNA gene. The primer set targeting the V3 region of bacterial 16 S rRNA gene consisted of GC-338f (5′-GCclamp-CACGGGGGGACTCCTACGGGAGGCAGCAG-3′) (GC clamp = CGCCCGCCGCGCGCGGCGGGCGGGGC GGGGGCACGGGGGG) and 518r (5′-ATTACCGCGGCTGCTGG-3′). The PCR amplification and DGGE were performed as per the conditions based on our previous studies48. After separation by DGGE, DNA of bands was extracted by using of Poly gel extraction kits (CWBiotech, Beijing, China). 16S rRNA gene fragments were amplified by PCR and then sequenced by ABI Prism 3730XL automated fluorescence sequencer (Applied Biosystems, Foster City, CA, USA). All sequences were compared to those in the GenBank database (http://www.ncbi.nlm.nih.gov). The nucleotide sequences reported in this paper have been deposited in the European Nucleotide Archive database under accession numbers LN649240 to LN649254 (16S rRNA gene DGGE band sequences).
Real-time qPCR test of degrading genes with SYBR green I
Three genes AlkB, Nah, and Phe involved in degradation of oil were quantified by real-time qPCR method. In order to minimize effects of environmental factors, numbers of copies of genes were normalized to total number of copies of 16S rRNA genes. Quantification of target genes by qPCR were carried out on a BioRad CFX96 (Hercules, USA) with a C1000 thermal cycler iCycler. Primers for quantification of target genes and related references were given in Supplementary Table S6. Each reaction was performed in a 25 μL volume containing 12.5 μL of qPCR super mix, 0.5 μL of passive reference dye, 0.2 µM of each primer, and 1 μL of DNA template. Cycling conditions were as follows: hold for 30 s at 94 °C followed by 40 cycles of denaturing at 94 °C for 5 s, annealing temperature of target gene primers were set as in Supplementary Table S6 for 15 s, extension at 72 °C for 10 s; hold at 55 °C for 30 s; melting curves were obtained at 55 to 95 °C at a 0.5% heating rate.
Data availability
Sequences data of amplicons from Illumina HiSeq have been deposited at DNA Data Bank of Japan database (http://www.ddbj.nig.ac.jp/) with accession numbers of SAMD00042819-SAMD00042826. The 16S rRNA gene DGGE band sequences reported in this paper have been deposited in the European Nucleotide Archive database (http://www.ebi.ac.uk/ena) under accession numbers LN649240 to LN649254. A supplement data of this article was represented in Supplemental Material. The Supplement related to this article is available online.
Statistical analyses
Statistical comparisons of metabolic gene abundances were conducted by SPSS 19.0 for Windows (IBM, Chicago, IL, USA) using nonparametric one-way ANOVA and Kruskal-Wallis Multiple Comparison Z test with Bonferroni adjustment. Redundancy analysis (RDA) were conducted by CANOCO 5.0 for Windows (Microcomputer Power Inc., Ithaca, NY).
Electronic supplementary material
Acknowledgements
This work was supported by (1) National Natural Science Foundation of China (41473070, 31270544), (2) The National Water Pollution Control and Treatment Science and Technology Major Project (2015ZX07203-011-06), (3) Natural Science Foundation of Tianjin (17JCQNJC07800). Prof. Giesy was supported by the program of 2012 “High Level Foreign Experts” (#GDT20143200016) funded by the State Administration of Foreign Experts Affairs, the P.R. China to Nanjing University and the Einstein Professor Program of the Chinese Academy of Sciences. He was also supported by the Canada Research Chair program.
Author Contributions
Q.L. and J.T. wrote the main manuscript text. R.G. prepared the Fig. 3. J.P.G. and J.T. prepared the Discussion and J.P.G. and J.T. edited the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14032-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McKew BA, et al. Efficacy of intervention strategies for bioremediation of crude oil in marine systems and effects on indigenous hydrocarbonoclastic bacteria. Environ Microbiol. 2007;9:1562–1571. doi: 10.1111/j.1462-2920.2007.01277.x. [DOI] [PubMed] [Google Scholar]
- 2.Liao JQ, Wang J, Jiang DL, Wang MC, Huang Y. Long-term oil contamination causes similar changes in microbial communities of two distinct soils. Appl Microbiol Biot. 2015;99:10299–10310. doi: 10.1007/s00253-015-6880-y. [DOI] [PubMed] [Google Scholar]
- 3.Jurelevicius D, Cotta SR, Peixoto R, Rosado AS, Seldin L. Distribution of alkane-degrading bacterial communities in soils from King George Island, Maritime Antarctic. Eur J Soil Biol. 2012;51:37–44. doi: 10.1016/j.ejsobi.2012.03.006. [DOI] [Google Scholar]
- 4.Yang Y, Wang J, Liao J, Xie S, Huang Y. Abundance and diversity of soil petroleum hydrocarbon-degrading microbial communities in oil exploring areas. Appl Microbiol Biot. 2014;99:1935–1946. doi: 10.1007/s00253-014-6074-z. [DOI] [PubMed] [Google Scholar]
- 5.Xu K, Tang Y, Ren C, Zhao K, Sun Y. Diversity and abundance of n-alkane-degrading bacteria in the near-surface soils of a Chinese onshore oil and gas field. Biogeosciences. 2013;10:2041–2048. doi: 10.5194/bg-10-2041-2013. [DOI] [Google Scholar]
- 6.Zhang J, Wang R, Du X, Li F, Dai J. Characterization of contamination, source and degradation of petroleum between upland and paddy fields based on geochemical characteristics and phospholipid fatty acids. J Environ Sci. 2012;24:1995–2003. doi: 10.1016/S1001-0742(11)60997-2. [DOI] [PubMed] [Google Scholar]
- 7.Lladó S, et al. Comparative assessment of bioremediation approaches to highly recalcitrant PAH degradation in a real industrial polluted soil. J Hazard Mater. 2013;248–249:407–414. doi: 10.1016/j.jhazmat.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Lee S-H, Oh B-I, Kim J-G. Effect of various amendments on heavy mineral oil bioremediation and soil microbial activity. Bioresource Technol. 2008;99:2578–2587. doi: 10.1016/j.biortech.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 9.Fang H, Cai L, Yu Y, Zhang T. Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge. Bioresource Technol. 2013;129:209–218. doi: 10.1016/j.biortech.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin BR, Nakatsu CH, Nies L. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microb. 2003;69:3350–3358. doi: 10.1128/AEM.69.6.3350-3358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurelevicius D, Alvarez VM, Peixoto R, Rosado AS, Seldin L. The use of a combination of alkB primers to better characterize the distribution of alkane-degrading bacteria. PLoS ONE. 2013;8:e66565. doi: 10.1371/journal.pone.0066565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki MT, Taylor LT, Delong EF. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microb. 2000;66:4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Y, et al. Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J. 2010;5:403–413. doi: 10.1038/ismej.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte LG, et al. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. Fems Microbiol Ecol. 2002;41:141–150. doi: 10.1111/j.1574-6941.2002.tb00975.x. [DOI] [PubMed] [Google Scholar]
- 15.Smits THM, Balada SB, Witholt B, van Beilen JB. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J Bacteriol. 2002;184:1733–1742. doi: 10.1128/JB.184.6.1733-1742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaghan AV, et al. Diversity of benzyl- and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures. Environ Sci Technol. 2010;44:7287–7294. doi: 10.1021/es1002023. [DOI] [PubMed] [Google Scholar]
- 17.Hamamure N, Fukui M, Ward DM, Inskeep WP. Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic, functional gene (alkB) and physiological analyses. Environ Sci Technol. 2008;42:7580–7586. doi: 10.1021/es800030f. [DOI] [PubMed] [Google Scholar]
- 18.Meynet P, et al. Effect of activated carbon amendment on bacterial community structure and functions in a PAH impacted urban soil. Environ Sci Technol. 2012;46:5057–5066. doi: 10.1021/es2043905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stallwood B, Shears J, Williams PA, Hughes KA. Low temperature bioremediation of oil-contaminated soil using biostimulation and bioaugmentation with a Pseudomonas sp. from maritime Antarctica. J Appl Microbiol. 2005;99:794–802. doi: 10.1111/j.1365-2672.2005.02678.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Tao S, Zhang N, Zhang DY, Li XQ. The effect of soil organic matter on fate of polycyclic aromatic hydrocarbons in soil: A microcosm study. Environ Pollut. 2010;158:1768–1774. doi: 10.1016/j.envpol.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Ji X, Ripp S, Layton A, Sayler G, Debruyn J. Assessing long term effects of bioremediation: soil bacterial communities 14 years after polycyclic aromatic hydrocarbon contamination and introduction of a genetically engineered microorganism. J Bioremed Biodeg. 2013;4:1–8. [Google Scholar]
- 22.Gadd GM. Metals, minerals and microbes: Geomicrobiology and bioremediation and bioremediation. Microbiology. 2010;156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- 23.Liao J, Wang J, Huang Y. Bacterial community features are shaped by geographic location, physicochemical properties, and oil contamination of soil in main oil fields of China. Microbial Ecol. 2015;70:380–389. doi: 10.1007/s00248-015-0572-0. [DOI] [PubMed] [Google Scholar]
- 24.Hamamura N, Olson SH, Ward DM, Inskeep WP. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl Environ Microb. 2006;72:6316–6324. doi: 10.1128/AEM.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abed RMM, Al-Sabahi J, Al-Maqrashi F, Al-Habsi A, Al-Hinai M. Characterization of hydrocarbon-degrading bacteria isolated from oil-contaminated sediments in the Sultanate of Oman and evaluation of bioaugmentation and biostimulation approaches in microcosm experiments. Int Biodeter Biodegr. 2014;89:58–66. doi: 10.1016/j.ibiod.2014.01.006. [DOI] [Google Scholar]
- 26.Bacosa HP, Suto K, Inoue C. Bacterial community dynamics during the preferential degradation of aromatic hydrocarbons by a microbial consortium. Int Biodeter Biodegr. 2012;74:109–115. doi: 10.1016/j.ibiod.2012.04.022. [DOI] [Google Scholar]
- 27.Mansur AA, et al. Assessing the hydrocarbon degrading potential of indigenous bacteria isolated from crude oil tank bottom sludge and hydrocarbon-contaminated soil of Azzawiya oil refinery, Libya. Environ Sci Pollut R. 2014;21:10725–10735. doi: 10.1007/s11356-014-3018-1. [DOI] [PubMed] [Google Scholar]
- 28.van Beilen JB, Wubbolts MG, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 29.Rentz JA, Alvarez PJJ, Schnoor JL. Benzo[a]pyrene degradation by Sphingomonas yanoikuyae JAR02. Environ Pollut. 2008;151:669–677. doi: 10.1016/j.envpol.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Witzig R, Junca H, Hans-Ju¨rgen Hecht, Pieper DH. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropyl benzene dioxygenases. Appl Environ Microb. 2006;72:3504–3514. doi: 10.1128/AEM.72.5.3504-3514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Wang L, Shao Z. Diversity and abundance of oil-degrading bacteria and alkane hydroxylase (alkB) genes in the subtropical seawater of Xiamen Island. Microbial Ecol. 2010;60:429–439. doi: 10.1007/s00248-010-9724-4. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn E, Bellicanta GS, Pellizari VH. New alk genes detected in Antarctic marine sediments. Environ Microbiol. 2009;11:669–673. doi: 10.1111/j.1462-2920.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 33.Li H, et al. Molecular detection, quantification and distribution of alkane-degrading bacteria in production water from low temperature oilfields. Int Biodeter Biodegr. 2013;76:49–57. doi: 10.1016/j.ibiod.2012.06.007. [DOI] [Google Scholar]
- 34.Wasmund K, Burns KA, Kurtboke DI, Bourne DG. Novel alkane hydroxylase gene (alkB) diversity in sediments associated with hydrocarbon seeps in the Timor Sea, Australia. Appl Environ Microb. 2009;75:7391–7398. doi: 10.1128/AEM.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arenghi FLG, Berlanda D, Galli E, Sello G, Barbieri P. Organization and regulation of meta cleavage pathway genes for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl Environ Microb. 2001;67:3304–3308. doi: 10.1128/AEM.67.7.3304-3308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nebe J, Baldwin BR, Kassab RL, Nies L, Nakatsu CH. Quantification of aromatic oxygenase genes to evaluate enhanced bioremediation by oxygen releasing materials at a gasoline-contaminated site. Environ Sci Technol. 2009;43:2029–2034. doi: 10.1021/es900146f. [DOI] [PubMed] [Google Scholar]
- 37.Baldwin BR, Nakatsu CH, Nies L. Enumeration of aromatic oxygenase genes to evaluate monitored natural attenuation at gasoline-contaminated sites. Water Res. 2008;42:723–731. doi: 10.1016/j.watres.2007.07.052. [DOI] [PubMed] [Google Scholar]
- 38.Mnif S, Sayadi S, Chamkha M. Biodegradative potential and characterization of a novel aromatic-degrading bacterium isolated from a geothermal oil field under saline and thermophilic conditions. Int Biodeter Biodegr. 2014;86(Part C):258–264. doi: 10.1016/j.ibiod.2013.09.015. [DOI] [Google Scholar]
- 39.Liang Y, Zhao H, Zhang X, Zhou J, Li G. Contrasting microbial functional genes in two distinct saline-alkali and slightly acidic oil-contaminated sites. Sci Total Environ. 2014;487:272–278. doi: 10.1016/j.scitotenv.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 40.Labud V, Garcia C, Hernandez T. Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere. 2007;66:1863–1871. doi: 10.1016/j.chemosphere.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Lisiecki P, et al. Biodegradation of diesel/biodiesel blends in saturated sand microcosms. Fuel. 2014;116:321–327. doi: 10.1016/j.fuel.2013.08.009. [DOI] [Google Scholar]
- 42.Tellechea FRF, Martins MA, da Silva AA, da Gama-Rodrigues EF, Martins MLL. Use of sugarcane filter cake and nitrogen, phosphorus and potassium fertilization in the process of bioremediation of soil contaminated with diesel. Environ Sci Pollut R. 2016;23:18027–18033. doi: 10.1007/s11356-016-6965-x. [DOI] [PubMed] [Google Scholar]
- 43.Sun W, et al. Microbial communities inhabiting oil-contaminated soils from two major oilfields in Northern China: Implications for active petroleum-degrading capacity. J Microbiol. 2015;53:371–378. doi: 10.1007/s12275-015-5023-6. [DOI] [PubMed] [Google Scholar]
- 44.Tao S, Xu F, Liu W, Cui Y, Coveney RM., Jr. A chemical extraction method for mimicking bioavailability of polycyclic aromatic hydrocarbons to wheat grown in soils containing various amounts of organic matter. Environ Sci Technol. 2006;40:2219–2224. doi: 10.1021/es051967b. [DOI] [PubMed] [Google Scholar]
- 45.Xie YW, et al. Ecogenomic responses of benthic communities under multiple stressors along the marine and adjacent riverine areas of northern Bohai Sea, China. Chemosphere. 2017;172:166–174. doi: 10.1016/j.chemosphere.2016.12.121. [DOI] [PubMed] [Google Scholar]
- 46.Luque de Castro MD, Priego-Capote F. Soxhlet extraction: Past and present panacea. J Chromatogr A. 2010;1217:2383–2389. doi: 10.1016/j.chroma.2009.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Rajakovic V, Aleksic G, Radetic M, Rajakovic L. Efficiency of oil removal from real wastewater with different sorbent materials. J Hazard Mater. 2007;143:494–499. doi: 10.1016/j.jhazmat.2006.09.060. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Tang J, Bai Z, Hecker M, Giesy JP. Distribution of petroleum degrading genes and factor analysis of petroleum contaminated soil from the Dagang Oilfield, China. Sci Rep. 2015;5:11068. doi: 10.1038/srep11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurav R, et al. Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environ Sci Pollut R. 2017;24:11392–11403. doi: 10.1007/s11356-017-8446-2. [DOI] [PubMed] [Google Scholar]
- 50.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 52.Sun J, Zhang Q, Zhou J, Wei Q. Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS ONE. 2014;9:e111744. doi: 10.1371/journal.pone.0111744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences data of amplicons from Illumina HiSeq have been deposited at DNA Data Bank of Japan database (http://www.ddbj.nig.ac.jp/) with accession numbers of SAMD00042819-SAMD00042826. The 16S rRNA gene DGGE band sequences reported in this paper have been deposited in the European Nucleotide Archive database (http://www.ebi.ac.uk/ena) under accession numbers LN649240 to LN649254. A supplement data of this article was represented in Supplemental Material. The Supplement related to this article is available online.