Abstract
The protein encoded by the TERMINAL FLOWER1 (TFL1) gene maintains indeterminacy in inflorescence meristem to repress flowering, and has undergone multiple duplications. However, basal angiosperms have one copy of a TFL1-like gene, which clusters with eudicot TFL1/CEN paralogs. Functional conservation has been reported in the paralogs CENTRORADIALIS (CEN) in eudicots, and ROOTS CURL IN NPA (RCNs) genes in monocots. In this study, long-term functional conservation and selective constraints were found between angiosperms, while the relaxation of selective constraints led to subfunctionalisation between paralogs. Long intron lengths of magnoliid TFL1-like gene contain more conserved motifs that potentially regulate TFL1/CEN/RCNs expression. These might be relevant to the functional flexibility of the non-duplicate TFL1-like gene in the basal angiosperms in comparison with the short, lower frequency intron lengths in eudicot and monocot TFL1/CEN/RCNs paralogs. The functionally conserved duplicates of eudicots and monocots evolved according to the duplication-degeneration-complementation model, avoiding redundancy by relaxation of selective constraints on exon 1 and exon 4. These data suggest that strong purifying selection has maintained the relevant functions of TFL1/CEN/RCNs paralogs on flowering regulation throughout the evolution of angiosperms, and the shorter introns with radical amino acid changes are important for the retention of paralogous duplicates.
Introduction
TERMINAL FLOWER1 (TFL1) is a member of the phosphatidylethanolamine-binding protein (PEBP) family. It represses flowering by counteracting the action of another PEBP protein, FLOWERING LOCUS T (FT), which promotes flowering1. The function of indeterminacy on shoot meristem of Antirrhinum majus suggests that the CENTRORADIALIS (CEN) gene is conserved and that its product is functionally identical to that of TFL1 2,3. TFL1 and CEN are paralogous genes with conserved functions that involve the formation of inflorescences4,5 and the maintenance of indeterminacy in inflorescent meristems6. Most eudicot species possess low or one copy of TFL/CEN in their genomes7. The monocot TFL1/CEN-like paralogous genes, named ROOTS CURL IN NPA (RCN1 and RCN2), also share the same function and are expressed in a similar pattern in rice, whereas another duplicated gene, RCN3, may be a non-functional chimera8. The TFL1-like gene was also found in the transition in inflorescent indeterminacy/determinacy in Phaseolus vulgaris 9. The natural variation of TFL1-like gene may also be related to evolutionary transition of inflorescence architecture10,11. It has been suggested that gymnosperms lack orthologues of FT and TFL1/CEN 7. From functional analysis of the homologous FT/TFL1-like gene in gymnosperm, the repressive function of TFL1/CEN/RCNs in flowering is known to be plesiomorphic12. The angiosperm TFL1/CEN/RCNs paralogs are the result of multiple gene duplications: (1) after the divergence between basal angiosperms (TFL1-like) and eudicots + monocots (TFL1/CEN/RCNs paralogs), (2) two-time duplications resulting in RCN1–3 in monocots, and (3) gene duplication causing the divergence of TFL1 and CEN in eudicots (phylogeny of angiosperms refers to Amborella Genome Project13, Fig. 1). The conserved function of TFL1/CEN/RCNs prevents redundancy or silencing by functional divergence14,15, which occurs by positive selection or through the relaxation of environmental constraints15.
Figure 1.
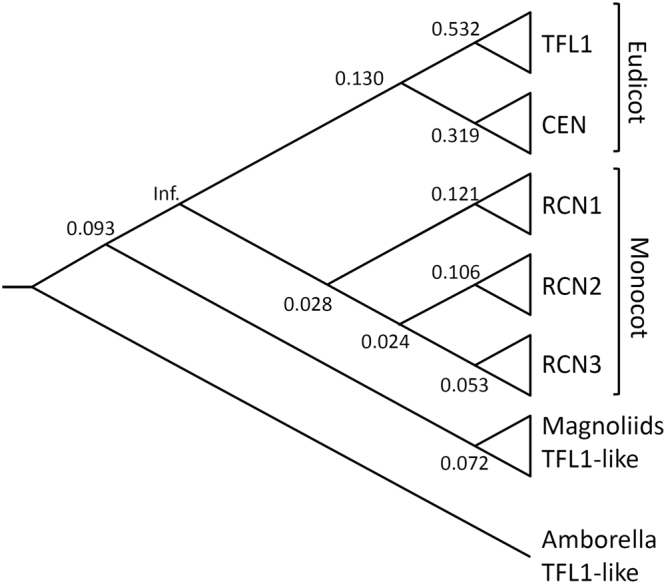
The hypothetical phylogenetic relationships of angiosperm TFL1/CEN/RCN paralogs. Values on the nodes indicate the ω of specific branches estimated under the free-ratio model, which suggest a pervasive purifying selection or selective constraints on the evolution of angiosperm TFL1/CEN/RCNs paralogs.
Different expression patterns of duplicated TFL1/CEN/RCNs genes in Arabidopsis 16, apple17, tomato18,19, and tobacco20 tissues have been reported. Such differential expression was suggested as complementary functions (subfunctionalisation)21,22. Following functional divergence, genes normally experience a phase free from selective constraints23. Because of the conserved properties of TFL1/CEN/RCNs paralogs, poor resolution of nucleotide phylogeny24 cannot explain their divergence. Nevertheless, a single reciprocally switched amino acid could cause functional interconversion between FT (flowering activator) and TFL (flowering repressor)25,26. Therefore, a few changes in the amino acid sequence can alter protein function to escape the redundancy of duplicates27. Therefore, determining radical amino acid changes between TFL1/CEN/RCNs paralogs (the type-II functional divergence of Gu28,29) could be useful for predicting their functional divergence after duplication.
The functional conservation and divergence of paralogous genes is not only reflected in coding sequences, but also in exon-intron structure. Structural divergence is prevalent in duplicated genes and leads to functionally divergent paralogs30. Variable intron lengths could be relevant to functional compensation in coexisting paralogs30 and provide heterogeneous regulatory functions in duplicate31–33. Highly expressed genes have longer introns than genes expressed at low levels33. Exon length was also suggested to be associated with molecular functions in flowering development cf.34. The Amborella trichopoda genome (http://www.amborella.org/)13 enables the comparison of gene structure and sequence variation in TFL1-like gene between basal angiosperms, and monocot and eudicot angiosperms. Comparisons of gene structure and intron lengths may enhance our understanding of evolution and its relevance among paralogs.
Genetic diversity among TFL1/CEN homologs played a key role in the diversification of flowering plants7,23, which was probably driven by heterogeneous selective pressures on different gene regions. For example, strong selective sweeps in coding regions, and balancing selection of promoters were detected in Arabidopsis 35. Furthermore, epistatic selection was identified through a QTL closely linked to the Arabidopsis TFL1 36. In addition, latitudinal gradients adaptation was also inferred by nonsynonymous polymorphisms of TFL1 37. However, there have been limited studies focussed on the effects of selective pressures on TFL1/CEN/RCNs paralog duplication, as well as the TFL1-like gene in basal angiosperms. These functionally conserved paralogous gene duplicates may be subject to strong purifying selection pressures that constrain redundant functions, such as the floral-regulatory paralogs SEPALLATA 1 (SEP1) and SEPALLATA 2 (SEP2), and SHATTERPROOF 1 (SHP1) and SHATTERPROOF 2 (SHP2)38. Selective constraints may be important in functionally redundant paralogous genes for buffering an organism’s phenotype against deleterious mutations39.
In this paper, a broad range of representative organisms from basal angiosperms, eudicots, and monocots were sampled to determine whether flowering plants exhibit divergent functions of TFL1/CEN/RCNs duplicates and how the selective pressure drove their evolution. General patterns of structural divergence in duplicated genes were analysed to represent the divergence/conservation of these paralogous genes. The aims of this research were to investigate (1) the evolution of intron variability in angiosperm TFL1/CEN/RCNs genes; (2) the effect of positive selection on angiosperm TFL1/CEN/RCNs coding sequences; and (3) the functional divergence between paralogs of angiosperm TFL1/CEN/RCNs, and thus infer the ancestral/derived type of TFL1/CEN/RCNs paralogs.
Results
Sequence length variation
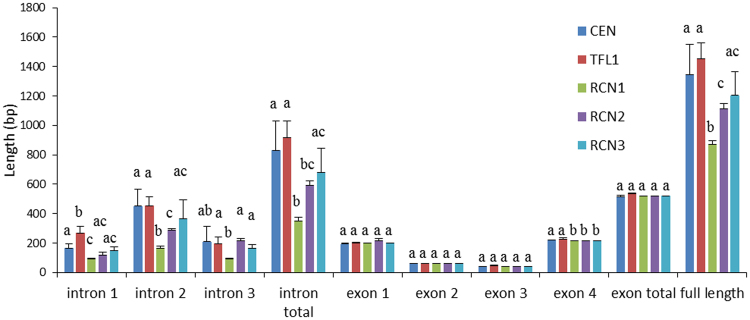
All sequences were confirmed as TFL1-like by the presence of histidine at the 92nd amino acid position (corresponding position at the 88th site of Arabidopsis)25. Only one copy for each Magnoliid species was obtained after amplification, and this result is consistent with only one TFL1/CEN/RCNs member in EST-library of basal angiosperm database (accession number: gnl|Liriodendron|b4_c119764, Ancestral Angiosperm Genome Project, http://ancangio.uga.edu/index.php). The sequences amplified from Magnoliid shown the best hit to TFL1/CEN/RCNs family (Nelumbo nucifera CEN-like protein 2, E-value: 5e−99–2e−92). Exon lengths of eudicot TFL1 and CEN, monocot RCN1, RCN2, and RCN3, and basal angiosperm (magnoliids + Amborella) TFL1-like gene range 519–609 bps, 447–531 bps, 522 bps, 522 bps, 522 bps and 516–522 bps, respectively. The length of introns from TFL1, CEN, RCN1, RCN2, RCN3, and basal angiosperm TFL1-like genes are 496–2048 bps, 320–3273 bps, 312–384 bps, 509–1007 bps, 510–643 bps, and 1444–3539 bps, respectively. Exon lengths were found to be constant and there were no significant differences between paralogs, with the exception of exon 4 between eudicots and monocots (Fig. 2). In contrast, the intron lengths were highly variable, and the monocot RCN1 was found to have relatively short but constant intron lengths compared with other paralogs. Furthermore, monocot RCNs had a higher number of intron length polymorphisms than eudicot TFL1/CEN (Fig. 2). Although only two TFL1-like full sequences were obtained from magnoliids, the synapomorphy of the long intron lengths of TFL1-like genes in Lauraceae and Magnoliaceae were confirmed by PCR (Additional file 1: Fig. S1). Analysis revealed that the monocot Sorghum bicolor has lost intron 1, and that exon 2 has merged with exon 1, and this sequence is removed when estimating the exon/intron length variation. The exon/intron structures and lengths are shown in Fig. 3 and Additional file 1: Table S2.
Figure 2.
Length polymorphisms of eudicot and monocot TFL1/CEN/RCN paralogs. Error bars represent one standard error. Different colors represent different TFL1/CEN/RCN lineages. Introns have greater length variation than exons, and the introns of monocot RCN1 are significantly shorter than other paralogs. Levels not connected by the same letter are significantly different based on Student’s t test.
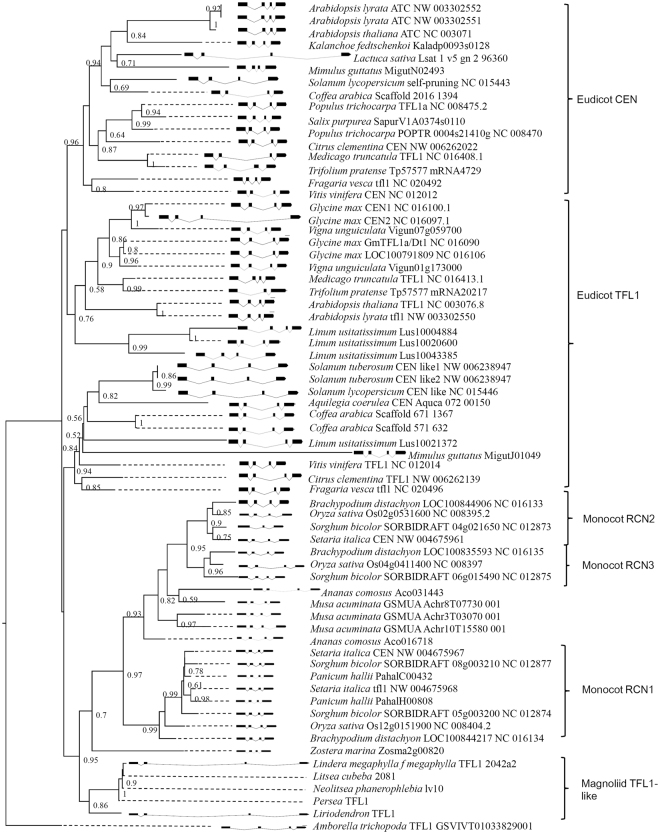
Figure 3.
Maximum likelihood tree and the exon-intron structure of angiosperm TFL1/CEN/RCN paralogs. Values of the nodes are bootstrapping supports for grouping. The bold boxes indicate the exon while the curves indicate the intron.
Correlation between intron lengths and conserved motifs
Twelve conserved motifs, which are identical to the motifs of putative cis-acting elements, were identified in noncoding regions (Additional file 1: Table S3), and the four-base motifs CAAT box and WRKY were abundant and both presence frequencies (0.0054 and 0.0064, respectively) are higher than those predicted by random occurrence (>1/256, p = 0.0245 and 0.0001, respectively) (Additional file 1: Table S3). The TFL1-like gene from magnoliids was found to have longer introns and more abundant cis-element to motifs. A strong and significant positive correlation between the number of cis-element motifs and intron length were found (R 2 = 0.711, p < 0.0001, Fig. 4), suggesting that noncoding regions in TFL1/CEN/RCNs paralogs are relevant to intron length.
Figure 4.
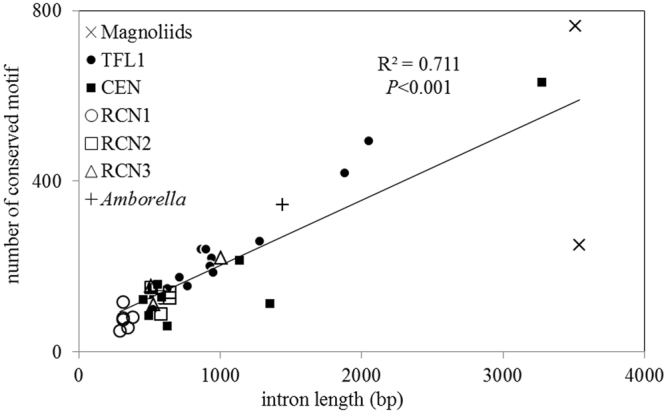
Significant positive correlation between the number of cis-acting elements and intron length. The correlation coefficient (R2) and significance of the correlation coefficient (P) were calculated. Intron length variations are listed in Additional file 1: Table S2. Types and locations of the putative cis-acting elements are listed in Additional file 1: Table S3.
Phylogenetic tree of angiosperm TFL1/CEN/RCNs paralogs
The phylogenetic tree of angiosperm TFL1/CEN/RCNs paralogs was reconstructed using amino acid sequences (Fig. 3) and was inconsistent with the hypothetical tree (Fig. 1 and Additional file 1: Fig. S2). The magnoliid TFL1-like gene was misgrouped with monocot RCNs (Fig. 3). The misgrouping for monocot and magnoliid paralogs was also revealed in the bush-like tree topology for basal lineages by Bayesian inference (Additional file 1: Fig. S3). The misgrouping of Magnoliid with monocot or eudicot is common in phylogenetic analysis using certain genes, which is probably due to combination of the relatively old age of these taxa and long branches attraction40–42. In contrast to the unresolved topology of basal lineages of eudicot and magnoliid paralogs, the monocot RCN paralogs were well grouped, with relatively high bootstrap supports in the ML and Bayesian trees (Fig. 3 and Additional file 1: Fig. S3). Furthermore, RCN2 and RCN3 are grouped together in both ML and Bayesian analyses (Fig. 3), implying that the duplication sequence in monocots is RCN1 and RCN2/3 followed by RCN2 and RCN3.
Positive selection analyses
To examine the effect of selective pressures on the angiosperm TFL1/CEN/RCNs paralogs, the ratio (ω) of missense (Ka) to silent mutation rates (Ks), an indicator of natural selection, was estimated. Likelihood ratio analysis revealed that the free ratio model was a better fit than the constant ratio model (2∆L = 97.8117, df = 61, P = 0.0004), suggesting a strong and pervasive purifying selection on angiosperm TFL1/CEN/RCNs paralogs (Fig. 1). To examine the grouping of magnoliid TFL1-like gene with eudicot TFL1/CEN and monocot RCNs (Table 1), the two and three ratio models were performed, and showed relaxation of selective constraints (ω 0 < ω 1, ω 2 < 1) for eudicot TFL1, CEN, monocot RCN2 and RCN3, and magnoliid TFL1-like gene, but strong purifying selection (ω 0 > ω 1) for the monocot RCN1 (Table 2). The two ratio model was a better fit for the evolution of monocot, eudicot and magnoliids TFL1/TFL1-like paralogs than three ratio models. This suggests that the grouping of eudicot and magnoliid TFL1/TFL1-like paralogs is a consequence of functional constraint and that both paralogs suffered different selective pressures for magnoliids TFL1/TFL1-like paralogs (Table 2).
Table 1.
Hypotheses and the corresponding scenarios for the grouping of eudicot TFL1 and magnoliid TFL1-like genes
| Hypotheses | Scenarios |
|---|---|
| 1.ω 1 = ω 2 ≤ 1 | Functional constraint hypothesis |
| 2.ω 1 = ω 2 > 1 | Synchronous selection hypothesis |
| 3.ω 1 ≠ ω 2 | Phylogenetic convergence driven by different selective pressures |
| 3.1.ω 1 > 1, ω 2 > 1 | Different strengths of positive selection on eudicot TFL1 and magnoliid TFL1-like |
| 3.2.ω 1 > 1, ω 2 ≤ 1 | Positive selection drives the convergence of eudicot TFL1 into magnoliid TFL1-like |
| 3.3.ω 2 > 1, ω 1 ≤ 1 | Positive selection drives the convergence of magnoliid TFL1-like into eudicot TFL1 |
| 3.4.ω 0 < ω 1 ≤ 1 | Relaxation of selective constraints for eudicot TFL1 |
| 3.5.ω 0 < ω 2 ≤ 1 | Relaxation of selective constraints for magnoliid TFL1-like |
| 3.6.ω 1 ≤ ω 0 ≤ 1 | Purifying selection on eudicot TFL1 |
| 3.7.ω 2 ≤ ω 0 ≤ 1 | Purifying selection on magnoliid TFL1-like |
ω 1, ω 2, and ω 0 are the Ka/Ks ratio of the branches of eudicot TFL1, magnoliid TFL1-like, and the other lineages (backgrounds), respectively. The ω 1 was also set for the eudicot CEN and monocot RCN1~3 for testing the same hypotheses.
Table 2.
Summary of the ω estimation and likelihood ratio test (2ΔL) between two-ratio (ω 0 ≠ ω 1 = ω 2) and three-ratio (ω 0 ≠ ω 1 ≠ ω 2) models.
| Hypothesis | np | lnL | 2ΔL | p | ω | Supporting hypothesis in Table 1 |
|---|---|---|---|---|---|---|
| TFL1 vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12594.3259 | ω 0 = 0.1044, ω 1 = ω 2 = 0.13221 | |||
| ω 1 ≠ ω 2 | 136 | −12593.9631 | 0.7256 | 0.3258 | ω 0 = 0.1021, ω 1 = 0.1361, ω 2 = 0.1678 | |
| CEN vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12597.2165 | ω 0 = 0.1173, ω 1 = ω 2 = 0.1072 | |||
| ω 1 ≠ ω 2 | 136 | −12597.2164 | 0.0002 | 0.9887 | ω 0 = 0.1054, ω 1 = 0.1193, ω 2 = 0.1674 | |
| RCN1 vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12596.0957 | ω 0 = 0.1170, ω 1 = ω 2 = 0.0891 | |||
| ω 1 ≠ ω 2 | 136 | −12595.6616 | 0.8682 | 0.2774 | ω 0 = 0.1140, ω 1 = 0.0841, ω 2 = 0.1665 | |
| RCN2 vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12597.6561 | ω 0 = 0.1137, ω 1 = ω 2 = 0.1164 | |||
| ω 1 ≠ ω 2 | 136 | −12597.5681 | 0.176 | 0.8708 | ω 0 = 0.1137, ω 1 = 0.1078, ω 2 = 0.1240 | |
| RCN3 vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12597.2484 | ω 0 = 0.1126, ω 1 = ω 2 = 0.1314 | |||
| ω 1 ≠ ω 2 | 136 | −12596.6628 | 1.1712 | 0.2052 | ω 0 = 0.1126, ω 1 = 0.1079, ω 2 = 0.1535 | |
| Eudicots vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 135 | −12592.2355 | ω 0 = 0.0915, ω 1 = ω 2 = 0.1255 | |||
| ω 1 ≠ ω 2 | 136 | −12592.0526 | 0.3658 | 0.5494 | ω 0 = 0.0916, ω 1 = 0.1085, ω 2 = 0.1264 | |
| Monocots vs. magnoliids | 1. Functional constraint hypothesis | |||||
| ω 1 = ω 2 | 65 | −12592.9536 | ω 0 = 0.1249, ω 1 = ω 2 = 0.0931 | |||
| ω 1 ≠ ω 2 | 66 | −12592.7766 | 0.354 | 0.5617 | ω 0 = 0.1249, ω 1 = 0.1073, ω 2 = 0.0917 | |
ω 1, ω 2, and ω 0 are the Ka/Ks ratio of the branches of the eudicot TFL1 (or eudicot CEN, monocot RCNs), magnoliid TFL1-like, and background lineages, respectively.
np: number of parameters
p: p-value obtained from fitted model using χ 2 test.
Evolutionary divergence between angiosperm TFL1/CEN/RCNs paralogs
The pairwise Ka/Ks ratio was calculated and plotted against Ks to reveal patterns of selection through time. No pairwise Ka/Ks > 1 were obtained suggesting that no positive divergent selection occurred between paralogs. Eudicot TFL1 and CEN were mostly distributed in the quadrant Ka/Ks < 1 and Ks > 1, indicating long-term purifying selection. The monocot RCNs and magnoliids TFL1-like genes were distributed in the quadrant Ka/Ks < 1 and Ks < 1. We divided this quadrant into two classes: (1) Ka/Ks > 0.097 (average Ka/Ks), suggesting the relaxation of selective constraints. This quadrant comprises the magnoliid TFL1-like and the monocot RCN2 and RCN3; and (2) Ka/Ks < 0.097, suggesting strong selective constraints or recent purifying selection, which comprised the monocot RCN1 (Fig. 5). This inference is consistent with the results of tests for selection hypotheses (Table 2).
Figure 5.
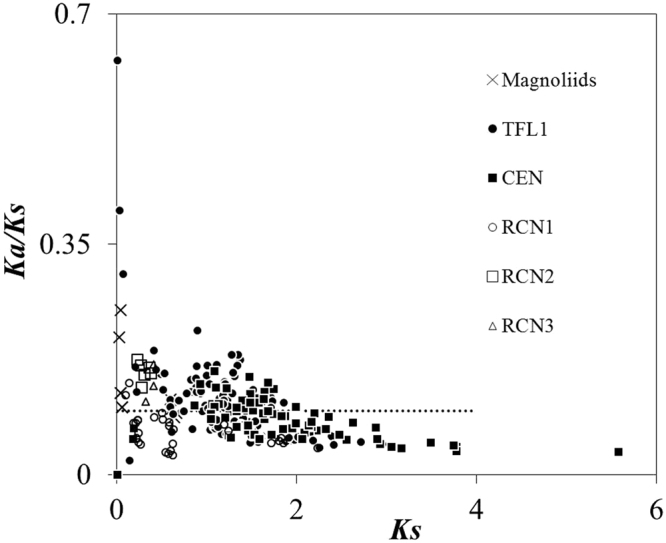
The Ka/Ks ratios against Ks values of pair comparisons of TFL1/CEN/RCN paralogous sequences within clades. The full and open symbols indicate the eudicot and monocot paralogous clades, respectively. The horizontal dotted line indicates the average Ka/Ks ratio (=0.0791) of all angiosperm TFL1/CEN/RCN paralogous sequences.
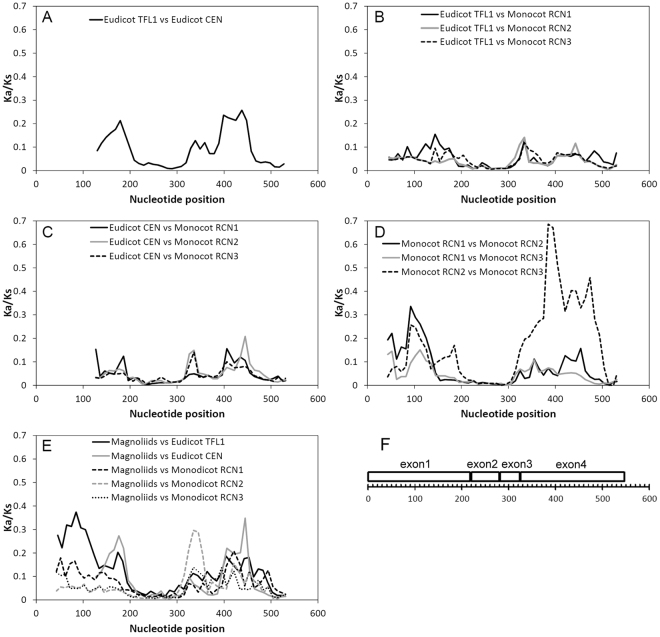
The sliding window analysis showed pairwise Ka/Ks < 1 in all regions among paralogs, with the greatest evolutionary divergence in exon 1 and exon 4 (Fig. 6). Relatively conserved regions in exon 2 and exon 3 indicate that these regions were subject to strong selective constraints. Relatively high Ka/Ks at exon 4 between the recently divergent monocot RCN2 and RCN3, indicate that this was subject to low selective pressures of constraining amino acid changes between RCN2 and RCN3 (Fig. 6D). Small Ka/Ks ratios between eudicot and monocot paralogs indicate functional conservatism divergence (Fig. 6B and C). Magnoliid TFL1-like gene was found to have a highly divergent exon 1 and exon 4 compared with the other paralogs (about 100th bp in TFL1, 180th and 450th bp in CEN, 340th bp in RCN2). This suggests that this gene was subject to different selective pressures than the eudicot and monocot paralogs, while the conservation of exon 2 and exon 3 suggests long-term and pervasive functional constraints on these genetic regions (Fig. 6E).
Figure 6.
Ka/Ks sliding windows of 50 nucleotides with a 10-bp step size between angiosperm TFL1/CEN/RCN paralogs. Comparisons (A) between eudicot TFL1/CEN paralogs, (B) between eudicot TFL1 and monocot RCNs, (C) between eudicot CEN and monocot RCNs, (D) between monocot RCN paralogs, and (E) between magnoliid TFL1-like, and eudicot and monocot TFL1/CEN paralogs. (F) The corresponding alignment positions of exons, revealing selective constraints on exon 2 and exon 3. The midposition of windows were listed in base pair (bp).
Radical functional divergence between angiosperm TFL1/CEN/RCNs paralogs
An in silico analysis of radical amino acid changes between duplicated genes was conducted for testing the functional divergence of TFL1/CEN/RCNs paralogs. Nonsignificant radical functional divergence, as estimated by the type-II functional divergence index (θII)43, was found between angiosperm TFL1/CEN/RCNs paralogs (Table 3). The proportion of fixed radical change between paralogs (F00,R) was zero between eudicot TFL1/CEN and paralogs of monocots and magnoliids, but more or less between paralogs between monocot RCNs and magnoliid TFL1-like genes (Table 3). This indicates that there is functional conservation of paralogs between eudicots and monocots, and functional specialization between paralogs within the eudicot and monocot species.
Table 3.
Summary of type-II functional divergence analysis for angiosperm TFL1/CEN/RCNs/TFL1-like paralogs.
| θII ± SE | p-value | aR/πR | GR | GC | F00,N | F00,R | F00,C | |
|---|---|---|---|---|---|---|---|---|
| TFL1/CEN | −0.181 ± 0.089 | 0.156 | 0.683 | 0.548 | 0.452 | 0.306 | 0 | 0 |
| TFL1/RCN1 | 0.060 ± 0.059 | 0.212 | −0.544 | 0.655 | 0.345 | 0.376 | 0 | 0 |
| TFL1/RCN2&3 | −0.132 ± 0.084 | 0.235 | −0.227 | 0.591 | 0.409 | 0.316 | 0.003 | 0 |
| TFL1/magnoliids | — | 1 | — | 1 | 0 | 0 | 0 | 0 |
| CEN/RCN1 | −0.111 ± 0.068 | 0.245 | −0.738 | 0.608 | 0.392 | 0.503 | 0.01 | 0.008 |
| CEN/RCN2&3 | −0.142 ± 0.074 | 0.253 | −0.072 | 0.570 | 0.430 | 0.376 | 0 | 0 |
| CEN/magnoliids | — | 1 | — | 1 | 0 | 0 | 0 | 0 |
| RCN1/RCN2&3 | −0.058 ± 0.053 | 0.177 | −0.184 | 0.457 | 0.543 | 0.503 | 0.010 | 0.006 |
| RCN1/magnoliids | — | 1 | — | 1 | 0 | 0 | 0 | 0 |
| RCN2&3/magnoliids | — | 1 | - | 1 | 0 | 0 | 0 | 0 |
θII, coefficient of type-II functional divergence (SE: standard error); p-value, significance test based on Z-score test to test the hypothesis of deviation of θII from zero; aR/πR: the ratio of radical change under functional divergence versus nonfunctional divergence; GR and GC, proportion of radical change and conserved change, respectively; F00,N, F00,R, and F00,C, proportion of none change, radical change, and conserved change of amino acids between clusters but no change within clusters, respectively.
Discussion
Exon length conservation and intron length variability
Exon length conservation implies constraints of gene functions among organisms34,44. Eudicot and monocot TFL1/CEN/RCNs are functionally conserved and the inflorescence architecture was determined by comparison with the model organisms Arabidopsis and rice45. Highly variable intron lengths and sequences of angiosperm CEN/RCNs/TFL1-like genes suggest absence of constraining reproductive function from noncoding regions. It is not known whether intron fragments have been gained or lost through evolution, due to poor or failed alignment in introns. However, we suspect that there was a gradual deletion throughout intron evolution because, generally, there are longer introns in basal angiosperms than in both eudicots and monocots (Fig. 3 and Additional file 1: Fig. S1). A deletion of this type in introns could be the result of recombination46 and may have contributed to the divergence and functional differentiation in this family of genes47. Intron lengths are positively correlated with the number of conserved motifs, which are identical to the putative transcription factor binding sites (R 2 = 0.711, P < 0.0001, Fig. 4). Furthermore, certain motifs in intron may stimulate gene expression48. Long introns with more conserved motifs could have a complicated folding structure, as well as alternative splicing sites that affect transcription, particularly for the basal angiosperms (such as Lauraceae, Magnoliaceae, and Amborella). Alternative splicing in TFL1/CEN paralogs was reported to influence terminal flowering and flowering time49. Formation of gene loops is also relevant to the activation or maintenance of Arabidopsis TFL1 expression45. Therefore, gene lengths are hypothesised to be a key factor affecting the expression efficiency of TFL1 orthologs.
TFL1 is targeted by several MADS-box genes, which have different functions during floral transition, and they coordinate the timing of flowering and floral development with TFL1 45, indicating that the TFL1 orthologs could have several protein binding sites. In addition, both AP1 and LFY can bind to the TFL1 locus and directly suppress TFL1 expression50–52. Suppression of TFL1 in inflorescence branching regulation by MADS-box genes also affects LFY and AP1 expression45. No AP1 binding sites (CArG box) or LFY binding sites were found in either eudicot TFL1 and monocot RCN1, but were present in basal angiosperms (Additional file 1: Table S3). This might suggest the functional relevance of long introns in the TFL1-like gene in basal angiosperms. In contrast, the short introns of eudicot and monocot paralogs could reflect their low expression33, which may facilitate the retention of duplicated genes and the conservation of their ancestral functions53.
Pervasive purifying selection and relaxation of selective constraints on eudicot and monocot TFL1/CEN/RCNs paralogs
It was suggested that the functions of angiosperm flowering development genes, have been conserved under selective constraint in eudicots and monocots34,45,54. Strong purifying selection of the TFL1 paralog with an average ω = 0.097 was inferred based on site-model analysis34, which is similar to the average pairwise Ka/Ks = 0.0791 estimated in our analysis (the horizontal dotted line in Fig. 5), and lower than the average ω of other floral-regulatory paralogs (SEP1 vs. SEP2 and SHP1 vs. SHP2, both ω = 0.1638). Nonsignificant radical functional divergence (θII) between paralogs supports the functional constraint hypothesis for angiosperm TFL1/CEN/RCNs paralogs (Table 3). However, in the phylogenetic analysis (Fig. 3 and Additional file 1: Fig. S3), low bootstrap values or posterior probabilities of basal lineages of the magnoliid TFL1-like and eudicot TFL1/CEN paralogs suggest a lack of fixed differences between paralogs. This indicates that multiple common polymorphisms are shared between clades or within-clade evolutionary constraints.
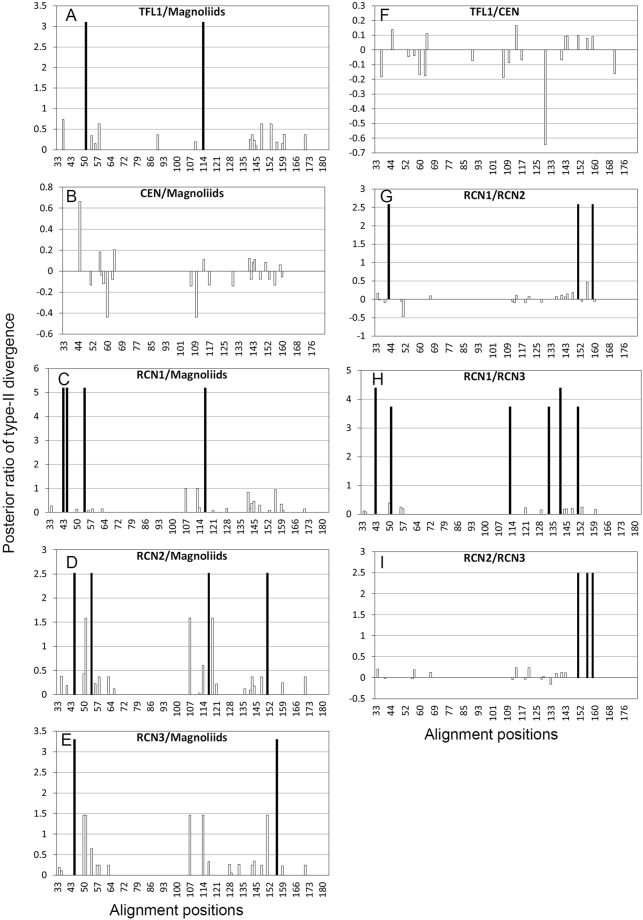
The pairwise Ka/Ks ratio and the sliding window analysis suggest that there were long-term selective constraints on eudicot TFL1 and CEN (Fig. 5), particularly on exon 2 and exon 3 of all angiosperm TFL1/CEN/RCNs paralogs (Fig. 6). Exon 2 and exon 3 are activator regions (ligand-binding site) of TFL1 25,55, and are highly conserved with no amino acid changes (Fig. 7). However, relatively higher pairwise Ka/Ks within monocot and magnoliid paralogs suggests that constraints were relaxed, particularly in exon 1 and exon 4 (Figs. 5 and 6), which is also supported by the analysis of site-specific radical functional change between paralogs of both monocots and magnoliids (Fig. 7). Residues 133–151 in exon 4 form an external loop, and this conformation determines the functional specificities of floral regulators55. Loss of the hydrogen bond between the external loop (exon 4) and the activator regions (exon 3) may be responsible for the functional conversion of activators of FT to floral repressors of TFL155. One radical change in RCN1/RCN2, RCN2/RCN3, RCN2/magnoliid TFL1-like, and three radical changes in RCN1/RCN3 within the external loop were estimated (Fig. 7), suggesting that the paralogous divergence occurred by relaxation of selective constraints, particularly between the monocot RCNs.
Figure 7.
Site-specific profile of type II functional divergence between angiosperm TFL1/CEN/RCN paralogs. Only the comparisons between the magnoliid TFL1-like and other paralogs, between eudicot TFL1 and CEN, and between monocot RCN1, RCN2, and RCN3 are shown. The full bars indicate the critical posterior ratio with a posterior probability >0.7.
Limited fixed radical differences (F00,R) could suggest the maintenance of ancestral function between paralogs of different taxa and imply that the eudicot TFL1/CEN and monocot RCNs do not fit the neo-functionalisation hypothesis of duplicate genes. Instead, the duplication could be a case of sub-functionalisation due to the relaxation of functional constraints, because one-third to a half frequency of radical change (GR) was detected (Table 3). The duplication-degeneration-complementation (DDC)56 and escape from adaptive conflict (EAC) models57 are commonly used to explain subfunctional divergence of duplicates and retention of duplicates58. The main difference between the DDC and EAC models is that the EAC puts more emphasis on positive selection for gene specialisation during or after duplication, while positive selection is not required for DDC59. In the case of eudicot TFL1/CEN and monocot RCNs, all duplicates retained plesiomorphic functionality with slight differences, by relaxation of selective constraints. However, no specific paralogs suffered positive selective pressures, suggesting a more likely evolutionary fit to DDC. The EAC hypothesis therefore, was rejected.
Relaxation of selective constraints and phylogenetic convergence of magnoliid TFL1-like genes
No duplication of TFL1-like genes was found in basal angiosperms. The constructed phylogenetic tree showed that the magnoliid TFL1-like genes are grouped with eudicot TFL1/CEN paralogs (Fig. 3 and Additional file 1: Fig. S3). This may suggest: (1) constraining ancestral functions of the eudicot TFL1 with the basal-angiosperm TFL1-like gene (functional constraints hypothesis), (2) identical selection pressures acted on both eudicot TFL1 and magnoliid TFL1-like genes synchronously (synchronous selection hypothesis), or (3) eudicot TFL1 and magnoliid TFL1-like genes evolved in parallel independently, resulting in phylogenetic convergence (phylogenetic convergence hypothesis). The LRTs for the two ratio and three ratio models showed the functional constraints between magnoliid TFL1-like and other paralogs (ω 1 = ω 2 ≤ 1) except the monocot RCN1 (ω 0 ≠ ω 1 ≠ ω 2) (Table 2). This indicates that (1) eudicot and monocot TFL1/CEN/RCNs could share ancestral polymorphisms and functions with TFL1-like gene of basal angiosperms, and (2) magnoliid TFL1-like and monocot RCN1 could functionally converge under heterogeneous evolutionary rates. The basic functions of TFL1/CEN/RCNs paralogs in magnoliids, eudicots, and monocots do not alter, but there is division of labour by small fractions of neutral or nearly neutral amino acid replacements, which is consistent with the functional divergence analysis (Table 3).
The long-term constrained evolution of floral development genes across divergent species was inferred by comparative analyses of 18 angiosperm species34. However, the evolutionary pattern of these genes in basal angiosperms, such as Amborella, Lauraceae, and Magnoliaceae, has not yet been investigated. Although the functional constraint hypothesis was supported between magnoliid TFL1-like genes and most other paralogs, the ω of foreground branches are larger than background lineages (Table 2), supporting the hypothesis of relaxation of constraints for flexing the non-duplicated magnoliid TFL1-like genes in shaping floral diversity inferred by both pairwise Ka/Ks (Fig. 5) and functional divergence analysis (Table 3). The relaxation of selective constraints was common for duplicated genes at the phase of early duplication that accelerated evolution of duplicated genes to escape from redundancy, while most gene duplicates were stochastically silenced with few survivors subsequently experiencing strong (10-fold efficiency) purifying selection14. Here, we provide at least two novel discoveries regarding the evolution of TFL1-like genes in basal angiosperms: (1) Lauraceae and Magnoliaceae TFL1-like genes are divergent from those of Amborella and are phylogenetically similar to the eudicot TFL1/CEN; (2) purifying selection prevailed over the magnoliid TFL1-like genes as well as the eudicot and monocot paralogs, but the unfixed paralogous radical replacement enabled their differentiation through the relaxation of selective constraints.
Conclusions
In this work, we inferred evolution and functional divergence of TFL1/CEN/RCN among 18 angiosperm species, including basal angiosperm species to elucidate the duplication history of TFL1/CEN/RCN genes. We found long-term retention of functionally redundant duplicates TFL1/CEN/RCNs in the angiosperm genomes. Based on the results of purifying selection on exon, radical amino acid changes and various intron lengths with cis-acting element analysis, the maintenance and conservation of their ancestral function could be explained by duplication-degeneration-complementation model. The ancestral function of TFL1/CEN/RCNs might be preserved and divided into each duplicates. Therefore, the strong selection pressure against removing any duplicates may cause the permanent establishment of duplicates during evolution of flowering plants. Consequently, these two duplicates together maintain the conservative mechanism in inflorescence architectures, and expansion of the PEBP gene members may be important factor for driving morphological divergence among angiosperms.
Intron length of TFL1 paralogs was various. TFL1 introns of basal angioserpm tend to have longer intron and more predicted cis-acting than monocot and eudicot. On the other hands, exon length was conserved with low amino acid substitution rate. These data suggest that strong purifying selection has maintained the relevant functions of TFL1/CEN/RCNs paralogs on flowering regulation throughout the evolution of angiosperms, and the shorter introns with radical amino acid changes are important for the retention of paralogous duplicates.
Methods
Data collection and phylogenetic tree reconstruction
The full lengths of angiosperm TFL1/CEN/RCNs genes were obtained from NCBI GenBank. Organisms without complete paralogs (e.g. only TFL1 of eudicot and RCN1 of monocot organisms) were excluded. Due to high similarity among PEBP gene family, many sequences named with TFL1 or CEN are belonged to FT/BFT/MFT. For preventing miss-inferring of phylogenetics of TFL1/CEN, we only included sequences which were previously identified as TFL1/CEN in our subsequent analysis e.g. ref.7. The TFL1 and CEN gene sequences from five eudicot species (Arabidopsis thaliana, A. lyrata [Brassicaceae], Citrus clementina [Rutaceae], Fragaria vesca [Rosaceae], Glycine max [Fabaceae], Medicago truncatula [Fabaceae], Populus trichocarpa [Salicaceae], Solanum lycopersicum [Solanaceae], Solanum_tuberosum [Solanaceae], Vitis vinifera [Vitaceae], Linum usitatissimum [Linaceae], Kalanchoe fedtschenkoi [Crassulaceae], Mimulus guttatus [Phrymaceae], Salix purpurea [Salicaceae], Trifolium pratense [Fabaceae], Vigna unguiculata [Fabaceae], Lactuca sativa [Asteraceae], Coffea arabica [Rubiaceae],), and RCN1–3 from four monocot species (Musa acuminata [Musaceae], Ananas comosus [Bromeliaceae], Zostera marina [Zosteraceae], Oryza sativa, Sorghum bicolor, Setaria italic, and Brachypodium distachyon, Panicum hallii [Poaceae]), and the TFL1-like gene from Amborella trichopoda (Amborellaceae) were obtained from GenBank. We also amplified complete TFL1-like sequences from two basal angiosperm species Lindear megaphylla (Lauraceae) and Liriodendron sp. (Magnoliaceae) using primers (MaLaTFL1-F1: 5′-ATGGCAAGAATGTTAGAGC-3′; MaLaTFL1-R1: 5′-CAACGTCTCCTNGCAGCTG-3′). Intron positions were rechecked based on the GT-AG rule. Exon-intron structures were drawn by Exon-Intron Graphic Maker (http://wormweb.org/exonintron). Exons of Litsea cubeba, Neolitsea phanerophlebia, Persea sp., and Michelia compressa (GenBank accession number: KY933631- KY933636) were also sequenced for coding region analyses. The identification of the exon sequences were conducted using BLAST. Sequences without best hit to TFL1/CEN/RCNs were discarded (eg. FT/BFT/MFT). The phylogenetic tree of TFL1/CEN/RCNs was reconstructed by exons using the Maximum likelihood method with the GTR+G model, gamma distribution (α = 0.46) for substitution rate among sites using PhyML 3.060. The tree bisection and reconnection (TBR) was adopted for tree rearrangement and fast bootstrap method aLRT was adopted for branch supports.
Conserved motifs in introns
Conserved motifs in introns were found by searching the database of plant cis-acting regulatory DNA elements, NEW PLACE61. From 212 types of predicted motifs like cis-acting elements, 12 putative functional cis-acting elements that have been reported to regulate the expression of TFL1/CEN/RCNs paralogs were identified (Additional file 1: Table S1)49,62–65 and the number of these putative cis-acting elements were calculated. Correlation between the number of cis-acting elements and the total intron length (i.e. intron1 + intron2 + intron3) was estimated.
Detection of positive selection on angiosperm TFL1/CEN/RCNs paralogs
To examine the effect of selective pressures, the ω ratio, which can be used for testing the gene neutrality hypothesis Ka/Ks (ω) = 1, was estimated by maximum likelihood approaches implemented in PAML 4.766. First, the ω under the free-ratio model was estimated, which allows varied ω on every branch. The likelihood ratio test (LRT) that calculates the 2× differences of log likelihood between constant-rate model and other evolutionary hypotheses (2ΔL) were used for evaluating the better fitted selective hypothesis by χ 2 test. Because TFL1-like genes in magnoliids were grouped with eudicot TFL1/CEN clades (Fig. 1), we hypothesized that (1) functional constraints between magnoliid TFL1-like and eudicot TFL1/CEN and (2) phylogenetic convergence was tested. To test these hypotheses, we estimated the ω and evaluated the goodness-of-fit of two-ratio model (ω 0 ≠ ω 1 = ω 2) and three-ratio model (ω 0 ≠ ω 1 ≠ ω 2) by LRT. ω 0 are the Ka/Ks ratio of background branches; ω 1 and ω 2 are Ka/Ks of foreground branches that allowed ω > 1, where ω 1 are the ω of eudicot TFL1/CEN or monocot RCNs paralogs and ω 2 are the ω of magnoliid TFL1-like genes. Detailed hypotheses and selection scenarios are listed in Table 1.
In addition, pairwise Ka/Ks comparisons between angiosperm TFL1/CEN/RCNs paralogs were calculated to examine the evolutionary divergence and represented by (1) the Ka/Ks against Ks plot and (2) sliding window analysis by DnaSP 5.067. The Ka/Ks against Ks plot helps to determine the degrees and relative times of paralogous divergence and the sliding windows provide details for clarifying the divergent regions from the regions under selective constraints.
Functional divergence between angiosperm TFL1/CEN/RCNs paralogs
Functional divergence between paralogs caused by radical amino acid changes was assessed by the type-II divergence function implemented in DIVERGE 3.043 with 500 bootstrap replications. Substitutions between amino acids with different radical biochemical properties (charge positive/negative, hydrophilic/hydrophobic) are classified as a radical change, and all others are classified as a conserved change. The Z-test was conducted to test the deviation of coefficients of type II functional divergence (θII) from zero, and value of θII greater than zero implied radical shifts in amino acid physiochemical properties after duplication. The fold of radical change related to non-functional change was calculated using the ratio of radical change under functional divergence versus nonfunctional divergence (aR/πR). The proportion of such as fixed radical change, conserved change and none change sites were also calculated. Site-specific estimation of posterior probability of radical changes was performed to assess the probable regions and shifts of biochemical properties between paralogous groups.
Availability of data and materials
The sequences have been submitted to GenBank with the accession number KY933631-KY933636.
Electronic supplementary material
Acknowledgements
We thank Li-Ping Ju for providing specimens, and the FuShan Botanical Garden, a long-term ecological research (LTER) site for providing samples. We also gratefully thank Chun-Neng Wang for his insightful comments on this paper. This research was financially supported by the Ministry of Science and Technology in Taiwan (MOST 105–2628-B-003-001-MY3 and MOST 105-2628-B-003-002-MY3) and was also subsidized by the National Taiwan Normal University (NTNU), Taiwan.
Author Contributions
P.C.L. conceived this study. B.H.H. and J.Y.C. collected materials. J.G. and Y.T.W. conducted experiments. B.H.H. and P.C.L. analysed the data. P.C.L. wrote the paper. J.G., B.H.H., and J.Q.L. critically reviewed the manuscript. All authors participated in the discussion and read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13645-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahn JH, et al. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley D, et al. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- 3.Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- 4.Zhang D, Yuan Z. Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 2014;65:553–578. doi: 10.1146/annurev-arplant-050213-040104. [DOI] [PubMed] [Google Scholar]
- 5.Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 7.Karlgren A, et al. Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 2011;156:1967–1977. doi: 10.1104/pp.111.176206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313X.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- 9.Repinski SL, Kwak M, Gepts P. The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis. TFL1. Theor. Appl. Genet. 2012;124:1539–1547. doi: 10.1007/s00122-012-1808-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen M-S, et al. Comparative transcriptome analysis between gynoecious and monoecious plants identifies regulatory networks controlling sex determination in Jatropha curcas. Front Plant Sci. 2016;7:1953. doi: 10.3389/fpls.2016.01953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaire, R. & Wilde, H. D. Natural allelic variation in blueberry TERMINAL FLOWER 1. Plant Genetic Resources, 1–9, 10.1017/s1479262116000435 (2016).
- 12.Klintenas M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O. Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol. 2012;196:1260–1273. doi: 10.1111/j.1469-8137.2012.04332.x. [DOI] [PubMed] [Google Scholar]
- 13.Amborella Genome Project. The Amborella Genome and the Evolution of Flowering Plants. Science 342, 1467, 10.1126/science.1241089 (2013). [DOI] [PubMed]
- 14.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 15.Hughes AL. The evolution of functionally novel proteins after gene duplication. P. Roy. Soc. B-Biol. Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 16.Mimida N, et al. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells. 2001;6:327–336. doi: 10.1046/j.1365-2443.2001.00425.x. [DOI] [PubMed] [Google Scholar]
- 17.Mimida, N. et al. Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus x domestica Borkh.). Plant Cell Physiol50, 394-412, doi:10.1093/pcp/pcp001 pcp001 [pii] (2009). [DOI] [PubMed]
- 18.Pnueli L, et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- 19.Shalit A, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. P. Natl. Acad. Sci. USA. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaya I, Ratcliffe OJ, Bradley DJ. Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell. 1999;11:1405–1417. doi: 10.1105/tpc.11.8.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarte S, et al. Sequence variation, differential expression, and divergent evolution in starch-related genes among accessions of Arabidopsis thaliana. Plant Mol. Biol. 2015;87:489–519. doi: 10.1007/s11103-015-0293-2. [DOI] [PubMed] [Google Scholar]
- 22.Huang, B.-H., Chen, Y.-W., Huang, C.-L., Gao, J. & Liao, P.-C. Imbalanced positive selection maintains the functional divergence of duplicated DIHYDROKAEMPFEROL 4-REDUCTASE genes. Sci Rep6, 39031, 10.1038/srep39031https://www.nature.com/articles/srep39031#supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 23.Krizek BA, Fletcher JC. Molecular mechanisms of flower development: An armchair guide. Nat Rev Genet. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- 24.Ballerini, E. S. & Kramer, E. M. In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front Plant Sci 2, 81, Artn 81 10.3389/Fpls.2011.00081 (2011). [DOI] [PMC free article] [PubMed]
- 25.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. P. Natl. Acad. Sci. USA. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Hans H, Christian J, Molina C. Mutations in single FT- and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front Plant Sci. 2014;5:282. doi: 10.3389/fpls.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seoighe C, Johnston CR, Shields DC. Significantly different patterns of amino acid replacement after gene duplication as compared to after speciation. Mol. Biol. Evol. 2003;20:484–490. doi: 10.1093/molbev/msg059. [DOI] [PubMed] [Google Scholar]
- 28.Gu X. Maximum-likelihood approach for gene family evolution under functional divergence. Mol. Biol. Evol. 2001;18:453–464. doi: 10.1093/oxfordjournals.molbev.a003824. [DOI] [PubMed] [Google Scholar]
- 29.Gu X. A simple statistical method for estimating Type-II (Cluster-Specific) functional divergence of protein sequences. Mol. Biol. Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- 30.Xu GX, Guo CC, Shan HY, Kong HZ. Divergence of duplicate genes in exon-intron structure. P. Natl. Acad. Sci. USA. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duester G, Jornvall H, Hatfield GW. Intron-dependent evolution of the nucleotide-binding domains within alcohol dehydrogenase and related enzymes. Nucleic Acids Res. 1986;14:1931–1941. doi: 10.1093/nar/14.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirby DA, Muse SV, Stephan W. Maintenance of pre-mRNA secondary structure by epistatic selection. P. Natl. Acad. Sci. USA. 1995;92:9047–9051. doi: 10.1073/pnas.92.20.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren XY, Vorst O, Fiers MWEJ, Stiekema WJ, Nap JP. In plants, highly expressed genes are the least compact. Trends Genet. 2006;22:528–532. doi: 10.1016/j.tig.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Mol. Biol. Evol. 2014;31:560–573. doi: 10.1093/molbev/mst223. [DOI] [PubMed] [Google Scholar]
- 35.Olsen KM, Womack A, Garrett AR, Suddith JI, Purugganan MD. Contrasting evolutionary forces in the Arabidopsis thaliana floral developmental pathway. Genetics. 2002;160:1641–1650. doi: 10.1093/genetics/160.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinig C, et al. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics. 2003;165:321–329. doi: 10.1093/genetics/165.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller SR, Levsen N, Ingvarsson PK, Olson MS, Tiffin P. Local selection across a latitudinal gradient shapes nucleotide diversity in Balsam poplar, Populus balsamifera L. Genetics. 2011;188:941–952. doi: 10.1534/genetics.111.128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore RC, Grant SR, Purugganan MD. Molecular population genetics of redundant floral-regulatory genes in Arabidopsis thaliana. Mol. Biol. Evol. 2005;22:91–103. doi: 10.1093/molbev/msh261. [DOI] [PubMed] [Google Scholar]
- 39.Gu X. Evolution of duplicate genes versus genetic robustness against null mutations. Trends Genet. 2003;19:354–356. doi: 10.1016/S0168-9525(03)00139-2. [DOI] [PubMed] [Google Scholar]
- 40.Leebens-Mack J, et al. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein Zone. Mol. Biol. Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- 41.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu X, et al. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013;30:1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- 44.Fu, G. C. L. & Lin, W. C. Identification of gene-oriented exon orthology between human and mouse. BMC Genomics 13, S10, doi:Artn S10 10.1186/1471-2164-13-S1-S10 (2012). [DOI] [PMC free article] [PubMed]
- 45.Liu C, et al. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Hu KJ. Intron exclusion and the mystery of intron loss. FEBS Lett. 2006;580:6361–6365. doi: 10.1016/j.febslet.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 47.Park KC, Kwon SJ, Kim NS. Why genes are in pieces? A genomics perspective. Genes Genom. 2008;30:429–437. [Google Scholar]
- 48.Rose AB, Carter A, Korf I, Kojima N. Intron sequences that stimulate gene expression in. Arabidopsis. Plant Mol. Biol. 2016;92:337–346. doi: 10.1007/s11103-016-0516-1. [DOI] [PubMed] [Google Scholar]
- 49.Tsaftaris A, et al. Isolation of a CENTRORADIALIS/TERMINAL FLOWER1 homolog in saffron (Crocus sativus L.): characterization and expression analysis. Mol. Biol. Rep. 2012;39:7899–7910. doi: 10.1007/s11033-012-1634-8. [DOI] [PubMed] [Google Scholar]
- 50.Kaufmann K, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 51.Moyroud E, et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winter CM, et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 53.Qian WF, Liao BY, Chang AYF, Zhang JZ. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends Genet. 2010;26:519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Kang S, Lee J, Lee MS, Seok C. Structural basis of functional conversion of a floral repressor to an activator: A molecular dynamics simulation study. B Korean Chem Soc. 2008;29:408–412. doi: 10.5012/bkcs.2008.29.2.408. [DOI] [Google Scholar]
- 56.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 58.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 60.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 61.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benlloch R, et al. Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J. 2011;67:1094–1102. doi: 10.1111/j.1365-313X.2011.04660.x. [DOI] [PubMed] [Google Scholar]
- 63.West AG, Shore P, Sharrocks AD. DNA binding by MADS-box transcription factors: A molecular mechanism for differential DNA bending. Mol. Cell. Biol. 1997;17:2876–2887. doi: 10.1128/MCB.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Randoux M, et al. Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot. 2012;63:6543–6554. doi: 10.1093/jxb/ers310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu JX, et al. Photoperiodic control of FT-like gene ClFT initiates flowering in Chrysanthemum lavandulifolium. Plant Physiol. Biochem. 2014;74:230–238. doi: 10.1016/j.plaphy.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 67.Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences have been submitted to GenBank with the accession number KY933631-KY933636.