Abstract
Amyotrophic lateral sclerosis (ALS) is a chronic neurodegenerative disease characterized by progressive degeneration of motor neurons. The pathogenesis of ALS remains largely unknown. RNA helicase DDX3 is a multifunctional protein involved in several steps of gene expression. Casein kinase 1ε (CK1ε) is an important signal molecule of Wnt signaling pathway and is closely related to neurite growth. However, the roles of DDX3 and CK1ε in the pathogenesis of ALS remain unclear. In this study, we first investigated the expression of DDX3 and CK1ε in the spinal cord of SOD1-G93A ALS transgenic mice using RT-PCR, Western blot and immunohistochemical technique. Results showed that the altered expression of DDX3 and CK1ε was found in the spinal cord of ALS mice. DDX3 and CK1ε positive cells were mainly distributed in the anterior horn of spinal cord and co-localized with neurons not with glial cells, suggesting that the altered expression of DDX3 and CK1ε was closely related to motor neuron degeneration of ALS. Moreover, we selected NSC34 cell line and transfected pEGFP-G93A-SOD1 plasmid to further examine the mechanism. Knockdown of DDX3 that uses small interfering RNA (siRNA) decreased the mRNA and protein levels of CK1ε significantly and inhibited neurite outgrowth of SOD1 mutant NSC34 cells in vitro. Co-immunoprecipitation kit confirmed that DDX3 could band with CK1ε in vivo. Our data suggested that DDX3 binding with CK1ε was closely related to motor neuron degeneration of ALS by affecting neurite outgrowth. Thus, elucidating the underlying mechanisms of ALS is crucial for future development of ALS treatments.
Keywords: Amyotrophic lateral sclerosis, SOD1-G93A transgenic mice, NSC34 cells, DDX3, CK1ε
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disease characterized by progressive muscle paralysis determined by the degeneration of motor neurons in spinal cord, brainstem and cerebral cortex [1,2]. The mechanisms that regulate the indication and/or the progression of motor neuron degeneration in this disease remain enigmatic, despite studies demonstrated the involvement of several altered signaling pathways, such as mitochondrial dysfunction, glutamate excitotoxicity, oxidative stress and neuroinflammation [2,3]. Cu/Zn superoxide dismutase 1 (SOD1) mutations are strongly associated with the pathogenesis of ALS. The most widely used transgenic mouse models of ALS are based on SOD1 mutations and recapitulate the key clinical and histopathological features of the disease [4]. The G93A mutation (glycine 93 changed to alanine) is frequently investigated as it was the first to be used in a transgenic mouse model of the disease [5,6].
DDX3 is a member of the DEAD/H box helicase family and is a multifunctional protein involved in several steps of gene expression, including transcription, translation and pre-mRNA processing. In addition, DDX3 is also involved in a variety of cellular biogenesis processes, including cell cycle regulation, cell differentiation, cell survival and apoptosis [7-9]. A study reported that DDX3 modulates neurite development via translationally activating an RNA regulation involved in Rac1 activation [10].
Recent evidence has identified that DDX3 is a novel regulator of the Wnt/β-catenin network, where it acts as a regulatory subunit of casein kinase 1ε (CK1ε). DDX3 binds CK1ε and directly stimulates its kinase activity in a Wnt-dependent manner [11,12]. Our previous study revealed that the altered expression of many Wnt signaling molecules was found in the spinal cord of SOD1-G93A ALS mice, indicating that the abnormalities of Wnt signaling pathway were closely related to the pathogenesis of ALS [13-16]. CK1ε is one of the important signal molecules of Wnt signaling pathway and is closely related to neurite growth [17]. CK1ε also plays an important role in the cleavage of amyloid precursor protein, which makes it an attractive target for the treatment of Alzheimer’s disease [18]. However, the role of DDX3 and CK1ε in the pathogenesis of ALS remains unclear.
In this study, we showed that the expression of DDX3 and CK1ε was altered in the spinal cords of SOD1-G93A ALS transgenic mice. We also provided evidence that DDX3 can band with CK1ε in vivo and knockdown of DDX3 not only decreased the mRNA and protein levels of CK1ε significantly but also reduced cell proliferation and inhibited neurite outgrowth of SOD1-G93A mutant NSC34 cells in vitro. These findings bring new insights into the mechanism of motor neuron degeneration and provide the basis for future therapeutic strategies of ALS.
Materials and methods
Mice and tissue preparations
The human SOD1-G93A transgenic mice (ALS mice) and wild-type mice (WT mice) were obtained from Jackson Laboratory (Bar Harbor, ME). The mice were genotyped using DNA isolated from a 0.5 cm piece of mouse tail as suggested by Jackson Laboratory genotyping protocol. Mice were killed at various ages of 95 (early stage), 108 (middle stage), or 122 days (end stage) and spinal cords were quickly dissected. Dissected samples were either stored in fresh frozen in liquid nitrogen for molecular biological analysis or immersion fixed in 4% paraformaldehyde for immunohistological analysis [13,14]. The following numbers of animals were used for the study: ALS mice at 95 days of age (n = 11), 108 days of age (n = 11) and 122 days of age (n = 11); among the eleven mice, four for RT-PCR, four for Western blot and three for immunohistological analysis; the same number of wild-type mice was used for the control. All animal experiments were performed according to protocols approved by the Animal Care and Use Committee of Weifang Medical University.
Cell lines and cell co-transfection
Cell culture was conducted essentially as described previously [19]. All reagents for cell culture were purchased from Gibco (Rockville, MD). NSC34, a mouse neuroblastoma N18TG2 and mouse embryonic spinal cord motor neuron hybrid cell line were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, plus 100 U/mL penicillin, 100 µg/mL streptomycin and cultured at 37°C in a humidified atmosphere with 5% CO2 for five to seven days before the cell treatment.
The human SOD1 plasmid pEGFP-G93A-SOD1 [20] were kindly provided by Professor Angelo Poletti (University of Milan, Italy). The small interfering RNA (siRNA) of DDX3 (si-DDX3) and control siRNA (Con) were purchased from Ribobio (China). The target sequence of DDX3 siRNA is 5’-CCTGAACTCTTCAGATAAT-3’. DDX3 siRNA and control siRNA were used at a final concentration of 50 nM. Co-transfection of NSC34 cells was performed using Lipofectamine™ 2000 (Thermo Fisher Scientific, USA) as previously described [19].
MTS proliferation and viability assay
The proliferation and survival of NSC34 cells were measured by MTS assay using the protocol recommended by CellTiter 96® AQueous One Solution Cell Proliferation Assay from Promega. In brief, NSC34 cells of experimental group on 96-well plate were co-transfected with DDX3 siRNA and pEGFP-G93A-SOD1 and the NSC34 cells of the control group were co-transfected with the control siRNA (Con) and pEGFP-G93A-SOD1. At 24, 48, 72, 96 and 120 h after co-transfection, 20 µl MTS reagent (5 mg/ml) was added to each well and kept for 1 h at 37°C. The plates were read by measuring the absorbance at 490 nm with the use of BioTek PowerWave Microplate Reader (BioTek, Winooski, VT, USA). The experiment was performed independently in triplicate. The cell proliferation curve was plotted at various time points.
RT-PCR
Total RNA from spinal cord tissue and NSC34 cells was extracted by homogenization in 1 ml TRIzol reagent (Life Technologies) following the manufacturer’s specifications. The amount and quality of RNA was determined using ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). Total RNA at 2 µg was reverse transcribed at 42°C for 1 h using random hexamers followed by PCR amplification using a pair of gene-specific primers and agarose gel running. PCR was performed in optimized conditions as follows: initial denaturation at 94°C for 3 min, annealing at 58°C for 30 s, extension at 72°C for 30 s followed by 30 cycles and a final extension step of 72°C for 5 min. The sequences of primers were synthesized as follows: mouse DDX3, forward: 5’-CAAGAAGTGCCTTCTTGGTTAGAG-3’; reverse: 5’-CTTGTTTGGCAGGGTGACCTA-3’ (331 bp); mouse β-actin, forward: 5’-GTCGT ACCAC AGGCATTGTG ATGG-3’; reverse: 5’-GCAAT GCCTG GGTAC ATGGT GG-3’ (493 bp); mouse CK1ε, forward: 5’-TTAGCGAGAAGAAGATGTCAACG-3’; reverse: 5’-TGGGAGAAGTATTGCCAGTTTG-3’ (411 bp); mouse β-actin, forward: 5’-CGTTGACATCCGTAAAGACC-3’; reverse: 5’-ACAGTCCGCCTAGAAGCAC-3’ (280 bp). The signal intensity of each RT-PCR product in the gel was measured by the Image-Pro Plus 6.0 analytic system. β-actin was used as a loading control.
Western blot
Western blot was performed as previously reported [19]. In brief, whole-cell lysates were prepared and protein concentrations were determined using Pierce™ BCA Protein Assay Kit (Thermo Scientific). Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto PVDF membranes (Millipore) and blotted with the following antibodies and dilutions: Rabbit anti-DDX3 (1:2000, Abcam) and Rabbit anti-CK1ε antibodies (1:800, Novus). Horseradish peroxidase-labeled anti-rabbit IgG antibody (1:10000) was purchased from Jackson ImmunoResearch Laboratories. The bands were visualized using enhanced chemiluminescence (ECL) detection kit (Thermo Scientific Pierce) by exposure to X-ray film (KODAK X-OMAT BT). To reprobe the blots, the membrane was stripped for 10 min in Restore Western blot buffer (Beyotime Biotechnology), blocked and reprobed with a mouse anti-GAPDH (1:2000; Proteintech Group) and horseradish peroxidase-labeled anti-mouse IgG antibody (1:10000, Jackson ImmunoResearch Laboratories). The bands were scanned and quantified by densitometric analysis using Image-Pro Plus 6.0 analytic system. GAPDH was used as a loading control.
Immunohistochemistry and immunofluorescence
Immunohistochemistry and immunofluorescence staining of spinal cord tissue slices were performed as described [19] by the following antibodies and dilutions: rabbit anti-DDX3 (1:200, Abcam), rabbit anti-CK1ε (1:100, Sigma), mouse anti-glial fibrillary acidic protein (GFAP) (1:300; Cell signaling technology) and mouse anti-β-tubulin III (1:200; R&D). Alexa Fluor 488-conjugated anti-mouse IgG (1:400; Jackson ImmunoResearch) and Cy3 conjugated anti-rabbit IgG (1:400; Jackson ImmunoResearch) were used as corresponding secondary antibodies. The images were obtained under fluorescence microscope (Olympus BX53, Japan).
Co-immunoprecipitation (Co-IP) kit
The spinal cord of SOD-G93A ALS transgenic mice and NSC34 cells, which were transiently transfected with pEGFP-G93A-SOD1, were harvested and lysed in ice cold lysis buffer (Thermo Scientific) with protease inhibitor cocktail (Sigma). After centrifugation at 13200 rpm for 10 min, supernatants were collected and protein concentrations were measured by Pierce™ BCA Protein Assay Kit. Part of the lysate was saved as the control “input”. Equal amounts of proteins were used for immunoprecipitation and incubated with 1 µg of rabbit anti-CKIε antibody (Novus) overnight with gentle rotation at 4°C and then added with 40 µl of protein G/A agarose beads (Thermo Scientific) for 3 h at 4°C. The beads were then washed with cell lysis buffer three times to obtain the bound proteins and were saved as “Co-IP”. The two groups of protein were assessed by Western blot analysis with the corresponding antibodies: rabbit anti-DDX3 (1:2000, Abcam) and rabbit anti-CK1ε (1:800, Novus).
Measurement of neurite outgrowth
NSC34 cells were co-transfected with pEGFP-G93A-SOD1 and DDX3 siRNA or pEGFP-G93A-SOD1 and control siRNA according to the method as described previously followed by treatment with 10 µmol/L retinoic acid (RA, Sigma-Aldrich) for two days to induce the neurite outgrowth. Cells in each group were observed under a microscope (Leica Microsystems CMS GmbH) at 0, 24 and 48 h after co-transfection. The images were obtained from five random fields. A neurite is defined as a cellular process as long or wide as the cell body [21]. Then image analysis was performed to measure the percentage of cells with neurite and the cell’s longest neurite. Data were obtained from three independent experiments.
Data analysis
All data were expressed as the means ± SD from at least three replicate experiments. Significant differences between two groups were analyzed by two-tailed Student’s t-test. Asterisks indicate statistically significant differences between the compared groups: *p < 0.05, **p < 0.01, ***p < 0.001.
Results
Altered expression of DDX3 in the spinal cord of SOD1-G93A ALS transgenic mice
To understand the expression of DDX3 in the spinal cord of SOD1-G93A ALS transgenic mice, we performed RT-PCR and Western blot analysis. The result showed that compared with wild-type mice, DDX3 mRNA in the spinal cord of ALS mice was all up-regulated significantly at the early stage (95 d), middle stage (108 d) and end stage (122 d) (p < 0.05, p < 0.01), but DDX3 protein remained unchanged at 95 d, up-regulated at 108 and 122 d (p < 0.01, p < 0.001) (Figure 1A-D).
Figure 1.
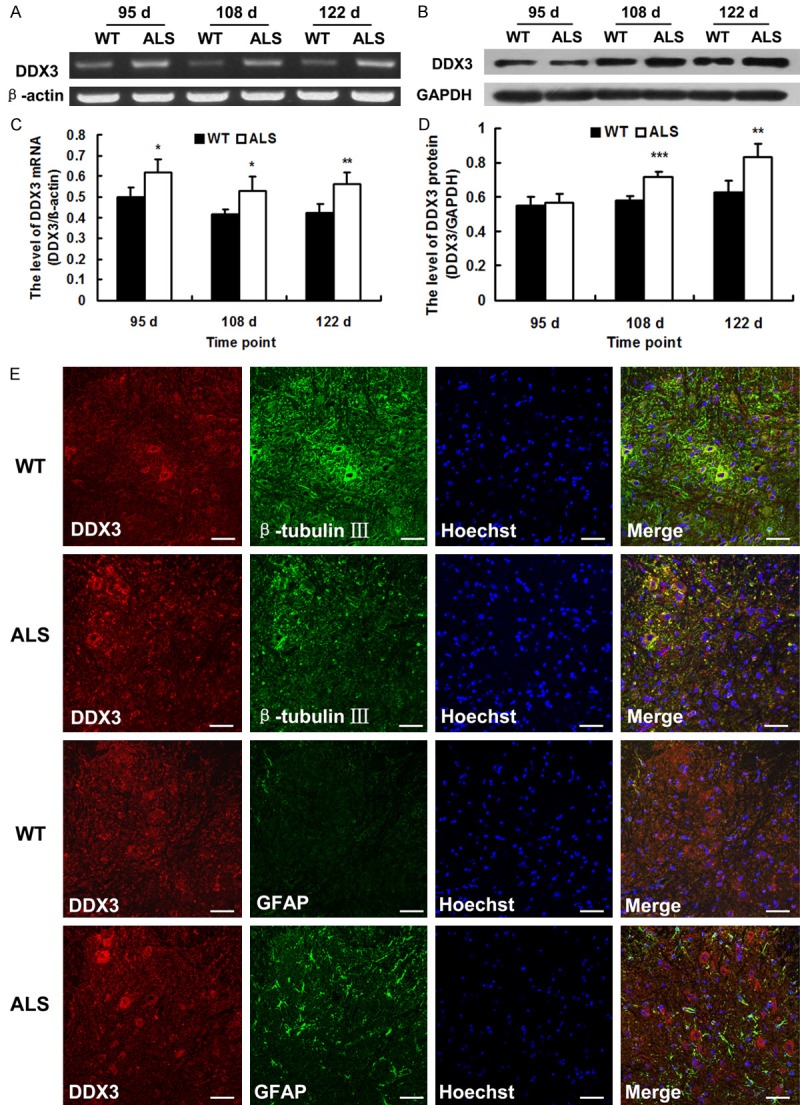
The altered expression of DDX3 in the spinal cords of SOD1-G93A transgenic mice (ALS) and wide-type (WT) mice. A. Representative RT-PCR images of DDX3. β-actin was used as loading control. B. Representative western blot images of DDX3 protein. GAPDH was used as loading control. C. The level of DDX3 mRNA as analyzed by RT-PCR (n = 4). D. The level of DDX3 protein as analyzed by western blot (n = 4). E. Confocal microscopy images showing the co-localization of DDX3 with β-tubulin III or GFAP. Representative confocal images were taken of gray matter of 108-day-old mice. Scale bar = 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001, vs. WT.
Double labeling was performed to further explore the localization of DDX3 with β-tubulin III for neurons or GFAP for mature astrocytes in the spinal cord of ALS mice. Results showed that DDX3 was co-localized with β-tubulin III and most of the double-positive cells were in the ventral horn where motor neuron degenerated. No DDX3/GFAP double positive cells were detected in both the ALS mice and wild-type mice (Figure 1E). Immunohistochemistry labeling was used to explore the distribution of DDX3 positive cells in the spinal cord of ALS transgenic mice. Most of the DDX3 positive cells were found to be distributed in the anterior horn of the gray matter. Nerve fibers in the white matter were also immunopositive. At 95 d, the immunoreactivity of DDX3 protein remains unchanged between the ALS mice and wild-type mice. At 108 and 122 d, the immunoreactivities of DDX3 protein in ALS mice were stronger than those in wild-type mice (Figure 2). All the data indicated that the expression of DDX3 was altered in the spinal cord of SOD1-G93A ALS transgenic mice, which may be closely associated with the degeneration of motor neuron.
Figure 2.
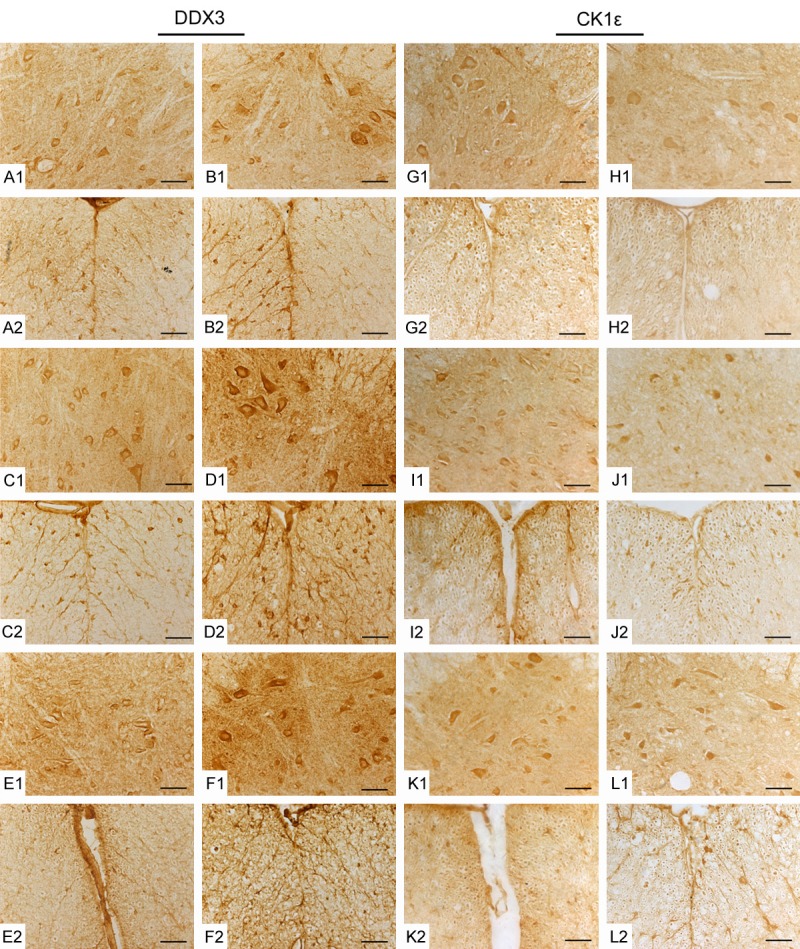
Immunohistochemical (IHC) staining showing the distribution of DDX3 and CK1ε in the spinal cords of ALS mice and wide-type (WT) mice. The representative images were taken of gray matter (GM) and white matter (WM) of the spinal cords at different stages. 95 d (A, B, G, H); 108 d (C, D, I, J); 122 d (E, F, K, L); GM (1); WM (2). WT (A, C, E, G, I, K); ALS (B, D, F, H, J, L). Scale bar = 50 μm.
Altered expression of CK1ε in the spinal cord of SOD1-G93A ALS transgenic mice
We detected the expression of CK1ε by RT-PCR, Western blot and immunohistochemistry labeling to test whether CK1ε expression is altered in the spinal cord of SOD1-G93A ALS transgenic mice. As shown in Figure 2, most of the CK1ε positive cells were also located in the anterior horn of the gray matter and the distribution of CK1ε was similar to that of DDX3. At 95, 108 and 122 d, the immunoreactivities of CK1ε protein in ALS mice were all weaker than those of age-matched wild-type mice (Figure 2). CK1ε mRNA in the spinal cord of ALS mice remained unchanged but CK1ε protein was all down-regulated significantly at 95, 108 and 122 d (p < 0.001) compared with wild-type mice as determined by RT-PCR and Western blot analysis (Figure 3A-D). CK1ε was expressed mainly in neurons not in astrocytes (Figure 3E). The altered expression of CK1ε was also closely related to the degeneration of motor neuron of ALS.
Figure 3.
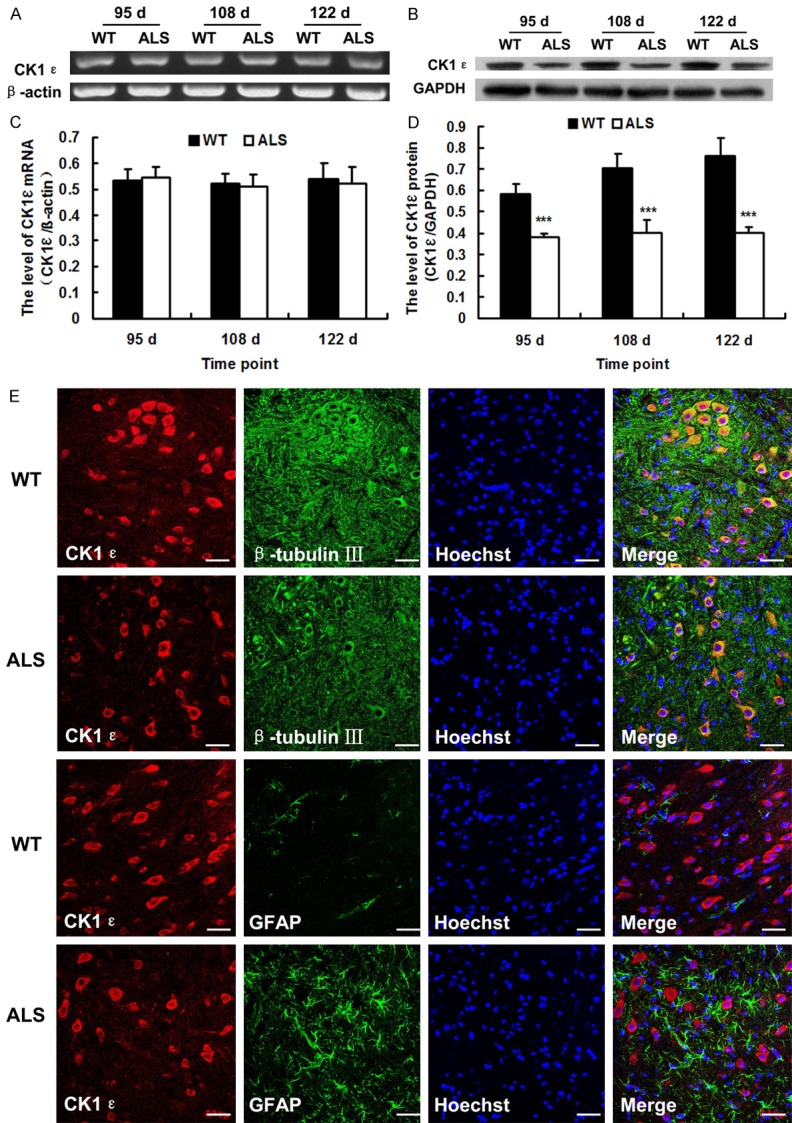
The altered expression of CK1ε in the spinal cords of SOD1-G93A transgenic mice (ALS) and wide-type (WT) mice. A. Representative RT-PCR images of CK1ε. β-actin was used as loading control. B. Representative western blot images of CK1ε protein. GAPDH was used as loading control. C. The level of CK1ε mRNA as analyzed by RT-PCR (n = 4). D. The level of CK1ε protein as analyzed by western blot (n = 4). E. Confocal microscopy images showing the co-localization of CK1ε with β-tubulin III or GFAP. Representative confocal images were taken of gray matter of 108-day-old mice. Scale bar = 50 μm. ***P < 0.001, vs. WT.
DDX3 can band with CK1ε in vivo and knockdown of DDX3 decreased the mRNA and protein levels of CK1ε significantly in vitro
Recent in vitro studies reveal that DDX3 functions as a regulatory subunit of CK1ε and DDX3 binding with CK1ε that can directly stimulate its kinase activity. DDX3 is a regulator of the Wnt-β-catenin network [11,12]. To assess whether similar mechanism was involved in the pathogenesis of ALS, we preformed co-immunoprecipitation kit to confirm the relationship between DDX3 and CK1ε using ALS mice and transfected mutate NSC34 cells in vitro and in vivo. Results showed that DDX3 can band with CK1ε in vivo but not in vitro (Figure 4A, 4B). The two proteins interact with each other in ALS tissue but not in SOD1 mutant NSC 34 cells. CK1ε protein level maybe low in NSC34 cells.
Figure 4.
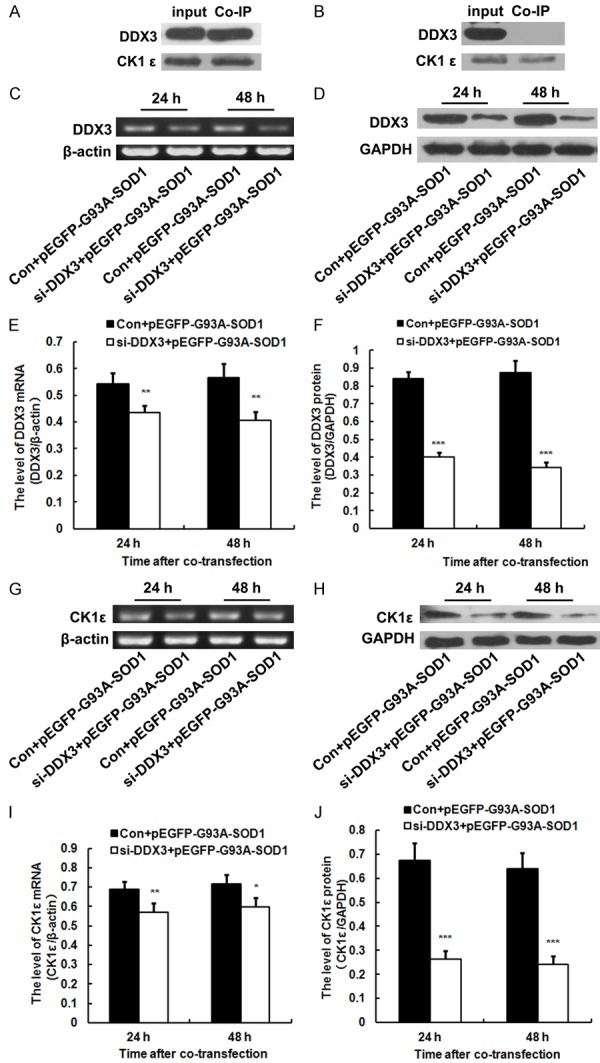
DDX3 can band with CK1ε in the spinal cord of ALS mice but not in NSC34 cells transfected with pEGFP-G93A-SOD1. Knockdown of DDX3 significantly decreased the mRNA and protein levels of CK1ε in vitro. NSC34 cells were co-transfected with DDX3 siRNA (si-DDX3) and pEGFP-G93A-SOD1 or control siRNA (Con) and pEGFP-G93A-SOD1. A. Co-immunoprecipitation kit confirmed that DDX3 can band with CK1ε in the spinal cord of ALS mice. CK1ε antibody was used to co-immunoprecipitate DDX3. Cell lysate (input) and immunoprecipitated products (Co-IP) were blotted with the DDX3 and CK1ε antibody respectively. B. Co-immunoprecipitation kit confirmed that DDX3 can not band with CK1ε in the NSC34 cells transfected with pEGFP-G93A-SOD1 at 48 h after transfection. Cell lysate (input) and immunoprecipitated products (Co-IP) were blotted with the DDX3 and CK1ε antibody respectively. C. Representative RT-PCR images of DDX3 in the co-transfected NSC34 cells. β-actin was used as loading control. D. Representative western blot images of DDX3 protein in the co-transfected NSC34 cells. GAPDH was used as loading control. E. RT-PCR analysis illustrating the knockdown of DDX3 (n = 4). F. Western blot analysis illustrating the knockdown of DDX3 protein. The amount of DDX3 was quantified and normalized against GAPDH (n = 4). G. Representative RT-PCR images of CK1ε in the co-transfected NSC34 cells. β-actin was used as loading control. H. Representative western blot images of CK1ε protein in the co-transfected NSC34 cells. GAPDH was used as loading control. I. RT-PCR analysis illustrating the down-regulation of CK1ε (n = 4). J. Western blot analysis illustrating the down-regulation of CK1ε protein. The amount of CK1ε was quantified and normalized against GAPDH (n = 4).
Given that DDX3 acts as a regulatory subunit of CK1ε, we also tested if DDX3 modulates the level of CK1ε in SOD1 mutant NSC34 cells. We used siRNA to knockdown DDX3 and subsequently validated the efficiency of DDX3 knockdown by RT-PCR and Western blot analysis first. Results showed that DDX3 mRNA and protein levels in the NSC34 cells co-transfected with DDX3 siRNA and pEGFP-G93A-SOD1 were dramatically decreased significantly compared with those co-transfected with control siRNA and pEGFP-G93A-SOD1 at 24 and 48 h after co-transfection, suggesting that a DDX3-targeting siRNA could efficiently deplete DDX3 in the transfected NSC34 cells (Figure 4C-F). DDX3 knockdown in SOD1 mutant NSC34 cells also significantly decreased the mRNA and protein levels of CK1ε at 24 and 48 h after co-transfection (Figure 4G-J).
Knockdown of DDX3 reduced the cell proliferation and inhibited the neurite outgrowth of SOD1 mutant NSC34 cells in vitro
We performed MTS assay to explore the role of DDX3 in cell proliferation of SOD1 mutant NSC34 cells and the results revealed that the OD values of NSC34 cells co-transfected with DDX3 siRNA and pEGFP-G93A-SOD1 were lower than those of NSC34 cells co-transfected with control siRNA and pEGFP-G93A-SOD1 at 48, 72 and 96 h after transfection (p < 0.05, p < 0.01) (Figure 5A). These data suggested that knockdown of DDX3 reduced cell proliferation and survival of SOD1 mutant NSC34 cells. DDX3 is indeed required for cell growth [22].
Figure 5.
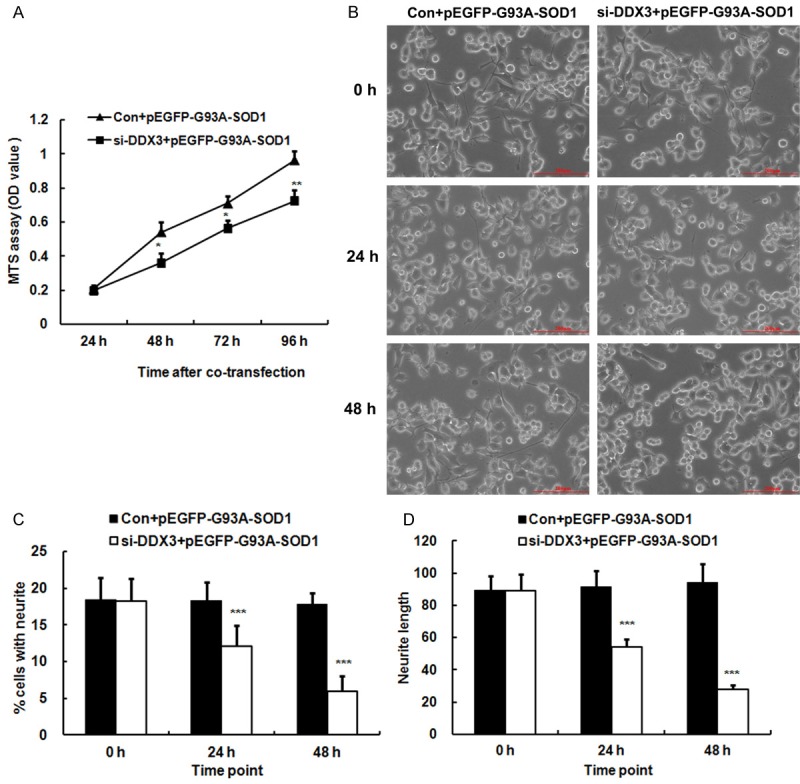
Knockdown of DDX3 reduced the cell proliferation and inhibited the neurite outgrowth of SOD1 mutant NSC34 cells in vitro. A. Growth curve of NSC34 cells co-transfected with DDX3 siRNA (si-DDX3) and pEGFP-G93A-SOD1 or control siRNA (Con) and pEGFP-G93A-SOD1 as determined by MTS assay at different time points. Each data point is the mean ± SD of three independent experiments. B. Representative images showing the neurites of NSC34 cells co-transfected with si-DDX3 and pEGFP-G93A-SOD1 or Con and pEGFP-G93A-SOD1 at different time points. Bar = 200 µm. C. Bar chart showing the percentage of cells having one or more neurites in the NSC34 cells co-transfected with si-DDX3 and pEGFP-G93A-SOD1 or Con and pEGFP-G93A-SOD1. Each data point is the mean ± SD of three independent experiments. D. Bar chart showing the lengths of the cell’s longest neurite in the NSC34 cells co-transfected with si-DDX3 and pEGFP-G93A-SOD1 or Con and pEGFP-G93A-SOD1. Each data point is the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, vs. Con and pEGFP-G93A-SOD1.
Recent study reported that CK1ε was closely related to neurite growth [17]. To determine whether knockdown of DDX3, which is a regulatory subunit of CK1ε, was associated with neurite growth, NSC34 cells were co-transfected with DDX3 siRNA and pEGFP-G93A-SOD1 and control siRNA pEGFP-G93A-SOD1 followed by treatment with 10 µmol/L RA for two days to induce the neurite outgrowth and then the length and number of cell projections were investigated. Consulting the length of the neurites against the cell body, both the number and length of neurites appeared to be apparently decreased after DDX3 inhibition. At 0, 24 and 48 h after co-transfection, we used a microscope to obtain the length and number of cell projections. The percentages of cells having one or more neurites in the NSC34 cells co-transfected with control siRNA and pEGFP-G93A-SOD1 were 18.44% ± 2.95%, 18.26% ± 2.55% and 17.79% ± 1.58% at 0, 24 and 48 h, whereas the percentages of cells having one or more neurites in the NSC34 cells co-transfected with DDX3 siRNA and pEGFP-G93A-SOD1 were 18.27% ± 2.98%, 12.09% ± 2.82% and 5.95% ± 2.10% at 0, 24 (p < 0.001) and 48 h (p < 0.001) (Figure 5B, 5C). Moreover, the lengths of the NSC34 cell’s longest neurite in control siRNA and pEGFP-G93A-SOD1 co-transfected cells were 89.31 ± 8.80 µm, 91.35 ± 10.28 µm and 94.06 ± 11.40 µm at 0, 24 and 48 h, whereas the lengths of the NSC34 cell’s longest neurite in DDX3 siRNA and pEGFP-G93A-SOD1 co-transfected cells were 88.84 ± 10.59 µm, 54.11 ± 5.07 µm and 28.01 ± 2.51 µm at 0, 24 (p < 0.001) and 48 h (p < 0.001) (Figure 5B, 5D). Knockdown of DDX3 inhibits the neurite outgrowth of SOD1 mutant NSC34 cells, suggesting that DDX3 is essential for neurite outgrowth.
Discussion
RNA helicases are a large family of proteins with a distinct motif, referred to as DEAD/H (Asp-Glu-Ala-Asp/His). The exact functions of all the DEAD/H box proteins are unknown. However, these proteins are associated with several aspects of RNA metabolism, including translation, ribosome biogenesis and pre-mRNA splicing [8]. In addition, DEAD/H box proteins play an important role in the regulation of nuclear-cytoplasmic transport and organellar gene expression. As a member of this RNA helicase family, DDX3 has been identified in a variety of cellular biogenesis processes, including cell cycle regulation, cellular differentiation, cell survival and apoptosis [23]. Knockdown of DDX3 expression reduces growth and proliferation, most likely by impeding the G1/S-phase transition of the cell cycle through cyclin D1 and cyclin E1 mRNA translation [22]. DDX3 has also been associated with cancer biogenesis [24-27]. At present, the research on DDX3 mainly focuses on the aspects of tumor and virus infection. Fewer studies on DDX3 is found on neural development and neurodegenerative diseases. Changes in the expression of DDX3 in the spinal cord of ALS have not yet been detected and the role and mechanisms of DDX3 in the pathogenesis remain uncertain.
In the present study, we first investigated the expression of DDX3 in the spinal cord of SOD-G93A ALS transgenic mice. Results showed that compared with wild-type mice, DDX3 protein in ALS mice was up-regulated at 108 and 122 d and DDX3 mRNA was all up-regulated significantly at 95, 108 and 122 d. Altered expression of DDX3 was found in ALS mice. DDX3 positive cells were mainly distributed in the anterior horn of spinal cord where motor neuron degenerated and co-localized with β-tubulin III-labeled neurons not in GFAP-labeled astrocytes, suggesting that the altered expression of DDX3 was closely related to motor neuron degeneration of ALS.
Chen et al. [10] reported that DDX3 affected the neurite outgrowth of both the cultured rat embryonic cortical neurons and N2A cells. The inhibition of DDX3 activity or expression in neonatal mice impaired dendritic outgrowth and spine formation of hippocampal neuron. These results indicate that DDX3 modulates neurite development. To assess the neuronal function of DDX3 in the pathogenesis of ALS, we investigated the effect of DDX3 on neuron outgrowth in vitro and found that the percentages of cells with neurite in DDX3 siRNA and pEGFP-G93A-SOD1 co-transfected NSC34 cells decreased significantly at 24 and 48 h after co-transfection compared with those in control siRNA and pEGFP-G93A-SOD1 co-transfected NSC34 cells. Moreover, the lengths of neurites in DDX3 siRNA and pEGFP-G93A-SOD1 co-transfected cells were also shorter than those in control siRNA and pEGFP-G93A-SOD1 co-transfected cells at 24 and 48 h after co-transfection. Data showed that DDX3 knockdown inhibits neurite outgrowth of SOD1 mutant NSC34 cells in vitro. DDX3 is essential for neurite outgrowth. The results of MTS assay showed that knockdown of DDX3 also reduced the cell proliferation and survival of SOD1 mutant NSC34 cells. All the data suggested that DDX3 is indeed required for cell growth.
Recent evidence has identified that DDX3 acts as an activator of CK1ε in the Wnt-β-catenin signaling pathway [11,12]. CK1ε belongs to a family of serine/threonine-specific kinases comprising seven members (α, β, γ1, γ2, γ3, δ and ε) with a highly conserved kinase domain and divergent C-terminal tails. CK1ε has been implicated in cell proliferation, migration, differentiation and cell cycle progression [28,29]. CK1ε also functions as important regulators of human health conditions such as neurodegenerative diseases, somatic tumor formation and pain [30,31].
Bischof et al. [17] reported that as one of the important signal molecules of Wnt signaling pathway, CK1ε was expressed in the growth cones of both mature retinal ganglion cells and PC12 cells and provided evidence that CK1ε activity was essential for neurite growth and extension. Studies revealed that CK1ε was expressed in all tissues, especially in the brain by Western blot and immunohistochemistry analyses [32]. CK1ε activity is also implicated in human neurodegenerative diseases [28]. An increase in CK1ε expression has been described in the human Alzheimer’s disease (AD) brain. Overexpression of active CK1ε leads to an increase in Aβ peptide production. Conversely, CK1-specific inhibitors significantly reduced endogenous Aβ peptide production, indicating that CK1ε represents a therapeutic target for the prevention of Aβ formation in AD [33]. Studies examining the role of CK1ε in ALS remain limited.
In this study, no significant change was observed in the level of CK1ε mRNA between wild-type mice and ALS mice, but CK1ε protein in the spinal cord of ALS mice was all down-regulated significantly at 95, 108 and 122 d compared with wild-type mice. Results obtained from double immunofluorescence labeling confirmed that CK1ε was expressed mainly in neurons not in astrocytes, indicating that the altered expression of CK1ε protein was also related to the degeneration of motor neuron of ALS. CK1ε mRNA remained unchanged, whereas CK1ε protein changed significantly because other regulatory mechanisms, such as the regulation of microRNA, may exist. The expression of microRNA was different in the pathogenesis of ALS [34]. The regulation of CK1ε by microRNA and its regulatory mechanism must be further explored. Our results showed that most of the CK1ε positive cells distributed in the ventral horn of gray matter where motor neuron degenerated and expressed in the β-tubulin III labeled neurons, suggesting that the change in expression of CK1ε may be closely related to ALS motor neuron degeneration.
The presence of a functional link between DDX3 and CK1ε in the progression of ALS was determined by co-immunoprecipitation experiments of endogenous protein. Results showed that DDX3 can band with CK1ε directly in vivo, consistent with the finding of Cruciat et al. [11]. He et al. [35] also found that DDX3 could function via the CK1ε/Dvl2 axis. The interaction of DDX3 with CK1ε was also confirmed by Co-IP of NSC34 cell transfected with pEGFP-G93A-SOD1, but unfortunately no interaction was detected between them. The reason may be related to the low content of endogenous protein in NSC34 cells.
Conclusions
Our study demonstrated that altered expression of DDX3 and CK1ε was found in the spinal cord of SOD1-G93A ALS transgenic mice. DDX3 binding with CK1ε was closely related to motor neuron degeneration of ALS by affecting neurite outgrowth. DDX3 may serve as a promising therapeutic target for the treatment of ALS. These findings may have important implications for developing therapeutic strategy treatment of motor neuron degeneration.
Acknowledgements
We thank Angelo Poletti (the University of Milan, Italy) for providing SOD1 plasmids. We also thank the key lab of neurological disease and regeneration & repair. This research project was supported by the National Natural Science Foundation of China (grant number 81401066 and 81271413), the Shandong Province Natural Science Foundation of China (grant number ZR2012HQ021, ZR2016HM60 and ZR2016HL20), the Shandong Province Science and Technology Development Program of China (grant number 2012GSF11827), the Shandong Provincial Education Department of China (grant number J11LF16), Shandong Medical and Health Science and Technology Development Program of China (grant number 2016WS0691 and 2016WS0666), the Science and Technology Development Program of Weifang in China (grant number 201301074) and the Shandong Province Taishan Scholar Project of China.
Disclosure of conflict of interest
None.
References
- 1.Bonafede R, Mariotti R. ALS pathogenesis and therapeutic approaches: the role of mesenchymal stem cells and extracellular vesicles. Front Cell Neurosci. 2017;11:80. doi: 10.3389/fncel.2017.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor JP, Brown RH Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirianni AC, Jiang J, Zeng J, Mao LL, Zhou S, Sugarbaker P, Zhang X, Li W, Friedlander RM, Wang X. N-acetyl-l-tryptophan, but not N-acetyl-d-tryptophan, rescues neuronal cell death in models of amyotrophic lateral sclerosis. J Neurochem. 2015;134:956–968. doi: 10.1111/jnc.13190. [DOI] [PubMed] [Google Scholar]
- 4.Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, Nicholson G, Ravits J, Shaw PJ, Swash M, Talbot K, Traynor BJ, Van den Berg LH, Veldink JH, Vucic S, Kiernan MC. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 6.Rosenblum LT, Shamamandri-Markandaiah S, Ghosh B, Foran E, Lepore AC, Pasinelli P, Trotti D. Mutation of the caspase-3 cleavage site in the astroglial glutamate transporter EAAT2 delays disease progression and extends lifespan in the SOD1-G93A mouse model of ALS. Exp Neurol. 2017;292:145–153. doi: 10.1016/j.expneurol.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tipassociated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14:188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soto-Rifo R, Ohlmann T. The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wiley Interdiscip Rev RNA. 2013;4:369–385. doi: 10.1002/wrna.1165. [DOI] [PubMed] [Google Scholar]
- 10.Chen HH, Yu HI, Tarn WY. DDX3 modulates neurite development via translationally activating an RNA regulon involved in Rac1 activation. J Neurosci. 2016;36:9792–9804. doi: 10.1523/JNEUROSCI.4603-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 12.Wrighton KH. Cell signalling: DDX3 in command of CK1epsilon. Nat Rev Mol Cell Biol. 2013;14:192. [PubMed] [Google Scholar]
- 13.Chen Y, Guan Y, Liu H, Wu X, Yu L, Wang S, Zhao C, Du H, Wang X. Activation of the Wnt/beta-catenin signaling pathway is associated with glial proliferation in the adult spinal cord of ALS transgenic mice. Biochem Biophys Res Commun. 2012;420:397–403. doi: 10.1016/j.bbrc.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Guan Y, Zhang Z, Liu H, Wang S, Yu L, Wu X, Wang X. Wnt signaling pathway is involved in the pathogenesis of amyotrophic lateral sclerosis in adult transgenic mice. Neurol Res. 2012;34:390–399. doi: 10.1179/1743132812Y.0000000027. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Guan Y, Chen Y, Zhang C, Shi C, Zhou F, Yu L, Juan J, Wang X. Expression of Wnt5a and its receptor Fzd2 is changed in the spinal cord of adult amyotrophic lateral sclerosis transgenic mice. Int J Clin Exp Pathol. 2013;6:1245–1260. [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Guan Y, Chen Y, Li X, Zhang C, Yu L, Zhou F, Wang X. Role of Wnt1 and Fzd1 in the spinal cord pathogenesis of amyotrophic lateral sclerosis-transgenic mice. Biotechnol Lett. 2013;35:1199–1207. doi: 10.1007/s10529-013-1199-1. [DOI] [PubMed] [Google Scholar]
- 17.Bischof J, Muller A, Fander M, Knippschild U, Fischer D. Neurite outgrowth of mature retinal ganglion cells and PC12 cells requires activity of CK1delta and CK1epsilon. PLoS One. 2011;6:e20857. doi: 10.1371/journal.pone.0020857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler C, Dinekov M, Gotz J. Granulovacuolar degeneration and unfolded protein response in mouse models of tauopathy and abeta amyloidosis. Neurobiol Dis. 2014;71:169–179. doi: 10.1016/j.nbd.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, Ju J, Pu L, Du H, Wang X. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 2015;7:1574–1587. [PMC free article] [PubMed] [Google Scholar]
- 20.Poletti A, Cattaneo E, Taroni F. The neurotoxicity of mutant proteins 20 years after the discovery of the first mutant gene involved in neurodegeneration. Foreword. Prog Neurobiol. 2012;97:53. doi: 10.1016/j.pneurobio.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Luo X, Guo Z, Xiong A, Dong H, Zhang Q, Liu C, Zhu J, Wang H, Yu N, Zhang J, Hong Y, Yang L, Huang J. Neuritin inhibits notch signaling through interacted with neuralized to promote the neurite growth. Front Mol Neurosci. 2017;10:179. doi: 10.3389/fnmol.2017.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai MC, Chang WC, Shieh SY, Tarn WY. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30:5444–5453. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariumi Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis and viral infection. Front Genet. 2014;5:423. doi: 10.3389/fgene.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Mao Y, Zhou J, Zhao Y, Cao Y, Chen X. Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am J Cancer Res. 2016;6:387–402. [PMC free article] [PubMed] [Google Scholar]
- 25.Wu DW, Lin PL, Cheng YW, Huang CC, Wang L, Lee H. DDX3 enhances oncogenic KRA sinduced tumor invasion in colorectal cancer via the betacatenin/ZEB1 axis. Oncotarget. 2016;7:22687–22699. doi: 10.18632/oncotarget.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heerma van Voss MR, Schrijver WA, Ter Hoeve ND, Hoefnagel LD, Manson QF, van der Wall E, Raman V, van Diest PJ. The prognostic effect of DDX3 upregulation in distant breast cancer metastases. Clin Exp Metastasis. 2017;34:85–92. doi: 10.1007/s10585-016-9832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen HH, Yu HI, Cho WC, Tarn WY. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34:2790–2800. doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 28.Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Piao S, Lee SJ, Xu Y, Gwak J, Oh S, Park BJ, Ha NC. CK1epsilon targets Cdc25A for ubiquitin-mediated proteolysis under normal conditions and in response to checkpoint activation. Cell Cycle. 2011;10:531–537. doi: 10.4161/cc.10.3.14757. [DOI] [PubMed] [Google Scholar]
- 30.Ghoshal N, Smiley JF, DeMaggio AJ, Hoekstra MF, Cochran EJ, Binder LI, Kuret J. A new molecular link between the fibrillar and granulovacuolar lesions of alzheimer’s disease. Am J Pathol. 1999;155:1163–1172. doi: 10.1016/S0002-9440(10)65219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai E, Kurihara T, Kouchi K, Saegusa H, Zong S, Tanabe T. Upregulation of casein kinase 1epsilon in dorsal root ganglia and spinal cord after mouse spinal nerve injury contributes to neuropathic pain. Mol Pain. 2009;5:74. doi: 10.1186/1744-8069-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utz AC, Hirner H, Blatz A, Hillenbrand A, Schmidt B, Deppert W, Henne-Bruns D, Fischer D, Thal DR, Leithauser F, Knippschild U. Analysis of cell type-specific expression of CK1 epsilon in various tissues of young adult BALB/c mice and in mammary tumors of SV40 T-Ag-transgenic mice. J Histochem Cytochem. 2010;58:1–15. doi: 10.1369/jhc.2009.954628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flajolet M, He G, Heiman M, Lin A, Nairn AC, Greengard P. Regulation of alzheimer’s disease amyloid-beta formation by casein kinase I. Proc Natl Acad Sci U S A. 2007;104:4159–4164. doi: 10.1073/pnas.0611236104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou F, Guan Y, Chen Y, Zhang C, Yu L, Gao H, Du H, Liu B, Wang X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int J Clin Exp Pathol. 2013;6:1826–1838. [PMC free article] [PubMed] [Google Scholar]
- 35.He TY, Wu DW, Lin PL, Wang L, Huang CC, Chou MC, Lee H. DDX3 promotes tumor invasion in colorectal cancer via the CK1epsilon/Dvl2 axis. Sci Rep. 2016;6:21483. doi: 10.1038/srep21483. [DOI] [PMC free article] [PubMed] [Google Scholar]


