ABSTRACT
Hanseniaspora uvarum (anamorph Kloeckera apiculata) is a predominant yeast on wine grapes and other fruits and has a strong influence on wine quality, even when Saccharomyces cerevisiae starter cultures are employed. In this work, we sequenced and annotated approximately 93% of the H. uvarum genome. Southern and synteny analyses were employed to construct a map of the seven chromosomes present in a type strain. Comparative determinations of specific enzyme activities within the fermentative pathway in H. uvarum and S. cerevisiae indicated that the reduced capacity of the former yeast for ethanol production is caused primarily by an ∼10-fold-lower activity of the key glycolytic enzyme pyruvate kinase. The heterologous expression of the encoding gene, H. uvarum PYK1 (HuPYK1), and two genes encoding the phosphofructokinase subunits, HuPFK1 and HuPFK2, in the respective deletion mutants of S. cerevisiae confirmed their functional homology.
IMPORTANCE Hanseniaspora uvarum is a predominant yeast species on grapes and other fruits. It contributes significantly to the production of desired as well as unfavorable aroma compounds and thus determines the quality of the final product, especially wine. Despite this obvious importance, knowledge on its genetics is scarce. As a basis for targeted metabolic modifications, here we provide the results of a genomic sequencing approach, including the annotation of 3,010 protein-encoding genes, e.g., those encoding the entire sugar fermentation pathway, key components of stress response signaling pathways, and enzymes catalyzing the production of aroma compounds. Comparative analyses suggest that the low fermentative capacity of H. uvarum compared to that of Saccharomyces cerevisiae can be attributed to low pyruvate kinase activity. The data reported here are expected to aid in establishing H. uvarum as a non-Saccharomyces yeast in starter cultures for wine and cider fermentations.
KEYWORDS: enology, genetic markers, chromosomes, ploidy, apiculatus yeast, glycolysis
INTRODUCTION
Alcoholic fermentation in the wine industry is generally attributed to the activity of the wine, beer, and baker's yeast Saccharomyces cerevisiae, which, in large-scale fermentations, is routinely added as a starter culture with a cell density of approximately 106 cells/ml (1). Even without the addition of such starter cultures, i.e., in spontaneous fermentations, S. cerevisiae usually dominates the fungal population after the first few days of vinification, explaining its importance for humankind over thousands of years (2, 3). Besides ethanol, S. cerevisiae produces several other compounds that determine the final wine quality, including glycerol, different esters, and fusel alcohols (4). Such compounds may be either desired or deleterious, frequently depending on their concentration. Acetate produced as a by-product of fermentation is generally regarded as unfavorable (5).
In contrast to its prevalence in the later stages of fermentation, S. cerevisiae displays a low abundance on the skin of grapes or in freshly prepared musts, with as few as one cell being found on 1,000 grapes (6). Instead, Hanseniaspora uvarum is the predominant yeast species and frequently constitutes more than 80% of the yeast population in must during early stages of fermentation (7, 8). H. uvarum is also still widely known for its imperfect form, Kloeckera apiculata, which led to the coining of the name “apiculate yeasts” due to its lemon-shaped cell morphology. It prevails within the first 48 h of fermentation, after which S. cerevisiae usually takes over and persists until the end of fermentation (9, 10). Initially, the decline of the H. uvarum population was attributed to its higher sensitivity to increasing ethanol concentrations during the course of fermentation. However, recent studies indicate that antimicrobial peptides secreted by S. cerevisiae are more important in reducing the population of non-Saccharomyces yeasts (11, 12). Nevertheless, H. uvarum forms a major part of the fungal microbiome in wine fermentations worldwide, including those in Austria (13), Brazil (14), China (15), France (16), Germany (17), Italy (18, 19), Portugal (20), Slovakia (21), and Spain (22). It should be noted that H. uvarum not only inhabits and ferments grapes but also resides on other fruits such as plums (23, 24) and apples, being a major factor in cider production (25, 26). This yeast is also found on more exotic substrates such as African coffee (27) and in chocolate production (28, 29). H. uvarum has further been suggested to be a useful agent in the biocontrol of molds such as Botrytis cinerea, one of the major plant pathogens (30). On the other hand, it is considered a spoilage yeast in orange juice, together with S. cerevisiae (31), and produces a killer toxin that is active against S. cerevisiae and Candida albicans (32, 33). These examples demonstrate the widespread nature, economical importance, and future potential of H. uvarum in food biotechnology.
Regarding vinification, different natural isolates of H. uvarum are known for their high capacities to form fruity esters but are also infamous for producing high levels of acetate and ethyl acetate (28, 34). Since during the first days of wine fermentation, H. uvarum can be present at cell densities similar to those of S. cerevisiae, even if the latter is added as a starter culture, its metabolism is expected to contribute significantly to the aroma profile and the final wine quality (5). In fact, after initial classifications as a spoilage yeast, different isolates have been tested for their performance in wine fermentations in mixed cultures with S. cerevisiae, with promising perspectives (35). In addition, a type strain of H. uvarum has been reported to be fructophilic, which could be of special importance regarding stuck fermentations attributed to an imbalance between glucose and fructose (19, 36).
Despite this obvious importance of H. uvarum in all kinds of fruit fermentations, data on its genetic makeup remained scarce. Thus, karyotyping approaches suggested the presence of 7 to 9 chromosomes, with high variability between different isolates (37, 38). As expected, H. uvarum also appears in deep-sequencing approaches addressing the microbiome in wine fermentations, since it dominates the yeast population and is readily identified by its ribosomal DNA (rDNA) sequences (39, 40). However, only a few genes have been isolated and sequenced, such as those encoding pyruvate decarboxylase or actin (40, 41). The mitochondrial genome of H. uvarum has an exceptional structure among fungi, as it is represented by a short, linear DNA molecule (42). Recently, two brief reports on whole-genome approaches have appeared for H. uvarum isolates, underlining the growing interest in this yeast, but with limited information regarding genome annotations (43) (GenBank accession number JPPO00000000).
Clearly, the determination of the physiological capacities of H. uvarum, with the future possibility of employing metabolic design to eliminate undesirable traits, would profit from more detailed genetic studies. We therefore initiated a genome-sequencing project with a readily available H. uvarum type strain. By applying next-generation sequencing approaches, an estimated 93% of the total genome sequence was deciphered, with >80% being assigned to contigs of >50 kb. From the annotation of these data and additional manual sequencing of specific PCR products, we identified more than 4,000 predicted protein-encoding genes, 3,043 of which were assigned putative functions by comparison with characterized yeast genomes. This included the structure of the highly repetitive rDNA repeats. We applied this information to identify genes coding for hexose transporters, the entire glycolytic pathway, and the fermentative pyruvate decarboxylase and alcohol dehydrogenases. Specific enzyme activities relevant for alcoholic fermentation were determined by using crude extracts of H. uvarum and S. cerevisiae. The functionality of genes encoding key enzymes of this pathway was confirmed by complementation studies with the respective S. cerevisiae deletion mutants.
RESULTS AND DISCUSSION
Selection of nonflocculating H. uvarum cells from type strain DSM2768.
Basic research on other yeast species, especially on the model yeasts S. cerevisiae and Kluyveromyces lactis, has profited from the use of small sets of laboratory strains after their original isolation from natural sources (44, 45). Therefore, we decided to start genetic work on H. uvarum using a readily available type strain (DSM2768; DMSZ [German Collection of Microorganisms and Cell Cultures], Braunschweig, Germany). As shown in Fig. 1A (left), cells of this strain tended to cluster in liquid cultures, which impedes the use of common microbiological and biochemical techniques such as the deduction of cell numbers from measurements of the optical density at 600 nm (OD600). In order to avoid this problem, a selection regime that involved inoculation in liquid yeast extract-peptone-dextrose (YEPD) medium, shaking overnight at 30°C, sedimentation without agitation for 1 h at room temperature, and reinoculation from the upper medium phase was applied. This was repeated more than 60 times over a period of several weeks, resulting in a strain producing exclusively nonflocculating single budding cells in liquid cultures (Fig. 1A, right), which retained the original cell size and shape and was used for all further investigations. Cells measured approximately 2 by 4 μm and are thus considerably smaller than those of S. cerevisiae, which are in the range of 5 by 7 μm. It should be noted that the smaller size of H. uvarum cells is also reflected in the relationship of the optical density of the cultures to live-cell platings. Thus, we found that an OD600 of 1 equals approximately 108 CFU of H. uvarum, as opposed to 107 CFU for S. cerevisiae. This should be taken into account when using H. uvarum for inoculations in cocultures with S. cerevisiae for experimental wine fermentations.
FIG 1.
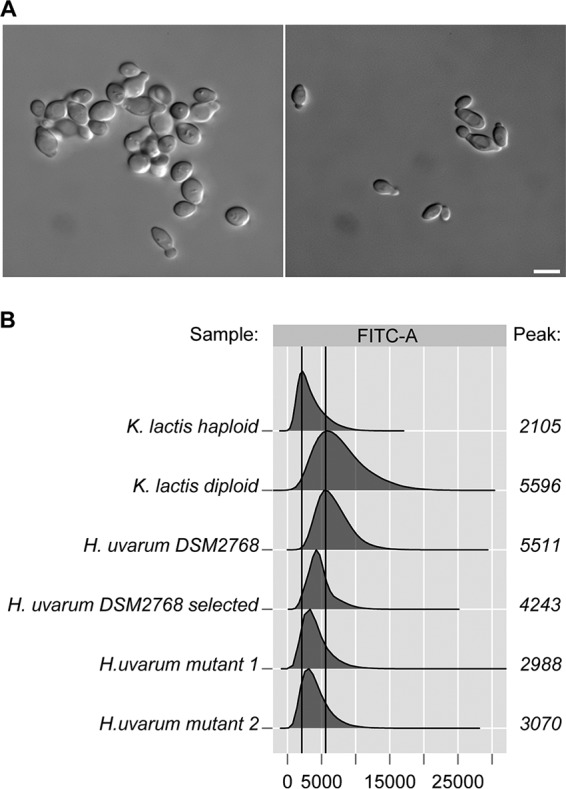
Counterselection of clustering in liquid cultures of H. uvarum type strain DSM2768. (A) Micrographs of cultures of the original H. uvarum type strain obtained from the stock culture (left) and after selection for nonclustering cells (right). Cells for image preparations were grown to logarithmic phase in YEPD medium. Bar, 2 μm. Note that clustering of the original strain was also observed by FACS analysis. (B) Flow cytometric analyses of DNA contents of different yeast strains. FACS analysis was performed on the yeast indicated and as described in Materials and Methods. The x axis corresponds to the fluorescence intensity of the DNA dye, and the number of cells is depicted on the y axis. One million cells were measured for each experiment. The peak for each distribution was calculated and plotted next to the corresponding graph. H. uvarum mutants 1 and 2 refer to Huura3 mutants selected on 5-FOA after prior treatment with nocodazole to induce aneuploidy. FITC, fluorescein isothiocyanate.
Examination of genome composition.
In order to assess the ploidy of the H. uvarum type strain employed here, we determined the DNA content per cell using Sybr green-stained G0 cells and flow cytometry. According to a recent study, this method provides the best alternative for estimations of the ploidy of yeast cells (46). As controls, haploid and diploid strains of the milk yeast Kluyveromyces lactis were employed, which should have a haploid genome size of 10.7 Mbp, i.e., slightly larger than that of H. uvarum, which is approximately 9 Mbp. As evident from Fig. 1B, the DNA content per cell for the type strain of H. uvarum more closely resembled that of the diploid control cultures than that of the haploids. Moreover, we were unable to select colonies that were resistant to 5-fluorootic acid (5-FOA), i.e., a mutation in the H. uvarum ura3 (Huura3) gene, after ethyl methanesulfonate (EMS) mutagenesis despite several attempts. Only after the prior induction of chromosome loss by nocodazole treatment, a method inspired by work on the diploid wine yeast Zygosaccharomyces bailii (47), were two Huura3 mutants obtained in presumed aneuploid derivatives (Fig. 1B). Taken together, these findings suggested that the genome of the H. uvarum type strain employed here is probably diploid. This assumption is further supported by the data from genome sequencing, where variants in 50% ± 5% of the reads were found in 10% of the annotated open reading frames (ORFs) of sequences with at least 20-fold read coverage. In addition, Fig. 2 shows that the mean of the fitted normal distribution of the percentage of all variant reads in open reading frames (n = 1,717) is very close to 50%. The fact that these mismatches are distributed over all large contigs assigned to the different chromosomes argues in favor of the yeast being entirely diploid rather than aneuploid for only some chromosomes.
FIG 2.
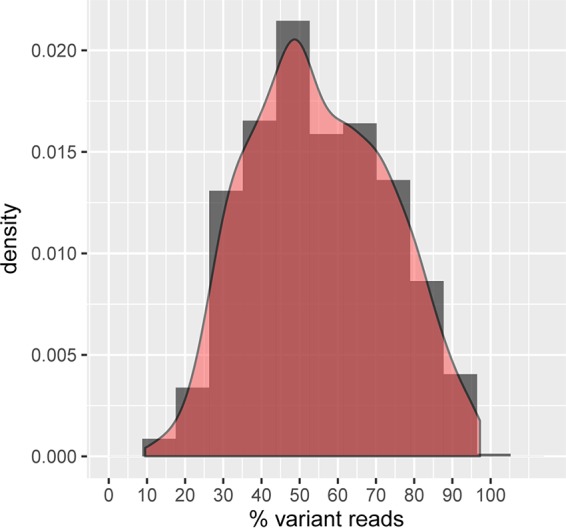
Reads in heterozygosity loci in the genome of the H. uvarum type strain. Shown is a histogram (dark gray) and a fitted normal distribution curve (red) of the percentage of reads displaying a variant in all loci showing heterozygosity (n = 1,717 loci). The peak at 50% variant reads suggests that the annotated genome is diploid.
Different natural isolates of H. uvarum were previously investigated for their chromosomal constitution by pulse-field gel electrophoresis (PFGE) and yielded different numbers of chromosomes and genome sizes (37). In order to check the adapted type strain described above for these parameters, we used FIGE (field inversion gel electrophoresis) separations. As exemplified in Fig. 3A, these analyses revealed that the type strain used here most likely contains 7 chromosomes, with chromosomes VI and VII having similar sizes, so they could not be separated (the genome analysis data presented below strongly indicate that two different chromosomes form this signal). The electrophoretic mobility in FIGE analyses is inversely proportional to the chromosome size; i.e., the larger chromosomes migrated faster. Based on this, all chromosomes detected add up to a genome size of ∼9 Mbp (Table 1). This correlates well with the 9.6-Mbp genome size calculated for another H. uvarum type strain (37). We further noticed a high degree of variation observed in karyotyping analyses of different natural isolates of H. uvarum with regard to the numbers of chromosomal bands and the total genome sizes, which we also found in our investigations of isolates from grapes from different locations in Germany and Spain (data not shown).
FIG 3.
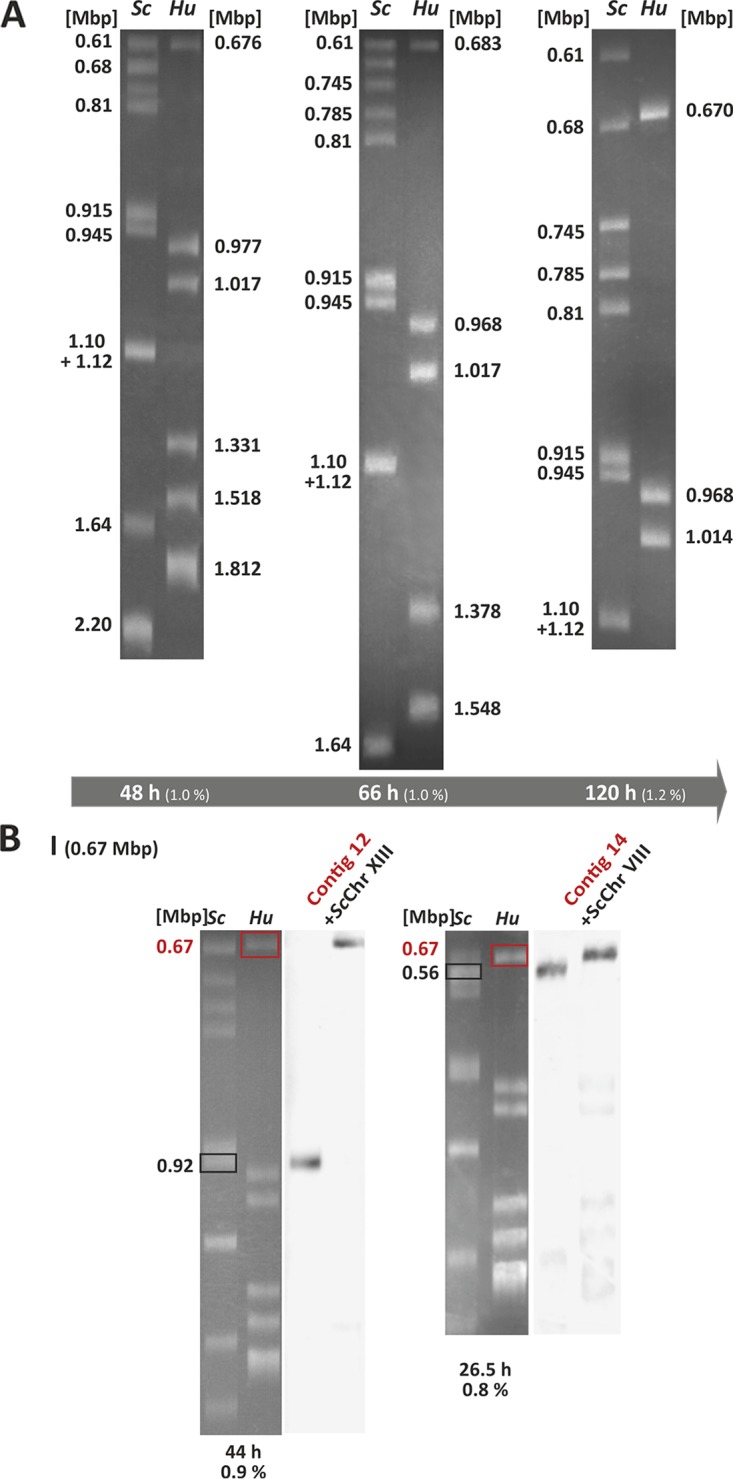
Karyotyping of H. uvarum. (A) FIGE analysis of the chromosomal constitution of the H. uvarum type strain. Chromosomal bands appearing after staining with ethidium bromide are shown, with three representative separation conditions (run times and agarose gel concentrations, as indicated at the bottom) being depicted. Sc, chromosomes from S. cerevisiae used as a size standard; Hu, chromosomes from H. uvarum. Numbers at the left and right indicate sizes in megabase pairs. (B) Example of Southern analysis for the assignment of contigs 12 and 14 to the smallest H. uvarum chromosome, chromosome I. DNA was stained with ethidium bromide (left) (white bands on a gray background). Gels were blotted and hybridized with a mixture of DIG-labeled probes with one probe for the indicated chromosomes of S. cerevisiae (right) (dark bands on a light background). Data from the entire set of experiments used to construct the genome map can be found in Fig. S1 in the supplemental material.
TABLE 1.
Chromosomes and sizes deduced from FIGE gels and sequence analysisb
| Chromosome | Contig | Size (Mbp) | Coverage (%) |
|---|---|---|---|
| I (0.67 Mbp) | 12 | 0.227 | |
| 14 | 0.215 | ||
| Sum | 0.44 | 66 | |
| II (0.97 Mbp) | 3 | 0.564 | |
| Sum | 0.56 | 58 | |
| III (1.02 Mbp) | 2 | 0.572 | |
| 4 | 0.412 | ||
| Sum | 0.98 | 97 | |
| IV (1.36 Mbp) | 7a | 0.345 | |
| 8 | 0.323 | ||
| 10 | 0.263 | ||
| 18 | 0.162 | ||
| Sum | 1.09 | 81 | |
| V (1.52 Mbp) | 6 | 0.355 | |
| 11 | 0.251 | ||
| 15 | 0.197 | ||
| 17 | 0.186 | ||
| Sum | 0.99 | 65 | |
| VI and VII (1.77 Mbp) | |||
| VI | 5 | 0.364 | |
| 9 | 0.309 | ||
| 13 | 0.226 | ||
| 16a | 0.193 | ||
| Sum | 1.09 | 62 | |
| VII | 1 | 0.998 | |
| rDNA | 0.298 | ||
| 25 | 0.110 | ||
| Sum | 1.41 | 80 |
Contigs 7 and 16 give an additional signal on chromosome I.
Percent coverage gives the estimated percentage of the sequence obtained for each chromosome.
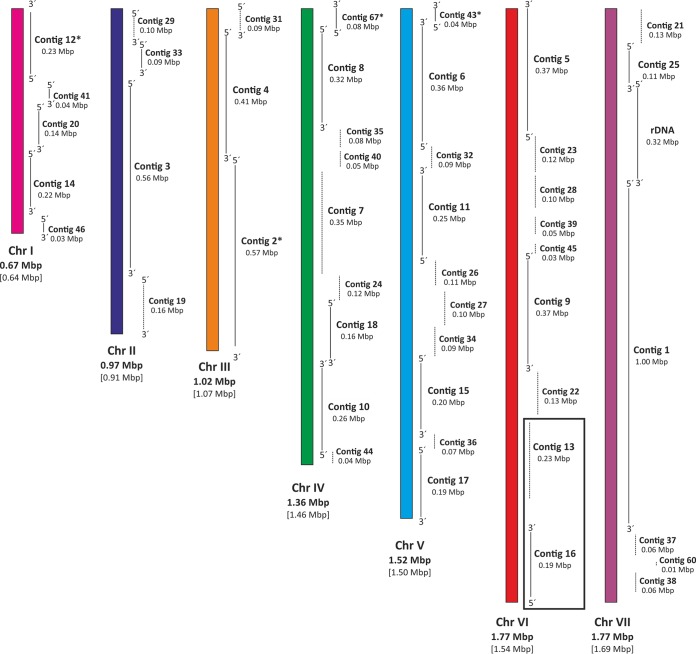
Gels used for the karyotyping analysis described above were further employed for Southern blot analyses to assign the larger contigs obtained by genome sequencing as described below to specific chromosomal bands. Mixtures of three PCR-generated probes from each contig were used for this purpose (Fig. 3B). All Southern data obtained are presented in Fig. S1 in the supplemental material. These results combined with results of synteny analyses of the genome sequence (see below) using the Yeast Gene Order Browser (YGOB) server (http://ygob.ucd.ie/) (48) allowed us to create an approximate chromosome map of H. uvarum (Fig. 4) (for detailed descriptions of these analyses, see reference 49). An indication of the validity of these analyses was provided by the finding that outward primers designed for contigs 13 and 16 employed for PCR on genomic DNA of H. uvarum revealed that they were indeed located next to each other and separated by only a small gap of approximately 900 bp (Fig. S2).
FIG 4.
Draft of a genome map for H. uvarum. Sequence data, annotations, data from synteny analyses, and data from the Southern analyses described in the text and in Fig. S1 in the supplemental material were combined to assemble a predicted order of large contigs on the seven chromosomes. Contigs and contig sizes are indicated. Note that the gaps between contigs are most likely not drawn to scale, due to the lack of sequence information.
Sequence and annotation of the H. uvarum genome.
In order to facilitate future genetic manipulations and gain more insight into the phylogenetic position of H. uvarum, we decided to take advantage of the ease of modern sequencing techniques. Since longer reads significantly simplify the assembly of de novo-sequenced genomes, we utilized a combination of long PacBio RS reads with reads from a GS FLX run used for error correction, as described in Materials and Methods. As judged from the calculations of chromosome separations reported above, an estimated 93% of the total genome sequence could thus be obtained. In summary, 360 contigs with a total size of >9.7 Mb now represent the annotated genome.
Regarding its phylogenetic relationships, genome sequence and synteny analyses indicated that H. uvarum belongs to the group of yeasts not having undergone a whole-genome duplication (WGD) and is most closely related to Kluyveromyces lactis, more distantly related to Ashbya gossypii (Eremothecium gossypii), and clearly distinct from the Saccharomyces sensu stricto group of yeasts (Fig. 5). Interestingly, this positions H. uvarum quite close to the whole-genome duplication event in phylogenetic trees. It should be noted that our genome sequence available in GenBank (accession number APLS01000000) was recently included in a highly detailed phylogenetic tree of yeast evolution, which supports these conclusions (50).
FIG 5.
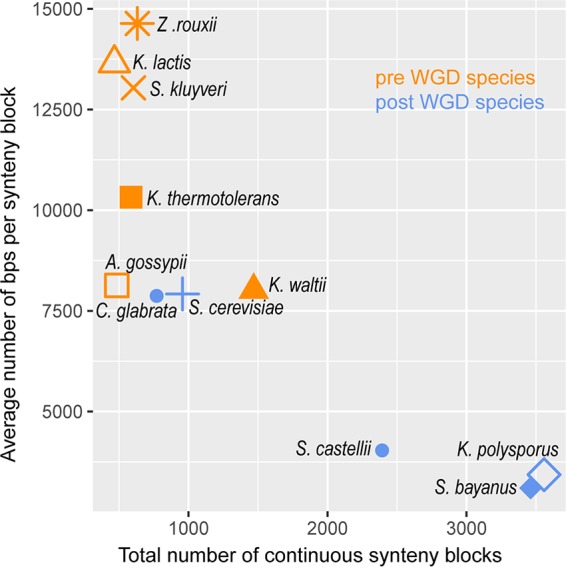
Synteny of the H. uvarum genome with the genomes of other yeast species. Relating the number of continuous synteny blocks to their average size, the annotated H. uvarum genome most closely resembles that of Kluyveromyces lactis and belongs to the group not having undergone a whole-genome duplication (pre- and post-WGD species). Genomes of the following species were obtained from YGOB (http://ygob.ucd.ie/) (48) (with YGOP file names and, if existent, alternative names of the species compared in parentheses): Zygosaccharomyces rouxii (Zrouxii_sequence.fsa), Kluyveromyces lactis (Klactis_sequence.fsa), Saccharomyces kluyveri (Lachancea kluyveri; Lkluyveri_sequence.fsa), Kluyveromyces thermotolerans (Lachancea thermotolerans; Lthermotolerans_sequence.fsa), Ashbya gossypii (Eremothecium gossypii; Egossypii_sequence.fsa), Candida glabrata (Cglabrata_sequence.fsa), Saccharomyces cerevisiae (Scerevisiae_sequence.fsa), Kluyveromyces waltii (Lachancea waltii; Lwaltii_sequence.fsa), Saccharomyces castellii (Ncastellii_sequence.fsa), Kluyveromyces polysporus (Vanderwaltozyma polyspora; Vpolyspora_sequence.fsa), and Saccharomyces bayanus (Suvarum_sequence.fsa).
With respect to genome composition, relating the size of chromosome VII with the assigned contigs and the size of a single rDNA repeat determined by our separate analyses of cloned PCR products (see Fig. S3A in the supplemental material), we calculated a repeat number of approximately 40 rDNA units to be present in the H. uvarum genome, which are located between contigs 1 and 25, presenting overlapping terminal sequences (Fig. 4). This is about 30% of the number of rDNA units reported for S. cerevisiae and may be attributed to the smaller size of H. uvarum cells (Fig. 1), which could explain the smaller number of ribosomes per cell.
Telomeres are also chromosomal structures that are difficult to assign by genome-sequencing approaches due to the large number of short nucleotide repeats. We found 28 small contigs with putative telomeric sequences. Their general composition is depicted in Fig. S3B. This number well exceeds the 14 telomeres expected from the presence of seven chromosomes and may be explained by the small sizes of the contigs on which these sequences are located, owing to their high degree of redundancy; i.e., several of these contigs may in fact constitute parts of the same chromosome ends.
As mentioned above, H. uvarum is the sexual form of anamorphic K. apiculata and should therefore undergo meiosis and produce spores (51). Despite several attempts with the H. uvarum type strain used here, we were unable to detect any hint of sporulation or sexual reproduction. In fact, genome analyses did not reveal the presence of a dimorphic mating type locus or of further silenced mating gene copies. Although the lack of these features could be assigned to the 7% of the genome not yet sequenced, alternative explanations are that sexual reproduction has been lost, at least in this type strain, or that the mating loci escaped detection by homology and synteny alignments due to high sequence divergence. In this context, it should be noted that a high degree of genetic variability was observed in numerous natural isolates of H. uvarum from wineries (52). Moreover, the loss of sexual reproduction may not be uncommon, as it is frequently found even in strains of S. cerevisiae used by fermentation industries (2).
The annotation of 3,010 protein-encoding genes also indicated a number of physiological capacities with implications for wine making, which are compiled in Tables S1 to S3 in the supplemental material. Thus, homologs of all genes encoding enzymes necessary for glucose fermentation, including putative hexose transporters, were found. However, as expected from a nonduplicated genome, less redundancy was observed, with a maximum of 8 putative hexose transporter homologs compared to 20 in the S. cerevisiae genome (53) (Table S1). Likewise, only one homolog each of the two hexokinase and two enolase genes present in S. cerevisiae could be identified in the H. uvarum genome. Regarding possible effects on wine quality, we were able to identify 28 genes in H. uvarum with putative functions in the production of aroma compounds, which include esterases and glycosidases but also overlap the subset involved in central carbohydrate metabolism in the production of glycerol and aldehydes (Table S2). Interestingly, at least six putative genes for xylose-degrading activities were identified, which are represented by only one homolog in either S. cerevisiae or Aspergillus nidulans, supporting the notion that H. uvarum contributes significantly to the production of aroma compounds in the process of vinification. Finally, survival in vineyards and in wine fermentations requires the presence of various stress response pathways in yeast cells (54), which we also detected in H. uvarum (Table S3). These include homologs of most components of the yeast cell wall integrity (CWI) pathway, again with less redundancy of components of the downstream mitogen-activated protein kinase (MAPK) pathway (55). Interestingly, H. uvarum seems to have a single isoform of protein kinase C (Pkc), which, as in S. cerevisiae, displays a prototypic structure of Pkc isoforms found in higher eukaryotes, including the regulatory homology domains mediating interactions with a small GTPase located near the N-terminal end (56, 57). In addition, homologs encoding proteins of the high-osmolarity glycerol (HOG) pathway (58) were identified. Although some intermediary components have not been annotated, the homologs detected indicate that the pathway, like its counterpart in S. cerevisiae, is bifurcated in the upstream signaling components. We also found several genes with putative functions in the oxidative stress response (59) (Table S3). Although transcription factors such as Yap1 and Skn7 did not appear in this search, the presence of detoxifying enzymes such as Sod1 and Gsh1/Gsh2 indicates that H. uvarum encounters and is well equipped to cope with oxidative stress. Together, these findings suggest that H. uvarum disposes of the necessary mechanisms to respond properly to stresses encountered in the vinification process.
Comparative investigation of fermentative capacities of H. uvarum and S. cerevisiae.
H. uvarum and other non-Saccharomyces yeasts that dominate the first stage of spontaneous wine fermentations have a limited capacity for alcohol production (60). We decided to address this question in more detail by determining the specific activities of all 12 enzymes involved in alcoholic fermentation in H. uvarum under different growth conditions and comparing them to those of a diploid laboratory strain of S. cerevisiae (HHD1). The latter is derived from the CEN.PK series (61), has a fermentation capacity and an aroma profile similar to those of commercial yeast strains, and is thus also suitable for spirit production (62).
As summarized in Table 2, most of the specific enzyme activities measured in extracts from cells grown in the presence of 1% glucose plus 1% fructose are in the same range for H. uvarum and S. cerevisiae; i.e., differences are <2-fold. This indicates a general conservation and similar capacities of the glycolytic pathways and the subsequent reactions leading to ethanol production in the two species. However, pyruvate kinase shows a striking difference, with H. uvarum having a >15-fold-lower specific activity than that of S. cerevisiae. This is especially interesting since pyruvate kinase catalyzes the second irreversible step specific for glycolysis after phosphofructokinase (PFK), thus being an ideal target to control metabolic flux. In fact, pyruvate kinase has been suggested to be a major determinant at the branching point of respiratory and fermentative sugar degradation (63). Of note, fructose-1,6-bisphosphate, the product of the PFK reaction, is a potent allosteric activator of pyruvate kinase in S. cerevisiae, thus connecting the two controlling steps of sugar degradation (64). We conclude that the reduced activity of pyruvate kinase in H. uvarum may be a major factor explaining the lower fermentative capacity of this yeast than of S. cerevisiae and also its classification as a Crabtree-negative yeast (65). It should be noted that specific pyruvate kinase activities are increased in both S. cerevisiae (approximately 2-fold) and H. uvarum (approximately 10-fold) when cells are grown in the presence of 20% as opposed to 2% sugar, i.e., under conditions similar to those of must fermentations (Fig. S4). Nevertheless, activities are still 3-fold lower in H. uvarum than in S. cerevisiae and are expected to decrease during the course of wine fermentations, as sugars are degraded. Although the specific activities of several other enzymes tested after growth with high sugar concentrations were also higher, the increases were similar for H. uvarum and S. cerevisiae, not affecting the ratios of <2-fold differences between the two species. The same is true for the generally lower specific activities measured after growth on 2% ethanol as a nonfermentable carbon source (Fig. S4).
TABLE 2.
Comparative analysis of specific enzyme activities relevant for alcoholic fermentation in H. uvarum and S. cerevisiaea
| Enzyme | Abbreviation | Mean sp act (mU/mg protein) ± SD |
Ratio of sp act of S. cerevisiae/H. uvarum | |
|---|---|---|---|---|
| Hanseniaspora uvarum | Saccharomyces cerevisiae | |||
| Hexokinase | Hxk | 525 ± 24 | 594 ± 38 | 1.1 |
| Phosphoglucose isomerase | Pgi | 1,192 ± 23 | 1,416 ± 118 | 1.2 |
| Phosphofructokinase | PFK | 361 ± 22 | 556 ± 84 | 1.5 |
| Aldolase | Fba | 420 ± 128 | 459 ± 87 | 1.1 |
| Triosephosphate isomerase | Tpi | 22,581 ± 2,847 | 26,804 ± 2,329 | 1.2 |
| Triosephosphate dehydrogenase | Tdh | 718 ± 305 | 937 ± 297 | 1.3 |
| Phosphoglycerate kinase | Pgk | 753 ± 157 | 1,033 ± 267 | 1.4 |
| Phosphoglycerate mutase | Gpm | 1,379 ± 243 | 1,987 ± 366 | 1.4 |
| Enolase | Eno | 1,251 ± 255 | 2,015 ± 326 | 1.6 |
| Pyruvate kinase | Pyk | 193 ± 61 | 3,263 ± 362 | 16.9 |
| Pyruvate decarboxylase | Pdc | 494 ± 169 | 707 ± 160 | 1.4 |
| Alcohol dehydrogenase | Adh | 1,727 ± 201 | 3,802 ± 699 | 2.2 |
Specific enzyme activities are given in milliunits per milligram of protein at 30°C. The means of data from at least three biological and three technical replicates for each enzyme and yeast species were determined, and standard deviations were calculated. Cells were grown in rich medium with 1% glucose and 1% fructose (YEPD medium plus fructose) prior to the preparation of crude extracts. For a comparison of specific activities after growth with higher sugar concentrations and with ethanol as a carbon source, see Fig. S4 in supplemental material.
From data obtained for a different H. uvarum isolate grown in chemostat cultures at different dilution rates, it was previously concluded that glucose degradation is limited by a step “before pyruvate formation,” which is in good agreement with our findings (66). Those authors determined the specific activities of a number of fermentative enzymes and those of pyruvate dehydrogenase and acetyl coenzyme A (acetyl-CoA) synthase, concluding that the low activity of the latter is a major cause of acetate production. It should be noted that the specific activities of the fermentative enzymes reported here differ considerably from those determined in the work cited above, which may be attributable to different assay conditions. Besides the use of batch cultures here, fructose-2,6-bisphosphate and fructose-1,6-bisphosphate, potent allosteric activators of phosphofructokinase and pyruvate kinase, respectively, for example, were apparently not added to mixtures in the enzymatic assays reported previously (66). Nevertheless, the aim of the present study was to compare specific enzyme activities of the H. uvarum type strain and S. cerevisiae. Since the same assay mixtures were used to assess activities in crude extracts from both species, it allowed us to assess the relative contribution of each step to alcoholic fermentation.
Functional analyses of genes encoding phosphofructokinase and pyruvate kinase.
The data described above suggested crucial differences in the activity of pyruvate kinase in H. uvarum compared to that in S. cerevisiae. These differences may be attributed to either different regulatory circuits governing PYK1 gene expression in the two yeast species or inherent properties of the enzyme itself. In order to distinguish between these possibilities, we decided to express the HuPYK1 gene in a pyk1 deletion mutant of S. cerevisiae. Indeed, HuPYK1 introduced on a CEN/ARS vector rescued the glucose-negative growth phenotype of the deletion strain VWH3B (pyk1::HIS3), indicating functional complementation. This was confirmed by enzyme assays of cultures grown in rich medium with 2% glucose, yielding specific pyruvate kinase activity of 465 ± 100 mU/mg protein, as determined by using two independent transformants carrying plasmid pJJH2144 and three technical replicates. This value is higher than the specific activity determined for the wild-type H. uvarum strain described above under similar growth conditions (193 ± 39 mU/mg protein) (Table 2) but only approximately 15% of that observed for transformants with a plasmid carrying the S. cerevisiae PYK1 (ScPYK1) gene (3,179 ± 270 mU/mg protein) or diploid wild-type strain HHD1 (3,087 ± 163 mU/mg protein). However, placement of the HuPYK1 open reading frame under the control of the ScPYK1 promoter yielded 1,982 ± 82 mU/mg protein when it was introduced into VWH3B (pyk1::HIS3), demonstrating that the low specific activity is probably due to a low level of gene expression rather than being determined by the structure of the heterologously produced enzyme.
Finally, the key glycolytic enzyme PFK of S. cerevisiae is a hetero-octamer, composed of four α- and four β-subunits, which are encoded by the genes PFK1 and PFK2, respectively (67). While mutations in either of these two genes do not prohibit growth on glucose, pfk1 pfk2 double mutants are glucose negative (68, 69). This has been attributed to the conservation of all catalytic and allosteric domains in each of the encoded subunits, retaining the activity of homooligomeric forms in vivo but not in vitro (70). For H. uvarum, we also identified two homologous genes in the annotated sequence and designated them HuPFK1 and HuPFK2 based on sequence alignments and synteny analyses. These genes encode proteins with deduced molecular masses of 103,428 Da and 85,351 Da, respectively. It should be noted that the annotated genome sequence of HuPFK1 is lacking an adenine nucleotide at position +442 relative to the correct ATG translation start codon, as confirmed by cloning and Sanger sequencing (GGAAGCCACCAAAAAGAAAAAAATTGCTG). Thus, the underlined sequence in fact contains five rather than four nucleotides, which leads to a considerable extension of the 5′ end of the predicted HuPFK1 open reading frame. For use in further studies, we therefore again obtained the entire coding sequences by Sanger sequencing of cloned PCR products and submitted the sequences of the genes under the accession numbers given in Materials and Methods.
When individually introduced on CEN/ARS vectors into an S. cerevisiae pfk1 pfk2 double-deletion mutant (HD114-8D [pfk1::HIS3 pfk2::HIS3]), either gene from H. uvarum complemented the glucose-negative phenotype, indicating that the subunits are functionally conserved and that they also exert catalysis in vivo. However, as for S. cerevisiae single-deletion mutants, PFK activity was below detectable levels in enzyme assays of crude extracts from strains carrying either of the HuPFK genes, even when introduced on multicopy vectors (Table 3). However, the introduction of both heterologous genes together on a CEN/ARS vector restored in vitro PFK activity in the double-deletion mutant, which amounted to approximately one-third of the specific activity determined for either the H. uvarum or S. cerevisiae wild-type strain. As expected, overproduction by the expression of the two genes from a multicopy vector yielded much higher activities. This indicates that PFK of H. uvarum is also an oligomeric enzyme and may form a hetero-octamer, like its counterparts in S. cerevisiae and K. lactis (71). The overall domain structure of the subunits, which retain both the catalytic and allosteric substrate binding sites, is consistent with the observation that sufficient glucose can be metabolized in vivo by either HuPfk1 or HuPfk2. However, as in S. cerevisiae, heterooligomerization seems to stabilize the enzyme structure, so in vitro activity can be observed only if both subunits are produced in one cell (70). The lower specific activity measured in the transformants with the two HuPFK genes carried on a CEN/ARS vector than in the wild-type strains of both species could be due to a lower expression level caused by the native HuPFK gene promoters in S. cerevisiae, analogous to the data presented above for HuPYK1. It should also be noted that heterooligomeric complexes between subunits of the two species either do not form or are not stable, since transformants of single Scpfk1 or Scpfk2 deletion mutants with the respective HuPFK genes did not restore measurable enzyme activity in crude extracts (data not shown).
TABLE 3.
Specific activities of phosphofructokinasea
| Strain (relevant genotype) | Plasmid | PFK gene(s) introduced | Mean sp act of PFK (mU/mg protein) ± SD |
|---|---|---|---|
| H. uvarum (HuPFK1 HuPFK2) | None | None | 413 ± 25 |
| HHD1 (PFK1 PFK2) | None | None | 326 ± 33 |
| HOD114-8D (pfk1::HIS3 pfk2::HIS3) | None | None | <1 |
| YCp111u | ScPFK1 | <1 | |
| pJJH2203 | ScPFK2 | <1 | |
| pJJH2200 | ScPFK1 ScPFK2 | 384 ± 27 | |
| pJJH2185 | HuPFK1 | <1 | |
| pJJH2208 | HuPFK1 | <1 | |
| pJJH1868 | HuPFK2 | <1 | |
| pJJH2209 | HuPFK2 | <1 | |
| pJJH1856new | HuPFK1 HuPFK2 | 63 ± 6 | |
| pJJH2211 | HuPFK1 HuPFK2 | 2,999 ± 232 |
Specific PFK activities were determined from at least three biological replicates and three technical replicates, with the exception of pJJH1856new, for which only two biological replicates were employed. Cells were grown in rich medium with 2% glucose.
The enzymatic determinations and data from sequence analyses were further substantiated by performing Western blot analyses using antiserum raised against ScPFK, which also detects the subunits of K. lactis PFK (72). As shown in Fig. 6, the HuPFK subunits migrate according to their deduced molecular weights, with the β-subunit encoded by HuPFK2 migrating much faster than its counterpart from S. cerevisiae. While all PFK subunits from different yeasts and fungi analyzed so far carry an N-terminal elongation of approximately 200 amino acids compared to their bacterial or mammalian homologs, this extension is missing in the β-subunit of H. uvarum. Determinations of PFK activities after controlled proteolysis, which cleaved off the N-terminal parts of the S. cerevisiae enzyme, demonstrated that these extensions are actually not required for catalysis (73). It is thus rather surprising that sequences encoding the N termini of PFK subunits should have been eliminated only during the evolution of the HuPFK2 gene and should not have occurred in other yeasts. With the growing number of reported genome sequences of other non-Saccharomyces wine-related yeasts, it will be interesting to see if this is indeed a single “evolutionary accident” confined to H. uvarum. If so, this would indicate a selective advantage of larger subunits whose biological significance remains to be elucidated.
FIG 6.
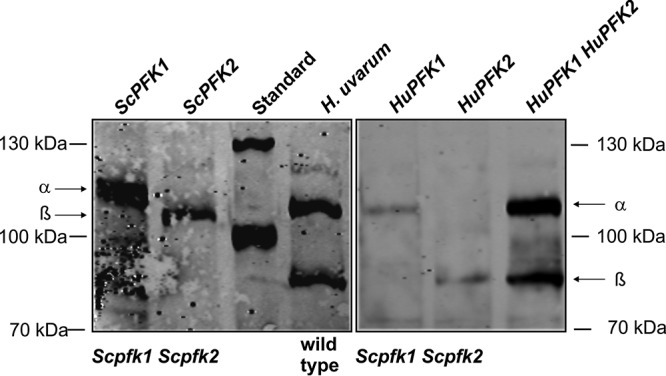
Western blot analysis of yeast phosphofructokinase. Total protein was prepared from strain HD114-8D (Scpfk1 Scpfk2) (see Materials and Methods for the complete genotype) carrying plasmids with the PFK genes indicated above the blots or from the adapted wild-type H. uvarum strain. Samples were separated by SDS-PAGE, blotted, and subjected to immunological detection with ScPFK antiserum as described in Materials and Methods. α and β refer to the PFK subunits encoded by PFK1 and PFK2 for S. cerevisiae (left) and H. uvarum (right). Plasmids employed were YCp111u (ScPFK1), pJJH2203 (ScPFK2), pJJH2208 (HuPFK1), pJJH2209 (HuPFK2), and pJJH2211 (HuPFK1 HuPFK2). Note that for reasons of detectability, ∼10-fold-larger amounts of protein were loaded for the samples with H. uvarum PFKs than for those containing the S. cerevisiae subunits, with the exception of the HuPFK1 HuPFK2 overproducer, for which only double the amount of total protein was loaded. HuPFK subunits produced from CEN/ARS vectors in the S. cerevisiae pfk1 pfk2 double-deletion strain, although barely detectable, confirmed the subunit sizes depicted here (data not shown).
Conclusions.
Here we present a comprehensive analysis of the genome of an H. uvarum type strain, including extensive annotation. This and other non-Saccharomyces yeasts are of growing interest for the production of fermented beverages, especially wine, due to their important contribution of desired aroma compounds (5). The genome sequence presented here provides the basis for future manipulations of the underlying pathways, e.g., for increasing the production of desired and reducing that of undesired metabolites. It also allows the assessment of physiological capacities. Thus, our studies indicate that in H. uvarum, pyruvate kinase activity could be a limiting step in alcoholic fermentation. This also has implications with regard to climate change and the corresponding increase in grape sugar content. Thus, cofermentations with non-Saccharomyces yeasts have been suggested as a measure to control increasing ethanol concentrations in wine (74). Although our functional studies of key glycolytic enzymes demonstrate the usefulness of the annotated genome sequence, they can be considered just a proof of principle. Further functional studies on enzymes involved in the production of aroma compounds, such as esterases or glycosylases, will follow. Moreover, this genome sequence provides the basis to study wine-related phenotypes, such as the capacity to flocculate and the signaling pathways involved in coping with environmental stress conditions during vinification (54). All these studies would greatly profit from the development of a system for the targeted genetic manipulation of H. uvarum, which is the most urgent subject to be addressed in future research on this yeast.
MATERIALS AND METHODS
Strains, plasmids, media, and culture conditions.
In this study, type strain DSM2768 of H. uvarum was obtained from the German Collection of Microorganisms. It is equivalent to ATCC 9774 (American Type Culture Collection). Presumably aneuploid derivatives with mutations in the HuURA3 gene were obtained by treatment with nocodazole prior to selection on medium containing 5-FOA. Haploid and diploid reference strains of Kluyveromyces lactis employed in fluorescence-activated cell sorter (FACS) analyses were type strains CBS2359 and KHO70, respectively (44). For the preparation of S. cerevisiae chromosomes as a size standard, strain BY4743 (Euroscarf, Frankfurt, Germany) was used. For comparisons of fermentative enzyme activities, we used diploid S. cerevisiae strain HHD1 (MATa/MATα ura3-52/URA3 LEU2/leu2-3,112) as a reference, which is a derivative of the CEN.PK series applied previously in fermentations for spirit production (62). HD114-8D (MATα ura3-52 leu2-3,112 his3-11,15 pfk1::HIS3 pfk2::HIS3) (75) served as a recipient strain for PFK gene expression, and VWH3B (MATα ura3-52 leu2-3,112 his3Δ1 trp1-289 pyk1::HIS3), also a derivative of the CEN.PK series, was used for the expression of genes encoding pyruvate kinase.
Standard procedures for the handling of yeast, Escherichia coli strain DH5α, and DNA were followed, as described previously (76). Genes from H. uvarum were amplified by PCR using the High Fidelity Taq polymerase kit from Roche (Mannheim, Germany) and cloned by restriction/ligation into yeast-E. coli shuttle vectors by either using natural restriction sites or adding the recognition motifs to the 5′ ends of the primer sequences, if required. Plasmids employed were based on the CEN/ARS vector YCplac33 (77) and the 2μm vector YEp352 (78). Genes with their flanking regions were amplified from genomic DNA preparations by PCR with appropriate primer pairs and cloned by conventional restriction/ligation. Sequences of oligonucleotides and the resulting plasmids are available upon request. Specifically, the PYK1 gene from S. cerevisiae was cloned as a BamHI/HindIII fragment into YCplac33 to yield pJJH2137. The gene from H. uvarum (HuPYK1) was also cloned as a BamHI/HindIII fragment into the same vector, yielding pJJH2144. To place the open reading frame of HuPYK1 between the promoter and terminator regions of ScPYK1, pJJH2145 was obtained by in vivo recombination using pJJH2137 as a recipient plasmid and a PCR fragment with HuPYK1 and the appropriate flanking regions for recombination. For phosphofructokinase clones, ScPFK1 was cloned as an SphI fragment into YCplac33 to yield YCp111u, and ScPFK2 was cloned as a SacI/SalI fragment to yield pJJH2203. The latter fragment was also inserted into YCp111u to give pJJH2200, which carries both ScPFK genes on the CEN/ARS vector. PFK genes from H. uvarum were obtained as follows. HuPFK1 was obtained as a SacI/HindIII fragment and cloned into YCplac33 (pJJH2185) and YEp352 (pJJH2208). HuPFK2 was obtained as a SacI/PstI fragment and cloned into YCplac33 (pJJH1868) and YEp352 (pJJH2209). Plasmids containing both HuPFK genes were obtained by cloning HuPFK2 as a SalI/SphI fragment into pJJH2208 to yield the multicopy plasmid pJJH2211 and by simultaneously cloning HuPFK1 as a BamHI/SacI fragment and HuPFK2 as a SacI/PstI fragment into YCplac33 linearized with BamHI/PstI to yield the CEN/ARS plasmid pJJH1856new.
Rich media were based on 1% yeast extract and 2% Bacto peptone (Difco) with 2% glucose (wt/vol) as a carbon source (YEPD), if not stated otherwise. Synthetic media were based on yeast nitrogen base with ammonium sulfate, supplemented with 2% glucose and a mixture of amino acids and bases as described previously (79), omitting compounds for plasmid selection as required. Mixtures of 10% (wt/vol) glucose plus 10% (wt/vol) fructose were used to mimic the initial stages of grape must fermentation, and 1% (wt/vol) each sugar was used for later stages.
Preparation of genomic DNA and sequencing.
The preparation of larger amounts of genomic DNA for sequencing of the H. uvarum genome was carried out with the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The resulting DNA was eluted in sterile, distilled water and showed an absorbance ratio at 260/280 nm of ≥1.8 and an absorbance ratio at 260/230 nm of ≥1.9.
LS454 sequencing using GS FLX Titanium technology (Roche, Basel, Switzerland) was done by Microsynth AG (Balgach, Switzerland), additional PacBio RS sequencing was performed by GATC Biotech (Constance, Germany), and data were combined to generate the final assembly. The assembly was performed by GATC Biotech using a hybrid approach for error correction of PacBio reads (80), with final assembly being performed by using Celera Assembler (May 2013 version).
Custom Sanger sequencing was performed by GATC Biotech (Cologne, Germany) with oligonucleotide primers obtained from Metabion (Munich, Germany).
Genome annotation and synteny analysis.
The main objective of the genome annotation was to accomplish high data reliability. As a first step, we therefore used the Yeast Genome Annotation Pipeline (YGAP) (81) (http://wolfe.ucd.ie/annotation/). This pipeline employs preexisting annotations of related species and also includes synteny information. YGAP resulted in a compilation of putative ORFs and a list of tRNA genes. The YGAP list contained 4,300 entries. We decided to further scrutinize this list since it also contained incomplete ORFs (e.g., those with missing start or stop codons). In addition, we found that automated annotation of intron-containing ORFs was not reliable, and we thus excluded all entries of ORFs that were continuous between a start codon and a stop codon. To this end, a list was generated from the assembly by using GetORF (part of the EMBOSS package) (82), and only entries present in the YGAP and GetORF lists were incorporated in further annotations. This resulted in the annotation of 3,010 protein-encoding sequences. Deduced tRNA genes were then added to the resulting list. Annotation of ribosomal DNA was performed with a BLAST search (83) using a sequence of H. uvarum rDNA as a query, which was obtained by the cloning of PCR-amplified DNA fragments and conventional Sanger sequencing. The sequence assembly and annotation were formatted according to GenBank guidelines. Further analyses of the data were performed by using software tools from the EMBOSS package and R statistical language v 3.3.2 (84). Synteny analysis was performed by using MUMmer version 3 (85). The SAMtools package (86) was used for variant analysis.
FIGE and Southern analysis.
Chromosome sizes in FIGE gels were determined by using LabImage 1D (Kapelan Bio-Imaging Solutions, Leipzig, Germany). A total of 25 gels obtained under various electrophoresis conditions were used to determine mean chromosome sizes. For Southern analyses, three different target sequences were chosen from each contig for PCR amplification with the oligonucleotides listed in Table 4. Probes for the detection of a number of S. cerevisiae chromosomes were also prepared by PCR with the oligonucleotides listed in Table 5. Strain BY4743, a diploid derivative of the S288C strain employed for the genome-sequencing project, was used as a source for the S. cerevisiae size standard (Euroscarf Collection, Frankfurt, Germany). Probes were prepared with the hexanucleotide digoxigenin (DIG) DNA labeling kit from Roche (Mannheim, Germany).
TABLE 4.
Oligonucleotides used to generate contig-specific probes used in Southern analysesa
| Contig | Forward primer | Reverse primer |
|---|---|---|
| 1 | TTCAATTCCGCAAGTGATGG | TATAGAGGCAGGTAACTGAG |
| CCATACATTTGCCAAGGTTC | CTTACCACCAAAGATCATTC | |
| CGCAAGCCAATGTACCAAAG | CACCGTTACACAACTACC | |
| 2 | AACGCTAACAATTCTACAAG | TTAGGCCCTATAGTCAAATC |
| ATATTGAAGTACCCGCAATG | TCTTGGTAACCATGACTAAC | |
| TCACCAACCACATCCAGTAG | AAGCATCTCTTCAACTGTAG | |
| 3 | ATTCTCCACTTAGGACTTAC | ATTCATCACCACCAGTATTG |
| GCATCAACTCGTCAAGATAG | GTAGACAGTGAGGTTGAAAG | |
| GTTCCTGTTCTGGTGAAATC | CTAGGCTCAGTCTCTACTTC | |
| 4 | TGTTCTCTGTTGGAGGATTC | GAGTTGCAAGATGCTGTTTC |
| CAACAACCAACTGGTTTCTC | AGCAGCTATGACTGCTAGAG | |
| TGCCGTTGATAGATATTCTG | ATGTAGATGCAGAATCTACC | |
| 5 | TTTCTACTTCGCTTAATTGC | TCGGTTACTTCAAGAAATGG |
| TGAACATGACCAGGAATTAG | TCCAGTATGAGGACATTTC | |
| AATTCGCTGTGGCAAATTGG | GTCAATCACCTTTGTTATCG | |
| 6 | TTGCATTAAACAGCGCAAAG | ACCAAATCCAACGACCAATG |
| TAAGACAGAGTCGTCTAGAG | AATTGCGGCTGATGAGTAAG | |
| TGGGTGCAAGAAATAGATGG | AACGTCTCCACTGCTCTATC | |
| 7 | AATCTGCTTCAGGCTGATAC | ATACCACTTCTTCTGGAATG |
| TCTTATCAGCCACATGTCAC | ATGGACTATTAGGCGATAAC | |
| CTTGCCTGGGTTGTTGATAG | TCCTTCCCTTAGAGAAGTTG | |
| 8 | TCTACCATAGCCAATCAAAG | CCAGACTTAGAGCTGATGAC |
| GCTTACAGCACCTAAAGATG | GGCTCAAATTTAGCACCATC | |
| TTGACATGTTCGAGGTGTTC | GATCCCTGCTTTACTTGAC | |
| 9 | AACTAAGGCAGTATCCAAAG | GGTTTAGCAGAATCTTTAGC |
| TGCCTACCTTAAATTCTCAC | GCAGCTGATCTATCACTTTG | |
| TGGAATAACGGAGTGAATAC | GTCATGTTCTTCACCATACC | |
| 10 | AAATGGCCGAAACCATTGGG | CTTGTGTTGTTGCTGAATAC |
| ATCATCGCCGTTATTAACAC | GGAAATTAATCCCTGACTTG | |
| CCTGCATTGTGAATTTCTTG | GCTATATCGCTTGAAGAAGG | |
| 11 | CGGCAATTTCGGAGTATTTC | TTGGTTCCCACAGACCAAAG |
| CATTGCTGCTGGTACTTATG | GCAATTCAGGTGCTCTATAC | |
| TTAGAGAACCCGCTGTTATC | TACCGTTGTTTGTAACAAAG | |
| 12 | GTCTGAAATATGTGCACTTG | CATGAACCAACTTGGAATAG |
| CAGAACTGATGCCTTTAATG | GGTATTTAGGCCACAGTATC | |
| AGAATCTGCTTCTGCTTCTG | CTTGGCCCATAAGTAGATTG | |
| 13 | AATCTGTTGCCTTGTGTTTG | GTATACCTGTGGAGATATGG |
| CAGCAACTTGCTCGTTATAC | ACTGATCCGTACATCGAAAG | |
| AGGTTGACCTTGAGTGAATG | ATCTGCTTTAGCCAACAAGG | |
| 14 | TAGAGTTACCACTGCTGTTC | CTGGTATTGCCGCCAGTAAC |
| GCCAACGTAGCACTTTCTTC | AAAGACCGTTTAATCAGATG | |
| TCAGATGGACCATTCCTTAC | ACTCATCATGACCATCTTC | |
| 15 | CTCTTTCACAAGCTTCTAAC | TGACTTGTTTGGCTTCATTC |
| AAAGTTTGCGGATGCTTATG | AACTTCATCAACAGGCTTAC | |
| CTACCCAGAAGAATTCAAAG | CCAAAGAACTTGTGGTAGTC | |
| 16 | GTTAGAGACGCTTTAAACAG | AAACGGTTCTAACACCCTTG |
| CTGACTTCCAGAAAGTATTG | TCTGCCTTTATATCCATTGC | |
| ACCCATGCTGTATCCCATTC | GTACAGTTGTACGAGCATTC | |
| 17 | TTTGGAATGGACCCATATTG | ATTGCTGAGTACCAATTGAC |
| TTGGTTTGAAGACACAAATG | GTTCCAAACAAGGCTCATAC | |
| TCTATTGGTGGAGCTGAATC | CTTGATAGCCTTGTGAATAG | |
| 18 | GTATTGACCAGGAACAATAC | ATGAGCATCAAATCCTCATC |
| ATATCAAGCCGTTAGTAAGC | AAAGTACCAGTGACTCTATC | |
| AAGGTTACTGGAGCTTTGTG | TGTAGATGATGCTGGTATTC | |
| 25 | ATATACCTGTCCATCCAAAG | AAGATCCTGGCGAATATGTC |
| TGCAGCAGATAACTCAGAAC | AACATTGCTTACAGTGTTAG | |
| CCAATGTAAGCACTGTATTC | TGCTTCTCTAGCCGCTTCTG |
Mixtures of three probes for each contig generated by the oligonucleotide pairs indicated were used for Southern analyses on blots from FIGE-separated genomic DNA to assign contigs to chromosomes. Sequences are all given in the 5′-to-3′ direction.
TABLE 5.
Oligonucleotides used to generate specific probes for S. cerevisiae chromosomes in Southern analysesa
| Gene | Forward or reverse primer | S. cerevisiae chromosome (size [Mb]) |
|---|---|---|
| BUD14 | gcgtcctgcagCTATTAAGAGCTGATGGAATCATCTTTCGAA | I (0.2) |
| CGTGGATAGCGCCGATAAGG | ||
| SLA1 | GAACCAGTTCACTGGTGGAG | II (0.81) |
| CTTAGGGTCGACTCCACCATTTC | ||
| TRP1 | GCGGCTTGCAGAGCACAGAGG | IV (1.61) |
| GTCTCCACACCTCCGC | ||
| GEA2 | gcgtacccgggAAACCACAGCCACATTAAC | V (0.61) |
| GGTGCCGTTGGAAATCACTG | ||
| PFK1 | ggatctcgagTCAATCTCAAGATTCATGCTACGG | VII (1.12) |
| gagcgctcgagTATTCAGTACCTGGAACG | ||
| GEA1 | gcgtcgcatgcGGTATAAATGCCACCGTCG | X (0.745) |
| CAGCCAAAGACCGCCAATTTGC | ||
| HXT1 | GGTACCATTGTTTTCCAGGCTGTCGG | VIII (0.56) |
| GCCGGTGAAGGTCAAGAACTAG | ||
| GPM1 | GAAGCCGCTAGAGCCGGTG | XI (0.68) |
| GGCAACAGCAGCGGCACCAGC | ||
| MID2 | GTGGAACGTTAAAGCACTCG | XII (1.9) |
| CCTCAAGTGCTGACTCATCTTCCC | ||
| rDNA | gcgcgaattcGCTAGTACCGATTGAATGGCTTAG | XII (1.9) |
| gcgcggatccGATGCGAGAACCAAGAGATCCG | ||
| PFK2 | ggatctcgagTACTGTTACTACTCCTTTTGTG | XIII (0.915) |
| cagcgctcgagTATTCAGTACCTGGAAC | ||
| WSC2 | GTGGAATATGCACCTAGATCTC | XIV (0.785) |
| CCACAAACCACACCTACTAC | ||
| Noncoding sequence | GCGTTTATTGTATCCCTTGAC | XV (1.09) |
| GGTAGATAGCTTGAGGCAC | ||
| GAL4 | GGTCTCCGCTGACTAGGGCAC | XVI (0.945) |
| CCCCCTCTATACACCAGGCTCC |
For S. cerevisiae chromosomes, probes corresponding to the genes indicated were obtained by PCR with the oligonucleotide pairs given. Lowercase type indicates sequences added to create restriction sites for cloning, which do not hybridize to the target DNA. Sequences are all given in the 5′-to-3′ direction.
Flow cytometry measurements of DNA content.
The fixation and staining of cells were performed according to methods described previously (46). Samples were measured on a BD FACSAria II flow cytometer (Becton Dickinson, Heidelberg, Germany). R statistical language v 3.3.2 (84) and flow cytometry packages from Bioconductor (87) were used for data analysis and the generation of graphics.
Determination of specific enzyme activities.
To obtain crude extracts for the determination of specific enzyme activities, yeast cells were washed and broken with glass beads, and the supernatant was obtained after 10 min of centrifugation in a microcentrifuge at 13,000 × g at 4°C as described previously (88). Specific activities were obtained by coupling the enzyme reactions in question to either NAD(P)H oxidation or reduction with ancillary enzymes, as described previously (89). Tests were performed with a Beckmann DU800 photometer, recording kinetics at 30°C and determining the slope after constant reaction rates were reached. As controls, kinetics were recorded without the presence of substrates prior to the start of the reactions. Buffers, concentrations of reagents, and ancillary enzymes employed are listed in Table 6. Protein concentrations in crude extracts were determined by the Micro-Biuret method, using bovine serum albumin as a standard (90).
TABLE 6.
Compositions of mixtures for determination of enzyme activitiesa
| Enzyme | Composition of assay mixture | Substrate (concn [mM]) |
|---|---|---|
| Hexokinase | IB, 0.4 mM NADP, 1 mM ATP, 10 mM MgCl2, 0.2 mM EDTA, 0.3 U/ml glucose-6-phosphate dehydrogenase | Fructose (5) |
| Phosphoglucose isomerase | IB, 0.4 mM NADP, 1 mM ATP, 10 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.3 U/ml glucose-6-phosphate dehydrogenase | Fructose-6-phosphate (5) |
| Phosphofructokinase | PB, 0.2 mM NADH, 1 mM ATP, 1 mM AMP, 10 mM MgCl2, 5 μM fructose-2,6-bisphosphate, 1 U/ml fructose-1,6-bisphosphate aldolase, 0.5 U/ml each of glycerol-3-phosphate dehydrogenase and triosephosphate isomerase | Fructose-6-phosphate (2.5) |
| Aldolase | PB, 0.2 mM NADH, 5 mM MgCl2, 30 mM KCl, 10 mM NH4Cl, 0.3 mM MnCl2, 0.2 mM Zn-acetate, 0.5 U/ml each of glycerol-3-phosphate dehydrogenase and triosephosphate isomerase | Fructose-1,6-bisphosphate (2) |
| Triosephosphate isomerase | IB, 0.2 mM NADH, 10 mM MgCl2, 0.1 mM EDTA, 0.5 U/ml glycerol-3-phosphate dehydrogenase | Glyceraldehyde-3-phosphate (5) |
| Triosephosphate dehydrogenase | PB, 0.2 mM NADH, 1 mM ATP, 10 mM MgCl2, 5 mM cysteine, 1 U/ml 3-phosphoglycerate kinase | 3-Phosphoglycerate (4) |
| Phosphoglycerate kinase | PB, 0.2 mM NADH, 10 mM ATP, 10 mM MgCl2, 100 mM KCl, 5 mM cysteine, 1 U/ml glyceraldehyde-3-phosphate dehydrogenase | 3-Phosphoglycerate (10) |
| Phosphoglycerate mutase | IB, 0.2 mM NADH, 1 mM ADP, 10 mM MgCl2, 0.5 U/ml enolase, 1 U/ml pyruvate kinase, 1 U/ml lactate dehydrogenase | 3-Phosphoglycerate (10) |
| Enolase | PB, 0.2 mM NADH, 1 mM ADP, 10 mM MgCl2, 100 mM KCl, 1 U/ml pyruvate kinase, 1 U/ml lactate dehydrogenase | 2-Phosphoglycerate (10) |
| Pyruvate kinase | PB, 0.2 mM NADH, 0.6 mM ADP, 10 mM MgCl2, 100 mM KCl, 1 mM fructose-1,6-bisphosphate, 1 U/ml lactate dehydrogenase | Phosphoenolpyruvate (10) |
| Pyruvate dehydrogenase | CB, 0.2 mM NADH, 10 mM MgCl2, 100 mM KCl, 1.7 mM cysteine, 1.6 mM thiamine pyrophosphate, 1 U/ml alcohol dehydrogenase | Pyruvate (5) |
| Alcohol dehydrogenase | PPB, 2 mM NAD, 1 mM glutathione | Ethanol (0.6) |
The following assay conditions were modified from those described previously: hexokinase, glucose isomerase, triosephosphate isomerase, phosphoglycerate kinase, and enolase (92); phosphofructokinase (93); aldolase (94); triosephosphate dehydrogenase; (95); phosphoglycerate mutase (96); pyruvate kinase (63); pyruvate dehydrogenase (97), and alcohol dehydrogenase (98). Ancillary enzymes and reagents were obtained from Roche (Mannheim, Germany), as far being as available in ammonium sulfate suspensions and used far beyond the indicated expiration date, partially due to the lack of current commercial resources. Alternatively, enzymes and substrates were obtained from Sigma-Aldrich. IB, 50 mM imidazole buffer, pH 7.0; PB, 50 mM sodium phosphate buffer, pH 7.0; CB, 50 mM sodium citrate buffer, pH 7.0; PPB, 85.5 mM sodium pyrophosphate buffer, pH 7.0.
Immunological detection of phosphofructokinase subunits.
Crude extracts prepared with glass beads as described above were obtained from cells grown in YEPD medium, boiled with loading buffer, and separated by SDS-polyacrylamide gel electrophoresis (PAGE) on gels with 7.5% acrylamide. A polyclonal antiserum raised against PFK from S. cerevisiae was employed for the immunological detection of H. uvarum PFK subunits, applied at a dilution of 1:10,000 (68). Since the reactivity against the heterologous enzyme is much weaker than that against S. cerevisiae PFK, approximately 250 μg of protein from crude extracts prepared from H. uvarum cells or S. cerevisiae pfk1 pfk2 double-deletion mutants transformed with either HuPFK1 or HuPFK2 carried on multicopy plasmids was loaded into each lane of the SDS-PAGE gel. For transformants carrying a multicopy plasmid with both heterologous PFK genes, only approximately 50 μg of protein was loaded. This concentration was also used for strains carrying endogenous ScPFK genes. Secondary anti-rabbit antibodies coupled to an infrared dye with fluorescence at 700 nm were then employed for the detection of PFK signals using the Odyssey imaging device as described previously (91). Contrast, brightness, and frames of the images were adjusted by using Corel PhotoPaint, which was applied only to the entire images shown in the figures and not to single bands.
Accession number(s).
The annotated genome sequence was submitted to GenBank (https://www.ncbi.nlm.nih.gov/GenBank/) under accession number APLS01000000. The two HuPFK sequences were obtained again independently and submitted under GenBank accession numbers MF509744 (HuPFK1) and MF509745 (HuPFK2).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by two consecutive grants from the Forschungsring des Deutschen Weinbaus (FDW) to J.J.H. A.-K.L. and F.J.B. were funded by grants from the FDW. We thank Josef Hermann (Veitshöchheim, Germany) for providing a natural isolate of H. uvarum and financial support for a second round of sequencing.
We also thank Andrea Murra for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01580-17.
REFERENCES
- 1.Boulton RB, Singleton VI, Bisson LF, Kunkee RE. 1999. Principles and practices of winemaking. Springer, New York, NY. [Google Scholar]
- 2.Chambers PJ, Pretorius IS. 2010. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep 11:914–920. doi: 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heard GM, Fleet GH. 1985. Growth of natural yeast flora during the fermentation of inoculated wines. Appl Environ Microbiol 50:727–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisson LF, Karpel JE. 2010. Genetics of yeast impacting wine quality. Annu Rev Food Sci Technol 1:139–162. doi: 10.1146/annurev.food.080708.100734. [DOI] [PubMed] [Google Scholar]
- 5.Jolly NP, Varela C, Pretorius IS. 2014. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14:215–237. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- 6.Mortimer R, Polsinelli M. 1999. On the origins of wine yeast. Res Microbiol 150:199–204. doi: 10.1016/S0923-2508(99)80036-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Garcia-Fernandez D, Mas A, Esteve-Zarzoso B. 2015. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front Microbiol 6:1156. doi: 10.3389/fmicb.2015.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zott K, Miot-Sertier C, Claisse O, Lonvaud-Funel A, Masneuf-Pomarede I. 2008. Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking. Int J Food Microbiol 125:197–203. doi: 10.1016/j.ijfoodmicro.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Capece A, Fiore C, Maraz A, Romano P. 2005. Molecular and technological approaches to evaluate strain biodiversity in Hanseniaspora uvarum of wine origin. J Appl Microbiol 98:136–144. doi: 10.1111/j.1365-2672.2004.02434.x. [DOI] [PubMed] [Google Scholar]
- 10.Fleet GH. 2003. Yeast interactions and wine flavour. Int J Food Microbiol 86:11–22. doi: 10.1016/S0168-1605(03)00245-9. [DOI] [PubMed] [Google Scholar]
- 11.Albergaria H, Francisco D, Gori K, Arneborg N, Girio F. 2010. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biotechnol 86:965–972. doi: 10.1007/s00253-009-2409-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Mas A, Esteve-Zarzoso B. 2015. Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int J Food Microbiol 206:67–74. doi: 10.1016/j.ijfoodmicro.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Lopandic K, Tiefenbrunner W, Gangl H, Mandl K, Berger S, Leitner G, Abd-Ellah GA, Querol A, Gardner RC, Sterflinger K, Prillinger H. 2008. Molecular profiling of yeasts isolated during spontaneous fermentations of Austrian wines. FEMS Yeast Res 8:1063–1075. doi: 10.1111/j.1567-1364.2008.00385.x. [DOI] [PubMed] [Google Scholar]
- 14.Bezerra-Bussoli C, Baffi MA, Gomes E, Da-Silva R. 2013. Yeast diversity isolated from grape musts during spontaneous fermentation from a Brazilian winery. Curr Microbiol 67:356–361. doi: 10.1007/s00284-013-0375-9. [DOI] [PubMed] [Google Scholar]
- 15.Li SS, Cheng C, Li Z, Chen JY, Yan B, Han BZ, Reeves M. 2010. Yeast species associated with wine grapes in China. Int J Food Microbiol 138:85–90. doi: 10.1016/j.ijfoodmicro.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Beltran G, Torija MJ, Novo M, Ferrer N, Poblet M, Guillamon JM, Rozes N, Mas A. 2002. Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst Appl Microbiol 25:287–293. doi: 10.1078/0723-2020-00097. [DOI] [PubMed] [Google Scholar]
- 17.Brysch-Herzberg M, Seidel M. 2015. Yeast diversity on grapes in two German wine growing regions. Int J Food Microbiol 214:137–144. doi: 10.1016/j.ijfoodmicro.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 18.Caruso M, Capece A, Salzano G, Romano P. 2002. Typing of Saccharomyces cerevisiae and Kloeckera apiculata strains from Aglianico wine. Lett Appl Microbiol 34:323–328. doi: 10.1046/j.1472-765X.2002.01090.x. [DOI] [PubMed] [Google Scholar]
- 19.Ciani M, Fatichenti F. 1999. Selective sugar consumption by apiculate yeasts. Lett Appl Microbiol 28:203–206. doi: 10.1046/j.1365-2672.1999.00505.x. [DOI] [PubMed] [Google Scholar]
- 20.Teixeira A, Caldeira I, Duarte FL. 2015. Molecular and oenological characterization of Touriga Nacional non-Saccharomyces yeasts. J Appl Microbiol 118:658–671. doi: 10.1111/jam.12727. [DOI] [PubMed] [Google Scholar]
- 21.Brezna B, Zenisova K, Chovanova K, Chebenova V, Krakova L, Kuchta T, Pangallo D. 2010. Evaluation of fungal and yeast diversity in Slovakian wine-related microbial communities. Antonie Van Leeuwenhoek 98:519–529. doi: 10.1007/s10482-010-9469-6. [DOI] [PubMed] [Google Scholar]
- 22.Clavijo A, Calderon IL, Paneque P. 2010. Diversity of Saccharomyces and non-Saccharomyces yeasts in three red grape varieties cultured in the Serrania de Ronda (Spain) vine-growing region. Int J Food Microbiol 143:241–245. doi: 10.1016/j.ijfoodmicro.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Janisiewicz WJ, Jurick WM II, Peter KA, Kurtzman CP, Buyer JS. 2014. Yeasts associated with plums and their potential for controlling brown rot after harvest. Yeast 31:207–218. doi: 10.1002/yea.3009. [DOI] [PubMed] [Google Scholar]
- 24.Vadkertiova R, Molnarova J, Vranova D, Slavikova E. 2012. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can J Microbiol 58:1344–1352. doi: 10.1139/cjm-2012-0468. [DOI] [PubMed] [Google Scholar]
- 25.Morrissey WF, Davenport B, Querol A, Dobson AD. 2004. The role of indigenous yeasts in traditional Irish cider fermentations. J Appl Microbiol 97:647–655. doi: 10.1111/j.1365-2672.2004.02354.x. [DOI] [PubMed] [Google Scholar]
- 26.Pando Bedrinana R, Querol Simon A, Suarez Valles B. 2010. Genetic and phenotypic diversity of autochthonous cider yeasts in a cellar from Asturias. Food Microbiol 27:503–508. doi: 10.1016/j.fm.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 27.Masoud W, Cesar LB, Jespersen L, Jakobsen M. 2004. Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast 21:549–556. doi: 10.1002/yea.1124. [DOI] [PubMed] [Google Scholar]
- 28.Batista NN, Ramos CL, Dias DR, Pinheiro AC, Schwan RF. 2016. The impact of yeast starter cultures on the microbial communities and volatile compounds in cocoa fermentation and the resulting sensory attributes of chocolate. J Food Sci Technol 53:1101–1110. doi: 10.1007/s13197-015-2132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Illeghems K, De Vuyst L, Papalexandratou Z, Weckx S. 2012. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PLoS One 7:e38040. doi: 10.1371/journal.pone.0038040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu HM, Guo JH, Cheng YJ, Liu P, Long CA, Deng BX. 2010. Inhibitory activity of tea polyphenol and Hanseniaspora uvarum against Botrytis cinerea infections. Lett Appl Microbiol 51:258–263. doi: 10.1111/j.1472-765X.2010.02888.x. [DOI] [PubMed] [Google Scholar]
- 31.Renard A, Gomez di Marco P, Egea-Cortines M, Weiss J. 2008. Application of whole genome amplification and quantitative PCR for detection and quantification of spoilage yeasts in orange juice. Int J Food Microbiol 126:195–201. doi: 10.1016/j.ijfoodmicro.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Radler F, Schmitt MJ, Meyer B. 1990. Killer toxin of Hanseniaspora uvarum. Arch Microbiol 154:175–178. doi: 10.1007/BF00423329. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt MJ, Poravou O, Trenz K, Rehfeldt K. 1997. Unique double-stranded RNAs responsible for the anti-Candida activity of the yeast Hanseniaspora uvarum. J Virol 71:8852–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciani M, Beco L, Comitini F. 2006. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int J Food Microbiol 108:239–245. doi: 10.1016/j.ijfoodmicro.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Tristezza M, Tufariello M, Capozzi V, Spano G, Mita G, Grieco F. 2016. The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front Microbiol 7:670. doi: 10.3389/fmicb.2016.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schütz M, Gafner J. 1993. Sluggish alcoholic fermentation in relation to alterations of the glucose-fructose ratio. Chem Mikrobiol Technol Lebensm 15:73–78. [Google Scholar]
- 37.Cadez N, Raspor P, de Cock AW, Boekhout T, Smith MT. 2002. Molecular identification and genetic diversity within species of the genera Hanseniaspora and Kloeckera. FEMS Yeast Res 1:279–289. doi: 10.1111/j.1567-1364.2002.tb00046.x. [DOI] [PubMed] [Google Scholar]
- 38.Esteve-Zarzoso B, Peris-Toran MJ, Ramon D, Quero A. 2001. Molecular characterisation of Hanseniaspora species. Antonie Van Leeuwenhoek 80:85–92. doi: 10.1023/A:1012268931569. [DOI] [PubMed] [Google Scholar]
- 39.Belda I, Ruiz J, Alastruey-Izquierdo A, Navascues E, Marquina D, Santos A. 2016. Unraveling the enzymatic basis of wine “flavorome”: a phylo-functional study of wine related yeast species. Front Microbiol 7:12. doi: 10.3389/fmicb.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurtzman CP, Robnett CJ. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 41.Holloway P, Subden RE. 1994. The nucleotide sequence and initial characterization of pyruvate decarboxylase from the yeast Hanseniaspora uvarum. Yeast 10:1581–1589. doi: 10.1002/yea.320101207. [DOI] [PubMed] [Google Scholar]
- 42.Pramateftaki PV, Kouvelis VN, Lanaridis P, Typas MA. 2006. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts. FEMS Yeast Res 6:77–90. doi: 10.1111/j.1567-1364.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- 43.Sternes PR, Lee D, Kutyna DR, Borneman AR. 2016. Genome sequences of three species of Hanseniaspora isolated from spontaneous wine fermentations. Genome Announc 4:e01287-16. doi: 10.1128/genomeA.01287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinisch JJ, Buchwald U, Gottschlich A, Heppeler N, Rodicio R. 2010. A tool kit for molecular genetics of Kluyveromyces lactis comprising a congenic strain series and a set of versatile vectors. FEMS Yeast Res 10:333–342. doi: 10.1111/j.1567-1364.2009.00604.x. [DOI] [PubMed] [Google Scholar]
- 45.Schacherer J, Ruderfer DM, Gresham D, Dolinski K, Botstein D, Kruglyak L. 2007. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS One 2:e322. doi: 10.1371/journal.pone.0000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delobel P, Tesniere C. 2014. A simple FCM method to avoid misinterpretation in Saccharomyces cerevisiae cell cycle assessment between G0 and sub-G1. PLoS One 9:e84645. doi: 10.1371/journal.pone.0084645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues F, Ludovico P, Sousa MJ, Steensma HY, Corte-Real M, Leao C. 2003. The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: a screening method for haploidization. Appl Environ Microbiol 69:649–653. doi: 10.1128/AEM.69.1.649-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langenberg AK. 2016. Genetische und physiologische Charakterisierung von Hanseniaspora uvarum. PhD thesis University of Osnabrück, Osnabrück, Germany. [Google Scholar]
- 50.Shen XX, Zhou X, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. 2016. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 6:3927–3939. doi: 10.1534/g3.116.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnett JA, Payne RW, Yarrow D. 2000. Yeasts: characteristics and identification. Cambridge University Press, Cambridge, England. [Google Scholar]
- 52.Albertin W, Setati ME, Miot-Sertier C, Mostert TT, Colonna-Ceccaldi B, Coulon J, Girard P, Moine V, Pillet M, Salin F, Bely M, Divol B, Masneuf-Pomarede I. 2015. Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front Microbiol 6:1569. doi: 10.3389/fmicb.2015.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boles E, Hollenberg CP. 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 54.Heinisch JJ, Rodicio R. 2017. Stress response in wine yeast. In König H, Unden G, Fröhlich J (ed), Biology of microorganisms on grapes, in must and wine, 2nd ed, in press Springer-Verlag, Berlin, Germany. [Google Scholar]
- 55.Jendretzki A, Wittland J, Wilk S, Straede A, Heinisch JJ. 2011. How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity. Eur J Cell Biol 90:740–744. doi: 10.1016/j.ejcb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 56.Schmitz HP, Heinisch JJ. 2003. Evolution, biochemistry and genetics of protein kinase C in fungi. Curr Genet 43:245–254. doi: 10.1007/s00294-003-0403-6. [DOI] [PubMed] [Google Scholar]
- 57.Schmitz HP, Lorberg A, Heinisch JJ. 2002. Regulation of yeast protein kinase C activity by interaction with the small GTPase Rho1p through its amino-terminal HR1 domain. Mol Microbiol 44:829–840. doi: 10.1046/j.1365-2958.2002.02925.x. [DOI] [PubMed] [Google Scholar]
- 58.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matallana E, Aranda A. 2017. Biotechnological impact of stress response on wine yeast. Lett Appl Microbiol 64:103–110. doi: 10.1111/lam.12677. [DOI] [PubMed] [Google Scholar]
- 60.De Benedictis M, Bleve G, Grieco F, Tristezza M, Tufariello M. 2011. An optimized procedure for the enological selection of non-Saccharomyces starter cultures. Antonie Van Leeuwenhoek 99:189–200. doi: 10.1007/s10482-010-9475-8. [DOI] [PubMed] [Google Scholar]
- 61.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WH, Klaassen P, Paddon CJ, Platt D, Kotter P, van Ham RC, Reinders MJ, Pronk JT, de Ridder D, Daran JM. 2012. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schehl B, Muller C, Senn T, Heinisch JJ. 2004. A laboratory yeast strain suitable for spirit production. Yeast 21:1375–1389. doi: 10.1002/yea.1189. [DOI] [PubMed] [Google Scholar]
- 63.Boles E, Schulte F, Miosga T, Freidel K, Schluter E, Zimmermann FK, Hollenberg CP, Heinisch JJ. 1997. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol 179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. 1998. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 6:195–210. doi: 10.1016/S0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 65.Rodicio R, Heinisch JJ. 2017. Carbohydrate metabolism in wine yeast. In König H, Unden G, Fröhlich J (ed), Biology of microorganisms on grapes, in must and wine, 2nd ed, in press Springer-Verlag, Berlin, Germany. [Google Scholar]
- 66.Venturin C, Boze H, Moulin G, Galzy P. 1995. Glucose metabolism, enzymic analysis and product formation in chemostat culture of Hanseniaspora uvarum. Yeast 11:327–336. doi: 10.1002/yea.320110405. [DOI] [PubMed] [Google Scholar]
- 67.Heinisch J, Ritzel RG, von Borstel RC, Aguilera A, Rodicio R, Zimmermann FK. 1989. The phosphofructokinase genes of yeast evolved from two duplication events. Gene 78:309–321. doi: 10.1016/0378-1119(89)90233-3. [DOI] [PubMed] [Google Scholar]
- 68.Heinisch J. 1986. Construction and physiological characterization of mutants disrupted in the phosphofructokinase genes of Saccharomyces cerevisiae. Curr Genet 11:227–234. doi: 10.1007/BF00420611. [DOI] [PubMed] [Google Scholar]
- 69.Breitenbach-Schmitt I, Schmitt HD, Heinisch J, Zimmermann FK. 1984. Genetic and physiological evidence for the existence of a second glycolytic pathway in yeast parallel to the phosphofructokinase-aldolase reaction sequence. Mol Gen Genet 195:536–540. doi: 10.1007/BF00341459. [DOI] [Google Scholar]
- 70.Arvanitidis A, Heinisch JJ. 1994. Studies on the function of yeast phosphofructokinase subunits by in vitro mutagenesis. J Biol Chem 269:8911–8918. [PubMed] [Google Scholar]
- 71.Bär J, Schellenberger W, Kopperschläger G. 1997. Purification and characterization of phosphofructokinase from the yeast Kluyveromyces lactis. Yeast 13:1309–1317. [DOI] [PubMed] [Google Scholar]
- 72.Heinisch J, Kirchrath L, Liesen T, Vogelsang K, Hollenberg CP. 1993. Molecular genetics of phosphofructokinase in the yeast Kluyveromyces lactis. Mol Microbiol 8:559–570. doi: 10.1111/j.1365-2958.1993.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 73.Kopperschläger G, Bar J, Stellwagen E. 1993. Limited proteolysis of yeast phosphofructokinase. Sequence locations of cleavage sites created by the actions of different proteinases. Eur J Biochem 217:527–533. doi: 10.1111/j.1432-1033.1993.tb18273.x. [DOI] [PubMed] [Google Scholar]
- 74.Ciani M, Morales P, Comitini F, Tronchoni J, Canonico L, Curiel JA, Oro L, Rodrigues AJ, Gonzalez R. 2016. Non-conventional yeast species for lowering ethanol content of wines. Front Microbiol 7:642. doi: 10.3389/fmicb.2016.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raben N, Exelbert R, Spiegel R, Sherman JB, Nakajima H, Plotz P, Heinisch J. 1995. Functional expression of human mutant phosphofructokinase in yeast: genetic defects in French Canadian and Swiss patients with phosphofructokinase deficiency. Am J Hum Genet 56:131–141. [PMC free article] [PubMed] [Google Scholar]
- 76.Rodicio R, Koch S, Schmitz HP, Heinisch JJ. 2006. KlRHO1 and KlPKC1 are essential for cell integrity signalling in Kluyveromyces lactis. Microbiology 152:2635–2649. doi: 10.1099/mic.0.29105-0. [DOI] [PubMed] [Google Scholar]
- 77.Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 78.Hill JE, Myers AM, Koerner TJ, Tzagoloff A. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 79.Zimmermann FK. 1975. Procedures used in the induction of mitotic recombination and mutation in the yeast Saccharomyces cerevisiae. Mutat Res 31:71–86. doi: 10.1016/0165-1161(75)90069-2. [DOI] [PubMed] [Google Scholar]
- 80.Bashir A, Klammer AA, Robins WP, Chin CS, Webster D, Paxinos E, Hsu D, Ashby M, Wang S, Peluso P, Sebra R, Sorenson J, Bullard J, Yen J, Valdovino M, Mollova E, Luong K, Lin S, LaMay B, Joshi A, Rowe L, Frace M, Tarr CL, Turnsek M, Davis BM, Kasarskis A, Mekalanos JJ, Waldor MK, Schadt EE. 2012. A hybrid approach for the automated finishing of bacterial genomes. Nat Biotechnol 30:701–707. doi: 10.1038/nbt.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Proux-Wera E, Armisen D, Byrne KP, Wolfe KH. 2012. A pipeline for automated annotation of yeast genome sequences by a conserved-synteny approach. BMC Bioinformatics 13:237. doi: 10.1186/1471-2105-13-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 83.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 84.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 85.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Breitenbach I, Schmitt HD, Heinisch J, Zimmermann FK. 1984. Yeast mutants without phosphofructokinase activity can still perform glycolysis and alcoholic fermentation. Mol Gen Genet 195:530–535. doi: 10.1007/BF00341458. [DOI] [Google Scholar]
- 89.Ciriacy M, Breitenbach I. 1979. Physiological effects of seven different blocks in glycolysis in Saccharomyces cerevisiae. J Bacteriol 139:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zamenhoff S. 1957. Preparation and assay of deoxyribonucleic acids from animal tissue. Methods Enzymol 3:702–704. [Google Scholar]
- 91.Kock C, Arlt H, Ungermann C, Heinisch JJ. 2016. Yeast cell wall integrity sensors form specific plasma membrane microdomains important for signalling. Cell Microbiol 18:1251–1267. doi: 10.1111/cmi.12635. [DOI] [PubMed] [Google Scholar]
- 92.Maitra PK, Lobo Z. 1971. A kinetic study of glycolytic enzyme synthesis in yeast. J Biol Chem 246:475–488. [PubMed] [Google Scholar]
- 93.Rodicio R, Strauss A, Heinisch JJ. 2000. Single point mutations in either gene encoding the subunits of the heterooctameric yeast phosphofructokinase abolish allosteric inhibition by ATP. J Biol Chem 275:40952–40960. doi: 10.1074/jbc.M007131200. [DOI] [PubMed] [Google Scholar]
- 94.Boles E, Zimmermann FK. 1993. Saccharomyces cerevisiae phosphoglucose isomerase and fructose bisphosphate aldolase can be replaced functionally by the corresponding enzymes of Escherichia coli and Drosophila melanogaster. Curr Genet 23:187–191. doi: 10.1007/BF00351494. [DOI] [PubMed] [Google Scholar]
- 95.McAlister L, Holland MJ. 1985. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem 260:15019–15027. [PubMed] [Google Scholar]
- 96.Heinisch JJ, Muller S, Schluter E, Jacoby J, Rodicio R. 1998. Investigation of two yeast genes encoding putative isoenzymes of phosphoglycerate mutase. Yeast 14:203–213. [DOI] [PubMed] [Google Scholar]
- 97.Schmitt HD, Zimmermann FK. 1982. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol 151:1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ciriacy M. 1975. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet 138:157–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.