ABSTRACT
The human microbiome contributes significantly to the genetic content of the human body. Genetic and environmental factors help shape the microbiome, and as such, the microbiome can be unique to an individual. Previous studies have demonstrated the potential to use microbiome profiling for forensic applications; however, a method has yet to identify stable features of skin microbiomes that produce high classification accuracies for samples collected over reasonably long time intervals. A novel approach is described here to classify skin microbiomes to their donors by comparing two feature types: Propionibacterium acnes pangenome presence/absence features and nucleotide diversities of stable clade-specific markers. Supervised learning was used to attribute skin microbiomes from 14 skin body sites from 12 healthy individuals sampled at three time points over a >2.5-year period with accuracies of up to 100% for three body sites. Feature selection identified a reduced subset of markers from each body site that are highly individualizing, identifying 187 markers from 12 clades. Classification accuracies were compared in a formal model testing framework, and the results of this analysis indicate that learners trained on nucleotide diversity perform significantly better than those trained on presence/absence encodings. This study used supervised learning to identify individuals with high accuracy and associated stable features from skin microbiomes over a period of up to almost 3 years. These selected features provide a preliminary marker panel for future development of a robust and reproducible method for skin microbiome profiling for forensic human identification.
IMPORTANCE A novel approach is described to attribute skin microbiomes, collected over a period of >2.5 years, to their individual hosts with a high degree of accuracy. Nucleotide diversities of stable clade-specific markers with supervised learning were used to classify skin microbiomes from a particular individual with up to 100% classification accuracy for three body sites. Attribute selection was used to identify 187 genetic markers from 12 clades which provide the greatest differentiation of individual skin microbiomes from 14 skin sites. This study performs skin microbiome profiling from a supervised learning approach and obtains high classification accuracy for samples collected from individuals over a relatively long time period for potential application to forensic human identification.
KEYWORDS: skin microbiome, human identification, forensic profiling, metagenomics, supervised learning
INTRODUCTION
The human microbiome plays a critical role in health, metabolism, and immune response (1) and can be influenced by numerous factors, including but not limited to genetics, geography, diet, and hygiene (2–4). Colonization of the human microbiome begins at birth and continues to change throughout development (5, 6), contributing an additional 5,000,000 genes from the gut microbiome alone (7) to the repertoire of human genes. Since unique genetic and environmental factors help shape the microbiome, the composition of the microbiome has the potential to be unique to its host individual. Features of the personal microbiome, such as strain-specific signatures (8, 9), which may be stable over time, make microbiome characterization potentially applicable to forensic human identification.
Current forensic human profiling methods typically utilize autosomal short tandem repeats (STRs) profiles to attribute forensic biological evidence to a suspect (or victim) (10). Often, evidentiary samples contain mixtures of human DNA from multiple sources or contain small amounts (i.e., low copy number) or degraded DNA, making interpretation of mixed or partial profiles difficult or inconclusive. In these cases, alternative methods may be used, such as sequencing high-copy-number markers (e.g., targeting the hypervariable regions of the mitochondrial genome [11, 12] or whole mitochondrial genomes [13]) or methods to enhance sensitivity of detection, including concentrating DNA extracts, increasing PCR cycles, or performing whole-genome amplification (14). The human microbiome is an example of another high-copy-number genetic marker, since microbial cells may be at ratios of 1:1 (15) to 10:1 (16) to human cells, and thus it is a potential target to complement partial or inconclusive STR profiles to increase resolution for human source attribution.
Recent studies have demonstrated the potential to use microbiome profiling for forensic identification, mainly using unsupervised methods to show that microbiome samples from touched objects resemble their respective donors (17–19). Few studies have addressed microbiome profiling from a supervised approach, i.e., for the purposes of classification. Franzosa et al. (8) used a nearest-neighbor classification approach using clade-specific markers and 1-kb genomic windows to identify strain-level metagenomic codes specific to individuals; however, this method could identify only <30% of individuals using skin microbiomes (i.e., anterior nares) sampled over a time interval of 30 to 300 days. Lax et al. (20) and Williams et al. (21) used random forests trained on operational taxonomic units (OTU) abundances of targeted 16S rRNA sequences for human identification. Although both approaches were highly accurate (96.3 and 97.3%, respectively), the samples were collected over short time intervals (<3 days or just a single time point, respectively) (20, 21), making their results less applicable to a typical forensics setting.
Individual-specific microbiome features with the greatest temporal stability (up to almost 3 years) include single-nucleotide variant (SNV) profiles of Propionibacterium acnes from the skin (9) and gene signatures (i.e., clade-specific markers and 1-kb genomic windows) from the gut microbiome (8). Strain-level signatures from shotgun sequencing provide far more depth of resolution than 16S rRNA based features, such as terminal restriction fragment length polymorphism profiles (18, 22, 23), OTU abundances (8, 19–21, 24, 25), and biological community distances (e.g., UniFrac distance) (17, 20). Nucleotide diversity of strains, which measures the strain-level heterogeneity of the microbial population, also has been shown to be greater between individuals than within the same individual (26). Thus far, features used for microbiome profiling at the strain level demonstrate the most success for differentiating individuals over time. However, a method has yet to be described that identifies differentiating features stable over reasonably long time intervals and applies appropriate measures on these markers to perform classification (i.e., via supervised learning) to attribute skin microbiome samples to their donors.
In this study, a novel approach is described to attribute skin microbiomes to their individual hosts with a high degree of accuracy and to identify genetic markers which may be well suited to individual skin microbiome differentiation. Unsupervised learning techniques were first evaluated to assess inter- versus intrasample variation across host microbiomes sampled across 14 body sites. To assess whether microbiomes could be used to be predictive of their host, two feature types capturing strain-level variation within shotgun metagenomes were compared using two supervised learning techniques. In particular, Propionibacterium acnes pangenome presence/absence features and the nucleotide diversities of clade-specific markers were used in conjunction with regularized multinomial logistic regression (RMLR) and 1-nearest-neighbor (1NN) classifiers to form predictions on host microbiomes based on samples separated by up to 3 years. Feature selection was then used to identify stable features which can be used to attribute skin microbiomes from multiple body sites to their respective hosts. This reduced set of markers was then evaluated to determine whether they could provide similar predictive power despite using much less information. The results from our classification algorithms were then formally compared to evaluate whether different body sites and different classification techniques significantly vary in their predictive capabilities.
RESULTS
Sample and shotgun metagenomic processing.
Publically available shotgun metagenomic data sets from Oh et al. (9) were used in this study. Briefly, the Oh et al. (9) data set consists of an extensive spatial and temporal sampling of skin microbiomes from 12 healthy individuals across 17 body sites (i.e., antecubital fossa [Ac], alar crease [Al], back [Ba], cheek [Ch], external auditory canal [Ea], forehead [Fh], hypothenar palm [Hp], inguinal crease [Ic], interdigital web [Id], manubrium [Mb], occiput [Oc], popliteal fossa [Pc], plantar heel [Ph], retroauricular crease [Ra], toenail [Tn], toe web space [Tw], and volar forearm [Vf]). Skin microbiome samples were collected at three different time points over a period of almost 3 years, sampled over long (ranging from 10 to 30 months) and short (ranging from 5 to 10 weeks) time intervals (9). In total, 2,446 fastq files from 585 samples, containing a total of 23 billion reads (mean of 39.3 million reads per sample) were downloaded from the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) (27). Data were preprocessed to remove sequencing adapters, to trim reads with a quality score of less than 20, to remove reads less than 50 bp in length, and to remove any human host-associated reads. A total of 12.6 billion quality-controlled reads (mean 21.5 million reads per sample) remained after read preprocessing. Several samples had substantially lower read depth after read preprocessing, and only individuals with samples from all three time points at a particular body site, with ≥10× average read depth across all shared markers, were included in the study (n = 381; see Table S1 in the supplemental material). Three body sites from the foot (i.e., plantar heel [Ph], toenail [Tn], and toe web space [Tw]) also were excluded from the study, since they only shared two to five markers among samples.
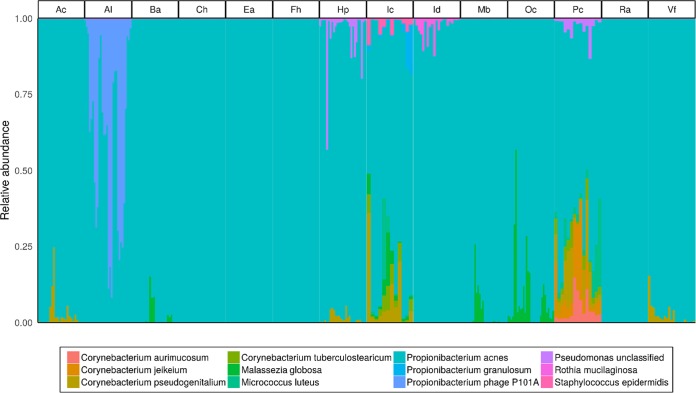
Taxonomic classification was performed using MetaPhlAn2 (28) to identify the core skin microbial species shared by all individuals, stable over time (i.e., present at each time point) to identify likely candidate species which may serve as forensically relevant targets. The core skin microbial taxa comprised of all shared species at a particular body site, together included 10 bacterial species (Corynebacterium aurimucosum, Corynebacterium jeikeium, Corynebacterium pseudogenitalium, Corynebacterium tuberculostearicum, Micrococcus luteus, Propionibacterium acnes, Propionibacterium granulosum, Pseudomonas sp., unclassified, Rothia mucilaginosa, and Staphylococcus epidermidis), 1 fungal species (Malassezia globosa), and 1 bacteriophage (Propionibacterium phage P101A) (Fig. 1). Propionibacterium acnes was the only species present in all samples at all body sites, ranging in average relative abundance from 35 to 89%, suggesting P. acnes may serve as an informative target species for forensic applications using skin microbiomes. Indeed, Oh et al. (9) previously reported that P. acnes strain single-nucleotide variant profiles are stable and individual-specific, and the known P. acnes pangenome (i.e., the composition of all core and accessory genes present from all known strains of a given species) reaches saturation from all P. acnes strains sampled across individuals (i.e., all genes from the P. acnes pangenome are present across all samples). Therefore, in this study, the findings of Oh et al. (9) are expanded upon and different features from P. acnes were evaluated as potential forensic targets in a supervised learning context.
FIG 1.
Proportional relative abundances of core skin microbiome taxa from 14 skin body sites. The core skin microbial taxa include prokaryotic, eukaryotic, and viral microbial species common to all samples (i.e., all individuals and time points) with ≥1% average relative abundance at each body site.
Propionibacterium acnes strain characterization and classification using P. acnes pangenome presence/absence features.
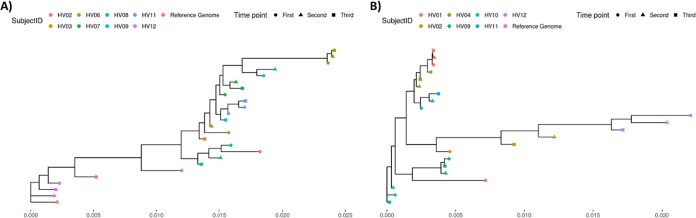
To further assess whether P. acnes may serve as a viable taxon for human identification, maximum-likelihood phylogenetic trees were constructed over 200 markers specific to the P. acnes pangenome using RAxML (29). Phylogenies of P. acnes clade-specific markers from each individual show that P. acnes strains tended to place samples from the same individuals at different time points within similar positions in the tree, though some exceptions are noted (Fig. 2).
FIG 2.
Maximum-likelihood phylogenies of P. acnes strains from all individuals and time points sampled from the antecubital fossa (Ac) (A) and cheek (Ch) (B). Phylogenetic trees were constructed using 200 P. acnes species-specific markers.
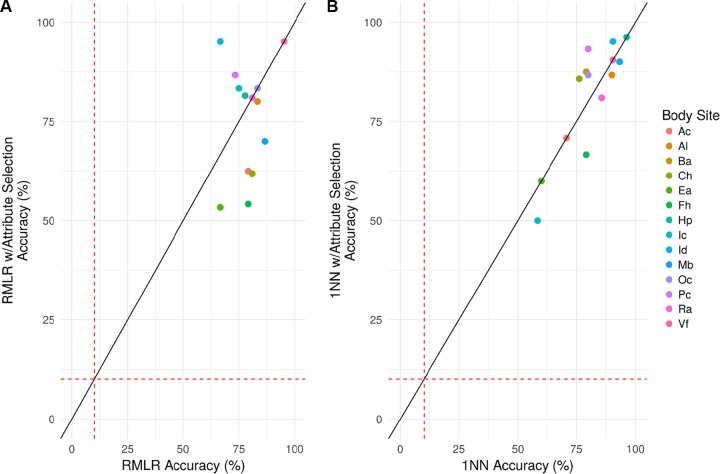
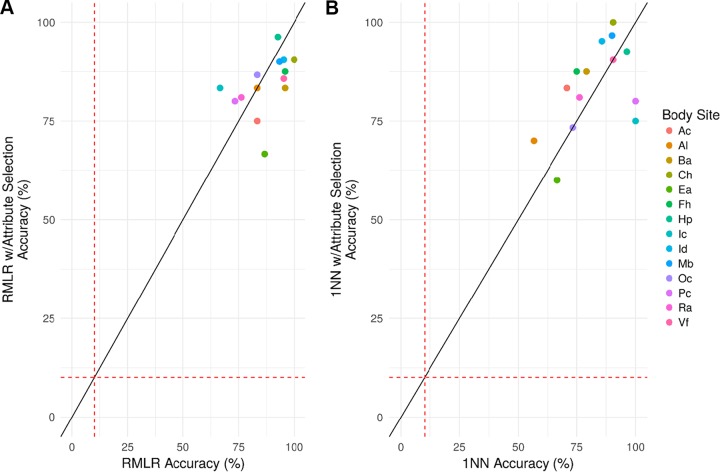
As previously reported, P. acnes strains across all samples reach pangenome gene saturation (9). Therefore, supervised learning using P. acnes pangenome gene presence/absence profiles was evaluated as a potential method for attributing skin microbiomes to their respective donors. P. acnes pangenome presence/absence profiles were constructed by aligning all P. acnes-associated reads to a database comprised of all known genes from 60 P. acnes genomes to determine the presence or absence of each gene within each sample. Presence/absence feature vectors, comprised of 551 (ear [Ea]) to 1,646 (manubrium [Mb]) features, were used to perform classification of host individuals across time points. In particular, regularized multinomial logistic regression (RMLR) and 1-nearest neighbor (1NN) classification (see Materials and Methods) were used to predict host individuals based on their microbiome signature taken at various time points. RMLR accuracies ranged from 66.67% at the ear (Ea) and interdigital web (Id) to 95.24% at the volar forearm (Vf) (4.67- to 9.52-fold higher accuracies than by random chance, respectively) with a mean accuracy of 79.40% (see Table S2 in the supplemental material). 1NN accuracies ranged from 58.33% at the inguinal crease (Ic) to 96.30% at the hypothenar palm (Hp) (3.21- to 12.52-fold higher accuracies than by random chance, respectively) with a mean accuracy of 80.71%. RMLR and 1NN classification also were evaluated on a reduced set of attribute selected markers (n = 9 to 39), with this subset of markers chosen to have similar predictive power as the sets from which they came (see Materials and Methods). The attribute-selected loci had nearly identical classification accuracies as classification using all markers collectively (Fig. 3).
FIG 3.
Classification accuracies of host individuals using P. acnes pangenome gene presence/absence features. (A and B) RMLR (A) and 1NN (B) classification, with (y axis) or without attribute selection (x axis), were used to attribute microbiomes from three time points (spanning >2.5 years) to their individual donor for 14 skin body sites. The red dashed lines represent the average predictive accuracies by random chance (10.1%).
Feature selection and classification of skin microbiomes using nucleotide diversities of stable clade-specific markers.
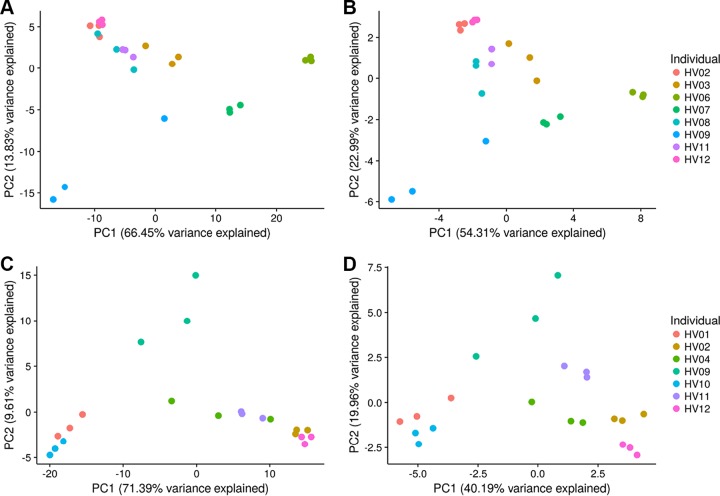
The nucleotide diversities of universal, stable clade-specific markers were evaluated as a novel feature for microbiome profiling of skin microbiomes for forensic applications. Nucleotide diversity was calculated for each clade-specific marker shared by all individuals and all time points for each body site. The number of clade-specific markers shared by all samples at each body site ranged from 239 (manubrium [Mb]) to 344 (popliteal fossa [Pc]) markers. Principal-component analysis (PCA) depicts less variation of nucleotide diversities of all shared markers between samples from the same individuals sampled at different times, than microbiomes from different individuals (Fig. 4). As represented in Fig. 4, greater variation (up to 20.85 percentage points more for the cheek [Ch]) was explained by the PCA using all shared features; however, marker reduction using feature selection (i.e., correlation-based feature subset selection, using the CfsSubsetEval evaluator in Weka [30]; see Materials and Methods) resolves overlapping clusters from different individuals to produce more defined boundaries around samples from the same individual, likely due to the reduction of redundant features contributing toward the same level of variation.
FIG 4.
PCA depicting the variance across skin microbiomes sampled from the antecubital fossa (Ac) (A) and cheek (Ch) (C) using the nucleotide diversity of shared clade-specific markers (242 and 252 markers, respectively) at each body site and using the nucleotide diversity from selected features using correlated feature selection to reduce the number of features to 27 markers at the Ac site (B) and 31 markers at the Ch site (D).
RMLR and 1NN classification were used to classify microbiome samples with respect to their individual donor in the same manner as the assessment of presence/absence markers. RMLR accuracies ranged from 66.67% at the inguinal crease (Ic) to 100% at the cheek (Ch) (3.67- to 10-fold higher accuracies than by random chance, respectively) with a mean accuracy of 87.21% (see Table S3 in the supplemental material). 1NN accuracies ranged from 56.67% at the alar crease (Al) to 100% at the inguinal crease (Ic) and popliteal fossa (Pc) (8.22- to 7-fold higher accuracies than by random chance, respectively) with a mean accuracy of 82.20%. RMLR and 1NN classification also were evaluated on a reduced set of attribute selected markers (n = 14 to 47), with this subset of markers chosen to have similar predictive power as the sets from which they came. The attribute-selected loci had nearly identical classification accuracies as classification using all markers collectively (Fig. 5).
FIG 5.
Classification accuracies of host individuals using nucleotide diversities of clade-specific markers shared by all individuals at each time point. (A and B) RMLR (A) and 1NN (B) classification, with (y axis) or without (x axis) attribute selection, were used to attribute microbiomes from three time points (spanning >2.5 years) to their individual donor for 14 skin body sites. The red dashed lines represent the average predictive accuracies by random chance (10.1%).
To assess whether our classification methods were robust to differences in time, 1NN classification accuracies, with and without attribute selection, were compared between the shortest time intervals (sampling collection time points 2 versus 3 [5 to 10 weeks]) and longest time intervals (sampling collection time points 1 versus 3 [>2.5 years]) at each body site. Microbiome samples collected 5 to 10 weeks apart could be attributed to their host individual with higher accuracy than microbiomes samples collected more than 10 to 30 months apart (see Fig. S1 in the supplemental material). Long time interval accuracies ranged from 30% at the alar crease (Al) to 100% at the popliteal fossa (Pc) and inguinal crease (Ic) with a mean accuracy of 69.52% (8.94-fold greater accuracy than by random chance). Short time interval accuracies ranged from 50% at the ear (Ea) to 100% at the forehead (Fh), inguinal crease (Ic), popliteal fossa (Pc), and volar forearm (Vf), with a mean accuracy of 85.85% (11.03-fold greater accuracy than by random chance) (see Fig. S1 in the supplemental material).
Feature selection identified 187 clade-specific markers from the following 12 clades that contributed the most to individual classification across all body sites: family level (n = 1; i.e., Propionibacteriaceae), species level (n = 10; i.e., Corynebacterium sp. strain HFH0082, Corynebacterium tuberculostearicum, Propionibacterium acnes, Propionibacterium humerusii, Propionibacterium sp. strain 434 HC2, Propionibacterium sp. strain 5 U 42AFAA, Propionibacterium sp. strain HGH0353, Propionibacterium sp. strain KPL1844, Propionibacterium sp. strain KPL1854, and Propionibacterium sp. strain KPL2008), and subspecies level (n = 1; i.e., Propionibacterium namnetense SK182B-JCVI) (see Table S4 in the supplemental material). These feature-selected markers only represent 3 of the 12 core skin microbiome species (see Fig. 1), indicating that both high- and low-abundance taxa contribute to stable features used for individual differentiation.
Assessing classifier accuracy.
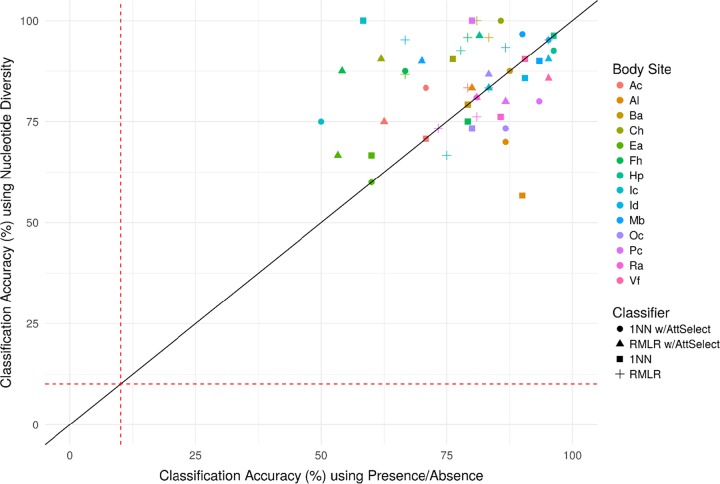
Several factors (indicated as italicized terms) may influence the probability of a correct classification (p) of a given classifier: accuracy varied substantially across body sites (BS), across feature vector type (diversity or presence/absence) (Type), and feature selection/classifier type (Classifier) may also impact p. Conditional binomial logistic regression was used to model log(p/1 − p) ∼ BS + Type + Classifier, controlling for intraindividual variation by stratifying on the (host) individual (see Materials and Methods). Several of the coefficients (log odds ratios) were statistically significant (see Table S5 in the supplemental material). In particular, the odds of an accurate classification are estimated to be 28% lower for presence/absence features than for nucleotide diversity (P < 0.01). Mean classification accuracies (p) were also contrasted between presence/absence and diversity (Fig. 6) across classifier types, and since most points are above the main diagonal (i.e., higher accuracy for diversity over presence/absence), further evidence is provided that presence/absence features are less individualizing than nucleotide diversity. RMLR and 1NN, both with and without attribute selection, did not significantly impact classification accuracy. Classification accuracies did, however, significantly vary across body sites. Compared to the occiput (Oc) body site (see Materials and Methods), which had medial classification accuracy, samples collected from the volar forearm (Vf) (P < 0.05), hypothenar palm (Hp) (P < 0.01), manubrium (Mb) (P < 0.001), and the check (Ch) (P < 0.05) had significantly higher odds of being classified correctly, and samples collected from the ear (Ea) (P < 0.001) had significantly lower odds for being classified correctly (see Table S5 in the supplemental material).
FIG 6.
Comparison of classification accuracies from RMLR and 1NN analyses, with or without attribute selection (AttSelect), using P. acnes pangenome presence/absence features and nucleotide diversities of stable clade-specific markers. The red dashed lines represent the average predictive accuracies by random chance (10.1%).
DISCUSSION
A novel approach is described for the attribution of skin microbiome samples to their individual donors with a high degree of accuracy. Microbiome samples were collected over a large time span (>2.5 years), and yet, classifier accuracies were high across a variety of body sites (see Tables S2 and S3 in the supplemental material). Of the body sites assessed, those that are likely of the greatest forensic relevance—the Mb body site (shirt) and the Hp body site (palm)—yield highly accurate rates of classification (97%/96%, respectively, using 1NN classification on nucleotide diversity), with odds ratios of 2.64 and 2.60, respectively, relative to that of a typical body site (occiput [Oc]) (see Table S5 in the supplemental material). This finding is somewhat unexpected for the hand, especially since it is likely the target of frequent recolonization from life's daily tasks and has been shown to contain relatively few (∼17%) shared phylotypes between different hands of the same individual (4). Lax et al. (20) observed similar classification accuracy (96.3%) when attributing microbiome samples from phone surfaces (i.e., touch samples from the hands and face) to their owners, when sampled from one time point for the majority of sample subjects and multiple time points over 2 days for 2 participants, whereas, when assessing classification accuracy for a skin site (i.e., anterior nares) over longer time intervals (i.e., 30 to 300 days), Fransoza et al. (8) were only able to differentiate <30% of the total number of individuals in the study. The methods reported here were used to attribute skin microbiomes to their hosts over long time intervals (>2.5 years) and obtain high classification accuracy for multiple skin body sites.
In this study, two different feature types were assessed with supervised learning (i.e., RMLR and 1NN) to differentiate skin microbiomes from different individuals. P. acnes pangenome presence/absence features were selected based on the stability of P. acnes strain-level signatures and pangenome saturation over time (9) and yielded high classification accuracies (up to 96.3%), likely due to high species abundance across multiple body sites allowing for greater genome coverage for characterization. Nucleotide diversity of shared clade-specific markers was selected as a feature type to capture population-level genetic variation of stable markers, since nucleotide diversity of strains has been shown to differ significantly between individuals from different geographical regions (26). Nucleotide diversity of stable markers yielded accuracies as high as 100% from the cheek (Ch), inguinal crease (Ic), and popliteal fossa (Pc) and contributed significantly greater (by an estimated 28%, with a 95% confidence interval of 10 to 43%) to classification accuracies than presence/absence features (P < 0.01) (see Table S5 in the supplemental material). This finding contrasts with those from Fransoza et al. (8), who showed that minimum cardinality sets of presence/absence features (i.e., 1-kb genomic window counts) are an ideal feature type for human identification. However, we demonstrate that presence/absence features do provide high classification accuracies (see Table S2 in the supplemental material), this feature type fails to capture additional genetic variation which significantly contributes to classification accuracy (i.e., nucleotide diversity) (see Table S5 in the supplemental material). Furthermore, presence/absence findings as inferred from shotgun sequencing data are likely susceptible to stochastic effects, increasing the likelihood that informative markers may drop out in highly diverse, poorly collected, or degraded samples, sample types typical in forensic settings, and further requires parameterization on what constitutes “absence.”
Attribute selection also was performed to evaluate classification performance using reduced subsets of features, selected to have similar predictive power as the full set of markers. Since attribute selection was performed using a correlation-based approach, features were selected independent of the classifier type (unlike features selected specific to a particular classifier; see, for example, reference 31), and thus the markers identified in this study are potentially informative for a wide range of supervised learning algorithms. Feature selection did not have a significant effect on classification accuracy (see Table S5 in the supplemental material), indicating that using an average of 24 markers reduced from 1,108 for presence/absence features and an average of 32 markers reduced from 263 clade-specific markers resulted in classification accuracies comparable to those achieved using full sets of features. Feature reduction helps eliminate markers which do not significantly contribute to microbiome classification (see Table S5 in the supplemental material), thus eliminating potential noise and redundancy in signal, and helps select for a reduced panel of candidate markers to be developed into a multiplex assay for targeted sequencing assays for microbiome characterization.
In this study, the nucleotide diversities of subsets of clade-specific markers were used to differentiate skin microbiomes samples from individuals sampled over relatively long time intervals with a high degree of accuracy. The main limitation within the study herein was sample size (n = 12 to 30 per body site). In this study, within a given body site only three intraindividual samples were available, which limits training. Larger sample sizes are needed to further validate the methods described here and to develop statistical models to incorporate the likelihood of microbiome classification to provide weight to similar or inclusionary comparisons. These results support future development of a robust and reproducible method for human identification using skin microbiomes. Since microbiomes likely do not have the same level of genetic stability as the human genome, identifying the most stable, personalizing features within microbiomes allows for further studies to more comprehensively assess the stability of these features and how these features contribute to classification accuracy using significantly larger population sample sets.
The study here does not address whether the data are applicable to real or mock forensic applications (e.g., touching an object and recovering deposited skin flora). That study cannot be performed as public data of this nature are not available. More importantly performing that study would be premature. For forensic applications informative targets will likely need to be enriched, as they are for current human identification methods. Our study has identified candidate markers that may be suitable to test forensically relevant samples, such as touched items which would tend to have low biomass and may be somewhat degraded. Targeted enrichment and sequencing using a panel of the most informative markers would provide an ideal solution for microbiome profiling for forensic identification to obtain high coverage at stable informative sites. A multiplex is being designed to empirically test these selected candidate markers for classification accuracy and sensitivity at various sites on the human skin, including the currently low informative foot region. Once assessed for performance, larger data sets (e.g., population studies) can be generated to enable statistical weighting and resolution comparisons with those of human identification forensic genetic marker systems. The field of microbial forensics has expanded from strictly focusing on biothreat attribution to include multiple areas of microbiome applications (32), and as such, future studies should consider method development as well as new statistical models to more accurately interpret microbiome data and establish standards and validation criteria before microbiome profiling can be actively used for investigative leads and attribution within the forensic scientific community.
MATERIALS AND METHODS
Public skin microbiome data set selection and download.
Skin microbiome shotgun metagenomic data sets, comprised of samples from 12 healthy individuals across 17 body sites and sampled at three different time points, were downloaded using the NCBI SRA Toolkit (27), using the program fastq-dump to download 2,446 fastq files (corresponding to 585 samples) from the SRA (27), under BioProject accession number PRJNA46333. Sample collection and sequencing methods are described by Oh et al. (9).
Metagenomic sequence data analysis.
Metagenomic data sets were preprocessed for read quality control, using (i) Cutadapt (33) to remove sequence adapters, trim reads with quality scores of <20, and remove reads of <50 bp; (ii) the Burrows-Wheeler alignment tool (34) to align and remove human host-associated reads; and (iii) Samtools v1.3.1 (35) to convert sorted .bam files to fastq format for downstream use. Taxonomic classification of skin microbiomes was performed using MetaPhlAn2 (28) using default parameters. Variant calls and associated coverage for aligned MetaPhlAn2 (28) markers shared by all samples at a particular body site were determined using Samtools mpileup (35). Only samples that met the following criteria for each body site were included in the study: ≥50× maximum coverage at any marker site within samples, ≥10× average coverage across all markers, and samples with all three time points for an individual (see Table S1 in the supplemental material). Three body sites from the foot (i.e., plantar heel [Ph], toenail [Tn], and toe web space [Tw]) also were excluded from the study, since they only shared two to five markers among samples.
A custom perl script was used to parse mpileup outputs and calculate nucleotide diversity (π) of each marker, with ≥5× coverage, shared by all individuals and time points for each body site. The nucleotide diversity (π) was calculated using the following equation:
where pi is the frequency of the reference base at the ith site in the nth base of the marker, as described in Nayfach and Pollard (26). Strain maximum-likelihood phylogenies of P. acnes were constructed using RAxML (29) as implemented in StrainPhlAn (36). Briefly, StrainPhlAn was used to generate sequence alignments using MUSCLE (37) from sequence reads aligned to 200 P. acnes markers from MetaPhlAn2 (28), and RAxML (29) was used to generate maximum-likelihood phylogenetic trees. The ggtree (38) and ggplot2 (39) R libraries using the “strainphlan_ggtree.R” script from https://bitbucket.org/biobakery/breadcrumbs was used to build the trees. Pangenome gene presence/absence profiles for P. acnes were generated using PanPhlAn (40), using the preprocessed “panphlan_pacnes16” database (https://bitbucket.org/CibioCM/panphlan/wiki/Pangenome%20databases).
Unsupervised learning, supervised learning, and attribute selection.
PCA was performed using the prcomp command in R. Statistical classification was performed in WEKA (30). Classification of individuals was performed by evaluating two data feature types: nucleotide diversity and pangenome gene presence/absence. Nucleotide diversity and pangenome feature vectors were created using a custom R script, which also removed any invariant features (defined as having a standard deviation <1e−6 across all samples). RMLR and 1NN classification using the Euclidean distance measure were used to perform classification, with all parameters set to their default values. Classification accuracy (i.e., the percentage of correctly classified samples in the data set) was assessed using leave-one-out cross-validation (i.e., n-fold cross-validation; n = sample size) so as to maximize the size of the training data set while mitigating the effects of overfitting. Thus, n sets each composed of n−1 individuals were used to train classifiers, and accuracies were assessed on the single “left out” individual, with the overall accuracies being the sums of the n correct and incorrect classifications. Attribute selection was performed by a correlation-based feature subset selection method, using the CfsSubsetEval evaluator in Weka (30), prior to each classification method, with default parameters and using leave-one-out cross validation. Upper and lower 95% confidence intervals were calculated for our estimates of classification accuracies using the binom.confint function from the binom R library (41) using the “asymptotic” method. All figures were created using the ggplot2 (39) and cowplot (42) R libraries unless stated otherwise. All custom scripts can be accessed online (https://github.com/SESchmedes/HIDskinmicrobiome).
Conditional binomial logistic regression.
Conditional binomial logistic regression was used to evaluate classifier accuracy, which models the log odds of a correct classification (p) as a linear function of the classifiers employed, the body site, and the feature vectors evaluated. In particular, log(p/1 − p) was modeled as a function of classifier type (1NN and RMLR, both with and without feature selection), the body site (column 1 of Table S1 in the supplemental material), and feature vector type, i.e., whether the classification was performed using presence/absence (encoded as 1) or diversity (encoded as 0). Since these measures were repeated within individuals, traditional binomial logistic regression would otherwise underestimate error terms. Instead conditional binomial logistic regression was used to account for the repeated measures design, using the host individual as a stratum, with the clogit function in R. As the body site independent variable, we chose the Oc body site as our reference category since it had medial marginal accuracy (rank 7 of 14), and the largest marginal sample size (n = 240).
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institute of Justice (award 2015-NE-BX-K006) and by a Texas Branch of the American Society for Microbiology 2014 Eugene and Millicent Goldschmidt Graduate Student Award.
We thank Jonathan L. King and David Warshauer for support and technical assistance. We also thank the authors of the Oh et al. (9) study for making their skin microbiome publically available, allowing us to perform the present study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01672-17.
REFERENCES
- 1.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fierer N, Hamady M, Lauber CL, Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A 105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. 2011. Diversity of the human skin microbiome early in life. J Invest Dermatol 131:2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, D Lieber A, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:1–14. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C. 2015. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A 112:E2930–E2938. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh J, Byrd AL, Park M, Kong HH, Segre JA. 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hares DR. 2015. Selection and implementation of expanded CODIS core loci in the United States. Forensic Sci Int Genet 17:33–34. doi: 10.1016/j.fsigen.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Wilson MR, DiZinno JA, Polanskey D, Replogle J, Budowle B. 1995. Validation of mitochondrial DNA sequencing for forensic casework analysis. Int J Legal Med 108:68–74. doi: 10.1007/BF01369907. [DOI] [PubMed] [Google Scholar]
- 12.Holland MM, Parsons TJ. 1999. Mitochondrial DNA sequence analysis: validation and use for forensic casework. Forensic Sci Rev 11:21–50. [PubMed] [Google Scholar]
- 13.King JL, LaRue BL, Novroski NM, Stoljarova M, Seo SB, Zeng X, Warshauer DH, Davis CP, Parson W, Sajantila A, Budowle B. 2014. High-quality and high-throughput massively parallel sequencing of the human mitochondrial genome using the Illumina MiSeq. Forensic Sci Int Genet 12C:128–135. doi: 10.1016/j.fsigen.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Budowle B, Eisenberg AJ, van Daal A. 2009. Validity of low copy number typing and applications to forensic science. Croat Med J 50:207–217. doi: 10.3325/cmj.2009.50.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacterial cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage DC. 1977. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 17.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. 2010. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A 107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goga H. 2012. Comparison of bacterial DNA profiles of footwear insoles and soles of feet for the forensic discrimination of footwear owners. Int J Legal Med 126:815–823. doi: 10.1007/s00414-012-0733-3. [DOI] [PubMed] [Google Scholar]
- 19.Meadow JF, Altrichter AE, Green JL. 2014. Mobile phones carry the personal microbiome of their owners. PeerJ 2:e447. doi: 10.7717/peerj.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lax S, Hampton-Marcell JT, Gibbons SM, Colares GB, Smith D, Eisen JA, Gilbert JA. 2015. Forensic analysis of the microbiome of phones and shoes. Microbiome 3:21. doi: 10.1186/s40168-015-0082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams DW, Gibson G. 2017. Individualization of pubic hair bacterial communities and the effects of storage time and temperature. Forensic Sci Int Genet 26:12–20. doi: 10.1016/j.fsigen.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Nishi E, Tashiro Y, Sakai K. 2014. Discrimination among individuals using terminal restriction fragment length polymorphism profiling of bacteria derived from forensic evidence. Int J Legal Med 129:425–433. doi: 10.1007/s00414-014-1092-z. [DOI] [PubMed] [Google Scholar]
- 23.Nishi E, Watanabe K, Tashiro Y, Sakai K. 2017. Terminal restriction fragment length polymorphism profiling of bacterial flora derived from single human hair shafts can discriminate individuals. Leg Med 25:75–82. doi: 10.1016/j.legalmed.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown G, Green JL, Bohannan BJ. 2015. Humans differ in their personal microbial cloud. PeerJ 3:e1258. doi: 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leake SL, Pagni M, Falquet L, Taroni F, Greub G. 2016. The salivary microbiome for differentiating individuals: proof of principle. Microbes Infect 18:399–405. doi: 10.1016/j.micinf.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Nayfach S, Pollard KS. 2015. Population genetic analyses of metagenomes reveal extensive strain-level variation in prevalent human-associated bacteria. bioRxiv doi: 10.1101/031757. [DOI] [Google Scholar]
- 27.Kodama Y, Shumway M, Leinonen R. 2012. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 29.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank E, Hall MA, Witten IH. 2016. The WEKA Workbench. Online appendix for “Data mining: practical machine learning tools and techniques.” http://www.cs.waikato.ac.nz/ml/weka/Witten_et_al_2016_appendix.pdf.
- 31.Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc 58:267–288. [Google Scholar]
- 32.Schmedes SE, Sajantila A, Budowle B. 2016. Expansion of microbial forensics. J Clin Microbiol 54:1964–1974. doi: 10.1128/JCM.00046-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:1. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 34.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 39.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 40.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, Truong DT, Tett A, Morrow AL, Segata N. 2016. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 41.Dorai-Raj S. 2014. binom: binomial confidence intervals for several parameterizations. R package version 1.1-1. https://cran.r-project.org/package=binom. [Google Scholar]
- 42.Wilke CO. 2016. cowplot: streamlined plot theme and plot annotations for “ggplot2.” R package version 0.7.0. https://cran.r-project.org/package=cowplot. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.