Sze et al. show that H3K4 methylation by Set1A is dispensable for ESC proliferation and self-renewal, but Set1AΔSET ESCs are unable to undergo normal differentiation. During differentiation, Set1A but not Mll2 functions as the H3K4 methylase on bivalent clusters and is required for their expression.
Keywords: COMPASS, Set1A, H3K4 methylation, pluripotency, self-renewal, differentiation
Abstract
Of the six members of the COMPASS (complex of proteins associated with Set1) family of histone H3 Lys4 (H3K4) methyltransferases identified in mammals, Set1A has been shown to be essential for early embryonic development and the maintenance of embryonic stem cell (ESC) self-renewal. Like its familial relatives, Set1A possesses a catalytic SET domain responsible for histone H3K4 methylation. Whether H3K4 methylation by Set1A/COMPASS is required for ESC maintenance and during differentiation has not yet been addressed. Here, we generated ESCs harboring the deletion of the SET domain of Set1A (Set1AΔSET); surprisingly, the Set1A SET domain is dispensable for ESC proliferation and self-renewal. The removal of the Set1A SET domain does not diminish bulk H3K4 methylation in ESCs; instead, only a subset of genomic loci exhibited reduction in H3K4me3 in Set1AΔSET cells, suggesting a role for Set1A independent of its catalytic domain in ESC self-renewal. However, Set1AΔSET ESCs are unable to undergo normal differentiation, indicating the importance of Set1A-dependent H3K4 methylation during differentiation. Our data also indicate that during differentiation, Set1A but not Mll2 functions as the H3K4 methylase on bivalent genes and is required for their expression, supporting a model for transcriptional switch between Mll2 and Set1A during the self-renewing-to-differentiation transition. Together, our study implicates a critical role for Set1A catalytic methyltransferase activity in regulating ESC differentiation but not self-renewal and suggests the existence of context-specific H3K4 methylation that regulates transcriptional outputs during ESC pluripotency.
The highly conserved COMPASS (complex of proteins associated with Set1) family of methylases implements methylation at Lys4 of histone H3 (H3K4) (Shilatifard 2012; Sze and Shilatifard 2016), a mark associated with transcriptionally active chromatin. The H3K4 methyltransferase Set1A is one of six COMPASS members identified in mammals and has consistently been shown to deposit bulk H3K4 dimethylation and trimethylation (H3K4me2/me3, respectively) across the genome (Wu et al. 2008; Ardehali et al. 2011; Mohan et al. 2011; Hallson et al. 2012; Bledau et al. 2014; Fang et al. 2016; Cao et al. 2017). Set1A is critical for early mouse embryonic development, as Set1A knockout results in early embryonic lethality at embryonic day 7.5 (E7.5) (Bledau et al. 2014). Defects exhibited in Set1A knockout embryos suggest that Set1A functions shortly after inner cell mass formation but before gastrulation (Bledau et al. 2014). Unlike any of its other COMPASS familial relatives (Bledau et al. 2014; Denissov et al. 2014; Cao et al. 2017; Dorighi et al. 2017), Set1A is essential for embryonic stem cell (ESC) viability: ESCs could not be derived from Set1A knockout blastocysts (Bledau et al. 2014; Fang et al. 2016), and homozygous Set1A knockout ESCs could not be generated via CRISPR/Cas9 (Cao et al. 2017). Depletion of Set1A hinders ESC proliferation and triggers their apoptosis (Bledau et al. 2014; Fang et al. 2016; Cao et al. 2017), affirming the importance of Set1A in ESC survival and identity. The involvement of Set1A in cellular differentiation has been explored in several cellular contexts. Set1A reportedly mediates hematopoietic lineage differentiation in culture (Deng et al. 2013) and B-cell development in vivo (Tusi et al. 2015). Our laboratory has also recently established a role for Set1A as a transcriptional activator of Hox gene expression during ESC differentiation (Cao et al. 2017). To date, we still have very limited knowledge of the role of Set1A in regulating ESC pluripotency; specifically, whether the function of Set1A in ESC maintenance is dependent on its methylase activity remains elusive.
Results and Discussion
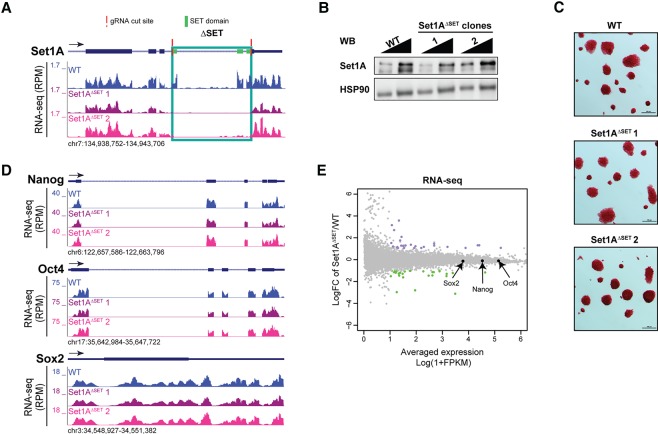
To investigate the role of the enzymatic activity compared with the rest of the Set1A protein in ESC pluripotency, we began by determining whether deletion of its C-terminal catalytic SET domain adversely affects ESC self-renewal and viability (Fig. 1; Supplemental Fig. S1). Using CRISPR/Cas9, we deleted an ∼1.7-kb endogenous genomic sequence coding for the SET domain of Set1A (Supplemental Fig. S1B,C). We identified two homozygous mutant clones containing the SET domain deletion of Set1A (Set1AΔSET) via PCR genotyping and further verified by Sanger sequencing and RNA sequencing (RNA-seq) (Fig. 1A; Supplemental Fig. S1B,C). Sanger sequencing of both the genomic DNA and mRNA revealed that these mutant clones harbor the SET domain deletion that would result in the introduction of an early stop codon, leading to protein truncation that also removes the post-SET domain (Supplemental Fig. S1A,C). Truncated Set1A protein stability is comparable with that of wild-type protein detected in the parental ESCs (Fig. 1B). Remarkably, Set1AΔSET ESCs retain their self-renewal characteristics: Mutant cells display a morphology strikingly similar to that of the wild-type cells, as evidenced by alkaline–phosphatase staining (Fig. 1C). Expression of the principal pluripotency factors Nanog, Oct4, and Sox2 is not altered in Set1AΔSET ESCs (Fig. 1D; Supplemental Fig. S1D), and further global analyses demonstrated that the overall transcriptome is indeed similar (Fig. 1E; Supplemental Table 1). Although deletion of Set1A was shown to perturb ESC pluripotency, our data show that the SET domain of Set1A is not required for ESC self-renewal, and the self-renewal function associated with Set1A requires a region of the protein upstream of the catalytic domain.
Figure 1.
Set1AΔSET does not perturb ESC self-renewal. (A) RNA-seq results confirm CRISPR/Cas9-mediated deletion (green box) of the genomic sequence coding for the SET domain of Set1A for two homozygous clones. (RPM) Reads per million. (B) Western blot (WB) of Set1A levels in wild-type (WT) and Set1AΔSET ESCs, with HSP90 as the loading control. Samples were loaded at a 1:2 ratio. (C) Representative images of alkaline–phosphatase staining of wild-type and Set1AΔSET ESC colonies. Bar, 100 µm. (D) RNA-seq tracks of pluripotency factors Nanog, Oct4, and Sox2 between wild-type ESCs and mutant clones. (E) MA plot comparing the global transcriptome between parental and Set1AΔSET ESCs. Expressions of pluripotency factors Nanog, Oct4, and Sox2 are indicated. (FPKM) Fragments per kilobase of exon per million.
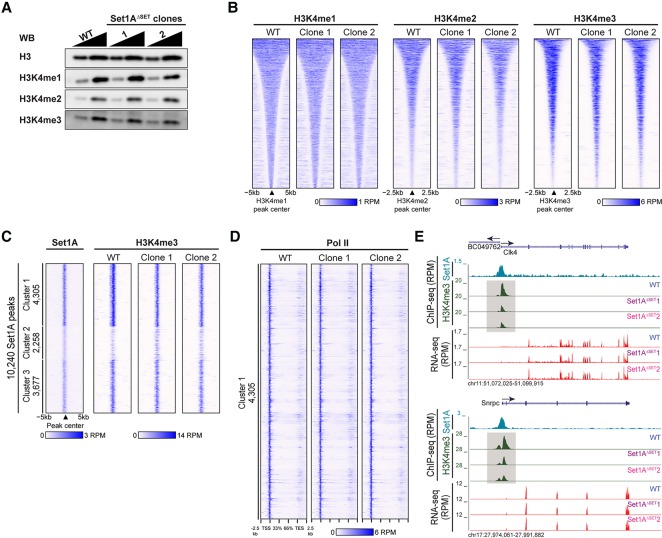
We assessed H3K4 methylation in the Set1AΔSET ESCs compared with that in parental wild-type cells. As observed by Western blot, bulk H3K4me1/me2/me3 levels are comparable between wild-type and mutant ESCs (Fig. 2A). To determine the changes in the levels and pattern of H3K4 methylation on chromosomes, ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analyses of global H3K4 methylation in wild-type and Set1AΔSET clones were performed, and these data further support our findings (Fig. 2B). To identify genomic sites with H3K4me3 changes specific to Set1A, we performed ChIP-seq of Set1A. A total of 10,240 Set1A-binding regions was characterized, with promoter localization being the predominant annotation as calculated by HOMER (Supplemental Fig. S2A). Examination of genome-wide Set1A occupancy revealed similar Set1A binding in wild-type and Set1AΔSET ESCs (Supplemental Fig. S2B). We subsequently centered H3K4me3 peaks from wild-type and Set1AΔSET ESCs at Set1A-binding regions. Specifically, we used K-means clustering to partition H3K4me3 occupancy at Set1A sites into three clusters for the parental cells and the Set1AΔSET clones (Fig. 2C). The first cluster, containing loci with the strongest Set1A binding, exhibits a detectable reduction in H3K4me3 peak occupancy in both Set1AΔSET mutants compared with wild-type cells, which is further illustrated in the differential heat map for H3K4me3 occupancy (Fig. 2C; Supplemental Fig. S2C; Supplemental Table 2). We examined the expression of the nearest genes for all three peak clusters and noted that cluster 1 coincides with the highest nearest gene expression (Supplemental Fig. S2D). Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al. 2010) analysis showed that cluster 1 peaks are present primarily at regions closest to transcription start sites (TSSs) (Supplemental Fig. S2E). Furthermore, despite the observed H3K4me3 decrease of cluster 1 peaks, there were no compelling changes in RNA polymerase II (Pol II) occupancy (Fig. 2D). Representative track examples in Figure 2E provide a clear visualization of H3K4me3 decrease at the promoters, although the accompanying RNA-seq data show no difference in the pattern of gene expression. We speculate the following: (1) Residual H3K4me3 at these specific loci is adequate to maintain gene expression, or (2) Set1AΔSET may be affecting recruitment of other H3K4 methyltransferases in the COMPASS family, which we plan to explore in future studies. We currently have reason to believe that Set1B, a close homolog of Set1A, does not compensate for Set1AΔSET ESCs sufficiently to sustain global H3K4me3 because Set1B overexpression was unable to rescue defects caused upon loss of Set1A in ESCs (Bledau et al. 2014). Nevertheless, the findings thus far communicate that the catalytic SET domain of Set1A and its associated H3K4me3 are dispensable for ESC survival and identity.
Figure 2.
Set1AΔSET in undifferentiated ESCs resulted in a decrease of H3K4me3 at specific sites. (A) Western blot comparing H3K4 methylation levels in Set1AΔSET with parental wild-type cells. H3 served as the loading control. Samples were loaded at a 1:2 ratio. (B) Heat maps of H3K4me1, H3K4me2, and H3K4me3 ChIP-seq occupancy levels in wild-type and Set1AΔSET cells. Occupancy levels were aligned to wild-type peaks sorted by decreasing peak width for each individual modification. (C) Set1A peaks (10,240) were called and partitioned into three groups by K-means clustering, and the corresponding H3K4me3 occupancy at the Set1A peaks was plotted for wild-type and mutant ESCs. (D) Pol II occupancy at the cluster 1 sites for wild-type and Set1AΔSET ESCs. (E) Genome browser track examples with corresponding RNA-seq. H3K4me3 and Set1A ChIP-seq tracks are shown. Decreases in H3K4me3 as a result of Set1AΔSET are highlighted in gray boxes.
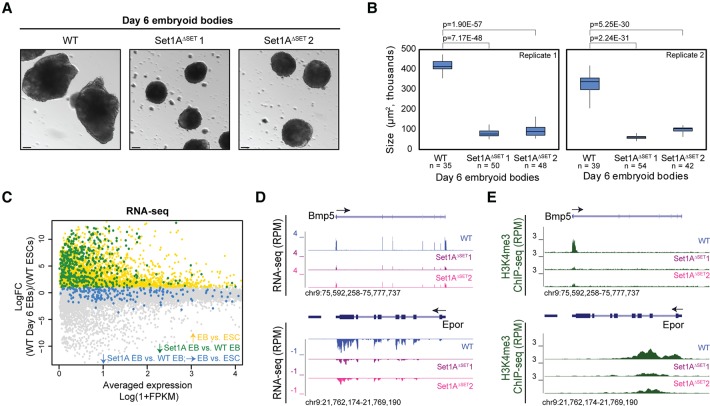
Set1A is required for early embryogenesis (Bledau et al. 2014); however, our data show that ablation of Set1A's catalytic activity does not perturb ESC self-renewal. Therefore, we investigated whether our mutant ESCs exhibit defective differentiation by culturing wild-type and Set1AΔSET ESCs to form embryoid bodies (EBs). By day 6 of the EB generation process, spheroids generated from Set1AΔSET ESCs were significantly smaller than those derived from wild-type cells (Fig. 3A,B). We performed RNA-seq of day 6 EBs to compare gene expression profiles of wild-type and Set1AΔSET EBs. Differential expression analyses revealed that 649 genes are significantly down-regulated in both mutant EBs compared with wild-type EBs (Supplemental Fig. S3A), suggesting that Set1AΔSET cells are undergoing defective differentiation. We subsequently identified that 447 of these 649 down-regulated genes are those that fail to be normally up-regulated during mutant Set1AΔSET EB formation (Fig. 3C; Supplemental Fig. S3B; Supplemental Table 3). Gene ontology (GO) annotation shows that these 447 genes are enriched for biological processes linked to tissue specification and development, especially an enrichment for genes involved primarily in mesoderm differentiation (Supplemental Fig. S3C). Track examples of Bmp5 and Epor, two genes involved in mesoderm differentiation, exhibit such diminished gene expression in both Set1AΔSET EBs compared with wild-type EBs (Fig. 3D; Supplemental Fig. S3D). Consistent with the notion that Set1AΔSET ESCs display impaired differentiation, we observed elevated expressions of Nanog, Oct4, and Sox2 in mutant EBs compared with wild type (Supplemental Fig. S3E,F), indicating that these pluripotency factors are not properly extinguished during Set1AΔSET EB formation, which may suggest a role for H3K4 methylation in transcriptional repression either directly or indirectly. To validate our findings that Set1AΔSET impairs ESC differentiation, we tested monolayer differentiation by culturing the cells in N2B27 medium without 2i/LIF (MEK inhibitor, GSK3 inhibitor, and LIF), which would promote ESC differentiation toward the neuronal lineage (Ying et al. 2003). 2i/LIF withdrawal in the first 2 d promoted exit from the self-renewal state in both wild-type and Set1AΔSET cells (Supplemental Fig. S3G). However, differentiation of Set1AΔSET cells was radically disrupted by day 3, for the Set1AΔSET clones were unable to further generate neurite projections as seen in wild-type cells (Supplemental Fig. S3G). We also scrutinized the effects on HoxA activation upon 24-h retinoic acid (RA) treatment of wild-type versus Set1AΔSET ESCs, since we had reported previously that HoxA gene activation is Set1A-dependent (Cao et al. 2017). Decreased expressions of Hoxa4, Hoxa5, and Hoxa7 genes were detected in Set1AΔSET cells compared with wild-type cells upon RA induction (Supplemental Fig. S3H), reinforcing both our previous findings and that Set1A SET domain-dependent H3K4me3 is key in differentiation (Cao et al. 2017). Our results thus far evince that Set1A's H3K4 trimethylase catalytic activity is required for normal ESC differentiation but is dispensable for ESC self-renewal, suggesting multiple roles for Set1A and its catalytic activity in ESC pluripotency.
Figure 3.
Set1AΔSET mutants exhibit defective EB differentiation. (A) Wild-type and Set1AΔSET ESCs were induced to form EBs. Emboss contrast images were taken at day 6 of EB formation. Bar, 100 µm. (B) Box plot contrasting the size (in square micrometers) of wild-type day 6 EBs to that of Set1AΔSET EBs. The number of EBs measured per sample per replicate is indicated. P-values were calculated using the Student's t-test. (C) MA plot of gene expression changes between undifferentiated wild-type ESCs and wild-type day 6 EBs. (Yellow) Up-regulated genes during differentiation; (blue) genes down-regulated in Set1AΔSET EBs not pertinent to differentiation; (green) genes down-regulated in Set1AΔSET EBs but activated during normal differentiation. Colors correspond to the categories illustrated in Supplemental Figure S3B. (D) Two genome browser track examples of genes Bmp5 and Epor down-regulated in Set1AΔSET EBs compared with wild-type EBs. (E) Example genome tracks of decreased H3K4me3 occupancy in Set1AΔSET EBs compared with wild-type EBs for the two genes shown in D.
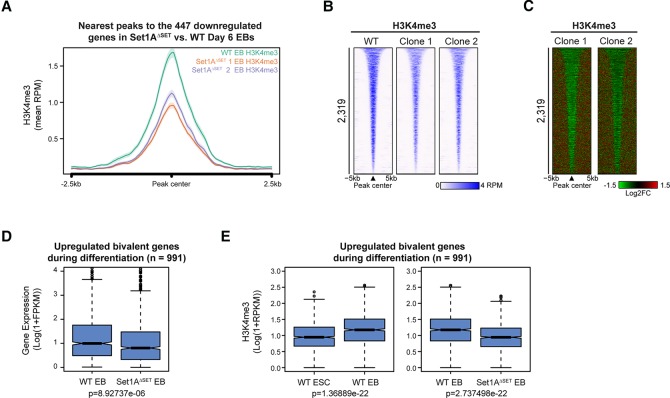
To further determine whether the enzymatic activity of Set1A regulates H3K4 methylation in Set1AΔSET EBs, we sought to ascertain H3K4me3 levels via ChIP-seq analyses. We noticed that reduced H3K4me3 occupancy in Set1AΔSET EBs is evident at Bmp5 and Epor (Fig. 3E) and therefore surveyed H3K4me3 levels at the 447 misregulated genes in Set1AΔSET EBs. Decreased H3K4me3 was observed in Set1AΔSET EBs compared with wild-type EBs (Fig. 4A), correlating with down-regulated expression of these genes in mutant EBs. To examine the extent of H3K4me3 changes attributed to Set1A activity, we first identified sites of differentially increased H3K4me3 occupancy during differentiation (Supplemental Fig. S4A). At the sites of increased H3K4me3 peaks during differentiation, we observed a compelling reduction of H3K4me3 occupancy in Set1AΔSET EBs compared with the occupancy in wild-type EBs (Fig. 4B,C; Supplemental Fig. S4B), in line with our findings that the catalytic activity of Set1A is necessary for proper stem cell differentiation.
Figure 4.
Set1A catalytic activity is required for H3K4me3 implementation during differentiation. (A) Metaplot of H3K4me3 levels for the 447 genes significantly down-regulated (as described in Fig. 3C and Supplemental Fig. S3B) in Set1AΔSET day 6 EBs relative to wild-type EBs. Peaks were centered at EB H3K4me3 peaks. (B) Heat map of H3K4me3 occupancy in wild-type and Set1AΔSET day 6 EBs. Peaks were centered at increased H3K4me3 peaks during differentiation (refer to Supplemental Fig. S4B). Peaks are rank-ordered by H3K4me3 peak width. (C) Log2 fold changes in H3K4me3 binding for peaks ordered in B. (D) Box plot gene expression analysis of the 991 bivalent genes activated during differentiation in mutant EBs versus wild-type EBs. The list of bivalent genes was from Galonska et al. (2015). P-value was determined by the Welch two-sample t-test. (E) Box plot representation of the levels of H3K4me3 peaks nearest to the 991 bivalent genes in wild-type ESCs versus wild-type EBs (left) and in wild-type EBs versus Set1AΔSET EBs (right). P-values were determined by the Welch two-sample t-test. (RPKM) Reads per kilobase of transcript per million mapped reads.
Another member of the COMPASS family of H3K4 methyltransferases, Mll2, deposits H3K4me3 at bivalent promoters in ESCs (Hu et al. 2013). However, upon differentiation of Mll2-depleted cells, increased gene expression and H3K4me3 were still observed at bivalent genes (Hu et al. 2013; Denissov et al. 2014), indicating that another enzyme might be responsible for activating and catalyzing H3K4me3 at these loci. We analyzed the expression of bivalent genes up-regulated during differentiation and found a significant reduction in their expression with a coincident decrease in H3K4me3 occupancy in Set1AΔSET EBs compared with wild-type EBs (Fig. 4D,E). These data suggest that Set1A catalytic activity is key in driving transcriptional activation of bivalently marked genes (which denote poised developmental regulators in pluripotent stem cells) (Bernstein et al. 2006; Barski et al. 2007) during cellular differentiation. Without Set1A methyltransferase activity, these regulators could not be expressed normally, rendering them unable to govern proper differentiation.
Taken together, our data demonstrate that while Set1A H3K4 methyltransferase activity is dispensable for ESC self-renewal and maintenance, it is essential for properly coordinating gene expression during ESC differentiation. Our findings substantially differ from a recent study that reported the negligible impact of catalytic-deficient Mll3 and Mll4, COMPASS relatives of Set1A, on ESC viability and transcriptional program (Dorighi et al. 2017). Unlike Mll3/Mll4 knockout ESCs, Set1A deletion in ESCs renders the cells nonviable (Bledau et al. 2014; Fang et al. 2016; Dorighi et al. 2017). Therefore, the revelation that the enzymatic activity of Set1A/COMPASS is not essential for ESC self-renewal is remarkable. On the basis of our findings, we propose the following: In self-renewing ESCs, TSSs at lineage-specific genes harbor low levels of H3K4me3 implemented by Mll2/COMPASS (Hu et al. 2013; Denissov et al. 2014); however, during differentiation, Set1A/COMPASS, potentially acting in concert with other coactivating complexes, promotes H3K4me3 deposition at these developmental genes, thus designating them for transcriptional activation. Our data collectively suggest that there is a potential functional switch between COMPASS family members (here, Mll2/COMPASS and Set1A/COMPASS) in stage-specific epigenetic regulation of transcriptional outputs. When we compared mRNA levels of Set1A and Mll2 in ESCs and day 6 EBs, we found a drop in Mll2 expression, while Set1A levels remained constant in the EB state, signifying that Set1A's functional activity in EBs is correspondingly more impactful in transcriptional regulation during differentiation (Supplemental Fig. S4C). Additionally, upon generating Mll2 knockout (Hu et al. 2017) day 6 EBs and assessing changes in H3K4me3 and gene expression at the same set of activated bivalent genes, we found that, unlike Mll2 knockout, there was a significant decrease in both H3K4me3 deposition and gene expression in Set1AΔSET EBs at the indicated bivalent genes (Supplemental Fig. S4D). The data suggest that Set1A but not Mll2 is responsible for H3K4 methylation and expression of bivalent genes during differentiation and support our proposal of a functional switch in transcriptional regulation between Mll2 and Set1A during the self-renewing-to-differentiation transition. In other words, our findings suggest that there might be a context-specific pattern of H3K4 methylation that directs transcriptional regulation during ESC self-renewal and differentiation.
Materials and methods
Antibodies
The following antibodies were generated in-house: anti-H3, anti-H3K4me1, anti-H3K4me2, anti-H3K4me3, and anti-Set1A. Anti-HSP90 was purchased from Santa Cruz Biotechnology (no. 7947), and anti-Rpb1 was purchased from Cell Signaling Technology (no. D8L4Y).
ESC culture, EB formation, and CRISPR/Cas9-mediated deletion
Mouse V6.5 ESCs were cultured in N2B27 medium supplemented with 2i/LIF as described previously (Ying et al. 2008). EBs were generated via the hanging drop method, where 1500 ESCs were cultured in 25 µL of EB differentiation medium on the lid of 150-mm culture plates for 6 d. EB differentiation medium was composed of DMEM supplemented with 15% FBS (Gemini Bio-Products), 1× GlutaMAX (Gibco), 1× MEM nonessential amino acids (Gibco), 1× β-mercaptoethanol (Gibco), and 1× penicillin–streptomycin (Gibco). N2B27 monolayer differentiation was performed as described previously (Ying et al. 2003). For RA-induced differentiation, ESCs were grown in N2B27 medium with 1 µM all-trans RA (Sigma) without 2i/LIF, and RA treatments were performed for 24 h. Generation of CRISPR single-guide RNA (sgRNA) targeting vectors and subsequent ESC transfection were performed as described (Cao et al. 2017). Targeted ESC clones were screened by PCR and confirmed by Sanger sequencing. CRISPR sgRNA oligo sequences used in this study are listed in Supplemental Figure S1C. The following primers were used to genotype CRISPR clones: Set1A wild-type forward (ACATGAGCCTGGAAAAGTGG), Set1A wild-type reverse (CATGTCCGCTACCATCTGTG), Set1AΔSET forward (TAATCGGGTGCTTTCTGAGC), and Set1AΔSET reverse (TAC AGGCTGGTACCCCAGGT).
Additional information on materials and methods is in the Supplemental Material.
Data availability
Next-generation sequencing data sets have been deposited at Gene Expression Omnibus with accession number GSE98988.
Supplementary Material
Acknowledgments
We are grateful to members of the Shilatifard laboratory for their critical feedback on this manuscript. We thank Dr. E.R. Smith for critical reading and editing of this manuscript. C.C.S. is supported in part by National Institutes of Health/National Cancer Institute (NIH/NCI) training grant T32CA09560. Studies in the Shilatifard laboratory regarding the role of the COMPASS family in development and cancer are supported by NCI’s Outstanding Investigator Award (R35CA197569).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.303768.117.
References
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 30: 2817–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bledau AS, Schmidt K, Neumann K, Hill U, Ciotta G, Gupta A, Torres DC, Fu J, Kranz A, Stewart AF, et al. 2014. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141: 1022–1035. [DOI] [PubMed] [Google Scholar]
- Cao K, Collings CK, Marshall SA, Morgan MA, Rendleman EJ, Wang L, Sze CC, Sun T, Bartom ET, Shilatifard A. 2017. SET1A/COMPASS and shadow enhancers in the regulation of homeotic gene expression. Genes Dev 31: 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Li Y, Liang S, Cui K, Salz T, Yang H, Tang Z, Gallagher PG, Qiu Y, Roeder R, et al. 2013. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet 9: e1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141: 526–537. [DOI] [PubMed] [Google Scholar]
- Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD II, Garcia BA, Adelman K, Wysocka J. 2017. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell 66: 568–576.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhang J, Zhang H, Yang X, Jin X, Zhang L, Skalnik DG, Jin Y, Zhang Y, Huang X, et al. 2016. H3K4 methyltransferase Set1a is a key Oct4 coactivactor essential for generation of Oct4 positive inner cell mass. Stem Cells 34: 565–580. [DOI] [PubMed] [Google Scholar]
- Galonska C, Ziller MJ, Karnik R, Meissner A. 2015. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell 17: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. 2012. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20: 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Cao K, Morgan MA, Mas G, Smith ER, Volk AG, Bartom ET, Crispino JD, Di Croce L, et al. 2017. Not all H3K4 methylations are created equal: Mll2/COMPASS dependency in primordial germ cell specification. Mol Cell 65: 460–475.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CC, Shilatifard A. 2016. MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb Perspect Med 6: a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusi BK, Deng C, Salz T, Zeumer L, Li Y, So CW, Morel LM, Qiu Y, Huang S. 2015. Setd1a regulates progenitor B-cell-to-precursor B-cell development through histone H3 lysine 4 trimethylation and Ig heavy-chain rearrangement. FASEB J 29: 1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data sets have been deposited at Gene Expression Omnibus with accession number GSE98988.