Abstract
Filoviruses, consisting of Ebola virus, Marburg virus and Cuevavirus, cause severe hemorrhagic fevers in humans with high mortality rates up to 90%. Currently, there is no approved vaccine or therapy available for the prevention and treatment of filovirus infection in humans. The recent 2013–2015 West African Ebola epidemic underscores the urgency to develop antiviral therapeutics against these infectious diseases. Our previous study showed that GPCR antagonists, particularly histamine receptor antagonists (antihistamines) inhibit Ebola and Marburg virus entry. In this study, we screened a library of 1220 small molecules with predicted antihistamine activity, identified multiple compounds with potent inhibitory activity against entry of both Ebola and Marburg viruses in human cancer cell lines, and confirmed their anti-Ebola activity in human primary cells. These small molecules target a late-stage of Ebola virus entry. Further structure-activity relationship studies around one compound (cp19) reveal the importance of the coumarin fused ring structure, especially the hydrophobic substituents at positions 3 and/or 4, for its antiviral activity, and this identified scaffold represents a favorable starting point for the rapid development of anti-filovirus therapeutic agents.
Keywords: Ebola, Marburg, Antiviral, Antihistamine, Histamine receptor antagonist
1. Introduction
Filoviruses are single-stranded, negative-sense RNA viruses with filamentous morphology, which cause severe hemorrhagic fevers in humans and nonhuman primates. Filoviruses consist of three members: Marburgvirus (MARV), Ebolavirus (EBOV), and Ceuvavirus (Kuhn et al., 2010). The first filovirus was recognized and named Marburg virus for its outbreak origin in Marburg, Germany in 1967. That outbreak led to 7 deaths among the 31 reported cases (Siegert et al., 1967). Since then, fatal filovirus outbreaks, particularly Ebolavirus, occurred sporadically, with mortality rates up to 90% (Feldmann and Geisbert, 2011). The latest 2013–2015 West Africa Ebola epidemic was the most widespread outbreak of EBOV in history and persisted for about two years. It led to 28,652 infections (including suspected, probable and confirmed cases) and 11,325 reported deaths from the 15,261 laboratory-confirmed cases with a mortality rate of 74% (as of April 13th, 2016 updated by the World Health Organization). Since there is no FDA-approved vaccine or therapy available against these viruses for humans, development of antiviral therapies is urgently needed to prevent and control future outbreaks and potential bioterrorism attacks.
Viral entry, the first step of the viral replication cycle, represents a potential target for antiviral drug development. Filovirus entry is mediated by a single viral glycoprotein (GP), which is composed of two subunits, GP1 and GP2 (Lee and Saphire, 2009; Manicassamy et al., 2005). GP1 mediates the initial attachment of virion to the host cell surface, likely by binding with heparan sulfate and other similar glycosaminoglycans (O’Hearn et al., 2015; Salvador et al., 2013), or C-type lectin family members like LSECtin (liver and lymph node sinusoidal endothelial cell C-type lectin), DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin), L-SIGN (liver/lymph node-specific ICAM-3 grabbing nonintegrin), mannose-binding lectin, and hMGL (human macrophage galactose- and N-acetylgalactosamine-specific C-type lectin) (Alvarez et al., 2002; Gramberg et al., 2005; Takada et al., 2004). The virus is then internalized via macropinocytosis and trafficked to late endosomes/lysosomes (Nanbo et al., 2010; Saeed et al., 2010), where the GP1 is trimmed by the cysteine proteases cathepsin B and L in the low pH environment (Brecher et al., 2012; Chandran et al., 2005; Schornberg et al., 2006). The mucin-like domain and the glycan cap of GP1 are removed and the receptor binding domain is exposed for the interaction with the internal EBOV receptor, Niemann-Pick C1 (NPC1), which triggers GP2 conformational change and leads to viral-endosomal membrane fusion (Carette et al., 2011; Cote et al., 2011; Gong et al., 2016; Wang et al., 2016).
We previously screened an FDA-approved drug library, identified several groups of GPCR antagonists as potent entry inhibitors, and revealed antihistamines as the most enriched GPCR antagonists (Cheng et al., 2015). In this report, we screened a library of 1220 antihistamine-like compounds (designed with ligand-based approach) and identified multiple compounds as potent inhibitors of filovirus entry. These small molecules exhibit potent inhibitory activity against both pseudotyped EBOV and MARV in adenocarcinomic human alveolar basal epithelial cells (A549). More importantly, the anti-Ebola activities of cp15 and cp19 were further validated with human primary cells including peripheral blood mononuclear cells (PBMC) derived macrophages and human foreskin fibroblasts (HFF). The compound cp19, which has an IC50 value of 3.4 μM in PBMC derived macrophages and an IC50 value of 1.2 μM in HFF against infectious EBOV infection, was further evaluated with structure-activity relationship (SAR) studies. Our results suggest that the coumarin fused ring structure of cp19 is important for its anti-filovirus activity and that the hydrophobic substituents at positions 3 and/or 4 confer an important interaction with its target.
2. Materials and methods
2.1. Cell culture
Human embryonic kidney cells (239T, ATCC# CRL-1573), human lung epithelial cells (A549, ATCC#CCL185) and Hela cells (ATCC#CCl-2) were cultured in DMEM (Cellgro) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 μg/mL of streptomycin and 100 units of penicillin (Invitrogen). The HFF-1 (SCRC-1041) cells was cultured in MEM supplemented with 10% FBS and 0.5 mM sodium pyruvate for limited amount of passages not exceeding 16. Human peripheral blood mononuclear cells (PBMCs) were isolated from human buffy coats obtained from healthy volunteers (New York blood center, Leukocytes E3752V00) and maintained in RPMI-1640 with GlutaMAX™ (Gibco) supplemented with 10% FBS, 10 mM Hepes, 100 units/mL penicillin and streptomycin. Cells were purified using ficoll columns from 50 mL of blood and cryopreserved at 2 × 106 cells/vial.
2.2. Generation of pseudovirions
The pseudovirions were created by the following plasmids: hemagglutinin (HA) and neuraminidase (NA), isolated from A/Goose/Qinghai/59/05 (H5N1) strain, Marburgvirus glycoprotein, Ebolavirus Zaire envelope glycoprotein, Lassa virus envelope glycoprotein and the HIV-1 proviral vector pNL4-3.Luc.R−E−(obtained through the NIH AIDS Research and Reference Reagent program). All four types of pseudovirions, HIV/MARV, HIV/AIV, HIV/EBOV, and HIV/LASV were produced by transient cotransfection of human 293T cells using a polyethylenimine (PEI)-based transfection protocol. Plasmids encoding MARV GP, hemagglutinin (HA)/neuraminidase (NA), EBOV GP, LASV GP and replication-defective HIV vector (pNL4-3.Luc.R−E−) were used for transient cotransfection into 293T producing cells. Six hours after transfection, cells were washed with phosphate-buffered saline (PBS) and 20 mL of fresh media was added to each plate (150 mm). Twenty-four hours post-transfection, the supernatants were collected and filtered through 0.45 μm pore size filter (Nalgene). The pseudovirion stocks were stored at 4 °C prior to use.
2.3. Compound library and chemical reagents
The histamine receptor antagonist library of 1220 compounds was obtained from OTAVA chemicals (Vaughan, Canada). On each 384-well plate, 320 unique compounds were arrayed at 5 mg/mL in DMSO from column 3 to 22, leaving columns 1, 2, 23 and 24 with DMSO. The positive control, an HIV reverse transcriptase inhibitor Azidothymidine (AZT, Sigma), was solubilized at 10 mM in DMSO and used at a final concentration of 5 μM for the screen. Benztropine mesylate (Santa Cruz Biotech) was dissolved in DMSO at 10 mM. All chemicals were stored at −80 °C until use.
2.4. High-throughput screen
The antihistamine library was screened in 384-well plates with a final concentration of 6.25 μg/mL in 0.125% DMSO (v/v) to identify Marburg virus (MARV) entry inhibitors. Low-passage A549 cells were seeded at the density of 1000/well in 384-well plates 24 h before infection. In the presence of compounds, A549 cells were infected by HIV/MARV or HIV/LASV pseudotyped virus, which contained luciferase reporter gene. Plates were incubated for an additional 48 h and infection was quantified by the luciferase activity of infected A549 cells using the neolite Reporter Gene Assay System (PerkinElmer). Virus alone with DMSO was used as the negative control; and virus with 5 μM AZT, an HIV reverse transcriptase inhibitor, was used as the positive control. Data was normalized by plate median and the criterion of average > 80% inhibition in duplicate plates was applied for picking hits.
The selected active compounds were then reformatted into new 384-well plates and tested against HIV/MARV, HIV/EBOV, HIV/LASV or HIV/AIV at 6.25 μg/mL in 0.125% DMSO (v/v) to confirm the primary results and to identify filovirus-specific hits. The cytotoxicity of hit compounds was examined using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) in the A549 cells treated the same way as for antiviral screen. The signals in the 16 negative control wells (DMSO) were averaged and used to data normalization.
The hit compounds were serial diluted for IC50 and CC50 evaluation. IC50 and CC50 values were determined by fitting the dose-response curves with four-parameter logistic regression in GraphPad.
2.5. Time-of-addition experiment
A549 cells were incubated with HIV/EBOV at 4 °C for 1 h to allow virus attachment to the cells. Virus was removed and cells were washed with cold PBS twice before fresh media was added. The temperature was shifted to 37 °C to trigger virus entry. At different time points of virus entry, cp15 (10 μg/mL), cp19 (10 μg/mL), benztropine (25 μM), or AZT (1 μM) was introduced to assess their impact on virus entry. Triplicate wells were used for each treatment at each time point. Control infected cell cultures were treated with drug vehicle (DMSO) only. Virus infection was measured 48 h post-infection as described above.
2.6. Infectious virus assays
Experiments using live authentic Ebola (Kikwit) and Marburg (Ci67) were performed in biosafety level 4 (BSL-4) facilities at the United States Army Medical Research Institute for Infectious disease (USAMRIID). Different plaque forming units (pfu) were used for different viruses and even same virus and different cell lines to achieve similar “observed” or detectable infection rate in 48 h of incubation.
2.6.1. EBOV infection assay of A549 and HFF-1 cells
Two hours after compound treatment, 384-well plates containing A549 cells or human foreskin fibroblasts (HFF-1) were transferred to the BSL-4 suite and infected with Ebola/Kikwit at a multiplicity of infection of 2.0 pfu per cell for A549 cells (calculated for 6000 cells/well, assuming one complete round of replication at 15–20 h after cell seeding) and 24 pfu per cell for HFF-1 cells (calculated for the cell density at the time of seeding) in 10 μL of media. The assay plates were incubated in a tissue culture incubator for 48 h before infection was terminated by fixing the samples in formalin solution prior to immunostaining.
2.6.2. EBOV human macrophage infection assay
Seven days before assays were performed, PBMC cells were placed in culture dishes for differentiation in the presence of 5 ng/mL GM-CSF and 50 μM BME. Media was changed about every 2 days and in 5–7 days attached cells were harvested with EDTA/PBS and scrapping and were plated at 30,000 cells/well in 100 μL media 24 h prior to compound treatment and infection. About 2 h after compound treatment, plates were transferred into the BSL-4 suite and the cells were infected with 7 pfu per cell of Ebola/Kikwit in 10 μL of media and incubated for 1 h. The inoculum was removed and the media was replaced with fresh media containing diluted compounds. At 48 h post-infection virus replication was terminated by fixation in formalin.
2.6.3. Marburg Hela infection assay
Two hours after compound treatment, plates containing Hela cells were transferred to the BSL-4 suite and infected with Marburg (Ci67) virus strain at a multiplicity of infection of 1 pfu per cell (calculated for 4000 cells/well, assuming one complete round of replication at 15–20 h after cell seeding). The assay plates were incubated in a tissue culture incubator for 48 h. Infection was terminated by fixing the samples in formalin solution prior to immunostaining.
2.6.4. Immunostaining assay
Immunostaining was used to visualize infection in cells treated with unmodified authentic EBOV and MARV virus isolates. After formalin fixation steps all wells on the assay plates were treated with viral antigen specific monoclonal antibody (mm 6D8 anti-GP for EBOV and mm 9G4 anti-GP for MARV) at 1 to 1000 dilution for 1 h and followed staining with Dylight488 conjugated anti-mouse IgG (Thermo) (1–1000 dilution) in blocking buffer containing 3% BSA in PBS. All wells also were stained with Draq5(-Biostatus) for nuclei and cytoplasm detection. Five images from each well on the assay plate were acquired on the Opera confocal imaging plate reader (Perkin Elmer) using 10× Air objective. Signal from anti-virus staining was detected at 488 nm emission wavelength and signal for nuclei and cytoplasm at 640 nm and used for cell count and masking correct location of the viral staining. Image analysis is performed using PE Acapella algorithms. The amount of infected cells were calculated for five images in each well and converted into present of infection. The plate based normalization was done using average of negative control (16 wells) and positive control (16 wells) for each plate using Gene data analytical software. Analysis of the dose response curve in order to determine EC50 was performed using GeneData software by applying the Levenberg-Marquardt algorithm (LMA) for curve fitting strategy.
3. Results
3.1. Screening an antihistamine chemical library for anti-filovirus entry inhibitors
We previously screened an FDA-approved drug library (Prestwick) and identified numerous GPCR antagonists as potent anti-filovirus entry inhibitors (Cheng et al., 2015). Since many of the anti-filovirus inhibitors identified belonged to antihistamine groups (9 out of 34 H1 receptor antagonists were effective, shown in Fig. 1a), we reasoned that screening antihistamine-focused chemical libraries could allow the identification of novel anti-filovirus compounds with more diversified scaffolds, which could be further optimized to develop anti-filovirus therapies.
Fig. 1. High-throughput screen of anithistamine library.
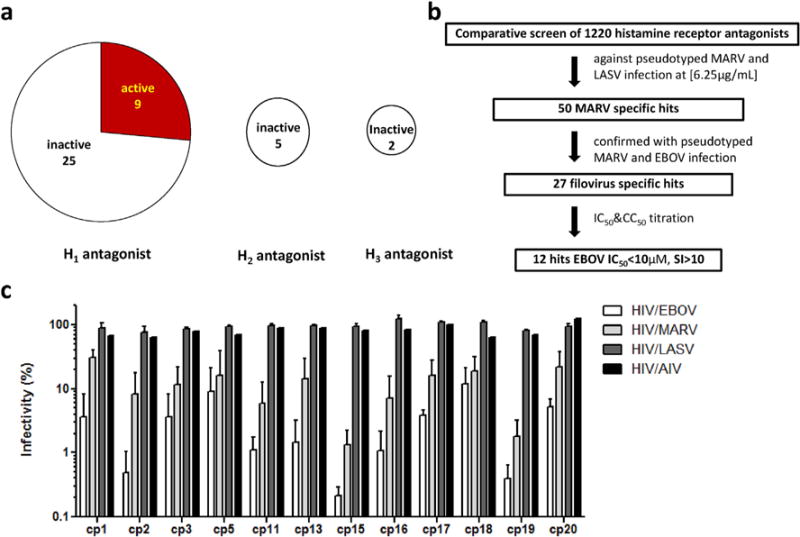
(a) Antihistamines represent a potential reservoir for anti-filovirus drug development. A category analysis of drugs from Prestwick library shows 9 out of 34 histamine H1 receptor antagonists exhibit inhibitory activity against filovirus infection. (b) An antihistamine chemical library of 1220 compounds was screened against pseudotypted MARV and LASV at a final compound concentration of 6.25 μg/mL. Fifty compounds showing >80% inhibition against MARV and <30% inhibition against LASV were picked up for confirmation. The filovirus specific hits were further validated by various pseudovirions including HIV/EBOV, HIV/MARV, HIV/LASV and HIV/AIV. The top 27 filovirus-specific hits were further evaluated by dose-response titrations for both pseudotyped filovirus infection and cell toxicity. Twelve compounds with IC50<10 μM, SI>10 were selected for authentic filovirus validation. (c) Top 12 hits specifically inhibit entry of pseudovirions HIV/EBOV and HIV/MARV but not HIV/LASV and HIV/AIV at 6.25 μg/mL. Data are means ± SEM from three independent experiments for HIV/EBOV, HIV/MARV and HIV/LASV (n = 3); data are mean for HIV/AIV from one experiment (n = 1). The statistical analysis is reported in Supplementary Table 2.
We adapted an HIV-based pseudotyping system to study the entry of highly pathogenic viruses like EBOV and MARV in a biosafety level 2 (BSL-2) facility and designed a comparative screen protocol for drug discovery (Wang et al., 2014). With this protocol, a focused library (OTAVA Ltd.) containing 1220 small molecules with predicted antihistamine activity was screened to identify filovirus-specific entry inhibitors. The workflow of the screen is shown in Fig. 1b. The antihistamine library was screened at a final compound concentration of 6.25 μg/mL against HIV/MARV pseudovirion and counter-screened against HIV/LASV pseudovirion (as specificity controls) to identify MARV entry-specific inhibitors (Z′ was used to assess screen quality, Supplementary Table 1). From the primary screen, we obtained 50 MARV specific hits showing >80% inhibition against HIV/MARV but <30% inhibition against HIV/LASV. These compounds were then screened again at 6.25 μg/mL with HIV/MARV, HIV/EBOV, HIV/AIV and HIV/LASV pseudovirions for confirmation. The top 27 confirmed hits with >80% inhibition against both HIV/MARV and HIV/EBOV but <30% inhibition against HIV/AIV and HIV/LASV were picked for further characterization. To prioritize the lead compounds, we performed dose-response titrations to assess the compounds’ antiviral activity and cell-toxicity. Among them, 12 compounds demonstrated the 50% inhibitory concentrations (IC50 values) less than 10 μM in pseudotyped filovirus infection assay and the 50% cytotoxic concentrations (CC50) > 50 μM in CellTiter-Glo® luminescent cell viability assay, with the selectivity index (CC50/IC50) ratios greater than 10. The antiviral effects of these 12 compounds against EBOV, MARV, LASV and AIV pseudovirions at 6.25 μg/mL are shown in Fig. 1c.
3.2. Validation of selected antihistamine-like small molecules
To validate the efficacy of these 12 antihistamine-like small molecules against authentic EBOV isolates, we further evaluated their ability to inhibit replication of infectious Ebola virus strain Ebola/Kikwit in tissue-cultured A549 cells under BSL-4 containment conditions. Seven compounds were confirmed against infectious EBOV infection with IC50 < 10 μM and selectivity index >10 (Table 1).
Table 1.
Anti-filovirus activities of validated compounds.
| Hit ID | Structure | Pseudovirus
|
Infectious EBOV
|
||||
|---|---|---|---|---|---|---|---|
| IC50, μM
|
CC50, μM | IC50, μM | CC50, μM | SI | |||
| EBOV | MARV | ||||||
| cp3 |
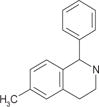
|
6.9 ± 1.4 | 9.1 ± 1.0 | 156.9 ± 12.5 | 5.2 ± 0.7 | 223 | 43 |
| cp5 |
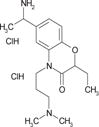
|
4.1 ± 1.3 | 9.0 ± 1.8 | >264.3 | 4.0 ± 0.6 | 264 | 66 |
| cp11 |
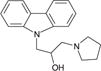
|
3.8 ± 0.7 | 5.6 ± 0.7 | 74.9 ± 19.4 | 7.5 ± 1.3 | 85 | 11 |
| cp13 |
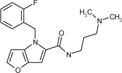
|
6.4 ± 0.9 | 9.2 ± 1.7 | >291.2 | 8.7 ± 0.9 | 150 | 17 |
| cp15 |
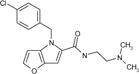
|
1.8 ± 0.7 | 5.1 ± 1.6 | 50.3 ± 2.7 | 5.2 ± 0.2 | 80 | 16 |
| cp16 |
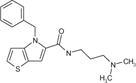
|
3.8 ± 0.9 | 6.9 ± 0.9 | 77.8 ± 14.0 | 5.1 ± 0.1 | 146 | 28 |
| cp19 |
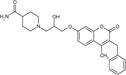
|
2.4 ± 0.3 | 4.8 ± 0.4 | 48.0 ± 4.3 | 3.7 ± 1.5 | 71 | 19 |
Abbreviations: CC50, 50% cytotoxic concentration; EBOV, Ebolavirus; IC50, 50% inhibitory concentration; MARV, Marburgvirus; SI, selectivity index. Data are means ± SD from at least two independent experiments.
Two compounds, cp15 and cp19, were picked for further characterization. These two compounds showed typical S-shaped dose-response inhibition against both EBOV and MARV infections. Compound cp15 displayed an IC50 value of 1.8 μM against EBOV pseudovirion infection, and to a little less extent, an IC50 value of 5.1 μM against MARV pseudovirion infection (Fig. 2a). For infectious EBOV, cp15 exhibited a closely comparable IC50 value of 5.2 μM in A549 cells and a CC50 value of 80 μM (Fig. 2b, Supplementary Fig. 1). This compound also had a strong inhibitory activity against infectious MARV infection with an IC50 value of 7.7 μM and a CC50 value of 128 μM in Hela cells (Fig. 2c, Supplementary Fig. 2). Very similar to cp15, cp19 inhibited pseudotyped EBOV and infectious EBOV efficiently, with IC50 values of 2.4 μM and 3.7 μM, respectively (Fig. 2d and e); c19 inhibited MARV infection to a similar extent, with IC50 values of 4.8 μM against pseudovirion and 3.7 μM against authentic MARV isolate (Fig. 2d and f).
Fig. 2. Anti-filovirus activities of cp15 and cp19.
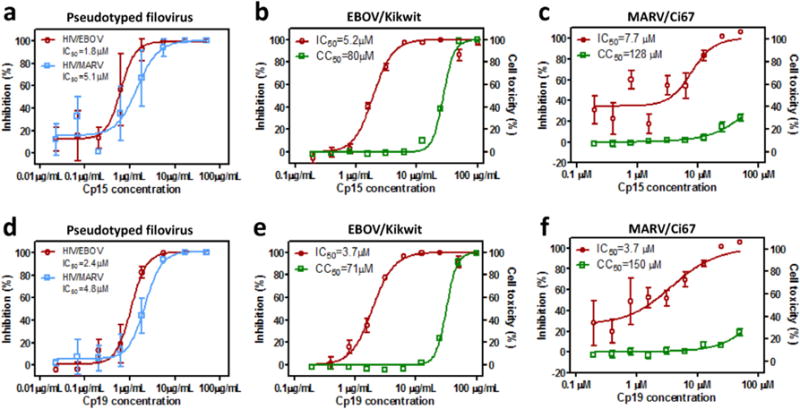
The in vitro dose-response curves of cp15 is shown in (a) against HIV/EBOV (red) and HIV/MARV (blue) pseudovirion infections in A549 cells, (b) against infectious EBOV/Kikwit infection (red) in A549 cells or in cell toxicity assay (green), and (c) against infectious MARV/Ci67 infection (red) in Hela cells or in cell toxicity assay (green). The in vitro dose-response curves of cp19 is shown in (d) against EBOV (red) and MARV (blue) pseudovirion infections in A549 cells, (e) against infectious EBOV/Kikwit infection (red) in A549 cells or in cell toxicity assay (green), and (f) against infectious MARV/Ci67 infection (red) in Hela cells or in cell toxicity assay (green). The data are mean ± SD for pseudoviruses from two independent experiments (n = 6); the data mean ± SD for infectious filovirus from two independent experiments (n = 4). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further confirm the anti-filovirus efficacy of cp15 and cp19, they were tested in two types of human primary cells: PBMC derived macrophages and HFFs. As shown in Fig. 3, both cp15 and cp19 inhibited authentic EBOV infection in these cells as effectively as in A549 cell line. Compound cp15 shows an IC50 value of 3.4 μM in macrophages (Fig. 3a) and an IC50 value of 5.5 μM in HFFs (Fig. 3c); cp19 exhibited an IC50 value of 3.4 μM in macrophages (Fig. 3b) and an IC50 value of 1.2 μM in HFFs (Fig. 3d). Both compounds showed little toxicity in the primary cells at 50 μM, which was the highest compound concentration tested.
Fig. 3. Anti-EBOV activities of cp15 and cp19 in macrophages and HFF cells.
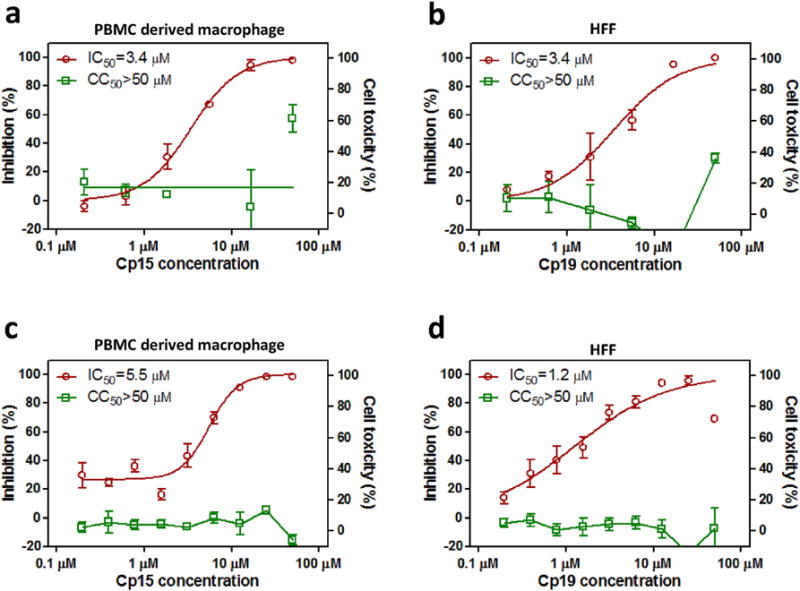
Dose-response titrations were evaluated in human PBMC derived macrophages against infectious EBOV/Kikwit infection (red) or in cell toxicity assay (green) for cp15 (a) and cp19 (b). Dose-response titrations were evaluated in human foreskin fibroblasts (HFF) against infectious EBOV/Kikwit infection (red) or in cell toxicity assay (green) for cp15 (c) and cp19 (d). Data are means ± SD (n = 2) for macrophage test and means ± SD (n = 4) for HFF test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Both cp15 and cp19 inhibit EBOV infection at a late stage of virus entry
In our previous study, we found that the GPCR antagonists (e.g., benztropine) inhibit filovirus entry at a post-attachment step (Cheng et al., 2015). To investigate whether cp15 and cp19 also block the post-binding steps (e.g., fusion), a “time-of-addition experiment” was performed with HIV/EBOV pseudovirions in the presence cp15 or cp19 using benztropine mesylate and zidovudine (AZT) as controls. HIV/EBOV virion was incubated with A549 cells at 4 °C at time point −1 h for 1 h. The cells were washed to remove unbound virions and the temperature was shifted to 37 °C to trigger virus entry. Compounds were introduced at various time points to evaluate their antiviral effects. As expected, benztropine started to lose its inhibitory effect when it was added at 2 h post entry and beyond, while AZT (an HIV reverse transcriptase inhibitor) were still effective even when it was introduced at a very late time points (>8 h). Similar to benztropine, cp15 and cp19 showed strong inhibition if they were added −1 h, 0 h and 1 h post-virus entry (Fig. 4). That these two compounds were still effective when they were introduced at 1 h post entry indicates that they act at a post-attachment step; that they lost their inhibitory ability much earlier than AZT suggests that they block infection of EBOV prior to viral/endosome membrane fusion.
Fig. 4. Compounds cp15 and cp19 inhibit EBOV infection at late entry step.
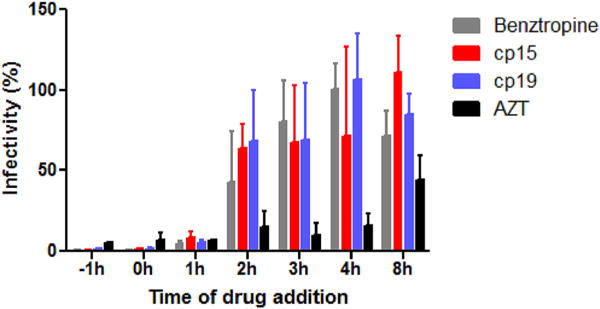
Pseudotyped HIV/EBOV was incubated with A549 cells at 4 °C at the −1 h time point. After 1 h of incubation, the unbound virus was removed and temperature was shifted to 37 °C to trigger virus internalization. Benztropine (25 μM), AZT (1 μM), cp15 (10 μg/mL) or cp19 (10 μg/mL) was introduced at different time points of virus infection, and the compounds’ effects on viral infection are shown (means ± SD (n = 3)). The statistical analysis is reported in Supplementary Table 3.
3.4. Preliminary structure activity relationship studies
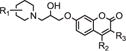
Compound cp19 consists of a piperidine ring and a coumarin ring linked by a 4-atom unit. To study the function of the piperidine and coumarin rings antiviral activity, we purchased and evaluated an additional 10 structurally-related compounds against the HIV/EBOV pseudovirion in A549 cells. In Table 2 is shown the structure of our coumarin-based anti-Ebola compound cp19 also referred to as CBS1129 and its activity in the pseudovirus assay. Also shown are anti-Ebola activites for ten close analogs, where modifications have been made in two locations on cp19/CBS1129 (Table 2).
Table 2.
Coumarin-based compound CBS1129/cp19 and related compounds as anti-Ebola agents.
| Compound No. |
 Compound structurea |
IC50 valueb μM |
CC50 valuec μM |
SI value | ||
|---|---|---|---|---|---|---|
| R1 | R2 | R3 | ||||
| CBS1129 | CONH2 | CH3 | CH2C6H5 | 2.4 | 56 | 23 |
| CBS1130 | CONH2 | C6H5 | H | 3.4 | 50.5 | 15 |
| CBS1131 | 2- CH3 | -(CH2CH2CH2)- | 9.7 | >100 | >10 | |
| CBS1132 | 4- CH3 | -(CH2CH2CH2)- | 25.7 | >100 | >4 | |
| CBS1133 | 3- CH3 | -(CH2CH2CH2)- | 35.2 | >100 | >3 | |
| CBS1134 | H | -(CH2CH2CH2)- | 42.7 | >100 | >2 | |
| CBS1135 | H | CH3 | H | >100 | >100 | NA |
| CBS1136 | 2- CH3 | CH3 | H | >100 | >100 | NA |
| CBS1137 | 3- CH3 | CH3 | H | >100 | >100 | NA |
| CBS1138 | 4- CH3 | CH3 | H | >100 | >100 | NA |
| CBS1139 | 2-CH2CH3 | CH3 | H | >100 | >100 | NA |
Compounds CBS1131–CBS1139 were evaluated as their hydrochloride salts.
Pseudovirus (HIV/EBOV) assay results.
These values were determined in A549 cells.
Our preliminary SAR analysis shows that certain modifications in the coumarin fused ring structure of cp19 are tolerated. Thus, the aromatic ring can be moved from the 3-position of the coumarin ring (CBS1129) to the 4-position (CBS1130) without a significant dimunition of activity. Replacing the benzyl group in CBS1129 and the phenyl group in CBS1130 with a trimethylene fusion at positions 3 and 4 diminishes (but does not eliminate) activity from 2- to 20-fold, as shown with compounds CBS1131–CBS1134; note that the substitution pattern in the piperidine ring is simultaneously changing with these compounds as well. The change in piperidine substitution with this set of four compounds shows that changes in this portion of the molecule are tolerated for activity, and that with methyl substitution, the 2-position is optimal. Interestingly, when the coumarin ring is substituted with a single methyl group at the 4-position, activity is lost, as shown with compounds CBS1135–CBS1139. This change suggests that relatively large hydrophobic substituents are required at the 3- or 4-positions or both positions for biological activity, and that this may be due to a hydrophobic binding pocket interaction that is necessary for activity. It can be concluded that (1) appropriate substitution of the coumarin ring is necessary for biological activity; (2) substituents on the piperidine ring can be varied; and (3) optimization of the substitution patterns on both the coumarin and piperidine portions of the molecule need to be systematically optimized (Fig. 5).
Fig. 5.
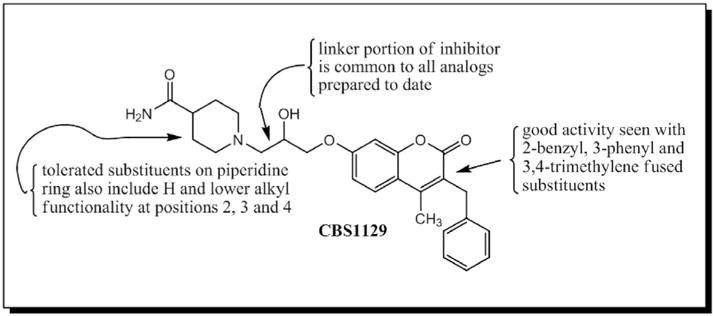
Structure activity relationship (SAR) developed for cp19/CBS1129.
4. Discussion
Previously, we showed that several groups of G protein-coupled receptor (GPCR) antagonists were capable of potently inhibiting the entry of numerous strains of both EBOV and MARV (Cheng et al., 2015). Since roughly 40% of prescription drugs target GPCR receptors and many of these are antagonists (Stevens et al., 2013), this finding provides us with a series of FDA-approved GPCR antagonists with potential anti-filovirus properties. Based on function-enrichment analysis of the GPCR antagonists originally identified in our screen, antihistamines emerged as the most enriched GPCR drug class. Therefore, we decided to screen an antihistamine-focused library containing 1220 compounds and 12 compounds had IC50s < 10 μM and SIs>10 μM against both pseudotyped HIV/MARV and HIV/EBOV. The most potent compounds, cp15 and cp19, showed excellent entry inhibition against both infectious EBOV and MARV; as such, we believe they represent excellent scaffolds for developing novel anti-filovirus therapeutics targeting filovirus entry.
Filovirus entry is a multistep process consisting of attachment and internalization of the virus on the cell surface, trafficking of this viral particle through the endosome system, and membrane fusion of the virus with the endosome/lysosome in the later phase (Bhattacharyya et al., 2012). In the time-of-addition assay, both compound cp15 and cp19 retained their full antiviral effect when they were introduced 1 h after virus entry began but lost their activity much faster than the reverse transcriptase inhibitor AZT, suggesting that these compounds target a post-attachment, pre-fusion step in the virus entry process (Fig. 4). The acidic pH environment in the endosome and the general endosomal trafficking pathways are not likely to be affected by these two compounds because the pH-dependent entry of other pseudotyped viruses, including avian influenza virus H5N1 and Lassa virus, which also go through the endosomal system, were not affected by these two compounds (Fig. 1c). Benztropine and toremifene, two late-stage entry filovirus inhibitors, have been recently reported to be able to destabilize the fusion loop of the Ebola GP protein by binding to a hydrophobic cavity between the GP1 and GP2 subunits, preventing the GP mediated membrane fusion step (Zhao et al., 2016). Based on our observations that cp15 and cp19 have antiviral activities similar to benztropine and that they lose their antiviral activities in a comparable time course to that of benztropine in the time-of-addition assay, it is quite possible that cp15 and cp19 are mechanistically similar to benztropine. In addition, we have recently obtained the structures of Ebola GP complexed with three different antihistamines (unpublished data), which shows that the antihistamines bind to the same pocket as toremifene does, indicating that antihistamines act by a similar mechanism as toremifene and benztropine. Though antihistamines and antihistamine-like compounds cp15 and cp19 are likely to act through preventing Ebola GP mediated fusion via directly binding to the GP protein, other possible mechanisms of action may exist. Recently, many cationic amphiphilic drugs (CADs) have been reported to be able to inhibit Ebola virus infection at a late entry stage (Fan et al., 2017; Johansen et al., 2015; Shoemaker et al., 2013; Sun et al., 2017). CADs are a wide group of chemicals with structure features of a hydrophobic aromatic ring system and a hydrophilic side-chain containing an ionizable amine. They inhibit Ebola virus entry via diverse mechanisms like preventing lysosomal calcium release and/or inhibiting acid sphingomyelinase activity, but these mechanisms require further elucidation. All but one of our compounds are structurally related to tertiary amines whose reported pKa values are >8. We believe the compounds described in this report may well fall into the CAD classification. There is a possibility that antihistamine-like compounds cp15 and cp19 may act similarly as CADs. Also, these compounds may inhibit Ebola virus entry through multiple mechanisms including binding to the GP proteins and inhibiting acid sphingomyelinase activity and lysosomal calcium release to prevent GP mediated membrane fusion both directly and indirectly.
We performed preliminary structure-activity relationship (SAR) studies on the coumarin-based anti-Ebola agents (Fig. 5). The scaffold consists of three parts: a piperidine ring, a coumarin ring and a linker between the two rings. The linker portion has remained constant in all of the molecules that we have evaluated to date. We think that the composition of this linker portion of the molecule may be important to the activity of the compounds and there are many opportunities for changing this part of the molecule. Different substituents on the piperdine ring were tested and some of them (hydrogen; e.g. no substituents; and lower alkyl) are tolerated. From a series of 2-methyl (CBS1131), 3-methyl (CBS1132) and 4-methyl (CBS1133) compounds that we have evaluated, it appears that the methyl substituent at the 2-position confers significantly more activity than it does at the 3- or 4-positions. The coumarin fused ring system is clearly an important part of the molecule for inhibitory activity. We have learned that if only a single methyl substituent is retained at the 4-position of the coumarin ring, as in compounds CBS1135–CBS1139, activity is totally lost. A benzyl substituent at the 3-position (and a methyl group at the 4-position), which is the substitution pattern in hit compound CBS1129, confers good inhibitory activity, as does a single phenyl group at the 4-position (CBS1130), or trimethylene fusion at positions 3 and 4 (CBS1131–CBS1134). Thus, it appears that our active compounds contain hydrophobic substituents at positions 3 and/or 4, and they may be binding into a hydrophobic pocket, such as the cavity previously described between the GP1 and GP2 subunits, as their primary site of activity.
A few compounds have been described recently targeting Ebola virus entry. Nonspecific cathepsin inhibitors (E64D and leupeptin) and specific cathepsin B/L inhibitors (CA-074, FY-DMK, CID23631927, K11777, and a series of compounds based on epoxysuccinate esters or epoxysuccinate propylamide scaffolds) have been developed to block Ebola virus entry (Barrientos and Rollin, 2007; Gnirss et al., 2012; Kaletsky et al., 2007; Nyakatura et al., 2015; Shah et al., 2010; van der Linden et al., 2016; Zhou et al., 2015). However, due to the dispensability of cathepsins B and L in EBOV replication in vivo, the mechanistic importance of these protease inhibitors, and whether they will provide any therapeutic benefit, is quite questionable (Marzi et al., 2012; Rhein and Maury, 2015). Compounds targeting the NPC1-Ebola GP interaction (compound 3.47, MBX2254 and MBX2270) have also been identified. Because of the relatively large interface between the NPC1 receptor and Ebola GP, it remains unclear if these compounds can be optimized to reach a potent antiviral effect (Basu et al., 2015; Cote et al., 2011). The compounds with predicted antihistamine activity we have identified represent a new class of chemicals for further anti-filovirus drug development. Firstly, our data suggests that antihistamine targets Ebola GP which is an attractive target for drug development. Secondly, the preliminary SAR analysis has given us specific direction for further compound optimization. Thirdly, GPCR drugs including antihistamines have been intensively studied and we have now assembled a significant base of knowledge that will allow us to rapidly optimize selected compounds for in vivo study.
In conclusion, we have identified two novel small molecules with predicted antihistamine activity, which have potent anti-filoviral properties and likely work in a similar fashion as benztropine and toremifene. Preliminary SAR studies revealed important information regarding the antiviral activity of compound 19 and point to further optimization studies that we will undertake to improve the molecule to provide a novel therapeutic agent for filovirus infections. More importantly, we have shown that GPCR antagonist libraries represent a vast repertoire of untapped anti-filovirus compounds.
Supplementary Material
Acknowledgments
We thank Julie Tran at USAMRIID for her help in purification of monoclonal antibodies for immunostaining assays.
This research was supported by National Institutes of Health (USA) grants AI77767 to L.R. USAMRIID efforts were supported by the Joint Science and Technology Office for Chemical and Biological Defense (JSTO-CBD) of the Defense Threat Reduction Agency (DTRA).
Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.antiviral.2017.06.015.
References
- Alvarez CP, Lasala F, Carrillo J, Muniz O, Corbi AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos LG, Rollin PE. Release of cellular proteases into the acidic extracellular milieu exacerbates Ebola virus-induced cell damage. Virology. 2007;358:1–9. doi: 10.1016/j.virol.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Basu A, Mills DM, Mitchell D, Ndungo E, Williams JD, Herbert AS, Dye JM, Moir DT, Chandran K, Patterson JL, Rong L, Bowlin TL. Novel small molecule entry inhibitors of ebola virus. J Infect Dis. 2015;212(Suppl 2):S425–S434. doi: 10.1093/infdis/jiv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Mulherkar N, Chandran K. Endocytic pathways involved in filovirus entry: advances, implications and future directions. Viruses. 2012;4:3647–3664. doi: 10.3390/v4123647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Lear-Rooney CM, Johansen L, Varhegyi E, Chen ZW, Olinger GG, Rong L. Inhibition of ebola and Marburg virus Entry by G Protein-coupled receptor antagonists. J Virol. 2015;89:9932–9938. doi: 10.1128/JVI.01337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Du X, Zhang J, Zheng H, Lu X, Wu Q, Li H, Wang H, Shi Y, Gao G, Zhou Z, Tan DX, Li X. Selective inhibition of Ebola entry with selective estrogen receptor modulators by disrupting the endolysosomal calcium. Sci Rep. 2017;7:41226. doi: 10.1038/srep41226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirss K, Kuhl A, Karsten C, Glowacka I, Bertram S, Kaup F, Hofmann H, Pohlmann S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology. 2012;424:3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Qian H, Zhou X, Wu J, Wan T, Cao P, Huang W, Zhao X, Wang X, Wang P, Shi Y, Gao GF, Zhou Q, Yan N. Structural insights into the niemann-pick C1 (NPC1)-Mediated cholesterol transfer and ebola infection. Cell. 2016;165:1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T, Hofmann H, Moller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH, Becker S, Munch J, Pohlmann S. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LM, DeWald LE, Shoemaker CJ, Hoffstrom BG, Lear-Rooney CM, Stossel A, Nelson E, Delos SE, Simmons JA, Grenier JM, Pierce LT, Pajouhesh H, Lehar J, Hensley LE, Glass PJ, White JM, Olinger GG. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med. 2015;7:290ra289. doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of ebola virus GP1 in viral entry. J Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Reinheckel T, Feldmann H. Cathepsin B & L are not required for ebola virus replication. PLoS Negl Trop Dis. 2012;6:e1923. doi: 10.1371/journal.pntd.0001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macro-pinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyakatura EK, Frei JC, Lai JR. Chemical and structural aspects of ebola virus entry inhibitors. ACS Infect Dis. 2015;1:42–52. doi: 10.1021/id500025n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hearn A, Wang M, Cheng H, Lear-Rooney CM, Koning K, Rumschlag-Booms E, Varhegyi E, Olinger G, Rong L. Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J Virol. 2015;89:5441–5449. doi: 10.1128/JVI.03689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhein BA, Maury WJ. Ebola virus entry into host cells: identifying therapeutic strategies. Curr Clin Microbiol Rep. 2015;2:115–124. doi: 10.1007/s40588-015-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador B, Sexton NR, Carrion R, Jr, Nunneley J, Patterson JL, Steffen I, Lu K, Muench MO, Lembo D, Simmons G. Filoviruses utilize glycosaminoglycans for their attachment to target cells. J Virol. 2013;87:3295–3304. doi: 10.1128/JVI.01621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Wang T, Kaletsky RL, Myers MC, Purvis JE, Jing H, Huryn DM, Greenbaum DC, Smith AB, 3rd, Bates P, Diamond SL. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol Pharmacol. 2010;78:319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Schornberg KL, Delos SE, Scully C, Pajouhesh H, Olinger GG, Johansen LM, White JM. Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One. 2013;8:e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Shu HL, Slenczka W, Peters D, Muller G. On the etiology of an unknown human infection originating from monkeys. Dtsch Med Wochenschr. 1967;92:2341–2343. doi: 10.1055/s-0028-1106144. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, Rosen H, Wuthrich K. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, He S, Martinez-Romero C, Kouznetsova J, Tawa G, Xu M, Shinn P, Fisher EG, Long Y, Motabar O, Yang S, Sanderson PE, Williamson PR, Garcia-Sastre A, Qiu X, Zheng W. Synergistic drug combination effectively blocks Ebola virus infection. Antivir Res. 2017;137:165–172. doi: 10.1016/j.antiviral.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Fujioka K, Tsuiji M, Morikawa A, Higashi N, Ebihara H, Kobasa D, Feldmann H, Irimura T, Kawaoka Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden WA, Schulze CJ, Herbert AS, Krause TB, Wirchnianski AA, Dye JM, Chandran K, Bogyo M. Cysteine cathepsin inhibitors as anti-ebola agents. ACS Infect Dis. 2016;2:173–179. doi: 10.1021/acsinfecdis.5b00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. Ebola viral glycoprotein bound to its endosomal receptor niemann-pick C1. Cell. 2016;164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cheng H, Ratia K, Varhegyi E, Hendrickson WG, Li J, Rong L. A comparative high-throughput screening protocol to identify entry inhibitors of enveloped viruses. J Biomol Screen. 2014;19:100–107. doi: 10.1177/1087057113494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ren J, Harlos K, Jones DM, Zeltina A, Bowden TA, Padilla-Parra S, Fry EE, Stuart DI. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature. 2016;535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R, Jr, Nunneley JW, Barnard D, Pohlmann S, McKerrow JH, Renslo AR, Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


