Abstract
Cellular FLICE-like inhibitory protein (c-FLIP, gene symbol CFLAR) was first identified as a negative regulator of death receptor-mediated apoptosis in mammals. To understand the ubiquity and diversity of the c-FLIP protein subfamily during evolution, c-FLIP orthologs were identified from a comprehensive range of vertebrates, including birds, amphibians, and fish, and were characterized by combining experimental and computational analysis. Predictions of three-dimensional protein structures and molecular phylogenetic analysis indicated that the conserved structural features of c-FLIP proteins are all derived from an ancestral caspase-8, although they rapidly diverged from the subfamily consisting of caspases-8, -10, and -18. The functional role of the c-FLIP subfamily members is nearly ubiquitous throughout vertebrates. Exogenous expression of non-mammalian c-FLIP proteins in cultured mammalian cells suppressed death receptor-mediated apoptosis, implying that all of these proteins possess anti-apoptotic activity. Furthermore, non-mammalian c-FLIP proteins induced NF-κB activation much like their mammalian counterparts. The CFLAR mRNAs were synthesized during frog and fish embryogenesis. Overexpression of a truncated mutant of c-FLIP in the Xenopus laevis embryos by mRNA microinjection caused thorax edema and abnormal constriction of the abdomen. Depletion of cflar transcripts in zebrafish resulted in developmental abnormalities accompanied by edema and irregular red blood cell flow. Thus, our results demonstrate that c-FLIP/CFLAR is conserved in both protein structure and function in several vertebrate species, and suggest a significant role of c-FLIP in embryonic development.
Abbreviations: CARD, caspase-recruitment domain; CASc, Caspase, interleukin-1 β converting enzyme homologs; c-FLIP, cellular FLICE-like inhibitory protein; CHX, cycloheximide; DED, death effector domain; EGFP, enhanced green fluorescent protein; FADD, Fas-associated death domain protein; MO, morpholino oligonucleotide; NF-κB, Nuclear factor-kappa B; ODC, ornithine decarboxylase; PCR, polymerase chain reaction; TRAF2, tumor necrosis factor receptor-associated factor 2; tubα6, tubulin α6; RT-PCR, reverse transcription-polymerase chain reaction
Keywords: Apoptosis, Caspase-8, Embryogenesis, Evolution, NF-κB, Pseudocatalytic triad
Highlights
-
•
c-FLIP/CFLAR is specific to the vertebrate lineage.
-
•
A rapid evolutionary divergence of the c-FLIP subfamily members has occurred.
-
•
The pseudocatalytic triad is formed by evolutionarily conserved amino acids.
-
•
Vertebrate c-FLIP proteins work on both apoptotic inhibition and NF-κB activation.
-
•
c-FLIP proteins play a developmental role during embryogenesis of frog and fish.
1. Introduction
Apoptotic cell death is essential for multicellular organisms. In the apoptosis process, a family of cysteine proteases known as caspases are activated and work as executors to induce the proteolytic cleavage of many critical proteins leading cells to suicide [1]. The activation of caspases converges from two major signaling pathways: the extrinsic signaling pathway initiated by ligation of the cell surface death receptors such as Fas (also called APO-1/CD95) and the intrinsic signaling pathway triggered by cytochrome c release from mitochondria into the cytosol by apoptotic stimuli [2], [3]. The Fas-mediated pathway is well characterized as the representative extrinsic pathway. Oligomerization of Fas by its natural ligand or an agonistic antibody recruits an adapter molecule, Fas-associated death domain protein (FADD) to the death domain within the intracellular region. The initiator caspase, caspase-8 (CASP8), which contains tandem death effector domain (DED) motifs and a protease domain, caspase, interleukin-1 β converting enzyme homologs (CASc), in turn associates with FADD by way of an interaction between their respective DEDs. In the complex composed of Fas-FADD-CASP8, CASP8 progresses through an auto-processing step and transforms to an active form. Activated CASP8 subsequently processes and activates downstream molecules, resulting in cell death.
Cellular FLICE-like inhibitory protein (c-FLIP; also called CFLAR, CLARP, FLAME-1, I-FLICE, Casper, CASH, and Usurpin, gene symbol CFLAR) is a negative regulator of Fas-induced apoptosis [4], [5], [6], [7], [8], [9], [10], [11]. Mammalian c-FLIP exists as two alternatively spliced isoforms: c-FLIP(L) (long form of c-FLIP) is homologous to CASP8, except it lacks critical amino acids for proteolytic activity [8]; c-FLIP(S) (short form of c-FLIP) consists of only two DED motifs. In humans, a third isoform, c-FLIP Raji (c-FLIP(R)) was also identified [12]. These isoforms are able to bind directly to CASP8 by homophilic interactions through their DED domains, and thereby regulate the activation of CASP8 and apoptosis [12], [13]. c-FLIP can also induce Nuclear Factor-kappa B (NF-κB) activation [14], [15]. In previous studies, we and other groups have reported that both CFLAR and CASP8 genes, along with their homologous genes caspase-10 (CASP10) and caspase-18 (CASP18), localize to the same chromosomal region in marsupials, birds, reptiles, and amphibians [16], [17], [18], [19]. Therefore, it is likely that these four genes are derived from an ancestral gene by duplication during evolution. Interestingly, the CASP18 gene is deleted in eutherian mammals, and the Casp10 gene is further deleted in rodents [16], [17], [18]. However, the CFLAR gene as well as the CASP8 gene is found from fish to mammals, suggesting that both are requisite in vertebrates.
In this study, we show both the conservation of the three-dimensional structure of c-FLIP proteins in vertebrates and the functional universality in regulation of apoptosis and NF-κB signaling. Furthermore, we clarify the action of c-FLIP proteins in embryos of non-mammalian vertebrates, pointing to an important role during embryonic development. Based on these results, we propose the uniqueness of c-FLIP relative to its paralogous molecules, as well as its conserved function through vertebrate evolution.
2. Materials and methods
2.1. Animals, cell lines, and reagents
Adult wild-type Xenopus laevis were purchased from Hamamatsu Seibutsukyozai Co. (Shizuoka, Japan). In vitro fertilization of X. laevis eggs was performed as previously described [20]. Fertilized embryos were dejellied in 3% cysteine hydrochloride, washed several times in water and used for several experiments. Developing embryo stages were determined according to the scheme described by Nieuwkoop and Faber [21]. Wild-type zebrafish Danio rerio AB and RW lines were kept in a light and temperature controlled facility and maintained at optimal breeding conditions [22]. Embryos prepared by breeding were staged according to Kimmel et al. [23]. Human cervical carcinoma HeLa cells and embryonal kidney HEK293 cells were cultured in Dulbecco’s Modified Eagle’s medium with 10% fetal calf serum. An agonistic anti-human Fas antibody, CH-11, was prepared as previously described [24]. Anti-Flag (M2, Sigma-Aldrich, St. Louis, MO), anti-HA (HA124, Nacalai Tesque, Kyoto, Japan), anti-Myc (9E10, Roche Diagnostics GmbH, Mannheim, Germany), anti-actin (MAB1501R, Chemicon International Inc., Temecula, CA), and HRP-conjugated anti-mouse IgG (Cell Signaling Technology, Danvers, MA, USA) antibodies and anti-FLAG M2 affinity gel (Sigma-Aldrich) were purchased.
2.2. DNA sequencing
To search for non-mammalian orthologs of the human CFLAR gene in the NCBI DNA database, the TBLASTN program [25] was used, and four candidate EST clones from chicken, African clawed frog, medaka, and stickleback were identified and obtained from distributors as described in our previous study [26]. The nucleotide sequence of these clones was confirmed on both strands using TaqDyeDeoxyterminator Cycle sequencing (Applied Biosystems Inc., Foster City, CA) on automated DNA sequencers (3130XL Genetic Analyzer, Applied Biosystems Inc.).
2.3. Database search
The following web sites were used to access nucleotide and protein sequence databases: NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi), Ensembl (http://www.ensembl.org/index.html), Xenbase (http://www.xenbase.org/entry/), and ZFIN (http://zfin.org/action/blast/blast). Proteins related to known c-FLIP and its paralogous proteins were identified using TBLASTN [27]. Domain searches were performed using either Simple Modular Architecture Research ToolSMART (SMART) (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) or the Pfam database (http://pfam.sanger.ac.uk/search). Multiple sequence alignments were performed using Clustal W (http://www.genome.jp/tools/clustalw/).
2.4. Molecular phylogenetic analysis
To construct a molecular phylogenetic tree, we collected published or predicted amino acid sequences of c-FLIP and its paralogs, CASP8, CASP10, CASP18, and CARD (caspase-recruitment domain)-casp8 from several databases for mammals [human (Homo sapiens), mouse (Mus musculus), opossum (Monodelphis domestica), pig (Sus scrofa), and rat (Rattus norvegicus)], birds [chicken (Gallus gallus) and turkey (Meleagris gallopavo)], reptiles [anole lizard (Anolis carolinensis) and alligator (Alligator mississippiensis)], amphibian [African clawed frog (X. laevis)], fish [catshark (Scyliorhinus canicula), coelacanth (Latimeria chalumnae), medaka (Oryzias latipes), skate (Leucoraja erinacea), spotted gar (Lepisosteus oculatus), stickleback (Gasterosteus aculeatus), zebrafish (D. rerio), and lamprey (Petromyzon marinus)], and chordates [ascidian (Ciona intestinalis) and lancelet (Branchiostoma floridae)] (Table A1). For phylogenetic reconstruction, the amino acid sequences were multiply aligned using MAFFT [28] and manually inspected. Based on the unambiguously aligned amino acid positions in the protease-like/protease (CASc*/CASc) domain, the phylogenetic tree was constructed by Randomized Axelerated Maximum Likelihood (RAxML) [29] using the GAMMA-WAG amino acid substitution model.
2.5. Comparative modeling of non-mammalian c-FLIP proteins and comparison with human c-FLIP
To build structural models of a CASc* domain of non-mammalian c-FLIP proteins, we used the crystal structure of human c-FLIP, 3H13 [30], which has more than 25% sequence identity with other c-FLIPs, as a template. The models were built using the Molecular Operating Environment (MOE) software package (MOE version 2012.10, Chemical Computing Group Inc., Tokyo), based on the alignments calculated by FORTE, a profile–profile comparison method for protein structure prediction [31], and were superimposed onto the structure of human c-FLIP.
2.6. Construction and transfection of plasmids
To express chicken, African clawed frog, zebrafish, and medaka c-FLIP proteins in mammalian cell lines, plasmid constructs pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, pME18S-Flag/DrFLIP, and pME18S-Flag/OlFLIP were generated by fusion of the respective CFLAR polymerase chain reaction (PCR)-amplified coding sequences from EST clones of these species and the Flag-tag sequence, followed by insertion into a mammalian expression vector, pME18S [32]. The pCMV-Flag/HsFLIP plasmid was generated by in-frame inserting human CFLAR cDNA into pCMV-Tag2B (Agilent Technologies, Santa Clara, CA). The pExpress1-GaFLIP plasmid, which carries full-length stickleback cflar cDNA, was obtained from Thermo Fisher Scientific Open Biosystems (Huntsville, AL) and used for the expression of stickleback c-FLIP. The pCS2-XlFLIP(DEDs) plasmid was generated by inserting a DNA fragment amplified by PCR into an expression vector, pCS2. The pME18S-HA/hFADD plasmid was generated by inserting the human FADD coding sequence fused with an HA-tag sequence into pME18S, and pME18S-Myc/TRAF2 was generated by inserting the mouse Traf2 coding sequence fused with a Myc-tag sequence. The pCAG-Venus plasmid, which was used to distinguish between transfected and untransfected cells, and the pCAG-p35 plasmid, which was used to inhibit caspase activation in transfected cells, have been previously described [33], [34]. Transfection of plasmid DNAs into HeLa and HEK293 cells was performed using Lipofectamine and PLUS Reagent or LTX Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions.
2.7. Immunoblotting and immunoprecipitation analyses
To detect human, chicken, African clawed frog, and zebrafish c-FLIP proteins in mammalian culture cells, HeLa cells were transfected with pCMV-Flag/HsFLIP, pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, pME18S-Flag/DrFLIP, or pME18S, cultured for 48 h, harvested, and suspended in lysis buffer containing 50 mM Tris–HCl (pH 7.5), 20 mM MgCl2, 0.5% Nonidet P-40, 150 mM NaCl, and the protease inhibitor cocktails (Nacalai Tesque). After removing cell debris by centrifugation, the cell lysates were added to Laemmli sample buffer, denatured by boiling, resolved by SDS-PAGE, and probed by immunoblotting with anti-Flag and anti-actin antibodies, respectively. After incubation with a secondary HRP-conjugated anti-mouse IgG antibody, immune complexes were visualized with ImmobilonTM Western chemiluminescent reagent (Millipore Corporation, Billerica, MA), using a luminescent image analyzer (LAS-3000, Fujifilm, Tokyo, Japan).
To detect the physical association of non-mammalian c-FLIP proteins with human FADD or mouse TRAF2 in cells, HEK293 cells were transfected with plasmids encoding non-mammalian Flag/c-FLIP and either plasmid pME18S-HA/hFADD or pME18S-Myc/TRAF2. In the case of transfection with pME18S-HA/hFADD, the plasmid pCAG-p35 was co-transfected to prevent cell death of FADD-expressing cells. Two days after transfection, cell lysates were prepared from transfected cells, incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4 °C overnight, and the beads were washed 5 times with lysis buffer. The immunoprecipitated proteins were eluted with Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting with the antibodies indicated in the figure legends.
2.8. Cytotoxicity assays
For cytotoxicity assays, HeLa cells were transiently transfected with pCMV-Flag/HsFLIP, pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, pME18S-Flag/DrFLIP, pME18S-Flag/OlFLIP, pExpress1-GaFLIP, or pME18S in conjunction with pCAG-Venus. At 48 h after transfection, cells were stimulated with 250 ng/ml of CH11 and 5 μg/ml of cycloheximide (CHX). After an additional 8 h incubation, cells were fixed in PBS containing 3.7% formaldehyde, washed with PBS, and observed by phase-contrast and fluorescence microscopy (DMIRE2, Leica Microsystems, Wetzlar, Germany).
2.9. Reporter assays for detection of NF-κB activation
The ability of non-mammalian c-FLIP proteins to activate NF-κB was examined by a Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Briefly, the plasmids carrying the non-mammalian CFLAR cDNAs were transfected with either reporter plasmid pTAL-Luc or pNFκB-Luc in conjunction with another reporter plasmid pRL-TK into HEK293 cells. After 48 h cultivation, cells were lysed in lysis buffer and the cell lysates were assayed for the activities of firefly and sea pansy luciferases, using a multilabel counter (ARVO-SX, PerkinElmer Life Science-Wallac Oy, Turku, Finland).
The NF-κB activation by non-mammalian c-FLIP proteins was also examined with another reporter plasmid, pCMV-EGFP/p65 [35]. By co-transfecting pCMV-EGFP/p65 with plasmids carrying the non-mammalian CFLAR cDNAs, the cellular localization of an EGFP/p65 protein was evaluated by microscopy after 48 h cultivation.
2.10. Reverse transcription-polymerase chain reaction (RT-PCR) analyses
Total RNAs were isolated from X. laevis embryos collected at various stages and RT-PCR was performed as described previously [36]. Briefly, first-strand cDNAs were amplified with two sets of primers: primers (forward: 5’-GCCGAGCGATATGTGGTTAGAA-3’, reverse: 5’- GCTTCAACTGGGATTTTAGGCG-3’) for the detection of CFLAR transcripts, primers (forward: 5’-CAGCTAGCTGTGGTGTGG-3’ and reverse: 5’-CAACATGGAAACTCACACC-3’) for the detection of ornithine decarboxylase (ODC) transcripts as an internal control. PCR consisted of an initial denaturation at 94 °C for 3 min followed by 20 (CFLAR) or 18 (ODC) cycles of 94 °C for 1 min, 57 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 3 min. Amplified PCR products were analyzed by 7.5 % acrylamide-gel electrophoresis.
Total RNAs were also isolated from zebrafish embryos collected at various stages as described previously [16]. For RT-PCR analysis, 2 μg of each RNA sample was used for first-strand cDNA synthesis with an oligo-dT primer on Ready-To-Go RT-PCR beads (Amersham Biosciences, Arlington Heights, IL), according to the manufacturer's recommendations. First-strand cDNAs were amplified with two sets of primers: primers (forward: 5’-GAAGCGCTATGATTTACTACGC-3’, reverse: 5’-TTTTGGACACTTGGTTGCGTT-3’) for the detection of cflar transcripts, and primers (forward: 5’-CTGTTGACTACGGAAAGAAGT-3’, reverse: 5'-TATGTGGACGCTCTATGTCTA-3') for tubulin α6 (tubα6) transcripts as an internal control. PCR consisted of an initial denaturation at 96 °C for 3 min followed by 35 cycles of 94 °C for 45 s, 58 °C for 60 s, 72 °C for 90 s, and a final extension at 72 °C for 5 min.
2.11. Microinjection of synthetic mRNA into Xenopus embryos
To express a truncated form of c-FLIP in X. laevis embryos, capped mRNA was synthesized using a template DNA, pCS2-XlFLIP(DEDs), with the mMESSAGE mMACHINE Kit (Ambion, Austin, TX), and purified by passing through a Sephadex G-50 column (GE Healthcare, Arlington Heights, IL) as described previously [37]. Microinjection of synthetic mRNA into embryos was performed at the four-cell stage. The injected embryos were statically cultured at 17 °C overnight in 3% Ficoll and 0.1×Steinberg’s solution [460 nM Tris–HCl (pH 7.2), 5.8 mM NaCl, 67 nM KCl, 34 nM Ca(NO3)2, 83 nM MgSO4, and 0.1 mg/ml Kanamycin], then transferred in 0.1×Marc’s Modified Ringers [0.5 mM HEPES (pH 7.8), 10 mM NaCl, 0.2 mM KCl, 0.2 mM CaCl2, 0.1 mM MgSO4, and 0.1 mM EDTA] and incubated at 23 °C until the proper stages for experiments.
2.12. Microinjection of morpholino oligonucleotide into zebrafish embryos
The antisense morpholino oligonucleotide (MO) for the coding sequence containing the start ATG codon of the zebrafish cflar gene (5’-CCAGAGTAGAAAATCCATCTGCCAT-3’) and the control MO carrying the reverse sequence (5’-TACCGTCTACCTAAAAGATGAGACC-3’) were obtained from GeneTools LLC (Philomath, OR) and dissolved at a concentration of 500 mM in 1×Danieau’s buffer [5 mM Hepes (pH 7.6), 58 mM NaCl, 0.7 mM KCl, 0.6 mM Ca(NO3)2, and 0.4 mM MgSO4]. One nanoliter of this solution or 1×Danieau’s buffer was injected into one- or two-cell embryos, which were prepared as described [38], before allowing the embryos to develop at 23 °C.
2.13. Statistical analyses
Data are presented as the means and standard deviations from at least three independent experiments. Significant differences between treatment groups were evaluated by calculating P values based on Student’s t-test.
3. Results
3.1. Isolation of the CFLAR/c-FLIP genes from non-mammalian vertebrates and the assessment of similarities
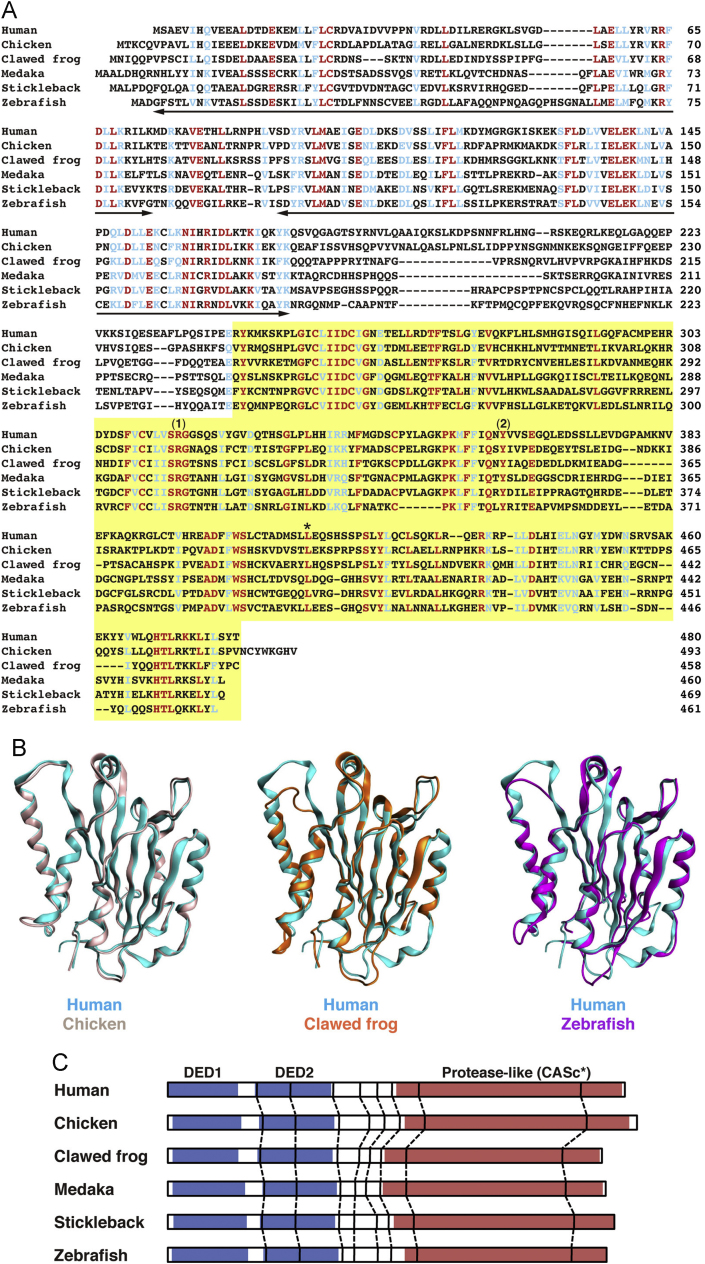
To investigate the distribution of genes orthologous to human CFLAR in animals, the NCBI DNA database was searched using TBLASTN. Candidate EST clones in the chicken (G. gallus), African clawed frog (X. laevis), medaka (O. latipes), and stickleback (G. aculeatus) were identified. Sequencing of these EST clones led to the determination of the complete open reading frames encoding 493, 458, 460, and 469 amino acids, respectively. Each of the predicted amino acid sequences exhibited the highest similarity to the long form of human c-FLIP in a reciprocal BLASTP analysis.3 As shown in Fig. 1A, the multiple alignment of amino acid sequences of these proteins with human and zebrafish c-FLIPs verified high similarity in two DED motifs and an inactive CASc (CASc*) domain. Therefore, these proteins are non-mammalian orthologs of c-FLIP.
Fig. 1.
Analysis of the protein structures of c-FLIP proteins. (A) Multiple alignment of amino acid sequences of human, chicken, African clawed frog, medaka, stickleback, and zebrafish c-FLIP proteins. Identical and similar amino acids in all family members are indicated by red and blue, respectively. The two bold lines and a yellow box indicate the DED motif and the protease-like CASc* domain, respectively. The numbers (1) and (2) shown above the sequences indicate the crucial arginine and tyrosine amino acid residues, which are the cause of the deactivation of the protease activity [8], and an asterisk indicates the conserved leucine residue. (B) Structural superposition of human c-FLIP and non-mammalian c-FLIP proteins. Structural models of the CASc* domain of chicken (light pink), clawed frog (orange), and zebrafish (magenta) c-FLIP proteins were computationally generated and superimposed with the CASc* domain of human c-FLIP (cyan, PDB ID: 3H13). (C) Comparison of the exon-intron organization of the CFLAR genes. The splice junction sites of the CFLAR/c-FLIP genes in vertebrates are indicated by vertical lines. Their positions were defined by the comparison of the respective genomic and cDNA sequences from the species listed in Table A2. The regions corresponding to two DED motifs and a CASc* domain are indicated by blue and red boxes, respectively.
To assess the structural similarity between non-mammalian c-FLIP proteins and human c-FLIP, three-dimensional structural models were constructed and compared with the crystal structure of human c-FLIP (Fig. 1B). The models showed that the core structural elements of non-mammalian CASc* domains comprise six β-sheets and five α-helices. When these structural models were superimposed with a crystal structure of the human CASc* domain, they showed a resemblance of the overall fold. Thus, the prediction of the spatial structure indicates that the tertiary structure of the c-FLIP family members is conserved from fish to mammals, and is consistent with the analysis of the primary amino acid sequence (see below).
In the CFLAR-paralogous CASP8 gene, new intron insertions have occurred in the coding region during teleost evolution [26]. In addition, both Casp10 and CASP18 paralogous genes have been lost during the evolution of various mammalian lineages [18], [19]. These observations suggest that the CFLAR genes might have diverged during evolution. To define the genomic structure of the CFLAR genes encoding c-FLIP proteins in vertebrates, we examined the genomic sequences published in the NCBI, Ensembl, and Xenbase genomic databases, and identified genomic sequences, which cover the CFLAR cDNA sequence, from chicken, African clawed frog, medaka, stickleback, and zebrafish (Table A2). Comparisons of these genomic and corresponding cDNA sequences clarified the similar organization of the CFLAR genes in these five animals. The CFLAR genes consist of nine to eleven exons. In every case, however, the CFLAR coding region basically covered nine exons (Fig. 1C and Table A2). Based on these data, we confirmed that the splice junction sites of the vertebrate CFLAR genes are well conserved in both the second DED motif and a CASc* domain, although they vary in position within the non-functional hinge region (Fig. 1C). Consequently, the organization of the CFLAR genes is evolutionarily conserved and maintained in the four living classes of vertebrates, excluding unexamined reptiles. Taken together, our results suggest that c-FLIP proteins retain a relatively unchanged structure during vertebrate evolution.
3.2. Molecular phylogenetic analysis of c-FLIP with its paralogous proteins
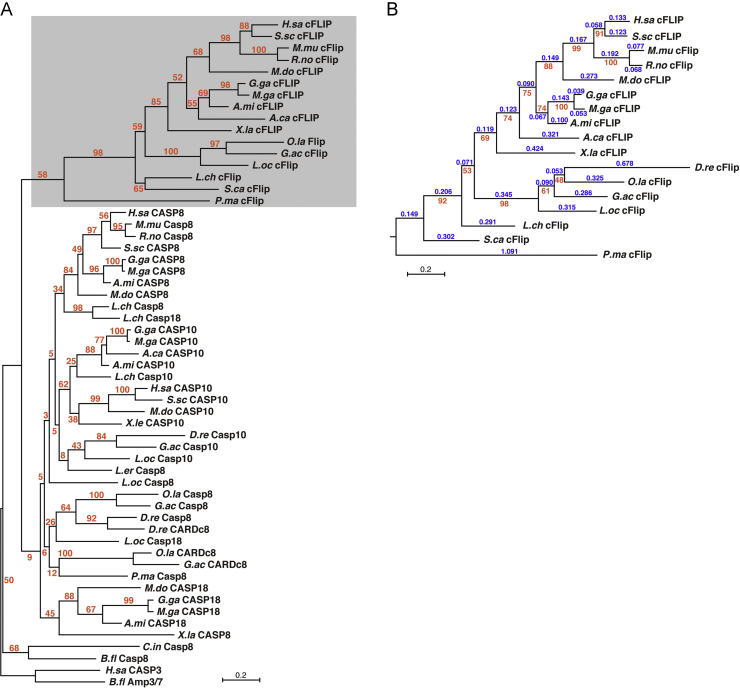
In previous studies, we proposed that the CFLAR/c-FLIP, CASP8, CASP10, and CASP18 genes diverged from the ancestral gene by gene duplication during evolution of early vertebrates, and the CARD-casp8 (tentatively named) gene in the fish lineage is also a diverged gene because it is located next to the casp8 gene, with sequence similarity in the protease domain [16], [18]. To better understand the evolutionary conservation of c-FLIP proteins in vertebrates, we constructed a molecular phylogenetic tree composed of the CASc*/CASc domains of c-FLIP and its paralogous proteins, CASP8, CASP10, CASP18, and CARD-casp8. For construction of the tree, we included additional c-FLIP protein sequences from various primitive fishes, the lamprey (P. marinus, Cyclostomata), the catshark (S. canicula, Chondrichthyes), the spotted gar (L. oculatus, Actinopterygii), and the coelacanth (L. chalumnae, Sarcopterygii) predicted from the genome and transcriptome databases, as listed in Table A1. In addition to known amino acid sequences, we also searched for unreported homologs in a set of sequence databases to identify new members in the caspase-8 superfamily (Table A1). The assembled molecular phylogenetic tree clearly indicated that the c-FLIP subfamily forms a unique clade distinct from the other subfamilies consisting of CASP8, CASP10, CASP18, and CARD-casp8 (Fig. 2A), suggesting that the c-FLIP subfamily members are independently evolving. In addition, the longer branch lengths of the c-FLIP subtree suggest that the amino acid changes in c-FLIP have quickly progressed compared with the other paralogous proteins after the divergence of the c-FLIP clade from the other subfamilies (Fig. 2A and B). Furthermore, comparison of the species divergence between CASc* and CASc domains revealed that the molecular evolutionary rate of c-FLIP is 1.7-fold higher than that of CASP8 when were analyzed in human, mouse, and chicken (Table 1). Thus, c-FLIP seems to have evolved in its own particular way compared to the other paralogous molecules in vertebrates.
Fig. 2.
Molecular phylogenetic analysis of c-FLIP and its paralogous proteins. (A) A molecular phylogenetic tree of c-FLIP and its paralogs, CASP8, CASP10, CASP18, and CARD-casp8 was constructed by RAxML, based on an alignment of the protease-like/protease (CASc*/CASc) domains (145 amino acid sites without mismatches including insertion/deletion (indel)). Human CASP3 and amphioxus CASP3-like AmphiCASP3/7 proteins were used as an out-group for rooting the tree. Numbers (in orange) at internal branches indicate the percentage of support of that branching pattern based on a bootstrap method with 100 replicates (bootstrap probabilities). The scale bar indicates the evolutionary distance of 0.2 amino acid substitutions per site. (B) A molecular phylogenetic tree restricted to the c-FLIP subfamily proteins. The tree was constructed by RAxML, based on an alignment of the c-FLIP CASc* domains (164 amino acid sites) derived from 17 animal species including zebrafish. The number (in blue) above each branch shows branch length, while the number (in orange) beneath each internal branch shows bootstrap probability. The scale bar indicates the evolutionary distance of 0.2 amino acid substitutions per site. The identification numbers of the c-FLIP subfamily and its paralogous proteins shown in the tree are listed in Table A1. Abbreviations of the species: A.ca, A. carolinensis (anole lizard); A.mi, A. mississippiensis (alligator); B.fl, B. floridae (lancelet); D.re, D. rerio (zebrafish); G.ac, G. aculeatus (stickleback); G.ga, G. gallus (chicken); H.sa, H. sapiens (human); L.ch, L. chalumnae (coelacanth); L.er, L. erinacea (little skate); L.oc, L. oculatus (spotted gar); M.do, M. domestica (opossum); M.ga, M. gallopavo (turkey); M.mu, M. musculus (mouse); O.la, O. latipes (medaka); R.no, R. norvegicus (rat); S.ca, S. canicula (catshark); S.sc, S. scrofa (pig); X.la, X. laevis (African clawed frog)
Table 1.
Comparison of evolutionary distances (Poisson correction) and molecular evolutionary rates of protease-like/protease (CASc#/CASc) domains.
| compared species | CASP8 | CASP10 | c-FLIP |
|---|---|---|---|
| human–mouse | 0.239 (1.00) | 0.253# (1.06) | 0.434 (1.82) |
| human–chicken | 0.392 (1.00) | 0.533 (1.36) | 0.688 (1.76) |
| mouse–chicken | 0.460 (1.00) | 0.592# (1.29) | 0.733 (1.59) |
| Average | 1.24 | 1.72 |
Evolutionary distances were calculated for highly homologous 179 amino acid sites without mismatches including insertion/deletion (indel). Each value in parentheses is relative molecular evolutionary rate normalized by the evolutionary distance of CASP8.
For CASP10, the amino acid sequence data of the naked mole rat (Heterocephalus glaber) was used instead of the mouse because the Casp10 gene has been lost in mice.
3.3. The evolutionarily conserved amino acid residues of c-FLIP family proteins
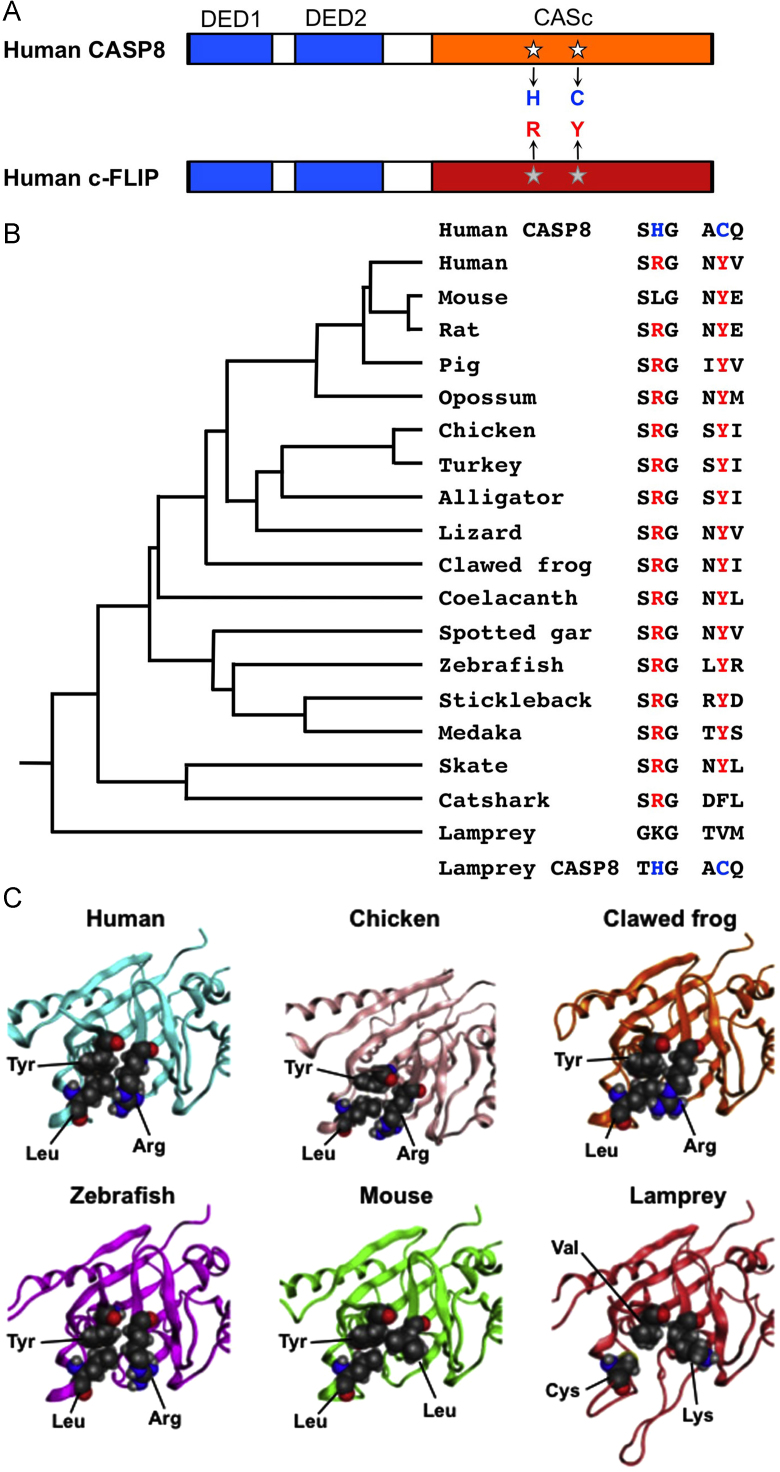
Both histidine (H) and cysteine (C) residues in the CASc of caspases form a catalytic dyad and are necessary for catalysis as a cysteine protease (Fig. 3A) [39]. These two amino acid residues were conserved in the active site of all CASP8, CASP10, CASP18, and CARD-casp8 proteins.4 By comparing amino acid sequences, both arginine (R) and tyrosine (Y) residues (R315 and Y360 in human c-FLIP), located in the inactive CASc* domain of c-FLIP proteins, replace the histidine and cysteine residues in the other subfamilies (Fig. 3A) [8], and are preserved in most animals examined, except lamprey, catshark, and mouse (Fig. 3B and Fig. A1). The lamprey and catshark, which belong to the Cyclostomata and Chondrichthyes, respectively, exhibited no conservation. Therefore, these two amino acids were likely conserved after evolving in bony vertebrates including bony fish and tetrapods (Actinopterygii and Sarcopterygii). Furthermore, the substitution from arginine to leucine has uniquely occurred in the mouse because the rat, which is a closely allied species to the mouse, has retained the arginine residue. The time of divergence between these two species is estimated at 23–33 million years ago [40], [41]. As it was thought that the arginine and tyrosine residues are crucial for the function of c-FLIP, the structural arrangement surrounding these amino acid residues was examined by computational analysis. For this analysis, the leucine residue (L413 in human) was also examined since it is conserved throughout vertebrates (Fig. A1). Modeling verified that these three residues, arginine, tyrosine, and leucine, are capable of forming a “pseudocatalytic triad” (Fig. 3C). By contrast, mouse and lamprey c-FLIP proteins may be unable to form such a firm structure (Fig. 3C). Thus, with the exception of mouse, c-FLIP proteins have evolutionarily conserved the spatial arrangement of amino acids required not only for the skeletal structure, but also for the triad formation, while they nevertheless show characteristic differences from the other subfamily members including CASP8, CASP10, CASP18, and CARD-casp8.
Fig. 3.
The specified amino acid substitutions of the CASc⁎ domain and their evolutionary conservation. (A) A schematic diagram of human CASP8 and c-FLIP proteins. Two DED motifs, a CASc protease domain, and a CASc* protease-like domain are indicated by boxes, respectively. White stars indicate the position of the essential amino acid residues, histidine (H) and cysteine (C) for the catalytic dyad formation in CASP8, whereas the gray stars indicate arginine (R) and tyrosine (Y) of c-FLIP positionally corresponding to two amino acid residues of CASP8, and are also coincident with positions (1) and (2), as shown in Fig. 1A. (B) A summary of critical amino acid residues of c-FLIP proteins conserved in vertebrates. Both the arginine (R) and tyrosine (Y) residues are evolutionarily conserved in most bony vertebrates. In the mouse c-Flip protein, the arginine residue is exceptionally changed to leucine. In the fish lineage, a phenylalanine (F) residue instead of tyrosine is present in the catshark whereas both lysine (K) and valine (V) residues are present at these two critical positions in the lamprey. A taxonomic tree of the species shown at the left was generated based on the previous study [70]. (C) Closeup views of the pseudocatalytic triad of c-FLIP proteins. The arginine, tyrosine, and leucine residues, which are shown in spheres, in human, chicken, clawed frog, and zebrafish c-FLIP proteins are brought close together in the three-dimensional shape forming a triad. In the lamprey, c-FLIP includes lysine, valine, and cysteine residues instead of the conserved amino acids, resulting in the failure of the triad formation. As mouse c-FLIP has replaced the arginine residue with leucine (L), there is little interaction with another leucine. The gray, red, and blue spheres indicate C, O, and N atoms, respectively.
3.4. Non-mammalian c-FLIP proteins play a role as negative regulators of apoptosis
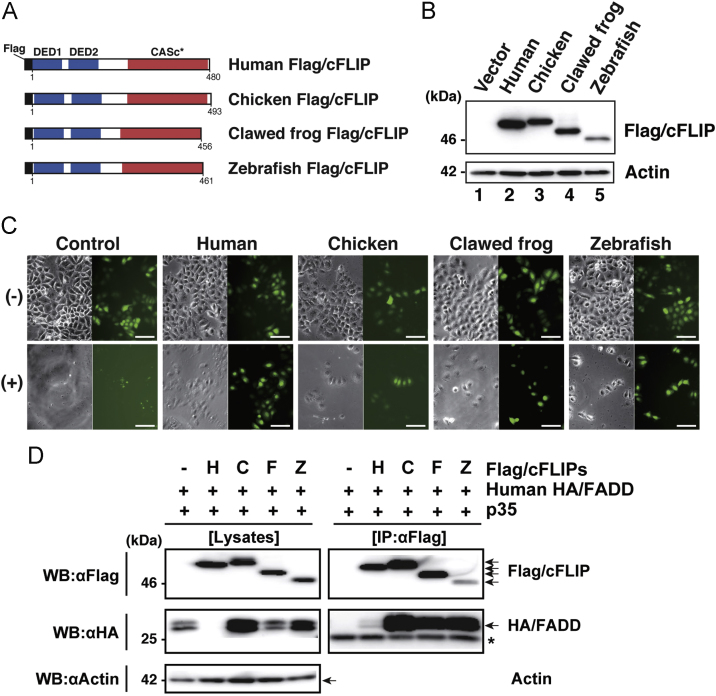
c-FLIP plays a role as a regulator of CASP8 activation mediated through death receptors [42]. We examined the anti-apoptotic activity of non-mammalian c-FLIP proteins in human HeLa cells. First, the ectopic expression of chicken, African clawed frog, and zebrafish c-FLIP proteins in transfected HeLa cell was confirmed by immunoblot analysis. To detect products in cultured cells, each c-FLIP was tagged with the Flag epitope (Fig. 4A). Proteins with the predicted size were detected (Fig. 4B). We then determined their ability to protect cells from death receptor-mediated apoptotic induction by examining the viability of transfectants subjected to Fas-ligation. Control cells expressing only Venus, which is a variant of yellow fluorescent protein [43], were sensitive to Fas-induced apoptosis, resulting in no Venus-positive cells after apoptotic stimuli (Fig. 4C). In contrast, chicken, frog, or zebrafish c-FLIP-expressing HeLa cells survived even in the presence of an agonistic anti-Fas antibody, as evidenced by the detection of only Venus-positive cells, while untransfected cells underwent apoptosis in the same culture dish. Similar results were observed in cells expressing medaka or stickleback c-FLIP (Fig. A2). These data indicated that non-mammalian c-FLIP proteins as well as human c-FLIP are able to protect mammalian cells from Fas-mediated apoptotic induction. FADD is a pro-apoptotic adapter molecule responsible for recruiting CASP8, CASP10, or c-FLIP into the death receptor complex through homotypic DED–DED interactions [44], [45] in response to extrinsic apoptotic stimuli. We investigated the interaction between non-mammalian c-FLIP proteins and human FADD by coimmunoprecipitation and immunoblot analyses. Human FADD was detected in the c-FLIP complex precipitated with an anti-Flag antibody (Fig. 4D), suggesting that non-mammalian c-FLIP proteins as well as human c-FLIP are able to interact with FADD. Taken together, these results suggest that c-FLIP proteins derived from birds, amphibians, and fish functionally act as anti-apoptotic molecules in the extrinsic apoptotic pathway by interacting with pro-apoptotic molecules.
Fig. 4.
Assessment of the anti-apoptotic activity of the c-FLIP subfamily proteins. (A) The protein structure of Flag-tagged non-mammalian c-FLIP proteins consisting of two DED motifs and a CASc* protease-like domain. (B) Immunoblot analysis of non-mammalian c-FLIP proteins. The empty vector, pCMV-Flag/HsFLIP, pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, or pME18S-Flag/DrFLIP were transiently transfected into HeLa cells. After culturing for 48 h, transgene products were analyzed by immunoblotting with an anti-Flag antibody. (C) Cytological analysis of transfectants expressing non-mammalian c-FLIP proteins. HeLa cells expressing Venus with or without human, chicken, clawed frog, and zebrafish c-FLIP were incubated in the presence (lower panels) or absence (upper panels) of anti-Fas antibody and CHX for 8 h, and examined for viability by monitoring Venus-positive cells. Both phase-contrast and fluorescence images in the same field were captured under the microscope. Scale bars represent 100 μm. (D) Co-immunoprecipitation and immunoblot analysis of physical interactions between non-mammalian c-FLIP proteins and human FADD. Human HEK293 cells were co-transfected with pME18S-HA/hFADD in conjunction with pME18S empty vector, pCMV-Flag/HsFLIP, pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, or pME-Flag/DrFLIP. Baculovirus p35 was introduced into all transfected cells to prevent cell death. After 2 days of cultivation, transfected cells were harvested and lysed in lysis buffer. The cell lysates were immunoprecipitated with an anti-FLAG M2 affinity gel. Coimmunoprecipitates and aliquots of cell lysates were examined by immunoblot analysis with anti-Flag, anti-HA, and anti-actin antibodies, respectively. An asterisk indicates immunoglobulin light-chain. Abbreviations: H, human; C, chicken; F, clawed frog; Z, zebrafish; IP, immunoprecipitation; WB, western immunoblotting.
3.5. Non-mammalian c-FLIP proteins function as NF-κB activators
It was previously reported that human c-FLIP induces NF-κB activation [14], [15]. To investigate whether non-mammalian c-FLIP proteins are also able to induce NF-κB activation, we examined their potential in transfected cells with the dual-luciferase reporter assay system (Fig. 5A, B). Exogenous expression of either chicken or clawed frog c-FLIP proteins increased the luciferase activity in transfected HEK293 cells (Fig. 5A). This indicated that NF-κB acquired transcriptional activity in the presence of the exogenous c-FLIP proteins. By changing the transfection reagent, we re-examined whether zebrafish c-FLIP is able to induce NF-κB activation. In these experiments, c-FLIP proteins derived from zebrafish and the other two fish, medaka and stickleback, increased the luciferase activity in transfected cells (Fig. 5B). To confirm NF-κB activation by non-mammalian c-FLIP proteins, the intracellular localization of p65 (RelA), an NF-κB subunit, was checked in non-mammalian c-FLIP-expressing cells. We co-transfected a plasmid, pEGFP/p65 encoding a fusion protein, EGFP/p65, consisting of enhanced green fluorescent protein (EGFP) and p65, with plasmids encoding non-mammalian c-FLIP proteins into HEK293 cells. The EGFP/p65 protein translocates to the nucleus when the NF-κB signaling pathway is activated [35]. As shown in Fig. 5C and D, EGFP/p65 nuclear localization increased in non-mammalian c-FLIP-expressing cells relative to control cells. In this assay system, human c-FLIP also stimulated the nuclear translocation of EGFP/p65. Furthermore, the interaction between tumor necrosis factor receptor-associated factor 2 (TRAF2) and non-mammalian c-FLIP proteins was examined. Previous studies have shown that TRAF2 plays a crucial role in NF-κB activation [46], and human c-FLIP associates with TRAF2 [47]. We confirmed their interactions by coimmunoprecipitation and immunoblot analyses (Fig. 5E). TRAF2 proteins were detected in the precipitated complex. Taken together, these results suggest that non-mammalian c-FLIP proteins have the ability to induce NF-κB activation in human cells.
Fig. 5.
Functional analyses of non-mammalian c-FLIP proteins on NF-κB activation ability. (A, B) Enzymatic analysis of NF-κB activation induced by non-mammalian c-FLIP proteins. The empty vector or plasmids carrying c-FLIP were transiently co-transfected with pNFκB-Luc and pRL-TK into HEK293 cells, and cultured for 48 h. NF-κB activation was analyzed by measuring enzyme activities of dual luciferases produced in transfected cells using a luminometer. Data are presented as the means and standard deviations of samples counted from three independent experiments. The statistically-significant difference between two groups was evaluated by Student’s t-test. (C) Cytological analysis of a NF-κB component, p65, in cells expressing c-FLIP. HEK293 cells were transiently transfected with an empty vector or plasmids carrying c-FLIP together with pEGFP/p65, and cultured for 24 h. The localization of EGFP/p65 proteins in transfected cells was analyzed by fluorescence microscopy. Typical patterns of subcellular localization of EGFP/p65: EGFP/p65 normally localizes in the cytoplasm (upper panels), but it translocates into the nucleus when the NF-κB signaling pathway undergoes activation (lower panels) [35]. Scale bars indicate 20 μm. (D) A summary of cytological analyses on the translocation of EGFP/p65. Within each field, positive cells (dark gray) and negative cells (light gray) for EGFP/p65 translocation were counted under the fluorescent microscope and percentages of total were calculated. N indicates the total number of transfected cells examined in four independent experiments. (E) Co-immunoprecipitation and immunoblot analysis of physical interactions between non-mammalian c-FLIP proteins and mouse TRAF2. HEK293 cells were transfected with either pCMV-Flag/HsFLIP, pME18S-Flag/GgFLIP, pME18S-Flag/XlFLIP, pME18S-Flag/DrFLIP, or control vector in combination with pME18S-Myc/TRAF2. Forty-eight hours after transfection, cells were lysed and the c-FLIP complex was immunoprecipitated from whole cell lysates with an anti-Flag antibody. Samples were analyzed alongside aliquots of the cell lysates by immunoblotting with anti-Flag, anti-Myc, and anti-actin antibodies, respectively. An asterisk indicates immunoglobulin light-chain. Abbreviations: H, human; C, chicken; F, clawed frog; Z, zebrafish; IP, immunoprecipitation; WB, western immunoblotting.
3.6. Involvement of c-FLIP in embryonic development
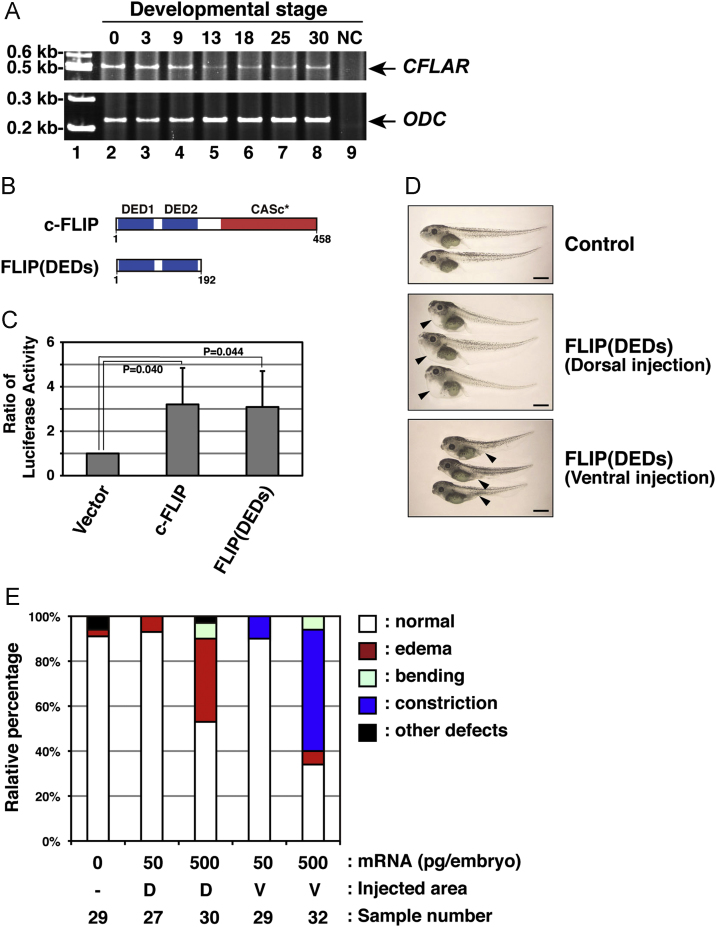
In the mouse, c-FLIP is critical for embryonic development. c-FLIP deficiency leads to embryonic lethality due to irregular development of the heart [48]. To more comprehensively understand the physiological roles of c-FLIP during embryonic development in vertebrates, we investigated the expression profiles of CFLAR/c-FLIP transcripts during embryogenesis of non-mammals. First we isolated total RNAs from embryos of African clawed frog (X. laevis) at various stages of development, and examined CFLAR mRNA by RT-PCR. In X. laevis embryos, CFLAR mRNA was detected in all samples prepared from stages 0, 3, 9, 13, 18, 25, and 30 (Fig. 6A). These data indicated that CFLAR mRNA exists in both maternal and zygotic transcripts during embryogenesis. To determine if CFLAR/c-FLIP is critical in embryogenesis, we generated a plasmid construct encoding a truncated form of c-FLIP, FLIP(DEDs), which contains only the two DED motifs (Fig. 6B), and synthesized mRNA using this plasmid as a template. The resulting truncated mutant protein, lacking the CASc* domain, was expected to dysregulate the function of full-length endogenous c-FLIP in organisms. In previous studies, discordant results have been reported, with one concluding that the amino-terminal half of human c-FLIP is a weaker NF-κB activator than full-length c-FLIP [14], whereas another study concluded that a truncated form contributed to higher NF-κB activation than full-length [49]. When an amphibian truncated form of c-FLIP was examined for its ability to induce NF-κB activation, this mutant induced NF-κB activation to the same extent as full-length c-FLIP in mammalian HEK293 culture cells (Fig. 6C). Following microinjection of mRNAs for FLIP(DEDs) into the four-cell-stage blastocysts, the morphology of embryos was examined after developing to stage 45. Injection into the equatorial areas of the two dorsal blastomeres caused abnormal development of embryos associated with thorax edema (Fig. 6D, middle panel). In contrast, abnormal embryos associated with scoop of the abdomen were observed after microinjection of the mRNA into the ventral area (Fig. 6D, lower panel). These phenotypes appeared in a dose-dependent manner (Fig. 6E). These data clearly indicate that irregular development of embryos occurs frequently following overexpression of a truncated form of c-FLIP. Thus, our results suggest that CFLAR/c-FLIP is involved in the early development of clawed frog embryos.
Fig. 6.
Irregular development of X. laevis embryos expressing a c-FLIP mutant. (A) The expression profile of African clawed frog CFLAR transcripts during embryogenesis. Total RNAs isolated from X. laevis embryos, which were collected at indicated stages (lanes 2–8), was analyzed by RT-PCR. PCR products amplified with primers specific for CFLAR (upper panel) and ODC (lower panel) were resolved by acrylamide-gel electrophoresis. Molecular weight markers were run in lane 1, and a negative control (NC) with no polymerase was run in lane 9. Arrows indicate the expected molecular weights of the CFLAR and ODC PCR products, respectively. (B) The structure of a truncated c-FLIP mutant, FLIP(DEDs), consisting of only two DED motifs. (C) Effect of a FLIP(DEDs) mutant on NF-κB activation. The empty vector or plasmids encoding intact or truncated forms of clawed frog c-FLIP were transiently co-transfected with pNFκB-Luc and pRL-TK into HEK293 cells. After 48 h in cell culture, NF-κB activation was analyzed by measuring enzyme activities of dual luciferases produced in transfected cells using a luminometer. Data are presented as the means and standard deviations of samples counted from three independent experiments. The statistically-significant difference between two groups was evaluated by Student’s t-test. (D) Morphological analysis of embryos expressing FLIP(DEDs). X. laevis embryos were injected without (upper panel) or with mRNA encoding FLIP(DEDs), at the equatorial area of two dorsal (middle panel) or two ventral (lower panel) blastomeres at the four-cell-stage. Images of the developing embryos were acquired at stage 45. Arrowheads indicate the edema and abdominal constriction of the injected embryos, respectively. Scale bars indicate 1 mm. (E) A summary of the phenotypic data presented in (D). Embryos displaying edema (red), abdominal constriction (dark blue), bending (light blue), and other defects (black) were counted under the microscope. Data represent the percentages calculated from five independent experiments. Abbreviations: D, dorsal; V, ventral.
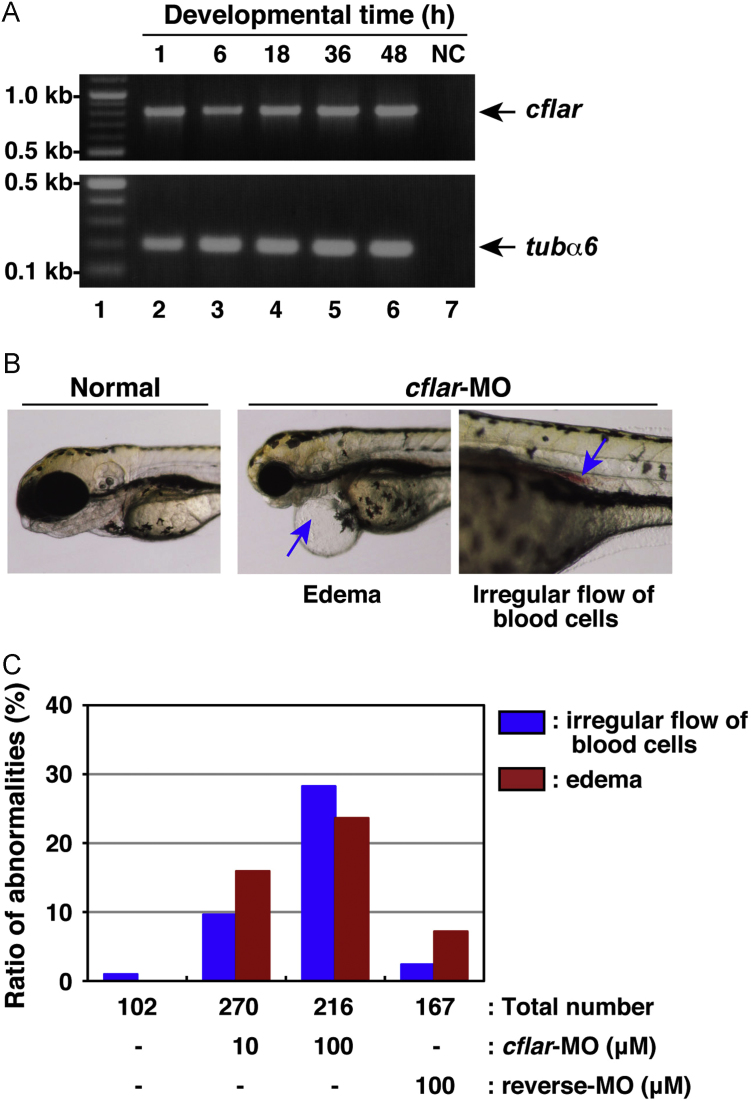
We also investigated the function of the fish c-Flip protein in embryonic development. The expression profile of cflar/c-Flip transcripts during embryogenesis in zebrafish was examined (Fig. 7A). During development, cflar transcripts were detected in all samples prepared from the cleavage, gastrula, segmentation, pharyngula, and hatching periods. These data indicate that cflar mRNA exists in both maternal and zygotic transcripts during embryogenesis of fish. To understand the physiological role of cflar/c-Flip in fish embryos, we examined the effects of an antisense morpholino oligonucleotide specific to cflar/c-Flip, cflar-MO, in the developing embryos. We injected cflar-MO into the fertilized eggs and examined the morphology of embryos during development. We observed abnormal development of the injected embryos in a dose-dependent manner, resulting in thorax edema and the delayed velocity of red blood cell flow or the clogging of red blood cells (Fig. 7B). We statistically confirmed these abnormalities (Fig. 7C). Thus, the deregulation of c-FLIP expression derailed embryonic development, suggesting the involvement of c-FLIP in embryogenesis.
Fig. 7.
Developmental anomalies in zebrafish embryos with knockdown of cflar transcripts. (A) The expression profile of zebrafish cflar transcripts during embryogenesis. Total RNAs isolated from embryos, which were collected at 1, 6, 18, 36, and 48 h after fertilization (lanes 2–6), was analyzed by RT-PCR. PCR products amplified with primers specific for cflar (upper panel) and tubulin α6 (tubα6) (lower panel) were resolved by agarose-gel electrophoresis. Molecular weight markers were run in lane 1, and a negative control (NC) with no template DNA was run in lane 7. Arrows indicate the expected positions of the cflar and tubα6 PCR products, respectively. (B) Morphological analysis of developing embryos subjected to morpholino oligonucleotide (MO) injection. Fertilized eggs were injected with an antisense MO for cflar, cflar-MO and their development was monitored under the microscope. Images of the developing embryos were acquired at three days later. The resulting abnormal phenotypes consisted of edema (middle panel) and a cluster of blood cells in the vessel (right panel), indicated by an arrow. (C) A summary of the phenotypic data presented in (B). Embryos displaying the edema (red) or irregular flow of red blood cells (blue) were counted under the microscope. Data represent the percentages calculated from four independent experiments.
4. Discussion
In this study, in order to understand the ubiquity and diversity of the c-FLIP/CFLAR subfamily members during vertebrate evolution, we identified and characterized non-mammalian c-FLIP proteins. The three-dimensional structure analysis showed a similar skeletal structure in vertebrate c-FLIP proteins. In addition, the arginine and tyrosine residues located in the pseudocatalytic triad of the CASc* domain are conserved in most bony vertebrates. Non-mammalian c-FLIP proteins functionally retain their potential as both apoptosis-regulating molecules and inducers of NF-κB activation. These results provide evidence that c-FLIP proteins are evolutionarily conserved in protein structure and function across vertebrate species. This prompts the hypothesis that these conserved functions were already established in the fish ancestor. On the other hand, the phylogenetic analysis verified that, compared with paralogous proteins such as CASP8, c-FLIP proteins have a faster substitution rate of amino acids in the CASc*/CASc domains, although they have preserved several amino acids that might be critical for maintenance of the structure and functionality. Consequently, c-FLIP, which arose at an early stage of vertebrate evolution, still might be continuing its molecular evolution while keeping the core structures.
In our previous studies, we proposed that the CFLAR, CASP8, CASP10, and CASP18 genes have been tandemly-duplicated from a single ancestral gene in the course of vertebrate evolution [16], and we and another group showed that both CFLAR and CASP8 genes invariably exist in vertebrates, whereas the other two genes, CASP10 and CASP18 have been lost in some mammals [16], [17], [18]. The main function of c-FLIP seems to be the regulation of CASP8 activation, but it is restricted to the vertebrate lineage. As supporting evidence for such a lineage specificity, no orthologs of c-FLIP have been reported in the fruit fly Drosophila melanogaster and ascidian Ciona intestinalis databases, while CASP8 exists in both animals [50], [51]. To understand the appearance and the acquisition of the functional properties of c-FLIP in the process of evolution, the investigation of jawless fish, such as lamprey and hagfish, is helpful because lamprey c-FLIP protein has not yet been characterized with regard to its functionality, and it clearly exhibits differences in the protein structure relative to other c-FLIP proteins.
c-FLIP was previously identified as a regulator of death receptor-mediated apoptosis. Whereas the deficiency of these death receptors causes no irregular embryogenesis [52], c-FLIP-deficient mice died at mid-gestation during embryogenesis due to irregular development of the heart [48]. This suggests that c-FLIP functions during embryonic development independent of death receptor-mediated signal transduction. A recent study reported that c-FLIP is indispensable for limited activation of CASP8 during embryonic development of rodents [53]. Therefore, c-FLIP and CASP8 act in a coordinated manner during embryogenesis. Apart from their respective roles in apoptosis, these proteins suppress necroptotic cell death mediated by receptor-interacting serine/threonine kinases 1 and 3 (RIPK1 and RIPK3) are involved in [53], [54], [55]. Necroptosis is a form of regulated or programmed necrosis and its signaling pathway has been defined [55], [56], [57], [58], [59], [60], [61]. When CASP8 is inactive and hence unable to control RIPK1, the necroptotic signaling pathway is activated: RIPK1 is free to interact with and phosphorylate RIPK3, and activated RIPK3 phosphorylates and activates an effector molecule, mixed lineage kinase domain-like (MLKL), resulting in cell death. These core components required for necroptosis are also detectable in non-mammalian vertebrates by searching the database.5 It is thus likely that a major developmental role of vertebrate c-FLIP in conjunction with CASP8 may be to jointly antagonize the induction of necroptotic cell death, although the molecular mechanism of the c-FLIP and CASP8 coordination still remains elusive [55], [60], [61], [62]. Interestingly, we noticed that the phenotypes of clawed frog embryos expressing a truncated mutant of c-FLIP are morphologically similar to those of embryos exogenously expressing a protease-deficient mutant of CASP10β [63]. This observation suggests that a CASP8 paralog, CASP10 also works with c-FLIP in developing embryos. Thus, c-FLIP might have pivotal roles not only for regulation of apoptosis associated with CASP8 activation but also for development and/or proliferation.
It has been reported that c-FLIP is required for heart development during mouse embryogenesis [48]. In humans, the expression of c-FLIP was detected in endothelial cells of the coronary arteries [64]. In addition, human umbilical vein endothelial cells (HUVECs) and dermal and pulmonary microvascular endothelial cells (MVECs) have been reported to express c-FLIP [65], [66]. In our study, both clawed frog embryos expressing a mutant FLIP(DEDs) and zebrafish embryos with down-regulated cflar expression exhibited edema during development (Fig. 6, Fig. 7). Edema is thought to be swelling caused by fluid leakage from the vascular system [67]. Both lines of evidence strongly suggest that c-FLIP plays a role in angiogenesis.
5. Conclusions
We demonstrated the universal functionality of c-FLIP proteins during vertebrate evolution. In vertebrates, the c-FLIP subfamily members commonly function as a negative regulator of the extrinsic apoptosis associated with CASP8 activation, and as an inducer for NF-κB activation. Although these physiological actions of c-FLIP proteins might be the result of the deficiency of protease activity, the specific amino acid substitutions in the catalytic dyad of the caspase domain and their conservation during evolution under selective pressure are striking, suggesting the possibility that c-FLIP has acquired a new function, not merely the loss of the enzyme activity, as was proposed previously [68]. Previous studies have shown that regardless of cell death, c-FLIP functions as a modulator of the Wnt signaling pathway [69], and an activator of the Erk signaling pathway [15]. Therefore, future studies should be directed at investigating the molecular mechanisms by which c-FLIP acts as a new functional molecule with interactions apart from caspase signaling.
Acknowledgments
The authors are grateful to N. Inohara (University of Michigan), J. Inoue (University of Tokyo), A. Miyawaki (RIKEN), J.A. Schmid (Medical University of Vienna), and K. Nomura (Kyushu University) for providing the zebrafish cflar, TRAF2, and Venus cDNAs, a EGFP/p65 reporter plasmid, and a FAD-II monoclonal antibody, and Y. Satou, M. Furutani-Seiki, A. Momoi, H. Yamashita, and S. Sakata for their technical assistance. We also thank to J.A. Hejna (Kyoto University) for critical reading of the manuscript. The complete sequence of the stickleback caspase-10 cDNA (GenBank Accession Number: HQ285318) has been deposited in the GenBank database. This work was supported in part by Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Data not shown
Data not shown
Data not shown
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.005.
Appendix A. Supplementary material
Fig. A1.
Multiple alignment of partial amino acid sequences in the CASc* domain of c-FLIP proteins. The conserved arginine (R), tyrosine (Y), and leucine (L) residues are shown in red. The assignments of α-helix and β-sheet secondary structure elements are based on a previous report (see Ref. [30]).
Fig. A2.
Cytological analysis of transfectants expressing fish c-FLIP proteins. HeLa cells expressing Venus with or without medaka and stickleback c-FLIP proteins were incubated in the presence (lower panels) or absence (upper panels) of anti-Fas antibody and CHX for 8 h, and examined for viability by monitoring Venus-positive cells. Both fluorescence and phase-contrast images in the same field were captured under the microscope. Scale bars represent 100 μm.
Supplementary material
Supplementary material
Supplementary material
References
- 1.Thornberry N.A., Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.C., Cullen S.P., Martin S.J. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 3.Galluzzi L., Vitale I., Abrams J.M., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., Dawson T.M., Dawson V.L., El-Deiry W.S., Fulda S., Gottlieb E., Green D.R., Hengartner M.O., Kepp O., Knight R.A., Kumar S., Lipton S.A., Lu X., Madeo F., Malorni W., Mehlen P., Nunez G., Peter M.E., Piacentini M., Rubinsztein D.C., Shi Y., Simon H.U., Vandenabeele P., White E., Yuan J., Zhivotovsky B., Melino G., Kroemer G. Molecular definitions of cell death subroutines: recommendations of the nomenclature committee on cell death. Cell Death Differ. , 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goltsev Y.V., Kovalenko A.V., Arnold E., Varfolomeev E.E., Brodianskii V.M., Wallach D. CASH, a novel caspase homologue with death effector domains. J. Biol. Chem. 1997;272:19641–19644. doi: 10.1074/jbc.272.32.19641. [DOI] [PubMed] [Google Scholar]
- 5.Han D.K., Chaudhary P.M., Wright M.E., Friedman C., Trask B.J., Riedel R.T., Baskin D.G., Schwartz S.M., Hood L. MRIT, a novel death-effector domain-containing protein, interacts with caspases and BclXL and initiates cell death. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11333–11338. doi: 10.1073/pnas.94.21.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu S., Vincenz C., Ni J., Gentz R., Dixit V.M. I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1- and CD- 95-induced apoptosis. J. Biol. Chem. 1997;272:17255–17257. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 7.Inohara N., Koseki T., Hu Y., Chen S., Nunez G. CLARP, a death effector domain-containing protein interacts with caspase-8 and regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10717–10722. doi: 10.1073/pnas.94.20.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J.L., Schroter M., Burns K., Mattmann C., Rimoldi D., French L.E., Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 9.Rasper D.M., Vaillancourt J.P., Hadano S., Houtzager V.M., Seiden I., Keen S.L., Tawa P., Xanthoudakis S., Nasir J., Martindale D., Koop B.F., Peterson E.P., Thornberry N.A., Huang J., MacPherson D.P., Black S.C., Hornung F., Lenardo M.J., Hayden M.R., Roy S., Nicholson D.W. Cell death attenuation by 'Usurpin', a mammalian DED-caspase homologue that precludes caspase-8 recruitment and activation by the CD-95 (Fas, APO-1) receptor complex. Cell Death Differ. 1998;5:271–288. doi: 10.1038/sj.cdd.4400370. [DOI] [PubMed] [Google Scholar]
- 10.Shu H.B., Halpin D.R., Goeddel D.V. Casper is a FADD- and caspase-related inducer of apoptosis. Immunity. 1997;6:751–763. doi: 10.1016/s1074-7613(00)80450-1. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasula S.M., Ahmad M., Ottilie S., Bullrich F., Banks S., Wang Y., Fernandes-Alnemri T., Croce C.M., Litwack G., Tomaselli K.J., Armstrong R.C., Alnemri E.S. FLAME-1, a novel FADD-like anti-apoptotic molecule that regulates Fas/TNFR1-induced apoptosis. J. Biol. Chem. 1997;272:18542–18545. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 12.Golks A., Brenner D., Fritsch C., Krammer P.H., Lavrik I.N. c-FLIPR, a new regulator of death receptor-induced apoptosis. J. Biol. Chem. 2005;280:14507–14513. doi: 10.1074/jbc.M414425200. [DOI] [PubMed] [Google Scholar]
- 13.Krueger A., Baumann S., Krammer P.H., Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol. Cell. Biol. 2001;21:8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W.H., Johnson H., Shu H.B. Activation of NF-kappaB by FADD, Casper, and caspase-8. J. Biol. Chem. 2000;275:10838–10844. doi: 10.1074/jbc.275.15.10838. [DOI] [PubMed] [Google Scholar]
- 15.Kataoka T., Budd R.C., Holler N., Thome M., Martinon F., Irmler M., Burns K., Hahne M., Kennedy N., Kovacsovics M., Tschopp J. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr. Biol. 2000;10:640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 16.Sakata S., Yan Y., Satou Y., Momoi A., Ngo-Hazelett P., Nozaki M., Furutani-Seiki M., Postlethwait J.H., Yonehara S., Sakamaki K. Conserved function of caspase-8 in apoptosis during bony fish evolution. Gene. 2007;396:134–148. doi: 10.1016/j.gene.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhart L., Ballaun C., Hermann M., Vandeberg J.L., Sipos W., Uthman A., Fischer H., Tschachler E. Identification of novel mammalian caspases reveals an important role of gene loss in shaping the human caspase repertoire. Mol. Biol. Evol. 2008;25:831–841. doi: 10.1093/molbev/msn012. [DOI] [PubMed] [Google Scholar]
- 18.Sakamaki K., Satou Y. Caspases: evolutionary aspects of their functions in vertebrates. J. Fish Biol. 2009;74:727–753. doi: 10.1111/j.1095-8649.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamaki K., Imai K., Tomii K., Miller D.J. Evolutionary analyses of caspase-8 and its paralogs: deep origins of the apoptotic signaling pathways. Bioessays. 2015;37:767–776. doi: 10.1002/bies.201500010. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A., Thies R.S., Yamaji N., Song J.J., Wozney J.M., Murakami K., Ueno N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10255–10259. doi: 10.1073/pnas.91.22.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieuwkoop P.D., Faber J. North Holland Publishing Co.; Amsterdam: 1967. Normal Table of Xenopus laevis (Daudin) [Google Scholar]
- 22.Westerfield M. University of Oregon Press; Eugene: 1994. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 23.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 24.Yonehara S., Ishii A., Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J. Exp. Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gertz E.M., Yu Y.K., Agarwala R., Schaffer A.A., Altschul S.F. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamaki K., Shimizu K., Iwata H., Imai K., Satou Y., Funayama N., Nozaki M., Yajima M., Nishimura O., Higuchi M., Chiba K., Yoshimoto M., Kimura H., Gracey A.Y., Shimizu T., Tomii K., Gotoh O., Akasaka K., Sawasaki T., Miller D.J. The apoptotic initiator Caspase-8: its functional ubiquity and genetic diversity during animal evolution. Mol. Biol. Evol. 2014;31:3282–3301. doi: 10.1093/molbev/msu260. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., BLAST Gapped. and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 30.Yu J.W., Jeffrey P.D., Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8169–8174. doi: 10.1073/pnas.0812453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomii K., Akiyama Y. FORTE: a profile–profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- 32.Sakamaki K., Miyajima I., Kitamura T., Miyajima A. Critical cytoplasmic domains of the common beta subunit of the human GM-CSF, IL-3 and IL-5 receptors for growth signal transduction and tyrosine phosphorylation. EMBO J. 1992;11:3541–3549. doi: 10.1002/j.1460-2075.1992.tb05437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakamaki K., Takagi C., Yoshino J., Yokota H., Nakamura S., Kominami K., Hyodo A., Takamune K., Yuge M., Ueno N. Transgenic frogs expressing the highly fluorescent protein Venus under the control of a strong mammalian promoter suitable for monitoring living cells. Dev. Dyn. 2005;233:562–569. doi: 10.1002/dvdy.20350. [DOI] [PubMed] [Google Scholar]
- 34.Sakamaki K., Takagi C., Kominami K., Sakata S., Yaoita Y., Kubota H.Y., Nozaki M., Yonehara S., Ueno N. The adaptor molecule FADD from Xenopus laevis demonstrates evolutionary conservation of its pro-apoptotic activity. Genes Cells. 2004;9:1249–1264. doi: 10.1111/j.1365-2443.2004.00802.x. [DOI] [PubMed] [Google Scholar]
- 35.Birbach A., Gold P., Binder B.R., Hofer E., de Martin R., Schmid J.A. Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. J. Biol. Chem. 2002;277:10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- 36.Goda T., Takagi C., Ueno N., Rnd1 Xenopus. and Rnd3 GTP-binding proteins are expressed under the control of segmentation clock and required for somite formation. Dev. Dyn. 2009;238:2867–2876. doi: 10.1002/dvdy.22099. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T.S., Takagi C., Hyodo A.C., Ueno N. Suppression of head formation by Xmsx-1 through the inhibition of intracellular nodal signaling. Development. 2001;128:2769–2779. doi: 10.1242/dev.128.14.2769. [DOI] [PubMed] [Google Scholar]
- 38.Momoi A., Yoda H., Steinbeisser H., Fagotto F., Kondoh H., Kudo A., Driever W., Furutani-Seiki M. Analysis of Wnt8 for neural posteriorizing factor by identifying Frizzled 8c and Frizzled 9 as functional receptors for Wnt8. Mech. Dev. 2003;120:477–489. doi: 10.1016/s0925-4773(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 39.Denault J.B., Salvesen G.S. Caspases: keys in the ignition of cell death. Chem. Rev. 2002;102:4489–4500. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 40.Adkins R.M., Gelke E.L., Rowe D., Honeycutt R.L. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol. Biol. Evol. 2001;18:777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- 41.Nei M., Xu P., Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozturk S., Schleich K., Lavrik I.N. Cellular FLICE-like inhibitory proteins (c-FLIPs): fine-tuners of life and death decisions. Exp. Cell Res. 2012;318:1324–1331. doi: 10.1016/j.yexcr.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 44.Boldin M.P., Goncharov T.M., Goltsev Y.V., Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 45.Muzio M., Chinnaiyan A.M., Kischkel F.C., O'Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J.D., Zhang M., Gentz R., Mann M., Krammer P.H., Peter M.E., Dixit V.M. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death–inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 46.Tada K., Okazaki T., Sakon S., Kobarai T., Kurosawa K., Yamaoka S., Hashimoto H., Mak T.W., Yagita H., Okumura K., Yeh W.C., Nakano H. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J. Biol. Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 47.Kataoka T., Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol. Cell. Biol. 2004;24:2627–2636. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh W.C., Itie A., Elia A.J., Ng M., Shu H.B., Wakeham A., Mirtsos C., Suzuki N., Bonnard M., Goeddel D.V., Mak T.W. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 49.Golks A., Brenner D., Krammer P.H., Lavrik I.N. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappaB activation. J. Exp. Med. 2006;203:1295–1305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aravind L., Dixit V.M., Koonin E.V. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291:1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- 51.Terajima D., Shida K., Takada N., Kasuya A., Rokhsar D., Satoh N., Satake M., Wang H.G. Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death Differ. 2003;10:749–753. doi: 10.1038/sj.cdd.4401223. [DOI] [PubMed] [Google Scholar]
- 52.Wajant H. Death receptors. Essays Biochem. 2003;39:53–71. doi: 10.1042/bse0390053. [DOI] [PubMed] [Google Scholar]
- 53.Oberst A., Dillon C.P., Weinlich R., McCormick L.L., Fitzgerald P., Pop C., Hakem R., Salvesen G.S., Green D.R. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaiser W.J., Upton J.W., Long A.B., Livingston-Rosanoff D., Daley-Bauer L.P., Hakem R., Caspary T., Mocarski E.S. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silke J., Rickard J.A., Gerlic M. The diverse role of RIP kinases in necroptosis and inflammation. Nat. Immunol. 2015;16:689–697. doi: 10.1038/ni.3206. [DOI] [PubMed] [Google Scholar]
- 56.Holler N., Zaru R., Micheau O., Thome M., Attinger A., Valitutti S., Bodmer J.L., Schneider P., Seed B., Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 57.Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G.D., Mitchison T.J., Moskowitz M.A., Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 58.Sun L., Wang X. A new kind of cell suicide: mechanisms and functions of programmed necrosis. Trends Biochem. Sci. 2014;39:587–593. doi: 10.1016/j.tibs.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Vanden Berghe T., Linkermann A., Jouan-Lanhouet S., Walczak H., Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 60.Justus S.J., Ting A.T. Cloaked in ubiquitin, a killer hides in plain sight: the molecular regulation of RIPK1. Immunol. Rev. 2015;266:145–160. doi: 10.1111/imr.12304. [DOI] [PubMed] [Google Scholar]
- 61.Newton K. RIPK1 and RIPK3: critical regulators of inflammation and cell death. Trends Cell Biol. 2015;25:347–353. doi: 10.1016/j.tcb.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 62.Silke J., Strasser A. The FLIP side of life. Sci. Signal. 2013;6:pe2. doi: 10.1126/scisignal.2003845. [DOI] [PubMed] [Google Scholar]
- 63.Kominami K., Takagi C., Nozaki M., Kurata T., Kitayama A., Nozaki M., Sawasaki T., Kuida K., Endo Y., Manabe N., Ueno N., Sakamaki K. The initiator caspase, caspase-10β, and the BH-3-only molecule, Bid, demonstrate evolutionary conservation in Xenopus of their pro-apoptotic activities in the extrinsic and intrinsic pathways. Genes Cells. 2006;11:701–717. doi: 10.1111/j.1365-2443.2006.00983.x. [DOI] [PubMed] [Google Scholar]
- 64.Imanishi T., McBride J., Ho Q., O'Brien K.D., Schwartz S.M., Han D.K. Expression of cellular FLICE-inhibitory protein in human coronary arteries and in a rat vascular injury model. Am. J. Pathol. 2000;156:125–137. doi: 10.1016/S0002-9440(10)64712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alladina S.J., Song J.H., Davidge S.T., Hao C., Easton A.S. TRAIL-induced apoptosis in human vascular endothelium is regulated by phosphatidylinositol 3-kinase/Akt through the short form of cellular FLIP and Bcl-2. J. Vasc. Res. 2005;42:337–347. doi: 10.1159/000086599. [DOI] [PubMed] [Google Scholar]
- 66.Stefanescu R., Bassett D., Modarresi R., Santiago F., Fakruddin M., Laurence J. Synergistic interactions between interferon-gamma and TRAIL modulate c-FLIP in endothelial cells, mediating their lineage-specific sensitivity to thrombotic thrombocytopenic purpura plasma-associated apoptosis. Blood. 2008;112:340–349. doi: 10.1182/blood-2007-10-119552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vestweber D. Molecular mechanisms that control endothelial cell contacts. J. Pathol. 2000;190:281–291. doi: 10.1002/(SICI)1096-9896(200002)190:3<281::AID-PATH527>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 68.Adrain C., Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol. 2012;13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- 69.Naito M., Katayama R., Ishioka T., Suga A., Takubo K., Nanjo M., Hashimoto C., Taira M., Takada S., Takada R., Kitagawa M., Matsuzawa S., Reed J.C., Tsuruo T. Cellular FLIP inhibits beta-catenin ubiquitylation and enhances Wnt signaling. Mol. Cell. Biol. 2004;24:8418–8427. doi: 10.1128/MCB.24.19.8418-8427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hedges S.B., Dudley J., Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material