Summary
Indoor Air Quality (IAQ) in libraries is influenced by the presence of specific factors which can impact on both paper storage as well as people health. Microclimatic conditions induce and support a biodiversity pattern involving environmental and anthropic microorganisms. We used a multidisciplinary monitoring model to characterize microflora biodiversity by Next Generation Sequencing (NGS). Biodiversity indexes were adapted to evaluate anthropic vs environmental pollution by combining Shannon mean index (H), species representativeness (EH), human/environmental pollution ratio (SA) to better characterize the NGS output and acquire synthetic information on Indoor Air Microbial Biodiversity (IAMB). Results indicate a frequently low microbial load (IGCM/m3 < 1000) characterized by different species (n = 102), including several cellulose metabolizing bacteria. Workers and visitors appeared a relevant source of microbial contamination. Air biodiversity assayed by NGS seems a promising marker for studying IAQ.
Key words: library, indoor air quality, microflora, NGS, monitoring
Introduction
Indoor air quality (IAQ) is recognized as a key factor for human health, due to the fact that people spend 80%- 90% of their time in indoor environments [1-4]. Biological, physical and chemical agents can affect the characteristics of the indoor atmosphere [5]. Biological agents relevant to health are widely heterogeneous, including pollen, allergens, plants spores, bacteria, fungi, algae and some protozoa. Moreover, activities like talking, sneezing, coughing, can generate airborne biological particulate, and this is considered a main factor contributing to the buildup and spread of airborne microbial contamination [6, 7]. The exposure to microbial pollution can be associated with respiratory symptoms, allergies, asthma and immunological reactions [8]. Microclimatic conditions, as humidity, temperature and ventilation rates, together with geometry of building, influence individual comfort and environmental health [9-11]. Volatile compounds (VOCs) can be released from building materials, furnishings, office machinery, or cleaning products [12]. Libraries represent an exemplificative model of sensitive indoor environments such as hospitals, schools, museums, laboratories, drug or food factories, where IAQ impacts on both people health and materials, products or procedures. During the last decades, studies on specific indoor environments have identified risk sources and threshold levels for different pollutants, proposing targeted control measures. Even the role of the structure itself has been investigated and analysis of the Sick Building Syndrome has been considered for the implications on the health of workers, dwellers, as well as for the preservation of materials and procedures [13-15]. However, the combined effects of biological agents and chemical mixtures still require further investigation, and innovative tools are needed to improve quality of indoor air [16, 17]. When it happens to specific facilities, as libraries, archives and schools, it needs further consideration to identify most appropriate markers to assess indoor air quality for people, paper materials and activities. Libraries are indoor facilities where air quality is influenced by both environmental and anthropic microorganisms, interacting with chemical and physical pollutants.
Microclimatic conditions influence the growth of several microorganisms (e.g. Aspergillus, Penicillium, Trichoderma, Alternaria, Rhizopus) that can attack library collections, eventually favoring the biodeterioration processes [3]. Traditional cultural methods or classical molecular approaches cannot address a comprehensive microflora analysis [18, 19]. The limit is mainly due to technical restrictions in growing, detecting or classifying the environmental species as well as in providing an overall view of the whole microflora pattern in that setting. The rapid diffusion of Next Generation Sequencing (NGS) and new bioinformatic tools are overcoming this problem, offering a wider approach to microflora biodiversity, at increasingly affordable costs. Starting from the microflora DNA (mfDNA) it became possible to detect simultaneously different species even from very complex matrices [20, 21]. NGS satisfies the need for qualitative and quantitative information on microbial complexity, disclosing innovative solutions for prevention and hygiene. Since the strategy is based on analysis of genomic properties, the method is independent from the knowledge of the microorganisms or their culture offering new potentials as already shown for other DNA based techniques [22-24]. Moreover, even unknown or uncultivable species can be identified, quantified and characterized following an established phylogenetic approach, by amplification of rDNA, massive sequencing and bioinformatics analysis.
Following a traditional approach, previous studies have already investigated the effects of biological and chemical conditions on paper conservation, outlining also issues related to the health of library users, such as students, visitors and employed personnel [12, 26, 27]. However, most of these studies focused on independent categories of risk factors, such as chemical agents [12, 26, 28, 29], physical [30-32], without adopting an integrated approach. The characterization of microbial air contamination using single culture-based sampling not only provides a limited understanding of the phenomenon, but also restrains a comprehensive evaluation of IAQ and the implementation of corrective actions [33]. Analysis of microbial biodiversity in indoor air (IAMB) can play an important role in characterizing specific environments, identifying possible biological risks and acquiring information for the anthropic component.
In the present study, environmental samplings have been simultaneously performed in order to obtain a wider overview focused on biological issues and integrated with microclimate and environmental parameters. Moreover, for the first time, an advanced approach for microorganism identification was adopted through NGS, with the final aim of typing the indoor microflora and acquiring information on the impact of anthropic and environmental microflora on IAQ.
Air microflora is influenced by environmental conditions and may play a role on human health as well as on safety of library resources, especially when storing valuable ancient books. NGS seems to offer an extraordinary and promising opportunity for making microflora biodiversity as a new marker for monitoring specific indoor environments.
Materials and methods
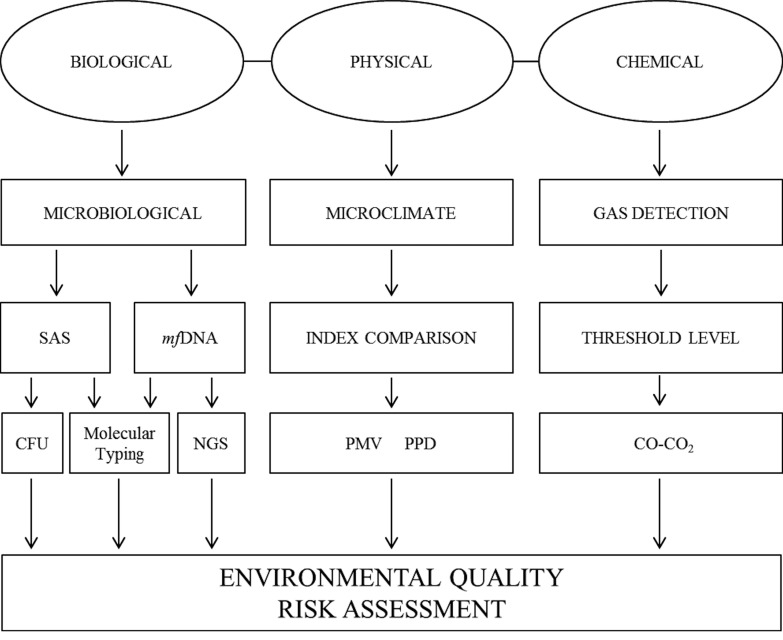
In order to evaluate IAQ, three different kinds of libraries were chosen: Site I, a university library; Site II, an old scientific library; Site III, a digital library. Several environmental samplings were performed following an integrated approach (Figure 1), to the aim of acquiring an overview on microclimate, chemical and biological parameters. Biological monitoring was performed by a novel passive method adapted for DNA extraction and by classical active sampling techniques (SAS). Microorganisms were characterized by both culture-based methods and by molecular techniques based on mfDNA analysis.
Fig. 1.
Flowchart with panel of sampling techniques and integrated monitoring approach.
STUDY DESIGN
Environmental sampling was performed to provid an overview of microclimate and microbiological parameters in these facilities and to show the feasibility of an integrated approach for IAQ surveillance. Biological monitoring was performed with different sampling techniques (active and passive) and methods (classical microbiology and DNA techniques). Statistical analysis was performed to determine the distribution of indoor mesophilic, psichrophilic bacteria and fungi concentrations in association with independent variables, such as the buildings age, light exposure, air exchange rate and ventilation systems. Similarly, the indoor mesophilic, psichrophilic and fungi concentration have been correlated with the relative humidity, the external and internal temperature.
STUDY AREA
Three scientific libraries in the city of Rome (Italy), were selected as exemplificative of different typologies. The first site (Site I) is a "university library" founded in 1994, consisting of a single reading room with desks for students, computers and a loft with shelves for the books storage. The documentary print heritage consists in over 5000 monographs and 2000 periodicals, providing access to about 2000 electronic journals and scientific databases. The library also has books storage room, located in the same building, in the basement. The second site (Site II) was founded in 1930s and represents a centralized reference, specialized in scientific documentation. The print documents legacy consists in more than 9000 periodicals and about 200000 monographs including publications of national and international institutes, pharmacopoeias, official documents, texts of health legislation and grey literature. This library also collects thousands of electronic journals in full text, and accesses to biomedical databases. It also stores historic 16th-18th century dated books, and precious documents. The library is organized on three levels, where the books are stored in shelves both fixed and mobile. The library has also a book storage room, located in an independent building, and divided into two rooms. The third one (Site III) is a "digital library": it was founded in 1930 and in 2005 was moved in an independent building and organized as an electronically accessible library, associated to a hospital and research centre. The collection currently amounts to about 10000 online magazines and different local databases. It offers a reading room with 15 PC workstations intended to health consulting and health education activities open to visitors, including both patients and professionals; in the same building the upper floor houses the management offices. Libraries have an usual access of about 10-50 visitors per day, employing 3-6 workers dedicated to reading rooms, office and management activities.
In Site I, sampling points (n = 2) were: the reading room and the book storehouse. In Site II (n = 7): Flat A Room 1, Flat A Room 2, Flat B room 3, Flat B room 4, Reading room, Book storehouse 1, Book storage 2. Site III samplings (n = 2): the reading room and the manager office. The samplings were performed in the period ranging from September 2014 to July 2015.
Daily averages of outdoor temperature data were obtained from a monitoring station at an average distance of about 10 km from the library sites. For each library, site elevation, proximity to agricultural/industrial processes or major arterial roadways was verified also by the support of satellite geographic maps (Google Map) to exclude major air pollution sources. Building characteristics were considered, including construction materials, restorations, heating and ventilation systems. Signs of moisture, water damage or fungal growth on building materials were assessed visually and annotated in the sampling form.
PHYSICAL AND CHEMICAL PARAMETERS
The instrument HD32.3 DataLogger® (Delta Ohm s.r.l.), conforms to ISO 7730, 7726, 27243, 7933, 11079, 8996 (Supplementary Table, S1), was adopted for microclimate analysis. The unit has three probes, thermohygrometric, anemometer and globe thermometer to determine the discomfort indices, PMV (Predicted Mean Vote) and PPD (Percentage of Person dissatisfied). The PMV is an individual wellness index. It is a mathematical function which gives as result a numerical value in the range between -3 (feeling too cold) and +3 (feeling too hot), where 0 represents the thermal comfort. The PPD expresses the percentage of dissatisfied people in a particular environment. Through DeltaLog10® software (Delta Ohm s.r.l.) the data were downloaded. During sampling, the instrument was positioned in the sites center, for 15 minutes, with 15 sec intervals in measurement at the worker's chest height.
Carbon dioxide (CO2) and carbon monoxide (CO) quantification was also performed by HD21AB17 Datalogger ® (Delta Ohm s.r.l). It has a single probe which can simultaneously measure the level of CO2, CO and atmospheric pressure. The instrument can also measure the temperature, the relative humidity and calculate the dew point, the wet bulb temperature, absolute humidity, mixing ratio and enthalpy. Through DeltaLog10® software (Delta Ohm s.r.l.) data were downloaded and analyzed. The tool has been positioned in the room centre at the worker's chest height and the measures were performed for 15 minutes, with 15 seconds of intervals between each measure. We referred to the European standard EN 13779-2008 on the Ventilation for non-residential buildings to evaluate the CO2 levels and to the European directive 2008/50/ EC to evaluate the CO levels.
MICROBIOLOGICAL AIR SAMPLING
Active sampling
The microbiological air sampling was performed by SAS® (Surface Air System, VWR International, LLC, Radnor, USA), a plate impact active sampler, slot type, using Petri dishes Ø 55mm. The air aspiration volume was 180 L for each sampling. We used two culture media types: TSA (Tryptic Soy Agar - Oxoid, Germany) for bacteria, SDA (Sabouraud Dextrose Agar - Oxoid, Germany) supplemented with Chloramphenicol® (Oxoid, Germany) for molds and yeasts. The TSA and SDA plates were incubated both at 37°C (for 48 hours) and at 22°C (for 72 hours) respectively. For each plate, the calculation of the CFU/m3 was obtained as follows: CFU/ m3 = (MPN/plate x 1000) / air volume (L). MPN (Most Probable Number)/plate was obtained by comparing the CFU of each plate with the conversion table, in attachment to the device manual.
In addition, from each TSA plate, colonies were collected by sterile loop, in a 1.5 ml tube containing 200 μl of sterile water. From each SDA plate, representative colonies of each fungal species were collected by sterile loop in a 1.5 ml tube, containing 200 μl of sterile water. All tubes were centrifuged at maximum speed (16000 g for 5 minutes). The supernatant was removed and the pellet was frozen at - 4°C. The DNA extraction by pellets was performed with GenElute® Bacterial Genomic DNA Kit, following the manufacturer's instruction.
Passive sampling
In Site II, a tissue-based passive sampling approach was developed and tested to collect bacteria and extract DNA. Sterilized cotton tissues (size 5 cm2) were placed for one month on the shelves of Flat A Room 1, Flat A Room 2, Flat B Room 3, Flat B Room 4, and named respectively T1, T2, T3, T4. The collection was followed by direct DNA extraction. Briefly, a piece of 1 cm2 was cut from each tissue. After exposure, each sample was transferred in a 1.5 ml tube at room temperature and then frozen at -20°C for 20 minutes for cryofreezing fracturing. The samples were pretreated with glass beads® (Sigma Aldrich, USA) and 200 μl of Lysozyme Solution® (Sigma Aldrich, USA), adapting the protocol as previous described (Giampaoli 2012) In a second phase we followed the standard protocol procedure of GenElute® Bacterial Genomic DNA Kit (Sigma Aldrich, USA). The DNA extraction by pellets was performed with GenElute ® Bacterial Genomic DNA Kit, following the manufacturer's instruction.
DNA ANALYSIS
Fungal ITS PCR amplification and Sanger sequencing
DNA was amplified with primer designed for fungal ITS (Internal Transcribed Spacer) region [34]. The PCR amplification was performed in 25 μl reaction mixture consisting of 1× Taqmastermix® (Promega, USA), 1 μM of forward and reverse universal primers and template DNA on TechneTC-PLUS® thermalcycler (VWR International, LLC, Radnor, USA). Thermocycling conditions were the following: for 35 cycles (each cycle is 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 60 s), with an initial hot start (94 °C for 15 min) and a final extension (72 °C for 10 min). The purified PCR products were sequenced using AB Big-Dye 3.1® dye chemistry (Applied Biosystems). A total of 20 μL sequencing reactions contained 2 μL of cleaned PCR product, 1 μL of BigDye Terminator v3.1 Ready Reaction Mix, 2 μL of 5× Sequencing Buffer, 1.6 pmol of Forward or Reverse sequencing primer and PCR grade water. Sequencing reactions were performed for 25 cycles (each cycle is 96 °C for 30 s, 50 °C for 15 s, and 60 °C for 4 min in a GeneAmp PCR 9700® thermocycler (Applied Biosystems). After the cleanup step with the Performa® DTR Gel Filtration Cartridges (Edge Bio, Gaithersburg, MD, USA) the reaction was run on AB 3500 XL® automated DNA sequencers (Applied Biosystems).
PCR amplification and NGS sequencing
Samples were prepared according to the "16S Metagenomic Sequencing Library Preparation" guide (Part# 15044223 rev. A; Illumina, San Diego, CA, USA). The amplicon PCR has been performed using the following primers (containing overhang adapters): Ba27F 5'-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCCTGGCTCAG- 3', Ba338R 5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCTGCCTCCCGTAGGAGT- 3'[35]. Libraries have been quantified through PicoGreen ds- DNA quantitation assay (Thermo Fisher Scientific, Waltham, MA, USA) and validated on Bioanalyzer DNA 1000 chip (Agilent, Santa Clara, CA. USA). The sequencing was performed on MiSeq desktop sequencer (Illumina).
Analysis of sequence reads and IAQ evaluation by Microbial Biodiversity parameters
The sequence reads were analyzed in the cloud environment BaseSpace through the 16S Metagenomics app (version 1.0.1; Illumina®): the taxonomic database used is an Illumina-curated version of the May 2013, a release of the Greengenes Consortium Database (greengenes.secondgenome.com); the classification algorithm is an implementation of the Ribosomal Database Project (RDP) Classifier [36]. In order to acquire information on IAQ by Microbial Biodiversity (MB) a novel putative parameter (Indoor Air Microbial Biodiversity, IAMB) was assumed by composing the following indexes: Shannon mean index, Shannon's Equitability index and calculating and an Anthropogenic pollution index based on NGS reads. In particular, community richness and indoor air microbial biodiversity were computed through Shannon mean index (H) and Shannon's Equitability index (EH) using EstimateS software [37, 38]. Briefly, a cut-off corresponding to taxonomic species was applied to the NGS data, and the indices were calculated on the filtered matrix. Moreover, to verify the origin of air pollution we calculated the proportion of anthropogenic species SA compared to the total identified taxonomic units (Number of species of human source/total selected species). The combination of microbial load (H), species representativeness (EH), human/environmental pollution ratio (SA) was used to acquire information for IAMB.
STATISTICAL ANALYSES
Statistical analysis was performed by IBM SPSS statistical software, version 23.0 for Windows (SPSS, Chicago, IL). Continuous variables were reported as arithmetic mean, whereas categorical variables were reported as absolute values. A multiple linear regression equation was created for the outcome interesting of indoor mesophilic, psichrophilic bacteria and fungi concentration. All continuous variables were tested for a linear correlation with each other (Pearson r). Similarly, all categorical variables were tested for statistically significant relationships with other independent variables and choices made as for continuous variables.
Results
The comfort parameters were acceptable in all sampled sites, as shown in Table I: in Site I the value of PMV was 0.94 for the Reading room and 1.02 for the Books store. The PPD was similar for both environments with a percentage of the 25%. The CO2 concentration was 572.08 and 721.13 ppm respectively for Reading room and Books store. In Site II the PMV value was less than 1 for all environments. The maximum PPD value was detected in Flat B room 3 and in the Reading room (respectively of 18.47% and 13.58%); in the other environments was around the 5%. The CO2 concentration was around 550 ppm, with a lower value detected in the Reading room (445.97 ppm). In Site III, the PMV values were 0.35 and 0.88 respectively for the Reading room and the Manager office. The maximum PPD value was detected in the Manager office (21.37%), where the CO2 concentration was 614.21, whether 752.89 in Reading room.
Tab. I.
Evaluation of PMV, PPD, CO-CO2 at Sites I, Site II and Site III.
| Sampling site | PMV | PPD | CO2 | CO | |
|---|---|---|---|---|---|
| Site I | Reading room | 0.94 | 23.83 | 572.08 | 0 |
| Books storage | 1.02 | 27.35 | 721.13 | 0 | |
| Site II | Flat A room 1 | 0.03 | 5.06 | 560.87 | 0 |
| Flat A room 2 | 0.16 | 5.53 | 470.43 | 0 | |
| Flat B room 3 | 0.8 | 18.47 | 542.79 | 0 | |
| Flat B room 4 | 0.14 | 6.1 | 583.23 | 0 | |
| Flat C Reading room | 0.64 | 13.58 | 445.97 | 0 | |
| Books storage 1 | -0.52 | 11.01 | 442.03 | 0 | |
| Books storage 2 | -0.61 | 12.91 | 444.75 | 0 | |
| Site III | Reading room | 0.35 | 7.51 | 752.89 | 0 |
| Manager office | 0.88 | 21.37 | 614.21 | 0 |
SAS air sampling data were analyzed according to "American Conference of Governmental Industrial Hygienists" and reported in Table II [39]. The IGCM index (Global Index of Microbiological Contamination) was very low in Reading room and low in the Books storage of Site I. In Flat A room 2, Flat B room 3, Reading room and books storage of Site II was very low as in other environments. The values of Reading room of Site III and manager office were respectively intermediate and low. IAQ was estimated according to the "European Collaborative Action" [40], showing in the Reading room of Site I an intermediate level of bacterial pollution at 22°C (101-500 CFU/m3) and a low one at 37°C (80 CFU m3); in the Books storage, it was intermediate at both temperatures. The fungal pollution was lower for both environments. In Flat A room 1, Flat A room 2, Flat B room 3, Flat B room 4 of Site II bacterial pollution was intermediate at both temperatures, while in the Reading room it was low (50-100 CFU/m3) at 37°C and very low (< 50 CFU/m3) at 22°C; in the Books storage 1 and 2 it was intermediate at both temperatures. The fungal pollution was very low in Flat A room 1, Flat A room 2, Flat B room 3, Flat B room 4 sites; while it was low (< 100 CFU/m3) in the Books storage 1 and 2. In Site III a low bacterial pollution at 37°C was observed for both environments; at 22°C, the Reading room showed high bacterial pollution (500-2000 CFU/m3), while the Manager office an intermediate one (101-500 CFU/m3). The fungal pollution in the Reading room and in the Manager office was respectively 500-2000 CFU/m3 and 100-500 CFU/m3 (Supplementary Tables, S2-S3).
Tab. II.
Evaluation of IGCM, according to ACGIH [39], in Site I, Site II and Site III.
| Group ofmicrobes | Range of value (CFU/m3) | Pollution degree | Site I | Site II | Site III | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reading room | Books storage | Flat A room 1 | Flat A room 2 | Flat B room 3 | Flat B room 4 | Flat C Reading room | Books storage 1 | Books storage 2 | Reading room | Manager office | |||
| IGCM/m3 | < 500 | Very low | √ | √ | √ | √ | √ | ||||||
| < 1000 | Low | √ | √ | √ | √ | ||||||||
| > 1000 | Intermediate | √ | |||||||||||
| > 5000 | High | ||||||||||||
| > 10,000 | Very high | ||||||||||||
Table III shows the categorical variables for indoor mesophilic, psichrophilic and fungi microbial load used for statistical analysis: the buildings age, light exposure, air exchange rate and ventilation systems. Table IV shows the continuous variables with statistically significant relationship with indoor mesophilic, psichrophilic and fungi concentration. These variables have been correlated with the relative humidity, the external and internal temperature. Significant correlations were observed for mesophilic concentrations versus air exchange rate (+1; p < 0.05), psychrophilic concentrations and fungi concentration versus light exposure (+1; p < 0.05) and mean indoor relative humidity (respectively 0.831- p < 0.01; 0.848-p<0.05). Negative correlations were observed for fungi concentration versus air exchange rate (-1; p < 0.05) and ventilation system (-1; p < 0.01), air exchange rate versus psychrophilic concentrations (-1; p < 0.05), mesophilic concentrations versus light exposure (-1; p < 0.05) and mean indoor temperatures (-0.73; p < 0.01). Moreover, the table shows the results obtained for continuous variables with statistically significant relationship to indoor mesophilic, psichrophilic and fungi concentration in books storage. These variables have been correlated with the relative humidity, the external and internal temperature. Significant correlation was observed only for psichrophilic concentration compared to the three variables in all storage. In particular, mean indoor relative humidity showed a correlation coefficient of 0.999 (p < 0.01), while both mean indoor temperature and mean outdoor temperature showed a correlation coefficient of +1 (p < 0.05).
Tab. III.
Categorical variables for indoor mesophilic, psichrophilic and fungi concentration.
| Variable | N | Mesophilic bacteria (Mean CFU/m3) | Psichrophilic bacteria (Mean CFU/m3) | Fungi (mean CFU/m3) |
|---|---|---|---|---|
| Air exchange rate (a.c.h.) | ||||
| Yes | 4 | 160.8 | 397.5 | 219.5 |
| No | 7 | 198.6 | 255.7 | 88.3 |
| Building age (years) | ||||
| 11 | 1 | 71.5 | 748.0 | 559.0 |
| 22 | 1 | 208.5 | 250.0 | 44.5 |
| 82 | 1 | 210.4 | 197.7 | 41.3 |
| Ventilation system | ||||
| Natural | 2 | 235.5 | 695.0 | 417.0 |
| Artificial | 5 | 143.4 | 170.0 | 18.6 |
| None | 4 | 211.3 | 285.0 | 142.3 |
| Light exposure | ||||
| Yes | 6 | 138.7 | 360.3 | 196.5 |
| No | 5 | 240.2 | 243.6 | 63.4 |
Tab. IV.
Continuous variables with statistically significant relationship to indoor mesophilic, psichrophilic and fungi concentration.
| Mesophilic Bacteria (Mean CFU/m3) | Psichrophilic Bacteria (Mean CFU/m3) | Fungi (Mean CFU/m3) | ||
|---|---|---|---|---|
| Variable | Correlation Coefficent (Pearson r) | |||
| Others ambient | Building age | 0.629 | -0.685 | -0.624 |
| Air exchange rate | +1* | -1* | -1* | |
| Light exposure | -1* | +1* | +1* | |
| Ventilation system | -0.897 | -0.992 | -1** | |
| Mean indoor relative humidity | -0.424 | 0.831** | 0.848* | |
| Mean indoor temperature | -0.73** | -0.005 | 0.187 | |
| Mean outdoor temperature | -0.523 | 0.458 | 0.525 | |
| Books storage | Mean indoor relative humidity | 0.657 | 0.999** | -0.967 |
| Mean indoor temperature | 0.689 | +1* | -0.977 | |
| Mean outdoor temperature | 0.669 | +1** | -0.971 | |
Correlation is significant at the 0.05 level;
Correlation is significant at the 0.01 level.
Microbiological identification of environmental mycotic species was performed through DNA sequencing. The most of fungi isolates belonged to the Cladosporidium spp. and were found in all Sites. Two isolates belonged to Penicillium genus, in particular P. rivolii (Site I) and P. viticola (Site II). In Site I, one sample was identified as Cladosporidium uredinicola and one as Fusarium sp. Finally, in Site II only a sample belonged to Ascomycota genus (Supplementary Table, S4).
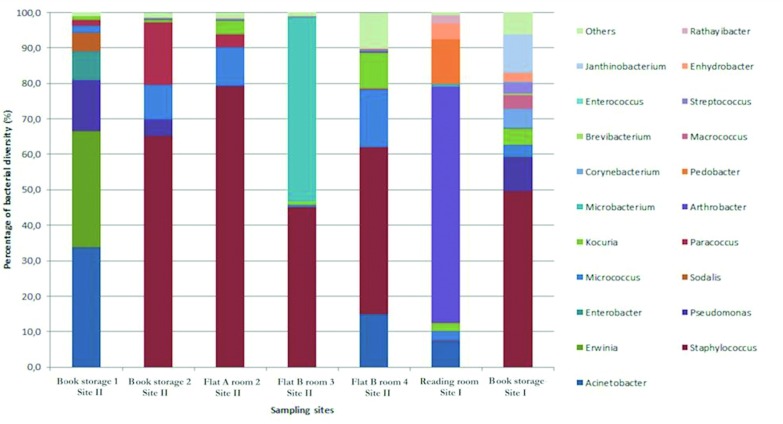
The relative abundance of bacterial diversity at the genus level was detected by NGS (Figure 2). A high abundance of Staphylococcus spp. was observed in most of the samples (65% in Books storage 2; 79% in Flat A Room 2; 45% in Flat B room 3; 47% Flat A Room 1 of Site II and 49% in Books storage of Site I). The most prevalent bacterial species identified among all the observed samples were Staphylococci, including S. aureus, S. epidermidis, and S. saprophyticus. Some other bacterial species were detected such as Acinetobacter (33% in Books storage 1 of Site II); Erwinia (32% in Books storage1 of Site II); Arthrobacter (66% in the Reading room of Site I), Microbacterium (52% in Flat B room 3 of Site II); Pseudomonas (14.4% and 4.7% respectively in Books storage 1 and 2 of Site II; 9.7% in Books storage of Site I); Janthinobacterium, Corynebacterium and Macrococcus (10.9%, 5.1% and 3.8% respectively in Books storage of Site I); Kocuria (3.8% and 10.3% in Flat A Room 2 and Room 1 of Site II respectively; 2.4% and 4.6% in Reading room and Books storage of Site I respectively); Sodalis (5.2% in Books storage1 of Site II); Pedobacter (12.5% in Reading room of Site I); Paracoccus (17.7%, 1.7% and 3.6% respectively in Books storage 1, 2 and Flat A Room 2 of Site II); Enterobacter (8.2% in Books storage1 of Site II). A total of 30 bacterial species were detected in all tissues sampled in this study.
Fig. 2.
Percentage of bacterial diversity at genus level of isolates sampled in Site I and II by NGS approach.
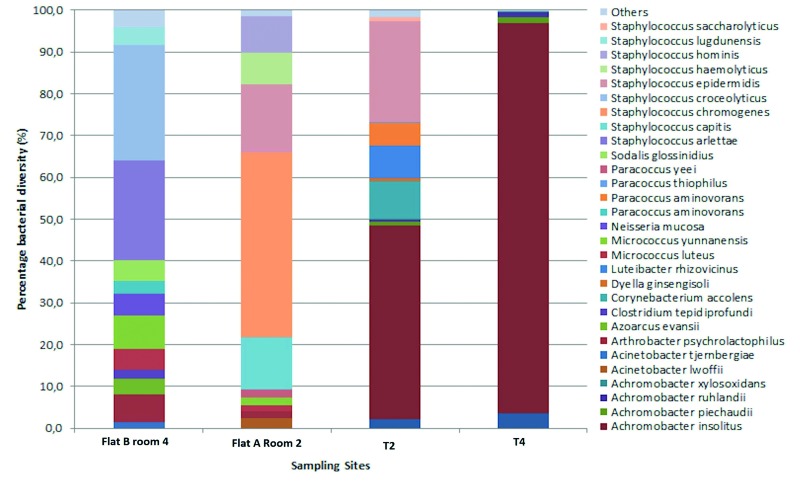
The predominant bacterial species in the analyzed surface samples were reported, considering their relative (cutoff > 1.0%) abundance (Figure 3). These species include Achromobacter insolitus (46.4% in T2 and 93.4% in T4) and Achromobacter arsenitoxydans (2.1% in T2 and 3.6% in T4). Only in T2 tissue the species detected were Corynebacterium accolens (9.1%), Luteibacter rhizovicinus (7.6%), Paracoccus aminovorans (5.6%) and Staphylococcus epidermidis (23.9%). Figure 3 also shows the comparison between the microflora which was found in tissues (T2 and T4), and bacterial isolates of the respective sampling areas (Flat A Room 2, Flat B room 4): T2 - Flat A Room 2 showed a matching, detecting Staphylococcus epidermidis in similar percentage (16.4% in Flat A Room 2; 23.9% in T2).
Fig. 3.
Representative distribution of bacterial diversity at species level of tissue samples and corresponding sampling points in Site II.
The bacterial community diversity was calculated on a total of 83 taxonomic units which fulfilled the cut off limit. The values of Shannon mean index (H) and Shannon's Equitability index (EH) are showed for each sample and compared with the control values for outdoor air and soil (Table V). In passive sampling (T1, T2, T3, T4), the H index was between 2.02 and 2.86 while the EH index was between 0.46 and 0.65. In active sampling (Site II: Flat B Room 3, Flat B Room 4, Flat A Room 2, Books storage1, Books storage2; Site I: Reading Room, Books store) the highest value of H index was 3.36 (Reading Room of Site I) and the lowest was 2.97 (Books storage 1 of Site II), same trend holds for EH index (0.67 and 0.76 respectively in Reading Room of Site I and Books storage1 of Site II). The distribution of anthropogenic niche (Table V) shows that passive sampling values were between 0 and 0.23, while the bacterial population in air samples as measured by active sampling provided a variety of human origin species between 0.62 and 0.89. For each indoor environment, the parallel consideration of the three indices provides information on the microflora biodiversity in the air (IAMB).
Tab. V.
Bacterial species distribution: results of Shannon diversity index richness (H), Evenness (EH) and Proportion of Anthropogenic species (SA) values.
| Samples | Shannon mean index (H) | Shannon's Equitability (EH) | Proportion of Anthropogenic species (SA) |
|---|---|---|---|
| T1 (Flat A Room 1 Site II) | 2.02 | 0.46 | 0.01 |
| T2 (Flat A Room 2 Site II) | 2.43 | 0.55 | 0.23 |
| T3 (Flat B Room 3 Site II) | 2.69 | 0.61 | 0.18 |
| T4 (Flat B Room 4 Site II) | 2.86 | 0.65 | 0.02 |
| Book storage 1 Site II | 2.97 | 0.67 | 0.62 |
| Book storage 2 Site II | 3.4 | 0.77 | 0.68 |
| Flat A Room 2 Site II | 3.28 | 0.74 | 0.82 |
| Flat B Room 3 Site II | 3.17 | 0.72 | 0.85 |
| Flat B Room 4 Site II | 3.2 | 0.72 | 0.89 |
| Book storage Site I | 3.34 | 0.76 | 0.73 |
| Reading Room Site I | 3.36 | 0.76 | 0.82 |
| Outdoor Air [44] | 4.38 | 0.98 | 0.05 |
| Soil [48] | 4.33 | 0.90 | 0.03 |
Discussion
Libraries represent one of those public places where monitoring IAQ is relevant not only for people's health, but also for the preservation of materials, in this case the paper of the books and their appropriate storage or restoration [12, 31]. Therefore, even if libraries do not represent settings at high occupational risk, they are exemplificative indoor environments to test monitoring strategies and improve air quality over the minimal requirements for occupational safety. Several studies regarding IAQ in libraries have been reported, focusing on the different categories of risk factors [12, 26, 29, 30, 42]. A monitoring program should include biological, physical and chemical parameters, following a "holistic method" to provide an extended vision of the phenomenon and an overall evaluation of IAQ.
In this study, we considered an integrated approach (Figure 1). Interestingly, both chemical and physical parameters are known to influence microflora composition, and, otherwise, each environmental microflora is typical of that ecological niche [24, 25, 42, 43]. Biodiversity analysis showed the presence of paper-related bacteria and allowed to define extent and diffusion of airborne microorganisms.
The collected data defined the safety of libraries air, the degree of comfort and wellness for users and workers. IAQ data comply with standards for non-industrial premises. The PPM and PMV indicators were acceptable for most sites, with some slight discrepancies related with external climatic conditions [30]. No main differences in PPM and PMV were observed for the different libraries, although these buildings have very different structures, use, age, light exposure suggesting a major role for the air conditioning/heating system.
Collected microclimatic and microbiological data suggested the presence of disadvantageous conditions for psychrophilic bacteria and fungi proliferation. Higher air exchange/ventilation rates correspond to a decrease in microbial load. Slight variations involved the psychrophilic count in Reading room of Site III that can be related to interference of ventilation and intense turnout of users, in accordance with previous reports [11, 41]. In storage room of Site II, the psychrophilic bacterial counts and fungal pollution are significantly higher than in the reading room, although the microclimate was acceptable even if visible molds were observable on walls, suggesting external factors or discontinuous indoor management. Overall mesophilic and psychrophilic growth was in line with data surveyed in other rooms within the same library, suggesting that similar disposition and typology of books, such as age, materials and storage conditions, may play a role. This observation agrees with statistical analysis showing a significant correlation with the relative humidity, external and internal temperature (Tab. IV).
Sequencing analysis by NGS allowed a comprehensive evaluation of microflora biodiversity, and the indoor distribution of microorganisms. The relative abundance of different bacteria in the indoor atmosphere is of high interest to evaluate IAQ. Airborne bacterial fluxes, indeed, represent an important way for microorganisms to colonize remote environments. In this context, the identification of levels and type of microbial pollution and analysis of its variations in space and time can contribute to implement IAQ assessment and dynamics[44]. A total of 110 ± 10 species were detected by NGS for each sampled point. To analyze this microflora complexity bioinformatic tools and several indices can be used [45-47]. The Shannon diversity (H) and Shannon equitability (EH) indices provide characterization of biodiversity, overcoming microbial richness. To evaluate microflora variations in indoor environments, we proposed the combined assessment of microbial load (H) and species representativeness (EH) parameters, adding a human/environmental pollution ratio (SA) to better characterize the NGS output and acquire synthetic information on Indoor Air Microbial Biodiversity (IAMB). As expected, the indices values were less rich in biodiversity than those reported for soil [48] or heavily polluted outdoor air [44] and the proportion of anthropogenic species SA compared to the total identified taxonomic units was higher (Tab. V). Passive sampling allowed acquisition of data for longer periods (1 month), showing a comparable trend for H and EH values but lower SA values (0.001-0.23) while the bacterial population collected by active techniques (60 seconds) showed a larger variety of human origin species, with SA values between 0.62 and 0.89. This can be explained considering the different sampling times, the drying effect of the tissue, the overall day/night exposure periods without visitors and the selective advantage of environmental resistant species respect to the survival capabilities of human anthropogenic mesophilic. Staphylococci were identified in the environments by 1 minute active sampling during working hours, but not in the corresponding 30 days passive sampling, except for Staphylococcus epidermidis that was identified in one site (T2 - Flat Room 2). In conclusion, there was a mismatch between the species detected by passive versus active sampling, therefore these microorganisms might be deposited, but were not cultivable on plate. However, in studying environmental microflora, plate culture may represent a kind of bias, because species are selected on the growth property we have available. The adoption of sampling and analysis systems strictly based on molecular methods without culture steps would bypass this limit, as shown by DNA analysis and NGS data.
In air samples analyzed by active techniques, the numerous sequences corresponding to anthropic bacteria (e.g. Micrococci, Paracocci, Staphylococci) suggested contaminations by people [49]. Besides, all the sampled libraries shared a similar microflora, including Micrococci ranging from 0.7% (Flat B room 3) to 10.9% (Flat A room 2) and Kocuria with a maximum of 10% in Flat A room 2. Amongst identified genera some as Corynebacterium, Macrococcus and Janthinobacterium were already reported as predominant flora in indoor air [50]. Furthermore, the presence of symbiotic (as Kocuria spp.) or phytopathogenic bacteria (as Erwinia and Sodalis) in the community illustrated the potential role of air in the dissemination of organisms interacting in different biological processes. The book storehouse in Site II showed a different scenario: although similar in terms of microclimate, fungal and psychrophilic count, a different mesophilic growth was observed, impacting on microflora biodiversity. Staphylococcus and Pseudomonas genera prevailed in the most accessible area (books storage 2), while Acinetobacter, Erwinia, Enterobacter and Sodalis were present only in the farthest part dedicated to long term storage (books storage 1). These environments selected a different microflora. Bacteria present in books storage 1, less adjacent to the external environment, are typical of paper and include species described papermaking industries [50]. Similarly, the presence of Pseudomonas spp. in the storehouse of Site I, can be related to the capability to grow on different carbon sources including cellulose [51-54]. Fungi presence in libraries and archives has been largely investigated, mainly in correlation with paper deterioration and possible adverse effects on workers due to the release of secondary metabolites [3, 32, 41]. Opportunistic pathogens were sporadically detected, as Fusarium genus that produces mycotoxins and secondary metabolites which can trigger allergic reactions. Most of the isolates collected in our study belong to the genera Penicillium and Cladosporidium, in line with previous studies [55, 56].
The proposed model and the microflora data by IAMB can provide a comprehensive evaluation of IAQ, suggesting whether a particular environment may fall within one of the different risk categories, including: i) the site is clean or poses acceptable risk and no further action is mandatory; ii) the site poses borderline risk levels and a further monitoring or follow up may be required; iii) the site is highly contaminated and remedial action is needed. Evaluation of the potential impact of the microflora biodiversity on IAQ depends on the specific environment, activities and procedures carried out in that context. Several guidelines and strategies are already available for air risk assessment and can be improved by the contribute of new molecular biology technologies.
Conclusions
A comprehensive evaluation of Indoor Air Quality requires a multidisciplinary approach and the identification of critical points. The analysis of microflora by Indoor Air Microbial Biodiversity is revealed as a valuable tool to assess IAQ and plan further corrective actions, based on reconstruction of contamination sources and routes. We can suggest that microflora biodiversity itself may represent an informative and valuable marker for IAQ, also when assayed by NGS. This NGS integrated approach may represent a promising tool for surveillance of IAQ in libraries and other occupational environments.
ACKNOWLEDGMENTS
Project was supported by grants P342013/15/INAIL and CDR2.RIC102013/IUSM. Authors are grateful to Manuela Camerino, Gaetana Cognetti, Rosalia Ferrara, Saverio Giampaoli, Franco Lufrani, Eugenio Sorrentino and Franco Toni, for participating in the preliminary discussion, sampling and in providing critical issues on safety and quality of libraries; Dr. Elena Scaramucci and Dr. Lucrezia Petruccioli for the editing.
Supplementary tables
S1.
Reference values of thermal comfort according to ISO 7730, 7726, 27243, 7933, 11079, 8996.
| PMV | PPD% | Evaluation of thermal comfort |
|---|---|---|
| 3 | 100 | Very hot |
| 2 | 75.7 | Hot |
| 1 | 26.4 | Slightly hot |
| 0.85 | 20 | Thermal environment within acceptable |
| -0,5 < PMV < 0,5 | < 10 | Thermal comfort |
| -0.85 | 20 | Thermal environment within acceptable |
| -1 | 26.8 | Slightly cold |
| -2 | 76.4 | Cold |
| -3 | 100 | Very cold |
S2.
Evaluation of air quality in Site I, Site II according to the sanitary standards for non-industrial premises [40].
| Group of microbes | Range of value (CFU/m3) | Pollution degree | Site I | Site II | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reading room | Books storage | Flat A room 1 | Flat A room 2 | Flat B room 3 | Flat B room 4 | Flat C Reading room | Books storage 1 | Books storage 2 | |||||||||||||
| 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | 37°C | 22°C | ||||
| Bacteria | < 50 | Very low | √ | ||||||||||||||||||
| 50-100 | Low | √ | √ | ||||||||||||||||||
| 101-500 | Intermediate | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | |||||
| 500 -2000 | High | ||||||||||||||||||||
| > 2000 | Very high | ||||||||||||||||||||
| Fungi | < 25 | Very low | √ | √ | √ | √ | √ | √ | √ | ||||||||||||
| < 100 | Low | √ | √ | ||||||||||||||||||
| < 500 | Intermediate | ||||||||||||||||||||
| < 2000 | High | ||||||||||||||||||||
| > 2000 | Very high | ||||||||||||||||||||
S3.
Evaluation of air quality in Site III according to the sanitary standards for non-industrial premises.
| Group of microbes | Range of value (CFU/m3) | Pollution degree | Site III | |||
|---|---|---|---|---|---|---|
| Reading room | Manager office | |||||
| 37°C | 22°C | 37°C | 22°C | |||
| Bacteria | <50 | Very low | ||||
| 50-100 | Low | √ | √ | |||
| 101-500 | Intermediate | √ | ||||
| 500-2000 | High | √ | ||||
| >2000 | Very high | |||||
| Fungi | < 25 | Very low | ||||
| <100 | Low | |||||
| < 500 | Intermediate | √ | ||||
| < 2000 | High | √ | ||||
| > 2000 | Very high | |||||
S4.
Type of fungi isolated from each site.
| Fungi isolate | Site I | Site II | Site III |
|---|---|---|---|
| Penicillium rivolii | + | ||
| Penicillium viticola | + | ||
| Cladosporidium sp. | + | + + | |
| Cladosporidium uredinicola | + | ||
| Fusarium sp. | + | ||
| Ascomycota genus | + |
References
- 1.Caselli M, Gennaro G, Saracino MR, Tutino M. Indoor contaminants from newspapers: VOC emission in newspaper stands. Environ Res. 2009;109:149–157. doi: 10.1016/j.envres.2008.10.011. doi:10.1016/j.envres. 2008.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Haleem Khan AA, Mohan Karuppayil S. Fungal pollution of indoor environments and its management. Saudi J Biol Sci. 2012;19:405–426. doi: 10.1016/j.sjbs.2012.06.002. doi: http://dx.doi.org/10.1016/j. sjbs.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalwasińska A, Burkowska A, Wilk I. Microbial air contamination in indoor environment of a university library. Ann Agric Environ Med. 2012;19:25–29. [PubMed] [Google Scholar]
- 4.Pegas PN, Alves CA, Evtyugina MG, Nunes T, Cerqueira M, Franchi M, Pio CA, Almeida SM, Freitas MC. Indoor air quality in elementary schools of Lisbon in spring. Environ Geochem Health. 2011;33:455–468. doi: 10.1007/s10653-010-9345-3. doi: 10.1007/s10653-010-9345-3. [DOI] [PubMed] [Google Scholar]
- 5.Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, Northcutt D, O'Connor TK, Womack AM, Brown GZ, et al. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air. 2014;24:41–48. doi: 10.1111/ina.12047. doi:10.1111/ina.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hospodsky D, Qian J, Nazaroff WW, Yamamoto N, Bibby K, Rismani-Yazdi H, Peccia J. Human occupancy as a source of indoor airborne bacteria. PLoS ONE. 2012;7:34867–34867. doi: 10.1371/journal.pone.0034867. doi:10.1371/ journal.pone.0034867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian J, Hospodsky D, Yamamoto N, Nazaroff WW, Peccia J. Size-resolved emission rates of airborne bacteria and fungi in an occupied classroom. Indoor Air. 2012;22:339–351. doi: 10.1111/j.1600-0668.2012.00769.x. doi:10.1111/j.1600-0668.2012.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. WHO. World Health Organization. 2009. Guidelines for indoor air quality: dampness and mould. Copenhagen, Denmark: World Health Organization; [Accessed on 24th November, 2013]. Available from: http://www. euro.who.int/__data/assets/pdf_file/0017/43325/E92645.pdf.
- 9.Graudenz GS, Oliveira CH, Tribess A, Mendes C, Jr, Latorre MR, Kalil J. Association of air conditioning with respiratory symptoms in office workers in tropical climate. Indoor Air. 2005;15:62–66. doi: 10.1111/j.1600-0668.2004.00324.x. doi: 10.1111/j.1600-0668.2004.00324.x. [DOI] [PubMed] [Google Scholar]
- 10.Rella R, Sturaro A, Vianello A. Incorrect installation and use of materials as the cause of a severe air pollution incident in a school building. Sci. Total Environ. 2014;487:255–259. doi: 10.1016/j.scitotenv.2014.04.031. doi:http:// dx.doi.org/10.1016/j.scitotenv.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Wamedo SA, Ede PN, Chuku A. Interaction between building design and indoor airborne microbial load in Nigeria. Asian J Biol Sci. 2012;5:183–191. doi: 10.3923/ajbs.2012.183.191. [Google Scholar]
- 12.Righi E, Aggazzotti G, Fantuzzi G, Ciccarese V, Predieri G. Air quality and well-being perception in subjects attending university libraries in Modena (Italy) Sci. Total Environ. 2002;286:41–50. doi: 10.1016/s0048-9697(01)00960-3. doi:10.1016/S0048-9697(01)00960-3. [DOI] [PubMed] [Google Scholar]
- 13.Brandt-Rauf PW, Andrews LR, Schwarz-Miller J. Sickhospital syndrome. J Occup Med. 1991;33:737–739. doi: 10.1097/00043764-199106000-00017. doi: 10.1097/00043764-199106000-00017. [DOI] [PubMed] [Google Scholar]
- 14.Burge PS. Sick building syndrome. Occup Environ Med. 2004;61:185–190. doi: 10.1136/oem.2003.008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauni R, Verbeek JH, Uitti J, Jauhiainen M, Kreiss K, Sigsgaard T. Remediating buildings damaged by dampness and mould for preventing or reducing respiratory tract symptoms, infections and asthma. Cochrane Database Syst Rev. 2015;25:2–2. doi: 10.1002/14651858.CD007897.pub3. doi: 10.1002/14651858.CD007897.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbritti G. Indoor environments, work and health. G Ital Med Lav Ergon. 2004;26:346–352. [PubMed] [Google Scholar]
- 17.Jantunen M, Oliveira Fernandes E, Carrer P, Kephalopoulos S. Promoting actions for healthy indoor air (IAIAQ) Luxembourg: European Commission Directorate General for Health and Consumers; 2011. [Google Scholar]
- 18.Kelley ST, Gilbert JA. Studying the microbiology of the indoor environment. Genome Biol. 2013;14:202–202. doi: 10.1186/gb-2013-14-2-202. doi:10.1186/gb-2013- 14-2-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanker A. Genome research in the cloud. OMICS. 2012;16:422–428. doi: 10.1089/omi.2012.0001. doi: 10.1089/omi.2012.0001. [DOI] [PubMed] [Google Scholar]
- 20.Giampaoli S, Berti A, Valeriani F, Gianfranceschi G, Piccolella A, Buggiotti L, Rapone C, Valentini A, Ripani L, Romano Spica V. Molecular identification of vaginal fluid by microbial signature. Forensic Sci Int Genet. 2012;6:559–564. doi: 10.1016/j.fsigen.2012.01.005. doi: 10.1016/j. fsigen.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Giampaoli S, Chillemi G, Valeriani F, Lazzaro D, Borro M, Gentile G, Simmaco M, Zanni G, Berti A, Romano Spica V. The SNPs in the human genetic blueprint era. N Biotechnol. 2013;30:475–484. doi: 10.1016/j.nbt.2012.11.015. doi: 10.1016/j.nbt.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Mansi A, Amori I, Marchesi I, Marcelloni AM, Proietto AR, Ferranti G, Magini V, Valeriani F, Borella P. Legionella spp. survival after different disinfection procedures: comparison between conventional culture, qPCR and EMA-qPCR. Microchem J. 2014;112:65–69. doi:http://dx.doi.org/10.1016/j.microc. 2013.09.017. [Google Scholar]
- 23.Valeriani F, Protano C, Gianfranceschi G, Cozza P, Campanella V, Liguori G, Vitali M, Divizia M, RomanoSpica V. Infection control in healthcare settings: perspectives for mfDNA analysis in monitoring sanitation procedures. BMC Infect Dis. 2016;16:394–394. doi: 10.1186/s12879-016-1714-9. doi: 10.1186/s12879-016-1714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valeriani F, Giampaoli S, Buggiotti L, Gianfranceschi G, Romano Spica V. Molecular enrichment for detection of S. aureus in recreational waters. Water Sci Technol. 2012;66:2305–2310. doi: 10.2166/wst.2012.435. doi:10.2166/wst.2012.435. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad I, Ahmad F, Pichtel J. Microbes and Microbial Technology: agricultural and environmental applications. New York: Ed. Springer; 2011. [Google Scholar]
- 26.Fenech A, Strlič M, Kralj Cigić I, Levart A, Gibson LT, Bruin G, Ntanos K. Volatile aldehydes in libraries and archives. Atmos Environ. 2010;44:2067–2073. doi:http://dx.doi.org/10.1016/ S0048-9697(96)05335-1. [Google Scholar]
- 27.Pasquarella C, Saccani E, Sansebastiano GE, Ugolotti M, Pasquariello G, Albertini R. Proposal for a biological environmental monitoring approach to be used in libraries and archives. Ann Agric Environ Med. 2012;19:209–212. [PubMed] [Google Scholar]
- 28.Fantuzzi G, Aggazzotti G, Righi E, Cavazzuti L, Predieri G, Francaschelli A. Indoor air quality in the university libraries of Modena, Italy. Sci Total Environ. 1996;193:46–56. doi: 10.1016/s0048-9697(96)05335-1. [DOI] [PubMed] [Google Scholar]
- 29.Lee CM, Kim YS, Nagajyoti PC, Park W, Kim KY. Pattern classification of volatile organic compounds in various indoor environments. Water Air Soil Pollut. 2011;215:329–338. doi: 10.1111/ j.1600-0668.1997.t01-2-00007.x. [Google Scholar]
- 30.Adam G, Pont U, Mahdavi A. Evaluation of thermal environments and indoor air quality in university libraries in Vienna. Adv Mat Res. 2014;899:315–320. doi:10.4028/www.scientific. net/AMR.899.315. [Google Scholar]
- 31.Borrego S, Guiamet P, Gomez de Saravia S, Batistini P, Garcia M, Lavin P, Perdomo I. The quality of air at archives and the biodeterioration of photographs. Int. Biodeterior. Biodegradation. 2010;64:139–145. doi: 10.1016/j.ibiod.2009.12.00. [Google Scholar]
- 32.Zielińska-Jankiewicz K, Kozajda A, Piotrowska M, Szadkowska-Stańczyk I. Microbiological contamination with molds in work environment in libraries and archive storage facilities. Ann Agric Environ Med. 2008;15:71–78. [PubMed] [Google Scholar]
- 33.Dacarro C, Picco AM, Grisoli P, Rodolfi M. Determination of aerial microbiological contamination in scholastic sports environments. J. Appl. Microbiol. 2003;95:904–912. doi: 10.1046/j.1365-2672.2003.02044.x. doi: 10.1046/j.1365-2672.2003.02044.x. [DOI] [PubMed] [Google Scholar]
- 34.White TJ, Bruns T, Lee S, Taylor J. PCR Protocols, a Guide to Methods and Applications: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 35.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One. 2013;8:47879–47879. doi: 10.1371/journal.pone.0047879. doi:http://dx.doi.org/10.1371/journal. pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;3:5261–5267. doi: 10.1128/AEM.00062-07. doi:10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colwell RK, Chao A, Gotelli NJ, Lin SY, Mao CX, Chazdon RL, Longino JT. Models and estimators linking individualbased and sample-based rarefaction, extrapolation, and comparison of assemblages. Journal of Plant Ecology. 2012;5:3–21. doi: https://doi.org/10.1093/jpe/rtr044. [Google Scholar]
- 38.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, et al. Introducing mothur: Opensource, platform-independent, communitysupported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;5:7537–7541. doi: 10.1128/AEM.01541-09. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. ACGIH. American Conference of Governmental Industrial Hygienists. Industrial Ventilation: A Manual of Recommended Practice. Cincinnati, OH; 1995.
- 40. CEC. Commission of the European Communities (EU). Indoor air quality and its impact on man. Report No. 12. Biological particles in indoor environments. Luxembourg; 1993.
- 41.Hayleeyesus SF, Manaye AM. Microbiological quality of indoor air university libraries. Asian Pac J Trop Biomed. 2014;1:312–317. doi: 10.12980/APJTB.4.2014C807. doi:10.12980/APJTB.4.2014C807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandi G, Sisti M, Paparini A, Gianfranceschi G, Schiavano GF, Santi M, Santoni D, Magini V, Romano-Spica V. Swimming pools and fungi: an environmental epidemiology survey in Italian indoor swimming facilities. Int J Environ Health Res. 2007;17:197–206. doi: 10.1080/09603120701254862. doi:10.1080/09603120701254862. [DOI] [PubMed] [Google Scholar]
- 43.Giampaoli S, Berti A, Di Maggio RM, Pilli E, Valentini A, Valeriani F, Gianfranceschi G, Barni F, Ripani L, Romano Spica V. The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci Int. 2014;240:41–47. doi: 10.1016/j.forsciint.2014.02.028. doi:http://dx.doi.org/10.1016/j.forsciint.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 44.Maron PA, Lejon DPH, Carvalho E, Bizet K, Lemanceau P, Ranjard L, Mougel C. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmos Environ. 2005;39:3687–3695. doi:http://dx.doi.org/10.1016/j.atmosenv.2005.03.002. [Google Scholar]
- 45.Begon M, Harper JL, Townsend CR. Ecology: from individuals to ecosystems. Oxford: Ed Blackwell Scientific Publications; 1986. [Google Scholar]
- 46.Gut IG. New sequencing technologies. Clin Transl Oncol. 2013;15:879–881. doi: 10.1007/s12094-013-1073-6. doi: 10.1007/s12094-013-1073-6. [DOI] [PubMed] [Google Scholar]
- 47.Jost L. The Relation between Evenness and Diversity. Diversity. 2010;2:207–232. doi:10.3390/d2020207. [Google Scholar]
- 48.Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 49.Rensburg JJ, Lin H, Gao X, Toh E, Fortney KR, Ellinger S, Zwickl B, Janowicz DM, Katz BP, Nelson DE, et al. The Human Skin Microbiome Associates with the Outcome of and Is Influenced by Bacterial Infection. MBio. 2015;6:01315–01315. doi: 10.1128/mBio.01315-15. doi:10.1128/mBio.01315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bajpai P. Pulp and Paper Industry: Microbiological Issues in Papermaking. 2015. Ed Elsevier. [Google Scholar]
- 51.Li TC, Ambu S, Mohandas K, Wah MJ, Sulaiman LH, Murgaiyah M. Bacterial constituents of indoor air in a high throughput building in the tropics. Trop Biomed. 2014;31:540–556. [PubMed] [Google Scholar]
- 52.Johansen C, Falholt P, Gram L. Enzymatic removal and disinfection of bacterial biofilms. Appl Environ Microbiol. 1997;63:3724–3728. doi: 10.1128/aem.63.9.3724-3728.1997. doi: 0099-2240/97/$04.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lessie TG, Phibbs PV., Jr. Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 54.Nandalal P, Somashekar RK. Prevalence of Staphylococcus aureus and Pseudomonas aeruginosa in indoor air flora of a district hospital, Mandya, Karnataka. J Environ Biol. 2007;28:197–200. [PubMed] [Google Scholar]
- 55.Cetinkaya Z, Fidan F, Unlu M, Hasenekoglu I, Tetik L, Demirel R. Assessment of indoor air fungi in Western-Anatolia, Turkey. Asian Pac J Allergy Immunol. 2005;23:87–92. [PubMed] [Google Scholar]
- 56.Lonc E, Kinga P, Kiewra D, Szczepańska A, Firling CE. Quantitative assessment of mycological air pollution in selected rooms of residential and dormitory housing facilities. Ann Parasitol. 2013;59:183–217. [PubMed] [Google Scholar]