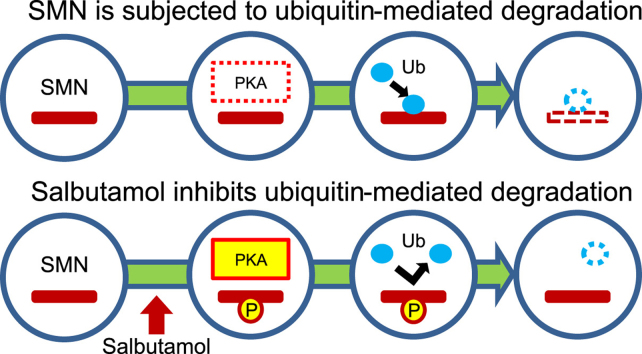
Abstract
Spinal muscular atrophy (SMA) is a common autosomal recessive neuromuscular disorder that is currently incurable. SMA is caused by decreased levels of the survival motor neuron protein (SMN), as a result of loss or mutation of SMN1. Although the SMN1 homolog SMN2 also produces some SMN protein, it does not fully compensate for the loss or dysfunction of SMN1. Salbutamol, a β2-adrenergic receptor agonist and well-known bronchodilator used in asthma patients, has recently been shown to ameliorate symptoms in SMA patients. However, the precise mechanism of salbutamol action is unclear. We treated SMA fibroblast cells lacking SMN1 and HeLa cells with salbutamol and analyzed SMN2 mRNA and SMN protein levels in SMA fibroblasts, and changes in SMN protein ubiquitination in HeLa cells. Salbutamol increased SMN protein levels in a dose-dependent manner in SMA fibroblast cells lacking SMN1, though no significant changes in SMN2 mRNA levels were observed. Notably, the salbutamol-induced increase in SMN was blocked by a protein kinase A (PKA) inhibitor and deubiquitinase inhibitor, respectively. Co-immunoprecipitation assay using HeLa cells showed that ubiquitinated SMN levels decreased in the presence of salbutamol, suggesting that salbutamol inhibited ubiquitination. The results of this study suggest that salbutamol may increase SMN protein levels in SMA by inhibiting ubiquitin-mediated SMN degradation via activating β2-adrenergic receptor-PKA pathways.
Keywords: Spinal muscular atrophy, Survival motor neuron gene, Salbutamol, Survival motor neuron protein, Ubiquitination
Graphical abstract
Highlights
-
•
Salbutamol increased SMN level in SMA fibroblast cells.
-
•
The increase of SMN is related to β2-adrenergic receptor-PKA pathways.
-
•
Salbutamol inhibits SMN ubiquitination in HeLa cells.
-
•
Salbutamol prevents ubiquitin-mediated SMN degradation via activated PKA pathways.
1. Introduction
Spinal muscular atrophy (SMA) is a common autosomal recessive neuromuscular disorder characterized by degeneration of α-motor neurons in the spinal cord, leading to muscular atrophy of the limbs and trunk. Unfortunately, SMA remains incurable. SMA is classified into three groups, depending on the age of onset and the achievement of motor milestones: SMA type I (Werdnig–Hoffman disease; severe form), SMA type II (intermediate form) and SMA type III (Kugelberg–Welander disease; mild form) [1].
The survival motor neuron genes SMN1 (telomeric SMN) and SMN2 (centromeric SMN) have been identified as candidate SMA-causing genes at chromosome 5q11.2–13.3 [2]. SMN1 and SMN2 are largely identical, but nucleotide +6 in the coding region is a C in SMN1 and a T in SMN2, though these are synonymous and do not result in an amino acid change. However, SMN1, but not SMN2, is now recognized as an SMA-causing gene, because SMN1 is completely deleted in more than 90% of SMA patients [3] and deleteriously mutated in the remaining patients [4].
Although the SMN1 homolog SMN2 encodes the same protein, it does not fully compensate for the loss or dysfunction of SMN1. The C-to-T transition alters the splicing pattern in SMN2 exon 7 [5], [6]. SMN1 exclusively produces full-length (FL) SMN1 transcripts, while SMN2 produces about 90% of exon 7-lacking (∆7) SMN2 transcripts and only about 10% of FL-SMN2 transcript [7]. The low levels of functional SMN protein produced by the FL-SMN2 transcript cannot fully compensate for the lack of functional SMN1-generated SMN protein. However, SMN2 is never lacking in SMA patients [2] and its copy number correlates inversely with disease severity; a higher SMN2 copy number may ameliorate the clinical phenotype [8]. SMN2 may thus partly compensate for the loss of SMN1 by modifying disease severity through the production of functional SMN protein.
Current treatment strategies for SMA have focused on enhanced production of SMN from SMN2, including by activation of SMN2 transcription and by modulation of exon 7 skipping [9]. Valproic acid, a histone deacetylase inhibitor, is the representative drug for the first of these strategies [10], [11], while ISIS-SMNRx, an antisense oligonucleotide masking a splicing suppressor in the SMN2 intron 7 [12], is used for the second strategy.
However, the β2-adrenergic receptor agonist, salbutamol, which is widely used as a bronchodilator in asthma patients, has recently been reported to alleviate symptoms in SMA patients [13], [14], [15], and Angelozzi et al. reported that salbutamol increased SMN protein levels in SMA fibroblast cells [16]. However, the mechanism whereby salbutamol increases SMN protein levels remains unclear. Angelozzi et al. suggested that salbutamol may increase SMN levels by increasing SMN2 transcripts [16]. Tiziano et al. also reported a significant and constant increase in FL-SMN2 transcripts in peripheral blood cells from SMA patients treated with salbutamol [17]. However, Burnett et al. found that the SMN complex was stabilized by activation of cyclic AMP (cAMP) and protein kinase A (PKA), with consequent increases in SMN protein levels [18], suggesting that salbutamol enhances SMN protein levels through other pathways.
To clarify the mechanism responsible for the salbutamol-induced increase in SMN protein levels, we treated SMA fibroblast cells lacking SMN1 and HeLa cells with salbutamol. We analyzed SMN2 transcript and SMN protein levels in SMA fibroblast cells, and assessed the effects of salbutamol on SMN protein ubiquitination in HeLa cells.
2. Materials and methods
2.1. Establishment of fibroblast cells and cell cultures
Skin tissue for primary culture of fibroblast cells was obtained from a 13-year-old girl with SMA type 3, who had complete absence of SMN1 but carried three copies of SMN2. This study was approved by the ethical committee of Kobe University, and informed consent was obtained from the patient and her parents.
Alteration of SMN2 transcript and SMN protein levels were investigated in SMA fibroblast cells. The fibroblast cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-low glucose (Sigma-Aldrich, St. Louis, MO, USA) containing 10,000 U/ml penicillin, 10 mg/ml streptomycin, 25 µg/ml amphotericin B (Fungizone; Life Technologies, Carlsbad, CA, USA), and 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich) in a 5% humidified CO2 atmosphere at 37 °C. Fibroblast cells were analyzed at 5–15 passages.
Ubiquitinated SMN was investigated in HeLa cells carrying two copies of SMN1 and two copies of SMN2. HeLa cells were used for co-immunoprecipitation (co-IP) experiments because of their ability to produce more SMN protein than fibroblast cells. HeLa cells were maintained in DMEM–high glucose (Sigma-Aldrich) containing 10,000 U/ml penicillin, 10,000 µg/ml streptomycin, 25 µg/mL amphotericin B (Life Technologies) and 8% heat-inactivated FBS in a 5% CO2 atmosphere at 37 °C.
2.2. Salbutamol treatment of fibroblast cells
Salbutamol sulfate was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Salbutamol solution 1 mM was freshly prepared in phosphate-buffered saline (PBS) before each use. To clarify if SMN2 transcript or SMN protein was induced by salbutamol, SMA fibroblast cells were incubated with final concentrations of salbutamol sulfate of 0.005, 0.05, and 0.5 µM, respectively. SMN2 transcript levels were measured at 0 m (pretreatment), 5 m, 15 m, 30 m, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h and 36 h. SMN protein levels were measured at 0 h (pretreatment), 1 h, 6 h, 8 h, 12 h, 24 h and 36 h.
The mechanism whereby salbutamol increased SMN protein levels was further investigated by adding PKA inhibitor 14–22 amide cell-permeable, myristoylated (Calbiochem; Darmstadt, Germany) (final concentration; 1 µM) and DUB inhibitor PR-619 (LifeSensors, Malvern, PA, USA) (final concentration; 10 µM), respectively, to SMA fibroblast cells with salbutamol (final concentration; 0.5 µM), and incubating for 36 h.
2.3. Reverse-transcription PCR analysis of salbutamol-treated fibroblast cells
Total RNA extraction and cDNA synthesis were conducted as described previously [19]. To evaluate transcript levels of the SMN genes, we amplified FL- and Δ7-SMN transcripts from cDNA of SMA fibroblast cells. Real-time quantitative reverse-transcription PCR was performed using a LightCycler 1.5 (Roche Diagnostics GmbH, Mannheim, Germany). cDNA amplification was carried out using FastStart DNA Master SYBR Green I (Roche Diagnostics GmbH).
The cDNA fragments containing exons 7 and 8 and exons 5 and 6/8 (flanking sequences of the exon boundary) represented FL- and Δ7-SMN, respectively. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous reference gene and the relative denominator for the SMN2 transcript. The amount of SMN2 transcript (relative to GAPDH) at 0 min (pretreatment time) was normalized to 1.0 in each dose group. The primer sequences and PCR conditions are listed in Table 1 (Supplementary material).
2.4. Western blotting analysis of salbutamol-treated fibroblast cells
Protein samples were prepared as described previously [19]. The protein samples were homogenized and subjected to western blotting. Briefly, the homogenized protein samples were electrophoresed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Cleveland, OH, USA) using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad, CA, USA). Immunostaining of the membranes was performed using the iBind Western System (Life Technologies). The following combinations of antibodies were used to detect SMN and tubulin: mouse anti-SMN antibody (BD Transduction Laboratories, Franklin Lakes, NJ, USA), ECL horseradish peroxidase (HP)-linked sheep anti-mouse IgG (GE Healthcare), rabbit polyclonal anti-beta tubulin antibody (Abcam, Cambridge, MA, USA), and ECL HP-linked donkey anti-rabbit IgG (GE Healthcare).
Chemiluminescent signals produced using Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare) were detected using a luminescent image analyzer LAS-3000 mini (Fujifilm, Tokyo, Japan). The signal intensity of the membrane was determined using Multi Gauge Version 3.0 software (Fujifilm). Western blotting experiments and signal-intensity analysis were repeated at least three times. Tubulin was used as an endogenous reference protein and as the relative denominator for the SMN protein. SMN protein levels (relative to tubulin) at 0 h (pretreatment time) in each dose group were normalized to 1.0.
2.5. Co-IP experiments of SMN complex in HeLa cells
We assessed the effect of salbutamol on SMN ubiquitination by co-IP experiments using HeLa cells. Our preliminary experiments showed that SMN expression level of HeLa cells was greater than that of human fibroblasts (data not shown). In addition, the tolerated dose of salbutamol in HeLa cells was higher than that in fibroblast cells (data not shown). Thus, we used HeLa cells for co-IP experiment and administered a higher dose of salbutamol to the HeLa cells.
HeLa cells were incubated with salbutamol sulfate (final concentration: 20 µM) for 24 h in the presence or absence of the proteasome inhibitor, MG132 (Wako Pure Chemical Industries, Ltd.) (final concentration; 5 µM).
Prior to co-IP, the cells were lysed in immunoprecipitation buffer (50 mM Tris–HCl pH 7.2, 100 mM NaCl, 5 mM EDTA pH 8.0, 2% Triton-X) and 2 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich) then sonicated for 7 s. After overnight incubation at −80 °C, the lysate was incubated with anti-SMN at 4 °C on a slowly rotating shaker for 2 h. The co-immunoprecipitate (SMN complex) was then isolated by adding protein G PLUS-Agarose beads (Santa Cruz Biotechnology, Inc., TX, USA). The beads were washed twice with PBS and once with the same immunoprecipitation buffer before adding to the lysate. After an additional 2 h rotation at 4 °C with the beads, the co-immunoprecipitate was collected by centrifugation, washed twice with PBS, and once with immunoprecipitation buffer. The pellet was eluted by boiling in 3× NuPAGE LDS Sample Buffer (Life Technologies Corporation) with β-mercaptoethanol.
The eluted co-IP samples were electrophoresed by 10% SDS-PAGE and blotted to PVDF membranes. Immunostaining was performed using iBind Western System (Life Technologies). Ubiquitinated SMN was detected using a mouse monoclonal anti-ubiquitin antibody (Sigma-Aldrich), mouse anti-SMN antibody, and ECL HP-linked sheep anti-mouse IgG. The chemiluminescent signal was determined as described above. The amount of ubiqtitinated SMN at the mock status (not treated with salbutamol) was normalized to 1.0 in each MG132 group (in the presence or absence of MG132). The experiments were repeated four times.
2.6. Statistical analysis
Statistical analysis was performed using Microsoft Excel with the add-in software Statcel3 (OMS Ltd., Tokyo, Japan). SMN2 transcript amounts were analyzed by two-way analysis of variance (ANOVA) with one observation per cell. Differences in SMN protein levels were analyzed by repeated measures two-way and one-way ANOVAs followed by post-hoc analyses with Tukey–Kramer's multiple-comparison test. Ubiquitinated SMN protein amounts were compared by Mann–Whitney test. p Values<0.05 were considered statistically significant.
3. Results
3.1. Salbutamol did not increase SMN2 transcript levels
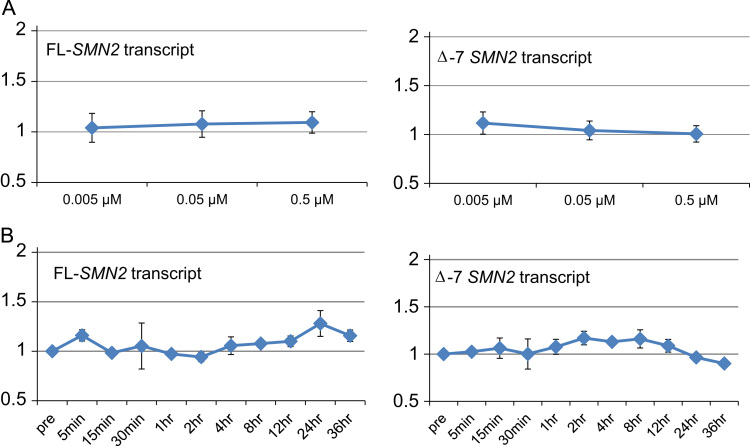
To test the possibility that salbutamol regulated the expression of SMN2 transcripts in SMA fibroblast cells, we assayed SMN2 transcript isoform levels using real-time quantitative PCR after treatment of cells with various concentrations of salbutamol (0.005, 0.05 and 0.5 µM) for time periods up to 36 h. The dose–response curve showed no significant differences in FL- and Δ7-SMN2 transcript levels in relation to salbutamol concentration (Fig. 1A). The time-course curve also showed no significant changes in FL- and Δ7-SMN2 transcript levels throughout the observation period (Fig. 1B).
Fig. 1.
FL- and Δ7-SMN2 transcript expression in salbutamol-treated SMA fibroblast cells. FL- and Δ7-SMN2 transcript levels were unaffected by different salbutamol concentrations and exposure time up to 36 h. Salbutamol concentrations used in this study were 0.005, 0.05 and 0.5 µM. Measurement time points were 0 m, 5 m, 15 m, 30 m, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h and 36 h. (A) Dose-response curve and (B) time-course curve.
3.2. Salbutamol increased SMN protein levels
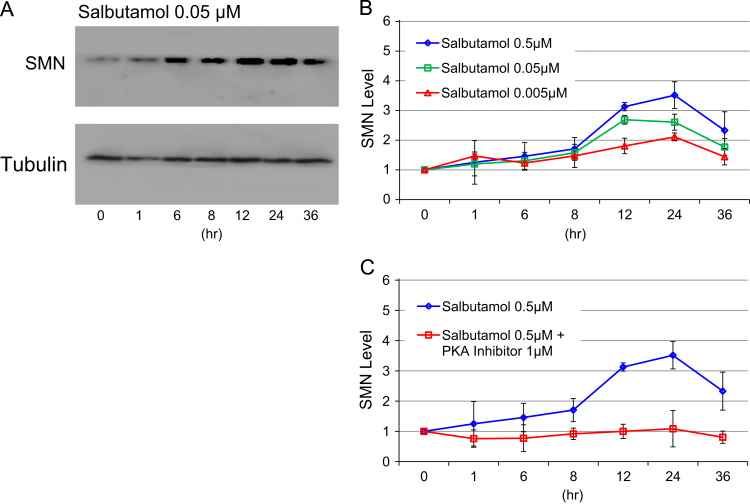
SMN protein levels were determined by western blotting at 0 h (pretreatment), 1 h, 6 h, 8 h, 12 h, 24 h and 36 h in the presence of 0.005, 0.05, and 0.5 µM salbutamol, respectively (Fig. 2A and B). SMN protein levels peaked at 12 h (with 0.05 µM salbutamol) or 24 h (with 0.005 and 0.5 µM salbutamol). The highest SMN protein levels with 0.005, 0.05, and 0.5 µM of salbutamol were 2.0-, 2.6- and 3.5-fold basal levels, respectively.
Fig. 2.
SMN protein expression in salbutamol-treated SMA fibroblast cells. Salbutamol concentrations used in this study were 0.005, 0.05 and 0.5 µM. Measurement time points were 0 h, 1 h, 6 h, 8 h, 12 h, 24 h and 36 h. (A) Western blotting analysis of SMN protein in SMA fibroblast cells. (B) Relative SMN protein levels in salbutamol-treated SMA fibroblast cells. Salbutamol increased SMN protein levels in dose- and time-dependent manners. (C) Effect of PKA inhibitor on SMN protein levels. PKA inhibitor, 14–22 amide cell-permeable, myristoylated, completely repressed the salbutamol (0.5 µM)-induced increase of SMN protein.
Salbutamol increased SMN protein levels in dose- and time-dependent manners. Two-way ANOVA showed significant differences for dose (0.005, 0.05, and 0.5 µM) (p<0.01) and time point (p<0.01), but no significant interaction between dose and time point. Post hoc analysis with Tukey–Kramer's multiple-comparison tests showed significant differences between 0.005 and 0.5 µM and between 0.05 and 0.5 µM, and between 0 h and 12 h, 0 h and 24 h, and 0 h and 36 h.
3.3. PKA inhibitor blocked salbutamol-induced increase in SMN protein
β2-adrenergic receptor binding to salbutamol activates PKA, which affects the downstream targets of the signaling pathways. To clarify the role of PKA pathways in the salbutamol-induced increase in SMN protein, we added a PKA inhibitor, 14–22 amide cell-permeable, myristoylated (final concentration: 1 µM), to the fibroblast cell culture medium together with salbutamol (final concentration: 0.5 µM) (Fig. 2C).
The PKA inhibitor completely repressed the salbutamol-induced increase in SMN. Two-way ANOVA showed significant differences in SMN protein levels in relation to PKA inhibitor (presence or absence) (p<0.01), time point (p<0.01), and PKA inhibitor×time point interaction (p<0.05). Post hoc analysis with Tukey–Kramer's multiple-comparison test showed significant differences between PKA inhibitor (presence) and PKA inhibitor (absence), and between 0 h and 12 h, and 0 h and 24 h. One-way ANOVA for PKA inhibitor (presence) followed by post hoc analysis with Tukey–Kramer's multiple-comparison test showed no significant increase in SMN protein levels during the observation period. These results suggested that the salbutamol-induced increase in SMN may be regulated by PKA pathways.
3.4. Salbutamol inhibited the ubiquitination of SMN protein
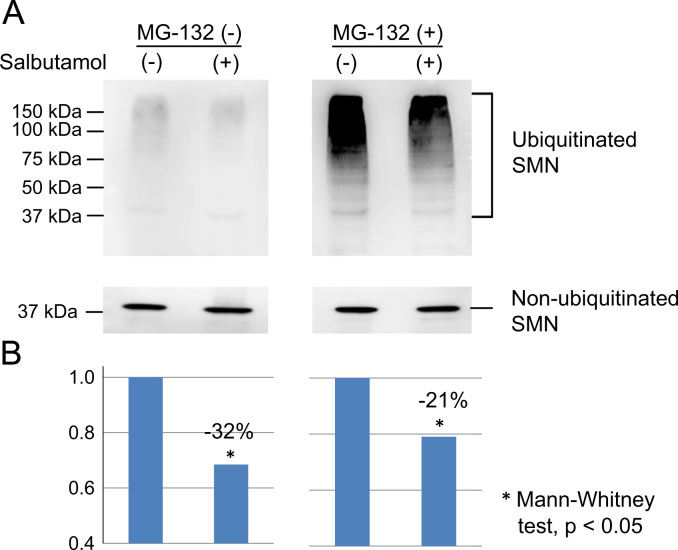
We investigated the effect of salbutamol on SMN ubiquitination by performing co-IP experiments in HeLa cells. Salbutamol reduced SMN ubiquitination in the presence or absence of the proteasome inhibitor MG132 (Fig. 3A and B). Quantification analysis showed that salbutamol significantly reduced SMN ubiquitination (Mann–Whitney test, p<0.05). The mean ubiquitinated SMN amount decreased by 20–30 percent after salbutamol treatment (Fig. 3B). These findings suggested that salbutamol may inhibit binding of ubiquitin to SMN.
Fig. 3.
Co-immunoprecipitation (co-IP) experiments in HeLa cells. Ubiquitinated SMN protein was determined by co-IP using anti-SMN antibody. (A) Ubiquitinated SMN extracted by co-IP. HeLa cells were incubated with salbutamol sulfate (20 µM) for 24 h in the presence or absence of the proteasome inhibitor, MG132 (5 µM). SMN ubiquitination was reduced in HeLa cells treated with salbutamol, in the presence or absence of MG132. (B) Quantification of ubiquitinated SMN levels. The amount of ubiquitinated SMN at the mock status (not treated with salbutamol) was normalized to 1.0 in each MG132 group (in the presence or absence of MG132). The mean ubiquitinated SMN amount decreased by 20–30 percent after salbutamol treatment.
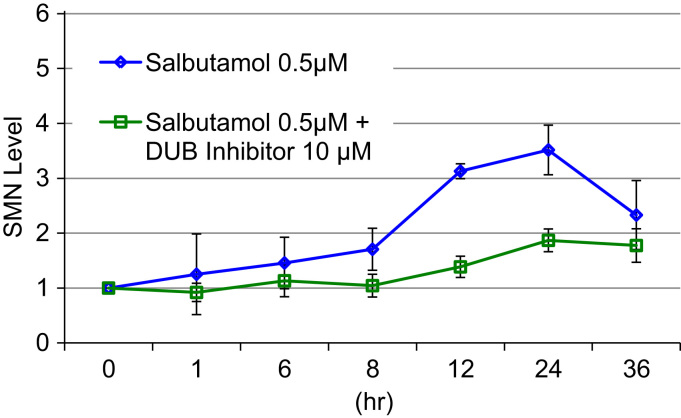
3.5. Deubiquitinase inhibitor hampered the salbutamol-induced increase in SMN protein
We hypothesized that salbutamol may inhibit ubiquitination during the process of SMN protein degradation. To test this hypothesis, we added a DUB inhibitor (PR-619, which facilitates ubiquitination) to the fibroblast cell culture medium (final concentration; 10 µM), together with salbutamol (final concentration; 0.5 µM) (Fig. 4).
Fig. 4.
Effect of deubiquitinase (DUB) inhibitor on SMN protein levels. Salbutamol concentration used in this study was 0.5 µM. Measurement time points were 0 h, 1 h, 6 h, 8 h, 12 h, 24 h and 36 h. The DUB inhibitor, PR-619, partially repressed the salbutamol -induced increase of SMN protein.
The DUB inhibitor partially repressed the salbutamol-induced increase in SMN. Two-way ANOVA showed significant differences in SMN protein levels in relation to the DUB inhibitor (presence or absence) (p<0.01) and time point (p<0.01), and DUB inhibitor×time point interaction (p<0.05). Post hoc analysis with Tukey–Kramer's multiple-comparison test showed significant differences between DUB inhibitor presence and absence, and between 0 h and 12 h, and 0 h and 24 h. One-way ANOVA for DUB inhibitor (presence) followed by post hoc analysis with Tukey–Kramer's multiple-comparison test showed a significant increase in SMN protein levels between 0 h and 12 h, 0 h and 24 h, and 0 h and 36 h, suggesting that the repression was incomplete. These results suggested that the salbutamol-induced increase of SMN may be related to SMN ubiquitination.
4. Discussion
In this study, we confirmed that salbutamol increased SMN protein levels in SMA fibroblast cells in accord with Angelozzi et al. [16]. However, Angelozzi et al. also found that FL-SMN2 transcript levels were increased by salbutamol, suggesting that salbutamol increased the production of SMN from SMN2. In contrast, we found no significant time- or dose-related changes in FL-SMN2 or Δ7-SMN2 transcript levels in response to salbutamol. This apparent discrepancy in terms of response of SMN2 transcripts to salbutamol may be attributable to the use of cells from a non-transcript-responder to salbutamol.
β2-adrenergic receptor agonists, including salbutamol, trigger elevation of cAMP and activate some PKA-signaling pathways. To examine the role of PKA-signaling pathways in our patient, we added a PKA inhibitor to the fibroblast cell culture medium, together with salbutamol, and found that the salbutamol-induced increase in SMN protein was completely blocked. This strongly suggested that salbutamol increased SMN protein levels via PKA-signaling pathways.
One of the PKA-signaling pathways may involve cAMP-response-element-binding (CREB) protein and cAMP-response elements (CREs) in gene promoters. The sequences of SMN1/2 promoter regions also includes CREs [20], [21]. The SMA fibroblast cells of Angelozzi et al. showed that salbutamol increased the production of FL-SMN2 transcript produced by SMN2 promoter activated through the PKA-signaling pathway. However, in some cases such as our patient, the effect of PKA signaling pathways on SMN2 promoter activation may be limited. Those patients may have some defects in the PKA pathway involving, CREB protein and CREs in the SMN2 promoter [22], and may not increase SMN2 transcripts. Our study suggested that there may be other mechanisms of salbutamol than activating the SMN2 promoter.
On the other hand, SMN protein is known to be degraded by the ubiquitin/proteasome system [23]. We confirmed the interaction between SMN and ubiquitin by co-IP with an anti-SMN antibody in HeLa cells. Then, we examined SMN ubiquitination levels before and after salbutamol treatment, with or without the proteasome inhibitor MG-132, and showed that salbutamol treatment partially blocked ubiquitination of SMN. This finding suggested that salbutamol inhibits ubiquitination of SMN, leading to inhibition of SMN degradation. To test this hypothesis, we added a DUB inhibitor, PR-619, to the fibroblast cells together with salbutamol, and found that the salbutamol-induced increase in SMN protein levels was blocked in the presence of the DUB inhibitor.
Our results raise the possibility that one of the PKA-signaling pathways may phosphorylate SMN and inhibit the ubiquitination of SMN. Phosphorylation by PKA-signaling pathway is known to inhibit ubiquitination of some proteins, such as catenin [24]. Burnett et al. has shown another possibility [18]. They demonstrated that PKA-signaling pathways increased the amount of SMN and non-SMN proteins (Gemin2, Gemin3 and Gemin5) in the SMN complex, suggesting that SMN stability may also be influenced by oligomerization and complex formation [18]. It may be summarized that salbutamol may increase SMN and non-SMN proteins in the SMN complex, facilitate and maintain SMN-complex formation, and affect SMN-complex function, thus promoting SMN-complex stability and function.
Salbutamol alleviates the symptoms in SMA patients [13], [14], [15]. The beneficial effects of salbutamol can be explained partly by prevention of SMN protein degradation. Kwon et al. found that the proteasome inhibitor bortezomib increased SMN protein levels in cultured cells and in peripheral tissues of SMA model mice. Furthermore, bortezomib improved motor function in SMN model mice [25]. Prevention of SMN degradation may thus represent a new treatment strategy for SMA.
This study was limited to the use of fibroblast cells from only one patient. Although salbutamol had no effect on SMN2 promoter activation in this patient's fibroblast cells, we were unable to detect this phenomenon in other SMA patients. However, the absence of SMN2 promoter activation in our patient allowed us to find the effect of salbutamol on SMN protein stabilization. In this study, we showed a new pathway of “salbutamol→PKA→X→inhibition of SMN ubiquitination” in one SMA subject. Further analyses using more SMA subjects are necessary to clarify the X factors.
In conclusion, this study demonstrated that salbutamol may increase SMN protein levels in SMA by inhibiting ubiquitin-mediated SMN degradation via activating β2-adrenergic receptor-PKA pathways. Prevention of SMN degradation may thus represent a new strategy for the treatment of SMA, and a better understanding of the SMN-degradation mechanism may afford new clues for the development of SMA therapeutics.
Acknowledgments
We are indebted to the SMA patient who participated in this study. We are also grateful to Ms. Mio Saegusa for her technical assistance. This study was supported in part by Grants-in-Aid from the Research Committee of Spinal Muscular Atrophy (SMA), Japan (research project number: H22-NANCHIIPPAN-012) and the Ministry of Education, Culture, Sports, Science and Technology, Japan (research project number: 25461549).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.10.012.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Zerres K., Davies K.E. 59th ENMC international workshop: spinal muscular atrophies: recent progress and revised diagnostic criteria. Neuromuscul. Disord. 1999;9:272–278. doi: 10.1016/s0960-8966(99)00016-4. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S., Bürglen L., Reboullet S. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Hahnen E., Forkert R., Marke C. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum. Mol. Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 4.Rochette C.F., Surh L.C., Ray P.N. Molecular diagnosis of non-deletion SMA patients using quantitative PCR of SMN exon 7. Neurogenetics. 1997;1:141–147. doi: 10.1007/s100480050021. [DOI] [PubMed] [Google Scholar]
- 5.Lorson C.L., Hahnen E., Androphy E.J. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 7.Jodelka F.M., Ebert A.D., Duelli D.M. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum. Mol. Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McAndrew P.E., Parsons D.W., Simard L.R. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nurpurta D.K., Lai P.S., Harahap N.F. Spinal muscular atrophy: from gene discovery to clinical trials. Ann. Hum. Genet. 2013;77:435–463. doi: 10.1111/ahg.12031. [DOI] [PubMed] [Google Scholar]
- 10.Brichta L., Holker I., Haug K. In vivo activation of smn in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 11.Sumner C.J., Kolb S.J., Harmison G.G. SMN mRNA and protein levels in peripheral blood Biomarkers for SMA clinical trials. Neurology. 2006;66:1067–1073. doi: 10.1212/01.wnl.0000201929.56928.13. [DOI] [PubMed] [Google Scholar]
- 12.Rigo F., Hua Y., Krainer A.R. Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 2012;199:21–25. doi: 10.1083/jcb.201207087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinali M., Mercuri E., Main M. Pilot trial of albuterol in spinal muscular atrophy. Neurology. 2002;59:609–610. doi: 10.1212/wnl.59.4.609. [DOI] [PubMed] [Google Scholar]
- 14.Pane M., Staccioli S., Messina S. Daily salbutamol in young patients with SMA type II. Neuromuscul. Disord. 2008;18:536–540. doi: 10.1016/j.nmd.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Morandi L., Abiusi E., Pasanisi M.B. Salbutamol tolerability and efficacy in adult type III SMA patients: results of a multicentric, molecular and clinical, double-blind, placebo-controlled study. Neuromuscul. Disord. 2013;23:738–852. [Google Scholar]
- 16.Angelozzi C., Borgo F., Tiziano F.D. Salbutamol increases SMN mRNA and protein levels in spinal muscular atrophy cells. J. Med. Genet. 2007;45:29–31. doi: 10.1136/jmg.2007.051177. [DOI] [PubMed] [Google Scholar]
- 17.Tiziano F.D., Lomastro R., Pinto A.M. Salbutamol increases SMN transcript levels in leukocytes of spinal muscular atrophy patients: relevance for clinical trial design. J. Med. Genet. 2010;12:856–858. doi: 10.1136/jmg.2010.080366. [DOI] [PubMed] [Google Scholar]
- 18.Burnett B.G., Muñoz E., Tandon A. Regulation of SMN protein stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harahap I.S.K., Saito T., San L.P. Valproic acid increases SMN2 expression and modulates SF2/ASF and hnRNPA1 expression in SMA fibroblast cell lines. Brain Dev. 2012;34:213–222. doi: 10.1016/j.braindev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Echaniz-Laguna A., Miniou P., Bartholdi D. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am. J. Hum. Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder S., Varadharaj S., Ghoshal K. Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene. J. Biol. Chem. 2004;279:14803–14811. doi: 10.1074/jbc.M308225200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Mack S.G., Cook D.J., Dhurjati P., Butchbach M.E.R. Systems biology investigation of cAMP modulation to increase SMN levels for the treatment of spinal muscular atrophy. PLoS ONE. 2014;9(12):e115473. doi: 10.1371/journal.pone.0115473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang H.C., Hung W.C., Chuang Y.J. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem. Int. 2004;45:1107–1112. doi: 10.1016/j.neuint.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Hino S., Tanji C., Nakayama K.I. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon D.Y., Motley W.W., Fischbeck K.H. Increasing expression and decreasing degradation of SMN ameliorate the spinal muscular atrophy phenotype in mice. Hum. Mol. Genet. 2011;20:3667–3677. doi: 10.1093/hmg/ddr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material