Abstract
DNA and histone modifications regulate transcriptional activity and thus represent valuable targets to reprogram the activity of genes. Current epigenetic therapies target the machinery that regulates these modifications, leading to global transcriptional reprogramming with the potential for extensive undesired effects. Epigenetic information can also be modified as a consequence of disrupting processive DNA replication. Here we demonstrate that impeding replication by small molecule-mediated stabilisation of G-quadruplex nucleic acid secondary structures triggers local epigenetic plasticity. We report the use of the BU-1 locus of chicken DT40 cells to screen for small molecules able to induce G-quadruplex-dependent transcriptional reprogramming. Further characterisation of the top hit compound revealed its ability to induce a dose-dependent inactivation of BU-1 expression in two steps, first loss of H3K4me3 and subsequently DNA cytosine methylation, changes that were heritable across cell divisions even after the compound was removed. Targeting DNA secondary structures thus represents a potentially new approach for locus-specific epigenetic reprogramming.
The ability to manipulate transcriptional activity in vivo has the potential to modify the development and progression of diverse pathological states. In particular, many cancers exhibit substantial dysregulation of gene expression associated with global changes in DNA methylation and histone modifications 1. There has been much recent interest in ‘epigenetic’ therapies that target histone methyl transferases, demethylases and deacetylases, which are commonly dysregulated in cancer cells. For instance, histone deacetylase inhibitors, such as Vorinostat or Phenylbutyrate, and DNA methyltransferase inhibitors, such as 5-Aza-CR (azacitidine), have proven useful for treating haematological malignancies and are currently in clinical use 2.
In vertebrate cells, it is becoming clear that the faithful transmission of epigenetic information, encoded in histone and DNA modifications, through DNA replication is important for the maintenance of gene expression patterns 3,4. Preservation of this information relies on the coupling of DNA synthesis with transfer of modified histones from the parental to the nascent daughter DNA strands, a process that requires an intricate, but as yet incompletely understood, network of histone chaperones associated with the replication fork 5,6. Under certain circumstances, replication impediments can lead to dysregulation of gene expression due to interruption of the recycling of parental histones 7–10. The ability to induce this phenomenon at discrete loci could be used to reprogram the transcriptional activity of certain genes.
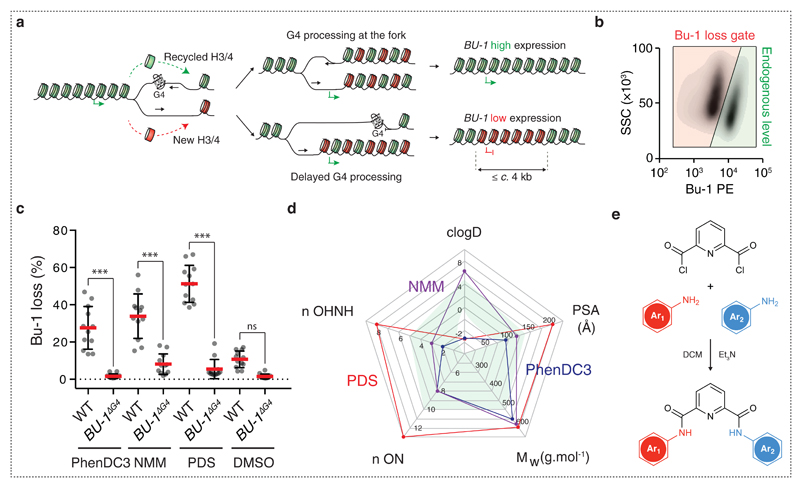
We have previously described a manifestation of this phenomenon in the BU-1 locus of chicken DT40 cells 9. We have shown that instability of BU-1 is induced in a replication-dependent manner by a DNA sequence that can form a replication-blocking G-quadruplex (G4) secondary structure. G4s form in G-rich DNA as a consequence of the ability of guanine to form thermodynamically stable Hoogsteen base-paired quartets 11. These structures present a significant impediment to DNA replication and are becoming appreciated as an important genomic feature 12–14. DT40 cells lacking key enzymes involved in G4 replication, for example the FANCJ helicase, REV1 DNA polymerase or PrimPol DNA primase/polymerase 8,9,15, or that are subjected to global replication stressors, such as hydroxyurea or aphidicolin 10, exhibit stochastic loss of normal BU-1 expression. We proposed that this results from localised uncoupling of the replicative helicase and polymerase on the leading strand resulting in correct DNA replication but a high frequency of loss of parental histone modifications. The resulting altered pattern of chromatin modifications is epigenetically inherited across cellular divisions and results in a permanent alteration in gene expression (Fig. 1a) 9.
Fig. 1. DT40 BU-1 gene expression is a sensitive screen for the in vivo activity of G4 ligands.
(a) Working model for replication-dependent loss of localised epigenetic marks 4,7,9. When the leading strand of a replication fork stalls at a G4 motif, histone recycling may be interrupted by the formation of a post-replicative gap resulting in loss of the parental epigenetic state for up to c. 4 kb downstream the G4 site. (b) Bu-1 loss variants. Flow cytometry for Bu-1 expression in WT DT40 cells treated with PDS showing gating for the wild type Bu-1high state (green region) and for Bu-1 expression variants (red region), which are quantitated as ‘percentage of Bu-1 loss’ in subsequent plots. (c) Induction of G4-dependent instability of Bu-1 expression with G4 ligands. WT or Bu-1ΔG4 DT40 were treated with either 5 µM PhenDC3, 2 µM NMM or 4 µM PDS starting from 5 cells for 7 days and then BU-1 level was monitored by flow cytometry. Red bar = median loss; whiskers = interquartile range; p-values for the comparison of Bu-1 loss distributions were calculated with Fisher’s tests of 5 windows spanning 20% BU-1 loss each, ***: p < 0.001, n.s: p = 1. (d) Radar chart reporting the physiochemical properties of NMM (purple), PhenDC3 (blue) and PDS (red). The green shaded area indicates the optimum range for each feature. Abbreviations (with optimum range) nOHNH = number of hydrogen bond donors (≤ 5); n ON = number of hydrogen bond acceptors (≤ 10); Mw = molecular mass (≤ 500 Da); PSA = polar surface area (80-140 Å2); clogD = partition coefficient (≤ 5) (e) Scheme for the creation of derivatives of PDS based on a pyridine-2,6-dicarboxamide scaffold. Details of Ar1 and Ar2 and the final compound library are given in Supplementary Methods.
Here we report the use of small molecules to induce heritable epigenetic changes in the BU-1 locus of DT40 cells through stabilisation of a single G4 motif. We exploit this phenomenon to design a sensitive assay to screen for small molecules capable of inducing G4-dependent transcriptional reprogramming of the BU-1 locus and hence of specific and effective in vivo G4 stabilisation. We identified several small molecules that are able to induce loss of BU-1 expression in a G4-dependent manner without significant perturbation of growth. We show that our lead compound, PDC12, induces the stochastic formation of variants with reduced Bu-1 expression in a dose-dependent manner over a wide concentration range. PDC12 triggers inactivation of the locus in two steps, first the loss of promoter H3K4me3, which subsequently leads to histone H3K9 and DNA cytosine methylation and complete shutdown of expression. Further, these changes persist after removal of the compound, showing that they are epigenetically heritable. This work demonstrates that stabilisation of DNA secondary structures by small molecules can be harnessed for high yield reprogramming of gene expression and suggests a potential new approach to epigenetic therapy.
Results
Expression of the BU-1 locus in wild type DT40 cells can be impaired by the use of G4-ligands
As previously reported 9, the stochastic loss of BU-1 expression under conditions of replication stress is dependent on a G4-forming sequence within the second intron of the gene (at 3.5 kb from the transcription start site (TSS) and subsequently referred to as the ‘+3.5 G4’). This G4 blocks leading strand replication of forks heading towards the TSS from the 3' end of the locus (Fig. 1a). This switching of expression of the locus away from the wild type Bu-1high state, which results from a reduction in transcription, can be readily monitored in individual cells using an antibody specific for surface Bu-1 protein coupled with flow cytometry 8,9 (Fig. 1b).
We first assessed the ability of known G-quadruplex ligands such as N-methyl mesoporphyrin IX (NMM) 16, PhenDC3 17 and Pyridostatin (PDS) 18 to induce Bu-1 loss variants (i.e. expression variants that have lost the wild type (WT) Bu-1high state; Fig. 1b) in WT DT40 19. Having determined the maximum dose of each compound that DT40 cells tolerate without any impact on doubling time, we set up a fluctuation analysis 9,20 for the emergence of Bu-1 expression variants in the presence of each ligand. A fluctuation analysis allows an estimate of the probability of a stochastic event per cell division when it is only possible to assess the prevalence of the event in a population. This is achieved by taking multiple parallel cultures through a defined number of cell divisions and examining the prevalence of the event, in this case the fraction of Bu-1 loss variants, at the end time point. Therefore, for each condition we started with 12 replicates of 5 cells per well in a 96 well plate and expanded these cells for 7 days, corresponding to approximately 14 cell divisions. In parallel with monitoring the proportion of Bu-1 loss variants in each well by flow cytometry, we recorded the final number of cells and consequently estimated the doubling time. As a control, we also treated cells in which the +3.5kb G4 had been deleted on both alleles 9, BU-1ΔG4 (Fig. 1c). We found that all three compounds generated Bu-1 loss variants in WT but not in BU-1ΔG4 cells, demonstrating that the stabilisation of this specific G4 is sufficient to induce stochastic loss of Bu-1 expression. PDS was then used to explore whether the fraction of cells exhibiting reduced BU-1 expression is related to dose. PDS was found to produce a linear dose-response for the frequency of Bu-1 loss variants at 7 days. However, it was not possible to generate a population with more than 50% Bu-1 expression variants due to acute toxicity of the compound at ≥ 5 µM (Supplementary Fig. 1).
In vivo screening for small molecules that cause G4-dependent epigenetic instability
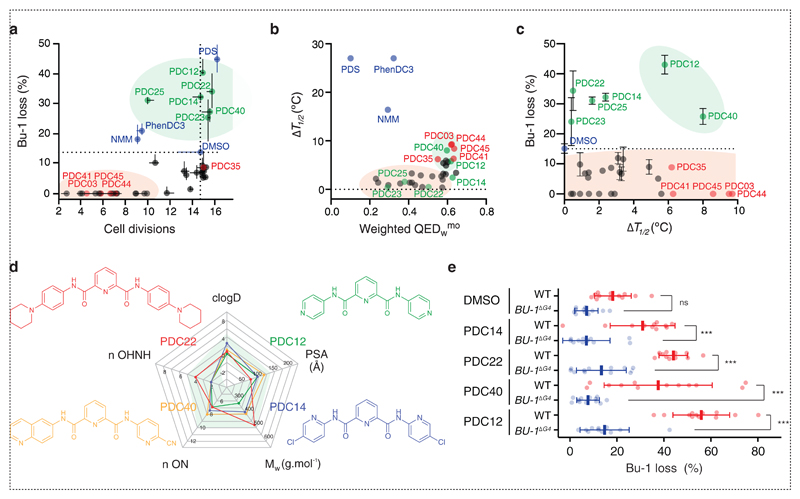
A sensitive assay that provides a read out for the explicit interaction of compounds with G4s in the genome of living cells enables the discovery of G4 targeted ligands with defined in vivo activity. Encouraged by the results presented above, we decided to employ BU-1 as a reporter locus to identify molecules capable of inducing G4-mediated transcriptional reprogramming and that exhibited an improved biological activity and therapeutic index. We first assessed the physiochemical properties of PDS, and other quadruplex ligands, using the “Lipinski rule of five” criteria (Fig. 1d) 21 that helps estimate a molecule’s ability to be absorbed by passive diffusion through cell membranes 22. We then postulated that decreasing the molecular weight, polar surface area, the number of hydrogen bond acceptors/donors and hydrophobicity of PDS should lead to small molecules combining the efficacy of PDS as a G-quadruplex ligand with increased bioavailability and wider therapeutic index (Supplementary Table 1). It is known that for small molecules a balance between cell permeability and potency must be struck, as low molecular weight and neutral compounds are generally more permeable whereas higher molecular weight compounds are generally more potent 23. Therefore, we designed and synthesised small molecules based on a pyridine-2,6-dicarboxamide (PDC) scaffold to generate a library of 32 low molecular weight, neutral compounds (Fig. 1e and Supplementary Methods). It is noteworthy that pyridine dicarboxamides of the same structural type as the PDC derivatives have been previously described as potential inhibitors of the action of telomerase via targeting the telomeric DNA G-quadruplex 24. Each PDC compound was screened for its effect on cell doubling time and the stability of BU-1 expression in a fluctuation analysis for loss of the Bu-1high state using WT DT40 cells. Each compound was assessed at two doses (5 µM and 10 µM), in triplicate and starting from 5 cells, as described above. After seven days of culture, the final viable cell number and percentage of Bu-1 expression variants was monitored by flow cytometry. The median percentage of Bu-1 loss variants in each culture is reported as a function of the estimated number of cell divisions (Fig. 2a). Compounds falling into the lower left quadrant (highlighted by the red area) are those that significantly inhibit growth while compounds in the lower right quadrant did not affect growth, but had no effect on Bu-1 expression. Ligands of interest (highlighted by the green area) appear in the upper right quadrant, as these molecules induced loss of Bu-1 expression without affecting cellular growth. At 5 µM, the compound that most potently induced loss of the Bu-1high expression state was PDS. However, at 10 µM, PDS became toxic, but several of the PDC derivatives (PDC12, 14, 22, 23, 25 and 40) were able to induce significant numbers of Bu-1 loss variants without appreciable toxicity (Fig. 2a).
Fig. 2. In vivo screen for new G4 quadruplex ligands.
(a) Effect of the PDC-derivatives on induction of Bu-1 loss variants and on cell proliferation after seven days. The plot shows the result of three independent repeats. Data presented on the plot were generated using 10 µM of the PDC derivatives and 5 µM of NMM, PhenDC3 or PDS. The green shaded area indicates compounds that were able to induced significant numbers of Bu-1 expression variants without significantly reducing cell growth. (b) Plot of induced thermal stabilisation of the BU-1 +3.5 G4 5’-d(GGGCTGGGTGGGTGCTGTCAAGGGCTGGG) as a function of the weighted quantitative estimate of drug likeness (QEDwmo) 22. Thermal stabilisation was monitored using CD spectroscopy of the BU-1 +3.5 G4 (2 µM) with 5 equivalents of the small molecule under test. (c) Plot of percentage of Bu-1 loss variants after exposure to each compound for 7 days plotted as a function of the induced thermal stabilisation of the +3.5 G4 motif. Horizontal and vertical bars, in panels a and c, represent standard errors among three repeats for the number of cell divisions and Bu-1 loss respectively. (d) Structure and physicochemical properties of the hits PDC12, 14, 22 and 40. (e) Induction of Bu-1 loss variants by the top four compounds requires the presence of the Bu-1 +3.5 G4. Repeat of the fluctuation analysis experiments shown in (a) for 10 µM over 7 days of PDC12, 14, 22 and 40 with DMSO as control on both WT and the Bu-1ΔG4 DT40 control. Bars = median loss; whiskers = interquartile range; p-values for the comparison of Bu-1 loss distributions were calculated with Fisher’s tests of 5 windows corresponding to 20% BU-1 loss, ns: non-significant, ***: p < 0.001, n.s: p = 1.
In a second step, we biophysically assessed the ability of each of these compounds to stabilise the Bu-1 G4 in vitro using Circular Dichroism (CD) spectroscopy-based thermal denaturation assays (to avoid fluorescence or UV-based experiments with which the small molecules themselves may interfere). Melting curves, followed at 263 nm, were recorded in the absence and presence of 5 equivalents (10 µM) of each of the small molecules and the stabilisation potency was extracted as the increase in melting temperatures (ΔT1/2, Supplementary Table 1). Taking into consideration the quantitative estimate of drug-likeness (QED) 22, (Supplementary Table 1), we identified the different compounds that displayed in vitro G4 stabilising properties and suitable theoretical physicochemical properties (Fig. 2b). The G4-stabilising molecules with weighted QED values > 0.5 (highlighted by the green area in Fig. 2b) represented the interesting hits from this biophysical screen. A plot showing the median number of Bu-1 loss variants generated in seven days as a function of ΔT1/2 (Fig. 2c) highlighted the six compounds, PDC12, 14, 22, 23, 25 and 40, as potential candidates for quadruplex-dependent reprogramming of BU-1 expression. We confirmed, using an alternative FRET-melting assay, the ability of each of the molecule to stabilise several unrelated quadruplex motifs in vitro alongside their ability to trigger loss of Bu-1 expression (Supplementary Fig. 2). Fig. 2d reports the structures of the four most potent molecules. It is noteworthy that these molecules all display improved physicochemical properties as compared as the parent molecule PDS. To confirm that these compounds acted directly on the +3.5 G4 motif, the fluctuation analysis was repeated by challenging the BU-1ΔG4 cells in parallel with wild type ones (Fig. 2e) with the four best small molecules. None of these ligands produced any significant effect on the expression stability of BU-1 when the +3.5 G4 motif was deleted. This demonstrated that the induction of BU-1 instability by these compounds was completely dependent on the presence of the G4 motif. Due to its higher apparent potency, PDC12 was selected for further investigation of BU-1 reprogramming.
PDC12-mediated BU-1 reprogramming is dose, G4 and replication dependent
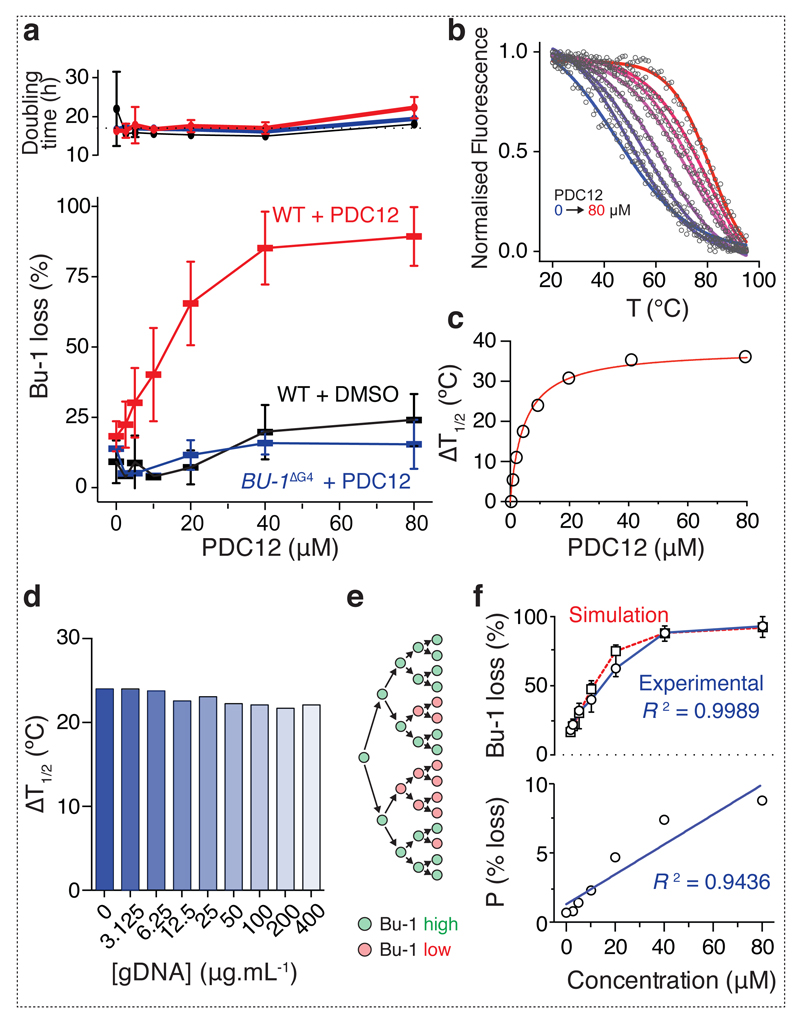
We next asked whether PDC12 was able to induce more complete reprogramming of BU-1 expression than PDS, which is limited by toxicity. A dose-response fluctuation analysis using the previous experimental design was performed and PDC12 was found to be active over a range of up to 80 µM without detectable toxicity (Fig. 3a), with a dose of 40 µM causing > 75% of all cells to lose wild type levels of Bu-1 expression, while BU-1ΔG4 exhibited no significant increase in the formation of Bu-1 loss variants above the background of the flow cytometry assay. We then used a FRET-melting assay to further assess the G4-binding properties and selectivity of PDC12 in vitro (Fig. 3b). Micromolar doses of PDC12 were found to stabilise the BU-1 G4 motif. We could demonstrate a significant thermal stabilisation (up to 35°C at 40 µM, Fig. 3c) of the +3.5 G4, and this was not affected by the presence of competing genomic DNA, demonstrating specificity of the compound for the BU-1 G4 (Fig. 3d). To further elucidate the mechanism by which PDC12 reprograms BU-1, we employed a Monte-Carlo simulation 9 to assess the correlation between the generation of Bu-1 loss variants and cell division. The simulation 9 is based on modelling the probability for each cell to irreversibly lose expression of wild type levels of Bu-1 at each cell cycle (Fig. 3e). Running the simulation with varying probabilities of loss of BU-1 expression per cell cycle allows the generation of a function that can be compared with the experimental data and used to compute the probabilities of loss for each concentration. The model was found to accurately recapitulate the kinetics with which Bu-1 loss variants are generated (R2 = 0.9989, Spearman correlation) (Fig. 3f) and allows an estimation of the per division probability of this event. For example, according to the model, the probability of conversion at 40 µM PDC12 is more than 7% per cell cycle. Interestingly, the probability of loss strongly correlates with the dose of the compound (R2 = 0.9436, Spearman correlation, Fig. 3f) supporting the idea that PDC12-induced loss of Bu-1 expression requires cell division. Together, these observations are consistent with the PDC12-mediated reprogramming of BU-1 locus being dependent on the small molecule dose, the BU-1 +3.5 G4 motif and DNA replication.
Fig. 3. G4- and PDC12-dependent induction of Bu-1 loss variants.
(a) Generation of Bu-1 loss variants as a function of PDC12 dose assessed by fluctuation analysis. Main graph: Each bar represents the median percentage of Bu-1 loss variants after 7 days in parallel cultures started from 5 Bu-1 positive cells (WT + PDC12 n= 24; WT + DMSO n = 12; Bu-1ΔG4 + PDC12 n = 12). Whiskers = interquartile range. Red: wild type cells treated with PDC12; black: wild type cells treated with DMSO; blue: Bu-1ΔG4 cells treated with PDC12. The top graph shows doubling time for each condition with mean and interquartile range. (b - d) PDC12 stabilizes the BU-1 +3.5 G4 in a concentration-dependent manner. (b) Thermal denaturation profiles of a dual-labelled BU-1 +3.5 G4 (200 nM) with an increasing concentration of PDC12 (from 0 to 80 µM, blue to red curves). The BU-1 +3.5 G4 was labelled with 6-FAM and TAMRA at its 5’ and 3’ ends (excitation at 494 nm and emission at 580 nm). (c) Stabilisation induced by PDC12 is reported as the concentration-dependent increase of the melting temperature (T1/2) of the G4/PDC12 complex. (d) PDC12 selectively stabilises the BU-1 +3.5 G4 as exemplified by a competition experiment with genomic DNA. PDC12 (10 µM) was incubated with a dual-labelled BU-1 +3.5 G4 (200 nM) and an increasing concentration of genomic DNA (from 0 to 400 µg.ml-1) and the melting temperatures of the G4/PDC12 complex were recorded as previously described. (e) Diagrammatic representation of the Monte Carlo computational model for emergence of Bu-1 loss variants dependent of cell divisions 9. (f) The upper graph shows a simulation of the generation of Bu-1 loss variants taking into account the experimental conditions used in this study (black squares and dotted red line) overlaid with the experimental data (black circles and solid blue line). The lower graph shows the correlation between probability of Bu-1 loss per division calculated using the Monte Carlo simulation and the results of the fluctuation analysis.
PDC12 induces repression of Bu-1 expression in two steps
To explore underlying mechanisms for the loss of Bu-1 expression in response to increasing PDC12 doses, five cells were expanded for seven days in the presence of 20 µM or 40 µM PDC12 or DMSO, then four replicate samples were pooled and expanded for a further seven days to allow recovery of sufficient material for expression and ChIP analysis.
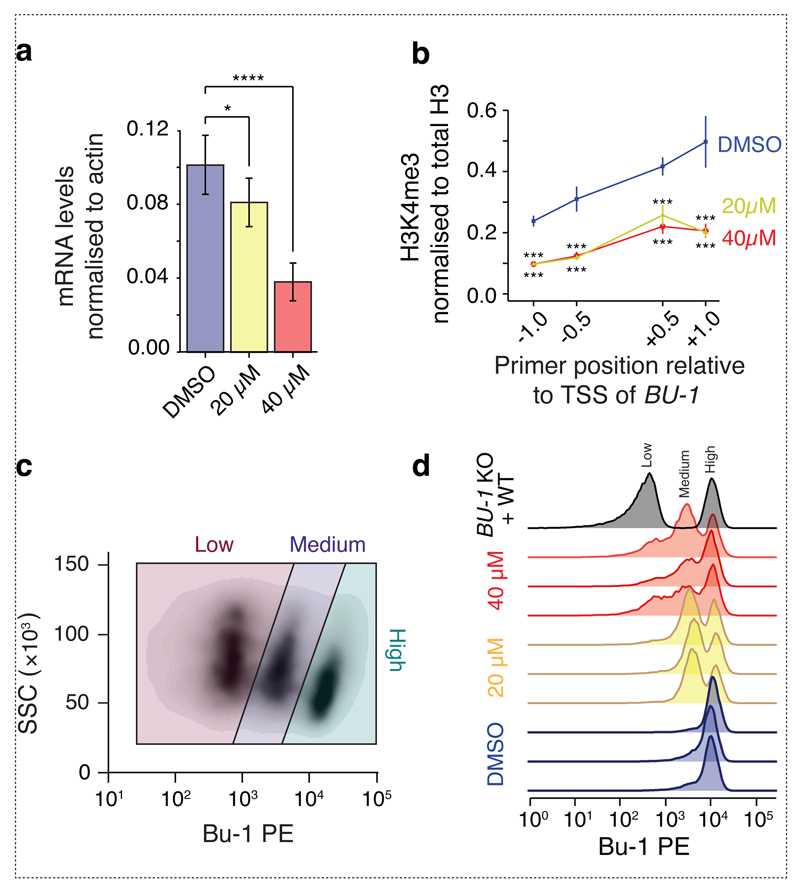
We first looked at BU-1 transcript levels. Exposure to PDC12 resulted in a dose-dependent reduction in BU-1 mRNA (Fig. 4a). Exposure of cells to some G4 ligands has been associated with the induction of DNA damage and the phosphorylation of H2Ax, a mechanism that has been linked to an acute reduction in gene expression 25. We therefore asked whether PDC12 induced a measurable DNA damage response by monitoring the induction of histone H2Ax and CHK1 S345 phosphorylation. It did not, even when cells were exposed to the highest dose (80 µM) used in these studies for two weeks (Supplementary Fig. 3). Further, the compound did not induce any change in the cell cycle (Supplementary Fig. 4). We were also unable to detect mutations of the +3.5 G4 motif suggesting that the formation of Bu-1 loss variants was not explained by mutation of the BU-1 locus around the G4 motif (Supplementary Fig. 5). Next, using chromatin immunoprecipitation, we monitored levels of H3K4me3, a histone modification associated with active transcription, around the TSS of BU-1. H3K4me3 was significantly reduced in the populations treated with both 20 µM and 40 µM PDC12 relative to the control (Fig. 4b). We also found a concomitant gain of H3K9me3 (Supplementary Fig. 6), consistent with our previous studies on mutants defective in G4 unwinding 8,9. However, there was no difference between the 20 µM and 40 µM conditions (Fig. 4b and Supplementary Fig. 6). This suggests that the additional repression of transcript production in cells treated with the higher dose of PDC12 may result from an additional mechanism. Examination of the distribution of expression of Bu-1 in the cells treated with 20 µM and 40 µM revealed that the latter exhibited an additional population with practically no expression of Bu-1 (Fig. 4c & d). This presence of a 'third', Bu-1low, state was reminiscent of the pattern of BU-1 expression in cells lacking the primase-polymerase, PrimPol 15. In this previous study, we showed that primpol Bu-1low cells had acquired DNA methylation at BU-1 in addition to the loss of H3K4me3 seen in the Bu-1medium population.
Fig. 4. PDC12 reprograms histone marks leading to reduced mRNA expression.
(a) Quantification of Bu-1 transcript levels by RT-qPCR as a function of PDC12 at 20 and 40 µM. Bu-1 transcript levels are normalised to actin B. Error bars show standard error of three biological replicates (each with three technical replicates). p-values computed with the Wilcoxon two-sided test, *: p < 0.05 and ***: p < 0.001. (b) ChIP for H3K4me3 around the BU-1 TSS normalised to total input and to total H3. Each point represents results from three biological replicates (each with three technical replicates) as previously described 9. ChIP for H3K9me3 and controls are shown in Supplementary Fig. 6. Bars represent standard error. p-values computed by the use of Wilcoxon two-sided test, ***: p < 0.001. (c) Representative flow cytometry scatter plot of WT DT40 cells treated with 40 µM of PDC12 for 14 days highlighting the gating for cells expressing high, medium and low level of Bu-1. Under this condition, PDC12 induces a third, Bu-1low expression state, not observed at seven days (Fig. 1b). (d) Bu-1 expression of cells used for the ChIP and qPCR experiments show in (a) and (b). Grey plot shows a control mix of WT Bu-1 positive DT40 and DT40 in which the BU-1 locus has been genetically disrupted. Triplicates have been performed for each condition (highlighted by different colours).
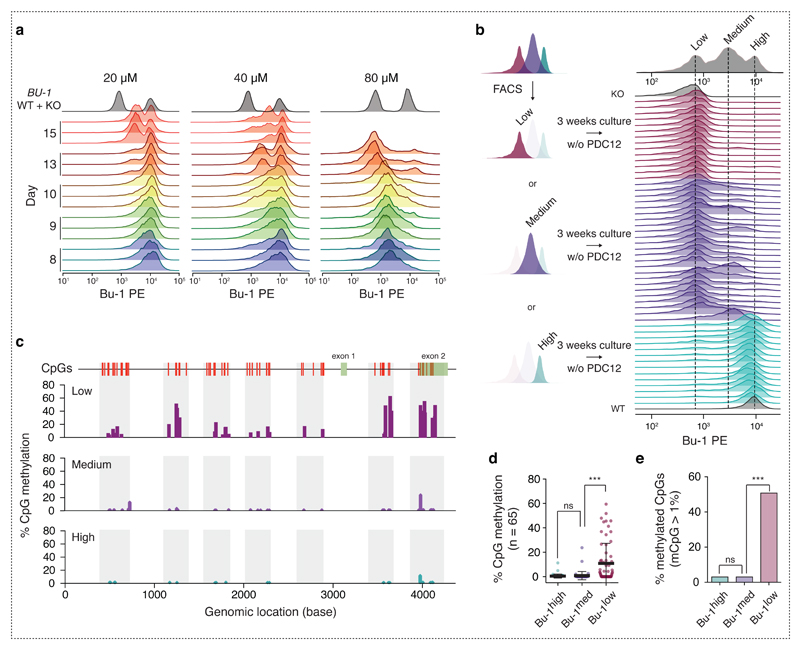
We therefore examined the way in which PDC12 induced the Bu-1medium and Bu-1low states in more detail. The appearance of Bu-1medium and Bu-1low populations is dependent on both the dose of drug and number of cell divisions (Fig. 5a). Thus, using 20 µM PDC12, a ratio of Bu-1medium: Bu-1high of 3 : 7 was achieved in 15 days. The same ratio was reached with 40 µM and 80 µM doses in just 10 and less than 8 days respectively. Consistent with our previous observations in PrimPol-deficient cells 15, the Bu-1low state required the prior establishment of a Bu-1medium population. The appearance of Bu-1low cells thus lagged behind the Bu-1medium population and they were only observed in significant numbers after 13 days in cells treated with 40 µM and 80 µM of the compound. Indeed, after this time, exposure to 80 µM PDC12 gave rise to an almost completely Bu-1low population.
Fig. 5. PDC12 induces irreversible loss of Bu-1 expression and subsequently CpG methylation.
(a) Bu-1 expression was found to depend on the duration and dose of PDC12 exposure. The flow cytometry profiles report three independent biological replicates of the change in Bu-1 expression after the indicated number of days (highlighted by different colours) and exposure to either 20, 40 or 80 µM PDC12. Grey plots show a control mix of WT Bu-1 positive DT40 and DT40 in which the BU-1 locus has been genetically disrupted. (b) The Bu-1high to Bu-1medium transition depends on PDC12, while the Bu-1medium to Bu-1low transition is spontaneous. Cells representative of a given expression state, after fourteen days in 40 µM PDC12, were sorted by FACS as depicted on the left hand-side. After three weeks in compound-free medium, several individual clones of each population, Bu-1high (light blue), Bu-1medium (dark blue) and Bu-1low (purple), were analysed by flow cytometry. (c - e) The irreversibility of the Bu-1low expression state can be explained by the DNA methylation of the BU-1 locus. (c) DNA methylation around the promoter and TSS of Bu-1low, Bu-1medium and Bu-1high populations. (d) Quantification of the percentage of methylation of the BU-1 locus. Black bar = median; whiskers = interquartile range. (e) Quantification of the percentage of methylated CpGs. p-values in (d) and (e) were computed by the use of Wilcoxon two-sided test, ns: non-significant, ***: p < 0.001.
The ability to control the formation of Bu-1 loss variants with PDC12 allowed us to assess the reversibility of each transition and the requirement for G4 stabilisation. We therefore selected a population that had been treated with 40 µM PDC12 for 16 days and that exhibited all three states (Fig. 5b). We sorted individual Bu-1high, Bu-1medium or Bu-1low cells into separate wells and expanded them for three weeks in the absence of PDC12. Bu-1 expression was then reassessed by flow cytometry (Fig. 5b). As expected, Bu-1high clones no longer generated Bu-1 loss variants in the absence of the compound. However, the majority of Bu-1medium clones (22 out of 24) spontaneously became Bu-1low, while none reverted to being Bu-1high. Bu-1low cells remained in this state.
To further characterise the cause of the irreversible Bu-1low state, we analysed CpG DNA methylation around the BU-1 locus. Genomic DNA of clones representing the three states was then extracted and subjected to bisulfite conversion. Seven regions around the BU-1 TSS were PCR amplified and sequenced in order to interrogate their CpG methylation status (Fig. 5c). While both Bu-1high and Bu-1medium cells had a very low level of CpG methylation of the BU-1 locus, Bu-1low cells exhibited a significant increase, suggesting formation of heterochromatin (Fig. 5d & e).
Together, our observations demonstrate that PDC12 exposure leads to the sustained methylation of the BU-1 promoter in a dose-dependent manner. Further, this expression state persists following removal of the G4 ligand demonstrating that the ligand has triggered an epigenetically heritable change in the expression state of the gene. Mechanistically, we show that this takes place in two steps. The first results from loss of H3K4me3 and is consistent with our previous model of epigenetic instability induced by localised G4-dependent replication arrest 8,9. This loss of H3K4me3 appears then to render the locus vulnerable to DNA methylation and total shut down of expression of the gene, but this second phase takes place without a requirement for the presence of PDC12.
Discussion
Here we report a robust and sensitive approach to evaluate small molecules for their ability to interact with genomic G4s. Using the extensively characterized BU-1 locus as reporter 8–10,15, we describe a sensitive in vivo screening platform that links a genomic G4 motif with a robust phenotypic readout in the generation of Bu-1 loss variants. By combining this in vivo assay with biochemical and computational approaches we have identified G4 interacting compounds with well-defined cellular intervention properties, while retaining low toxicity.
We show that the per division probability of a cell losing full expression of Bu-1 can be controlled by the dose of the G4 ligand used, suggesting that this assay monitors the in vivo potency of a specific ligand – G4 pair. It is noteworthy that the example of the +3.5 BU-1 G4 reveals that the ability of a compound to bind a G4 in vitro does not necessarily translate into in vivo potency. While there is a general trend linking the in vitro ability of the top hit compounds to stabilise G4s and their in vivo ability to induce G4 motif-dependent expression instability at BU-1 (Supplementary Fig. 2g), the correlation is not a strict one. This likely reflects the complexity of G4 formation and stabilisation in vivo and highlights the potential limitations of in vitro biophysical measurements. Since the cellular assay, which we describe here, is easily scalable it should be suitable for automation and allow the screening of larger chemical libraries. Coupled with the genetic tractability of DT40 cells this approach could be expanded to the study of other G4 or DNA structural sequences/motifs, inserted in place of the +3.5 BU-1 G4 allowing a more detailed understanding of the interaction of specific ligands with G4 targets in vivo.
While the use of a G4 ligand to induce gene expression changes is inevitably accompanied by the possibility that the observed effects are indirect, for example through inhibition of a G4-processing enzyme, the observation that chemically diverse compounds destabilise BU-1 expression in a manner requiring the +3.5 G4 motif (Fig. 1c and 2e) provides some confidence that they are indeed acting directly on the structure in vivo. There are a number of potential mechanisms by which G4 stabilisation can affect gene expression. Early studies by Hurley and colleagues demonstrated that the stabilisation of a G4 within the NHE III1 (nuclease hypersensitivity element) in the promoter of the c-Myc oncogene resulted in repression of transcription by preventing binding of key transcription factors 26, establishing a paradigm for direct transcriptional regulation by G4 stabilisation 27. G4 stabilisation has also been proposed to influence transcription indirectly by causing DNA damage, especially double strand breaks 25. The repair of double strand breaks in active genes results in transient transcriptional repression through heterochromatin formation 28,29. However, in neither case has the G4 stabilising compound has been shown to exert an epigenetically heritable effect on gene expression that persists even after removal of the compound. The data we present here shows that heritable and durable chromatin remodelling can be achieved by exploiting the ability of stabilised G4s to locally interrupt the propagation of epigenetic information through S phase.
We have taken advantage of the broad usable concentration range of PDC12 to shed further light on the step-wise mechanism by which the BU-1 locus is reprogrammed. In a first step, stabilisation of the BU-1 G4 results in the structure becoming a more significant impediment to replication. The observation that G4 ligands induce instability of expression of the BU-1 locus suggests that G4s actually form frequently during DNA replication, but are usually rapidly resolved. This may explain why some G4 motifs are associated with replication slow zones, for example the G4 within the chicken ρ-globin gene 30. We propose that the additional delay to replication imposed by the stabilisation of the structure leads to significant uncoupling of DNA synthesis and histone recycling leading to loss of the H3K4me3 normally found around the TSS of the gene, similar to cases where replication of the G4 is delayed 8,9.
The ability to control G4 stabilisation by removal of the ligand revealed that the subsequent appearance of DNA methylation, and further repression of expression, does not require the G4 to remain a replication impediment. The mechanism by which this DNA methylation is installed remains to be investigated. However, it may simply be a consequence of the loss of H3K4me3, which inhibits DNA methylation 31,32. H3K4me3 interferes with the activation of the de novo DNA methyltransferase DNMT3A by preventing the binding of the ADD domain of the enzyme to H3K4. Inability to bind H3K4 prevents the conformational change necessary to release the auto inhibition of the catalytic activity of the enzyme by the ADD domain 33. Thus, our data is consistent with the idea that replication-dependent maintenance of H3K4me3 at gene promoters not only represents a mechanism to facilitate stable, high level transcription but also prevents aberrant silencing by DNA methylation.
In summary, we demonstrate the principle of reprogramming the epigenetically determined transcriptional state of a locus through G4 stabilisation. Our observations suggest that targeting secondary structures in DNA to trigger replication-dependent reprogramming of key genes could potentially be harnessed to create epigenetic therapies.
Methods
DT40 cell culture and mutants
DT40 cells were cultured as previously described 34. Manipulation of the BU-1 locus, including the derivation of the BU-1Δ+3.5G4 line has been described previously 9,10.
G-quadruplex ligands
The small molecules Pyridostatin (PDS) and PhenDC3 were synthesised as previously reported 17,18. N-Methyl Mesoporphyrin IX (NMM) was purchased from Frontier Scientific (Catalogue No: NMM580). Detailed procedures for the synthesis of pyridine-2,6-dicarboxamide (PDC) derivatives and their characterization are provided in the Supplementary Methods. Physicochemical properties were predicted using ChemAxon Marvin (http://www.chemaxon.com).
Surface Bu-1 staining and analysis
Gene expression variation of the BU-1 locus in WT DT40 cells is reported for the BU-1A allele only, using a Bu-1A antibody (Santa Cruz 70447) coupled with Phycoerythrin (PE). As previously described,9 DT40 is an F1 hybrid expressing both Bu-1A and Bu-1B. All flow cytometry was carried using BD™ LSR II flow cytometer and analysed with FlowJoTm. Bu-1A expression has been gated using in Y-axis side scatter (SSC) and in X-axis Phycoerythrin (PE) fluorescence. Data was exported and further computation and plots performed using home-made R 35 scripts using the Beeswarm package 36. p-values were assessed by Fisher’s tests of 5 windows (corresponding to bins of 20%) as previously described 9.
Chromatin Immunoprecipitation (ChIP) and RT-qPCR
ChIP was performed as previously described 9. Total mRNA was extracted with TRIzol™ following the manufacturer's instructions. mRNA was converted into cDNA with the QuantiTect Reverse Transcription Kit (Qiagen cat nº 205311). Quantitative PCR was performed on a ViiA™ 7 Real-Time PCR System (ThermoFisher Scientific) using Power SYBR® Green PCR Master Mix (ThermoFisher cat nº 4367659). Ct thresholds were determined with the ViiA™ 7 software and the results analysed with custom R 35 scripts. Primers used are given in Supplementary Methods. For ChIP, quantitation was computed using the formula Quantitation of mRNA levels were computed using the formula E^(Ct(BU-1) – Ct(Actin)). E corresponds to primer efficiencies (Supplementary Methods).
Circular dichroism (CD) denaturation studies
CD experiments were conducted on a Chirascan Plus spectropolarimeter. Oligonucleotide solutions were prepared at a final concentration of 2 μM in 10 mM lithium cacodylate (pH 7.2) containing 1 mM of EDTA and 100 mM of KCl. The samples were annealed by heating at 95 °C for 10 min and slowly cooled to 20 °C. The small molecules were directly added to the oligonucleotide solutions from 10 mM DMSO stock solutions for a final concentration of 10 μM. Each sample was transferred to a quartz cuvette with 1 cm path length, covered with a layer of mineral oil, placed in the spectrophotometer and equilibrated at 5 °C for 10 min. Samples were then heated to 95 °C at a rate of 1 °C/min, with data collection every 1 °C. The CD signal at 263 nm was monitored and melting temperature (T1/2) values were extracted as the half-maximum increase in ellipticities.
Förster resonance energy transfer (FRET) melting experiments
Fluorescence experiments were conducted on a Varian Cary Eclipse spectrophotometer. A dual-labelled G4-BU-1 oligonucleotide, referred to as Fl-G4-BU-1, was used. The donor fluorophore was 6-carboxyfluorescein (FAM), and the acceptor fluorophore was 6-carboxytetramethylrhodamine (TAMRA). Oligonucleotide solutions were prepared at a final concentration of 200 nM in 10 mM lithium cacodylate (pH 7.2) containing 1 mM of EDTA and 100 mM of KCl. The samples were annealed by heating at 95 °C for 10 min and slowly cooled to 20 °C. PDC12 was directly added to the oligonucleotide solutions from a 10 mM DMSO stock solution. For competition experiments with genomic DNA, human placental DNA (Sigma) was directly added from a 1 mg.mL-1 solution. Each sample was transferred to a quartz cuvette with 1 cm path length, covered with a layer of mineral oil, placed in the spectrophotometer and equilibrated at 5 °C for 10 min. Samples were then heated to 95 °C at a rate of 1 °C/min, with data collection every 1 °C. FRET signals were monitored using an excitation wavelength of 483 nm and a detection wavelength of 533 nm. Melting temperature (T1/2) values were extracted as the half-maximum decrease in fluorescence.
Bisulfite sequencing
Bisulfite conversion was achieved using the TrueMethyl Seq kit (Cambridge Epigenetix), following the manufacturer’s instructions, starting with 1 μg of genomic DNA. After oxidation and bisulfite treatment, regions of interest from the BU-1 locus were amplified by PCR, using the VeraSeq Ultra DNA polymerase (Enzymatics) and bisulfite primers (Supplementary Methods). The amplicons were then blunt ended (NEBNext® End Repair Module) and dA-tailed (NEBNext® dA-Tailing Module), before ligation of Illumina adapters. Libraries were quantified using the KAPA Library Quantification Kits (Kapa Biosystems). Sequencing was performed on an Illumina NextSeq 500 with paired-end 300 cycle reads. Counts of converted and unconverted cytosine, i.e the methylation status, in the BU-1 locus were obtained after Trimming reads by seqtk (H. Li, https://github.com/lh3/seqtk/), alignment using Bismark 37 and Bowtie2 38. Data extracted from Bismark were further analysed and the Fig. generated with a home-made R 35 script.
Supplementary Material
Acknowledgments
The authors would like to thank Milena Stankovic for her help in genotyping PCR across BU-1 G4 and with the ChIP experiment. Maria Daly, Fan Zhang and Veronika Romashova in the LMB flow cytometry facility for cell sorting, Jake Grimmett and Toby Darling in LMB scientific computing for their help in massive sequencing analysis and Chris Lowe for proofreading the manuscript. Work in the Sale group is supported by a central grant to the LMB by the MRC (U105178808). SB is a Wellcome Trust Senior Investigator (grant no. 099232/z/12/z). The Balasubramanian group is supported by a European Research Council Advanced Grant (no. 339778) and receives core funding (C14303/A17197) and programme funding (C9681/A18618) from Cancer Research UK.
Footnotes
Data Availability Statement
All the data presented and analysed in this study are either included in this article or in the Supplementary Information, which includes the materials and methods used.
Author contribution
GG and PM designed, performed and analysed the experiments. BR analysed the impact of PDC12 on the DNA damage response. BCC and PM synthesised the G4 ligand library. AM performed the first screen of the library with GG. GG, PM & JES wrote the manuscript with contributions from all authors. JES and SB supervised the project.
Conflict of interest
The authors declare that they have no conflict of interest
References
- 1.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarkies P, Sale JE. Cellular epigenetic stability and cancer. Trends Genet. 2012;28:118–127. doi: 10.1016/j.tig.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Šviković S, Sale JE. The Effects of Replication Stress on S Phase Histone Management and Epigenetic Memory. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 6.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 7.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic Instability due to Defective Replication of Structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiavone D, et al. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014;33:2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papadopoulou C, Guilbaud G, Schiavone D, Sale JE. Nucleotide Pool Depletion Induces G-Quadruplex-Dependent Perturbation of Gene Expression. Cell Rep. 2015;13:2491–2503. doi: 10.1016/j.celrep.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Maizels N, Gray LT. The G4 Genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiavone D, et al. PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol Cell. 2016;61:161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 17.De Cian A, DeLemos E, Mergny J-L, Teulade-Fichou M-P, Monchaud D. Highly Efficient G-Quadruplex Recognition by Bisquinolinium Compounds. J Am Chem Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez R, et al. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J Am Chem Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba TW, Giroir BP, Humphries EH. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 20.Luria SE, Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 22.Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4:90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keserü GM, Makara GM. The influence of lead discovery strategies on the properties of drug candidates. Nat Rev Drug Discov. 2009;8:203–212. doi: 10.1038/nrd2796. [DOI] [PubMed] [Google Scholar]
- 24.Hittinger A, Caulfield T, Mailliet P, Bouchard H, Mandine E, Mergny J-L, Guittat, Riou J-F, Gomez D, Belmokhtar C. Patent application WO 2004072027. Chemical derivatives binding very specifically with G-quadruplex DNA structures and use thereof as a specific anti-cancer agent. 2004
- 25.Rodriguez R, et al. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prioleau MN, Gendron MC, Hyrien O. Replication of the Chicken β-Globin Locus: Early-Firing Origins at the 5' HS4 Insulator and the ρ- and βA-Globin Genes Show Opposite Epigenetic Modifications. Mol Cell Biol. 2003;23:3536–3549. doi: 10.1128/MCB.23.10.3536-3549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- 34.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Foundation for Statistical Computing. R: A language and environment for statistical computing. 2016 [Google Scholar]
- 36.Eklund A. Beeswarm: an add-on package for the R statistical environment. 2016 [Google Scholar]
- 37.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.