Abstract
We developed software tools to download, extract features, and organize the Cell Types Database from the Allen Brain Institute (ABI) in order to integrate its whole cell patch clamp characterization data into the automated modeling/data analysis cycle. To expand the potential user base we employed both Python and MATLAB. The basic set of tools downloads selected raw data and extracts cell, sweep, and spike features, using ABI’s feature extraction code. To facilitate data manipulation we added a tool to build a local specialized database of raw data plus extracted features. Finally, to maximize automation, we extended our NeuroManager workflow automation suite to include these tools plus a separate investigation database. The extended suite allows the user to integrate ABI experimental and modeling data into an automated workflow deployed on heterogeneous computer infrastructures, from local servers, to high performance computing environments, to the cloud. Since our approach is focused on workflow procedures our tools can be modified to interact with the increasing number of neuroscience databases being developed to cover all scales and properties of the nervous system.
Keywords: Computer simulation, Allen Brain Institute, database, NEURON, computational neuroscience, NeuroManager
1 Introduction
The systematic acquisition of experimental data in neuroscience is resulting in the emergence of many databases focusing on different scales and properties of the nervous system (ABI, 2017b; Autism Brain Imaging Data Exchange, 2017; Fox and Laird, 2017; George Mason University, 2017; International Neuroinformatics Coordinating Facility, 2017; SenseLab, 2017). While much effort has been invested in collecting, curating, and making the databases available, the rate limiting factor in their use can be interaction with users, which, as can be seen at the previously cited resources, typically involves a GUI–based selection process or manual download. Users must generate code to translate the data for their research environment and develop ad hoc strategies for the workflow of database access, download, cataloging and analysis, thus raising the barrier to database use and increasing the difficulty of sharing with other researchers. We have argued that automation of the workflow in computational neuro-science investigation can increase the efficiency of research, the preservation of provenance and the reproducibility of results (Stockton and Santamaria, 2016, 2015).
The Allen Brain Institute (ABI) (Grillner et al., 2016) provides and maintains the Cell Types Database (ABI–CTdb) (ABI, 2015a) of cell morphology and electrophysiological properties of mouse brain cells. Although ABI supports manual download like the other databases, they also support automated methods of search, selection, and retrieval, including a Python Software Development Kit (SDK) (ABI, 2017e). In this project we make use of the ABI automated features and develop software tools that facilitate the use of ABI–CTdb data when automating combined simulation and experimental work. We provide infrastructure to generate a local, specialized MySQL (My Structured Query Language) database of the electrophysiological data and extracted features; the user then can employ Python or MATLAB utilities to access the local database. We have also extended our workflow automation software (Stockton and Santamaria, 2016, 2015) to integrate the local database into the modeling/data analysis cycle within a single script. To support the investigation, we use a second MySQL database that associates input parameters with simulations, analyses, and comparisons, thus providing provenance information and post-investigation search capabilities. Overall, we provide tools to download, analyze and interact with the ABI–CTdb, providing flexibility and automation of daily research workflow. Given the standardization of database technology our approach in the ABI–CTdb can be extended to other databases (McDougal et al., 2017).
2 Methods
2.1 Using the Allen Brain Institute Cell Types Database
Public neuroscience databases are becoming a valuable tool in neuroscience (McDougal et al., 2017) and are a core part of the BRAIN Initiative (Bargmann et al., 2014). The ABI–CTdb is a publicly–accessible and actively maintained (ABI, 2017c) set of electrophysiological (ABI, 2015b), imaging (ABI, 2015c), and modeling data (ABI, 2015d, e). The database was gathered with a transparent, methodical protocol, and has a clear Application Programmer’s Interface (API) (ABI, 2017a) and several methods of access, including web page interaction (ABI, 2017b), RESTful1 interactions (ABI, 2017d), and an ABI–supplied Python Software Development Kit (SDK) (ABI, 2017e).
Electrophysiological data can be downloaded and is organized by donor (the animal), specimen (the cell), sweep (a stimulus–recording pair), and experiment (a trimmed version of the sweep that removes a leading subthreshold test pulse and most of the leading prestimulation data). The SDK includes Python code to extract a multitude of features at the cell, sweep, and spike levels (ABI, 2015b).
Our objective was to download data from the ABI–CTdb and construct an investigation–specialized local database called ”CellSurvey”, consisting of cell information, electrophysiological responses, and extracted features. We used the ABI Python SDK in combination with custom code in both Python and MATLAB Transfer to download data, perform feature extraction, and populate the local database.
2.2 Leveraging Python and MATLAB usage
Neuroscience researchers doing simulation and modeling often use Python as a programming medium (Antolík and Davison, 2013; Davison et al., 2009; Hines et al., 2009; Muller et al., 2015; Van Geit et al., 2016), whereas researchers on the experimental side often use MATLAB (Baek et al., 2016; Delorme and Makeig, 2004; Englitz et al., 2013; Felice et al., 2016; Lawhern et al., 2013; Schrouff et al., 2013; Shamlo et al., 2015; Vidaurre et al., 2011); there are exceptions to both as well as hybrids. On ModelDB as of February 28, 2016, there were 247 MATLAB models and 104 Python models hosted or linked to; NEURON models numbered 523 (McDougal et al., 2017). The ABI SDK, however, is written exclusively in Python. We used a combination of Python and MATLAB code to provide access to the ABI–CTdb from both languages. Both MATLAB and Python have a rich collection of utilities, which gives integrators the facilities for handling MySQL, HDF5/NWB, XML, and JSON using Python or MATLAB.2
2.3 Extending NeuroManager towards integrated simulation—analysis management
Our group has developed an object–oriented MATLAB program called NeuroManager which automates the workflow of simulation job submissions when using multiple heterogeneous computational resources (Stockton and Santamaria, 2017, 2016, 2015). Simulation characteristics are embodied in a Simulator, which has as its SimCore a simulator engine such as NEURON, MCell, or custom simulators written in any language (Teka et al., 2014, 2016). The results of the simulations are returned to the NeuroManager host in date/time–stamped directories. NeuroManager’s documentation includes examples that integrate MATLAB and Python. In this project we integrate the ABI Python SDK’s feature extraction code into the Simulator classes so that we extract spike train features remotely post–simulation, gaining the advantage of using the same feature extraction code for both experimental and simulated data. Then we extend NeuroManager with a persistent database to hold all investigation information, including input parameter vectors, simulator metadata, simulation results, and analysis results.
2.4 Using MySQL to build local experiment and simulation databases
A standard strategy in our software is to use intermediate forms that do simple translation between software units, such as XML or JSON files, which create hierarchical data representation within a text file. The standardized, universally–accessible formats provide a common language between software “cultures”. For example, the features extracted remotely are loaded into a JSON file that is downloaded for database insertion.
Another lingua franca is the relational database, a more complex structure than single text–file representations. MySQL (MySQL, 2017a) is an open–source implementation of relational database concepts that uses an extended version of the SQL (Structured Query Language) interface for constructing tables and inserting and retrieving data (ISO, 2017). MySQL allows multiple simultaneous users using a common specialized syntax to add and retrieve data in a controlled way, and has a large support base of compatible software, software utilities, education, and programmer expertise. In addition, both Python and MATLAB have utilities that reduce the amount of work required to work with a MySQL database. Here we use one MySQL database to hold ABI feature extractions, and another to hold all investigation data.
2.4.1 Separate or combined databases?
One of our original design concepts was that the Cell-Survey and Investigation databases should be combined. After all, with the addition of a few more tables, the data used in an investigation and the simulations produced therein could be found in the same place. The combined approach proved problematic and undesirable for many reasons:
In everyday use, there are many builds/rebuilds of the investigation database; in contrast there are few builds of the CellSurvey and they are seldom. Combining the databases would often cause a substantial amount of rework for no good reason.
We are very careful to ensure separation of experimental data and simulated data, and a read–only approach to the experimental data found in the Cell-Survey database helps ensure simulation activities do not corrupt experimental data.
As the number of tables in a database increases, the complexity of formulating a query increases. For practicality of creating queries easily and rapidly, it pays to keep the investigation database as trimmed as possible.
There may be situations and users that would like to utilize the CellSurvey database but not the investigation database. Keeping the two types of data separate enhances the modularity, flexibility, and coverage of the tools.
In general, the natures of the two databases and the feature extractions involved are fundamentally different and the two databases are focused on different purposes. The CellSurvey database is a local projection/reorganization of a subset of the entire ABI data that is intended to be easier to work with/use from the local research viewpoint (especially NeuroManager), and is static for everyday operations. In contrast, the investigation database is fluid; there are many resulting database snapshots and the researcher is continually re-evaluating and redoing the investigation approach. Additionally, the feature extraction represented in the Cell Survey database comes from several ABI sources (multi–sweep features, multi-stimulus features, etc), not just those sweep–limited features extracted using the few Python modules we use for the simulation features. The CellSurvey database feature extraction is intended to be a superset of the simulation feature extraction. The resulting feature extraction workflow for experimental data is different from that used for simulation data. For quality, however, we ensure that the code used to extract features from the simulation data is the same exact code as that used to extract the corresponding features from experimental data.
2.4.2 The MySQL Python Connector
The “MySQL Connector/Python” interface provides the ability to work with a MySQL database from within Python (MySQL, 2017b). The database must have been created previously using the mysql command line or an interface such as MySQL Workbench (MySQL, 2017c). The connector converts parameter values to/from MySQL and Python, provides secure TCP/IP connections, and does not require additional modules or libraries.
2.4.3 The MATLAB Database Toolbox
The MATLAB Database Toolbox (MathWorks, 2017) provides the ability to work with several database types from within MATLAB, including MySQL. For MySQL databases, MATLAB requires the user to have already set up a database (easily done through MySQL Workbench), and a “data source” manually using (on Windows) the Microsoft ODBC Data Source Administrator tool; this is launched by the Database Explorer app of the Database Toolbox.
3 Results
Here we describe our collection of Python, MATLAB, and MySQL tools to interact with the ABI–CTdb. The specific objectives of our work are to a) provide tools in Python and MATLAB to download the ABI–CTdb electrophysiology data; b) generate a local database of the data with ABI feature extractions; c) integrate the feature extraction tools of the ABI Python SDK into our NeuroManager software to provide single–script management of the modeling/data analysis cycle in neuroscience.
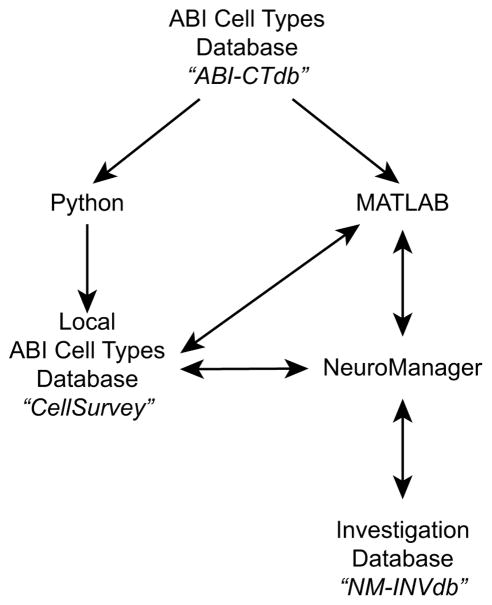
As illustrated in Figure 1, the user can use Python or MATLAB code to access and download the ABI–CTdb’s electrophysiological data and run SDK routines to extract features from the electrophysiological data. The user can also store them in a local database that we call “CellSurvey”, use a new extension of our NeuroManager software to run simulations and extract features from them, then compare the simulated and experimental data. The results of these interactions are stored in a second investigation database called “NM–INVdb”.
Fig. 1. Database and language independence.
The user can use Python or MATLAB code to access and download the ABI–CTdb’s electrophysiological data and run ABI’s Python SDK routines to extract features from the electrophysiological data. The user can also store them in a local MySQL database that we call “CellSurvey”, use a new extension of our NeuroManager software to run simulations and extract features from them, then compare the simulated and experimental data. The results of these interactions are stored in a second investigation database called “NM–INVdb”. For more details please see the User Guide
3.1 MATLAB classes for access to the online ABI Cell Types database
The first of our tools gives direct MATLAB access to the ABI electrophysiological data. We have created the “ABIApiML” package, consisting of classes ABICell-Data, ABISweep, and ABIExperiment. The classes access the cell’s electrophysiology summary, which is contained in a file in Neurodata Without Borders (NWB) format. NWB format (Neurodata Without Borders, 2016; NWB-CN Project, 2015) is a neurophysiology data format based on the HDF5.0 standard (Folk et al., 2011), and these MATLAB classes make use of the MATLAB hdf5 facilities (Mathworks, 2017) for accessing the downloaded file. The NWB file can be downloaded by choosing a cell using ABI’s web–based interface (ABI, 2017b) and pressing a download button manually; by using the ABI Python SDK as seen at ABI (2017e); or as a side effect of using our Python ABICellSurvey tool described in Section 3.2. Once the NWB file has been downloaded, the user can access the data contained therein through use of the ABIApiML classes and process them using MATLAB’s extensive computational facilities, or use the ABI Python SDK and process them using Python’s numerical facilities.
The ABICellData class represents a specific cell (specimen) in the NWB file and gives access to the cell’s webpage, collection information, subject characteristics, lists of experiments and sweeps, and the ability to create ABISweep or ABIExperiment instances from class methods.
The ABISweep class represents a specific sweep in the NWB file and gives access to a subset of the sweep’s information, experimental values such as capacitances, resistances, and time base, as well as the actual data of the sweep including stimulus time series, acquisition time series, and detected spike times.
The ABIExperiment class represents a specific experiment in the NWB file and gives access to the experiment’s basic information, including experiment description, associated sweep number, and start and stop times of the experiment.
A brief example is shown in Figure 2. A full example script is published on GitHub.
Fig. 2. ABIApiML example usage.
The ABIApiML classes give access to the ABI Cell Types electrophysiological data from within MATLAB
3.2 A Python tool to create a local MySQL database of extracted features
For extensive use of the features extracted from the ABI electrophysiological data, we have a Python tool that creates a specialized, local MySQL database of extracted features. A Python module called “ABI Cell Sur-vey” takes as input a list of specimen numbers and the name of a preexisting, possibly blank MySQL database (in this paper we call it the “CellSurvey” database), creates a set of tables that are designed for automated access, then employs the ABI SDK to populate the database with cell/sweep/experiment data and features extracted from the data. The module makes use of the ABI SDK’s CellTypesCache class to download the specimen data only if necessary, which makes the specimen’s NWB data file accessible to the other tools used here. Once the database has been built, the user can access it by issuing SQL commands through MySQL Workbench, through use of the Python or MATLAB database modules, or by using methods in the MATLAB class ABIFeatExtrData (see Section 3.3 below), developed as part of this suite.
The CellSurvey database is primarily designed to present extracted features based on specimen and experiment identifiers, and consists of five tables, named “donors”, “specimens”, “experiments”, “specimenFXs”, and “experimentFXs”. The donors, specimens, and experiments tables are simple and allow access to the sex of the animal that donated the cell specimen, associate the specimens with their experiments, and hold the sampling rate, stimulus type, and stimulus current associated with an experiment. The experimentFXs (experiment–level extracted features) and specimenFXs (specimen–level extracted features) tables hold a selected set of features extracted from the experiment and specimen, respectively, which we describe in detail in the next section. We did not design any statistics not provided by ABI, such as multi–specimen statistics, into the CellSurvey database since the choice of cells is directly related to the user’s investigation activities. Instead, the user would aggregate the statistics as part of an investigation script.
We have selected certain features for our own purposes, but the user is free to adapt the open–source code as necessary to extend the tables with additional ABI features. The CellSurvey database is easily reconstructed; the time required depends on how many specimens the user requests and how many of their nwb files have already been downloaded. More information can be found in the ABI Python SDK Electrophysiology code (ABI, 2017f) and the ABI whitepaper (ABI, 2015b). In addition, ABI (2017g) gives a compact high–level overview of the data.
Experiment–based features
At the experiment/sweep level, features are confined to those extractable from a single voltage waveform. The features we have selected for our purposes include:
number of spikes, if any;
first Inter–Spike Interval (ISI);
average ISI;
ISI coefficient of variation;
latency;
delay, if applicable, and its properties;
adaptation index;
average threshold for all the spikes in the train;
number of bursts, if any, and their properties;
number of pauses, if any, and their properties.
Specimen–based features
At the specimen level are features that require the cell’s full set of sweeps to acquire. The features we have selected for our purposes include:
all sweep–level characteristics of the “hero” sweep (the sweep with long-pulse stimulus at the minimum current level at which spikes first appear);
dendrite type;
average interspike voltage characteristics for each stimulus type such as fast trough voltage and time, slow trough voltage and time, trough voltage and time and sag fraction;
average max voltage value and time for each stimulus type;
average threshold voltage, current, and time for each stimulus type;
upstroke/downstroke ratio for each stimulus type;
average resting membrane voltage;
average input resistance;
and search–facilitating collectives such as has–spikes, has–bursts, and has–delays.
The local CellSurvey database can be accessed in several ways, depending on the user’s needs. The database is easy to browse manually using an interface such as MySQL Workbench using mouse clicks or by entering SQL commands. SQL–based queries also can be sent via database utilities such as the MySqL Python Connector or the MATLAB Database Toolbox. Figure 4 presents two examples of SQL commands that access the CellSurvey database. Finally, in Section 3.3 we describe a simple MATLAB class ABIFeatExtrData that pulls CellSurvey data out into MATLAB structs without the need for SQL.
Fig. 4. CellSurvey Database Queries.
The local MySQL CellSurvey Database holds features extracted from NWB files downloaded from the Allen Brain Institute Database website. We can use standard SQL queries like these to access the database. The INNER JOINs connect tables in the database, allowing disparate parts of the stored data to be accessed in a single query. The table design is straightforward and is easily seen in the code or by browsing the database with MySQL Workbench. Automating the SQL query employment using custom Python or MATLAB functions is straightforward and easily extended; we have provided a MATLAB wrapper class which is described in Section 3.3
3.3 A MATLAB class for access to the CellSurvey database
The class ABIFeatExtrData allows native MATLAB access to the local CellSurvey database instead of requiring MySQL queries such as those shown in the previous section. ABIFeatExtrData methods permit extraction of features based on specimen and experiment number, returning data in the MATLAB data structures called “structs”.
For example, the MATLAB snippet seen in Figure 5 uses ABIFeatExtrData to gather data from the Cell-Survey database about Specimen 484635029/Experiment 65, then inserts them plus other data into the investigation database represented by object invDB. The investigation database will be presented in the next section.
Fig. 5.
MATLAB access of CellSurvey database. The MATLAB class ABIFeatExtrData provides access to the local CellSurvey database of features extracted from the public online ABI Cell Types database
There are always tradeoffs in wrapping a sophisticated technology such as MySQL for a specific, simpler use. The wrapping action can simplify the user’s code and improve speed of adoption. However, it is impossible to foresee the user’s needs and the wrapping can induce strong false limitations in the user’s mind. Accordingly we have kept this class simple — it provides initial easy access, yet provides illustrative examples for the user to add new access methods. Both databases described in this paper use a small, easily learned subset of MySQL, and in our view it is more useful to the researcher to gain some facility with SQL queries than to try to wrap every possible neuroscience–related query and by so doing, induce an even greater learning task.
3.4 NeuroManager extension: Store salient aspects of a neuroscience investigation in a MySQL investigation database
NeuroManager is a research tool for neuroscientists to define models, programmatically adapt them to a set of input parameter vectors, and automatically submit the corresponding simulation jobs on a user–defined grid of heterogeneous computational resources. Extending NeuroManager to handle analyses allows us to place all aspects of a simulation/analysis–based investigation into a local MySQL database for inspection and retrieval. We have added support for an “investigation database” and developed support classes investigationDB and abiCompDB that enable exploring the input parameter space of a simulator in order to best match data from the ABI–CTdb. A session is a single NeuroManager use, which will have a set date/time stamp called the “sessionID”; this id becomes the identifier of the session in the database. A session may involve many serially–run SimSets.3 An investigation is composed of one or more sessions.
The investigation database is used by the NeuroManager host and not by remote computational resources. It stores all input parameter vectors, simulation results, and extracted features in a format that is accessible by users or other programs. Each simulation is stored with its own Simulation ID, its SimSet ID, and its Session ID, thus allowing reconstruction of the order in which the database was built. Full paths to raw data and results are provided in the simulationRuns table, allowing retrieval of simulation raw results as well as specialized plots developed remotely within the subclassed Simulator. The associations between input parameter set and simulation metadata, results, and analyses are preserved through the use of foreign keys.4 At the end of each session the current state of the investigation database is stored in the investigation directory using the session ID and can be reloaded into the database at any time using MATLAB or Python scripts or an application such as MySQL Workbench. At the beginning of a session, reloading of the most recent version of the database in the investigation directory simplifies adding to the database in a series of sessions, such as progressive refinement of parameter search spaces, or an explore stage followed by an exploit stage. Following an investigation, the database state is stored as a *.sql file, which can be shared, archived, or uploaded for publication.
The investigation database is single–simulation–based. Processing based upon multiple simulations, such as Monte Carlo or multi–specimen approaches, can be done through multiple queries or more advanced queries. Incorporating collective results such as these into the investigation database would be highly investigation–specific and thus we have excluded them from the design. Should the user find such an addition advantageous, the open–source code is easily modified and the existing code provides examples of table design/construction and data insertion, extraction and modification.
To fill–out the coverage of a neuroscience investigation, we have included simple parameter search classes and feature set feature ↔ set comparison classes as part of the extension and added comparisons to the investigation database. Except for feature extraction from simulation raw data output, which is done remotely as a part of each Simulator using Python and MATLAB scripts we provide in the tool set, analyses and comparisons are performed directly from the database. Queries of the investigation database can cover nearly any aspect of the investigation, such as (SQL omitted for brevity):
Show the simulation IDs for all simulations that had stimulus type “Long Square” and τ between 30 and 40 msec.
Show the simulation IDs for all simulations run on machine MyServer01.
Show all comparisons involving simulation “Point0010”.
Show comparisons, input parameters τm, α, and threshold, and identifying data for simulations with input parameter α equal to 1.2.
Show the input parameters, specimenID, sweepID, and simulation–identifying data for all comparisons with score1 < 0.15 and score2 < 0.4.
As part of the comparison class, we have included a MATLAB method called “visComparison” that plots the experimental and simulated voltage traces and current stimuli from any set of comparisons in the database. The new code, modifications to existing code, more details on design, and User Guide updates are published on the NeuroManager GitHub website (Stockton and Santamaria, 2017).
The new tools can be combined into a single, flexible MATLAB script in which the user selects ABI specimen/sweep combinations and automatically runs parameter search simulations against them. In this combined script, all experimental data is retrieved from the ABI database, all simulations and feature extractions are handled by NeuroManager on the designated machine set, and all metadata, simulation results, analyses, and comparisons are stored in the investigation database. We have included working example scripts in the User Guide, including one which uses the ABI data in conjunction with a NEURON model from ModelDB (McDougal et al., 2017; Miyasho et al., 2001).
4 Related work
There are several projects that are related to different aspects of our expanded NeuroManager suite, from the automated downloading of electrophysiological data to building databases for sorting and analysis. For example, the NeuroElectro project (Tripathy and Gerkin, 2015; Tripathy et al., 2014) hosts a online database of neuron electrophysiological properties automatically extracted from journal article tables. They provide a web–based interface and a RESTful API for access and contribution. These interface methods are also used by our Python and MATLAB objects that manage downloading, feature extraction, and creation of the CellSurvey database, and so it would be straightforward to expand our tools to accommodate the NeuroElectro resources.
We are aware of at least two portions of code on GitHub that access the ABI database using the ABI Python SDK. The first, called AIBS Cell Types,5 is composed of two iPython Notebook examples of performing accesses of both the ABI and NeuroElectro databases. The accesses seen in AIBS Cell Types are also handled by our CellSurvey database and utilities, which can be used for further modeling or analysis. The second code portion, aibs.py,6 gives access to sweep data and the rheobase sweep id of an ABI specimen through the NeuronUnit framework.7 Our tools automatically create a local MySQL subdatabase of ABI specimen and sweep data including sweep–by–sweep feature extraction, provide automatic access to that database, and enable feature extraction from simulations using the same ABI Python code; all of which are suitable for automated parameter space search using metaheuristic strategies.
Some computational neuroscience projects, including Neuronvisio and neuroConstruct, also tackle questions of automating computational neuroscience work-flow that include external data sources. Neuronvisio (Mattioni et al., 2012) acts as a workflow facilitator between ModelDB and the NEURON simulator, handling model selection, download, and manipulation, as well as results visualization. neuroConstruct (Gleeson et al., 2007), although the software does not connect directly to external libraries, automates workflow associated with the creation of networks of neurons, including extensive facilities for importing neuronal models/data in different formats, such as those output by Neurolucida8 or published on ModelDB.9
Four computational neuroscience projects also use an internal database or similar technology: PANDORA, the Neural Query System (NQS), Sumatra, and Mosaik. PANDORA (Günay, 2007, 2012; Günay et al., 2009) is a MATLAB toolbox that creates a custom database management system which provides commands and objects for accessing and manipulating raw electrophysiology data. In contrast to our work PANDORA does not use an SQL database with MATLAB–SQL layer and the MATLAB Database Toolbox; instead it uses a fully custom database and integrated object-oriented class methods to access database contents. Since PANDORA’s database is stored in MATLAB objects, there can be memory limitations; Günay et al. (2009) used an external MySQL database to hold electrophysiological data in bulk for transfer to PANDORA in chunks.
The NQS (Lytton, 2006) is a relational database tool for the NEURON simulator that embodies a subset of MySQL functionality, supplying an extension of the hoc language to create tables, insert data, and perform queries in a role similar to that of NeuroManager’s investigation database. NQS is compiled into the NEURON simulator as a module.
Sumatra (Davison, 2012) makes use of a local file repository managed by a version control system like Git (Git, 2017; Chacon, 2014) together with record stores handling by database functionality such as SQLite10 to keep track of both the user’s code versions and the data/metadata associated with simulations performed under the Sumatra capture mechanism. Sumatra uses an abstraction layer so that the user can use multiple types of database backend. The combination of the two allows the user to access the commit values for code that is submitted to the repository, enabling fine level provenance of each simulation. The computation record is available for later use and by other users.
Mosaik (Antolík and Davison, 2013) uses a datastore for holding raw data and simulation results and to support some query types. Mosaik preserves the datastore as an investigation product and the user can run additional analysis or visualization on the data therein. Although Mosaik uses lists of objects to form the datastore instead of formal database technology, the authors plan to use such in future work.
5 Discussion
In this work we presented a collection of tools to download, catalog, and extract data from the ABI-CTdb. In order to expand the potential user base we developed tools in Matlab and Python, the most used environments in neuroscience. We integrated this effort into our NeuroManager workflow automation software, allowing us to use the ABI–CTdb in the data analysis/simulation cycle while providing access to heterogeneous computational infrastructure, including local servers, high–performance computing clusters, and multiple cloud services. The existence of the ABI Python SDK provided us a framework for database interaction as well as the use of the SDK routines for extracting features from the electrophysiological data. The ability to combine all operations into a single MATLAB script simplifies the development of the workflow, clarifies the script, and enables collaboration and replication. Overall, our work provides a framework to incorporate other databases into automated simulation and data analysis workflow.
Since analyses and comparisons can be performed directly from the investigation database itself, the overall process has a modularity not seen in other approaches. This modularity facilitates 1) individual inspection, verification, and publication of simulation work and analyses; 2) redo of analyses without redoing simulations; 3) post–simulation reexamination of hypotheses; 4) investigation of multiple, possibly conflicting hypotheses independently and simultaneously; and post–investigation additions to the database. Even a manual exploration is facilitated by the database approach. For example, it is easy to use MySQL Workbench to create and submit a query, get the response within the Workbench as a table, then sort the table by clicking on the field titles. The investigation database gives flexibility and power in accessing the results of many simulations, either manually, automatically using a database connector or MATLAB toolbox, or through the use of the tools provided here. The approach is directly aligned with “Establish Platforms for Sharing Data” and “Validate and Disseminate Technology”, which are core principles of the BRAIN Initiative (Bargmann et al., 2014).
The use of standard database technology rather than custom software gives us immediate sophisticated database functionality, tested and maintained by external developers, together with substantial support software, easily obtained education, and substantial available expertise. This approach reduces our computational burden, provides protection against obsolescence, and enhances reproducibility and preservation of provenance.
The ABI’s substantial facilities for automatic interaction (website, RESTful interactions, and a Python SDK) make the CellTypes database attractive for use by automated workflows that can employ them. Recently, the Neuroscience Gateway (NSG, 2017a) also added RESTful support (NSG, 2017b). Although we used RESTful interactions minimally in the present project (instead we used the ABI SDK), we used them extensively in a previous work for accessing cloud resources (Stockton and Santamaria, 2016), and neuro-science databases may show increased public usage should they provide these types of interaction facilities.
Fig. 3. Creation of CellSurvey database.
The creation of the CellSurvey MySQL database from a list of desired specimens. The CSdbconfig module keeps user details out of the creation script. If a database called by the string held by databaseName does not exist, the module will create one. The manifest file is a JSON–format tool created and used by the ABI SDK for keeping track of which nwb files have been downloaded. resetDB is a flag which, if true, instructs the routine to clear all database tables and start again. verbose, if true, prints out a lot of supporting text to add in troubleshooting
Acknowledgments
National Science Foundation (NSF-DBI1451032), National Institutes of Health (NIH-G12MD007591) (for use of computational facilities at UTSA).
Footnotes
REST: Representational State
HDF5: Hierarchical Data Format Version 5; NWB: Neurodata Without Borders; XML: Extended Meta Language; JSON: Javascript Object Notation
A SimSet is a set of input parameter vectors, each of which is associated with a single simulation. A SimSet may represent many simulations, which will be scheduled to run in parallel on the Simulators in the Simulator Pool. Please see the NeuroManager documents for more details.
Each row in a table has a unique key; a foreign key in a row associates that row with a row in another table. In this way tables support the various characteristics of a thing in a database.
6 Conflict of Interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
7 Information Sharing Statement
The NeuroManager software, ABI Tools software, User Guides, and examples are available at GitHub11,12,13 under a UTSA open source license that permits free use for research purposes. MATLAB commercial software is available online,14 including student versions. Other software mentioned in this paper is freely available.
Contributor Information
David B. Stockton, Department of Biomedical Engineering, The University of Texas at San Antonio, San Antonio, TX 78249, USA
Fidel Santamaria, Department of Biology, The University of Texas at San Antonio, San Antonio, TX 78249, USA.
References
- Git Website. 2017 URL https://git-scm.com/
- ABI. Technical report. Allen Brain Institute; 2015a. Allen Cell Types Database - Overview. URL http://help.brain-map.org/download/attachments/8323525/CellTypesOverview.pdf?version=1&modificationDate=1456188760121. [Google Scholar]
- ABI. Technical report. Allen Brain Institute; 2015b. Allen Cell Types Database - Electrophysiology. URL http://help.brain-map.org/download/attachments/8323525/EphysOverview.pdf?version=1&modificationDate=1456188786670. [Google Scholar]
- ABI. Technical report. Allen Brain Institute; 2015c. Allen Cell Types Database - Morphology. URL http://help.brain-map.org/download/attachments/8323525/MorphOverview.pdf?version=1&modificationDate=1456525256645. [Google Scholar]
- ABI. Technical report. Allen Brain Institute; 2015d. Allen Cell Types Database - GLIF Models. URL http://help.brain-map.org/download/attachments/8323525/GLIFModels.pdf?version=1&modificationDate=1456188812960. [Google Scholar]
- ABI. Technical report. Allen Brain Institute; 2015e. Allen Cell Types Database - Biophysical Modeling - Perisomatic. URL http://help.brain-map.org/download/attachments/8323525/BiophysModelPeri.pdf?version=1&modificationDate=1456188760131. [Google Scholar]
- ABI. Allen Brain Institute Cell Types Database Application Programmer’s Interface. 2017a URL http://help.brain-map.org/display/celltypes/API.
- ABI. Allen Brain Institute Cell Types Webpage. 2017b doi: 10.1007/s12021-017-9337-x. URL http:/celltypes.brain-map.org. [DOI] [PMC free article] [PubMed]
- ABI. Allen Brain Atlas Portal - News and Updates. 2017c URL http://www.brain-map.org/announcements/index.
- ABI. Allen Brain Institute RESTful Model Access (RMA) 2017d URL http://help.brain-map.org/pages/viewpage.action?pageId=5308449.
- ABI. Allen Brain Institute Allen Brain Atlas Software Development Kit. 2017e URL http://alleninstitute.github.io/AllenSDK/
- ABI. Allen Brain Institute Software Development Kit Ephys Code Webpage. 2017f URL http://alleninstitute.github.io/AllenSDK/allensdk.ephys.html.
- ABI. Allen Brain Institute SDK Ephys Features. 2017g URL http://help.brain-map.org/display/celltypes/API#API-ephys_features.
- Antolík Ján, Davison Andrew P. Integrated workflows for spiking neuronal network simulations. Frontiers in Neuroinformatics. 2013;7(34):1–15. doi: 10.3389/fninf.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Brain Imaging Data Exchange. Autism Brain Imaging Data Exchange I – ABIDE I. 2017 URL http://fcon_1000.projects.nitrc.org/indi/abide/abide_I.html.
- Baek Kwangyeol, Shim Woo Hyun, Jeong Jaeseung, Radhakrishnan Harsha, Rosen Bruce R, Boas David, Frances-chini Maria, Biswal Bharat B, Kim Young R. Layer-specific interhemispheric functional connectivity in the somatosensory cortex of rats: resting state electrophysiology and fMRI studies. Brain Structure and Function. 2016;221(5):2801–2815. doi: 10.1007/s00429-015-1073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann Cornelia, Newsome William, Anderson A, Brown E, Deisseroth K, Donoghue J, MacLeish P, Marder E, Normann R, Sanes J, et al. Brain 2025: a scientific vision. Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Working Group Report to the Advisory Committee to the Director, NIH. 2014 URL https://www.braininitiative.nih.gov/2025/
- Chacon Scott. Pro Git 2. 2. Apress; 2014. [Google Scholar]
- Davison Andrew. Automated capture of experiment context for easier reproducibility in computational research. Computing in Science & Engineering. 2012;14(4):48–56. [Google Scholar]
- Davison Andrew P, Hines Michael L, Muller Eilif. Trends in programming languages for neuroscience simulations. Frontiers in Neuroscience. 2009;3(3):374–380. doi: 10.3389/neuro.01.036.2009. URL http://dx.doi.org/10.3389/neuro.01.036.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme Arnaud, Makeig Scott. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. Journal of Neuroscience Methods. 2004 Mar;134(1):9–21. doi: 10.1016/j.jneumeth.2003.10.009. URL http://dx.doi.org/10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Englitz Bernhard, Sorenson Mike D, Shamma Shihab A. MANTA — an open-source, high density electrophysiology recording suite for MATLAB. Frontiers in Neural. Circuits. 2013;7:69. doi: 10.3389/fncir.2013.00069. URL http://journal.frontiersin.org/article/10.3389/fncir.2013.00069/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice Carmelo J, Albarracín Ana L, Farfán Fernando D, Coletti Marcos A, Teruya Pablo Y. Electrophysiology for biomedical engineering students. Advances in Physiology Education. 2016;40:402–409. doi: 10.1152/advan.00073.2015. [DOI] [PubMed] [Google Scholar]
- Folk Mike, Heber Gerd, Koziol Quincey, Pourmal Elena, Robinson Dana. An overview of the HDF5 technology suite and its applications. Proceedings of the. EDBT/ICDT 2011 Workshop on Array Databases AD ’11, 36–47; New York, NY, USA. ACM; 2011. URL. http://doi.acm.org/10.1145/1966895.1966900. [DOI] [Google Scholar]
- Fox Peter, Laird Angie. BrainMap Website. 2017 URL http://brainmap.org/
- George Mason University. NeuroMorpho.Org. 2017 URL http://neuromorpho.org/index.jsp.
- Gleeson Padraig, Steuber Volker, Angus Silver R. neuroConstruct: A tool for modeling networks of neurons in 3d space. Neuron. 2007;54(2):219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner Sten, Ip Nancy, Koch Christof, Koroshetz Walter, Okano Hideyuki, Polachek Miri, Poo Muming, Sejnowski Terrence J. Worldwide initiatives to advance brain research. Nature Neuroscience. 2016;19(9):1118–1122. doi: 10.1038/nn.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günay Cengiz. PANDORA Neural Analysis Toolbox. 2007 URL https://senselab.med.yale.edu/simtooldb/
- Günay Cengiz. Plotting and Analysis for Neural Database Oriented Research Applications (PANDORA) Toolbox Users and Programmers Manual Rev 1293. 2012 URL https://senselab.med.yale.edu/SimToolDB/showTool.cshtml?tool=112112&.
- Günay Cengiz, Edgerton Jeremy R, Li Su, Sangrey Thomas, Prinz Astrid A, Jaeger Dieter. Database analysis of simulated and recorded electrophysiological datasets with PANDORA’s toolbox. Neuroinformatics. 2009 Jun;7(2):93–111. doi: 10.1007/s12021-009-9048-z. URL http://dx.html.doi.org/10.1007/s12021-009-9048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines Michael L, Davison Andrew P, Muller Eilif. NEURON and Python. Frontiers in Neuroinformatics. 2009;3(1) doi: 10.3389/neuro.11.001.2009. URL http://dx.doi.org/10.3389/neuro.11.001.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Neuroinformatics Coordinating Facility. INCF Website. 2017 URL https://www.incf.org/
- ISO. ISO/IEC 9075-X:2016 SQL Standards. 2017 URL https://www.iso.org/advanced-search/x/title/status/P/docNumber/9075.
- Lawhern Vernon, David Hairston W, Robbins Kay. DETECT: A MATLAB toolbox for event detection and identification in time series, with applications to artifact detection in EEG signals. PLOS ONE. 2013;8(4) doi: 10.1371/journal.pone.0062944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytton William W. Neural query system. Neuroinformatics. 2006;4(2):163–175. doi: 10.1385/NI:4:2:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathworks. MATLAB HDF5 Files Webpage. 2017 URL https://www.mathworks.com/help/matlab/hdf5-files.html.
- MathWorks. MATLAB Database Toolbox. 2017 URL https://www.mathworks.com/products/database.html.
- Mattioni Michele, Cohen Uri, Le Novere Nicolas. Neuronvisio: a graphical user interface with 3d capabilities for neuron. Frontiers in Neuroinformatics. 2012;6(20) doi: 10.3389/fninf.2012.00020. URL http://www.frontiersin.org/neuroinformatics/10.3389/fninf.2015.00024/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal Robert A, Morse Thomas M, Carnevale Ted, Marenco Luis, Wang Rixin, Migliore Michele, Miller Perry L, Shepherd Gordon M, Hines Michael L. Twenty years of ModelDB and beyond: building essential modeling tools for the future of neuroscience. Journal of Computational Neuroscience. 2017;42(1):1–10. doi: 10.1007/s10827-016-0623-7. URL http://dx.doi.org/10.1007/s10827-016-0623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasho T, Takagi H, Suzuki H, Watanabe S, Inoue M, Kudo Y, Miyakawa H. Low-threshold potassium channels and a low-threshold calcium channel regulate Ca2+ spike firing in the dendrites of cerebellar Purkinje neurons: a modeling study. Brain Research. 2001 Feb;891(1–2):106–115. doi: 10.1016/s0006-8993(00)03206-6. [DOI] [PubMed] [Google Scholar]
- Muller Eilif, Bednar James A, Diesmann Markus, Gewaltig Marc-Oliver, Hines Michael, Davison Andrew P. Python in neuroscience. Frontiers in Neuroinformatics. 2015;9:11. doi: 10.3389/fninf.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MySQL. MySQL website. 2017a URL https://www.mysql.com/
- MySQL. MySQL Connector/Python Developer Guide. 2017b URL https://dev.mysql.com/doc/connector-python/en/
- MySQL. MySQL Workbench. 2017c URL https://www.mysql.com/products/workbench/
- Neurodata Without Borders. NWB File Format Specification Version 1.0.3. 2016 URL https://github.com/NeurodataWithoutBorders/specification.
- NSG. Neuroscience Gateway Website. 2017a URL https://www.nsgportal.org/
- NSG. NSG REST Api (NSG-R) Website. 2017b URL https://www.nsgportal.org/guide.html.
- NWB-CN Project. Neurodata Without Borders —Computational Neuroscience Project. 2015 URL http://crcns.org/NWB.
- Schrouff J, Rosa MJ, Rondina JM, Marquand AF, Chu C, Ashburner J, Phillips C, Richiardi J, Mouro-Miranda J. PRoNTo: Pattern recognition for neuroimaging toolbox. Neuroinformatics. 2013 Jul;11(3):319–337. doi: 10.1007/s12021-013-9178-1. URL http://dx.doi.org/10.1007/s12021-013-9178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenseLab. ModelDB Website. 2017 URL https://senselab.med.yale.edu/ModelDB/default.cshtml.
- Shamlo Nima, Mullen Timothy, Kothe Christian, Su Kyung Min, Robbins Kay A. The PREP Pipeline: Standardized preprocessing for large-scale EEG analysis. Frontiers in Neuroinformatics. 2015;9(16) doi: 10.3389/fninf.2015.00016. URL http://www.frontiersin.org/neuroinformatics/10.3389/fninf.2015.00016/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton David, Santamaria Fidel. NeuroManager Website. 2017 URL https://github.com/SantamariaLab/NeuroManager.
- Stockton David B, Santamaria Fidel. Automating NEURON simulation deployment in cloud resources. Neuroinformatics. 2016 Sep; doi: 10.1007/s12021-016-9315-8. URL http://dx.doi.org/10.1007/s12021-016-9315-8. [DOI] [PMC free article] [PubMed]
- Stockton David Bruce, Santamaria Fidel. NeuroManager: A workflow analysis based simulation management engine for computational neuroscience. Frontiers in Neuroinformatics. 2015;9(24) doi: 10.3389/fninf.2015.00024. URL http://www.frontiersin.org/neuroinformatics/10.3389/fninf.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teka Wondimu, Marinov Toma M, Santamaria Fidel. Neuronal spike timing adaptation described with a fractional leaky integrate-and-fire model. PLoS Computational Biology. 2014 Mar;10(3):e1003526. doi: 10.1371/journal.pcbi.1003526. http://dx.doi.org/10.1371/journal.pcbi.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teka Wondimu, Stockton David Bruce, Santamaria Fidel. Power-law dynamics of membrane conductances increase spiking diversity in a Hodgkin–Huxley model. PLoS Computational Biology. 2016;12(3) doi: 10.1371/journal.pcbi.1004776. URL http://dx.doi.org/10.1371/journal.pcbi.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy Shreejoy J, Gerkin Richard C. NeuroElectro Project, 1915–1916. Springer New York; New York, NY: 2015. URL http://dx.doi.org/10.1007/978-1-4614-6675-8_477. [DOI] [Google Scholar]
- Tripathy Shreejoy J, Savitskaya Judith, Burton Shawn D, Urban Nathaniel N, Gerkin Richard C. Neuroelectro: a window to the world’s neuron electrophysiology data. Frontiers in neuroinformatics. 2014;8 doi: 10.3389/fninf.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Geit Werner, Gevaert Michael, Chindemi Giuseppe, Rössert Christian, Courcol Jean-Denis, Muller Eilif B, Schürmann Felix, Segev Idan, Markram Henry. BluePyOpt: Leveraging open source software and cloud infrastructure to optimise model parameters in neuroscience. Frontiers in Neuroinformatics. 2016;10 doi: 10.3389/fninf.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre Carmen, Sander Tilmann H, Schlögl Alois. BioSig: the free and open source software library for biomedical signal processing. Computational Intelligence and Neuroscience. 2011 doi: 10.1155/2011/935364. [DOI] [PMC free article] [PubMed] [Google Scholar]