Abstract
Rapidly proliferating tumors are exposed to a hypoxic microenvironment because of their density, high metabolic consumption, and interruptions in blood flow because of immature angiogenesis. Cellular responses to hypoxia promote highly malignant and metastatic behavior, as well as a chemotherapy-resistant state. To better understand the complex relationships between hypoxic adaptations and cancer progression, we studied the dynamic proteome responses of glioblastoma cells exposed to hypoxia via an innovative approach: quantification of newly synthesized proteins using heavy stable-isotope arginine labeling combined with accurate assessment of cell replication by quantification of the light/heavy arginine ratio of peptides in histone H4. We found that hypoxia affects cancer cells in multiple intertwined ways: inflammation, typically with over-expressed glucose transporter (GLUT1), DUSP4/MKP2, and RelA proteins; a metabolic adaptation with overexpression of all glycolytic pathway enzymes for pyruvate/lactate synthesis; and the EMT (epithelial-mesenchymal transition) and cancer stem cell (CSC) renewal with characteristic morphological changes and mesenchymal/CSC protein expression profiles. For the first time, we identified the vitamin B12 transporter protein TCN2, which is essential for one-carbon metabolism, as being significantly downregulated. Further, we found, by knockdown and overexpression experiments, that TCN2 plays an important role in controlling cancer cell transformation toward the highly aggressive mesenchymal/CSC stage; low expression of TCN2 has an effect similar to hypoxia, whereas high expression of TCN2 can reverse it. We conclude that hypoxia induces sequential metabolic responses of one-carbon metabolism in tumor cells. Our mass spectrometry data are available via ProteomeXchange with identifiers PXD005487 (TMT-labeling) and PXD007280 (label-free).
Hypoxia is a pervasive microenvironmental cellular stressor that plays a critical role in tissue inflammation and malignancy (1). The cellular response to hypoxia is mediated predominantly through hypoxia-inducible factor 1 (HIF-1α)1, a basic helix-loop-helix transcription factor that forms a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT, or HIF-1β) (2). Hypoxia promotes transformed cells to acquire invasive and mesenchymal characteristics, referred to as “type III” epithelial-mesenchymal transition (EMT) (3–10), which plays essential roles in cancer pathogenesis, including that of glioblastoma (11, 12).
Cells exposed to hypoxic stress respond with complex metabolic and transcriptional adaptation mechanisms (13). Hypoxia induces deficient mitochondria redox-oxidation cycles normally needed for energy production (14, 15). To compensate for this deficiency, anaerobic cells metabolize citric acid cycle intermediates and glucose, an adaptation historically known as the Warburg effect (16, 17). Mechanistically, HIF-1α activates transcription of the glucose transporter (GLUT)-1/3 (15, 18–21) and other major glycolytic pathway enzymes, while downregulating energy-consuming genes involved in DNA transcription and RNA translation, resulting in adaptive cell-cycle arrest (2). However, the proteome and metabolome of cells adapted to hypoxia have not been fully evaluated. In this study, we analyzed the hypoxic proteome by tandem mass tag (TMT) and label-free LC-MS/MS in U87 glioblastoma cells exposed for 5 days to hypoxia (1% O2) versus normoxic U87 cells. In addition to confirming the above-described pathways known to be regulated by hypoxia, we made the novel observation that hypoxia significantly downregulates the vitamin B12 transporter protein TCN2, which controls Met synthesis through the one-carbon metabolic pathway, resulting in a folate trap and arrest of cell replication. We have innovatively adapted the SILAC technique to TMT and label-free proteomics and the quantification of proteins in replicating cells in which light arginine (LR) was incorporated into the proteins, so that the effects from preexisting proteome of old cells, in which the arginines in proteins were originally labeled with heavy isotopes (13C615N4-arginine, denoted HR), can be minimized. An additional benefit of quantification starting with HR-labeled cells is that the cell proliferation rates can be readily obtained by calculation of the LR/HR ratios when proteins/peptides are analyzed by LC-MS.
In response to hypoxia, cells transition from an epithelial phenotype to a type III EMT mesenchymal phenotype consistent with an enhanced inflammatory and aggressive cancer status with cancer stem cell (CSC) properties. Hypoxia-induced inflammation and EMT seems to be cancer cell type- independent; they also occurred in glioblastoma U251 cells and nonsmall cell lung cancer (NSCLC) A549 cells among the few chosen for this study. We have demonstrated by knock-down and overexpression experiments that TCN2 plays an important role in regulating EMT and CSC transformations; a low level of TCN2 creates a phenotype similar to that of hypoxic cells with EMT and CSC properties, whereas high levels of TCN can reverse it, suggesting that TCN2 might be a potential activation target for the treatment of cancers. The role of TCN2 downregulation in hypoxia cells suggests that a blockage of the one-carbon metabolic pathway be induced by hypoxia, resulting in diminished consumption of Met essential for RNA and protein syntheses, but significant accumulation of cellular Ser and Gly. Because the cofactor of DNA/histone methyltransferases, S-adenosylmethionine (SAM), can be synthesized from Met through the Met cycle which is conjugated with the folate cycle to comprise the one-carbon metabolic pathway, studies on the biological mechanisms of cancer hypoxia should include metabolic regulation of the epigenome.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The purpose of this project was to identify the proteome under the stress of oxygen depletion in glioblastoma cells. We chose two typical glioblastoma cell lines, U87 and U251, for this study. Protein quantification was performed by both TMT-labeling and label-free (LFQ) approaches. The cell lysate from U87 cells was used for proteomics analysis and the cell lysates from both U87 and U251 cells were used for Westernblot analysis to verify the expression changes of selective proteins that were determined by proteomics as playing significant roles in identified cellular pathways. Served as two biological replicates, two sets of plates of U87 cells which were derived from the same originally seeding SILAC-modified cells were concurrently cultured; each set contains one plate (75-cm2) of control cells growing under normoxia (20% O2 in the incubator chamber) and five plates of “treated” cells growing under hypoxia (1% O2 in the incubator chamber) for varying duration from 1 up to 5 days. Typically, one plate of cells when harvested contained about 5 - 20 × 106 cells from which about 0.5 to 2 mg whole cell proteins were isolated. For TMT-labeling experiments, two analytical repeats (two separated preparation of TMT labeling of peptides followed by strong-cation-exchanging fractioning) were performed and at least three technical repeats (LC-MS/MS runs) were carried out on the QExactive instrument. For LFQ, the protein lysates from TMT-labeling were combined and then, after digestion, analyzed by three sequential LC-MS/MS runs (technical repeats). Protein identification and quantification was carried out by Protein Discoveror (PD) 1.4 using the intensities of the reporter ions of TMT-labeling peptides or by PEAKS using TIC peak areas of non-labeled peptides. Statistical analysis including FDR and p- or q-value calculation to determine the significance of protein identification and quantification was provided by PD and PEAKS; principal component analysis (PCA) and unsupervised hierarchical clustering analysis with heat map to group protein expression profiles, correlation analysis to evaluate the reproducibility between two biological repeats (sets) of proteomic data, were realized by the R programs. In addition, z-scores in IPA were used to predict activation or inhibition of targeting molecules or disease inclination and FDRs in STRING to determine significant KEGG pathways. Details in methods and procedures of statistical analysis are described in the following experimental subsection-Proteomics procedures.
Cell Culture and Treatment
U87 cells labeled with stable isotope-labeled (HR) arginine were prepared as described previously to ensure ∼ 98% incorporation of HR as determined by mass spectrometric analysis of histone peptides (22). The HR-enriched cells were stored in a liquid nitrogen tank until use. In this experiment, HR-enriched U87 cells were first cultured in Thermo Scientific SILAC MEM media (Ca# 88422, Grand Island, NY) minus l-lysine and l-arginine, 10% sera (Ca# 88440), and 1% antibiotic-antimycotic solution (Corning cellgroTM, Ca# 30-004-CI, Manassas, VA) for SILAC. The media were supplemented with 0.084 g/L HR (Thermo, Ca# 88434) and 0.146 g/L l-lysine (Sigma, Ca# L5501, St. Louis, MO). The cells were split into 12 NunclonTM cell culture dishes (∼2 × 106 cells in each 35 × 10 mm dish, Sigma Ca# D7804) when they reached confluence in the HR media. Then, they were grown in 12 ml regular (LR) EMEM (ATCC® 30–2003TM) with 10% fetal bovine serum (Sigma, Ca# F2442) and 1% Corning cellgro at 37 °C under either normoxic or hypoxic conditions. Twenty-four hours after seeding and every 24 h thereafter, two dishes of cells were moved to an incubator equilibrated with 1% O2 and 5% CO2 in N2 (Galaxy 170R incubator, New Brunswick, Eppendorf, VDR, Radnor, PA) until the last 2 dishes remained in normal atmosphere (20% O2 and 5% CO2) for an additional 24 h, which were treated as the normoxic cells and labeled in a reverse order as d0. The first two dishes taken from the normoxic incubator chamber to the hypoxic chamber were labeled as d5 because they had been grown in low oxygen for 5 days (d); the other dishes were labeled sequentially in decreasing order based on the number of days they were exposed to hypoxia. During this six-day period, the medium was changed once (on d3), and cells in all the dishes grew to fully or close to confluent by d6 except for the d5 hypoxic cells.
Examination of Cell Morphology
The cell morphology was examined under a phase contrast microscope, EVOS XL Core Cell Imaging System (Life Technologies). With Cell morphological changes were quantitatively measured by shape factors using the free open-source bio-image processing software ImageJ and Fiji (23); we typically used the aspect ratio (AR), a function of the largest diameter (dmax) and the smallest diameter (dmin) orthogonal to it (AR = dmin/dmax), and the circularity (fcirc), a function of the perimeter (p) and the area (A) (Fcirc = 4πA/p2). Normally, 50–100 cells from up to 10 randomly selected fields were measured. Elongated cells characteristic of EMT generally have small AR and large fcirc.
Proteomics Procedures
Sample Preparation
The cell pellet was lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Santa Cruz Biotechnology, Dallas, TX) supplemented with 1% Nonidet P40, PMSF (0.2 mm), and protease inhibitor mixture (Roche, one tablet per 10 ml). Samples were sonicated with a probe sonicator (Qsonica, model CL-188; 3 × 30 bursts at 40 Hz amplitude) and incubated on ice for 2 h. The cell lysate was centrifuged and the supernatant transferred into a clean tube. The protein concentration in the supernatant was determined by bicinchoninic acid assay (BCA). An aliquot of supernatant containing 300 μg of protein was treated by the addition of 200 mm TCEP solution (sigma) to a final TCEP concentration of 20 mm and then incubated in votex mixer at 50 °C for 30 min, followed by carboxymethylation with 25 mm iodoacetamide (final concentration) in the dark for 1 h. Four volumes of precooled acetone (−20 °C) was added to precipitate proteins at −20 °C overnight. The proteins were pelleted by centrifugation at 13,000 × g for 10 min and washed once with 1 ml precooled (−20 °C) acetone. After being air-dried overnight, the proteins were resuspended in 25 mm triethylammonium bicarbonate buffer (pH 7.8) and digested with trypsin (1 μg, Sigma) for ∼ 4 h till the solution became clear. The protein concentrations were measured again by BCA. About 60 μg of each protein sample was aliquoted and digested with trypsin at an enzyme ratio of 25:1 (by mass) overnight at 37 °C. To each digest, 0.8 mg of TMT-6 (Thermo-Fisher Scientific) reagent was added. After incubation for 2 h, 20 μl of 5% hydroxylamine was added to stop the labeling reaction. After all the samples labeled with individual TMT-6 reagents were combined, the solution was acidified with 1% formic acid (FA) and fractionated into six fractions with Thermo's strong cation-exchange (SCX) spin-columns with a KCl gradient from 0 to 0.5 m in 20% acetonitrile (ACN). The last elution solution was 5% ammonia hydroxide. Next, the fractions were dried under vacuum to remove ACN and then desalted by Thermo spin-columns with HypercarbTM. The ammonia-eluted fraction was acidified by 1% formic acid (FA) before desalting, and 0.8 ml of 60% ACN was used to elute peptides from the desalting column for each sample. The eluted peptides were vacuum-dried, reconstituted in 30 μl of 0.1% FA, and then subjected to LC-MS/MS analysis.
Mass Spectrometry Analysis
LC-MS/MS analysis was carried out on the Thermo QExactive mass spectrometer. Peptides were separated by online reverse-phase liquid chromatography (RPLC) using home-packed C18 capillary columns (15 cm long, 75 μm i.d., 3-μm particle size) with a 250-min gradient (solvent A, 0.1% FA in water; solvent B, 0.1% FA in ACN) from 5–30% solvent B. Approximately 2 μg of peptide sample was injected. The Orbitrap mass analyzer was set to acquire data at 35,000 resolution with Full Width at Half Maximum (FWHM) for the parent full-scan mass spectrum followed by data-dependent high collision-energy dissociation (HCD) MS/MS spectra for the top 15 most abundant ions acquired at 7500 resolution. For LFQ proteomics, the peptide digests without TMT labeling were analyzed by LC-MS/MS with the same instrumental acquisition settings except much shorter HPLC gradient, i.e. 100 min, was used.
Data Analysis
1. TMT-labeling Proteomics
Proteins were identified and quantified through the Proteome Discoverer 1.4 platform (Thermo) using Sequest HT (employing the Swiss-Prot Homo Sapiens database, release date 7/2015, containing 45,391 entries). The Sequest search parameters were: carbamidomethylation of cysteine and TMT-6 modification of peptide N terminus and lysine were set as fixed modifications and oxidation of methionine and deamination of asparagine and glutamine were set as variable modifications; trypsin was the protease selected and up to two missed cleavages were allowed. Mass tolerance for the precursor ions was 10 ppm, and for the MS/MS, 0.1 Da. Only peptides with a minimum length of four amino acids were considered, and peptides were filtered for a maximum false discovery rate (FDR) of 1%. PD1.4 searched the concatenation of the Homo sapiens database and a decoy database with the same size. FDR were estimated by the ratio of decoy # hits over target # hits among the total # hits, A.K. A., peptide spectrum matches (PSMs). At least one unique peptide with posterior error probability of less than 0.05 was accepted for quantification using the TMT reporter ions. The TMT proteomic raw and processed data have been deposited to the ProteomeXchange Consortium via the PRIDE (24) partner repository with the data set identifier PXD005487.
2. LFQ Proteomics
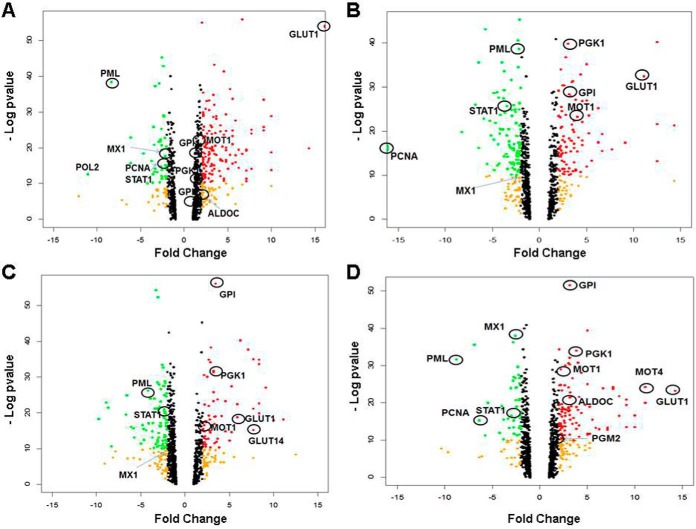
Proteins were identified by PEAK® DB to perform De Novo sequencing assisted database search against the Uniprot Homo Sapiens database with 88,760 entries of protein sequences (release date: 09/2014). Carbamidomethylation of cysteine was set as fixed modifications and oxidation of methionine and deamination of asparagine and glutamine were set as variable modifications. FDR were estimated by the ratio of decoy # hits over target # hits among PSMs. Maximum allowed FDR is 1%. Quantification of proteins was carried out by PEAKS® Q using peak areas in mass chromatograph (TIC) with retention time alignment and isotope summation. Significance of quantification (- 10xLog p value) was calculated based on three technical replicates and its relationship with the ratio or converted fold change of protein expression derived from the summation of the peak intensities of unique peptides (> = 1) was visualized by volcano graphics (Fig. 3). The LFQ QExactive-raw and PEEKS-peptides.pep (XML) data have been deposited to the ProteomeXchange with the data set identifier PXD007280.
Fig. 3.
Volcano plots of data from label free quantification of proteins. Dots representing proteins with fold change (compared with normoxia) > ± 2 expression are colored in orange; fold change > 2 and significance [- Log10(p value)] >10 are in red; fold change < 2 and significance >10 are in green. The upregulation zone included proteins involved in glycolysis and transportation of glucose and lactate; the downregulation zone included proteins involved in tumor suppression, anti-inflammation and replication, which are selectively circled and named. A-D, Hypoxia for 1–4 days.
Statistical Analysis
Multivariate statistics including hierarchical agglomerative clustering analysis, principal component analysis (PCA), and correlation coefficient analysis and their associated presentation graphics were performed in R, version Window Rx64, 3.4.1, as described previously (25) and in the following Results section. Ingenuity Pathway Analysis (IPA) was used to identify protein functions/pathways or upstream regulators significantly influenced by hypoxia. STRING was used for protein-protein interaction pathways and networks (PPIPNs) analysis using the list of proteins showing significant changes in expression under hypoxia. Significant STRING-predicted PPIPNs (FDR < 0.05) were visualized with Cytoscape 3.5.1 (26). Histograms of the correlation coefficients were calculated and visualized using the R program (Windows Rx64, 3.4.1) containing the custom-downloadable package “PerformanceAnalytics” (https://cran.r-project.org/web/packages/PerformanceAnalytics/PerformanceAnalytics.pdf) and its required subpackages “xts” (https://cran.r-project.org/web/packages/xts/vignettes/xts.pdf) and “zoo” (https://cran.r-project.org/web/packages/zoo/zoo.pdf). Calculation of q-values and local FDR (lfdr) from p value given by PEAKS 8 label-free quantification was based on the Benjamini and Hochberg method (27) using Window Rx64, 3.4.1containing the package qvalue (http://bioconductor.org/bioclite.R and http://qihub.com/jdstorey/qvalue).
Measurement of vitamin B12, glycine (Gly), serine (Ser), and methionine (Met)
Vitamin B12 was measured by ELISA using the Human VB12 ELISA Kit from Creative Diagnostics (Shirley, New York; ca#: DEIA4440V2) per the manufacturer's protocol. Ser, Gly, and Met were measured by GC-MS on an Agilent 5975C GC-MSD mass spectrometer using a protocol involving methanol extraction, derivatization with N-(t-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA) containing 1% tert-butyldimethylchlorosilane (TBDMCS, Sigma-Aldrich; ca#: 375934), and Selected-Ion-Monitoring (SIM) mass spectrometry analysis of Ser at m/z 390 Da, Gly at m/z 246, and Met at m/z 320 with their respective internal standards Ser+7 (13C3, 97–99%; D3, 97–99%; 15N, 97–99%; ca#: CDNLM-6813–0.25), Gly+5 (D5, 98%; ca#: DLM-280-PK), Met+3 (D3, 98%; ca#: DLM-431-PK) purchased from Cambridge Isotope Laboratories (Tewksburg, MA).
| Sequence (5′-3′) | Sequence (5′-3′) | |
|---|---|---|
| Primer Set | Forward | Reverse |
| hSNAI1 | GCGCTCTTTCCTCGTCAGG | GGGCTGCTGGAAGGTAAACTCT |
| hTCN2 | GGCCCTCACTGAGATGTGTG | CTGTGCAGGTAGAGGTCTTCC |
| hSOX2 | GCCGAGTGGAAACTTTTGTCG | GGCAGCGTGTACTTATCCTTCT |
| hZEB1 | GATGATGAATGCGAGTCAGATGC | GATGATGAATGCGAGTCAGATGC |
| hPPIA (Control) | CCCACCGTGTTCTTCGACATT | GGACCCGTATGCTTTAGGATGA |
Western Blot (WB) Analyses and Antibodies
Cell lysates containing ∼30 μg of total proteins in each sample were resolved on SDS-PAGE and transferred to nitrocellulose membranes, then probed with specific first antibodies followed by appropriate fluorophore-conjugated secondary antibodies (#929–70020) from LI-COR Biosciences (Lincoln, NE). The fluorescence signals were detected with a LI-COR Odyssey imaging system. Band intensities were quantified using Odyssey imaging software version 3.0. Anti-TCN2 (ab113873), β-Actin (ab8229), KDM3A (ab31739), SETD8 (ab107234) and UGT8 (ab84288) antibodies were purchased from Abcam (Cambridge, MA). Anti-Claudin (#4933) and anti-SOX2 (#3579) were from Cell Signaling Technology (Danvers, MA). Anti-MKP2 (MA5–17225) and anti-RelA/NFγB p65 (PA5–16545) were from ThermoFisher Scientific. Individual targeted proteins and the control (β-Actin) were probed on the same membrane.
qRT-PCR Analysis of Gene Expression
Total RNA was extracted using acid guanidinium phenol extraction (Tri Reagent; Sigma). Briefly, 1 μg of RNA was reverse-transcribed using SuperScript III in a 20-μl reaction mixture. The rest of the procedures were as described previously (28). Relative changes in gene expression were quantified using the ΔΔCT method using gene-specific primers (shown in the table below). Data shown are the fold-change in mRNA abundance normalized to cyclophilin. All Q-RT-PCR data presented are the mean ± S.D. from n = 3 experiments.
TCN2 Knock-down and Overexpression
TCN2 expression was knocked down (KD) in U87 cells by transfection of human transcobalamin II siRNA (sc-45320) with transfection reagents and medium purchased from Santa Cruz Biotechnology per the vendor's protocol. TCN2 was over-expressed in U87 cells under both normoxic and hypoxic conditions. The TCN2-pCMV3-SP-N-HA plasmid (pTCN2) and its negative control vector were from Sino Biological and the DNA amplified in E. coli. The DNA sequence of TCN2 was confirmed by next-generation sequencing to match NCBI RefSeq NM_000355.2 for TCN2. Equal amounts of pTCN2 and its negative control vector were electroporated into U87 cells. After 48 h of transfection, the cells were collected for extracting total RNA for qRT-PCR analysis of gene expression, and whole-cell extracts prepared for WBs of targeted proteins. WB/qRT-PCR analysis confirmed successful HA-TCN2 expression.
RESULTS
TMT Quantification of Proteins in Hypoxic U87 Cells
We employed a new approach, different from traditional SILAC technology, which started with cultured cells fully labeled with heavy isotope-containing arginine (HR) in their cellular proteins. Protein quantification was carried out using the peptides with incorporated unlabeled arginine after 6 days of incubation in regular EMEM medium containing light arginine, followed by use of the established TMT stable isotope labeling method. It appeared that more than 80% of heavy arginine was replaced by light arginine in cells growing at normal oxygen levels (normoxia) after 6 days of incubation under the described experimental conditions. The advantage of this method is that it not only quantifies the expression of newly synthesized proteins after cellular perturbations, but it also precisely monitors cell proliferation rates. These rates are determined by measuring LR/HR ratios in histone peptides in MS profiles, which enabled us to replace the less accurate cell-counting approach. Because a set of TMT-6 regents is just sufficient to label six samples (d0/normoxia and d1–d5 hypoxic cells), it was impossible to simultaneously label all of the samples more than once. Therefore, we performed the overall experiments twice. The TMT-labeled tryptic peptides were separated by SCX into seven fractions. Each fraction was run on LC-MS/MS at least three times.
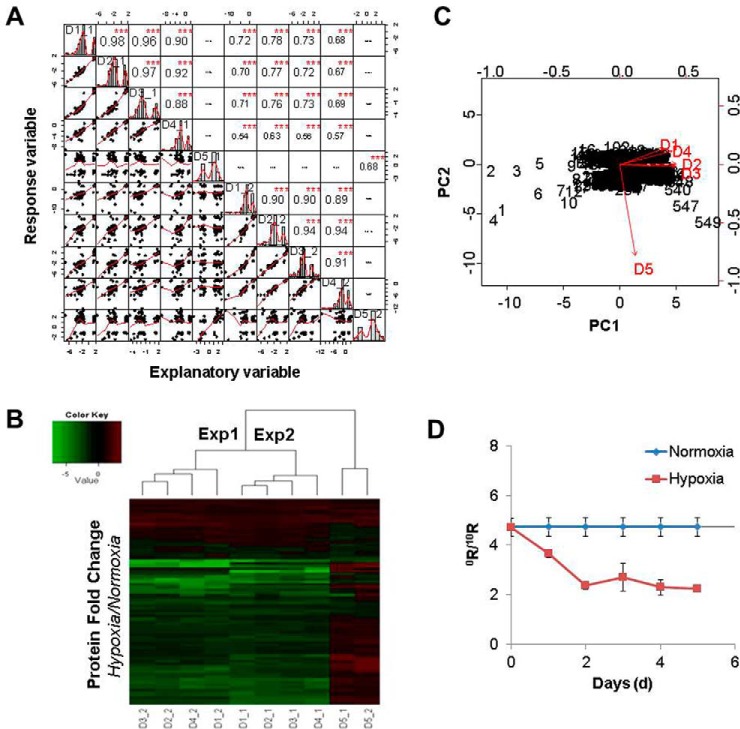
Experiment (Expt) 1 quantified 5180 unique proteins and Expt 2 quantified 4076 proteins. 2488 proteins were detected in both experiments, 80 of which were significantly changed in abundance (by more than ± 1.5 fold). This data set matrix was subjected to calculation of the correlation between the two biological experiments. The correlation coefficients (covariance) were calculated by the Pearson formula and visualized via a histogram. As indicated in Fig. 1A, nearly perfect intraday (d1–d4) correlations were observed for both Expts 1 and 2, because the majority of the correlation coefficients were larger than 0.9, shown as larger fonts in the diagonal histogram (Note: The maximum is 1.0, defined as identical between two separated subjects). This suggested that the hypoxic proteome was basically the same for the first 4 days, indicating that the effect of hypoxia was immediate. By contrast, the protein expression profile at d5 was clearly not correlated with that of the other 4 days, as the Pearson correlation coefficients were either close to zero or negative shown in almost invisible small fonts. The correlation between the two biological experiments was also significant, as most of the coefficients were larger than 0.7, except for d4 which was slightly lower (0.5 −0.6). Alternatively, as shown by a heat map, hierarchical agglomerative clustering analysis separates d5 from d1–4 as two major groups, whereas only small difference could be observed between the two subgroups of Expts 1 and 2, as judged by the color patterns (Fig. 1B). Thus, the reproducibility between the two experiments seemed satisfactory; we then merged the results from the two experiments, giving a total of 7527 unique proteins. In this mega-data set, there were a total of 549 proteins whose expression level changed significantly (by over 1.5-fold), of which 78 proteins were upregulated and 471 proteins were downregulated. PCA analysis (Fig. 1C) of these 549 proteins revealed the same result as shown by the heat map. These proteins with significantly changed expression were used for protein pathway and functionality analysis by Ingenuity Pathway Analysis (IPA). Hypoxic cells showed slower growth than normoxic cells, which could be seen visually by cell densities. However, the growth rates have been accurately measured by cell replication rates using the LR/HR ratio. As shown in Fig. 1D, for U87 cells growing under normoxia for 6 days, the LR/HR ratio of H4 peptide was 4.73, and the cell doubling time was calculated as 61 h (supplemental Table S6). When cells were grown under hypoxia for over 1d, their growth was arrested, because the LR/HR ratio of the H4 peptide had not changed at all from that of the previous day, when the cells were under normoxic conditions (supplemental Table S6).
Fig. 1.
Determination of quantification reproducibility by correlation analysis. A, Protein expression profile similarities were evaluated by correlation coefficients at consecutive 5 day time points and two experimental inter-groups. The distribution with fitted line of each variable (protein expression of each sample) is shown on the diagonal. Correlation coefficients were calculated by Pearson equation and visualized by the histogram display using the R program. Bottom-left half are linear regressions with dots representative of variables and red fitted lines (correlations) between every two data points (samples) and upright half are the calculated coefficients and asterisks denoting significance (p values). Better correlations are shown with larger fonts (identical is 1, whereas no correlation is 0 or negative.), *** = p < 0.0001. B, Heat map of protein expression calculated as fold changes compared with normoxic cells. Two experiments (Expts 1 and 2) were carried out for U87 cells growing under hypoxia for up to 5 days, and control cells under normoxia for the same period. C, Principle component analysis (PCA) of protein expression patterns for groups d1 to d5. The two most significant principal components, PC1 and PC2, representative for the 549 proteins (shown as numbers in the figure), are shown in the biplot. D, Ratios of newly synthesized histones (LR) to old histones (HR) in U87 cells growing under normxia and hypoxia were graphed on a 5-day time scale from the mass spectrometry total ion currents (TIC, area) of the MS/MS product ions (average of y5 and y6) of histone H4 peptides VFLENVILR/VFLENVIHR.
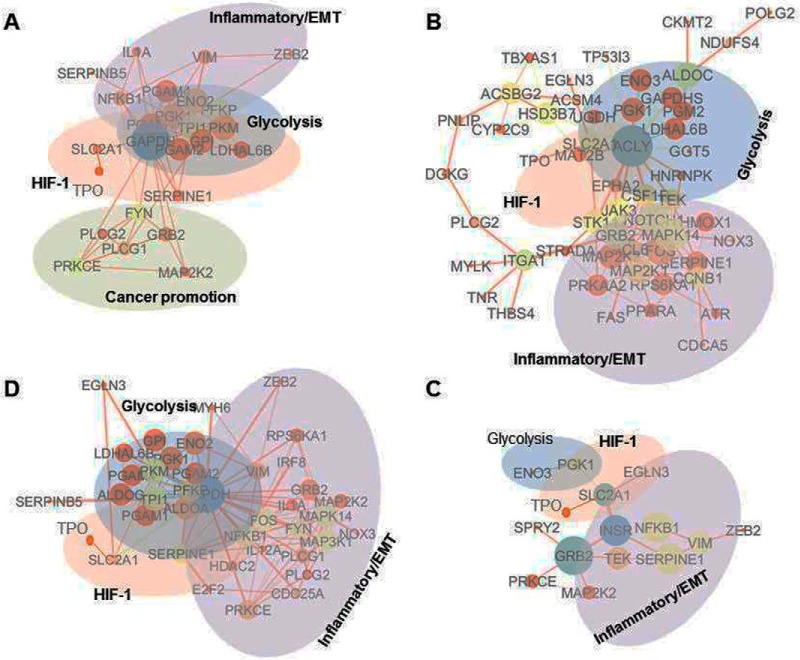
The proteins with significant expression changes were subjected to STRING pathway analysis. Proteins identified in the enriched biological process (GO) with FDR smaller than 0.05 were re-loaded to STRING for KEGG pathway analysis. The interaction network of the proteins was visualized by Cytoscape. Enriched KEGG pathways perturbed by hypoxia included glycolysis, HIF-1 signaling, carbon metabolism, and PI3K-Akt signaling/inflammation pathways (Fig. 2, supplemental Table S3).
Fig. 2.
Protein interaction networks of hypoxic proteome revealed by TMT proteomics. The analysis was done using protein interaction information based on the enriched KEGG pathways from the STRING database and visualized using Cytoscape. Each node represents a protein that is sized by the degrees of connections and colored based on betweenness centrality; each edge is sized by EgeBetweeness and colored based on its chromosome neighborhood. Protein fold changes were multiplied by the combined scores to form new Combined Scores. A–D are the protein interaction proteomes of 4 days of hypoxia from d1 to d4, respectively.
Label-free Quantification of Proteins in Hypoxic U87 Cells
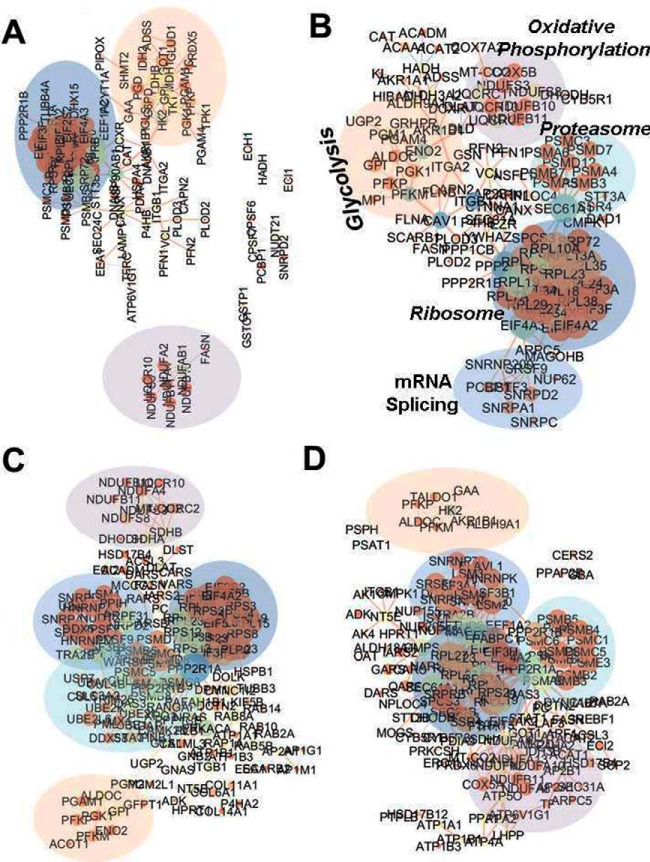
The combined two sets of peptide samples before TMT were analyzed by LC-MS/MS. In addition to the normoxic samples (d0), hypoxic samples for up to 5 days (d1–d5) were analyzed, by three injections each. A total of 18 LC-MS/MS raw data files were obtained. Because the TMT-measured protein expression profile at d5 was significantly different from those on d1–d4, the result at d5 was not presented again. Proteins with FDR smaller than 0.05 were considered real (supplemental Table S4). Supplemental Table S4 summarized the information on protein identification and quantitative results. For proteins whose identification was based on single unique peptides, their annotated spectra, which were sequentially numbered as appeared in supplemental Table S4, were provided as “Tandem MS spectra of single unique peptide for protein identification” in the Supplemental Materials. Based on supplemental Table S4, volcano plots (Fig. 3) were drawn to show the distribution of proteins with fold changes (x axis) of protein expressions in hypoxic cells relative to normoxic cells and the significance of the measurements (y axis with -Log p value). Noteworthy upregulated proteins indicated in the figure were glycolytic proteins (ALDOC, GPI, PGK1, PGM2) and the lactate/pyruvate transporter proteins MOT1/4 (MCT/14); downregulated proteins were Proliferation Cell Nuclear Antigen (PCNA, essential for replication), Promyelocytic Leukemia protein (PML, an oncogene suppressor protein), and proteins involved in the immune system with antiviral and anti-inflammatory function, including MX1 and STAT1. Further, STRING pathway analysis of proteins with significant expression changes indicated that glycolysis, carbon metabolism, proteasome, oxidative phosphorylation, and mRNA splicing were among the top enriched KEGG pathways as shown in supplemental Table S5 and Fig. 4, which was visualized by Cytoscape.
Fig. 4.
Protein interaction networks of hypoxic proteome revealed by LFQ. The analysis was done using protein interaction information based on the enriched KEGG pathways from the STRING database and visualized using Cytoscape. Each node represents a protein that is sized by the degrees of connections and colored based on betweenness centrality; each edge is sized by EgeBetweeness and colored based on its Chromosome Neighborhood. Protein fold changes were multiplied by the Combined Scores to form new Combined Scores. A–D are the protein interaction proteomes of 4 days of hypoxia from d1 to d4, respectively.
Hypoxia Promotes EMT and CSC Renewal
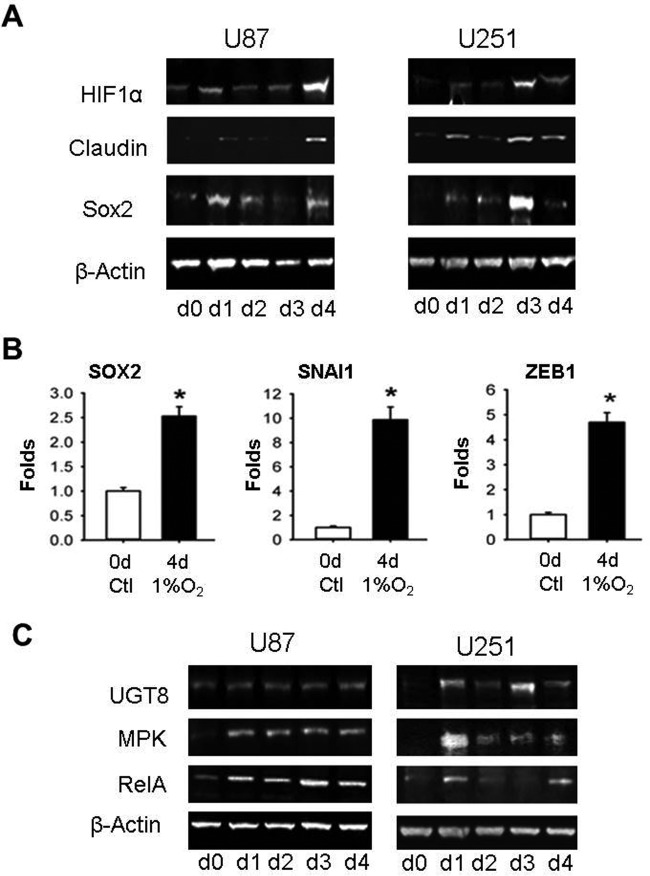
Surprisingly, the HIF-1α protein was not detected by our current proteomic approach, possibly because its expression in brain cells is not as high as we expected. Therefore, we turned to Western blot analysis using an anti-HIF-1α antibody. As shown in Fig. 5A, the expression of HIF-1α protein increased significantly on d1 to d4 for both U87 cells and U251 cells under hypoxia. The expression of HIF-1α was concurrent with that of GLUT1/SLC2A1; the latter is a HIF-1α-activating protein (29). Similar expression patterns were observed for the stem cell/cancer stem cell marker protein SOX2 and the EMT marker protein Claudin, an important component of tight junction complexes (Fig. 5A). Gene expression analysis by qRT-PCR revealed that mRNAs for SOX2 and two additional EMT markers, SNAI1 and ZEB1, were upregulated in hypoxic U87 cells (Fig. 5B), suggesting that overexpresssion of stem cell and EMT markers could be transcriptionally regulated by hypoxia. Promoter activation of both CSC and EMT marker genes by HIF-1α has previously been described in coronary endothelial cells and colorectal cancer cells (30–33), consistent with our findings here in glioblastoma cells. The hypoxic glioblastoma cells exhibited elongated morphologies characteristic of EMT, including increased Aspect Ratios (AR) and decreased Circularity (fcirc) (supplemental Fig. S1 and S2). Under the same experimental conditions as used for U87 cells, the small lung cancer A549 cells also underwent EMT under 5 days of hypoxia, showing the same cell phenotype and gene expression profiles of EMT as induced by TGFβ (supplemental Fig. S3). Thus, induction of EMT by hypoxia might be a common mechanism of metastatic cancer cells with CSC renewal and invasive/migratory properties, allowing them to disseminate and propagate to remote sites.
Fig. 5.
Hypoxia induces EMT. A, WB of HIF-1α, SOX2, and Claudin1 protein expression in hypoxic U87 and U251 cells. β-actin was used as the loading control. B, qRT-PCR analysis of SOX2, SNAI1 and ZEB1 at d5 under hypoxia. *p < 0.05, **p < 0.01, ***p < 0.001. C, WB of UGT8, MKP2, and RelA/p65 protein expression in hypoxic cells. β-actin was used as the loading control.
Our data also suggested that hypoxia triggers cellular transformation by upregulating the expression of ceramide UDP-galactosyltransferase (UGT8), a marker of invasiveness (34, 35), as shown in Fig. 5C. In addition, the inflammatory markers mitogen-activated protein kinase phosphatase (MKP2) and NF-κB p65/RelA (36) were also upregulated (Fig. 5C). Overexpression of MKP2 was previously detected in several cancer cell lines including liver, pancreatic, ovary, and breast (37–39). RelA is not only a transcriptional activator of Snail, one of the EMT/CSC markers (40, 41), but also one of the major components of the NF-κB signaling pathway activated in inflammation, immunity, and cancer (42, 43). IPA analysis of the d1–d4 TMT proteomics data set revealed several significantly altered pathways, including reduced cellular assembly and maintenance capability, decreased neuron migration, and downregulated cancer suppression gene p53 (z-scores are smaller than −2). By contrast, the pathway for forming solid cancer/tumor was significantly upregulated (z-scores are larger than 2; Table I), showing good agreement with the STRING analysis that identified several enriched inflammation pathways including MAPK, PI3K-Akt, mTOR, FoxO, and VEGF signaling pathways (Fig. 2, supplemental Table S3).
Table I. Activation z-score of major disease-related cell functionality (shown is hypoxia d4 data and not shown for d1-d3 are very similar).
| Cellular functions and diseases | Activation Z-score | p value -Log | |
|---|---|---|---|
| Cellular assembly | Microtubule dynamics | −3.24 | 4.29 |
| Organization of cytoskeleton | −3.43 | 3.95 | |
| Organization of cytoplasm | −3.53 | 3.52 | |
| Formation of cellular protrusions | −2.55 | 2.94 | |
| Neurological function | Migration of neurons | −2.40 | 1.8 |
| Cancer | Malignant solid tumor | 2.05 | 8.37 |
| Abdominal cancer | 2.07 | 7.47 | |
| Digestive organ tumor | 3.23 | 6.63 | |
| Gastrointestinal tract cancer | 2.11 | 6.05 | |
| Malfunction of genital organs | 2.39 | 3.39 | |
| Head and neck neoplasia | 2.19 | 5.48 | |
| Cancer suppression | p53 | −2.37 | 1.07 |
Hypoxia Upregulates Protein Expression in Glucose Transport and Glycolysis Pathways
All the proteins identified by TMT proteomics in the glucose transport and metabolic pathways were assembled and visualized graphically in a “pseudo WB” showing protein expression profiles by the degree of darkness of rectangles. As shown in Fig. 6, GLUT1 was upregulated by nearly 3-fold under 1% O2 on d1 and remained elevated throughout the experiment for 5 days. By contrast, GLUT3 was upregulated only 1.52 fold, from d4. The expression of nine key enzymes in the glycolytic pathway was incrementally increased with the number of days of hypoxia, reaching the maximum at d4 (Fig. 6) (44). Though not every protein identified by TMT was also identified by label-free analysis, the two proteomics experiments both identified glycolysis as one of the major pathways impacted by hypoxia (Fig. 2–4, supplemental Fig. S3 and S4). Upregulation of glycolysis compensates for the hypoxia-induced impairment of oxidative phosphorylation to provide energy for cells to survive. However, hypoxic cells' growth was arrested (Fig. 1D, supplemental Table S6). The switch between these two pathways was highlighted by label-free proteomics (Fig. 4).
Fig. 6.

MS pseudo-WB of hypoxia-regulated protein expression in glucose transportation and metabolic pathways. MS pseudo-WB is a metaphor for Western blot images, created by a heat map of the protein expression ratios acquired from quantitative proteomic analysis. Protein expression in normoxic cells (d0) was set at 1 and protein expression in hypoxic cells for up to 5 days (d1 - d5) were compared with that in the normoxic cells. Ratios ≤ 0.67 (or 1.5-fold) are considered to denote significant downregulation (white color), and ratios ≥ 1.5 are considered to show significant upregulation, which is visualized by the relative darkness of the bands.
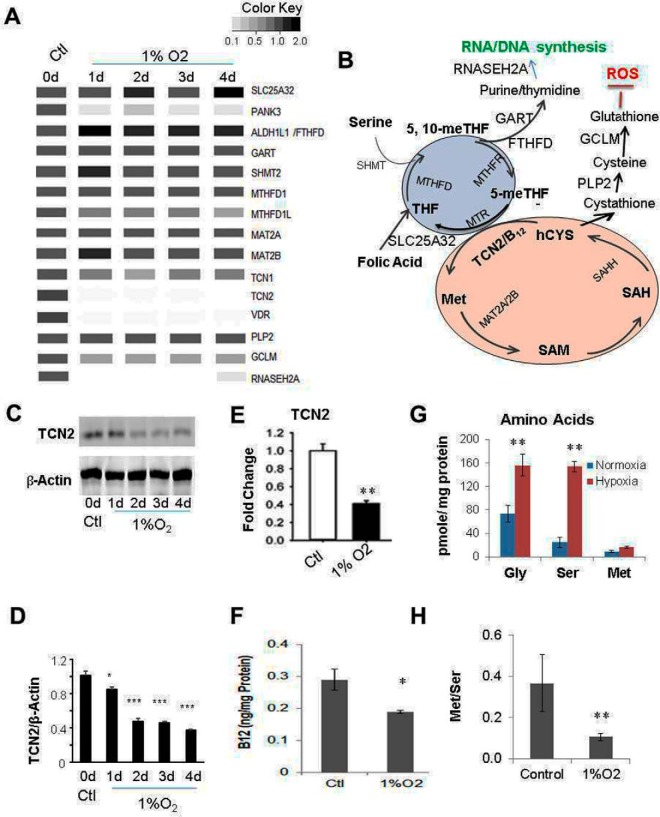
Hypoxia Downregulates TCN2 Expression Resulting in Folate-cycle Trap and the Arrest of Cell Growth
Met is used for the synthesis of proteins and SAM. It is normally taken up from extracellular nutrients, consumed, and recycled in the Met cycle, but also regenerated from Ser metabolism when a methyl group from 5-methyl-tetrahydrofolate (5-meTHF) is transferred to homocysteine, catalyzed by Met synthase, which requires vitamin B12 as a cofactor. Vitamin B12 from nutrients binds to intrinsic factor/transcobalamin III (TCN3) on the cell membrane before being taken up into the cells via receptor-mediated endocytosis. Vitamin B12 is further delivered to various cell compartments by its transport proteins haptocorrin (TCN1) and transcobalamin II (TCN2). Among the three transcobalamin proteins, TCN1 and TCN3 are less specific and have a lower turnover rate than TCN2, and the former two but not the latter bind to inactive B12 analogues. Hence, TCN2 is considered the best biomarker of active and available B12 (45). In our proteomics experiments, TCN2 was significantly downregulated under hypoxia right after d1, TCN1 was slightly downregulated, whereas TCN3 was not detected (Fig. 7A). Downregulation of TCN2 was further confirmed by WB (Fig. 7C, 7D) and RT-qPCR analysis (Fig. 7E). Without TCN2, there would be no vitamin B12 transported into the cells, resulting in its deficiency. As expected, vitamin B12 concentration measured by ELISA was significantly lower in hypoxic U87 cells (Fig. 7F). Because proteomic analysis revealed no significant changes in protein expression levels for most of the enzymes participating in the folate cycle (Fig. 7A, 7B), vitamin B12 deficiency because of downregulation of TCN2 by hypoxia might be the leading cause of the folate trap (methyl trap), because the reaction of methyl transfer from 5-me-THF to homocysteine to form Met is impeded without available vitamin B12 as the essential cofactor of Met synthase (MTR). Until 5-me-THF is demethylated, folate cannot participate in any other one-carbon transfer reactions. As a result, blocked Ser and Gly metabolism leads to their accumulation in the cells. Higher concentrations of Ser and Gly in hypoxic cells were confirmed by the measurement of cellular endogenous amino acid concentrations by GC/MS (Fig. 7G). Blocked Ser metabolism would cause decreased Met synthesis in the cell. However, the measured Met concentrations showed little change between normoxic cells and hypoxic cells (Fig. 7G), suggesting that depletion of Met from metabolic synthesis was largely compensated by its decreased consumption in protein synthesis resulting from the folate trap, as it stops the synthesis of purines and thymidines, the precursors of RNA and DNA, inducing the arrest of cell division. In this regard, the decreased Met/Ser ratio in hypoxic cells (Fig. 7H) would otherwise be a better barometer indicating the blocked one-carbon metabolic pathway in hypoxic cells.
Fig. 7.
Hypoxia impedes one-carbon metabolism by the vitamin B12-tranporter TCN2. A/B, MS pseudo-WB and locations of the proteins in the pathway. The most significant hypoxia-induced downregulated proteins were TCN2 (Transcobalamin II), RNASEH2A (Ribonuclease H2), PANK3 (pantothenate kinase 3, needed for CoA biosynthesis), VDR (vitamin D receptor), and GCLM (glutamate-cysteine ligase); less significantly downregulated proteins were TCN1 (Transcobalamin I), MTHFD1L (methylenetetrahydrofolate dehydrogenase 1-Like); no or slight change: SHMT2 (Ser hydroxymethyltransferase 2 (mitochondrial)), MTHFD (methylenetetrahydrofolate dehydrogenase 1), SCL25A32 (solute carrier family 25 (mitochondrial folate carrier), member 32), MAT2A/2B (Met adenosyltransferase 2A/2B), MTHFR (methylene tetrahydrofolate reductase), FTHFD (formyltetrahydrofolate dehydrogenase), GART (phosphoribosylglycinamide formyltransferase) and PLP2 (papain-like proteinase 2). C, Fluorescence image of WB of TCN2. D, Statistical analyses of three repeats of WB in C. E, mRNA of TCN2 in control and 4d under hypoxia by RT-qPCR. F, Vitamin B12 concentration in control (d0) and at 4d under hypoxia, by ELISA. G, Ser, Gly, and Met amino acid concentrations in control and at 4d under hypoxia, by GC/MS. H, Met/Ser calculated from G.
As shown in Fig. 7A, RNAseH2, a component of the heterotrimeric type II ribonuclease H enzyme, which is the major source of ribonuclease H activity in mammalian cells and endonucleolytically cleaves ribonucleotides essential for DNA replication, was dramatically downregulated in hypoxic cells. The arrest of cell replication results in decreased protein synthesis. As expected, the histone synthesis rate declined gradually in hypoxic cells compared with the normoxic cells growing in parallel according to the calculated light arginine incorporation rate (note: proteins in the seeded cells were fully labeled with heavy arginine) (Fig. 1D, supplemental Table S6), which could be used, on the other hand, as a surrogate of commonly used methods (using labeled nucleosides for measuring cell replication). It should be noted that the vitamin D receptor (VDR) was also significantly downregulated under hypoxia, indicating that hypoxia might also impair the homeostatic control of mineral metabolism, cell differentiation by VDR targeted gene expression, and functional activities responsible for growth hormones and immune responses (46). Taken together, impaired synthesis of proteins (except for the HIF-1α-activating genes/proteins), RNA/DNA, and possibly metabolites, could result in cell growth arrest coexisting with EMT (47), as slowed cell proliferation happened concurrently with morphologically visible EMT characterized by elongation (Fig. 1D, supplemental Fig. S1, S2). Slightly away from the major topic, as shown from the extension of Met metabolic cycle (Fig. 7B), the enzyme GCLM, which is responsible for conversion of cysteine into glutathione, was significantly downregulated, decreasing the cells' ability to combat reactive oxygen species (ROS), because the antioxidant glutathione is a ROS scavenger.
DISCUSSION
We have analyzed the proteome of glioblastoma, typically U87 cells, during 5 days of hypoxia. The quantification was carried out by both TMT-labeling and label-free LC-MS/MS. The two quantification methods have identified not only commonly shared enriched pathways, such as glycolysis. Consistent with the literature, a switch from oxidative phosphorylation to glycolysis is a major cancer cell response to hypoxia, shown by elevated glucose metabolism, high glucose consumption and increased glucose uptake (44, 48). Transport of glucose across the plasma membrane of mammalian cells is the first rate-limiting step in glucose metabolism, and is facilitated by glucose transporters (GLUT) and endocytosis. It has been widely accepted that increased GLUT levels and their transport activity together with an accelerated glycolysis rate contribute significantly to tumor, growth as historically indicated by the Warburg effect (49). GLUT has up to 14 isoforms (GLUT1-GLUT14), among which GLUT1, GLUT3, and GLUT4 have the highest affinity for glucose, allowing transport of glucose at a high rate under normal physiological conditions. GLUT4, which is sequestered in the intracellular compartments and is translocated to the cell membrane in the presence of insulin, assists in the transport of glucose by endocytosis/exocytosis (50). GLUT1 and GLUT3 are overexpressed in many malignant cell types, including glioblastoma. Of the 14 GLUT isoforms, our proteomic data indicate that GLUT1 is the most rapidly and highly upregulated protein under hypoxia before glycolysis (Figs. 3, 6, and 4S). The GLUT1 protein is one of the low-oxygen stress-induced or HIF-1α transcriptionally regulated proteins, which also include the HIF-1α stabilizer protein Thrombopoietin (TPO) (29, 51); these two proteins belong to the HIF-1α signaling pathway and were upregulated in hypoxic U87 cells as shown by TMT proteomics (Fig. 2). GLUT1 and GLUT3 also participate in vitamin C transport (supplemental Fig. S4). High vitamin C intake could partially compensate for oxygen deficiency- induced ROS and counter the adverse effects of low oxygen on demethylases (52). The TMT proteomics approach has quantified almost all of the proteins in the glycolysis pathway, which were all upregulated under hypoxia, accelerating the synthesis of pyruvate and its conversion into lactate that was consequently consumed for energy production in order to compensate for the loss from hypoxia-induced downregulation of oxidative phosphorylation in the Krebs cycle (Fig. 4S and 6). Beside the glycolytic enzymes, both TMT and label-free-proteomics have shown elevated expression of the monocarboxylate transporter 4 (MOT4), responsible for lactate transport (supplemental Table S1–S2, S4, Fig. 3), but small changes for MOT1 and lactate dehydrogenase A/B (simplified as LDHA/B), which are the two enzymes responsible for the conversion between pyruvate and lactate from either direction, demonstrating that lactate efflux in hypoxic cells is enhanced through glycolysis and transportation of lactate. Beside uptake from the cell culture medium, intercellular Ser levels could be regulated by its production through the Ser biosynthesis pathway, a branch from the main glycolysis pathway starting at 3-phospho-glycerate (3-P-G) (supplemental Fig. S4). Because the expression of the three proteins involved in Ser biosynthesis was inconsistent over 5 days of hypoxia and because only ∼10% of 3-P-G is metabolized to Ser as compared with pyruvate in cancer cells (53), we cannot predict how hypoxia affects Ser synthesis. Above all, our proteomic results support the long-standing Warburg theory that hypoxic cancer cells have large lactate production from glucose uptake and anaerobic glycolysis to maintain cell survival (54).
Although the glucose metabolism pathway in hypoxic cancer cells is accelerated to compensate for the loss of oxidative phosphorylation because of the low-oxygen stress for cell survival, overall cell proliferation is suppressed as evidenced by upregulation of the proteasome protein degradation pathway and downregulation of ribosome and mRNA splicing pathways, together with downregulation of the cell replication marker protein PCNA (Fig. 4). Decreased cell proliferation in hypoxic cells was further directly supported by the lower cell densities (supplemental Fig. S1 and S2), and decreased proliferation rates measured by arginine incorporation (LR/HR) in replicating cells (Fig. 1D and supplemental Table S6). We would again empathize that proteomic analysis starting with HR-labeled cells quantifies the proteome of replicating cells so that the effects from the preexisting proteome of old cells are minimized, and hence quantification accuracy is improved. This improved accuracy was more apparent when histone methylation from Ser metabolism was measured, which will be reported in a subsequent manuscript. In addition to the protein pathways regulating cell proliferation that were downregulated, the metabolic pathways essential for DNA and protein synthesis were also downregulated, indicating cross-talk between metabolites and proteins. It should be noted that when cells were switched to fresh medium at the end of d2, it took about a half hour for the hypoxic incubator to reach 1% O2, and extra time was possibly needed for the rebalancing of O2 in the medium; reoxygenation could have happened. This caused a slight resurgence in the proliferation rate (Fig. 1D and supplemental Table S4) and minor relaxation of the perturbed pathways at d3 (Fig. 2C, 3C). However, as compared with the 61-hour doubling time for normoxic cells (supplemental Table S6), the short-term re-oxygenation did not cause dramatic changes in the hypoxic proteome from d2 to d4 (Figs. 2 and 4).
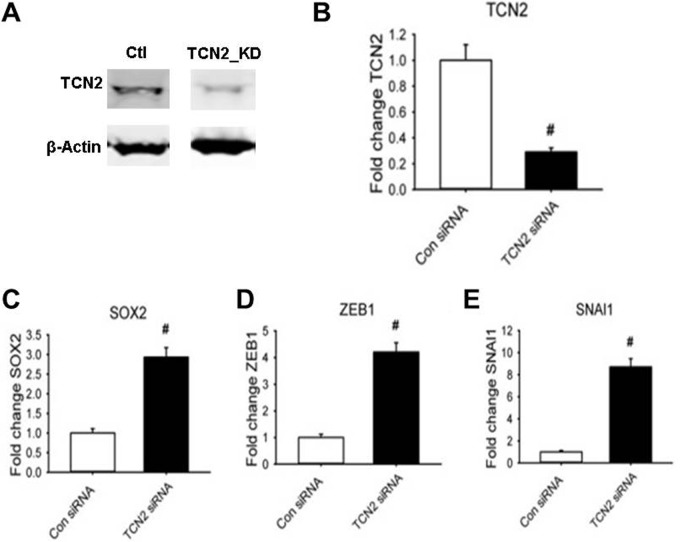
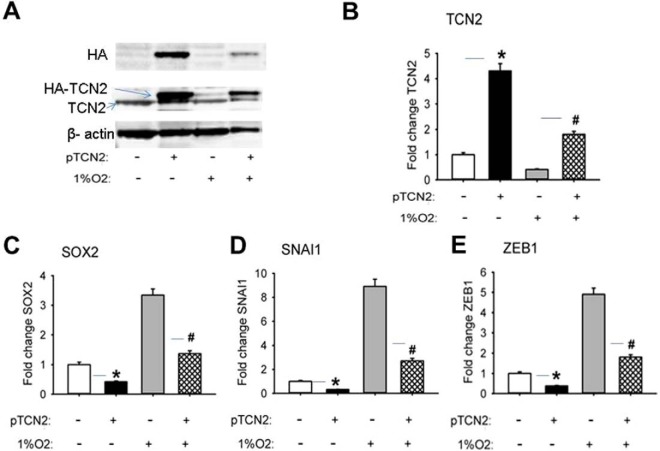
Cell growth arrest could directly result from the folate trap that is caused by the inactivation of Met synthase (MTR). MTR requires vitamin B12 as its cofactor. In hypoxic U87 cells, the cellular vitamin B12 concentration was found to be decreased (Fig. 7F). We further found that the cause of this decrease in vitamin B12 was likely the downregulation of TCN2, which is the major vitamin B12 transporter (Fig. 7A, 7C, 7D, 7E). The folate trap blocks the synthesis of the purine and thymidine substrates for RNA and DNA synthesis, and the synthesis of Met essential for protein synthesis. Because Met is not only the first amino acid of proteins before N-terminal Met excision in eukaryotic cells (AUG encoding Met is always the start code) but also one of the total 20 amino acids in the protein sequence, its deficiency blocks cell growth. The identification of downregulation of TCN2 in hypoxic cells led to our hypothesis that a hypoxia-induced decrease in TCN2 results in an intracellular B12 deficiency that sequentially causes blockage of the one-carbon metabolic pathway, cell growth arrest, EMT, and CSC renewal. To test this hypothesis, we measured the expression levels of several ESC and EMT markers (protein and/or mRNA) by WB and qRT-PCR in U87 cells in which TCN2 expression was either knocked down (KD) by transfection of TCN2 siRNA or overexpressed by electroporation of TCN2 cDNA. TCN2 was successfully knocked down by >80% at both the protein (Fig. 8A) and the mRNA level (Fig. 8B). As shown in Figs. 8C–8E, in TCN2 KD cells the mRNA level of the CSC marker SOX2 increased by 3-fold and the mRNA levels of the EMT markers ZEB1 and SNAI1increased by 4-fold and 8-fold, respectively, relative to the control. By contrast, in TCN2 over-expressing cells, the expression of both EMT and CSC marker proteins was inhibited, which blocked the hypoxic cells from EMT and CSC production (Fig. 9). These data strongly supported our hypothesis that TCN2 plays a critical role in regulating one-carbon metabolism and dictating cell transformation. Therefore, our identification of the role of TCN2 in cancer cells revealed a novel mechanism for understanding hypoxia biology.
Fig. 8.
Expression of CSC and EMT markers in TCN2 KD U87 cells. A, WB of TCN2 and β-actin (loading control). B, Fold change of TCN2 mRNA. C, Fold change of SOX2 mRNA. D, Fold change of SNAI1 mRNA. E, Fold change of ZEB1 mRNA. qPCR data (B–E) were normalized by the expression of cyclophinin. #: p < 0.01.
Fig. 9.
Expression of CSC and EMT markers in U87 cells over-expressed with TCN2 under 48 h of normoxia and hypoxia. A, WB of HA (the tag protein), TCN2, and β-actin (the loading control). B, Fold change of TCN2 mRNA. C, Fold change of SOX2 mRNA. D, Fold change of ZEB1 mRNA. E, Fold change of SNAI1 mRNA. qPCR data (B–E) were normalized by the expression of cyclophinin. */#: p < 0.01 (+pTCN2/-pTCN2).
A combination of proteomic analysis with mRNA analysis by qRT-PCR and protein expression analysis by WB illustrated the EMT progression of U87 cancer cells under hypoxic conditions. EMT occurred with recognizable mesenchymal morphology (supplemental Fig. S1 and S2) accompanied by overexpression of EMT markers including SNAI1, ZEB1/2, Claudin, and Vim at either the mRNA or protein level (Fig. 2, 4, and 5). Under hypoxia, cells responded to the low-oxygen stress by inflammation. In this regard, the proinflammatory molecule GLUT1/SLC2A1 (55, 56) (Figs. 2–4, 6, and (supplemental Fig. S4), inflammatory proteins including NF-κB p65/RelA (Fig. 5B) and proteins in the mitogen-activated protein kinase (MAPK) signal pathway including MKP2 (37, 38) and the PI3K/Akt/mTOR pathway were all overexpressed (supplemental Table S1). Concurrently with EMT, hypoxic cells were dedifferentiated with more CSC properties, which was supported by upregulation of the stem cell marker proteins SOX2 (Fig. 5A, 5B) and Vim (supplemental Table S1–S2, and S4). This feature was also consistent with previous physiological and biological discoveries obtained from studies in animal or human disease models (57, 58). The work presented here endorsed the consensus that hypoxia promotes EMT and CSC renewal, which are characteristic features of metastatic and aggressive cancer cells, in line with the downregulation of the anti-inflammatory proteins MX1 and STAT1 (Fig. 3), the downregulation of the oncogene suppressors p53 (Table I, Fig. 2B) and PML (Fig. 3), and upregulation of the cancer metastatic marker protein UGT8 (Fig. 5C) (34, 35).
In conclusion, cells growing under hypoxic stress experience metabolic and transcriptional adaptation (13). Hypoxia induces deficient mitochondria redox-oxidation cycles needed for energy production (14, 15). To compensate for this deficiency, cells adapt to the stress by anaerobic metabolism of citric acid cycle intermediates and glucose metabolism, historically known as the Warburg effect (54, 59, 60). This metabolic adaptation was previously seen in response to stem cell-like EMT in tumor cells in response to hypoxia (61, 62), albeit it is also required for normal human pluripotent stem cell maintenance and differentiation (63). Our current hypoxic experiments consistently identified EMT promotion and CSC renewal in hypoxic glioblastoma U87 cells and U251 cells. The same identification was achieved in human small lung cancer A549 cells (supplemental Fig. S3). However, no EMT characteristic morphological change was seen in normal lung epithelial cells under 1% O2 for over 15 days (data not shown). This suggested that hypoxia may cause type III EMT in all cancer cells, but is not sufficient to induce type I EMT in normal cells. Importantly, we have demonstrated that the metabolic adaptation of hypoxic cells was through downregulation of TCN2 protein, which is essential for delivering active and available vitamin B12 to the cells for their one-carbon metabolism. Reduced expression of TCN2 appeared to exhibit the same phenotype as that of hypoxic cells, especially with regard to the features of EMT and CSC (Fig. 8). By contrast, overexpression of TCN2 in hypoxic cells nullified the effects of hypoxia (Fig. 9). This suggests that TCN2 is not only important in regulating one-carbon metabolism, but also plays a significant role in cancer cell transformation.
The propagation of glioblastoma is closely linked to the microvascular proliferation in hypoxic areas of the brain, often because of the overexpression of HIF-1α-activating genes, including VEGF and GLUT1. Although 2D cell culture cannot fully mimic a tumor that grows in a spheroid 3D structure with a gradient of oxygen from normal at the outside surface to depleted at the center of a solid tumor, 2D culture allows effective analysis of hypoxia via gene/protein expression profiling to reveal global expression patterns and biological signaling pathways. For example, as indicated from our work, hypoxia itself suppressed cell proliferation, which was contrary to the perception that hypoxia induces proliferation in the tumor. However, the results of our proteomic analysis suggest that the blockage of one-carbon metabolism could be one of the causes of cell proliferation arrest, because one-carbon metabolic pathway provides essential fuels, such as amino acids, lipids, DNA/RNA and proteins needed for cells to grow. The identification of TCN2 as a potential therapeutic target for activating vitamin B12 delivery, reversing EMT, and controlling CSC lays the foundation for a novel preclinical study on the treatment for glioblastoma.
Because one-carbon metabolism is critical for maintaining Met homeostasis, and Met is required not only for cell proliferation but also to produce SAM, an essential cofactor of DNA and histone methylation, the current work promotes continuing studies that link the impact of hypoxia on one-carbon metabolism to the epigenome.
DATA AVAILABILITY
Mass spectrometry raw data was deposited via ProteomeXchange (http://www.proteomexchange.org) with identifiers IDs PXD005487 (TMT) and PXD007280 (LFQ). Processed data available as Table S1-S6 in the Supplemental.
Supplementary Material
Acknowledgments
We thank Dr. David Konkel for critically editing the manuscript.
Footnotes
Author Contributions: K.Z. partially performed and supervised H.T to carry out cell culture, mass spectrometry, and metabolite analysis experiments. H.T. carried out the mass spectrometry and vitamin B12 and SAM analyses. P.X. carried out WB experiments. B.M. did the cell morphology measurements. D.M. and J.H. performed GC/MS analysis of amino acids. Y.C. supervised P.X. for WB experiments. B.T. carried out TCN2 overexpression and RT-qRT-PCR analysis of TCN2 and ESC/EMT markers. J.S., A.B. and L.S. provided conceptual advice. K.Z. wrote the manuscript.
* This work was supported in part by the National Institutes of Health, National Cancer Institute (CA184097) and in part by the start-up funds provided to K. Z. by the Department of Pharmacology and Toxicology, University of Texas Medical Branch School of Medicine.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HIF-1
- hypoxia inducible factor
- GLUT
- glucose transporter
- TMT
- tandem mass tag
- EMT
- epithelial mesenchymal transition
- CSC
- cancer stem cell
- SAM
- S-adenosylmethionine
- PCA
- principal component analysis
- AR
- aspect ratio.
REFERENCES
- 1. Vaupel P., and Mayer A. (2007) Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 26, 225–239 [DOI] [PubMed] [Google Scholar]
- 2. Greijer A. E., van der Groep P., Kemming D., Shvarts A., Semenza G. L., Meijer G. A., van de Wiel M. A., Belien J. A., van Diest P. J., and van der Wall E. (2005) Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 206, 291–304 [DOI] [PubMed] [Google Scholar]
- 3. Bao B., Azmi A. S., Ali S., Ahmad A., Li Y., Banerjee S., Kong D., and Sarkar F. H. (2012) The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness. Biochim. Biophys. Acta 1826, 272–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Higgins D. F., Kimura K., Bernhardt W. M., Shrimanker N., Akai Y., Hohenstein B., Saito Y., Johnson R. S., Kretzler M., Cohen C. D., Eckardt K. U., Iwano M., and Haase V. H. (2007) Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J. Clin. Investig. 117, 3810–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marie-Egyptienne D. T., Lohse I., and Hill R. P. (2013) Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: potential role of hypoxia. Cancer Lett. 341, 63–72 [DOI] [PubMed] [Google Scholar]
- 6. Salnikov A. V., Liu L., Platen M., Gladkich J., Salnikova O., Ryschich E., Mattern J., Moldenhauer G., Werner J., Schemmer P., Buchler M. W., and Herr I. (2012) Hypoxia induces EMT in low and highly aggressive pancreatic tumor cells but only cells with cancer stem cell characteristics acquire pronounced migratory potential. Plos One 7, e46391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsai Y. P., and Wu K. J. (2012) Hypoxia-regulated target genes implicated in tumor metastasis. J. Biomed. Sci. 19, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoo Y. G., Christensen J., Gu J., and Huang L. E. (2011) HIF-1alpha mediates tumor hypoxia to confer a perpetual mesenchymal phenotype for malignant progression. Sci. Signal. 4, pt4. [DOI] [PubMed] [Google Scholar]
- 9. Zhang L., Huang G., Li X., Zhang Y., Jiang Y., Shen J., Liu J., Wang Q., Zhu J., Feng X., Dong J., and Qian C. (2013) Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1alpha in hepatocellular carcinoma. BMC Cancer 13, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q., Bai X., Chen W., Ma T., Hu Q., Liang C., Xie S., Chen C., Hu L., Xu S., and Liang T. (2013) Wnt/beta-catenin signaling enhances hypoxia-induced epithelial-mesenchymal transition in hepatocellular carcinoma via crosstalk with hif-1alpha signaling. Carcinogenesis 34, 962–973 [DOI] [PubMed] [Google Scholar]
- 11. Ortensi B., Setti M., Osti D., and Pelicci G. (2013) Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res. Therapy 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ismail-Beigi F. (1993) Metabolic regulation of glucose transport. J. Membrane Biol. 135, 1–10 [DOI] [PubMed] [Google Scholar]
- 13. Cairns R. A., Harris I. S., and Mak T. W. (2011) Regulation of cancer cell metabolism. Nat. Rev.. Cancer 11, 85–95 [DOI] [PubMed] [Google Scholar]
- 14. Giaccia A. J., Simon M. C., and Johnson R. (2004) The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Develop. 18, 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Semenza G. L. (2011) Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 [DOI] [PubMed] [Google Scholar]
- 16. Guo X., Shi M., Sun L., Wang Y., Gui Y., Cai Z., and Duan X. (2011) The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 58, 153–157 [DOI] [PubMed] [Google Scholar]
- 17. Kristian T. (2004) Metabolic stages, mitochondria and calcium in hypoxic/ischemic brain damage. Cell Calcium 36, 221–233 [DOI] [PubMed] [Google Scholar]
- 18. Humar R., Kiefer F. N., Berns H., Resink T. J., and Battegay E. J. (2002) Hypoxia enhances vascular cell proliferation and angiogenesis in vitro via rapamycin (mTOR)-dependent signaling. FASEB J. 16, 771–780 [DOI] [PubMed] [Google Scholar]
- 19. Mimeault M., and Batra S. K. (2013) Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J. Cell. Mol. Med. 17, 30–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heddleston J. M., Li Z., McLendon R. E., Hjelmeland A. B., and Rich J. N. (2009) The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 8, 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mobasheri A., Richardson S., Mobasheri R., Shakibaei M., and Hoyland J. A. (2005) Hypoxia inducible factor-1 and facilitative glucose transporters GLUT1 and GLUT3: putative molecular components of the oxygen and glucose sensing apparatus in articular chondrocytes. Histol. Histopathol. 20, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 22. Sowers J. L., Mirfattah B., Xu P., Tang H., Park I. Y., Walker C., Wu P., Laezza F., Sowers L. C., and Zhang K. (2015) Quantification of histone modifications by parallel-reaction monitoring: a method validation. Anal. Chem. 87, 10006–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicholas D., Tang H., Zhang Q., Rudra J., Xu F., Langridge W., and Zhang K. (2015) Quantitative proteomics reveals a role for epigenetic reprogramming during human monocyte differentiation. Mol. Cell. Proteomics 14, 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robles M. S., Cox J., and Mann M. (2014) In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Gen. 10, e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y., and Hochberg Y. (1995) Controllin gthe false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. B 57, 289–300 [Google Scholar]
- 28. Tian B., Yang J., and Brasier A. R. (2012) Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol. Biol. 809, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C., Pore N., Behrooz A., Ismail-Beigi F., and Maity A. (2001) Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 276, 9519–9525 [DOI] [PubMed] [Google Scholar]
- 30. Fuxe J., and Karlsson M. C. (2012) TGF-beta-induced epithelial-mesenchymal transition: a link between cancer and inflammation. Seminars Cancer Biol. 22, 455–461 [DOI] [PubMed] [Google Scholar]
- 31. Mathieu J., Zhang Z., Zhou W., Wang A. J., Heddleston J. M., Pinna C. M., Hubaud A., Stadler B., Choi M., Bar M., Tewari M., Liu A., Vessella R., Rostomily R., Born D., Horwitz M., Ware C., Blau C. A., Cleary M. A., Rich J. N., and Ruohola-Baker H. (2011) HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 71, 4640–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu X., Tan X., Tampe B., Sanchez E., Zeisberg M., and Zeisberg E. M. (2015) Snail is a direct target of hypoxia-inducible factor 1alpha (HIF1alpha) in hypoxia-induced endothelial to mesenchymal transition of human coronary endothelial cells. J. Biol. Chem. 290, 16653–16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang W., Shi X., Peng Y., Wu M., Zhang P., Xie R., Wu Y., Yan Q., Liu S., and Wang J. (2015) HIF-1alpha promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PloS One 10, e0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dziegiel P., Owczarek T., Plazuk E., Gomulkiewicz A., Majchrzak M., Podhorska-Okolow M., Driouch K., Lidereau R., and Ugorski M. (2010) Ceramide galactosyltransferase (UGT8) is a molecular marker of breast cancer malignancy and lung metastases. Br. J. Cancer 103, 524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Owczarek T. B., Suchanski J., Pula B., Kmiecik A. M., Chadalski M., Jethon A., Dziegiel P., and Ugorski M. (2013) Galactosylceramide affects tumorigenic and metastatic properties of breast cancer cells as an anti-apoptotic molecule. Plos One 8, e84191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cornell T. T., Fleszar A., McHugh W., Blatt N. B., Le Vine A. M., and Shanley T. P. (2012) Mitogen-activated protein kinase phosphatase 2, MKP-2, regulates early inflammation in acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 303, L251–L258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haagenson K. K., and Wu G. S. (2010) Mitogen activated protein kinase phosphatases and cancer. Cancer Biol. Ther. 9, 337–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keyse S. M. (2008) Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 27, 253–261 [DOI] [PubMed] [Google Scholar]
- 39. Wang H. Y., Cheng Z., and Malbon C. C. (2003) Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 191, 229–237 [DOI] [PubMed] [Google Scholar]
- 40. Jiang R., Li Y., Xu Y., Zhou Y., Pang Y., Shen L., Zhao Y., Zhang J., Zhou J., Wang X., and Liu Q. (2013) EMT and CSC-like properties mediated by the IKKbeta/IkappaBalpha/RelA signal pathway via the transcriptional regulator, Snail, are involved in the arsenite-induced neoplastic transformation of human keratinocytes. Arch. Toxicol. 87, 991–1000 [DOI] [PubMed] [Google Scholar]
- 41. Tian B., Li X., Kalita M., Widen S. G., Yang J., Bhavnani S. K., Dang B., Kudlicki A., Sinha M., Kong F., Wood T. G., Luxon B. A., and Brasier A. R. (2015) Analysis of the TGFbeta-induced program in primary airway epithelial cells shows essential role of NF-kappaB/RelA signaling network in type II epithelial mesenchymal transition. BMC Genomics 16, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoesel B., and Schmid J. A. (2013) The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., and Karin M. (2008) NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature 453, 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robin E. D., Murphy B. J., and Theodore J. (1984) Coordinate regulation of glycolysis by hypoxia in mammalian cells. J. Cell. Physiol. 118, 287–290 [DOI] [PubMed] [Google Scholar]
- 45. Behrooz A., and Ismail-Beigi F. (1997) Dual control of glut1 glucose transporter gene expression by hypoxia and by inhibition of oxidative phosphorylation. J. Biol. Chem. 272, 5555–5562 [DOI] [PubMed] [Google Scholar]
- 46. Pike J. W., and Meyer M. B. (2010) The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3). Endocrinol. Metab. Clin. North Am. 39, 255–269, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lovisa S., LeBleu V. S., Tampe B., Sugimoto H., Vadnagara K., Carstens J. L., Wu C. C., Hagos Y., Burckhardt B. C., Pentcheva-Hoang T., Nischal H., Allison J. P., Zeisberg M., and Kalluri R. (2015) Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 21, 998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Macheda M. L., Rogers S., and Best J. D. (2005) Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cellular Physiol. 202, 654–662 [DOI] [PubMed] [Google Scholar]
- 49. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 50. Hou J. C., and Pessin J. E. (2007) Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr. Opinion Cell Biol. 19, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoshida K., Kirito K., Yongzhen H., Ozawa K., Kaushansky K., and Komatsu N. (2008) Thrombopoietin (TPO) regulates HIF-1alpha levels through generation of mitochondrial reactive oxygen species. Int. J. Hematol. 88, 43–51 [DOI] [PubMed] [Google Scholar]
- 52. Blaschke K., Ebata K. T., Karimi M. M., Zepeda-Martinez J. A., Goyal P., Mahapatra S., Tam A., Laird D. J., Hirst M., Rao A., Lorincz M. C., and Ramalho-Santos M. (2013) Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amelio I., Cutruzzola F., Antonov A., Agostini M., and Melino G. (2014) Serine and glycine metabolism in cancer. Trends Biochem. Sci. 39, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vander Heiden M. G., Cantley L. C., and Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freemerman A. J., Johnson A. R., Sacks G. N., Milner J. J., Kirk E. L., Troester M. A., Macintyre A. N., Goraksha-Hicks P., Rathmell J. C., and Makowski L. (2014) Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 289, 7884–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macintyre A. N., Gerriets V. A., Nichols A. G., Michalek R. D., Rudolph M. C., Deoliveira D., Anderson S. M., Abel E. D., Chen B. J., Hale L. P., and Rathmell J. C. (2014) The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metabolism 20, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lopez-Novoa J. M., and Nieto M. A. (2009) Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 1, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ricciardi M., Zanotto M., Malpeli G., Bassi G., Perbellini O., Chilosi M., Bifari F., and Krampera M. (2015) Epithelial-to-mesenchymal transition (EMT) induced by inflammatory priming elicits mesenchymal stromal cell-like immune-modulatory properties in cancer cells. Br. J. Cancer 112, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Solaini G., Baracca A., Lenaz G., and Sgarbi G. (2010) Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta 1797, 1171–1177 [DOI] [PubMed] [Google Scholar]
- 60. Denko N. C. (2008) Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 8, 705–713 [DOI] [PubMed] [Google Scholar]
- 61. Otto A. M. (2016) Warburg effect(s) -a biographical sketch of Otto Warburg and his impacts on tumor metabolism. Cancer Metab. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cuyas E., Corominas-Faja B., and Menendez J. A. (2014) The nutritional phenome of EMT-induced cancer stem-like cells. Oncotarget 5, 3970–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shiraki N., Shiraki Y., Tsuyama T., Obata F., Miura M., Nagae G., Aburatani H., Kume K., Endo F., and Kume S. (2014) Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metabolism 19, 780–794 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry raw data was deposited via ProteomeXchange (http://www.proteomexchange.org) with identifiers IDs PXD005487 (TMT) and PXD007280 (LFQ). Processed data available as Table S1-S6 in the Supplemental.