Abstract
Asthma prevalence has been on the increase, especially in North America compared to other continents. However, the prevalence of asthma differs worldwide and in many countries the prevalence of asthma is stable or decreasing. This highlights the influence of environmental exposures, such as allergens, air pollution, and environmental microbiome, on disease etiology and pathogenesis. The epigenome may provide the unifying mechanism that translates the influence of environmental exposures to changes in gene expression, respiratory epithelial function, and immune cell skewing that are hallmarks of asthma. In this review, we will introduce the concept of the environmental epigenome in asthma, summarize previous publications of relevance to this field, and discuss future directions.
Keywords: asthma, DNA methylation, epigenetics, nasal epithelium, microbiome, air pollution, allergens
Asthma remains an important public health problem in the United States and worldwide because of high morbidity and inadequate disease control. Recent data from the International Study of Asthma and Allergies in Childhood (ISAAC) demonstrate geographical differences in asthma prevalence, suggesting that the environment and epigenetics play a key role in this disease. 1 While asthma is more problematic in children, this disease also affects the health of adults. The dynamic and unique biological responses that are triggered by allergens and air pollutants have proven difficult to predict and prevent. This review will focus on the epigenetic mechanisms that regulate the response to environmental exposures and are critical to primary and secondary prevention of asthma.
Etiology of asthma
While inheritance, 2–5 parent-of-origin, 6–8 general environment, 9–17 immunization,18 in utero exposures, 19–24 and Th2 immunity 25 play important roles in the etiology of asthma, there is no unifying mechanism accounting for these etiologic events.
Asthma concordance in MZ twins is only ~50%, 26 and the heritablitity of this disease is 0.40–0.85. 27, 28 Initial genome Wide Association Studies (GWAS) identified three putative susceptibility loci (5q22.1 near TMEM232, 11q13.5 near C11orf30, and the HLA region) associated with asthma risk, 3, 29 but the scope of these studies was limited. Moffatt et al. reported that genetic variants of ORMDL3 contribute to the risk of childhood asthma. 30 Recently, meta-analysis of GWAS identified seven asthma genetic risk loci (HLA-DQ, IL33, ORMDL3/GSDMB, IL1RL1/IL18R1, IL2RB, SMAD3, and TSLP 31, 32) and ten loci near TLR6, C11orf30, STAT6, SLC25A46, HLA-DQB1, IL1RL1, LPP, MYC, IL-2, and HLA-B that influence allergic sensitization. 33 In aggregate, approximately 50 replicated genes have been identified from association studies, several genes by linkage and fine mapping, and one gene identified by GWAS. However, the effect estimates are modest (odds ratios between 0.5 and 1.5), and it has been estimated that these gene variants predict less than 10% of the heritability of asthma. 34
Allergens and Asthma
Allergic sensitization is a critical risk factor for childhood asthma, conferring a 4–20 fold increase in the risk of developing disease. 10, 35 Both indoor (molds, house dust mites, cockroaches, rodents, and pets) and outdoor (pollens from trees, grass, and weeds) allergens are critical environmental triggers of asthma. 36–38 Seminal studies in this field have clearly demonstrated a role of the environment in the development of asthma; exposure to house dust mite, cat and dog allergen early in life and continuing throughout childhood determined the course of chronic airway hyper-responsiveness and impairment of lung function, 35 sensitization to dog, cat, or Alternaria was associated with increased bronchial responsiveness in children with mild to moderate asthma, 39 children who were both allergic to cockroach and exposed to high levels of this allergen had significantly more asthma-related hospitalizations, days of wheezing, missed school days, and nights with lost sleep, 40 and mouse sensitization/exposure was associated with acute care visits, decreased lung function, fraction of exhaled nitric oxide levels, and bronchodilator reversibility. 41 Pet allergens may also be protective in some settings although the evidence for this is mixed. 36, 42
Air pollution and Microbial Factors and the Severity of Asthma
Air pollutants are known to exacerbate the symptoms of asthma and may also play a role in the initiation of this disease. 43–45 In most urban areas, and increasingly suburban areas, components of traffic-related emissions are a major source of air pollution. However, air pollution represents a complex exposure with inorganic and organic components. Particulate matter (PM) carries both environmental pollutants such as polycyclic aromatic hydrocarbons (PAHs) formed during the incomplete combustion of fossil fuels and oil products, 46 and immune stimulating agents such as pollens, endotoxin and fungal spores. PAHs are widespread environmental contaminants formed as a result of incomplete combustion of organic materials and are a particularly toxic component of air pollution. 47, 48 PAHs in PM such as phenanthrene or other components of diesel exhaust can also directly enhance IgE production in vitro. 49 Endotoxin, a structural component of membranes of Gram negative bacteria, is ubiquitous in urban and suburban environments both in homes/daycares/schools 50–53 and is a component of air pollution. 54, 55 In addition to endotoxin, children who grow up in rural and inner city areas are exposed to many other aerosolized microorganisms and their toxins. 56, 57 Interestingly, germ-free (GF) mice are more prone to a Th2 phenotype and allergic airway disease, and this risk can be reduced by exposure to a diverse microbial flora. 58 Moreover, mono-colonization with a common intestinal bacterium Bacteroides fragilis restores Th2/Th1 balance using a mechanism dependent on stimulation of Th1 CD4+ T cell proliferation by a zwitterionic (neutral molecule with positive and negative charges) coat polysaccharide (ZPS) that directly activates CD4+ T cells, 59, 60 and can also be potent Treg cell inducers. 61 ZPSs have been characterized in commensals from multiple body sites including Streptococcus pneumoniae in the upper respiratory tract, and ZPSs from different organisms have similar biological properties and can display immune cross-reactivity. 59, 62, 63 Interestingly, the prevalence of Staphylococcus aureus (which produce ZPS and is present in bedding/household dust) was inversely associated with asthma. 64, 65
Introduction to Epigenetics
Epigenetic mechanisms control expression levels of genes without changing DNA sequence. Hypermethylation of cytosines within CpG islands in gene promoters leads to gene silencing and hypomethylation leads to active transcription. 66, 67 More recent studies have demonstrated that methylation of less CpG dense regions near islands (‘shores’)68,69 and within gene bodies 70, 71 is also important in regulation of gene expression and alternative splicing, and that the relationship between methylation and transcription is more complex. Further adding to this complexity is the presence of methylation marks in non-CpG context 72, 73 and presence of 5-hydroxymethylcytosine, which may be a mark of demethylation.74 Finally, the most recent data from international consortia (ENCODE75, FANTOM576, and Roadmap Epigenomics77) identify enhancers as critical regions involved in regulation of gene expression in addition to promoters. Promoters and enhancers are also characterized by the presence of a number of histone modifications, with histone methylation and acetylation being the most common.78, 79 Acetylation of lysine 27 on the histone H3 (H3K27ac) is one of the most informative single histone modifications. It is known to mark active enhancers and is also enriched at active promoters. Among histone methylation marks, H3K4me1 is present at poised and active enhancers while H3K4me3 marks poised or active promoters. Although non-coding RNAs such as micro RNAs (miRNAs) and long intergenic noncoding RNAs (lncRNAs) are sometimes viewed as a part of the epigenome as they are involved in regulation of gene expression,80 they will not be discussed in the current review and are reviewed elsewhere.81
Environment and the Epigenome
While some of the epigenetic marks are heritable (such as imprinted loci82, for example) and genome-wide studies demonstrate a genetic component to inter-individual variation in DNA methylation 83–86 and histone modification profiles, 83–85 epigenetic marks are also strongly influenced by the environment.86 Epigenetic processes translate environmental exposures associated with disease risk into regulation of chromatin, which shapes the identity, gene expression profile, and activity of specific cell types that participate in disease pathophysiology.87 Both DNA methylation and histone modifications are mutable and dynamic, responding to the environment, disease and aging.67, 86, 88 These alterations may persist for the life of a cell, demonstrate transgenerational inheritance, or may be altered prenatally and postnatally, influencing gene expression differentially throughout life.89, 90 We have previously proposed that epigenetic marks may be the missing link that connects environmental exposures in genetically predisposed individuals to transcriptional changes associated with development of asthma.91
Asthma Epigenetics
Epigenetic mechanisms, as a cause of asthma,91 build on our current knowledge about the etiology of asthma: non-Mendelian3 and parent-of-origin inheritance,6 influence of direct9 and in utero92 exposures, and a strong component of immune regulation.25 Our early work in mice showed that in utero exposure to high methyl donor diet resulted in an increase in airway inflammation (eosinophil recruitment and concentrations of IL-4 and IL-13), increase in serum IgE, a skewing of the lymphocytes toward a Th2 phenotype, and hypermethylation of Runx3, a transcription factor involved in regulation of Th2 immunity (unpublished data). It is well established that epigenetic mechanisms regulate expression of transcription factors and cytokines important in T cell differentiation (Th1, Th2, and Tregs).93–100 Another animal study contributed to our knowledge of the critical role of DNA methylation in allergic airway disease by showing that global DNA demethylation agent 5-aza-2′-deoxycytidine (5-AZA) prevented Th2 skewing and rebalanced Th1/Th2, and used adoptive transfer experiments to demonstrate that 5-AZA treated CD4+ T cells protect against allergic airway disease.101
Early studies in human cohorts demonstrated an association of DNA methylation in a few candidate genes in peripheral blood102, and buccal103, 104 and nasal105 cells with asthma phenotypes but did not elucidate the role of DNA methylation in the control of gene expression. Our work in African American inner city children identified 81 differentially methylated regions (DMRs) in peripheral blood mononuclear cells (PBMCs) associated with allergic asthma.106 Methylation changes in PBMCs are modest (median 1.3%; range 0.02%–3.1%) but consistent with the majority of DMRs hypomethylated in asthma. Several immune genes were hypomethylated in asthma, including IL-13, RUNX3, and TIGIT. Hypo- and hypermethylated genes were associated with increased and decreased gene expression respectively (P<0.6×10−11). We further explored the relationship between DNA methylation and gene expression using an integrative analysis and identified additional candidates relevant to asthma (IL-4 and ST2). Our group also contributed to a study that identified replicated associations, in three independent cohorts, between IgE and low methylation at 36 loci.107 Genes annotated to these loci encode known eosinophil products, and also implicate phospholipid inflammatory mediators, specific transcription factors, and mitochondrial proteins. We confirmed that methylation at these loci differed significantly in isolated eosinophils from subjects with and without high IgE levels. The top three loci accounted for 13.5% of IgE variation, explaining 10 fold higher variance than that derived from large genome-wide association studies (GWAS). A recent publication by another group identified asthma-specific enhancers in primary CD4+ T cells, marked by gaining the histone H3K4me2 mark during Th2 cell development.108
In the nasal epithelia of inner city African American children, we identified substantial (median 9.5%, range: 2.6–29.5% methylation change) methylation changes, both in the form of single CpG methylation (differentially methylated probes[DMPs]) and regions (DMRs) that are associated with their disease and changes in gene expression.109 The magnitude of the methylation changes observed in nasal epithelia of these asthmatic children approaches that reported in malignancies.110, 111 60% of genes that are differentially expressed in the asthmatic nasal epithelium have significant associations between DNA methylation and gene expression; these include asthma genes (ALOX15, CAPN14, POSTN), genes involved in inflammation and immunity, cell adhesion, extracellular matrix, obesity and autophagy, and epigenetic regulators, among others. 30% of the genes we identified were also found in an IL-13 DNA methylome signature of cultured airway epithelial cells of asthmatics, additionally demonstrating the relevance of our findings to allergic asthma. Collectively, these studies underscore the importance of the epigenome as a modifier of transcriptional profiles associated with asthma and demonstrate the potential utility of profiling nasal epithelia in the context of studying environmental exposures in asthma.
Nasal Epithelium as a Biosensor of the Environment with Relevance to the Disease Process
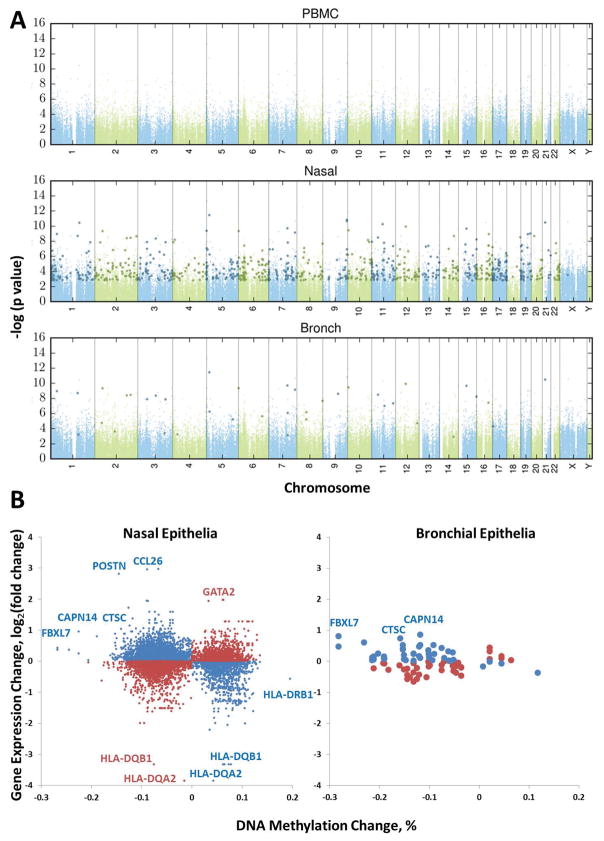
The nasal and airway epithelium is the primary interface with the respirable environment, interacts with air pollution,43 allergens112–114 and other environmental stimuli,115 and directs airway inflammatory, immune, and regenerative responses to these exposures. Gene expression profiles of the asthmatic airway epithelium have identified genes associated with exposure to endotoxin,116 house dust mite allergen,116 cigarette smoke,117 asthma,117, 118 and disease subtypes.119 Importantly, it has been shown that the nasal epithelium is a reasonable proxy for the airway epithelium; nasal airway transcriptome mirrors the bronchial airway transcriptome and reflects asthma status as well as Th2-related subphenotypes of disease.120 Our more recent work demonstrates that nasal epithelia capture disease activity seen in the lung airway epithelia but that there are many more significant associated DNA methylation changes in the nasal epithelia (Figure 1), suggesting an important role for the environment in influencing these epigenetic changes121 and the need to understand environmental exposures that are driving these changes.122 While the solubility and particle size of ambient air pollutants123 and allergens114 are key determinants of deposition and response, the nasal epithelium is the most proximal portion of the airway and may represent the only portion of the airway that comes in contact with relevant components of the environment that trigger airway responses. As such, nasal epithelium is constantly exposed to ozone, endotoxin, allergens, and other toxins from air pollution, functions to filter air pollution particles by the process of mucociliary clearance, and is an active component of both the immune and respiratory systems.
Figure 1.
Asthma-associated DNA methylation and gene expression changes in nasal epithelia of non-Hispanic White nonsmoker allergic asthmatics. (A) Differentially methylated single-CpG probes (DMPs) in bronchial and nasal epithelia, but not PBMCs, are associated with asthma after controlling for age, gender, technical variables, and batch effects in Caucasian adult subjects. Manhattan plot of the false discovery rate (FDR) adjusted p-values (q-values) for disease status (asthma/control) from the tissue-specific linear model. Probes with q<0.05 in the tissue-specific linear model are highlighted by darker larger symbols. (B) DNA methylation changes are associated with changes in gene expression in nasal and bronchial epithelia. Expression changes of genes nearest DMPs from part A. X-axis methylation difference is represented by the mean % methylation difference in asthma subjects compared to controls; y-axis expression difference is represented by the mean fold change in asthma subjects compared to controls (on the log2 scale). The blue symbols represent hypomethylated genes that were associated with increased gene expression as well as some hypermethylated genes associated with decreased gene expression. The red symbols represent methylation changes that were not associated with expected gene expression differences. Reprinted with permission of the American Thoracic Society from Yang et al.121 Copyright © 2017 American Thoracic Society. The American Journal of Respiratory and Critical Care Medicine is an official journal of the American Thoracic Society.
Epigenetic Changes Associated with Exposure to Air Pollution
Air pollution influences the peripheral blood epigenome among adults.124–135 These studies have shown that both short125, 127, 133 and long126, 129 term exposures to PM impact DNA methylation, and includes genes in innate immunity (TLR4, TLR2)125, 136 and asthma (HLA-DOB, HLA-DPA1, CCL11, CD40LG, ECP, FCER1A, FCER1G, IL9, IL10, IL13, MBP).129 One study has also demonstrated the effect of PM10 and PMBC on 5-hydroxymethylcytosine134. Similarly, cigarette smoke exposure has profound influence on DNA methylation among adult smokers137, 138 and as second hand smoke exposure (ETS) in childhood139, 140 and in utero,141–143 and also affects innate immune genes (CD14).139 Diesel exhaust particle exposure (DEP) has been associated with DNA methylation changes in peripheral blood144 and lung epithelial cells.145 DNA methylation in adults146 and children both as a result of direct147 and in utero148, 149 exposure is also influenced by PAHs, a bi-product of incomplete combustion of organic materials in airborne pollution. In children, PAH exposure has been associated with increased methylation of IFNG148 and the Treg transcription factor FOXP3 as well as impaired Treg function.147
Epigenetic Changes Associated with Exposure and Sensitization to Allergens
Very few studies to date have examined the relationship of allergen exposure and sensitization to changes in epigenetic marks. Sensitization to several allergens (tree, grass, house dust mite, and ragweed) has been associated with changes in DNA methylation in peripheral blood of older adults150 and in bronchial epithelial cells of adults in controlled exposure settings.145 CD4+ T cells isolated from ex vivo grass pollen extract stimulated PBMCs of patients with seasonal allergic rhinitis have extensive DNA methylation and gene expression changes compared to control patients.151 Decreased DNA methylation in the CD14 promoter has been associated with increased CD14 expression in placentas of mothers living on a farm compared with mothers not living on a farm 152 and methyl-CpG-binding protein Mbd2 has been shown to control Th2 induction by dendritic cells.153 No studies have been performed on specific indoor allergens.
A few studies have begun to understand the complex relationship of multiple exposures. DEP exposure in combination with allergen resulted in hypermethylation of the Th1 cytokine Ifng and hypomethylation of the Th2 cytokine Il4 in mice.154 In human airway epithelial cells, controlled exposure to allergen alone, diesel exhaust alone, or allergen and diesel exhaust together (coexposure) led to significant changes in only 7 CpG sites genome-wide at 48 hours.145 However, when the same lung was exposed to allergen and diesel exhaust but separated by approximately 4 weeks, significant changes in more than 500 sites were observed. These findings suggest that specific exposures can prime the lung for changes in DNA methylation induced by a subsequent insult.
Epigenetic changes and the Microbiome
The microbiome represents the multitude of microbes (bacteria, archaea, microbial eukaryotes such as fungi, and viruses) that live in the environment and that also inhabit our bodies. Exposure to a greater diversity or unique repertoire of microbes via bedding or household dust, 64, 155,65 birth by vaginal rather than cesarean section,156 or relatively restricted exposure to antibiotics in early life157–159 have all been associated with decreased incidence of childhood asthma in epidemiological studies. Furthermore, germ-free (GF) mice (i.e. mice born and raised in sterile isolators devoid of microbes) have increased airway resistance, increased total bronchoalveolar lavage fluid cell numbers, eosinophilia, and proinflammatory cytokine production, and higher serum IgE levels compared with conventional specific-pathogen-free (SPF) mice in an ovalbumin (OVA)–driven allergic-asthma mouse model.160 The relationship between microbial exposures and asthma susceptibility is particularly strong in early life. For instance, in humans, the largest reduction in risk of developing respiratory allergies with exposure to a farming environment is seen among those who are exposed prenatally and continuously thereafter.155 Furthermore, colonization of neonatal but not adult GF mice with a conventional microbiota protected them from OVA-driven allergic asthma.160
The mechanisms that mediate the influence of early-life microbial exposures on asthma/immune phenotypes are largely not understood, but microbially-mediated modification to the epigenome may in some cases provide this “missing link.” We are only beginning to understand the extent to which the microbiome exerts influence on host gene expression via modification to the epigenome. Different gut (fecal) microbiota compositions in humans have been associated with distinctive DNA methylomes in blood. 161 Experiments with GF mice have also indicated a link; for instance the methylation level of the TLR4 gene was significantly lower in Intestinal Epithelial Cells (IECs) of the large intestine of GF compared to conventional mice.162 Finally, in vitro experiments have allowed for exploration of how different microbes may affect the epigenome; immature enterocytes exposed to probiotic and pathogenic bacteria showed over 200 regions of differential DNA methylation, with decreased DNA methylation of genes associated with cytoskeleton/actin remodeling and cell adhesion functions after exposure to pathogenic gram-negative bacteria (Klebsiella spp.).163 Interestingly, fetal epithelial cells were more sensitive to these pathogenic-specific changes than adult epithelial cells.163
The mechanisms by which specific microbes may affect the epigenome are not well known but include via their metabolic activity. For instance, the short-chain-fatty-acid (SCFA) butyrate, a key product of the microbial fermentation of dietary fiber in the gut, induces the differentiation of colonic Treg cells and this was associated with enhanced histone H3 acetylation in the promoter and conserved non-coding sequence regions of the Foxp3 locus.164 It has also been proposed that microbiota can exert influence by altering the availability of chemical donors for DNA or histone modifications,165 for instance various probiotic bacteria have the ability to produce/consume folate and can affect both plasma and fecal folate levels following oral consumption.166 The relationship between the epigenome and microbiome can also “go both ways”, with differential epigenetic marks also affecting which microbes colonize hosts; for instance, epigenetic alterations induced by treating pregnant mice with dexamethasone were associated with altered composition of the gut microbiome of their offspring.163
Supporting a specific importance for epigenome modifications in mediating early microbe exposures and development of asthma, one study showed epigenome modification as a driver of the protection conferred by early colonization of GF mice in an OVA–driven allergic-asthma mouse model.160 Specifically, the accumulation of invariant natural killer T (iNKT) cells in the colonic lamina propria and lung in 8 week old GF mice, 160 was linked to increased expression of the mRNA for CXCL16, a ligand of chemokine receptor CXCR6 on iNKT cells. A region 5′ of the Cxcl16 gene that contains five potential CpG sites was hypermethylated in the colon and lungs, but not in other tissues such as the spleen and liver, in GF compared to SPF conditions. Since this observation was made by comparing GF mice to those colonized with a diverse microbiota, the specific bacteria and molecular factors that mediate this epigenetic modification remain to be discovered.
The complex interplay between early life microbial exposures and those that occur over time in the state of the epigenome and development of asthma susceptibility/exacerbations is not well understood. Given the high magnitude of methylation changes in genes differentially expressed in the asthmatic nasal epithelium, 109 it is interesting to consider that the nasal cavity is home to a complex and poorly understood community of microorganisms. Since mucous in the nasal cavity captures PM, some component on the nasal microbiome certainly represents the “respirable” microbiome (i.e. microbes that have been deposited on inhaled particles); the nasal microbiome thus can in part can be considered to be an “exposure” and in part a community that have adapted to inhabit and in some cases closely interact with the host.
Prior culture-independent studies of nasal microbiome have focused on bacteria in the nasal vestibule (e.g. the anterior nares) and nasal cavity, and have found the genera Corynebacterium, Propionibacterium, and Staphylococcus (all common skin bacteria) to be among the most frequent colonizers167, 168 with a relatively low prevalence of Propionibacterium that can use sebum as a growth substrate in the nasal cavity compared to vestibule.167 Specific microbial taxa169 and their expressed genes170 in the nasal cavity differ significantly between asthmatics and controls. Asthma-associated nasal microbes are also associated with differential expression of host genes such as increased expression of mediators of inflammation169, 170 and apoptosis.169
Future Perspective
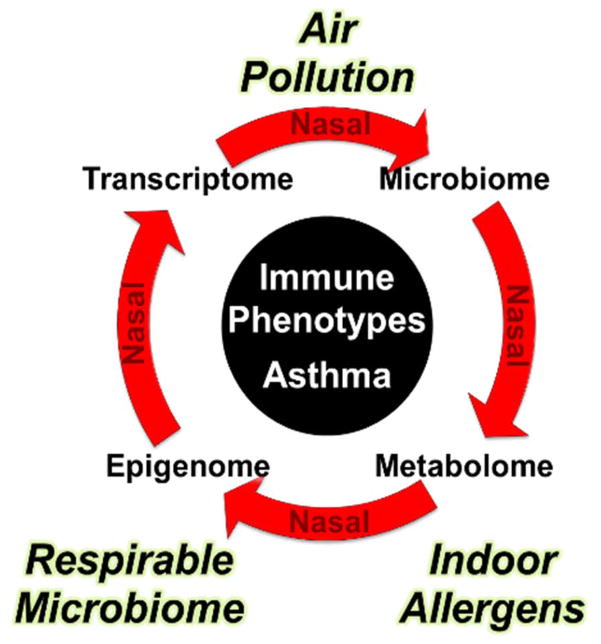
A major goal of this field is to understand the complex interaction of the microbiome, indoor allergens, and air pollution with the dynamic biological responses in the nares that predispose toward a Th2 phenotype and place individuals at risk of asthma (Figure 2). These environmental factors are likely to influence the epigenome differently based on genetic variants of the host. Isolated studies focused on genetics, epigenetics, pathobiology, and the microbiota have so far provided only a partial picture regarding disease risk and its mechanistic basis. A comprehensive and integrated approach to asthma will: 1) establish the basic molecular profiles to develop novel molecular insights into disease etiology and clinical severity/extent; 2) produce environmental and biological signatures that will create a roadmap for primary and secondary prevention of asthma; and 3) provide the rationale and targets/biomarkers for intervention in this disease that remains a significant public health problem, especially among children. While other tissues/cell types may be valuable for studies focused on immunology of asthma, the nasal epithelium is the ideal surrogate tissue for studies of environmental asthma as it is easily accessible in children and adults, has substantial epigenetic and gene expression changes associated with asthma, and appears to be an excellent proxy for the bronchial epithelium.
Figure 2.
Conceptual approach to integrative analysis of exposures and the epigenome/transcriptome in asthma. Sophisticated network approaches will be required to assess how environmental exposures (air pollution, allergens, microbiome), in the context of genetic susceptibility, interact to lead to changes in the epigenome and transcriptome, and ultimately to Th2 cell skewing and specific disease phenotypes in asthma. Our ability to predict, prevent, and control asthma will be substantially advanced by understanding the complex interactive relationships of the microbiome, allergens, and air pollutants with the dynamic biological responses in the airways that predispose toward a Th2 phenotype.
The integrative approach would require the use of systems biology to understand how different elements of the environment, in the context of genetic susceptibility, interact to lead to changes in the epigenome and transcriptome, and ultimately to Th2 cell skewing and specific disease phenotypes. Systems biology generally refers to a process of identifying networks of molecular pathways based on the evidence from multiple datasets and has been reviewed in the context of asthma.171
Findings from integrative studies will identify elements of exposures that have the most prominent effect on the epigenome/transcriptome. It is likely that exposures will be different depending on the geographical region but also that the host will respond differently to them based on their genetics and epigenetics, suggesting that personalized intervention will be needed, which aligns well with the goals of personalized or precision medicine.172 Importantly, we view epigenetic marks influenced by exposures as biomarkers of disease that would allow us to treat patients early in the course of the disease but may also provide specific treatments for different asthma endotypes173 and difficult-to-control asthma.174 Developing a greater understanding of how exposures to modifiable factors such as the microbiome may dynamically affect the epigenome of key immune and or asthma-associated genes may pave to way to novel intervention strategies.
These approaches will also prioritize genes that have epigenetic marks influenced by multiple exposures as the most relevant candidates for therapeutic intervention. DNA methylation changes have been shown to drive tumor formation and malignant progression,175 and as such have established basic mechanisms for disease pathogenesis and targets for intervention in cancer. DNA methyltransferase (DNMT) inhibitors have been approved for the treatment of myelodysplastic syndrome176, 177 and are in clinical trials for treatment of solid tumors.178, 179 While currently available DNMT inhibitors lack specificity for gene(s) of interest, locus-specific therapies are currently being developed.175, 180
Our ability to predict, prevent, and control asthma will be substantially advanced by understanding the complex interactive relationships of the microbiome, indoor allergens, and air pollutants with the dynamic biological responses in the airways that predispose toward a Th2 phenotype.
Abbreviations
- 5-AZA
5-aza-2′-deoxycytidine
- CpG
cytosine followed by guanine in DNA sequence
- DEP
diesel exhaust particle
- DMP
differentially methylated probe or position
- DMR
differentially methylated region
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- GF
germ free
- GWAS
genomewide association
- IEC
intestinal epithelial cells
- iNTK
invariant natural killer
- miRNA
micro RNA
- ncRNA
non-coding RNA
- OVA
ovalbumin
- PAH
poluaromatic hydrocarbon
- PBMC
peripheral blood mononuclear cells
- PM
particulate matter
- RNA
ribonucleic acid
- SCFA
short-chain-fatty-acid
- SNP
single nucleotide polymorphism
- SPF
specific pathogen free
- Th
T helper cells
- ZPS
zwitterionic coat polysaccharide
References
- 1.Mallol J, Crane J, von Mutius E, Odhiambo J, Keil U, Stewart A. The International Study of Asthma and Allergies in Childhood (ISAAC) Phase Three: a global synthesis. Allergol Immunopathol (Madr) 2013;41:73–85. doi: 10.1016/j.aller.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 3.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 4.March ME, Sleiman PM, Hakonarson H. The genetics of asthma and allergic disorders. Discov Med. 2011;11:35–45. [PubMed] [Google Scholar]
- 5.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–46. e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Cookson WO. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy. 1998;28(Suppl 1):56–61. doi: 10.1046/j.1365-2222.1998.0280s1056.x. discussion 5–6. [DOI] [PubMed] [Google Scholar]
- 7.Cookson WO, Young RP, Sandford AJ, Moffatt MF, Shirakawa T, Sharp PA, et al. Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet. 1992;340:381–4. doi: 10.1016/0140-6736(92)91468-n. [DOI] [PubMed] [Google Scholar]
- 8.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001;29:175–8. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 9.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–84. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ. Exposure to house-dust mite allergen (Der p I) and the development of asthma in childhood. A prospective study [see comments] N Engl J Med. 1990;323:502–7. doi: 10.1056/NEJM199008233230802. [DOI] [PubMed] [Google Scholar]
- 11.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 Variants and Smoking Exposure in Early-Onset Asthma. N Engl J Med. 2008 doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 12.Oh SS, Tcheurekdjian H, Roth LA, Nguyen EA, Sen S, Galanter JM, et al. Effect of secondhand smoke on asthma control among black and Latino children. J Allergy Clin Immunol. 2012;129:1478–83. e7. doi: 10.1016/j.jaci.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan-Yeung M, Malo JL. Occupational asthma. N Engl J Med. 1995;333:107–12. doi: 10.1056/NEJM199507133330207. [DOI] [PubMed] [Google Scholar]
- 14.Samet JM, Lambert WE. Epidemiologic approaches for assessing health risks from complex mixtures in indoor air. Environ Health Perspect. 1991;95:71–4. doi: 10.1289/ehp.919571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folkerts G, Busse WW, Nijkamp FP, Sorkness R, Gern JE. Virus-induced airway hyperresponsiveness and asthma. Am J Respir Crit Care Med. 1998;157:1708–20. doi: 10.1164/ajrccm.157.6.9707163. [DOI] [PubMed] [Google Scholar]
- 16.Gereda J, Leung D, Thatayatikom A, Streib J, Price M, Klinnert M, et al. Relation between house-dust endotoxin exposure, type 1 T-cell development, and allergen sensitisation in infants at high risk of asthma. Lancet. 2000;355:1680–3. doi: 10.1016/s0140-6736(00)02239-x. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz DA. Inhaled endotoxin, a risk for airway disease in some people. Respir Physiol. 2001;128:47–55. doi: 10.1016/s0034-5687(01)00264-x. [DOI] [PubMed] [Google Scholar]
- 18.Shirakawa T, Enomoto T, Shimazu SI, Hopkin JM. The inverse association between tuberculin responses and atopic disorder. Science. 1997;275:77–9. doi: 10.1126/science.275.5296.77. [DOI] [PubMed] [Google Scholar]
- 19.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest. 2005;127:1232–41. doi: 10.1378/chest.127.4.1232. [DOI] [PubMed] [Google Scholar]
- 20.Henderson AJ, Newson RB, Rose-Zerilli M, Ring SM, Holloway JW, Shaheen SO. Maternal Nrf2 and gluthathione-S-transferase polymorphisms do not modify associations of prenatal tobacco smoke exposure with asthma and lung function in school-aged children. Thorax. 2010;65:897–902. doi: 10.1136/thx.2009.125856. [DOI] [PubMed] [Google Scholar]
- 21.Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:660–2. doi: 10.1513/pats.200907-065DP. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Pinkerton KE. Air pollutant effects on fetal and early postnatal development. Birth Defects Res C Embryo Today. 2007;81:144–54. doi: 10.1002/bdrc.20097. [DOI] [PubMed] [Google Scholar]
- 23.Clark NA, Demers PA, Karr CJ, Koehoorn M, Lencar C, Tamburic L, et al. Effect of early life exposure to air pollution on development of childhood asthma. Environ Health Perspect. 2010;118:284–90. doi: 10.1289/ehp.0900916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzsimon N, Fallon U, O’Mahony D, Loftus BG, Bury G, Murphy AW, et al. Mothers’ dietary patterns during pregnancy and risk of asthma symptoms in children at 3 years. Ir Med J. 2007;100(suppl):27–32. [PubMed] [Google Scholar]
- 25.Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol. 2010;10:838–48. doi: 10.1038/nri2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Skadhauge LR, Backer V. Increase in the heritability of asthma from 1994 to 2003 among adolescent twins. Respir Med. 2011;105:1147–52. doi: 10.1016/j.rmed.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Los H, Postmus PE, Boomsma DI. Asthma genetics and intermediate phenotypes: a review from twin studies. Twin Res. 2001;4:81–93. doi: 10.1375/1369052012191. [DOI] [PubMed] [Google Scholar]
- 28.Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy. 2006;36:1382–90. doi: 10.1111/j.1365-2222.2006.02512.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 31.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC, et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–6. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss ST, Silverman EK. Pro: Genome-wide association studies (GWAS) in asthma. Am J Respir Crit Care Med. 2011;184:631–3. doi: 10.1164/rccm.201103-0485ED. [DOI] [PubMed] [Google Scholar]
- 35.Illi S, von Mutius E, Lau S, Niggemann B, Gruber C, Wahn U, et al. Perennial allergen sensitisation early in life and chronic asthma in children: a birth cohort study. Lancet. 2006;368:763–70. doi: 10.1016/S0140-6736(06)69286-6. [DOI] [PubMed] [Google Scholar]
- 36.Ahluwalia SK, Matsui EC. The indoor environment and its effects on childhood asthma. Curr Opin Allergy Clin Immunol. 2011;11:137–43. doi: 10.1097/ACI.0b013e3283445921. [DOI] [PubMed] [Google Scholar]
- 37.Baldacci S, Maio S, Cerrai S, Sarno G, Baiz N, Simoni M, et al. Allergy and asthma: Effects of the exposure to particulate matter and biological allergens. Respir Med. 2015;109:1089–104. doi: 10.1016/j.rmed.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 38.von Mutius E. Environmental factors influencing the development and progression of pediatric asthma. J Allergy Clin Immunol. 2002;109:S525–32. doi: 10.1067/mai.2002.124565. [DOI] [PubMed] [Google Scholar]
- 39.Nelson HS, Szefler SJ, Jacobs J, Huss K, Shapiro G, Sternberg AL. The relationships among environmental allergen sensitization, allergen exposure, pulmonary function, and bronchial hyperresponsiveness in the Childhood Asthma Management Program. J Allergy Clin Immunol. 1999;104:775–85. doi: 10.1016/s0091-6749(99)70287-3. [DOI] [PubMed] [Google Scholar]
- 40.Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, et al. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. N Engl J Med. 1997;336:1356–63. doi: 10.1056/NEJM199705083361904. [DOI] [PubMed] [Google Scholar]
- 41.Ahluwalia SK, Peng RD, Breysse PN, Diette GB, Curtin-Brosnan J, Aloe C, et al. Mouse allergen is the major allergen of public health relevance in Baltimore City. J Allergy Clin Immunol. 2013;132:830–5. e1–2. doi: 10.1016/j.jaci.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fretzayas A, Kotzia D, Moustaki M. Controversial role of pets in the development of atopy in children. World J Pediatr. 2013;9:112–9. doi: 10.1007/s12519-013-0412-6. [DOI] [PubMed] [Google Scholar]
- 43.Guarnieri M, Balmes JR. Outdoor air pollution and asthma. Lancet. 2014;383:1581–92. doi: 10.1016/S0140-6736(14)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann JK, Balmes JR, Bruckner TA, Mortimer KM, Margolis HG, Pratt B, et al. Short-term effects of air pollution on wheeze in asthmatic children in Fresno, California. Environ Health Perspect. 2010;118:1497–502. doi: 10.1289/ehp.0901292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng YY, Rull RP, Wilhelm M, Lombardi C, Balmes J, Ritz B. Outdoor air pollution and uncontrolled asthma in the San Joaquin Valley, California. J Epidemiol Community Health. 2010;64:142–7. doi: 10.1136/jech.2009.083576. [DOI] [PubMed] [Google Scholar]
- 46.Brandt HC, Watson WP. Monitoring human occupational and environmental exposures to polycyclic aromatic compounds. Ann Occup Hyg. 2003;47:349–78. doi: 10.1093/annhyg/meg052. [DOI] [PubMed] [Google Scholar]
- 47.Somers CM, Yauk CL, White PA, Parfett CL, Quinn JS. Air pollution induces heritable DNA mutations. Proc Natl Acad Sci U S A. 2002;99:15904–7. doi: 10.1073/pnas.252499499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.EPA U. National Air Quality and Emissions Trends Report. Research Triangle Park, NC: US Environmental Protection Agency; 2003. [Google Scholar]
- 49.Tsien A, Diaz-Sanchez D, Ma J, Saxon A. The organic component of diesel exhaust particles and phenanthrene, a major polyaromatic hydrocarbon constituent, enhances IgE production by IgE-secreting EBV-transformed human B cells in vitro. Toxicol Appl Pharmacol. 1997;142:256–63. doi: 10.1006/taap.1996.8063. [DOI] [PubMed] [Google Scholar]
- 50.Michel O, Kips J, Duchateua J, Vertongen F, Robert L, Collet H, et al. Severity of asthma is related to endotoxin in house dust. Am J Respir Crit Care Med. 1996;154:1641–6. doi: 10.1164/ajrccm.154.6.8970348. [DOI] [PubMed] [Google Scholar]
- 51.Milton D, Johnson D, Park J. Environmental endotoxin measurement: interference and sources of variation in the limulus assay of house dust. Am Ind Hyg Assoc J. 1997;58:861–7. doi: 10.1080/15428119791012199. [DOI] [PubMed] [Google Scholar]
- 52.Rullo VE, Rizzo MC, Arruda LK, Sole D, Naspitz CK. Daycare centers and schools as sources of exposure to mites, cockroach, and endotoxin in the city of Sao Paulo, Brazil. J Allergy Clin Immunol. 2002;110:582–8. doi: 10.1067/mai.2002.127511. [DOI] [PubMed] [Google Scholar]
- 53.Gereda JE, Leung DY, Liu AH. Levels of environmental endotoxin and prevalence of atopic disease. JAMA. 2000;284:1652–3. doi: 10.1001/jama.284.13.1652. [DOI] [PubMed] [Google Scholar]
- 54.Bonner JC, Rice AB, Lindroos PM, O’Brien PO, Dreher KL, Rosas I, et al. Induction of the lung myofibroblast PDGF receptor system by urban ambient particles from Mexico City. Am J Respir Cell Mol Biol. 1998;19:672–80. doi: 10.1165/ajrcmb.19.4.3176. [DOI] [PubMed] [Google Scholar]
- 55.Villarreal-Calderon A, Acuna H, Villarreal-Calderon J, Garduno M, Henriquez-Roldan CF, Calderon-Garciduenas L, et al. Assessment of physical education time and after-school outdoor time in elementary and middle school students in south Mexico City: the dilemma between physical fitness and the adverse health effects of outdoor pollutant exposure. Arch Environ Health. 2002;57:450–60. doi: 10.1080/00039890209601437. [DOI] [PubMed] [Google Scholar]
- 56.von Mutius E. Asthma and allergies in rural areas of Europe. Proc Am Thorac Soc. 2007;4:212–6. doi: 10.1513/pats.200701-028AW. [DOI] [PubMed] [Google Scholar]
- 57.Roy SR, Schiltz AM, Marotta A, Shen Y, Liu AH. Bacterial DNA in house and farm barn dust. J Allergy Clin Immunol. 2003;112:571–8. doi: 10.1016/s0091-6749(03)01863-3. [DOI] [PubMed] [Google Scholar]
- 58.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzianabos AO, Onderdonk AB, Rosner B, Cisneros RL, Kasper DL. Structural features of polysaccharides that induce intra-abdominal abscesses. Science. 1993;262:416–9. doi: 10.1126/science.8211161. [DOI] [PubMed] [Google Scholar]
- 60.Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol. 2010;28:107–30. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 61.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–58. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 62.Stingele F, Corthesy B, Kusy N, Porcelli SA, Kasper DL, Tzianabos AO. Zwitterionic polysaccharides stimulate T cells with no preferential V beta usage and promote anergy, resulting in protection against experimental abscess formation. J Immunol. 2004;172:1483–90. doi: 10.4049/jimmunol.172.3.1483. [DOI] [PubMed] [Google Scholar]
- 63.Neff CP, Rhodes ME, Arnolds KL, Collins CB, Donnelly J, Nusbacher N, et al. Diverse Intestinal Bacteria Contain Putative Zwitterionic Capsular Polysaccharides with Anti-inflammatory Properties. Cell Host Microbe. 2016;20:535–47. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 65.Pakarinen J, Hyvarinen A, Salkinoja-Salonen M, Laitinen S, Nevalainen A, Makela MJ, et al. Predominance of Gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environ Microbiol. 2008;10:3317–25. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- 66.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 67.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 68.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 71.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–42. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 72.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–6. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 75.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu AH, Babineau DC, Krouse RZ, Zoratti EM, Pongracic JA, O’Connor GT, et al. Pathways through which asthma risk factors contribute to asthma severity in inner-city children. J Allergy Clin Immunol. 2016;138:1042–50. doi: 10.1016/j.jaci.2016.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 80.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 81.Pua HH, Ansel KM. MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol. 2015;36:101–8. doi: 10.1016/j.coi.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lawson HA, Cheverud JM, Wolf JB. Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet. 2013;14:609–17. doi: 10.1038/nrg3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–2. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kilpinen H, Waszak SM, Gschwind AR, Raghav SK, Witwicki RM, Orioli A, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–7. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–9. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2009. [Google Scholar]
- 88.Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–83. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. J Allergy Clin Immunol. 2012;130:1243–55. doi: 10.1016/j.jaci.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharma S, Chhabra D, Kho AT, Hayden LP, Tantisira KG, Weiss ST. The genomic origins of asthma. Thorax. 2014;69:481–7. doi: 10.1136/thoraxjnl-2014-205166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–52. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.White GP, Hollams EM, Yerkovich ST, Bosco A, Holt BJ, Bassami MR, et al. CpG methylation patterns in the IFNgamma promoter in naive T cells: variations during Th1 and Th2 differentiation and between atopics and non-atopics. Pediatr Allergy Immunol. 2006;17:557–64. doi: 10.1111/j.1399-3038.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 95.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- 96.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 97.Webster RB, Rodriguez Y, Klimecki WT, Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J Biol Chem. 2007;282:700–9. doi: 10.1074/jbc.M609501200. [DOI] [PubMed] [Google Scholar]
- 98.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–79. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A. 2007;104:17052–7. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 101.Brand S, Kesper DA, Teich R, Kilic-Niebergall E, Pinkenburg O, Bothur E, et al. DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. J Allergy Clin Immunol. 2012;129:1602–10. e6. doi: 10.1016/j.jaci.2011.12.963. [DOI] [PubMed] [Google Scholar]
- 102.Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, et al. DNA hypomethylation at ALOX12 is associated with persistent wheezing in childhood. Am J Respir Crit Care Med. 2012;185:937–43. doi: 10.1164/rccm.201105-0870OC. [DOI] [PubMed] [Google Scholar]
- 103.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med. 2011;184:191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuriakose J, Rosa MJ, Perzanowski M, Miller R. Bronchial nitric oxide flux may be better associated with inducible nitric oxide synthase promoter methylation. Am J Respir Crit Care Med. 2012;185:460–1. doi: 10.1164/ajrccm.185.4.460. author reply 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baccarelli A, Rusconi F, Bollati V, Catelan D, Accetta G, Hou L, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics. 2012;4:91–100. doi: 10.2217/epi.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. 2015;136:69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liang L, Willis-Owen SA, Laprise C, Wong KCC, Davies GA, Hudson TJ, et al. An Epigenome-Wide Association Study of Total Serum Immunoglobulin E Concentration. Nature. 2015;520:4. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol. 2014;15:777–88. doi: 10.1038/ni.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gandhi VD, Davidson C, Asaduzzaman M, Nahirney D, Vliagoftis H. House dust mite interactions with airway epithelium: role in allergic airway inflammation. Curr Allergy Asthma Rep. 2013;13:262–70. doi: 10.1007/s11882-013-0349-9. [DOI] [PubMed] [Google Scholar]
- 113.Gandhi VD, Vliagoftis H. Airway epithelium interactions with aeroallergens: role of secreted cytokines and chemokines in innate immunity. Front Immunol. 2015;6:147. doi: 10.3389/fimmu.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lambrecht BN, Hammad H. Allergens and the airway epithelium response: gateway to allergic sensitization. J Allergy Clin Immunol. 2014;134:499–507. doi: 10.1016/j.jaci.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 115.Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med. 2012;18:684–92. doi: 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]
- 116.Yang IV, Tomfohr J, Singh J, Foss CM, Marshall HE, Que LG, et al. The clinical and environmental determinants of airway transcriptional profiles in allergic asthma. Am J Respir Crit Care Med. 2012;185:620–7. doi: 10.1164/rccm.201108-1503OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–63. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choy DF, Modrek B, Abbas AR, Kummerfeld S, Clark HF, Wu LC, et al. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. J Immunol. 2011;186:1861–9. doi: 10.4049/jimmunol.1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang IV, Richards A, Davidson EJ, Stevens AD, Kolakowski CA, Martin RJ, et al. The Nasal Methylome: A Key to Understanding Allergic Asthma. Am J Respir Crit Care Med. 2017;195:829–31. doi: 10.1164/rccm.201608-1558LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126:453–65. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peden DB. Effect of pollutants in rhinitis. Curr Allergy Asthma Rep. 2001;1:242–6. doi: 10.1007/s11882-001-0011-9. [DOI] [PubMed] [Google Scholar]
- 124.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bellavia A, Urch B, Speck M, Brook RD, Scott JA, Albetti B, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. 2013;2:e000212. doi: 10.1161/JAHA.113.000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, et al. Air pollution and gene-specific methylation in the Normative Aging Study: association, effect modification, and mediation analysis. Epigenetics. 2014;9:448–58. doi: 10.4161/epi.27584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo L, Byun HM, Zhong J, Motta V, Barupal J, Zheng Y, et al. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ Mol Mutagen. 2014;55:322–35. doi: 10.1002/em.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hou L, Zhang X, Zheng Y, Wang S, Dou C, Guo L, et al. Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ Mol Mutagen. 2014;55:256–65. doi: 10.1002/em.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sofer T, Baccarelli A, Cantone L, Coull B, Maity A, Lin X, et al. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics. 2013;5:147–54. doi: 10.2217/epi.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tarantini L, Bonzini M, Tripodi A, Angelici L, Nordio F, Cantone L, et al. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup Environ Med. 2013;70:418–25. doi: 10.1136/oemed-2012-101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Prins S, Koppen G, Jacobs G, Dons E, Van de Mieroop E, Nelen V, et al. Influence of ambient air pollution on global DNA methylation in healthy adults: a seasonal follow-up. Environ Int. 2013;59:418–24. doi: 10.1016/j.envint.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 132.Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–40. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–6. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen P, Piaggi P, Traurig M, Bogardus C, Knowler WC, Baier LJ, et al. Differential methylation of genes in individuals exposed to maternal diabetes in utero. Diabetologia. 2017;60:645–55. doi: 10.1007/s00125-016-4203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 136.Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, et al. Air Pollution and Markers of Coagulation, Inflammation, and Endothelial Function: Associations and Epigene-environment Interactions in an Elderly Cohort. Epidemiology. 2012 doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–82. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Buro-Auriemma LJ, Salit J, Hackett NR, Walters MS, Strulovici-Barel Y, Staudt MR, et al. Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum Mol Genet. 2013;22:4726–38. doi: 10.1093/hmg/ddt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Munthe-Kaas MC, Bertelsen RJ, Torjussen TM, Hjorthaug HS, Undlien DE, Lyle R, et al. Pet keeping and tobacco exposure influence CD14 methylation in childhood. Pediatr Allergy Immunol. 2012;23:747–54. doi: 10.1111/pai.12021. [DOI] [PubMed] [Google Scholar]
- 140.Kohli A, Garcia MA, Miller RL, Maher C, Humblet O, Hammond SK, et al. Secondhand smoke in combination with ambient air pollution exposure is associated with increasedx CpG methylation and decreased expression of IFN-gamma in T effector cells and Foxp3 in T regulatory cells in children. Clin Epigenetics. 2012;4:17. doi: 10.1186/1868-7083-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nicole W. Pattern of clues: evidence of distinct DNA methylation in newborns of smoking women. Environ Health Perspect. 2012;120:a402. doi: 10.1289/ehp.120-a402a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–31. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol. 2014;11:71. doi: 10.1186/s12989-014-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clifford RL, Jones MJ, MacIsaac JL, McEwen LM, Goodman SJ, Mostafavi S, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. 2017;139:112–21. doi: 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 146.Alegria-Torres JA, Barretta F, Batres-Esquivel LE, Carrizales-Yanez L, Perez-Maldonado IN, Baccarelli A, et al. Epigenetic markers of exposure to polycyclic aromatic hydrocarbons in Mexican brickmakers: a pilot study. Chemosphere. 2013;91:475–80. doi: 10.1016/j.chemosphere.2012.11.077. [DOI] [PubMed] [Google Scholar]
- 147.Hew KM, Walker AI, Kohli A, Garcia M, Syed A, McDonald-Hyman C, et al. Childhood exposure to ambient polycyclic aromatic hydrocarbons is linked to epigenetic modifications and impaired systemic immunity in T cells. Clin Exp Allergy. 2014 doi: 10.1111/cea.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. Maternal exposure to polycyclic aromatic hydrocarbons and 5′-CpG methylation of interferon-gamma in cord white blood cells. Environ Health Perspect. 2012;120:1195–200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sordillo JE, Lange NE, Tarantini L, Bollati V, Zanobetti A, Sparrow D, et al. Allergen sensitization is associated with increased DNA methylation in older men. Int Arch Allergy Immunol. 2013;161:37–43. doi: 10.1159/000343004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nestor CE, Barrenas F, Wang H, Lentini A, Zhang H, Bruhn S, et al. DNA methylation changes separate allergic patients from healthy controls and may reflect altered CD4+ T-cell population structure. PLoS Genet. 2014;10:e1004059. doi: 10.1371/journal.pgen.1004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Slaats GG, Reinius LE, Alm J, Kere J, Scheynius A, Joerink M. DNA methylation levels within the CD14 promoter region are lower in placentas of mothers living on a farm. Allergy. 2012;67:895–903. doi: 10.1111/j.1398-9995.2012.02831.x. [DOI] [PubMed] [Google Scholar]
- 153.Reynolds SD, Rios C, Wesolowska-Andersen A, Zhuang Y, Pinter M, Happoldt C, et al. Airway Progenitor Clone Formation Is Enhanced by Y-27632-Dependent Changes in the Transcriptome. Am J Respir Cell Mol Biol. 2016;55:323–36. doi: 10.1165/rcmb.2015-0274MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102:76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.von Mutius E, Radon K. Living on a farm: impact on asthma induction and clinical course. Immunol Allergy Clin North Am. 2008;28:631–47. ix–x. doi: 10.1016/j.iac.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 156.Tollanes MC, Moster D, Daltveit AK, Irgens LM. Cesarean section and risk of severe childhood asthma: a population-based cohort study. J Pediatr. 2008;153:112–6. doi: 10.1016/j.jpeds.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 157.Wang JY, Liu LF, Chen CY, Huang YW, Hsiung CA, Tsai HJ. Acetaminophen and/or antibiotic use in early life and the development of childhood allergic diseases. Int J Epidemiol. 2013;42:1087–99. doi: 10.1093/ije/dyt121. [DOI] [PubMed] [Google Scholar]
- 158.Muc M, Padez C, Pinto AM. Exposure to paracetamol and antibiotics in early life and elevated risk of asthma in childhood. Adv Exp Med Biol. 2013;788:393–400. doi: 10.1007/978-94-007-6627-3_53. [DOI] [PubMed] [Google Scholar]
- 159.Kuo CH, Kuo HF, Huang CH, Yang SN, Lee MS, Hung CH. Early life exposure to antibiotics and the risk of childhood allergic diseases: an update from the perspective of the hygiene hypothesis. J Microbiol Immunol Infect. 2013;46:320–9. doi: 10.1016/j.jmii.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 160.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012 doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kumar H, Lund R, Laiho A, Lundelin K, Ley RE, Isolauri E, et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio. 2014:5. doi: 10.1128/mBio.02113-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Takahashi K, Sugi Y, Nakano K, Tsuda M, Kurihara K, Hosono A, et al. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J Biol Chem. 2011;286:35755–62. doi: 10.1074/jbc.M111.271007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics. 2016;11:205–15. doi: 10.1080/15592294.2016.1155011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 165.Nagy-Szakal D, Kellermayer R. The remarkable capacity for gut microbial and host interactions. Gut Microbes. 2011;2:178–82. doi: 10.4161/gmic.2.3.16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118–34. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013;14:631–40. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 169.Castro-Nallar E, Shen Y, Freishtat RJ, Perez-Losada M, Manimaran S, Liu G, et al. Integrating microbial and host transcriptomics to characterize asthma-associated microbial communities. BMC Med Genomics. 2015;8:50. doi: 10.1186/s12920-015-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Perez-Losada M, Castro-Nallar E, Bendall ML, Freishtat RJ, Crandall KA. Dual Transcriptomic Profiling of Host and Microbiota during Health and Disease in Pediatric Asthma. PLoS One. 2015;10:e0131819. doi: 10.1371/journal.pone.0131819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bunyavanich S, Schadt EE. Systems biology of asthma and allergic diseases: a multiscale approach. J Allergy Clin Immunol. 2015;135:31–42. doi: 10.1016/j.jaci.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Weiss ST. New approaches to personalized medicine for asthma: where are we? J Allergy Clin Immunol. 2012;129:327–34. doi: 10.1016/j.jaci.2011.12.971. [DOI] [PubMed] [Google Scholar]
- 173.Levy BD, Noel PJ, Freemer MM, Cloutier MM, Georas SN, Jarjour NN, et al. Future Research Directions in Asthma. An NHLBI Working Group Report. Am J Respir Crit Care Med. 2015;192:1366–72. doi: 10.1164/rccm.201505-0963WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pongracic JA, Krouse RZ, Babineau DC, Zoratti EM, Cohen RT, Wood RA, et al. Distinguishing characteristics of difficult-to-control asthma in inner-city children and adolescents. J Allergy Clin Immunol. 2016;138:1030–41. doi: 10.1016/j.jaci.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yu DH, Waterland RA, Zhang P, Schady D, Chen MH, Guan Y, et al. Targeted p16(Ink4a) epimutation causes tumorigenesis and reduces survival in mice. J Clin Invest. 2014;124:3708–12. doi: 10.1172/JCI76507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–82. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 177.Saba HI. Decitabine in the treatment of myelodysplastic syndromes. Ther Clin Risk Manag. 2007;3:807–17. [PMC free article] [PubMed] [Google Scholar]
- 178.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 179.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–6. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]