Abstract
Diapause is a programmed dormancy that allows organisms to tolerate predictable periods of unfavorable conditions by temporarily halting development and reducing metabolism. Diapause is widespread among insects and is crucial for allowing organisms to coordinate their growth and reproduction with favorable environmental conditions. While the adaptive significance of diapause is well-understood, the molecular mechanisms underpinning diapause remain unresolved. We performed high-throughput sequencing to investigate the role of microRNAs in the diapause of the Asian tiger mosquito, Aedes albopictus. We first investigate miRNAs in diapause induction by characterizing maternally-provisioned miRNAs in mature oocytes of Ae. albopictus under diapause-inducing and diapause-averting conditions. Second, we investigate miRNAs in diapause maintenance by characterizing miRNAs in diapause and non-diapause pharate larvae. We identified 162 miRNAs, 152 previously known and 10 putatively novel. We identified no differentially abundant miRNAs in mature oocytes and seven differentially abundant miRNAs in pharate larvae. The predicted targets of differentially abundant miRNAs include genes affecting several processes related to diapause maintenance including ecdysone regulation, immune response, lipid metabolism, and regulation of development. Our results suggest that Ae. albopictus does not maternally-provision a unique set of miRNAs during diapause induction but miRNAs are a component of diapause maintenance in this species.
Keywords: diapause maintenance, diapause induction, microRNA (miRNA) regulation, Aedes albopictus
Introduction
All temperate insects face seasonally recurring environmental stress. For these species, overwinter survival requires a behavioral response, such as migrating to a warmer climate, and/or a physiological adaptation like diapause (Saunders, 2009). Diapause is an alternative life-history strategy characterized by a period of developmental arrest in response to a token cue (i.e., photoperiod) detected in advance of unfavorable conditions. Diapause involves a progression through four ecophysiological stages (Koštál, 2006). In the first stage, induction, the organism perceives the token cue(s) that initiate the diapause program. In the second stage, preparation, the organism undergoes physiological and developmental changes in preparation for entry into diapause. The third stage, maintenance, is characterized by reduced metabolism, developmental arrest, and a suite of physiological modifications to facilitate survival including increased stress tolerance, elevated immunity, and downregulation of the cell cycle (Goto et al., 2001; Denlinger, 2002; King & MacRae, 2015; Denlinger & Armbruster, 2016). Finally, during the fourth stage, termination, the organism becomes competent to resume growth and development. Diapause enables insects to maximize their fitness by synchronizing growth and reproduction with periods of favorable environmental conditions (Tauber et al., 1986). While the adaptive significance of diapause is well-established, the underlying molecular mechanisms regulating this complex phenotype are less clear.
The Asian tiger mosquito, Aedes albopictus (Skuse), is an emerging model for investigating the molecular underpinnings of diapause. Ae. albopictus is a highly invasive species and diapause has facilitated its establishment throughout temperate regions (Benedict et al., 2007; Urbanski et al., 2012). In Ae. albopictus, diapause is under maternal control; pupal and adult females detect short (autumnal) photoperiods as a token cue and respond by producing diapause offspring which overwinter as pharate larvae within the chorion of the egg (Mori & Oda, 1981; Denlinger & Armbruster, 2016). However, the mechanisms by which the mother induces diapause in offspring remain elusive. Diapause eggs are more cold tolerant and desiccation resistant than non-diapause eggs (Sota & Mogi, 1992; Hanson & Craig, 1994, 1995; Urbanski et al., 2010). Also, diapause eggs are larger and contain ~40% more surface hydrocarbons and ~30% more total lipids than non-diapause eggs (Urbanski et al., 2010; Reynolds et al., 2012). In addition to these physiological changes, transcriptome sequencing has identified thousands of coding genes that are differentially expressed throughout the four stages of diapause (Poelchau et al., 2011, 2013a,b; Huang et al., 2015).
Small, non-coding RNAs (sncRNAs) have been shown to regulate a wide range of developmental and physiological processes (Hussain et al., 2016). Regulatory sncRNAs are divided into three main classes according to their biogenesis pathway: small-interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), and microRNAs (miRNAs) (Lucas et al., 2013). siRNAs are ~21nt long double-stranded RNA molecules (dsRNA) which originate either endogenously from sequences in the genome or exogenously from foreign dsRNA (Hussain et al., 2016). piRNAs are ~24-31nt long and canonically guide cleavage of mobilized transposable elements in germline cells (Ishizu et al., 2012). Additionally, piRNAs can regulate the expression of protein coding genes (Iwasaki et al., 2015). miRNAs are ~22nt long and synthesized in pairs from a hairpin precursor (pre-miRNA) (Bartel, 2004). Members of a miRNA pair are denoted with either a -3p or -5p suffix corresponding to the arm of the hairpin precursor from which it was synthesized (Desvignes et al., 2015). miRNAs post-transcriptionally regulate gene expression by guiding an Argonaute protein complex to silence target mRNAs containing perfect or near-perfect complementarity to a 7nt seed region in the miRNA (Bartel, 2009). The short seed sequence allows for a single miRNA to target and affect the translation of dozens or even hundreds of genes (Lewis et al., 2005; Baek et al., 2008).
Several studies have implicated miRNAs in the regulation of diapause and diapause-associated physiological traits. Genes encoding proteins involved in miRNA biogenesis and function are differentially expressed during diapause maintenance in the flesh fly, Sarcophaga bullata (Reynolds et al., 2013). Specifically, Loquacious, encoding a protein necessary for miRNA maturation, and Argonaute-1, encoding a protein in the miRNA-induced silencing complex, are upregulated in diapause pupae of S. bullata (Reynolds et al., 2013). Furthermore, ten miRNAs were found to be differentially abundant during pupal diapause in S. bullata and subsequent in silico target prediction suggests these miRNAs could regulate development, stress tolerance, and metabolism (Reynolds et al., 2017). miRNAs also contribute to maintenance and termination of the dauer arrest in Caenorhabditis elegans, a diapause-like state in worms (Karp et al., 2011; Karp & Ambros, 2012). miRNAs are also known to regulate numerous pathways associated with diapause physiology such as metabolism, development, cell signaling, growth, apoptosis, cell division, and immunity (Huang et al., 2011; Asgari, 2012; Ebert & Sharp, 2012; Biggar & Storey, 2015; Lucas et al., 2015). Finally, miRNAs are known to be maternally provisioned in Drosophila melanogaster eggs (Votruba, 2011; Soni et al., 2013). Given the differential expression of genes involved in miRNA function and biogenesis, the role of miRNAs in regulation of diapause-related pathways, and the potential for miRNA transgenerational signaling, we hypothesized that miRNAs could be an important component of diapause regulation in Ae. albopictus.
In this study, we utilized next-generation sequencing to identify novel miRNAs in Ae. albopictus. We then analyzed the next-generation sequencing data to quantify and compare the abundance of miRNAs present in mature oocytes of adult females reared under diapause-averting and diapause-inducing photoperiod as well as miRNAs of pharate larvae in early diapause maintenance relative to developmentally-matched non-diapause pharate larvae. Finally, we predicted target sites for differentially abundant miRNAs and identified diapause-related functions of miRNA target genes.
Results
Diapause incidence
Diapause incidence was measured in replicates from both the mature oocyte (induction) and pharate larvae (maintenance) experiments (Fig. 1) to confirm that short-day (SD) photoperiod conditions had induced diapause and that long-day (LD) photoperiod conditions had induced non-diapause. In the mature oocyte experiment, diapause incidence was 96.7–98.4% in two SD photoperiod replicates and 16.1–18.0% in two LD photoperiod replicates. In the pharate larvae experiment, diapause incidence was 93.5–98.9% in four SD photoperiod replicates and 2.6–12.1% in four LD photoperiod replicates (Table S1).
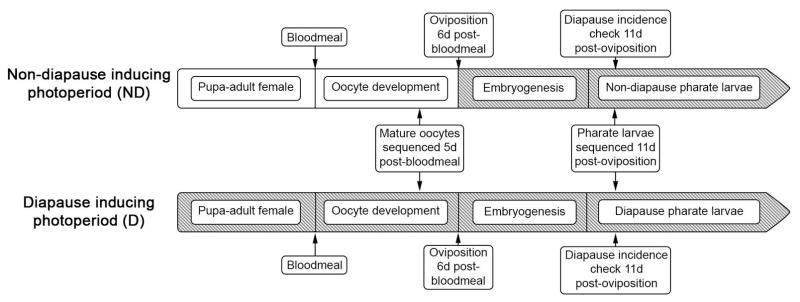
Figure 1.
Overview of experimental design. Unshaded portions of the timelines indicate exposure to long-day photoperiod (16h light:8h dark). Shaded portions of the timelines indicate exposure to short-day photoperiod (8h light:16h dark).
RNA libraries and read mapping
All reads generated in this project are available in the NCBI Short Read Archive with the accession numbers SRR5168334-SRR5168339 (mature oocytes) and SRR5168326-SRR5168333 (pharate larvae). Six mature oocyte libraries and eight pharate larvae libraries were sequenced, cleaned, and size sorted producing a total of 12.9 million cleaned reads (Table 1). The read lengths obtained in these two experiments were significantly different (Mann-Whitney U = 1.23 x 105, p < 0.001; Table S2). Mature miRNA length reads (20–22nt) comprised 17.7% of the mature oocyte libraries compared to 46.7% in the pharate larvae libraries. Mature piRNA length reads (24-31nt) comprised 58.4% of the mature oocyte libraries and just 15.9% of the pharate larvae libraries.
Table 1.
Summary of miRNA sequencing, cleaning, and mapping.
| Sample | Number of Reads | ||||
|---|---|---|---|---|---|
|
| |||||
| Sequenced | Cleaned Reads | Mapped to Genome | Mapped to Reference miRNAs | Mapped to Putative Novel miRNAs | |
| All Samples | 57,432,813 | 12,895,405 | 7,303,656 | 3,348,144 | 57,254 |
| Mature Oocytes | |||||
| Diapause 1 | 1,964,986 | 530,966 | 265,757 | 78,529 | 2,401 |
| Diapause 2 | 2,470,716 | 1,008,763 | 472,872 | 88,760 | 14,497 |
| Diapause 3 | 2,943,828 | 704,207 | 355,739 | 113,985 | 4,624 |
| Non-diapause 1 | 2,360,151 | 818,099 | 379,917 | 48,049 | 7,710 |
| Non-diapause 2 | 2,738,026 | 1,815,875 | 868,614 | 84,265 | 14,907 |
| Non-diapause 3 | 2,387,982 | 1,529,625 | 752,327 | 99,268 | 13,259 |
| Pharate Larvae | |||||
| Diapause 1 | 4,374,433 | 1,131,877 | 786,366 | 554,405 | 1,303 |
| Diapause 2 | 5,241,029 | 895,362 | 579,820 | 391,158 | 1,066 |
| Diapause 3 | 5,023,940 | 948,284 | 657,767 | 490,837 | 1,333 |
| Diapause 4 | 6,195,237 | 804,382 | 546,724 | 387,633 | 1,204 |
| Non-diapause 1 | 5,413,799 | 599,212 | 379,394 | 249,619 | 569 |
| Non-diapause 2 | 4,870,514 | 789,399 | 484,299 | 283,740 | 585 |
| Non-diapause 3 | 4,949,601 | 650,623 | 395,246 | 248,802 | 614 |
| Non-diapause 4 | 6,498,571 | 668,731 | 378,814 | 229,094 | 568 |
To identify putatively novel miRNAs that have not previously been annotated for Ae. albopictus, reads from all 14 libraries across both experiments were combined, mapped to the Ae. albopictus genome, and scored with miRDeep2. Ten putatively novel miRNA precursors had miRDeep2 scores >4, hairpin loops in the known size range for Ae. albopictus, presented at least ten mature reads for each arm, and appeared in at least two libraries (Table S3). Eight of the predicted novel miRNA precursors were identified in both the mature oocyte and pharate larvae experiments. No putative novel miRNA precursors were exclusively identified in the mature oocyte libraries and two putative novel miRNA precursors were exclusively identified in the pharate larvae libraries. One novel miRNA precursor appeared only in diapause libraries and no novel miRNA precursors appeared in only non-diapause libraries (Table S3). Each precursor miRNA produces two distinct mature miRNAs; these mature miRNAs were searched in miRBase (Kozomara & Griffiths-Jones, 2014) to identify Dipteran orthologs. Five of the predicted novel mature miRNAs have seed regions identical to known Dipteran miRNAs, four in Aedes aegypti and one in D. melanogaster. All 20 predicted novel mature miRNA sequences were added to the set of previously annotated Ae. albopictus miRNAs (Skalsky et al., 2010; Gu et al., 2013) to produce a final reference set of 257 mature miRNAs (Table S4).
Next, each of the libraries was mapped to the Ae. albopictus genome and read counts were obtained for each miRNA in the reference set. For the mature oocytes, 48.3% of cleaned reads aligned to the genome and of these aligned reads, 19.1% mapped to miRNAs in the reference set. For the pharate larvae, 64.9% of cleaned reads aligned to the genome and of these aligned reads, 67.6% mapped to miRNAs in the reference set (Table 1). In total, 162 mature miRNAs were identified: 89 in both mature oocytes and pharate larvae, 12 only in the mature oocytes, and 61 only in the pharate larvae.
MicroRNA analyses
The distance between the miRNA expression patterns of each library was visualized in an MDS plot (Fig. S1). Variation within each treatment group (diapause, non-diapause) and stage (mature oocytes, pharate larvae) was estimated by the biological coefficient of variation (BCV) in edgeR. BCV was higher in the mature oocyte libraries (diapause: 0.796, non-diapause: 0.351) than in the pharate larvae libraries (diapause: 0.233, non-diapause: 0.182).
Next, miRNAs were tested for differential abundance between diapause and non-diapause conditions for oocytes and pharate larvae. No miRNAs had significant differences in abundance between diapause and non-diapause mature oocytes (Fig. 2a). However, three miRNAs had absolute log2-fold changes >2 (aal-miR-1890-1-3p, aal-miR-957-3p, and aal-miR-92a-5p).
Figure 2.
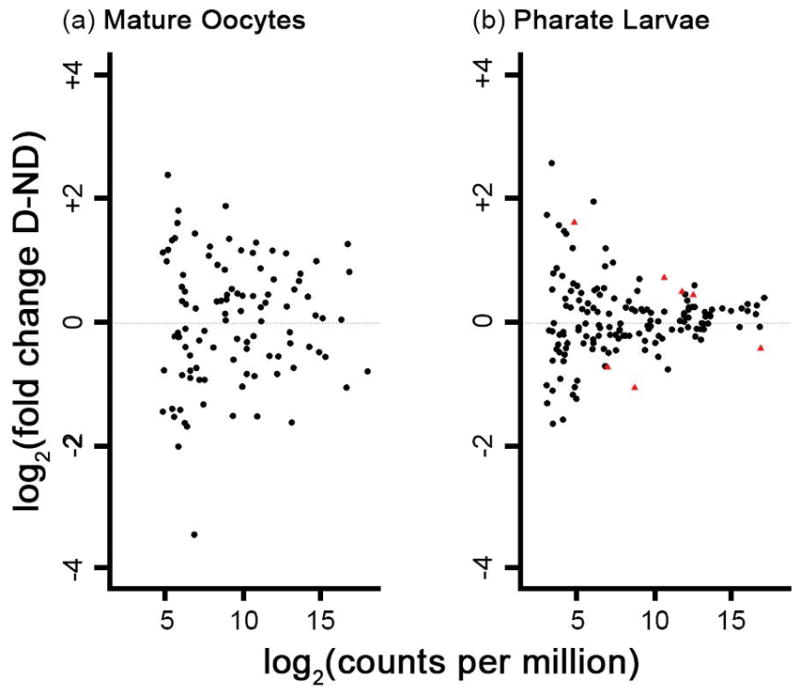
Log2(fold change) of miRNA abundance under diapause relative to non-diapause conditions versus abundance log2(counts per million) following TMM-normalization for (a) mature oocytes and (b) pharate larvae. Each point represents a single miRNA. MicroRNAs with higher abundance under diapause conditions have positive fold change values. Red triangles indicate significant differences in miRNA abundance between conditions (Fisher’s exact test p<0.05 following Benjamini-Hochberg correction).
Seven miRNAs were differentially abundant between diapause and non-diapause conditions in pharate larvae (Fig. 2b). Four miRNAs were over-represented and three miRNAs were under-represented under diapause relative to non-diapause conditions. The absolute values of log2-fold changes ranged from 0.41–1.58 (Table 2). All seven differentially abundant miRNAs were from the set previously annotated in Ae. albopictus (Skalsky et al., 2010; Gu et al., 2013). Fold change values measured during follow-up quantitative reverse-transcription PCR (qRT-PCR) experiments were strongly correlated to those obtained in the RNA-Seq analysis (r = 0.93, Table S5, Figure S2).
Table 2.
MicroRNAs with significant differences in abundance in diapause relative to non-diapause pharate larvae.
| miRNA | log2-FC | p-value | FDR |
|---|---|---|---|
| aal-miR-283-5p | +0.41 | 6.78e-04 | 0.035 |
| aal-bantam-5p | +0.68 | 7.20e-04 | 0.035 |
| aal-miR-2942-3p | −1.08 | 9.33e-04 | 0.035 |
| aal-miR-286b-3p | +0.46 | 1.09e-03 | 0.035 |
| aal-miR-282-5p | +1.58 | 1.12e-03 | 0.035 |
| aal-miR-14-5p | −0.75 | 1.87e-03 | 0.046 |
| aal-miR-1-3p | −0.45 | 2.07e-03 | 0.046 |
MicroRNA target prediction
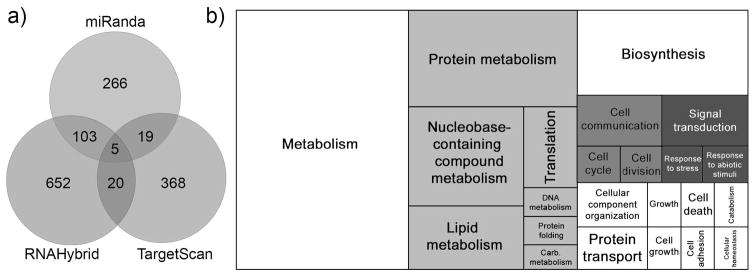
miRNA target prediction was used to characterize the potential downstream effects of regulation by differentially abundant miRNAs at the pharate larvae stage (early diapause) using three target prediction algorithms (Fig. 3a). The first algorithm, TargetScan, predicted 546 target sites in 412 3′ UTRs. RNAHybrid, predicted 801 target sites across 780 3′ UTRs and miRanda predicted 444 targets in 393 3′ UTRs. In total, 147 3′UTRs were predicted to contain miRNA target sites by at least two algorithms (Fig. 3a). Three of these 3′UTRs were predicted to be targeted by two miRNAs identified as differentially abundant and seven of 3′UTRs were predicted to contain more than one target site for a single differentially abundant miRNA (Table S6).
Figure 3.
(a) Count of 3′ UTR miRNA targets predicted by three algorithms for differentially abundant miRNAs. Targets predicted by at least two algorithms were retained for further investigation. (b) GO-Slim2 terms for predicted targets of differentially abundant miRNAs. The size of the square represents how frequently the term appears in annotations of genes with 3′UTRs targeted by differentially abundant miRNA. Groups of shaded squares are semantically similar subsets of terms as determined by REVIGO (SimRel score > 0.5). Predicted miRNA targets, target GO-Slim2 terms, and algorithms identifying particular miRNA-target pairs are detailed in Table S7.
GO-Slim2 annotations for genes containing predicted target sites were clustered by semantic similarity and sized according to frequency among the predicted target set (Fig. 3b). Changes in metabolism are a key feature of diapause and GO terms related to metabolism appear frequently within the set especially lipid, nucleobase, and protein metabolism.
Discussion
Diapause is a programmed dormancy that allows insects to anticipate and persist through periods of poor environmental conditions (Koštál, 2006). In the photoperiodically-induced diapause of Ae. albopictus, the pupal and adult female is photosensitive but developmental arrest occurs in the pharate larvae; the nature of the trans-generational signal is unknown (Denlinger & Armbruster, 2016). miRNAs can be maternally provisioned (Votruba, 2011; Soni et al., 2013) suggesting they may play a role in this trans-generational signaling. In S. bullata, genes encoding proteins involved in miRNA biogenesis and function undergo coordinated changes in expression during diapause preparation and initiation (Reynolds et al., 2013) and miRNAs are differentially abundant during diapause maintenance (Reynolds et al., 2017). Thus, we hypothesized that maternally-provisioned miRNAs in mature oocytes may be a component of the transgenerational diapause signal in Ae. albopictus. Furthermore, diapause maintenance requires coordinated regulation of diverse physiological processes including: developmental arrest, reduced metabolism, and increased immunity and stress tolerance (Denlinger, 2002). Because an individual miRNA can alter the translation of dozens or hundreds of genes via post-transcriptional regulation (Baek et al., 2008; Asgari, 2012; Lucas et al., 2015), individual miRNAs have the potential to underlie broad regulatory changes like those observed in diapause. Accordingly, we further hypothesized that key elements of the Ae. albopictus diapause phenotype may be regulated by differential abundance of miRNAs in pharate larvae during developmental arrest.
Putative novel miRNAs
miRDeep2 analysis of mature oocytes and pharate larvae small RNA samples led to the prediction of ten novel miRNA precursors (Table S3). miRNAs are often expressed in a stage-specific manner (Li et al., 2009; Gu et al., 2013) and no previous study has characterized Ae. albopictus miRNAs from either mature oocytes or pharate larvae. Two novel miRNA precursors were unique to pharate larval stage (Table S3).
Mature Oocyte sncRNAs
We characterized maternally-provisioned miRNAs by sequencing small RNA from mature oocytes dissected from virgin females reared under SD (diapause-inducing) or LD (diapause-averting) photoperiod. Relatively few miRNA-length reads were recovered (17.7% of cleaned reads; Table S2). In total, 101 miRNAs were present across six mature oocyte replicates. Contrary to our expectations, none of these miRNAs were differentially abundant between SD and LD photoperiods (Fig. 2). This result implies that maternally-provisioned miRNAs do not significantly contribute to the maternal regulation of the egg diapause in Ae. albopictus.
Intriguingly, 58.4% of the reads from mature oocytes were 24-31nt long, the expected size range of piRNAs (Table S2). This result is consistent with evidence from S. bullata that genes involved in piRNA biogenesis and function are upregulated during the photosensitive stage prior to diapause (Reynolds et al., 2013). Canonically, piRNAs are involved in transposon defense but recent work demonstrates that these molecules can also regulate the expression of protein coding genes (Iwasaki et al., 2015). In Ae. aegypti, just 19% of putative piRNAs are predicted to target transposable elements suggesting that non-canonical piRNA functions like targeting of protein coding genes are prevalent (Arensburger et al., 2011). Preliminary analysis of piRNAs in Ae. albopictus predicted that approximately 20% of piRNAs map to protein-coding genes (Liu et al., 2016). Thus, it seems likely that protein-coding genes may be regulated, at least to some extent, by piRNAs in this species. However, the preliminary annotation of Ae. albopictus piRNAs is less robust than for miRNAs (Liu et al., 2016), and thus we did not attempt to quantify differential abundance of piRNAs associated with diapause in this study.
Pharate Larvae miRNAs
We characterized miRNAs in early diapause maintenance by sequencing small RNAs from pharate larvae in either early diapause or non-diapause. In contrast to the low proportion of miRNAs in the mature oocytes, 46.7% of cleaned reads in the pharate larvae RNA samples were in the miRNA size range (Table S2) and 67.7% of genome-aligned reads mapped to reference miRNAs (Table 1). 150 miRNAs were identified across a total of eight diapause and non-diapause replicates. Seven miRNAs were differentially abundant in early diapause pharate larvae; four were up-regulated and three were down-regulated (Fig. 2, Table 2). A recent study identified ten differentially abundant miRNAs during the pupal diapause of S. bullata (Reynolds, et al., 2017) including miR-1-3p, one of the differentially abundant miRNAs identified in our study. In contrast to our results, miR-1-3p was more abundant in S. bullata during diapause relative to non-diapause, suggesting independent mechanisms of miRNA regulation of diapause in these two species. The seven differentially abundant miRNAs in the pharate larvae of Ae. albopictus have been implicated in processes associated with diapause including increased immunity, lipid accumulation and developmental regulation.
Differentially Abundant miRNAs in Pharate Larvae
miR-283-5p and miR-1-3p were both differentially expressed in pharate larvae and have been linked to immune response in Ae. albopictus. miR-283-5p was upregulated and miR-1-3p was absent in an Ae. albopictus cell line after exposure to Chikungunya virus (Shrinet et al. 2014). Similarly, in our study miR-283-5p was upregulated and miR-1-3p was downregulated in diapause pharate larvae. Enhanced immune function is thought to be an important component of diapause (Chernysh et al., 2000; Ragland et al., 2010), and we hypothesize that differential abundance of these two miRNAs may contribute to increased immunity during diapause.
miR-14-5p was downregulated during diapause in pharate larvae and has previously been linked to regulation of lipid metabolism. Knockout of the mir-14 precursor in D. melanogaster reduces lipid metabolism and increases lipid accumulation in the form of triglycerides and diglycerides (Xu et al., 2003). Previous work in Ae. albopictus found that diapause eggs have approximately 30% more lipids than non-diapause pharate eggs, due at least in part to upregulation of triglyceride storage and downregulation of lipid catabolism during diapause preparation (Reynolds et al., 2012). Thus, the diapause-associated down-regulation of miR-14-5p in our data is consistent with that evidence suggesting that differential abundance of this miRNA could contribute to the accumulation of lipids.
Developmental arrest is a hallmark of diapause and thought to be regulated in most insects by some combination of two hormones: juvenile hormone (JH) and ecdysone (Denlinger et al., 2012). For example, high levels of ecdysone initiate and maintain the pharate larval diapause of Lymantria dispar (Lee & Denlinger, 1997). In contrast, low levels of ecdysone maintain egg diapause in Locusta migratoria (Tawfik et al., 2002), larval diapause in Omphisa fuscidentalis (Singtripop et al., 1999), and pupal diapause in Mamestra brassicae (Endo et al., 1997). In Ae. albopictus, ecdysone signaling genes are differentially expressed in diapause-induced mature oocytes and embryos (Poelchau et al., 2011, 2013a). During diapause maintenance, genes related to JH binding and metabolism, but not ecdysone, are differentially expressed (Poelchau et al., 2013b). These previous findings implicate differential ecdysone signaling in Ae. albopictus diapause preparation but not diapause maintenance.
Our study provides evidence that post-transcriptional regulation of ecdysone-related genes by miRNAs could be involved in early diapause maintenance in Ae. albopictus pharate larvae. Two miRNAs linked to ecdysone regulation, bantam-5p and miR-282-5p, were overexpressed in diapause pharate larvae (Table 2). In D. melanogaster, bantam negatively regulates ecdysone production in the prothoracic gland and strong overexpression significantly extends time to pupation (Boulan et al., 2013). miR-282-5p negatively regulates the rutabaga gene in D. melanogaster (Vilmos et al., 2013). Rutabaga is most often associated with adult learning and memory functions (Livingstone et al., 1984) but mutation of this gene has also been found to delay larval molts by limiting the production of active ecdysone (Venkatesh et al., 2001). Together, these functional studies in D. melanogaster suggest that bantam-5p and miR-282-5p may negatively regulate ecdysone synthesis to suppress larval development.
Finally, miR-2942-3p and miR-286b-3p were differentially abundant in diapause pharate larvae and both have been linked to developmental transitions in Ae. albopictus (Puthiyakunnon et al., 2013). Knockdown of miR-2942-3p in Ae. albopictus larvae significantly reduces the rate of eclosion from pupa to adult (Puthiyakunnon et al., 2013). In our study, miR-2942-3p was downregulated in the diapause pharate larvae, consistent with the hypothesis that reduced levels of miR-2942-3p impede developmental transitions. Similarly, knockdown of miR-286b-3p in Ae. albopictus embryos delayed hatching and reduced hatching success (Puthiyakunnon et al., 2013). Delayed hatching is a primary component of diapause in Ae. albopictus, therefore we expected miR-286b-3p to be downregulated in our diapause pharate larvae. In contrast to this expectation, miR-286b-3p was upregulated under diapause. In some cases, both overexpression and knockdown of a particular miRNA generate similar mutant phenotypes (e.g. Hansen et al., 2013; Luo & Sehgal, 2012). Thus, one possible explanation is that an intermediate abundance of miR-286b-3p is required for normal hatching. The differential abundance of these two miRNAs related to developmental transitions further support the notion that differentially abundant miRNAs play a role in regulating the suppression of development during early diapause maintenance in Ae. albopictus.
miRNA Target Characterization
To characterize additional downstream regulatory effects, we identified putative gene targets for each of the seven differentially abundant miRNAs in pharate larvae. 147 genes were predicted to contain target sites for the differentially abundant miRNAs by at least two algorithms (Fig. 3a, Table S7). Among these targets, 58 genes were annotated with the GO-Slim2 term metabolism (Fig. 3b, Table S7). Of particular interest, four metabolism gene targets were annotated as components of lipid metabolism. Lipids are the primary energy source for most diapause insects (Hahn & Denlinger 2011) and altered lipid metabolism is a critical component of the diapause phenotype in Ae. albopictus (Reynolds et al., 2012; Poelchau et al., 2013b). One of the predicted miRNA target genes (AALF015949) is an ortholog of the D. melanogaster gene Hormone-sensitive lipase (HSL). HSL expression is induced by starvation and mobilizes triglycerides from intracellular lipid storage droplets for lipolysis (Bi et al., 2012). miRNA targeting of the HSL gene could potentially help fine-tune starvation response during diapause to conserve lipids. Triglyceride lipases were previously found to be downregulated during Ae. albopictus diapause preparation (Reynolds et al., 2012) and upregulated during early diapause maintenance (Poelchau et al., 2013b). Our results suggest that post-transcriptional targeting by miRNAs could provide another level of regulation for lipid metabolism in Ae. albopictus diapause.
We identified five putative target genes related to cell cycle regulation and apoptosis, two pathways typically downregulated during diapause (Hwang et al., 2005; Koštál et al., 2009; MacRae, 2010; Poupardin et al., 2015). In Ae. albopictus, differential expression of genes regulating the cell cycle and apoptosis have been previously observed in diapause-destined mature oocytes and early in diapause preparation (Poelchau et al., 2011, 2013a) but not during diapause maintenance (Poelchau et al., 2013b). Thus, this study provides the first support for regulation of these pathways during the diapause maintenance of Ae. albopictus.
Conclusions
This study is the first to directly investigate changes in miRNA abundance as a mechanism of diapause regulation in Ae. albopictus. We found that maternally-provisioned miRNAs did not differ between diapause-induced and diapause-averting mature oocytes suggesting that miRNAs do not act as the transgenerational signal for diapause induction in Ae. albopictus. We found seven differentially abundant miRNAs in diapause pharate larvae which are implicated in a wide variety of diapause-associated traits including developmental arrest, stress tolerance, immunity, and shifts in metabolism. Collectively, these results suggest that miRNAs may play an important role in orchestrating physiological processes during early diapause maintenance of Ae. albopictus.
Experimental Procedures
Insect rearing and tissue preparation
An overview of the experimental design for this study is provided in Figure 1. For both experiments (mature oocytes = diapause induction, pharate larvae = diapause maintenance), a laboratory colony of Ae. albopictus from Manassas, Virginia, USA (38.6°N, 77.4°W) was reared as described previously (Armbruster & Hutchinson, 2002; Armbruster & Conn, 2006) and briefly summarized below.
Mature oocytes
Samples of mature oocyte tissue were generated by rearing F7 Ae. albopictus larvae under LD photoperiod (16h light:8h dark) at 21°C, 80% humidity. Larvae were fed a near-optimal diet consisting of a slurry of dog food and brine shrimp (Armbruster & Hutchinson, 2002; Armbruster & Conn, 2006). Upon pupation, three replicate adult cages were established under SD photoperiod (8h light:16h dark) to induce diapause and three replicate adult cages were established under LD photoperiod to induce direct development (non-diapause). Each cage consisted of approximately 30 females. Cages were lined with a moist filter paper to help maintain humidity and females were provided organic raisins for sugar-feeding.
Virgin females were allowed to blood feed to repletion on a human host 5–8d post-eclosion. The Georgetown University Institutional Review Board (IRB) determined that mosquito blood feeding was not human research and thus did not require IRB approval. However, the blood feeding protocol was approved by the Georgetown University Occupational Health and Safety Committee. Five days post-blood meal, females from each cage were briefly anesthetized with CO2 and transferred to a 1.5mL tube. Tubes were snap frozen in liquid nitrogen and stored at -80°C. Mature oocytes, indicated by a clearly visible surface exochorion pattern (Clements, 1992), were dissected directly into RNA Later (Thermo-Fisher, Waltham, Massachusetts) from at least 13 females per cage and stored at 4°C overnight prior to RNA extraction (see below).
Diapause incidence
For both the mature oocyte and pharate larvae experiments, eggs from adult cages were collected to quantify the diapause incidence under SD and LD conditions. For the mature oocyte experiment, four mixed sex (50 male: 50 female) adult cages were established in parallel to the female-only cages used to produce mature oocyte tissue. Two cages were placed under SD photoperiod to induce diapause and two cages were placed under LD photoperiod to induce non-diapause. All females were allowed to blood feed to repletion 5–8d post-eclosion. Starting 4d post-blood meal, females were provided with a brown oviposition cup half-filled with deionized water and lined with unbleached paper towel. Eggs on the paper towel were collected after 24h, maintained for 48h, then gently air dried and stored at 21°C and 80% relative humidity in a sealed Tupperware container. For the pharate larvae experiment, diapause incidence was measured using approximately 100 eggs removed from egg papers prior to snap freezing samples for RNA extraction (see below).
Diapause incidence in both experiments was measured as described previously (Urbanski et al., 2012). Briefly, at 11d post-oviposition, eggs on paper towels were stimulated to hatch by submersion in water. The number of larvae hatched was recorded for each replicate and the egg papers were re-dried. This process was repeated a second time. Remaining unhatched eggs were bleached to clear the chorion (Trpiš, 1970) and the number of fully developed, unhatched embryos was counted. Finally, diapause incidence was calculated as (no. embryonated unhatched eggs)/(no. hatched eggs + no. embryonated unhatched eggs).
Pharate larvae
Samples of pharate larvae tissue were generated by rearing F12 Ae. albopictus larvae under LD conditions as described above. Upon pupation, four replicate adult cages were established under SD conditions to produce eggs with pharate larvae entering diapause and four replicate adult cages were established under LD conditions to produce eggs with non-diapause pharate larvae. Each cage consisted of approximately 90 females and 90 males.
Females were allowed to blood feed to repletion 14–21d post-eclosion as described above. Although female age varied by seven days within replicates prior to blood feeding, this variation is unlikely to impact the small RNA composition of samples as the developmental trajectory of oocytes is most strongly affected by time since blood feeding rather than female age (Clements, 1992). Five days post-blood meal, females were provided a brown oviposition cup half-filled with deionized water and lined with unbleached paper towel. Eggs were collected, dried, and stored as described above. All eggs, including those produced by females reared under LD photoperiod, were stored under SD photoperiod. Exposure of eggs to an SD photoperiod does not induce diapause in Ae. albopictus (Mori & Oda, 1981) and this approach was taken to minimize potential environmental effects of storage in different incubators. Eleven days post-oviposition, eggs were gently brushed from the unbleached paper towels into a 1.5mL tube, snap frozen in liquid nitrogen and stored at −80°C until RNA extraction.
RNA extraction and sequencing
RNA was extracted from both mature oocytes and pharate larvae by homogenizing tissue in TRI Reagent (Sigma-Aldrich, St. Louis, Missouri, USA) and performing a phenol-chloroform extraction followed by an isopropanol precipitation according to the manufacturer’s directions. Residual DNA was removed from each sample using TurboDNA Free (Ambion, Austin, Texas). RNA integrity and concentration were assessed on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Individually-barcoded sequencing libraries were prepared according to the TruSeq RNA sample preparation kit (Illumina, San Diego, California, USA) at the University of Maryland Institute for Genome Science. All six barcoded mature oocyte samples (3 diapause, 3 non-diapause) were pooled and run on a single Illumina MiSeq lane. All eight barcoded pharate larvae samples (4 diapause, 4 non-diapause) were pooled and then split evenly on two Illumina MiSeq lanes.
Read Cleaning
Raw reads were processed in several steps prior to analysis. First, raw reads were screened using SSAHA2 (Ning et al. 2004) to remove tRNA (NCBI accession AY072044.1), rRNA (NCBI accession L22060.1), adapter and primer sequences. Remaining reads were trimmed to their longest continuous sequence with a quality score >30 at each base and reads <18nt were removed using the DynamicTrim and LengthSort scripts, respectively (Cox et al., 2010)
miRNA annotation and differential abundance
In order to identify putatively novel miRNAs, reads obtained across all experiments and treatments were pooled and aligned to the Ae. albopictus genome (Chen et al., 2015) using miRDeep2 (Friedländer et al., 2008, 2012). Then, the 110nt region around each alignment was excised and scored for miRNA-like properties based on two criteria: [1] if the excised region can be folded into a stable hairpin precursor structure and [2] if reads aligned to the excised region match expected pre-miRNA splice sites. Next, alignments and structures were randomly permuted to estimate a true-positive rate across a range of miRDeep2 scores. Predicted miRNAs were considered putative novel miRNA precursors if they met the following four criteria: [1] achieved a miRDeep2 score >4 (corresponding to a true positive rate of 76±8%); [2] the length of the precursor miRNA hairpin loop was within the range of hairpin loop lengths observed in known Ae. albopictus miRNAs (16–145bp); [3] produced at least 10 mature reads from each arm across our 14 combined libraries; and [4] appeared in at least two separate libraries. The 20 mature miRNAs (derived from 10 precursor miRNAs) meeting these criteria were added to 237 previously annotated Ae. albopictus miRNAs (Skalsky et al., 2010; Gu et al., 2013) to produce a final reference set (Table S4).
Next, replicate libraries from each experiment and treatment were individually mapped to genome by using miRDeep2 and read counts for each mature miRNA in the final reference set were obtained. For each experiment, read counts from all replicate libraries were TMM-normalized using the edgeR package (Robinson et al., 2010) and only mature miRNAs present in at least half the libraries were retained. A distance matrix of miRNA expression patterns was visualized in a multi-dimensional scaling (MDS) plot using PlotMDS in the limma package (Ritchie et al., 2015). Variation within experimental treatment groups was quantified with the biological coefficient of variation (BCV) using the edgeR package. Differential abundance of miRNAs in diapause compared to non-diapause was determined at both the oocyte and pharate larval stage using the ExactTest function in the edgeR package. MicroRNAs were considered differentially abundant if they had a Benjamani-Hochburg corrected p-value <0.05.
qRT-PCR validation
qRT-PCR was used to validate our RNA-Seq results at the pharate larval stage. The six miRNAs selected for validation include two miRNAs with significantly greater abundance during diapause (aal-miR-283-5p and aal-bantam-5p), two miRNAs with significantly lower abundance during diapause (aal-miR-2942-3p and aal-miR-1-3p), and two miRNAs with no significant difference in abundance during diapause (aal-miR-133-3p and aal-miR-315-5p). Therefore, the results of the qRT-PCR test the correspondence of qRT-PCR and RNA-Seq across the range of fold change values obtained during our RNA-Seq experiment.
At least 460 eggs were collected from six biological replicates (three SD conditions; three LD conditions) and snap frozen 11dpov as described above. Total RNA was extracted from each biological replicate as described above for the RNA-Seq experiments. Mature miRNA was reverse transcribed with the miScript II RT Kit with HiSpec buffer (Qiagen, Germantown, Maryland, USA) per the manufacturer’s directions.
Relative abundance for each target miRNA was measured using the miScript SYBR Green PCR Kit (Qiagen, Germantown, Maryland, USA) on a Bio-Rad CFX96 Touch (Bio-Rad, Hercules, California, USA). Cycling parameters were 94°C for 15 min followed by 40 cycles of 94°C for 15s, 55°C for 30s, and 70°C for 30s. The miScript system utilizes a miRNA-specific forward primer and a proprietary universal reverse primer which binds to a sequence affixed to the -3p end of the mature miRNA during the reverse transcription process. For each target miRNA, we ordered one miRNA-specific miScript Primer Assay (Qiagen, Germantown, Maryland, USA). These primers were designed to align to the most abundant isomer of a particular miRNA in the RNA-Seq data (primer sequences are provided in Table S5).
For each target miRNA, the number of threshold cycles (Ct) was determined by averaging across three technical replicates per biological replicate. Relative abundance was calculated using the 2−ΔΔCt method with the geometric mean of Ct values for aal-miR-10-5p and aal-miR-87-3p as a reference. Finally, we calculated the Pearson correlation coefficient for fold change values determined in this qRT-PCR experiment versus fold change values obtained during the RNA-Seq experiment.
Target prediction of differentially abundant miRNAs
The potential regulatory functions of differentially abundant miRNAs were investigated by identifying miRNA target sites using three algorithms with different methods of target prediction: miRanda (Enright et al., 2003), RNAHybrid (Krüger & Rehmsmeier, 2006), and TargetScan (Lewis et al., 2005). Only targets predicted by at least two of the three algorithms were retained as putative targets. Although somewhat conservative, this strategy is likely to reduce false positives and produce a high confidence set of targets (Witkos et al., 2011). miRNA target sites are disproportionately located in the 3′ UTR region of mRNAs (Hausser & Zavolan, 2014), therefore 6,438 genes with annotated 3′ UTR regions in the Ae. albopictus genome were scanned for miRNA target sites.
miRanda
MiRanda uses a two-phase scoring algorithm to identify miRNA targets. First, potential targets for differentially abundant miRNAs are identified by aligning complementary miRNAs and 3′ UTRs. Alignments are scored by a position-weighted algorithm which favors matches in the seed region (miRNA nucleotide positions 2–8). Second, the free energy of each alignment is calculated to predict if interactions are thermodynamically favorable. MiRanda predictions were generated with the following settings: score cutoff 140, energy cutoff −20, gap open penalty −9, gap extension penalty −4.
RNAHybrid
RNAHybrid predicts targets by predicting the free energy of interaction between miRNAs and 3′UTR sequences. For each differentially abundant miRNA, the RNACalibrate module was run to estimate a null distribution for expected free energy of interaction for that miRNA against a random set of target sequences. Calibration output was subsequently used to run the main RNAHybrid module with the following settings: binding required in miRNA positions 2–7, p-value < 0.1, maximum target sequence length 100000, energy cutoff −20.
TargetScan
TargetScan identifies targets first by sequence alignment between differentially abundant miRNAs and 3′UTRs. Alignments are then checked for target conservation in a set of related species; in our analysis, conservation was identified by comparison to annotated 3′ UTR regions in three mosquito genomes: Ae. aegypti, Culex pipiens, and Anopheles gambiae (AegL3.3, CpipJ2.2, AgamP4.3, respectively, from VectorBase) as well as D. melanogaster (FB2016_03 from FlyBase). These four 3′UTR sets were aligned in a pairwise fashion to the Ae. albopictus 3′UTRs. For each species, the best hit for each 3′UTR was retained to generate a multiple sequence alignment file of 3′UTR putative orthologs. Next, putative orthologs for each differentially abundant miRNA were identified for each of the four Dipteran species using the sequence search tool on miRBase (Kozomara & Griffiths-Jones, 2014). Finally, this information on putative orthologous miRNAs and targets was provided to TargetScan and the algorithm was run with the default settings to identify conserved miRNA-target sites. Targets with 7nt or 8nt seed matches and conservation of nucleotides in the target seed region in at least one other species were retained.
Gene Ontology (GO) classification and visualization
To characterize the biological processes potentially affected by differentially abundant miRNAs, gene models predicted as miRNA targets by at least two algorithms were classified according to their associated Gene Ontology (GO) terms. GO terms were downloaded from VectorBase for each putative target. This list of GO terms was then consolidated to a simplified set of GO-Slim2 terms via the online CateGOrizer tool (Hu et al., 2008) to provide a concise overview of biological processes for target genes. During this process, each GO-Slim2 term retains a count of how often sub-terms appear in the initial set.
GO-Slim2 terms and associated frequency counts were visualized in REVIGO (Supek et al., 2011), which organizes GO terms by semantic similarity. The underlying semantic similarity algorithm, SimRel, uses the parent-child hierarchy of GO terms to assign similarity scores to pairs of GO terms based on the hierarchal distance between the pairs and how frequently each term appears overall in a given database. Pairs of GO terms which are closer together in the GO term hierarchy and appear less frequently receive higher similarity scores. Groups are formed when sets of GO terms receive similarity scores above a user-defined cutoff. Similarity scores >0.5 are considered functionally related (Schlicker et al., 2006) and 99% of randomly generated GO term pairs score <0.53 (Supek et al., 2011). Accordingly, the cutoff for similarity grouping was set to 0.5. D. melanogaster was chosen as a comparable GO term database for algorithm calculations.
Supplementary Material
Acknowledgments
We thank the University of Maryland Institute of Genome Sciences for performing the sequencing and Drs. Xin Huang and Julie Reynolds for technical assistance. We also thank Mr. Austin Garner, Mr. Kevin Martin, and two anonymous reviewers for valuable feedback on this manuscript. This work was supported in part by the National Institutes of Health (grant no. R15 AI111328 to PAA). The funding body did not have role in the study design, collection, analysis, or interpretation of the data or in writing the manuscript.
Footnotes
Conflict of interests: None
Data availability: All Illumina raw read sequence files are available from the NCBI Short Read Archive under BioProject accession PRJNA360961.
References
- Arensburger P, Hice RH, Wright JA, Craig NL, Atkinson PW. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genomics. 2011;12:606. doi: 10.1186/1471-2164-12-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster PA, Conn JE. Geographic variation of larval growth in North American Aedes albopictus (Diptera: Culicidae) Ann Entomol Soc Am. 2006;99:1234–1243. [Google Scholar]
- Armbruster PA, Hutchinson RA. Pupal mass and wing length as indicators of fecundity in Aedes albopictus and Aedes geniculatus (Diptera: Culicidae) J Med Entomol. 2002;39:699–704. doi: 10.1603/0022-2585-39.4.699. [DOI] [PubMed] [Google Scholar]
- Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2012;43:388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MQ, Levine RS, Hawley WA, Lounibos LP. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector Borne and Zoonotic Dis. 2007;7:76–85. doi: 10.1089/vbz.2006.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Xiang Y, Chen H, Liu Z, Gro S, Ku RP. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125:3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- Biggar KK, Storey KB. Insight into post-transcriptional gene regulation: stress-responsive microRNAs and their role in the environmental stress survival of tolerant animals. J Exp Biol. 2015;218:1281–1289. doi: 10.1242/jeb.104828. [DOI] [PubMed] [Google Scholar]
- Boulan L, Martín D, Milán M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol. 2013;23:473–478. doi: 10.1016/j.cub.2013.01.072. [DOI] [PubMed] [Google Scholar]
- Chen XG, Jiang X, Gu J, Xu M, Wu Y, Deng Y, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci USA. 2015;112:E5907–E5915. doi: 10.1073/pnas.1516410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernysh SI, Gordja NA, Simonenko NP. Diapause and immune response: induction of antimicrobial peptides synthesis in the blowfly, Calliphora vicina R.-D. (Diptera: Calliphoridae) Entomol Sci. 2000;3:139–144. [Google Scholar]
- Clements AN. The Biology of Mosquitoes. Chapman and Hall; London: 1992. [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denlinger DL. Regulation of diapause. Annu Rev Entomol. 2002;47:93–122. doi: 10.1146/annurev.ento.47.091201.145137. [DOI] [PubMed] [Google Scholar]
- Denlinger DL, Armbruster PA. Molecular physiology of mosquito diapause. In: Raikhel AS, editor. Advances In Insect Physiology. Elsevier; New York: 2016. pp. 329–361. [Google Scholar]
- Denlinger DL, Yocum GD, Rinehart JP. Hormonal control of diapause. In: Gilbert LI, editor. Insect Endocrinology. Academic Press; London: 2012. pp. 430–463. [Google Scholar]
- Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, et al. MiRNA nomenclature: a view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015;31:613–626. doi: 10.1016/j.tig.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for nncroRNAs in conferring robustness to ciological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Fujimoto Y, Kondo M, Yamanaka A, Watanabe M, Weihua K, et al. Stage-dependent changes of the prothoracicotropic hormone (PTTH) activity of brain extracts and of the PTTH sensitivity of the prothoracic glands in the cabbage armyworm, Mamestra brassicae, before and during winter and aestival pupal diapause. Zoolog Sci. 1997;14:127–133. [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, MacKowiak SD, Li N, Chen W, Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Sekine Y, Outa H, Hujikura M, Suzuki K. Relationships between cold hardiness and diapause, and between glycerol and free amino acid contents in overwintering larvae of the oriental corn borer, Ostrinia furnacalis. J Insect Physiol. 2001;47:157–165. doi: 10.1016/s0022-1910(00)00099-8. [DOI] [PubMed] [Google Scholar]
- Gu J, Hu W, Wu J, Zheng P, Chen M, James AA, et al. miRNA genes of an invasive vector mosquito, Aedes albopictus. PloS One. 2013;8:e67638. doi: 10.1371/journal.pone.0067638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn DA, Denlinger DL. Energetics of insect diapause. Annu Rev Entomol. 2011;56:103–121. doi: 10.1146/annurev-ento-112408-085436. [DOI] [PubMed] [Google Scholar]
- Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. MiRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct. 2013;218:817–831. doi: 10.1007/s00429-012-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Craig GB. Cold acclimation, diapause, and georgraphic origin affect cold hardiness in eggs of Aedes albopictus (Diptera: Culcidae) J Med Entom. 1994;31:192–201. doi: 10.1093/jmedent/31.2.192. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Craig GB. Aedes albopictus (Diptera: Culicidae) eggs: field survivorship during northern Indiana winters. J Med Entom. 1995;32:599–604. doi: 10.1093/jmedent/32.5.599. [DOI] [PubMed] [Google Scholar]
- Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions – beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- Hu ZL, Bao J, Reecy JM. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J Bioinform. 2008;9:108–112. [Google Scholar]
- Huang X, Poelchau MF, Armbruster PA. Global transcriptional dynamics of diapause induction in non-blood-fed and blood-fed Aedes albopictus. PLoS Negl Trop Dis. 2015;9:e0003724. doi: 10.1371/journal.pntd.0003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- Hussain M, Etebari K, Asgari S. Functions of small RNAs in mosquitoes. Adv In Insect Phys. 2016;51:189–213. [Google Scholar]
- Hwang JS, Go HJ, Goo TW, Yun EY, Choi KH, Seong Sl, et al. The analysis of differentially expressed novel transcripts in diapausing and diapause-activated eggs of Bombyx mori. Arch Insect Biochem Physiol. 2005;59:197–201. doi: 10.1002/arch.20057. [DOI] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Karp X, Ambros V. Dauer larva quiescence alters the circuitry of microRNA pathways regulating cell fate progression in C. elegans. Development. 2012;139:2177–2186. doi: 10.1242/dev.075986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Hammell M, Ow MC, Ambros V. Effect of life history on microRNA expression during C. elegans development. RNA. 2011;17:639–651. doi: 10.1261/rna.2310111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AM, MacRae TH. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 2015;60:59–75. doi: 10.1146/annurev-ento-011613-162107. [DOI] [PubMed] [Google Scholar]
- Koštál V. Eco-physiological phases of insect diapause. J Insect Physiol. 2006;52:113–27. doi: 10.1016/j.jinsphys.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Koštál V, Simunkova P, Kobelkova A, Shimada K. Cell cycle arrest as a hallmark of insect diapause: changes in gene transcription during diapause induction in the drosophilid fly, Chymomyza costata. Insect Biochem Mol Biol. 2009;39:875–883. doi: 10.1016/j.ibmb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Denlinger DE. A role for ecdysteroids in the induction and maintenance of the pharate first instar diapause of the gypsy moth, Lymantria dispar. J Insect Physiol. 1997;43:289–296. doi: 10.1016/s0022-1910(96)00082-0. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li S, Mead EA, Liang S, Tu Z. Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics. 2009;10:581. doi: 10.1186/1471-2164-10-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Dong Y, Gu J, Puthiyakunnon S, Wu Y, Chen XG. Developmental piRNA profiles of the invasive vector mosquito Aedes albopictus. Parasit Vectors. 2016;9:524. doi: 10.1186/s13071-016-1815-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Sziber PP, Quinn WG. Loss of calcium calmodulin responsiveness in adenylate-cyclase of rutabaga, a Drosophila learning mutant. Cell. 1984;37:205–215. doi: 10.1016/0092-8674(84)90316-7. [DOI] [PubMed] [Google Scholar]
- Lucas KJ, Myles KM, Raikhel AS. Small RNAs: a new frontier in mosquito biology. Trends Parasitol. 2013;29:295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KJ, Zhao B, Liu S, Raikhel AS. Regulation of physiological processes by microRNAs in insects. Curr Opin Insect Sci. 2015;11:1–7. doi: 10.1016/j.cois.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol Life Sci. 2010;67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Oda T. Studies on the egg diapause and overwintering of Aedes albopictus in Nagasaki. J Tropl Med. 1981;23:79–90. [Google Scholar]
- Poelchau MF, Reynolds JA, Denlinger DL, Elsik CG, Armbruster PA. A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation. BMC Genomics. 2011;12:619. doi: 10.1186/1471-2164-12-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. Deep sequencing reveals complex mechanisms of diapause preparation in the invasive mosquito, Aedes albopictus. Philos Trans R Soc Lond B BIol Sci. 2013a;280:20130143. doi: 10.1098/rspb.2013.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelchau MF, Reynolds JA, Elsik CG, Denlinger DL, Armbruster PA. RNA-Seq reveals early distinctions and late convergence of gene expression between diapause and quiescence in the Asian tiger mosquito, Aedes albopictus. J Exp Biol. 2013b;216:4082–4090. doi: 10.1242/jeb.089508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupardin R, Schöttner K, Korbelová J, Provazník J, Doležel D, Pavlinic D, et al. Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genomics. 2015;16:720. doi: 10.1186/s12864-015-1907-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyakunnon S, Yao Y, Li Y, Gu J, Peng H, Chen X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasit Vectors. 2013;6:230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland GJ, Denlinger DL, Hahn DA. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc Natl Acad Sci USA. 2010;107:14909–14914. doi: 10.1073/pnas.1007075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JA, Clark J, Diakoff SJ, Denlinger DL. Transcriptional evidence for small RNA regulation of pupal diapause in the flesh fly, Sarcophaga bullata. Insect Biochem Mol Biol. 2013;43:982–989. doi: 10.1016/j.ibmb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Reynolds JA, Poelchau MF, Rahman Z, Armbruster PA, Denlinger DL. Transcript profiling reveals mechanisms for lipid conservation during diapause in the mosquito, Aedes albopictus. J Insect Physiol. 2012;58:966–973. doi: 10.1016/j.jinsphys.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JA, Peyton JT, Denlinger DL. Changes in microRNA abundance may regulate diapause in the flesh fly Sarcophaga bullata. Insect Biochem Mol Biol. 2017;84:1–14. doi: 10.1016/j.ibmb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D. Photoperiodism in insects: migration and diapause responses. In: Nelson RJ, Denlinger DL, Somers DE, editors. Photoperiodism: The Biological Calendar. Oxford University Press; Oxford: 2009. pp. 218–258. [Google Scholar]
- Schlicker A, Domingues FS, Rahnenführer J, Lengauer T. A new measure for functional similarity of gene products based on gene ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinet J, Jain S, Jain J, Bhatnagar RK, Sunil S. Next generation sequencing reveals regulation of distinct Aedes microRNAs during Chikungunya Virus development. PLoS Negl Trop Dis. 2014;8:e2616. doi: 10.1371/journal.pntd.0002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singtripop T, Wanichacheewa S, Tsuzuki S, Sakurai S. Larval growth and diapause in a tropical moth, Omphisa fuscidentalis Hampson. Zoolog Sci. 1999;16:725–733. [Google Scholar]
- Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K, Choudhary A, Patowary A, Singh AR, Bhatia S, Sivasubbu S, et al. MiR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013;41:4470–4480. doi: 10.1093/nar/gkt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota T, Mogi M. Survival time and resistance to desiccation of diapause and non-diapause eggs of temperate Aedes (Stegomyia) mosquitos. Entomol Exp Appl. 1992;63:155–161. [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber MJ, Tauber CA, Masaki S. Seasonal Adaptations of Insects. Oxford University Press; Oxford: 1986. [Google Scholar]
- Tawfik AI, Tanaka Y, Tanaka S. Possible involvement of ecdysteroids in embryonic diapause of Locusta migratoria. J Insect Physiol. 2002;48:743–749. doi: 10.1016/s0022-1910(02)00099-9. [DOI] [PubMed] [Google Scholar]
- Trpiš M. A new bleaching and decalcifying method for general use in zoology. Can J Zool. 1970;48:892–893. [Google Scholar]
- Urbanski JM, Benoit JB, Michaud MR, Denlinger DL, Armbruster PA. The molecular physiology of increased egg desiccation resistance during diapause in the invasive mosquito, Aedes albopictus. Philos Trans R Soc Lond B BIol Sci. 2010;277:2683–2692. doi: 10.1098/rspb.2010.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski J, Mogi M, O’Donnell D, DeCotiis M, Toma T, Armbruster PA. Rapid adaptive evolution of photoperiodic response during invasion and range expansion across a climatic gradient. Am Nat. 2012;179:490–500. doi: 10.1086/664709. [DOI] [PubMed] [Google Scholar]
- Venkatesh K, Siddhartha G, Joshi R, Patel S, Hasan G. Interactions between the inositol 1,4,5-trisphosphate and cyclic AMP signaling pathways regulate larval molting in Drosophila. Genetics. 2001;158:309–318. doi: 10.1093/genetics/158.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilmos P, Bujna Á, Szuperák M, Havelda Z, Várallyay É, Szabad J, et al. Viability, longevity, and egg production of Drosophila melanogaster are regulated by the miR-282 microRNA. Genetics. 2013;195:469–480. doi: 10.1534/genetics.113.153585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votruba S. MicroRNAs in the Drosophila Egg and Early Embryo. University of Toronto; 2011. [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical Aspects of microRNA Target Prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.