Cancer cells can rewire genetic and epigenetic regulatory networks to promote cell proliferation and evade the immune system. Using a focused CRISPR/Cas9 genetic screen, Cuellar et al. identify a novel role for the SETDB1 histone methyltransferase in regulating the antiviral response in AML cells via the suppression of transposable elements.
Abstract
A propensity for rewiring genetic and epigenetic regulatory networks, thus enabling sustained cell proliferation, suppression of apoptosis, and the ability to evade the immune system, is vital to cancer cell propagation. An increased understanding of how this is achieved is critical for identifying or improving therapeutic interventions. In this study, using acute myeloid leukemia (AML) human cell lines and a custom CRISPR/Cas9 screening platform, we identify the H3K9 methyltransferase SETDB1 as a novel, negative regulator of innate immunity. SETDB1 is overexpressed in many cancers, and loss of this gene in AML cells triggers desilencing of retrotransposable elements that leads to the production of double-stranded RNAs (dsRNAs). This is coincident with induction of a type I interferon response and apoptosis through the dsRNA-sensing pathway. Collectively, our findings establish a unique gene regulatory axis that cancer cells can exploit to circumvent the immune system.
Introduction
Dysregulation of the epigenome can lead to global and local changes in gene expression (Dawson and Kouzarides, 2012), transforming normal cells and providing license to proliferate, metastasize, curb cell death, and evade the host immune system (Hanahan and Weinberg, 2011). Cancer genome sequencing efforts have focused on defining tumor-specific changes in unique coding regions and structural variants, as well as noncoding mutations that impact transcription factor–binding sites or the activity of functional RNAs (Mwenifumbo and Marra, 2013). However, evidence has begun to accumulate suggesting that deregulation of transposable elements (TEs), which comprise at least 45% of the human genome, may allow for tumor evolution through integration into and disruption of tumor suppressor loci or introduction of active promoter and enhancer elements within oncogenes (Lander et al., 2001; Haubold and Wiehe, 2006).
In the human and murine germlines, TEs, which include long terminal repeat (LTR)–containing endogenous retroviruses (ERVs), non-LTR–containing long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs), are thought to be marked for transcriptional silencing via H3K9 trimethylation (H3K9me3) and/or DNA methylation (Kassiotis and Stoye, 2016). Indeed, loss of these marks in embryonic stem cells by depletion of distinct lysine methyltransferases, DNA methyltransferases, or their cofactors leads to up-regulation of TEs and, either through direct or indirect effects, a host of developmental regulatory and TE-neighboring genes (Karimi et al., 2011). Suppression of TEs via specific histone modification is likely necessitated by the unique, DNA-hypomethylated state observed throughout distinct stages of early development (Ehrlich, 2002). H3K9me3 levels decrease in committed cells, such as mouse embryonic fibroblasts and neural precursor cells, compared with embryonic stem cells, and the primary mechanism of TE suppression in somatic cells is thought to be proximal DNA methylation driven by DNA methyltransferases (Mikkelsen et al., 2007; Kassiotis and Stoye, 2016).
Though genomic alterations induced by mobilized TEs may permit cellular transformation, their unrestrained expression can induce an immune response or, potentially, genomic instability, either of which could reduce cancer cell survival (Rodić et al., 2015; Zeller et al., 2016). Indeed, recent studies have found this phenomenon to be a potential vulnerability in cancer cells, as desilencing of endogenous retroelements with broad DNA methylation inhibitors can trigger increased expression of TE-specific double-stranded RNA (dsRNA) and induction of IFN through the RNA-sensing innate immune network (Chiappinelli et al., 2015; Roulois et al., 2015). Dysregulated expression of specific ERVs has recently been reported for a host of cancer subtypes, and it was speculated that increased ERV levels correlate with tumor immunity (Rooney et al., 2015). Furthermore, expression of TEs, particularly ERVs, can produce immunogenic peptides, which may sensitize cells harboring these antigens to immunoediting or the host to autoimmune disorders directed toward cells expressing these antigens (Young et al., 2013). Although TEs are tightly regulated by a combination of epigenetic and small RNA-based mechanisms, activation of these elements is thought to contribute to normal somatic cell diversity (Yang and Kazazian, 2006; Erwin et al., 2016). Accordingly, an emerging body of data suggests that normal tissues strike a balance between the advantageous and detrimental effects of TE activity (Kassiotis and Stoye, 2016). How this process is maintained, and whether cancer cells coopt the underlying mechanisms to enhance their survival, has yet to be fully established.
Acute myeloid leukemia (AML) represents the most common form of myeloid leukemia in adults, and alterations in epigenetic modifiers are known to contribute to myeloid malignancies (Plass et al., 2008; Shih et al., 2012; Sasca and Huntly, 2016). For instance, the H3K4 methyltransferase gene MLL exhibits translocation in 4–7% of AMLs. In addition, a high frequency of mutations are seen in genes that impact DNA methylation, including TET2 (7–23%), which converts 5-methylcytosine to 5-hydroxymethylcytosine (the first product in the active DNA demethylation process), IDH1 and IDH2 mutations that can impair TET2 activity (15–33%), and the de novo DNA methyltransferase DNMT3A (12–22%; Figueroa et al., 2010; Ito et al., 2011; Shih et al., 2012). Direct targeting of epigenetic modifiers may provide a therapeutic benefit in leukemia, and strategies to inhibit the activities of DOT1L, an H3K79 methyltransferase, and KDM1A (LSD1), a lysine demethylase, are being tested in the clinic (Daigle et al., 2011; Harris et al., 2012). Here, we identify SETDB1, an H3K9 methyltransferase, as a critical regulator of cell survival in human AML lines. SETDB1 expression is elevated in primary patient AML samples, and loss of SETDB1 triggers rapid desilencing of TEs, including ERVs, LINEs, and satellite repeats, despite only modest global reduction in H3K9me3 levels. The immediate effect of SETDB1 disruption in sensitive AML cell lines is a type I IFN antiviral response and apoptosis through a cytosolic dsRNA-sensing pathway. This is likely to take place through a concomitant increase in TE-derived overlapping sense and antisense transcripts. Our work supports a new model whereby SETDB1 plays a critical role in regulating the innate immune response via the suppression of endogenous repetitive elements, thus contributing to immune evasion and a prosurvival, oncogenic cell state.
Results
Identification of SETDB1 as a critical regulator of AML cell survival through a CRISPR/Cas9 genetic screen
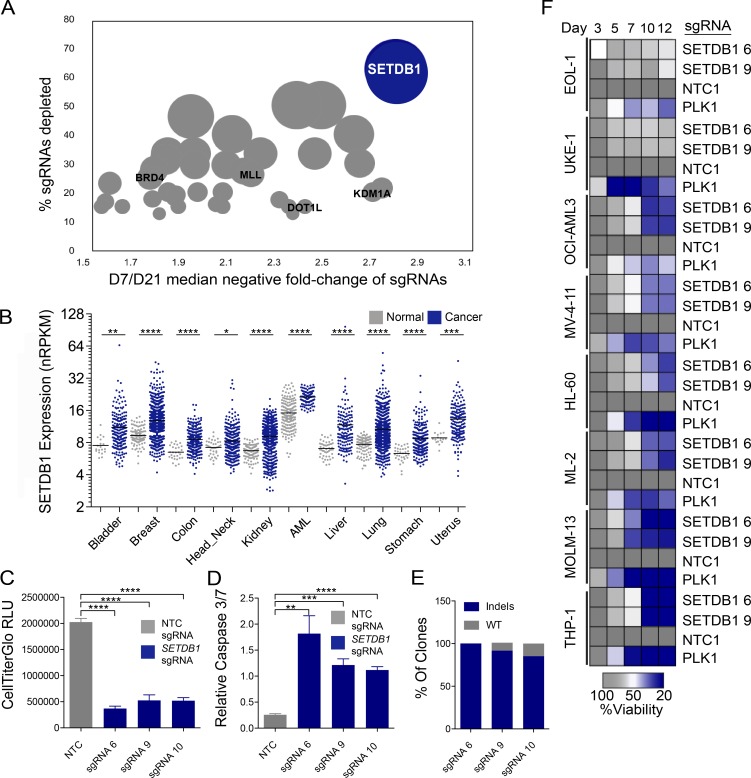
To explore epigenetic vulnerabilities in AML, we developed a loss-of-function (LOF) cell viability screen in the human p53-deficient, MLL-AF9 translocation–driven THP-1 AML cell line, which we engineered to stably express Streptococcus pyogenes Cas9 (Tsuchiya et al., 1980; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013). In parallel, we generated a custom pooled, lentivirus-based single-guide RNA (sgRNA) expression library that targets ∼350 known human epigenetic and transcriptional modifiers. THP-1–Cas9 cells were infected with the sgRNA pool at 1,000-fold representation (Fig. S1 A), and comparative analysis of sgRNA abundance at both an early (day 7, reference sample) and late (day 21) time point was performed via high throughput sequencing. Confirming the utility of our approach, we observed the depletion of sgRNAs specific to known regulators of AML cell growth or viability, including KDM1A (LSD1), DOT1L, MLL, and BRD4 (Fig. 1 A), as well as positive control sgRNAs targeting known essential genes (MCL1 and PLK1). Interestingly, the top hit in our screen, as determined by the fraction of depleted sgRNAs per gene and overall fold change related to sgRNA depletion, was SETDB1 (Fig. 1 A). An analysis of The Cancer Genome Atlas (TCGA) and other publicly available datasets suggests that SETDB1 expression is elevated in AML relative to normal blood cells and that SETDB1 is overexpressed or amplified across a broad range of malignancies (Fig. 1 B; Rodriguez-Paredes et al., 2014; Sun et al., 2014, 2015; Fei et al., 2015; Liu et al., 2015). However, a molecular mechanism related to SETDB1-dependent maintenance of AML cell growth and viability has yet to be defined, and we selected this target for additional follow-up studies.
Figure 1.
A CRISPR/Cas9 genetic screen identifies SETDB1 as a critical regulator of the leukemic cell survival that is overexpressed in many cancers. (A) Median negative fold changes of sgRNA abundance at day 21 versus day 7, ranked by percentage of sgRNAs depleted for each gene in THP-1–Cas9 cells. n = 3 biological replicates for the day 7 reference and n = 2 biological replicates for day 21. The size of each circle corresponds to the fraction of depleted sgRNAs/target. Select regulators of AML cell growth are highlighted in black text. Significant fold changes were calculated with DESeq2. (B) Compiled data from TCGA RNA-seq. SETDB1-normalized RPKM (reads per kilobase of transcript per million mapped reads) values in cancer and normal tissues. Student’s t tests were performed. (C and D) Viability and Caspase 3/7 levels, as measured by Caspase-Glo, in THP-1–Cas9 cells after 7 d of treatment with three different SETDB1-specific sgRNAs or an NTC sgRNA. Mean relative light units (RLU) are shown. n = 3 experiments. Error bars represent standard deviation. Student’s t tests were performed. (B–D) *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001. (E) Indel frequency at listed sgRNA target sites. Genomic DNA derived from THP-1–Cas9 cells 7 d after sgRNA infection. (F) Arrayed target validation screen in eight Cas9-stable AML lines. Mean viability at each day is shown. Screen performed in duplicate.
To validate and further examine the SETDB1 LOF phenotype in THP-1–Cas9 cells, we performed viability and apoptosis assays on cells exposed to three distinct SETDB1-specific sgRNAs or a nontargeting control (NTC) sgRNA (Fig. 1, C and D). SETDB1-targeting sgRNAs depleted SETDB1 protein, induced apoptosis in standard 2D tissue culture conditions, and reduced colony formation in 3D culture (Fig. 1, C and D; and Fig. S1, B–D). Analysis of transduced cell populations show that the SETDB1-specific sgRNAs introduced frame-shifting alleles (indels) with high frequency, and the proportion of deleterious mutations was consistent with penetrance of the apoptotic phenotype associated with each sgRNA (Fig. 1 E). To rule out the possibility that SETDB1 disruption synergizes with DNA damage–triggered cell death caused by Cas9, we performed an inducible shRNA-mediated depletion of SETDB1 mRNA (Wang et al., 2015; Aguirre et al., 2016; Munoz et al., 2016). Consistent with the knockout (KO) phenotypes, SETDB1 knockdown resulted in reduced protein levels, loss of viability, and induction of apoptosis markers (Fig. S2, A–D). To determine whether there is a general requirement for SETDB1 in AML cell survival, we engineered seven additional human AML cell lines to express Cas9 protein and treated these with the two distinct SETDB1 sgRNAs, an NTC sgRNA, or a positive control sgRNA targeting the essential gene PLK1. Like THP-1 cells, markedly reduced cell growth was observed in MOLM-13, ML-2, HL-60, OCI-AML-3, and MV-4-11 cells upon treatment with SETDB1-specific sgRNAs, whereas the UKE-1 and EOL-1 lines were insensitive (Fig. 1 F). Our results are consistent with recent CRISPR/Cas9 screens that identified a cell line–dependent, but perhaps AML-specific, requirement for SETDB1 (Hart et al., 2015; Tzelepis et al., 2016; Wang et al., 2017). Although reduced viability of SETDB1-depleted liver cancer cell lines has been shown to be driven by p53 mutation status, we could not identify an overt genetic signature that could explain sensitivity to SETDB1 loss in the evaluated AML lines (Fei et al., 2015). Therefore, we sought a deeper understanding of the molecular responses triggered upon loss of SETDB1 in distinct AML cell line contexts.
Disruption of SETDB1 leads to induction of viral response genes
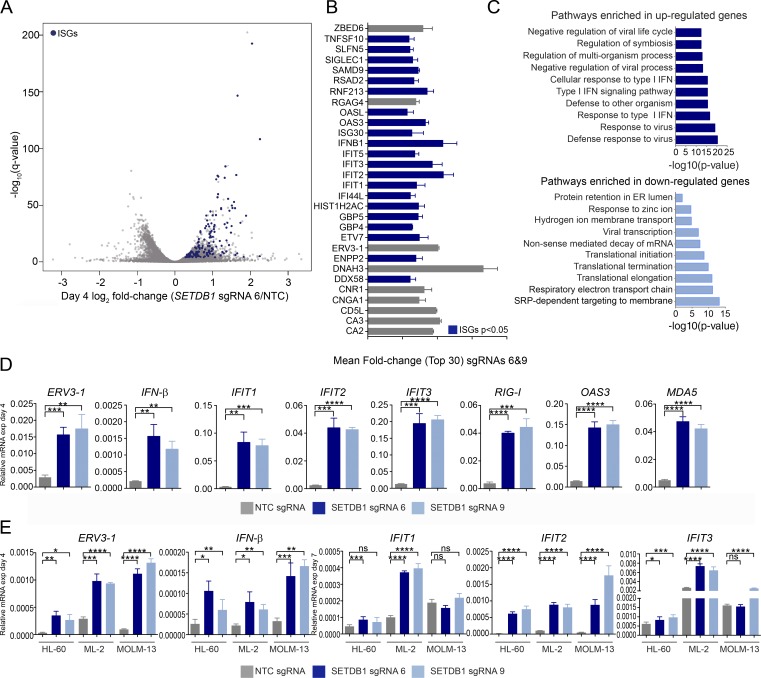
To gain insight into the molecular mechanisms by which SETDB1 provides a survival advantage in AML, we generated individual SETDB1 mutant THP-1 lines with two distinct SETDB1-specific sgRNAs and compared their transcriptome changes relative to that of an NTC sgRNA–treated line. Total mRNA was collected from these cells and analyzed by RNA-seq at 4 and 7 d after transduction with the gene-specific or control sgRNAs. Though analyses were performed on both day 4 and 7 samples, here we focus on the early time point (day 4) to study gene expression changes before appreciable cell death and collapse of the cell population, which occurs by day 7. RNA-seq profiling of SETDB1 sgRNA 6– and sgRNA 9–treated cells, relative to NTC sgRNA, showed similar gene expression across these populations (Fig. S1 E). In addition, the RNA-seq reads derived from SETDB1 sgRNA–treated cells exhibited a nearly uniform presence of mutated alleles at the expected sgRNA target sites, confirming gene disruption (Fig. S3 A). RNA-seq data from SETDB1 mutant cells at day 4 demonstrated a robust activation of IFN-stimulated genes (ISGs) with both target-specific sgRNAs (Fig. 2, A and B). Accordingly, panther gene ontology analysis revealed that type I IFN signaling and antiviral response genes were identified as the most up-regulated pathways in these cells (Fig. 2, A–C; Mi et al., 2017). The most significant down-regulated pathways include genes that regulate gene expression (translation) and viral transcription. We validated the RNA-seq data by quantitative real-time PCR (qRT-PCR) analysis of the SETDB1 mutant THP-1 lines, which confirmed increased expression of IFN-β and several ISGs, including IFIT1–3, RIG-I (DDX58), OAS3, and MDA5 (IFIH1; Fig. 2 D). ISG expression remained elevated at day 7 (Fig. S2 I). To exclude the possibility that DNA damage induced expression, ISG induction was further validated in THP-1 lines that express SETDB1-specific shRNAs (Fig. S2, E–G; Härtlova et al., 2015; Pépin et al., 2016). Consistent with the SETDB1 sgRNA–treated THP-1 cells, disruption of SETDB1 in three additional AML lines led to up-regulation of IFN-β at day 4 and increased ISG expression by day 7. Notably, though all three cell lines sensitive to SETDB1 mutation up-regulated IFN-β and IFIT2, they exhibited differential baseline and induced ISG expression levels (Fig. 2 E). Together, these results suggest that SETDB1 represses, directly or indirectly, type I IFN induction.
Figure 2.
Loss of SETDB1 leads to the induction of viral response genes. (A) Volcano plot of RNA-seq gene expression changes in THP-1–Cas9 cells after 4 d of treatment with representative SETDB1-specific sgRNA relative to NTC (DESeq2 log2 fold changes vs. significance −log10 [q-value], and n = 3 biological replicates). ISGs are highlighted in blue. (B) Bar plot of top 30 induced genes (ISGs are in blue) in THP-1–Cas9 cells after treatment with two SETDB1-specific sgRNAs (6 and 9) at day 4. Mean fold change was calculated using the fold changes for each SETDB1-specific sgRNA relative to NTC, and n = 3 biological replicates. (C) Panther gene ontology analysis shows most significantly enriched biological processes after SETDB1 disruption at day 4 (genes up-regulated/down-regulated ≥1.5-fold in both cells treated with SETDB1 sgRNAs 6 and 9 were used for the analysis). (D) Taqman qRT-PCR validation of RNA-seq data on day 4, showing relative expression of IFN-inducible transcripts after treatment with SETDB1 sgRNAs or an NTC control sgRNA. n = 3 experiments, and Student’s t tests were performed. (E) Taqman qRT-PCR data on day 4 showing relative expression of IFN-β and ERV3-1 as well as ISGs at day 7 after treatment of three different SETDB1 disruption-sensitive AML lines with SETDB1-specific or NTC sgRNAs. n = 3 experiment, and Student’s t tests were performed. Error bars represent standard deviation. (D and E) ns, not significant, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P ≤ 0.0001.
Loss of SETDB1 leads to a reduction of H3K9me3 at repetitive loci
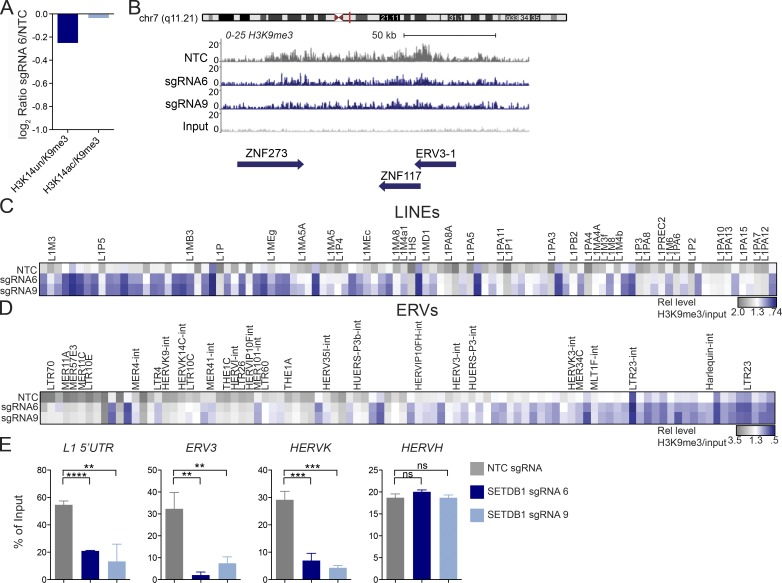
SETDB1 has been characterized as a transcriptional repressor, mediated by its deposition of the H3K9me3 mark on target loci, and a broad range of effects on global H3K9me3 levels has been observed upon loss of SETDB1 in distinct cellular contexts (Collins et al., 2015; Tchasovnikarova et al., 2015). To determine whether the gene expression changes in SETDB1 KO cells could be attributed to widespread or selective loss of H3K9me3, we first performed a histone mass spectrometry analysis on THP-1 cells treated for 6 d with either NTC or SETDB1 sgRNAs. This analysis revealed a modest loss of global H3K9me3 (Fig. 3 A). Interestingly, we observed that loss of H3K9me3 occurred only on unmodified H3K14, a neighboring mark, which may be reflective of SETDB1-mediated H3K9me3 occurring in regions of the genome that are transcriptionally inactive and H3K14 hypoacetylated (Karmodiya et al., 2012).
Figure 3.
Loss of SETDB1 leads to a modest reduction in H3K9me3. (A) Histone mass spectrometry analysis of H3K9me3 levels in THP-1–Cas9 cells after 6 d of treatment with SETDB1 sgRNA 6 or NTC (log2 ratios of sgRNA 6 over NTC are shown). (B) Density plot showing H3K9me3 levels over the ERV3-1/ZNF117 locus as well as the neighboring ZNF273 gene (levels of H3K9me3 in THP-1–Cas9 cells are shown after 6 d of treatment with control NTC sgRNA or distinct SETDB1-specific sgRNAs. Y axis is 0–25 and region shown is ∼150 kb. (C) Low input nChIP-seq heat map showing LINE-1 elements with a ≥1.2-fold reduction in H3K9me3 after 6 d of treatment with sgRNA 6, sgRNA 9, or NTC. For each sgRNA, the relative enrichment = fractional H3K9me3 reads/nonimmunoprecipitated chromatin reads (input/background). Highlighted genes were up-regulated at days 4, 7, or both days in SETDB1 mutant RNA-seq datasets. (D) As in C; heat map showing ERV elements with a ≥1.2-fold reduction in H3K9me3 after 6 d of treatment with sgRNA 6, sgRNA, 9 or NTC. (E) ChIP-PCR validation of ChIP-seq data from day 6, showing percentage of input of cells treated with NTC or SETDB1-specific sgRNAs for L1 LINE 5′UTR, HERV-K-rev, ERV3-1-exon, and HERV-H-pregag (n = 3 experiments. Student’s t tests were performed). All error bars represent standard deviation. ns, not significant, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To identify specific genetic loci that were depleted of H3K9me3 after SETDB1 disruption, we performed H3K9me3 chromatin immunoprecipitation (ChIP)–seq on cells treated with separate SETDB1-specific sgRNAs versus an NTC sgRNA control for 6 d. H3K9me3 ChIP-seq revealed that loss of this mark occurred at KRAB zinc finger genes, which are known to be repressed by SETDB1 and are linked to ERV silencing (Schultz et al., 2002; Jacobs et al., 2014). H3K9me3 levels were unchanged across loci specific to the ISGs or innate immune sensors up-regulated in our RNA-seq data, suggesting that induction of these genes was not directly attributable to reduced H3K9me3 at these positions (Fig. 3 B and Fig. S4, A and B). Refined analysis of our H3K9me3 ChIP-seq data, focusing on the repetitive genome, determined that loss of this mark occurs over repetitive loci, in accordance with the known role for SETDB1 in modifying H3K9 in repetitive parts of the genome (Fig. 3, C and D; Criscione et al., 2014). These findings were further validated by ChIP-qPCR, examples of which are shown in Fig. 3 E.
Loss of SETDB1 leads to induction of retro-TEs
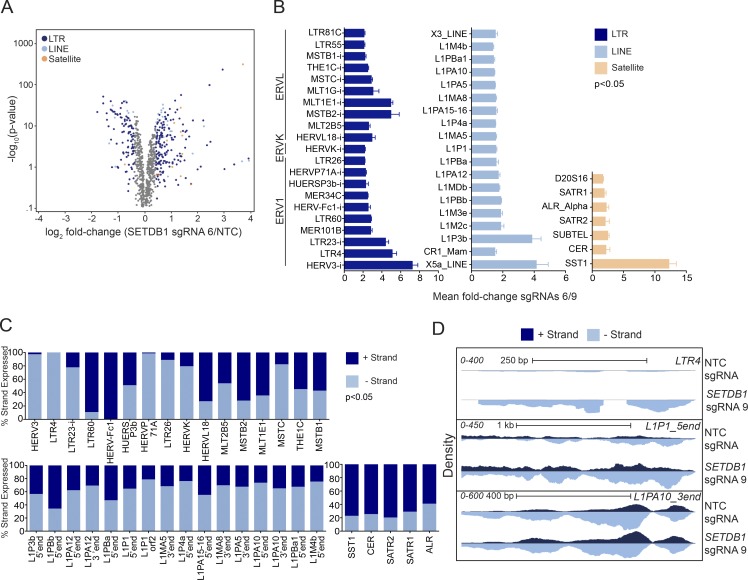
Increased ERV expression has been observed after the deletion of SETDB1 in developing cells, including embryonic stem cells and B cells, although ERV suppression has not been reported as a function of SETDB1 in cancer cell lines (Matsui et al., 2010; Karimi et al., 2011; Collins et al., 2015; Tchasovnikarova et al., 2015). RNA-seq analysis of SETDB1-disrupted cells revealed striking up-regulation of the ERV ERV3-1 in THP-1, as well as three additional AML cell lines treated with SETDB1-specific sgRNAs or SETDB1-targeting shRNAs (Fig. 2, B, D, and E; and Fig. S2, H and I). Notably, repetitive elements may be hypomethylated in cancers, enabling them to mobilize and potentially destabilize the genome (Howard et al., 2008; Ehrlich, 2009). Therefore, we hypothesized that TEs are suppressed by SETDB1 in THP-1 cells, and removing this block would induce expression of TE transcripts, thus contributing to the observed antiviral response. ERV3-1 bears a unique nucleotide sequence and can be distinguished from ERVs within the same class. Although this transcript is listed in the standard genomic annotations (e.g., Ensembl) and evaluated in standard RNA-seq toolkits, such as that used for our initial expression analyses, retrotransposons can be present in the genome at many copies and thus are masked out (repeat masked) of the standard genome annotations as being multimappers or low-complexity sequences. Therefore, we re-analyzed our RNA-seq data using a pipeline that enables alignment of repetitive (repeat masked) reads (Criscione et al., 2014). Using this approach, we found that many LTR-containing ERV subfamilies and non-LTR elements, including LINEs and satellite repeats, were consistently up-regulated in SETDB1 sgRNA–treated cells but not in the NTC controls (Fig. 4, A and B). Some LTRs are down-regulated, possibly because of induction of KRAB-ZNFs. Notable for their coevolution with and ability to silence ERVs, KRAB-ZNF genes are known to be enriched for the H3K9me3 mark (Schultz et al., 2002; Jacobs et al., 2014). These data suggest SETDB1 plays a critical role in repressing retro-TEs in AML cells.
Figure 4.
Loss of SETDB1 leads to the induction of TEs. (A) An RNA-seq volcano plot depicting expression changes of TEs in THP-1–Cas9 cells at day 7 after treatment with a representative SETDB1-specific sgRNA versus NTC (n = 3 for all samples, EdgeR GLM log2 fold change). (B) Bar plots showing statistically significant (P < 0.05) EdgeR GLM fold changes of TEs in SETDB1 sgRNA–treated THP-1–Cas9 cells (mean fold change of sgRNAs 6 and 9) over NTC at day 7 (n = 3 biological replicates, and error bars represent standard deviation). (C) Bar plots derived from strand-specific RNA-seq of SETDB1 or NTC sgRNA–treated THP-1–Cas9 cells at day 7. Up-regulated TEs are shown as a percentage of reads derived from 5′ or 3′ transcribed products. In B and C, "i" denotes "internal." (D) Density plots showing an example of a unidirectionally transcribed TE (LTR4), a bidirectionally transcribed element (L1P1_5′end), and a second bidirectionally transcribed element in which only one strand is increased after treatment with an SETDB1-specific sgRNA for 7 d (L1PA10_3′end).
It has previously been shown that both LINEs and ERVs exhibit bidirectional transcription and that these overlapping transcripts can pair to form dsRNAs (Dunn et al., 2006; Faulkner et al., 2009; Liu et al., 2016). Therefore, we reasoned that an increase in the homeostatic levels of dsRNAs, formed by expressed TEs, could trigger the IFN response we observed in SETDB1-disrupted AML cell lines. To determine whether there was an increase in overlapping sense and antisense transcripts in the up-regulated TEs, we performed strand-specific RNA sequencing of THP-1–Cas9 cells treated with an NTC or a representative SETDB1-specific sgRNA. The most up-regulated TE subfamily members (genes shown in Fig. 4 B) were aligned to the Dfam consensus sequence (Hubley et al., 2016). Our analysis, which is consistent with previously published work, shows that whereas a few ERVs are unidirectionally transcribed (e.g., LTR4; Fig. 4 C), many of the ERV subfamily members and all LINE-1 and satellites surveyed exhibited increased concurrent sense and antisense transcription (e.g., L1P1 and L1PA10) upon SETDB1 disruption (Fig. 4, C and D; Walsh et al., 1998; Domansky et al., 2000; Kanellopoulou et al., 2005; Dunn et al., 2006; Yang and Kazazian, 2006; Cruickshanks et al., 2013; Liu et al., 2016). Interestingly, for many of these elements, the ratio of sense to antisense transcripts did not change (Fig. S3, B–D). To visualize whether the increase in transcription from both orientations occurred over the same regions for each element, thus increasing the probability that they could pair and form dsRNAs, we generated RNA-seq read density plots. As predicted, an increased yield of overlapping transcripts was observed at select loci. Representative examples of uni- and bidirectionally transcribed TEs, induced upon SETDB1 disruption, are shown in Fig. 4 D.
SETDB1 KO leads to induction of dsRNAs
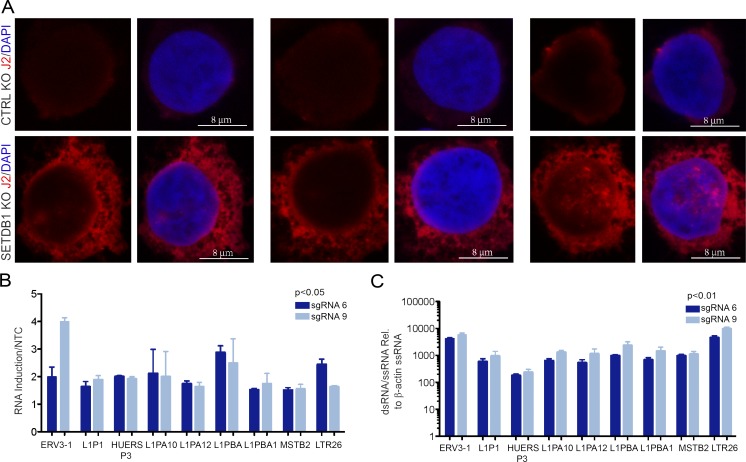
To experimentally determine directly whether there was an increase in the abundance of dsRNAs in cells after depletion of SETDB1, we stained THP-1 cells treated with either SETDB1- or CD81-specific synthetic gRNAs with the J2 antibody, which detects dsRNA (Weber et al., 2006). In these experiments, the CD81 gRNA was used to confirm CRISPR/Cas9 activity after gRNA nucleofection and control for the possibility that DNA breaks themselves may induce dsRNA expression. We observed induction of dsRNAs in cells that have lost SETDB1 (as verified based on the mutation status of SETDB1 from an aliquot of the cells used for image analysis; see Fig. 6 C) but not in the control cells, consistent with our stranded RNA-seq data (Fig. 5 A). We then sought to determine whether this increase in dsRNA content was correlated with up-regulation of dsRNA derived from derepressed TEs. To test this, we performed an RNase protection assay (Roulois et al., 2015). In brief, total RNA was collected from THP-1 cells treated with distinct SETDB1-specific or NTC sgRNAs, and the RNA was digested with a cocktail of RNase A and T1 to remove single-stranded species. TE regions predicted to generate dsRNAs, based on RNA-seq density plots (Fig. 4 D), were amplified and compared with a highly expressed mRNA (β-actin, ACTB) via qRT-PCR (Fig. 5 B). As shown, TE regions with overlapping sense and antisense transcripts were both up-regulated after SETDB1 disruption and were >100–1,000-fold resistant to RNase digestion compared with single-stranded RNA, suggesting the evaluated TE loci can produce stable dsRNAs (Fig. 5 C).
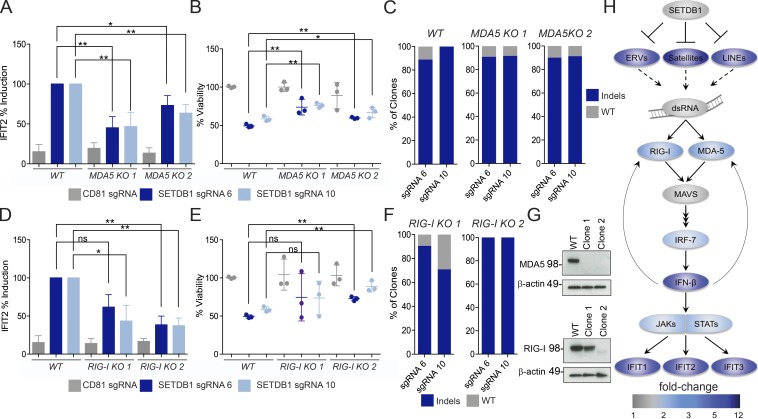
Figure 6.
IFN response and cell death in SETDB1 mutant cells are dependent on the viral sensing machinery. (A) Induction of IFIT2 expression after SETDB1 disruption or control gene (CD81) disruption, in THP-1–Cas9 cells (WT) or two MDA5 KO THP-1–Cas9 clones. Total RNA was analyzed 5 d after nucleofection of cells with synthetic SETDB1 gRNAs or control gRNA targeting CD81. (B) Cell viability at day 5 after nucleofection for cells and conditions described in A. (C) Indel rate at noted target sites for THP-1–Cas9 cells for cells and conditions described in A. Genomic DNA analyzed at day 5 after nucleofection. (D) Induction of IFIT2 expression after targeting SETDB1 or control gene (CD81) disruption in THP-1–Cas9 cells (WT) or RIG-I hypomorphic (clone 1) and KO (clone 2) THP-1–Cas9 clones. Total RNA was analyzed 5 d after nucleofection of cells with synthetic SETDB1 gRNAs or control gRNA targeting CD81. (E) Cell viability at day 5 after nucleofection for cells and conditions described in D. (A, B, D, and E) n = 3 experiments, and Student’s t tests were performed. (F) Indel rate at noted target sites for THP-1–Cas9 cells for cells and conditions described in D. Genomic DNA analyzed at day 5 after nucleofection. (G) Western blot analysis of MDA5, RIG-I, and β-actin in THP-1–Cas9 WT or clonal lines after overnight stimulation with IFN-β. (H) Model of SETDB1 function, based on ingenuity analysis 4 RNA-seq data 4 d after SETDB1 disruption (as shown in Fig. 2), as well as incorporating TEs into map. Color bar denotes fold change compared with NTC-treated cells. All error bars represent standard deviation. (A, B, D, and E) ns, not significant, P > 0.05; *, P < 0.05; **, P < 0.01.
Figure 5.
Loss of SETDB1 leads to the induction of dsRNAs. (A) Confocal microscopy images of representative THP-1–Cas9 cells 5 d after nucleofection with synthetic gRNAs targeting CD81 (negative control, top) or SETDB1 (target sequence 6, bottom). Cells were stained with J2 antibody to label dsRNA and DAPI to mark nuclei. (B) qRT-PCR validated expression of bidirectionally transcribed LINEs and ERVs in THP-1–Cas9 cells after 5 d of treatment with SETDB1 sgRNAs or an NTC control sgRNA (n = 3 biological replicates). (C) RNase protection assay showing expression of dsRNAs in THP-1–Cas9 cells after 5 d of treatment with synthetic SETDB1-specific gRNAs after digestion of total RNA with RNase A/T1. ssRNA, single-stranded RNA. Enrichments are relative to NTC control sgRNA (n = 3 biological replicates, and Student’s t tests were performed). Error bars represent standard deviation.
Cell death induced by SETDB1 disruption is dependent on the viral sensing machinery
Because we observed concordant loss of H3K9me3 and up-regulated transcription of TE loci, in SETDB1 disrupted cells, we reasoned that one possible trigger of the IFN antiviral response could be the increased output of aberrant, TE-specific transcripts and their subsequent detection by the nucleic acid–sensing innate immune receptors. To determine whether this machinery was responsible for inducing type I IFN and, consequently, cell death, we used CRISPR to knock out cytosolic RNA sensors of the innate immune system, including IFIH1 (MDA5), DDX58 (RIG-I), and MAVS, followed by disruption of SETDB1. MDA5, MAVS, and RIG-I–specific sgRNA expression conferred protection of viability in the presence of the SETDB1-specific sgRNAs (Fig. S5 A). The loss of MDA5 and MAVS in the combinatorial sgRNA-treated cells was confirmed by FACS analysis of THP-1–Cas9 cells treated with two different sgRNAs/gene (Fig. S5, B and C). RIG-I disruption was confirmed by sequencing the targeted loci for mutations, which demonstrated that >99% disruption occurred with two separate sgRNAs (Fig. S5 D).
As shown in Fig. 2 (D and E), expression of the ISG, IFIT2, provided a sensitive, robust readout of the IFN response within distinct SETDB1-disrupted AML lines. To evaluate whether activation of the cytosolic sensors contributed to IFIT2 induction, we generated single-cell THP-1–Cas9 KO clones for MDA5 as well as KO and hypomorphic clones for RIG-I by way of nucleofected gRNAs (Fig. 6). Western blotting was performed on each clone after overnight stimulation with IFN-β, and mutations were confirmed by sequencing (Fig. 6 G). IFIT2 induction and the negative effects on viability were significantly reduced when KO clones were nucleofected with two distinct gRNAs targeting SETDB1 compared with control THP-1–Cas9 cells (Fig. 6, A, B, D, and E). These effects were less pronounced in the RIG-I hypomorphic clone relative to the complete KO. SETDB1 gRNA activities were confirmed by DNA sequencing of the target sites, which show ∼70–100% disruptive mutation rates across the treated cell populations (Fig. 6, C and F). Together, the combination of expression changes and functional data provided in these studies suggests a complex mixture of RNAs become expressed in the absence of SETDB1, which triggers a cytosolic, nucleic acid–sensing cascade and IFN-mediated cell death (Fig. 6 H).
Discussion
Using an unbiased LOF screen and cross validation in human AML cell lines, we uncovered a novel axis for stimulating an innate immune response and reducing cell viability through the disruption of SETDB1. Specifically, the mutation of SETDB1 leads to rapid desilencing of TEs, including ERVs, satellite repeats, and LINEs, many of which exhibit bidirectional transcription. Expression of these elements is coincident with an increase in dsRNA content, activation of the cytosolic RNA–sensing pathway, and IFN-induced apoptosis (Fig. 6 H).
Intriguingly, the means by which SETDB1 silences TEs may differ between cancerous or normal adult tissues and the germline, owing to cell type– or context-specific differences in SETDB1-dependent H3K9me3 deposition. Similar to studies in developmental contexts, SETDB1 KO in THP-1 cells resulted in modest global depletion of H3K9me3, and increased transcription was likely caused by localized turnover of this mark at TE loci (Collins et al., 2015; Koide et al., 2016). Furthermore, despite their presence in our library, we did not observe the dropout of sgRNAs targeting HP1 family members or TRIM28/KAP1, factors known to be required for TE silencing in embryonic cell types (Rowe et al., 2013). This is consistent with the retrovirus-silencing properties of SETDB1 that occur independent of its known germline-associated cofactors (Fig. 1 A; Maksakova et al., 2011; Tchasovnikarova et al., 2015).
Here, we have defined a unique avenue for inducing apoptosis in AML contexts and identified a previously uncharacterized role for SETDB1 in suppressing innate immunity by limiting the overall abundance of TE expression in cancer cells. Because SETDB1 is amplified and/or overexpressed in many human cancers and its expression is induced upon lethal chemotherapeutic drug exposure, further work to determine whether up-regulation of this gene is a common mechanism to evade the host innate immune response, thus acting as a cancer cell immunity cloak, should be pursued (Guler et al., 2017). Additionally, while we and others have found that the KO and knockdown of SETDB1 reduces AML cell viability in vitro and in vivo, it remains to be seen whether chemical inhibition of this enzyme is feasible and can induce a similar molecular phenotype (Shi et al., 2015; Wang et al., 2015, 2017; Koide et al., 2016; Tzelepis et al., 2016). The impact of TE expression may extend beyond the cell-intrinsic growth defects evaluated in our study. Indeed, TE-derived antigens have been considered as targets of antitumor immunotherapy, and induction of these elements via inhibition of DNA methylation has been linked to sensitization of cells to checkpoint blockade in preclinical settings (Cherkasova and Selyatitskaya, 2013; Chiappinelli et al., 2015; Roulois et al., 2015; Kassiotis and Stoye, 2016).
Moreover, repeat-associated dsRNAs have been shown to trigger production of siRNAs and silencing by an RNAi-dependent mechanism (Yang and Kazazian, 2006). Curiously, several components of the RNAi machinery, including DICER1, AGO1, and AGO4, are up-regulated in SETDB1-mutated THP-1 cells (unpublished data). Therefore, in light of evidence that the KO of DICER leads to an accumulation of repeat-associated dsRNAs and induction of IFN in both mouse and human cultured cells, it is tempting to speculate that SETDB1 contributes to a potential network of overlapping mechanisms aimed at silencing inappropriate somatic TE transcription (Murchison et al., 2005; Yang and Kazazian, 2006).
Materials and methods
CRISPR library generation
A panel of ∼350 known or predicted epigenetic regulators was identified, and ∼15–25 sgRNAs were designed to target each gene. sgRNAs were designed to target coding regions common to the most isoforms, with the 5′ end of the CDS favored to maximize disruption. sgRNAs with the best off-target score based on the number and location of mismatches were used. Selected sgRNAs were formatted into a standard expression context (Mali et al., 2013) and cloned en masse into pLKO (SHC-201; Sigma-Aldrich) by Cellecta, Inc., creating what we term the Epi300 sgRNA library. Illumina sequencing confirmed that >99% of the designated sgRNAs were represented in the cloned plasmid library.
CRISPR/Cas9 sgRNA pooled screen and analysis
THP-1 cells (ECACC; Sigma-Aldrich) were transduced with a lentiviral vector (pLenti7.3; ThermoFisher) engineered to coexpress a human codon–optimized S. pyogenes Cas9 along with emerald GFP (emGFP). emGFP-expressing cells were collected after fluorescence-activated cell sorting with a flow cytometer (FACSAria; BD). 20 × 106 THP-1–Cas9–emGFP cells per biological replicate were seeded into a 50-ml conical tube containing 8 µg/ml polybrene and Epi300 sgRNA viral pool (MOI 0.3). Cells were dispensed into six 6-well plates and spin transduced for 45 min at 1,800 rpm at 20°C. The next day, cells were pooled and transferred to flasks (T-175; Corning). 2 d after transduction, the cells were placed under selection with media containing 1 µg/ml puromycin. Cells were maintained at a minimum of 10 × 106 cells at all times to maintain over 100× representation. Reference samples were taken at day 7, and competitive growth was assessed at day 21. At each time point, 10 × 106 cells were pelleted and flash frozen. Genomic DNA was prepared with a blood and tissue kit (DNeasy; Qiagen) and quantified using a fluorimeter (Qubit; ThermoFisher). To determine the sgRNAs that were enriched or depleted, we amplified sgRNAs from genomic DNA by PCR. The mass of DNA required to maintain representation of each sgRNA was determined by multiplying the number of cells with integrants by nanogram/genome. PCR was performed with 500 ng DNA per PCR reaction with polymerase (Phusion; NEB). Primer sequences were forward, 5′-TCTTGTGGAAAGGACGAGGTACCG-3′, and reverse, 5′-CTACTATTCTTTCCCCTGCACTGT-3′. ∼221 bp PCR products were isolated. Next-generation sequencing libraries were prepared with the TruSeq Nano DNA library (Illumina) with 5% PhiX spiked in or NuGen Ovation low-complexity kits followed by PCR amplification for six cycles. Samples were sequenced on a MiSeq system (Illumina) with reagent kit v3 for 150 cycles. Raw FASTQ files were aligned with the genomic short-read nucleotide alignment program (GSNAP), and differential analysis was performed using DESeq2 (Love et al., 2014; Wu et al., 2016).
TCGA data
For TCGA analysis, the data were based in part on data generated by the TCGA Research Network. Genotype-Tissue Expression project expression data for normal blood samples were obtained from https://www.genome.gov/gtex/.
Arrayed sgRNA transduction
As detailed in the CRISPR/Cas9 sgRNA pooled screen and analysis section, MV-4-11 (ATCC), MOLM-13 (DSMZ), OCI-AML-3 (DSMZ), UKE-1 (Coriell Institute), HL-60 (ATCC), ML-2 (DSMZ), and EOL-1 (DSMZ) cells were transduced with pLenti7.3-Cas9 and selected based on emGFP expression. Individual sgRNAs were cloned into pLKO, and lentivirus was prepared in 96-well plates. For each cell line, 35 × 105 Cas9-emGFP cells were seeded into round-bottom 96-well plates and transduced with virus in 8 µg/ml polybrene. Cells were spin transduced for 30 min at 1,800 rpm at 20°C. Screen was performed in duplicate. Viability was measured every 2–3 d for a total of 18 d using CellTiter-Glo (Promega). For caspase detection, the Caspase-Glo 3/7 assay kit (Promega) was used, and data were normalized to viability derived from CellTiter-Glo assays.
Inducible SETDB1 shRNA cell line generation
Individual shRNAs targeting human SETDB1 were designed using the DSIR algorithm. The Ren.713 non-targeting control was used in a previous study (Fellmann et al., 2013). All targeting sequences were converted into 125-nt DNA oligonucleotides, annealed, and cloned into the XhoI–EcoRI restriction sites of the lentiviral, doxycycline-inducible shRNA expression vector pMinDUCERv1-tRFP-miRE-ET2P, enabling expression of optimized miR-30–based shRNAs along with a turboRFP (Evrogen) marker.
shRNA oligo sequences were SETDB1_3_FOR, 5′-TCGAGAAGGTATATTGCTGTTGACAGTGAGCGATAGCTGAGACACCAAACGTCATAGTGAAGCCACAGATGTATGACGTTTGGTGTCTCAGCTAGTGCCTACTGCCTCGGACTTCAAGGGGCTAG-3′; SETDB1_3_REV, 5′- AATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGCACTAGCTGAGACACCAAACGTCATACATCTGTGGCTTCACTATGACGTTTGGTGTCTCAGCTATCGCTCACTGTCAACAGCAATATACCTTC-3′; SETDB1_4_FOR, 5′-TCGAGAAGGTATATTGCTGTTGACAGTGAGCGAACCTGATAGTCAGCATGCGAATAGTGAAGCCACAGATGTATTCGCATGCTGACTATCAGGTGTGCCTACTGCCTCGGACTTCAAGGGGCTAG-3′; SETDB1_4_REV, 5′-ATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGCACACCTGATAGTCAGCATGCGAATACATCTGTGGCTTCACTATTCGCATGCTGACTATCAGGTTCGCTCACTGTCAACAGCAATATACCTTC-3′; Ren.713_FOR, 5′-TCGAGAAGGTATATTGCTGTTGACAGTGAGCGAAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTGTGCCTACTGCCTCGGACTTCAAGGGGCTAG-3′; and Ren.713_REV, 5′-AATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGCACAGGAATTATAATGCTTATCTATACATCTGTGGCTTCACTATAGATAAGCATTATAATTCCTTCGCTCACTGTCAACAGCAATATACCTTC-3′.
THP-1 cells (ECACC; Sigma-Aldrich) were transduced with lentiviral particles as described in the CRISPR/Cas9 sgRNA pooled screen and analysis section. After transduction, stable, transgenic cells were selected with 1 µg/ml puromycin. Stable lines were treated with 500 ng/ml doxycycline to induce the expression of shRNAs for various assays.
RNA-seq and analysis
THP-1–Cas9 cells were transduced with either SETDB1 sgRNA 6 or 9 or an NTC sgRNA and collected at various time points. Total RNA was isolated with the RNAeasy micro kit (Qiagen). The concentration of total RNA samples was determined using a NanoDrop 8000 (Thermo Scientific). The integrity of RNA samples was determined using a bioanalyzer (2100; Agilent Technologies). 0.5 µg of total RNA was used as an input material for library preparation using an RNA sample preparation kit v2 (TruSeq; Illumina). For stranded RNA-seq, 100 ng of total RNA for each sample was used for library preparation with a TruSeq Stranded total RNA library prep kit. The size of the libraries was confirmed using a 2200 TapeStation and high-sensitivity screen tape (D1K; Agilent Technologies), and their concentration was determined via a qPCR library quantification kit (KAPA). The libraries were multiplexed and then sequenced on an HiSeq2500 system (Illumina); we generated 30 M single-end 100-bp reads for TruSeq RNA libraries and 30 M paired-end 75-bp reads for TruSeq Stranded RNA libraries. Resultant FASTQ files were aligned to GSNAP, and differential gene expression analysis was performed with DESeq2 after removing genes with less than five reads in any of the samples (Love et al., 2014; Wu et al., 2016).
TE analysis
RNA-seq and ChIP-seq FASTQ files were aligned to the human genome (hg19) using Bowtie version 0.12.9 (Langmead et al., 2009). Unique mapping and multimapping reads were separated and analyzed using RepEnrich (; Criscione et al., 2014). Differential enrichment analysis was performed with EdgeR GLM on the fractional counts data (Robinson et al., 2010). Library size was determined based on Bowtie, (reads processed) − (reads that failed to align).
Bidirectional transcript analysis
Consensus sequences for elements for up-regulated TEs were downloaded from the Dfam database (Hubley et al., 2016). Stranded RNA-seq data were then aligned to consensus sequences using BWA-MEM and quantified using HTseq-count (Li, 2013; Anders et al., 2015). Reads were visualized using the Integrative Genomics Viewer (IGV) genome browser and a custom pseudogenome that was generated from the Dfam consensus sequences (Robinson et al., 2011; Thorvaldsdóttir et al., 2013).
ChIP-seq and analysis
Native ChIP-seq on SETDB1 mutant cells (generated with SETDB1 sgRNA 6 or 9) or NTC-treated cells was performed as described in Brind’Amour et al. (2015). 10% of 300 µl MNase-digested chromatin was removed and saved as input/unenriched chromatin, and the rest was immunoprecipitated with an H3K9me3 antibody (49-1008; ThermoFisher). 30 µl of input chromatin was decross linked in the presence of 5 µl of 5 M NaCl and 2 µl proteinase K at 65°C overnight on a thermocycler. The input was then purified over a column (minElute; Qiagen) in 20 µl TE buffer (Qiagen). 50-bp single-end reads derived from immunoprecipitated and input samples were generated on a HiSeq2500 system. Reads were aligned to hg19 with Bowtie 2.2.4 and processed with MACS2 (Zhang et al., 2008; Langmead et al., 2009). Peak calling was performed with both broad- and narrow-peak functions using a bandwidth of 150, tag size of 50, and q-value cut-off of 0.05. The RepEnrich pipeline was used to determine H3K9me3 levels over TEs. This pipeline generates fractional counts for each element. Fractional counts are generated by quantifying reads that map exclusively to a single repetitive subfamily in addition to counting reads that map to multiple subfamilies using a fractional value (1/N), with N defined as the number of repetitive element subfamilies to which that read maps, as previously described (Criscione et al., 2014). The fractional counts generated by RepEnrich for each sgRNA were normalized to fractional counts from the input sample to generate the relative levels shown in the heat maps in Fig. 3 (relative value = SETDB1 sgRNA 6/9 or sgRNA NTC H3K9me3 fractional counts/input fractional counts).
qRT-PCR assays: ChIP-qPCR assay and Taqman assays
The ChIP-PCR primer sequences were L1 5′UTR forward, 5′-ACGGAATCTCGCTGATTGCTA-3′; and reverse, 5′-AAGCAAGCCTGGGCAATG-3′; HERVH-pregag forward, 5′-TTGCTCACACAAAGCCTGTT-3′; and reverse, 5′-GGGATTGATCTCCCAAGG-3′; HERVK-Rev forward, 5′-AGTTGCCATCCACCAAGAAG-3′; and reverse, 5′-CGATGGTTGCTGTCTCTTCA-3′; and ERV3-exon forward, 5′-GCAAGTTAACTCTCCTACTGGG-3′; and reverse, 5′-TTCACACTAACCGCCTCTTC-3′. A starting input of 10% was used for qRT-PCR analysis. PCR was quantified via SYBR-green (bimake.com). Data were analyzed using the percent input method (100 × 2adjusted input – Ct (IP)). Input was adjusted by taking Ct input − 3.32. For Taqman qRT-PCR experiments, total RNA was isolated with RNeasy micro kits (Qiagen), including an on-column DNase digestion step. Reverse transcription was performed with 250–1,000 ng of total RNA (N8080234; Applied Biosystems). All Taqman qPCRs were run with 5 µl cDNA (diluted 1:5 first), 5 µl of 2× Taqman Universal master mix (4304437; Applied Biosystems), and 0.5 µl probe per reaction. Taqman gene expression probes were obtained from ThermoFisher, ACTB Hs00357333_g1, ERV3-1 Hs04184598_s1, GAPDH Hs02758991_g1, IFN-β Hs01077958_s1, IFIT1 Hs03027069_s1, IFIT2 Hs01922738_s1, IFIT3 Hs01922752_s1, MDA5 (IFIH1) Hs00223420_m1, MAVS Hs00920075_m1, OAS3 Hs00196324_m1, RIG-I (DDX58) Hs01061436_m1, RPL36 Hs03006033_g1, and SETDB1 Hs01048361_m1.
J2 antibody staining
THP-1–Cas9 cells nucleofected with CD81- or SETDB1-specific gRNAs were attached to slides using a cytospin centrifuge at 800 rpm for 5 min after 5 d of treatment. Cells were fixed with 3% paraformaldehyde in PBS for 10 min and permeabilized in 0.5% Triton X-100 in PBS for 2 min. Slides were treated with 3% hydrogen peroxide solution to quench endogenous peroxidase activity for 60 min followed by blocking for 60 min with blocking buffer (B40913; ThermoFisher). Slides were incubated with the J2 primary antibody (10010200; Scicons) at 1:200 in PBS overnight at 4°C. The next day, the slides were processed with the Tyramide SuperBoost kit (B40913; ThermoFisher) according to the manufacturer’s protocol. Images were acquired using a confocal microscope (SP8; Leica) equipped with a 100× oil immersion lens (HC PL APO CS2 100×/1.4 oil), a pinhole of 0.5, and a Z step size of 0.19 µm, displayed as maximum intensity projections. DAPI was excited with a 405-nm laser diode at 2% and detected from 410 to 483 nm, and the tyramide Alexa Fluor 555–amplified J2 staining was excited using a white light laser at 50% output at 553 nm at 2% and acquired from 558 to 633 nm using HyD detectors in standard mode with 5× line averaging.
Histone purification and mass spectrometry
Core histones (H2A, H2B, H3, and H4) were purified from frozen cell pellets by acid extraction, ion exchange, and perchloric acid precipitation using a commercial kit (Histone Purification Mini kit; 40026; Active Motif) according to the manufacturer’s instructions. The purified histones were resuspended in deionized distilled water to a final concentration of 0.5–1.0/µl and stored at −80°C until use. 2 µg aliquots of endogenous histones was mixed with equal amounts of purified stable isotope–labeled core histones, purified from PC9 cells, and grown in media supplemented with 13C6,15N2 lysine and 13C6,15N4 arginine that served as internal standards. Samples were prepared for mass spectrometry by propionylation of lysines, digestion with trypsin, and derivatization of peptide N termini with phenyl isocyanate as previously described (Maile et al., 2015). Histone peptides were quantified by capillary reverse phase liquid chromatography nanoelectrospray tandem mass spectrometry on a hybrid quadrupole-orbitrap mass spectrometer (Q-Exactive HF; ThermoFisher) in a parallel reaction monitoring experiment. Quantitative data on 40 distinct posttranslational modifications of histones H3 and H4 in 78 combinations were extracted via Skyline software and normalized via internal standards as previously described (Vinogradova et al., 2016).
Viral sensor sgRNA and FACS assays
Cells were transduced with viral sensor sgRNAs (sgRNAs are in Table 2). 2 d after transduction, cells were subjected to a second transduction with SETDB1-specific sgRNAs or an NTC control sgRNA. Cells were selected with 1 µg/ml puromycin (viral sensor sgRNAs) and 5 µg/ml blasticidin (SETDB1 or NTC sgRNAs). Cells were harvested for FACS analysis 14 d after the second transduction. In brief, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and then washed with 1 ml PBS. Cells were permeabilized with PBS + Tween 0.1% for 15 min. Primary antibodies were used at a 1:100 dilution (MAVS; ab31334; Abcam; MDA5; ab69983; Abcam) in 150 µl FACS buffer (PBS with 0.5% BSA and 2 mM EDTA) at 4°C for 30 min. The cells were washed twice with 1 ml FACS buffer. The secondary was Alexa Fluor goat anti–rabbit IgG (H + L; A11008; ThermoFisher) and was used at a 1:500 dilution in 150 µl FACS buffer. Cells were washed twice with 1 ml FACS buffer and analyzed on a LSRII analyzer (BD).
Table 2. sgRNA sequences used for lentiviral vectors and Alt-R crRNA generation.
| Gene | sgRNA sequences |
|---|---|
| SETDB1 sgRNA 6 | 5′-TGGAAGTCCCGAGTTGAGG-3′ |
| SETDB1 sgRNA 9 | 5′-TGGTGGAAGTCCCGAGTTG-3′ |
| SETDB1 sgRNA 10 | 5′-CCACTCTTGAGCAGTACCA-3′ |
| NTC (Luciferase) | 5′-GCATGCGAGAATCTCACGC-3′ |
| PLK1 | 5′-GTTGTCCTCGAAAAAGCCG-3′ |
| MAVS sgRNA 1 | 5′-GGATTCCTTGGGATGGCTC-3′ |
| MAVS sgRNA 2 | 5′-CTGGAGTCCTCCTCTGACC-3′ |
| MAVS sgRNA 3 | 5′-CAGCCTCACACCATCCCGT-3′ |
| MAVS sgRNA 4 | 5′-GAGACACAGGCCCACGGGA-3′ |
| MAVS sgRNA 5 | 5′-CAGGGTCAGTTGTATCTAC-3′ |
| MDA5 sgRNA 1 | 5′-TAGCGGAAATTCTCGTCTG-3′ |
| MDA5 sgRNA 2 | 5′-GCCTGCATGTTCCCGGAGG-3′ |
| MDA5 sgRNA 3 | 5′-ACTGCCTGCATGTTCCCGG-3′ |
| MDA5 sgRNA 4 | 5′-CATGAGCGTTCTCAAACGA-3′ |
| MDA5 sgRNA 5 | 5′-AGAAATGGTATCGTGTTAT-3′ |
| RIG-I sgRNA 1 | 5′-AGCCTTCCAGGATTATATC-3′ |
| RIG-I sgRNA 2 | 5′-GATTATATCCGGAAGACCC-3′ |
| RIG-I sgRNA 3 | 5′-AACAACAAGGGCCCAATGG-3′ |
| RIG-I sgRNA 4 | 5′-GATCAGAAATGATATCGGT-3′ |
| RIG-I sgRNA 5 | 5′-ATTTCTGCTGTTCATACAC-3′ |
| CD81 sgRNA 2 (control) | 5′-GTTGGCTTCCTGGGCTGCTA-3′ |
Western blotting
To detect SETDB1 protein, 5–10 × 106 THP-1–Cas9 cells treated with the respective sgRNAs were lysed in 150 µl radio-immunoprecipitation assay buffer plus protease inhibitors (4693159001; Roche). Samples were placed on ice and allowed to incubate for 30 min (with vortexing briefly every 5 min). Samples were then sonicated with a Bioruptor for 11 cycles of 30 s on and 30 s off. Samples were spun down at 13,000 rpm for 10 min at 4°C. 50–100 µg of protein was prepared in 1× SDS loading buffer (ThermoFisher) and 1× reducing agent (ThermoFisher), heated to 70°C for 10 min, and loaded into 4–12% Bis-Tris gels (ThermoFisher). MOPS buffer was chilled to 4°C, and gels were run at 75 V for ∼3 h. Gels were transferred to polyvinylidine fluoride membranes using an iblot2 (ThermoFisher) for 7 min at 23 V. Membranes were blocked with 5% nonfat milk diluted in TBS with 0.1% Tween 20 (TBS-T) for 1 h and were rinsed briefly three times with TBS-T and incubated with primary antibodies overnight in TBS-T with 5% nonfat milk. Blots were washed three times for 15 s and then three times for 10 min in PBS-T and then incubated with secondary antibodies in TBS-T with 5% nonfat milk. Blots were washed as previously described, and detection was performed with Supersignal Femto (Pierce). The antibodies were 1:500 rabbit anti-SETDB1 (PA5-29101; ThermoFisher), 1:2,000 mouse antihistone H3 (3638; CST), 1:10,000 antimouse HRP (1721011; BIO-RAD), and 1:10,000 antirabbit HRP (1721019; BIO-RAD). To detect MDA5 and RIG-I, 1 × 106 THP-1 cells were lysed in 150 µl radio-immunoprecipitation assay buffer plus protease inhibitors. Samples were placed on ice and allowed to incubate for 30 min (with vortexing briefly every 5 min). Samples were spun down at 13,000 rpm for 10 min at 4°C. 10–20 µg of protein was loaded into 4–12% Bis-Tris gels (ThermoFisher) and run at 200 V in MOPS buffer for 50 min. Gels were transferred to polyvinylidine fluoride membranes using an iblot2 with program P0. Samples were blocked, washed, and developed as described earlier in this section. Antibodies were 1:1,000 rabbit anti-MDA5 (ALX-210-935; Enzo), 1:1,000 rabbit anti–RIG-I (ag-20b-0009; Adipogen), and 1:1,000 mouse anti–β-actin (47778; Santa Cruz Biotechnology).
RNase protection assay
RNase protection assays were performed as described in Roulois et al. (2015) with modifications. Total RNA from THP-1–Cas9 cells transduced with SETDB1 sgRNA 6 or 9 or NTC control sgRNA was purified with TRIzol (ThermoFisher) according to the manufacturer’s protocol. 3 µg of total RNA was digested with RNase A/T1 mix (EN0551; ThermoFisher) with 3 µl of enzymes in 150 µl RNase protection buffer (10 mM Tris-HCl, pH7.5, 300 mM NaCl, and 5 mM EDTA). Control samples were mock digested without the addition of RNases. RNA was digested for 30 min at 37°C, followed by the addition of 5 µg glycogen and purification with TRIzol. Reverse transcription was performed as described in the qRT-PCR assays section, and 5 µl cDNA (prediluted to 1:5 in water) was used for SYBR-green qPCRs. Thermocycling conditions were 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and then a melt curve of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s, according to the manufacturer’s protocol. dsRNA enrichment was determined by normalizing to β-actin. Primers were designed based on Dfam consensus sequences for each element and focused on predicted regions of dsRNAs inferred from our stranded RNA-seq data. Primer sequences are listed in Table 1.
Table 1. Primer sequences for TE qPCRs.
| Element | Forward primer | Reverse primer |
|---|---|---|
| HERV3 | 5′-CCTGCTCTAGTCACCCTGGA-3′ | 5′-CTTCCCTGATGATTACTCAAGC-3′ |
| L1P1_5′end | 5′-TGCCCTAAAAGAGCTCCTGA-3′ | 5′-TGTTTTTGCAGTGGCTGGTA-3′ |
| HUERSP3b | 5′-TCAAATTGTTTCTTCTCGCCTA-3′ | 5′-GGTTCAGTTTGCAGCACCAT-3′ |
| L1PA10_5′end | 5′-TGGAACCAAGTTGGAAAACA-3′ | 5′-TTGGCCTGTCTTGCTAGGTT-3′ |
| L1PA12_5′end | 5′-TCCATGAAAACTTCCCCAAC-3′ | 5′-TTCTCTGTATTTCCTGAATTTGACTG-3′ |
| L1Pba_5′end | 5′-CTTTGCAGACACTCCCCAGT-3′ | 5′-GGTCTAGCCACCCAGCAG-3′ |
| L1Pba1_5′end | 5′-TGGGTGAGGCCTGTGACT-3′ | 5′-TGCTGAGTCATGCAGGTTGT-3′ |
| MSTB2 | 5′-CTGCAGAACCATGAGCTAAAT-3′ | 5′-AACAGAATACCTGAGACTGGGTAA-3′ |
| LTR26 | 5′-ATGCAGTTTCCACATCCTGA-3′ | 5′-ATTGGGGTCATTGATTGGTC-3′ |
Methocult assay
THP-1–Cas9 cells were transduced with sgRNAs in 96-well plates as described in the Arrayed sgRNA transduction section. 2 d after transduction, cells were selected with 1 µg/ml puromycin. 5 d after transduction and before appreciable cell death, 1,000 cells were seeded into 1.1 ml methocult H4535 in 35-mm plates. Colonies were counted using a colony counter (GelCount; Oxford Optronix) after 28 d of growth.
Indel analysis
To identify the mutations generated with SETDB1-, RIG-I–, or MDA5-specific sgRNAs, cells were transduced or nucleofected with sgRNAs or gRNAs, respectively. Guide sequences are described in Table 2. After 5–7 d of treatment, genomic DNA was isolated with a DNeasy blood and tissue kit. Targeted loci were amplified via the following primer sets: SETDB1, forward, 5′-TCTCCTGGCCAAGTCTTTTC-3′, and reverse, 5′-TCAACAATGACCTGCAGAGG-3′; RIG-I, sgRNA1 targeted loci, forward, 5′-CTCGGAAAATCCCTGCTTTC-3′, and reverse, 5′ RIG-I; sgRNA3 targeted loci, forward, 5′-AGTGGCTTGGTGAAGAATGG-3′, and reverse, 5′-TTCCCCAGCTTTGAACCTAATGCAGATTCTTTTGTTGGATG-3′. MDA5 sgRNAs 1 and 4 targeted loci were amplified with the same primers: forward, 5′-CGTCATTGTCAGGCACAGAG-3′, and reverse, 5′-ACAGTTCCTCCTCCATGCAC-3′. PCR thermocycling used 2× Taq Mastermix (NEB) at 98°C for 30 s, 40 cycles at 98°C for 10 s, 54°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min and 4°C. Resultant PCR products were gel-isolated, TOPO cloned into pCR2.1 (ThermoFisher), and sequenced.
Nucleofection assay
To generate THP-1–Cas9 RIG-I, MDA5, and SETDB1 KO cells, nucleofections were performed with Alt-R gRNAs from IDT with the P4 Primary Cell 4D-Nucleofector X kit S (V4XP-4032; Lonza). 10 nM gene-specific CRISPR RNAs (crRNAs) and tracrRNAs (1072534; IDT) were reconstituted to 100 µM in IDT duplexing buffer. tracrRNA and crRNA were mixed 1:1 and hybridized (95°C for 5 min followed by cooling to 4°C). For each nucleofection, 2 × 105 THP-1–Cas9 cells were spun down and resuspended in 20 µl P4 primary cell buffer with supplement added. 4 µl of hybridized tracrRNA/crRNAs was added to each cuvette, and cells were layered on top, mixed, and allowed to incubate for 5 min at room temperature. Cells were nucleofected with protocol CM-138 using the Amaxa 4D system (Lonza). After nucleofection, 100 µl of prewarmed medium (RPMI + 10% FBS) was added to the cuvette and immediately pipetted into 24-well plate containing 1 ml of medium. For phenotypic assays, cells were allowed to incubate for 5 d before dilution to generate single-cell clones. For single-cell clone generation, cells were sorted with a FACSAria Fusion cell sorter 1 d after nucleofection crRNA sequences (Table 2).
Data deposition
The sequences reported in this paper have been deposited at the Gene Expression Omnibus as a SuperSeries and can be found under accession no. GSE103411.
Online supplemental material
Fig. S1 depicts a cartoon overview of the screen, verification of SETDB1 protein knockdown with sgRNAs, colony growth assays of THP-1 cells treated with SETDB1 sgRNAs, and a correlation plot showing the RNA-seq expression changes between two different SETDB1 sgRNAs used after 4 d of treatment. Fig. S2 depicts shRNA validation of SETDB1 KO phenotypes, including verification of protein knockdown with SETDB1 shRNAs, qRT-PCR validation of mRNA knockdown, viability and apoptosis data after SETDB1 knockdown, qRT-PCR analysis of ISG induction after SETDB1 knockdown, and additional time point analysis demonstrating that ISGs and an ERV are induced after SETDB1 KO. Fig. S3 depicts the verification of SETDB1 disruption mediated by CRISPR/Cas9 via an IGV view of the RNA-seq data and depiction of TE expression as a percentage of each strand expressed to assess bidirectional transcription of TEs after treatment with SETDB1 sgRNAs. Fig. S4 depicts ChIP-seq data from SETDB1 mutant cells across select IFIT and zinc-finger genes. Fig. S5 depicts the assessment of cell death after SETDB1 mutation and its dependence on viral sensing genes.
Supplementary Material
Acknowledgments
We wish to acknowledge Nobuhiko Kayagaki and Soren Warming for critical reading of the manuscript. We would also like to thank Amy Heidersbach, Clark Ho, Jingli Zhang, Gulfem Guler, Charles Tindell, Honglin Chen, Yuxin Liang, and Mark McCleland for their helpful input and technical assistance.
All authors were employed by Genentech, Inc., a member of the Roche group, during the preparation of the manuscript. Thus, the authors declare competing financial interests.
Author contributions: T.L. Cuellar and B. Haley designed experiments and wrote the manuscript with input from all authors. T.L. Cuellar, A.-M. Herzner, X. Zhang, Y. Goyal, J.M. Doerr, V. Janakiraman, and S. Chaudhuri performed the experiments. T.L. Cuellar, C. Watanabe, and B. Haley developed the CRISPR sgRNA library, and T.L. Cuellar developed the screening methods. T.L. Cuellar, A.-M. Herzner, X. Zhang, Y. Goyal, J.M. Doerr, B.A Friedman, and S. Durnick analyzed and interpreted data. M. Classon contributed intellectual expertise on chromatin biology. V. Janakiraman, S. Chaudhuri, J. Stinson, and Z. Modrusan provided expertise on next-generation sequencing, RNA-seq, and ChIP-seq. D. Arnott and T.K. Cheung performed and analyzed the histone mass spectrometry experiments.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- ChIP
- chromatin immunoprecipitation
- crRNA
- CRISPR RNA
- dsRNA
- double-stranded RNA
- emGFP
- emerald GFP
- ERV
- endogenous retrovirus
- H3K9me3
- H3K9 trimethylation
- ISG
- IFN-stimulated gene
- KO
- knockout
- LINE
- long interspersed nuclear element
- LOF
- loss-of-function
- LTR
- long terminal repeat
- NTC
- nontargeting control
- qRT-PCR
- quantitative real-time PCR
- sgRNA
- single-guide RNA
- TE
- transposable element
References
- Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B., et al. . 2016. Genomic copy number dictates a gene-independent cell Response to CRISPR/Cas9 targeting. Cancer Discov. 6:914–929. 10.1158/2159-8290.CD-16-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind’Amour J., Liu S., Hudson M., Chen C., Karimi M.M., and Lorincz M.C.. 2015. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6:6033. [DOI] [PubMed] [Google Scholar]
- Cherkasova A.P., and Selyatitskaya V.G.. 2013. Corticosteroid hormones and angiotensin-converting enzyme in the dynamics of chronic granulomatous inflammation. [In Russian]. Patol. Fiziol. Eksp. Ter. 2:26–31. [PubMed] [Google Scholar]
- Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. . 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 162:974–986. (published erratum appears in Cell 2017. 169:361) 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Kyle K.E., Egawa T., Shinkai Y., and Oltz E.M.. 2015. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. USA. 112:8367–8372. 10.1073/pnas.1422187112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione S.W., Zhang Y., Thompson W., Sedivy J.M., and Neretti N.. 2014. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics. 15:583 10.1186/1471-2164-15-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks H.A., Vafadar-Isfahani N., Dunican D.S., Lee A., Sproul D., Lund J.N., Meehan R.R., and Tufarelli C.. 2013. Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res. 41:6857–6869. 10.1093/nar/gkt438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Majer C.R., Sneeringer C.J., Song J., Johnston L.D., Scott M.P., Smith J.J., Xiao Y., et al. . 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 20:53–65. 10.1016/j.ccr.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M.A., and Kouzarides T.. 2012. Cancer epigenetics: from mechanism to therapy. Cell. 150:12–27. 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Domansky A.N., Kopantzev E.P., Snezhkov E.V., Lebedev Y.B., Leib-Mosch C., and Sverdlov E.D.. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472:191–195. 10.1016/S0014-5793(00)01460-5 [DOI] [PubMed] [Google Scholar]
- Dunn C.A., Romanish M.T., Gutierrez L.E., van de Lagemaat L.N., and Mager D.L.. 2006. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 366:335–342. 10.1016/j.gene.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Ehrlich M. 2002. DNA methylation in cancer: too much, but also too little. Oncogene. 21:5400–5413. 10.1038/sj.onc.1205651 [DOI] [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics. 1:239–259. 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin J.A., Paquola A.C., Singer T., Gallina I., Novotny M., Quayle C., Bedrosian T.A., Alves F.I., Butcher C.R., Herdy J.R., et al. . 2016. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 19:1583–1591. 10.1038/nn.4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., et al. . 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41:563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Fei Q., Shang K., Zhang J., Chuai S., Kong D., Zhou T., Fu S., Liang Y., Li C., Chen Z., et al. . 2015. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat. Commun. 6:8651 10.1038/ncomms9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y., et al. . 2013. An optimized microRNA backbone for effective single-copy RNAi. Cell Reports. 5:1704–1713. 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. . 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 18:553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler G.D., Tindell C.A., Pitti R., Wilson C., Nichols K., KaiWai Cheung T., Kim H.J., Wongchenko M., Yan Y., Haley B., et al. 2017. Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell. 32:221–237. [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Harris W.J., Huang X., Lynch J.T., Spencer G.J., Hitchin J.R., Li Y., Ciceri F., Blaser J.G., Greystoke B.F., Jordan A.M., et al. . 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 21:473–487. (published erratum appears in Cancer Cell 2016. 6:856) 10.1016/j.ccr.2012.03.014 [DOI] [PubMed] [Google Scholar]
- Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., et al. . 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 163:1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Härtlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. . 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Haubold B., and Wiehe T.. 2006. How repetitive are genomes? BMC Bioinformatics. 7:541 10.1186/1471-2105-7-541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G., Eiges R., Gaudet F., Jaenisch R., and Eden A.. 2008. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 27:404–408. 10.1038/sj.onc.1210631 [DOI] [PubMed] [Google Scholar]
- Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F., and Wheeler T.J.. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44:D81–D89. 10.1093/nar/gkv1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., and Zhang Y.. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 333:1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs F.M., Greenberg D., Nguyen N., Haeussler M., Ewing A.D., Katzman S., Paten B., Salama S.R., and Haussler D.. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 516:242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., and Doudna J.. 2013. RNA-programmed genome editing in human cells. eLife. 2:e00471 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., and Rajewsky K.. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19:489–501. 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M.M., Goyal P., Maksakova I.A., Bilenky M., Leung D., Tang J.X., Shinkai Y., Mager D.L., Jones S., Hirst M., and Lorincz M.C.. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 8:676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., and Tora L.. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 13:424 10.1186/1471-2164-13-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G., and Stoye J.P.. 2016. Immune responses to endogenous retroelements: taking the bad with the good. Nat. Rev. Immunol. 16:207–219. 10.1038/nri.2016.27 [DOI] [PubMed] [Google Scholar]
- Koide S., Oshima M., Takubo K., Yamazaki S., Nitta E., Saraya A., Aoyama K., Kato Y., Miyagi S., Nakajima-Takagi Y., et al. . 2016. Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood. 128:638–649. 10.1182/blood-2016-01-694810 [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. International Human Genome Sequencing Consortium . 2001. Initial sequencing and analysis of the human genome. Nature. 409:860–921. (published erratum appears in Nature 2001. 411:720) 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. arXiv:1303.3997 (Preprint posted March 16, 2013)
- Liu L., Kimball S., Liu H., Holowatyj A., and Yang Z.Q.. 2015. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget. 6:2466–2482. 10.18632/oncotarget.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Ohtani H., Zhou W., Ørskov A.D., Charlet J., Zhang Y.W., Shen H., Baylin S.B., Liang G., Grønbæk K., and Jones P.A.. 2016. Vitamin C increases viral mimicry induced by 5-aza-2′-deoxycytidine. Proc. Natl. Acad. Sci. USA. 113:10238–10244. 10.1073/pnas.1612262113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maile T.M., Izrael-Tomasevic A., Cheung T., Guler G.D., Tindell C., Masselot A., Liang J., Zhao F., Trojer P., Classon M., and Arnott D.. 2015. Mass spectrometric quantification of histone post-translational modifications by a hybrid chemical labeling method. Mol. Cell. Proteomics. 14:1148–1158. 10.1074/mcp.O114.046573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I.A., Goyal P., Bullwinkel J., Brown J.P., Bilenky M., Mager D.L., Singh P.B., and Lorincz M.C.. 2011. H3K9me3-binding proteins are dispensable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenetics Chromatin. 4:12 10.1186/1756-8935-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., and Shinkai Y.. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 464:927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., and Thomas P.D.. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. . 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 448:553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz D.M., Cassiani P.J., Li L., Billy E., Korn J.M., Jones M.D., Golji J., Ruddy D.A., Yu K., McAllister G., et al. . 2016. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 6:900–913. 10.1158/2159-8290.CD-16-0178 [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., and Hannon G.J.. 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 102:12135–12140. 10.1073/pnas.0505479102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenifumbo J.C., and Marra M.A.. 2013. Cancer genome-sequencing study design. Nat. Rev. Genet. 14:321–332. 10.1038/nrg3445 [DOI] [PubMed] [Google Scholar]
- Pépin G., Ferrand J., Höning K., Jayasekara W.S., Cain J.E., Behlke M.A., Gough D.J., G Williams B.R., Hornung V., and Gantier M.P.. 2016. Cre-dependent DNA recombination activates a STING-dependent innate immune response. Nucleic Acids Res. 44:5356–5364. 10.1093/nar/gkw405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C., Oakes C., Blum W., and Marcucci G.. 2008. Epigenetics in acute myeloid leukemia. Semin. Oncol. 35:378–387. 10.1053/j.seminoncol.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N., Steranka J.P., Makohon-Moore A., Moyer A., Shen P., Sharma R., Kohutek Z.A., Huang C.R., Ahn D., Mita P., et al. . 2015. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21:1060–1064. 10.1038/nm.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M., Martinez de Paz A., Simó-Riudalbas L., Sayols S., Moutinho C., Moran S., Villanueva A., Vázquez-Cedeira M., Lazo P.A., Carneiro F., et al. . 2014. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 33:2807–2813. 10.1038/onc.2013.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney M.S., Shukla S.A., Wu C.J., Getz G., and Hacohen N.. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., Han H., Liang G., Jones P.A., Pugh T.J., et al. . 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 162:961–973. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.M., Friedli M., Offner S., Verp S., Mesnard D., Marquis J., Aktas T., and Trono D.. 2013. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 140:519–529. 10.1242/dev.087585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasca D., and Huntly B.J.. 2016. Independence of epigenetic and genetic diversity in AML. Nat. Med. 22:708–709. 10.1038/nm.4136 [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., and Rauscher F.J. III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919–932. 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang E., Milazzo J.P., Wang Z., Kinney J.B., and Vakoc C.R.. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33:661–667. 10.1038/nbt.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih A.H., Abdel-Wahab O., Patel J.P., and Levine R.L.. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 12:599–612. 10.1038/nrc3343 [DOI] [PubMed] [Google Scholar]
- Sun Q.Y., Ding L.W., Xiao J.F., Chien W., Lim S.L., Hattori N., Goodglick L., Chia D., Mah V., Alavi M., et al. . 2015. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J. Pathol. 235:559–570. 10.1002/path.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wei M., Ren S.C., Chen R., Xu W.D., Wang F.B., Lu J., Shen J., Yu Y.W., Hou J.G., et al. . 2014. Histone methyltransferase SETDB1 is required for prostate cancer cell proliferation, migration and invasion. Asian J. Androl. 16:319–324. 10.4103/1008-682X.122812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchasovnikarova I.A., Timms R.T., Matheson N.J., Wals K., Antrobus R., Göttgens B., Dougan G., Dawson M.A., and Lehner P.J.. 2015. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 348:1481–1485. 10.1126/science.aaa7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., and Tada K.. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer. 26:171–176. 10.1002/ijc.2910260208 [DOI] [PubMed] [Google Scholar]
- Tzelepis K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O.M., Mupo A., Grinkevich V., Li M., Mazan M., et al. . 2016. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Reports. 17:1193–1205. 10.1016/j.celrep.2016.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova M., Gehling V.S., Gustafson A., Arora S., Tindell C.A., Wilson C., Williamson K.E., Guler G.D., Gangurde P., Manieri W., et al. . 2016. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12:531–538. 10.1038/nchembio.2085 [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., and Bestor T.H.. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116–117. 10.1038/2413 [DOI] [PubMed] [Google Scholar]
- Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., and Sabatini D.M.. 2015. Identification and characterization of essential genes in the human genome. Science. 350:1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., and Sabatini D.M.. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 168:890–903.e15. 10.1016/j.cell.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S.B., Hartmann R., and Paludan S.R.. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Reeder J., Lawrence M., Becker G., and Brauer M.J.. 2016. GMAP and GSNAP for genomic sequence alignment: Enhancements to speed, accuracy, and functionality. Methods Mol. Biol. 1418:283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- Yang N., and Kazazian H.H. Jr. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13:763–771. 10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- Young G.R., Stoye J.P., and Kassiotis G.. 2013. Are human endogenous retroviruses pathogenic? An approach to testing the hypothesis. BioEssays. 35:794–803. 10.1002/bies.201300049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., and Gasser S.M.. 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48:1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.