ABSTRACT
Understanding interactions between tumor and the host immune system holds great promise to uncover biomarkers for targeted therapies and clinical outcomes. However, systematical analysis of immune signatures in esophageal squamous cell carcinoma (ESCC) remains largely unstudied. In this study, immune signatures containing 708 immune related genes were curated from mRNA microarray data with tumor and paired normal tissues from 119 ESCC patients. Differential expression and survival analysis were performed with validations from Human Protein Atlas and an independent cohort of 110 ESCC patients by immunohistochemistry staining. We identified a total of 186 significantly dysregulated genes in ESCC, including downregulated genes SPINK5, IL1RN and upregulated genes SPP1 and PLAU, which were further confirmed in Human Protein Atlas data. Moreover, nine immune related genes (ABL1, ATF2, ATG5, C6, CD38, HMGB1, ICOSLG, IL12RB2 and PLAU) were significantly associated with patients' overall survival, among which, prognostic model was built including three independent factors ABL1, CD38 and ICOSLG. Validation by immunohistochemistry staining suggested that combination with tumor infiltrated CD4+ and CD8+ T lymphocytes would yield higher performance in distinguishing cases as high or low risk of unfavorable prognosis. In summary, we profiled the immune status in ESCC and established predictive and prognostic factors for ESCC, which could reflect immune disorders within tumor microenvironments and independently distinguish patients with a high risk of reduced survival, providing novel predictive and therapeutic targets for ESCC patients in the future.
KEYWORDS: ABL1, CD38, esophageal squamous cell carcinoma, ICOSLG, immune, prognosis, tumor-infiltrated lymphocyte
Abbreviations
- ABL1
C-abl oncogene 1, Non-receptor Tyrosine Kinase
- ATF2
Activating Transcription Factor 2
- ATG5
Autophagy Related 5
- AUC
Area Under Curve
- CD4
CD4 molecule
- C6
Complement Component 6
- CD8
CD8 molecule
- CD38
CD38 molecule
- ESCC
Esophageal Squamous Cell Carcinoma
- GO
Gene Ontology
- HMGB1
High Mobility Group Box 1
- ICOSLG
Inducible T-cell Co-stimulator Ligand
- IHC
Immunohistochemistry
- IL12RB2
Interleukin 12 Receptor, Beta 2
- IL1RN
Interleukin 1 Receptor Antagonist
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PLAU
Plasminogen Activator, Urokinase
- ROC
Receiver-operator Characteristic
- SPINK5
Serine Peptidase Inhibitor, Kazal type 5
- SPP1
Secreted Phosphoprotein 1
- TIL
Tumor Infiltrating Lymphocyte
Introduction
Esophageal carcinoma (EC) is the eighth most frequently diagnosed malignancies,1 which ranks as the sixth leading cause of cancer death worldwide.2 Over 70% of worldwide cases of EC occur in China and 95% is esophageal squamous cell carcinoma (ESCC), with an estimated 478,000 new cases and 375,000 new deaths in 2015.3,4 Despite advances in multidisciplinary treatment of ESCC, the prognosis of ESCC patients remains poor, with a 5-year survival rate ranging from 10% to 25%.3,5 Predictive and prognostic markers may benefit clinical decision making and provide novel insights into underlying mechanisms and biologic behaviors of ESCC.
Molecular profiles of tumor cells and cancer-related cells within their microenvironments represent promising candidates for predictive and prognostic biomarkers.6,10 Despite vigorous efforts have been made with major breakthroughs in high-throughput genomic technologies,11,12 translational implications suffer from inconsistent results due to heterogeneity in different cancer types, patient cohort and treatment strategies.13-15 The immune evasion, a strategy used by tumor cells to evade a host's immune response to maximize their probability to continue growing, is one of the hallmarks of human cancer.16 Immune disorders in tumor is regarded as a promoting factor during tumorigenesis and development.17,18 Immune responses stimulated by tumor antigens, which are supposed to eradicate tumor cells, are even subjugated to provide proper microenvironment for tumor growth.17 With intensive efforts made to elucidate the interactions between the tumor and the immune system,19-21 remarkable success has been achieved in cancer immunotherapy in treating advanced tumors, but only applicable to a substantial fraction of patients while others either are not suitable or failed to respond.21
Considering the poor outcome after standard treatment and few targeted therapeutics in ESCC, immunotherapy, a promising additional approach, is currently under intensive investigation. Meanwhile, several immune-related parameters, mainly tumor infiltrating lymphocytes, have been reported for predicting ESCC patient prognosis,18,22,23 further suggesting distinct immune status has profound influence on outcome of ESCC patients. Therefore, systematically investigation of the immune phenotype within the ESCC microenvironment is great needed to better understand the complex antitumor response and guide effective immunotherapies in ESCC.
Methods and materials
Patient samples
For patient cohort in microarray study, paired tumor and adjacent normal tissues from 119 ESCC patients were collected between December 2005 and December 2007; for patient cohort in immunohistochemistry (IHC) study, tumor tissues from 110 ESCC patients were collected between January 1998 and January 2003. All patients had surgically proven primary ESCC and underwent surgery at National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. The clinical and pathological information for patients in both cohorts was listed in Table 1. Samples were obtained with informed consent, and the study was approved by the medical ethics committee of National Cancer Center/Cancer Hospital.
Table 1.
Clinical and pathologic characteristics of ESCC patients in microarray and tissue array.
| Characteristics | Microarray Cohorts(N = 119) | Tissue array Cohorts(N = 110) | P value* | |
|---|---|---|---|---|
| Age | Median(range) | 59 (41–75) | 61 (28–85) | 0.439 |
| Gender | Male | 98 (82.4%) | 72 (65.5%) | 0.006 |
| Tobacco use | Yes | 80 (67.2%) | 73 (66.4%) | 0.890 |
| Alcohol use | Yes | 74 (62.2%) | 62 (56.4%) | 0.446 |
| Tumor location | Upper | 14 (11.8%) | 8 (7.3%) | 0.466 |
| Middle | 69 (58.0%) | 70 (63.6%) | ||
| Low | 36 (30.3%) | 32 (29.1%) | ||
| Tumor grade | Well | 23 (19.3%) | 8 (7.3%) | 0.011 |
| Moderately | 64 (53.8%) | 58 (52.7%) | ||
| Poorly | 32 (26.9%) | 44 (40.0%) | ||
| T stage | T1 | 8 (6.7%) | 3 (2.7%) | 0.242 |
| T2 | 20 (16.8%) | 17 (15.5%) | ||
| T3 | 62 (52.1%) | 81 (73.6%) | ||
| T4 | 29 (24.4%) | 9 (8.2%) | ||
| N stage | N0 | 54 (45.4%) | 66 (60.0%) | 0.037 |
| N1 | 65 (54.6%) | 44 (40.0%) | ||
| TNM stage | I | 6 (5.0%) | 3 (2.7%) | 0.016 |
| II | 47 (39.5%) | 66 (60.0%) | ||
| III | 66 (55.5%) | 41 (37.3%) |
Statistical analysis was performed between the microarray cohort and the tissue array cohort.
Microarray data curation
Microarray data, which was deposited in Gene Expression Omnibus (GEO) under accession number GSE53625, were processed as described previously.24 In brief, mRNA expression data in 119 paired tumor-normal samples were extracted by quantile normalization and were then log 2-scaled transformed. For genes with more than one probes, mean expression were calculated and used in the study. A total of 708 immunology-related human genes, curated from nCounter® PanCancer Immune Profiling Panel (NanoString), were then implemented as candidate genes in this studies, detailed annotations for these 708 genes were listed in Supplementary Table S1.
Bioinformatics analysis
Differentially expression analysis was tested by row t-test by genefilter package (Version 1.56.0) with P values adjusted by Benjamini-Hochberg (BH) method. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment for the dysregulated genes were analyzed by ClueGO25 (Version 2.3.2) within Cytoscape (Version 3.2). In searching for survival associated genes, rbsurv (Version 2.32.0) in R (Version 3.3.1) were used, univariate and multivariate COX regression were then applied. To compare the ability of the prognostic predictors, survivalROC package (Version 1.0.3) in R, which allows for time dependent ROC curve estimation with censored data,26 was used to generate the area under the curve (AUC) of the receiver-operator characteristic (ROC) curve for each parameters.
Immunohistochemistry (IHC) analysis
For validation of differentially expressed genes, IHC data of ESCC tissues and normal esophageal tissues were downloaded from Human Protein Atlas27 available from www.proteinatlas.org. For validation of prognostic genes, tissue arrays containing tumor tissues from 110 ESCC patients were constructed for IHC staining. Three to 5 μm thick ESCC tissues were consecutively cut, subsequently dewaxed and rehydrated through graded alcohols. Slides were immunohistochemically stained according to the manufacturer's instructions. Antibodies for identification of protein expression of ABL1 (abcam, #ab15130), CD38 (abcam, #ab108403), ICOSLG (abcam, #ab189052), CD4 (abcam, #133616) and CD8 (abcam, #ab4055) were used in this study. Quantitative evaluation were performed using Aperio pathology workstation (Aperio). The proportion of cells with positive staining was measured automatically. The results were carried out blindly to the clinical data.
Statistical analysis
Differentially expressed genes were compared with student t-test and adjusted P value less than 0.001 and fold change bigger than 2 was considered significantly dysregulated. The survival curves were compared using Kaplan-Meier method and log-rank test. Comparisons of gene expression with different clinicopathologic features were tested by Student t test for two groups, One-Way ANOVA test for more than two groups, and cohort clinicopathologic featurs were compared by χ2 tests or Wilcoxon tests. All tests were two sided, and a P value of less than 0.05 was considered as statistical significance unless stated otherwise. Data were analyzed using R (version 3.3.1).
Results
Differentially expressed immune signatures in ESCC
Normalized mRNA expression of ESCC and adjacent normal tissues were obtained from a total of 119 ESCC patients. Immune signatures containing a panel of 708 immune related genes were then extracted and classified into five groups based on GO annotations (Fig. 1 Panel A). Overview of expression of all the 708 immune related genes in the ESCC cohort was presented in Fig. 1 Panel A, with diverse expression patterns between tumor and normal tissues. Next, differentially expressed genes were tested and defined as adjusted P value less than 0.001 with fold change bigger than 2. As results, a total of 186 immune related genes, among which 99 genes were upregulated while 87 genes were downregulated, were significantly dysregulated (P<0.001, fold change>2) in ESCC tissues compared with paired normal tissues (Fig. 1 Panel B, Supplementary Table S1). The most significant upregulated and downregulated genes were SPP1 and SPINK5 (Fig. 1 Panel B), respectively. Moreover, main of these dysregulated immune related genes belonged to general immune response processes (Fig. 1 Panel C) since half of the panel were in this group (Fig. 1 Panel A) and there seemed no obvious trends of enrichment of upregulated or downregulated genes in one individual group (Fig. 1 Panel C). The detailed results of all the 708 genes were provided in Supplementary Table S1.
Figure 1.
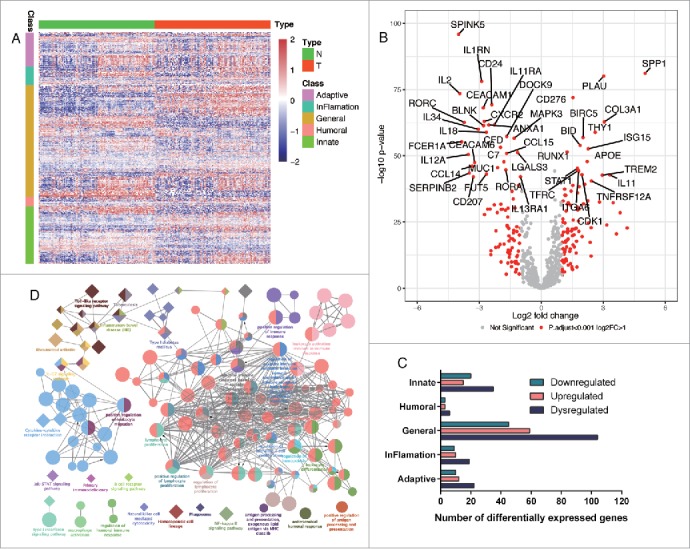
Differentially expressed immune signatures in ESCC and enriched GO and KEGG. Panel A. expression of immune signature containing 708 immune related genes in ESCC tissues (T) and paired normal tissues (N) were shown as heatmap. Generally, these genes were classified into five groups: adaptive, inflammation, general, humoral, and innate immune response genes. Panel B. Significantly downregulated and upregulated genes in ESCC were shown as volcano plot. Red dots represent significantly dysregulated genes which were defined as adjusted P value less than 0.001 and fold change bigger than 2. Panel C. Distribution of significantly dysregulated genes among the five classes. Major of the dysregulated genes were enriched in general immune response and upregulated or downregulated genes seemed to be equally divided in each class. Panel D. Significantly enriched GO and KEGG. Circles represent enriched GO while diamonds represent enriched KEGG. Objects with the same color belongs to the same group with labels in the same color by the side. Interactions between GO and or KEGG were lined up otherwise were placed alone.
Interestingly, GO enrichment analysis revealed that these dysregulated immune signatures were enriched in diverse immune processes, including T cell activation, leukocyte activation involved in immune response, cytokine-cytokine receptor interaction and positive regulation of lymphocyte proliferation (Fig. 1 Panel D) while IL-17, Jak-STAT and NF-kappa B signaling pathway were significantly enriched in the KEGG analysis (Fig. 1 Panel D). The detailed evolvement of the specific genes in each enriched immune processes or pathways were provided as Supplementary Table S2. Taken together, dysregulated immune signatures in ESCC might be a reflection of dynamic and heterogeneous immune responses in tumor microenvironment of ESCC.
Prognostic immune signatures in ESCC
Before studying the prognostic values of immune signatures, univariate survival tests were conducted to assess the relationship between clinical parameters and outcome in this ESCC cohort. As shown in Table 2, N stage (HR = 2.21, 95%CI: 1.35-3.62) was significantly associated with overall survival (P = 0.002). The results of this preliminary assessment indicated that the survival data for the ESCC cohort were informative and appropriate for use in the further analysis.
Table 2.
Univariate and multivariate COX regression analyses results in microarray data.
| Univariate analyses |
Multivariate analyses* |
|||
|---|---|---|---|---|
| Variables | HR (95%CI) | P value | HR (95%CI) | P value |
| ABL1 | 2.36 (1.47–3.81) | <0.001 | 2.44 (1.40–4.27) | 0.001 |
| ATF2 | 2.11 (1.24–3.61) | 0.006 | 1.18 (0.61–2.26) | 0.626 |
| ATG5 | 1.65 (1.03–2.62) | 0.035 | 1.06 (0.62–1.79) | 0.842 |
| C6 | 1.59 (1.00–2.53) | 0.049 | 1.34 (0.77–2.35) | 0.301 |
| CD38 | 0.50 (0.31–0.80) | 0.003 | 0.42 (0.25–0.68) | <0.001 |
| ICOSLG | 2.01 (1.15–3.50) | 0.014 | 2.01 (1.09–3.71) | 0.025 |
| IL12RB2 | 0.62 (0.39–0.98) | 0.039 | 0.65 (0.38–1.10) | 0.107 |
| HMGB1 | 1.99 (1.25–3.17) | 0.003 | 1.50 (0.87–2.60) | 0.145 |
| PLAU | 1.71 (1.08–2.74) | 0.023 | 1.57 (0.91–2.69) | 0.101 |
| N stage | 2.21 (1.35–3.62) | 0.002 | 1.51 (0.87–2.61) | 0.143 |
Factors included in the multivariate model were nine prognostic genes and N stage in the univariate analysis listed by the left.
Survival analysis were then conducted using rbsurv (version 2.32.0) in R (version 3.3.1), which is designed to select survival associated genes by robust likelihood-based survival modeling with microarray data. As results, nine genes (ABL1, ATF2, ATG5, C6, CD38, HMGB1, ICOSLG, IL12RB2 and PLAU), showed significant association with overall survival in the ESCC cohort (Table 2). Next, we separated the cohort into high expression and low expression groups (mean cut) based on expression of these nine genes, respectively. Consistently, Kaplan-Meier plots revealed that all these nine genes were significantly associated with patients' outcome (Fig. 2 Panel A-I). Among these nine survival associated genes, ABL1, ATF2, ATG5, C6, HMGB1, ICOSLG and PLAU were adversely prognostic genes while CD38 and IL12RB2 were favorably prognostic genes in ESCC cohort (Fig. 2 Panel A-I).
Figure 2.
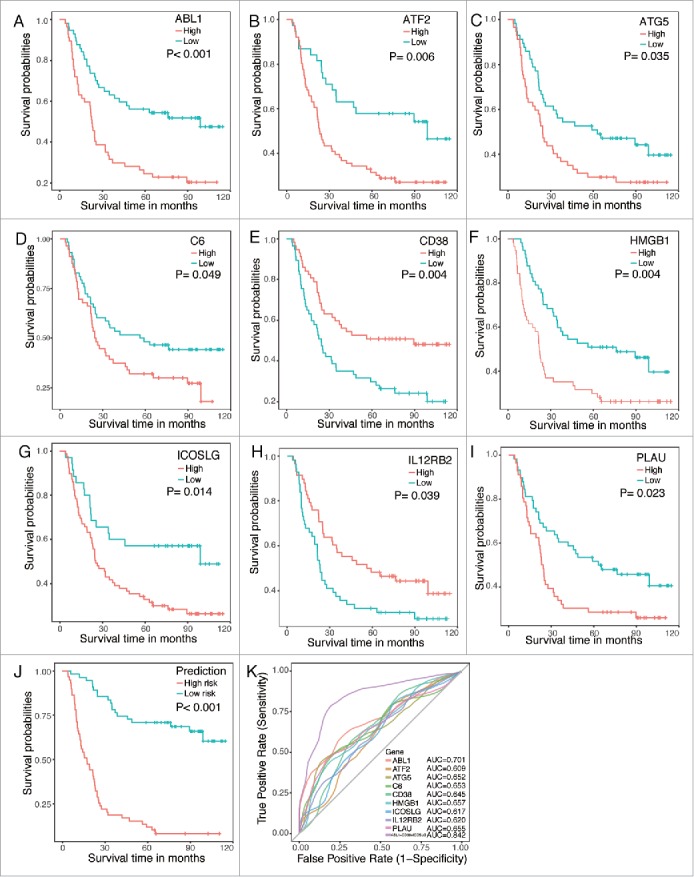
Kaplan-Meier plots and ROC curves of the survival associated genes in the microarray data. Panel A to I. Kaplan-Meier plots of the nine survival associated genes. Patients were divided into high expression (red line) and low expression (blue line) based on their gene expression by mean cut. Panel J. Kaplan-Meier plot of the prognostic predictor built with three independent factors ABL1, CD38 and ICOSLG. Patients were divided into high risk (red line) and low risk (blue line) by the prediction of the predictor. Panel K. ROC curves of each parameters with AUC scores.
To move any genes that might not be independent factors for ESCC patients, multivariate Cox regression with prognostic model construction was applied to these nine candidate immune related genes together with N stage. As shown in Table 2, ABL1, CD38 and ICOSLG were potential independent factors in the regression. Therefore, we built the final prognostic predictors with these three genes and ROC curves were also applied to compare the efficiency of these predictive model and genes (Fig. 2 Panel J). The AUC of the ROC for this predictor model was 0.842, which was much higher than other individual genes (Fig. 2 Panel K), indicating that the final predictor indeed showed much better performance in distinguishing good or poor survival in ESCC patients (Fig. 2 Panel K).
IHC validation of differentially expressed immune signatures
To further validate differentially expressed immune related genes generated by microarray data analysis, we chose the top2 most significantly downregulated and upregulated genes, which were SPINK5, IL1RN and SPP1, PLAU, respectively, as candidates. Representative images of IHC staining of ESCC tissues and normal esophageal tissues were obtained from Human Protein Atlas and shown in Fig. 3. The fraction of samples with antibody staining/protein expression level high, medium, low, or not detected were provided by the blue-scale color-coding. For significantly downregulated genes SPINK5 and IL1RN, 6 out of 9 samples (66.7%) showed high expression of SPINK5 and 3 out of 9 samples (33.3%) showed medium expression of SPINK5 in the normal esophageal tissues while majority of ESCC tissues (9/11, 81.8%) showed undetectable expression of SPINK5 (Fig. 3 Panel A); Similarly, all of the 4 normal esophageal tissues showed high expression of IL1RN while over half of the ESCC tissues showed undetectable or low expression of IL1RN (Not detectable: 3/7, 42.6%; Low: 1/7, 14.3%) (Fig. 3 Panel B). For significantly upregulated genes SPP1 and PLAU, 3 out of 7 samples (42.6%) showed medium expression of SPP1 while expression of SPP1 was low in all of the 3 normal esophageal samples (Fig. 3 Panel C); Consistently, all of the 3 ESCC samples show low expression of PLAU while both of the normal samples was not detectable (Fig. 3 Panel D). Taken together, although statistical analysis suffered from small cohorts, obvious trends could be seen that IHC staining results were largely consistent with microarray data analysis.
Figure 3.
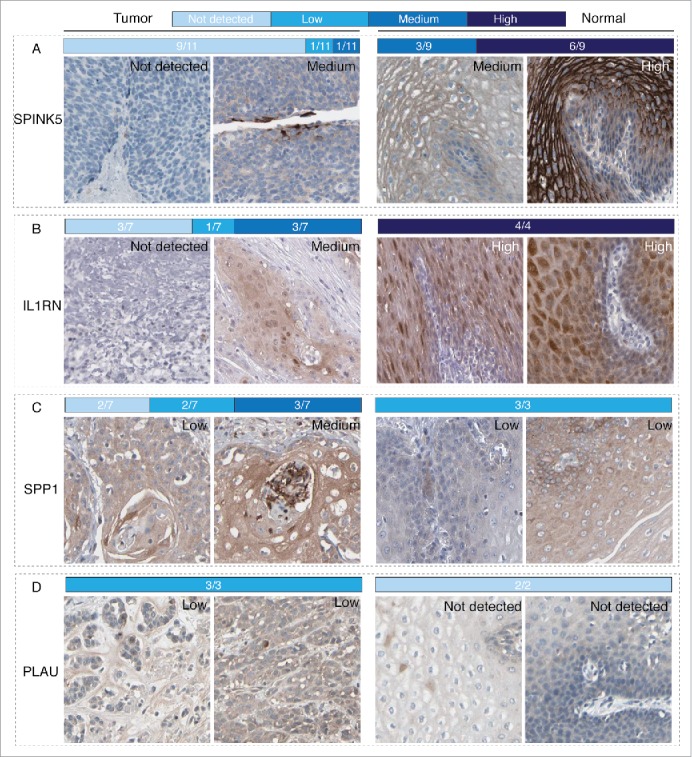
IHC validations of the top2 downregulated and upregulated genes in ESCC. Representative images for expression of each genes in ESCC tissues and normal esophageal tissues were shown with the fraction of samples with antibody staining/protein expression level high, medium, low, or not detected were provided by the blue-scale color-coding. Panel A and B. Expression of SPINK5 and IL1RN, which were top2 downregulated genes in the microarray data, in ESCC tissues were obviously lower than that in normal esophageal tissues. Panel C and D. Expression of SPP1 and PLAU, which were top2 upregulated genes in the microarray data, in ESCC tissues were obviously higher than that in normal esophageal tissues.
IHC validation of independent prognostic factors
In the validation of survival associated immune signatures in ESCC based on microarray data, independent prognostic factors ABL1, CD38 and ICOSLG were chosen as candidates. Besides, we also detected CD4+ and CD8+ T cells in ESCC tissues since previous studies indicate that tumor-infiltrating immune cells could predict the outcome of ESCC patients.18,28 A total 110 ESCC patients were enrolled as independent validation and patients were divided into two groups based on ratios of the positive regions in the ESCC slides (Fig. 4). Consistently, ABL1, CD38 and ICOSLG were significantly associated with patient survival (Fig. 5 Panel A-C). Patients with high ABL1 and ICOSLG expression had significantly worse prognosis while high expression of CD38 predicted favorable prognosis in ESCC patients (Fig. 5 Panel A-C). Similarly, we also built the predictor based on these three genes and the model performed well with much more higher AUC of ROC than individual genes (Fig. 5 Panel F and H). Moreover, tumor infiltration lymphocytes were also significantly associated with patients overall survival. As shown in Fig. 5 Panel E and F, CD4+ and CD8+ T lymphocytes were favorable prognostic factors in ESCC. Then, we tested if combination of tumor infiltration lymphocytes with the three-gene predictor would yield better performance (Fig. 5 Panel G). The AUC of ROC for this combination was 0.740 which was bigger than 0.724 for the three-gene predictor itself (Fig. 5 Panel H). Taken together, our results demonstrated that combination of the three-gene predictor and tumor infiltration lymphocytes was more powerful in distinguishing good or poor outcome of ESCC patients.
Figure 4.
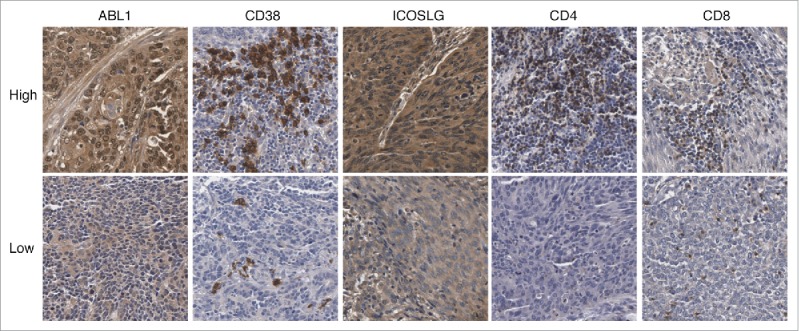
Representative images of expression of ABL1, CD38, ICOSLG, CD4 and CD8 in ESCC tissues by IHC staining.
Figure 5.
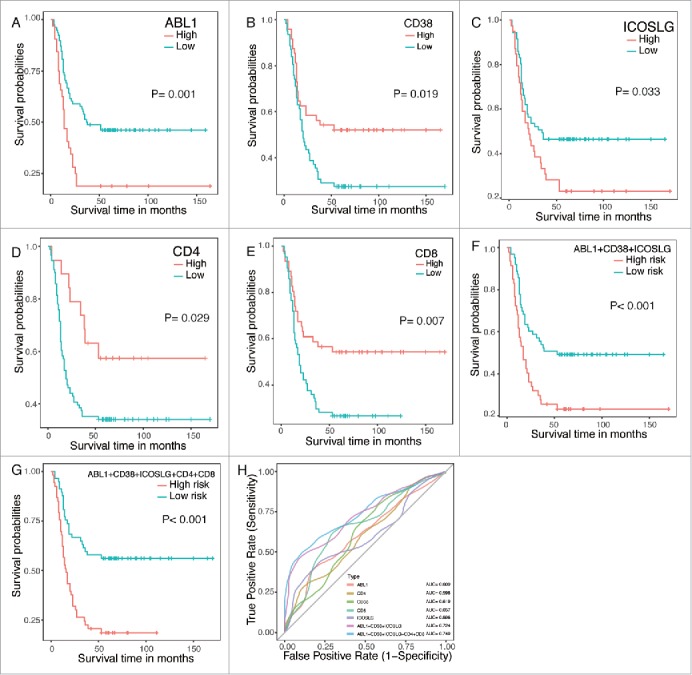
IHC validations of prognostic factors in ESCC. The independent prognostic factors ABL1, CD38 and ICOSLG were chosen as candidates in the validations. CD4+ and CD8+ T lymphocytes were also detected as controls. Panel A to E. Kaplan-Meier plots of each gene with red line represent high expression group while blue lines represent low expression group. Panel F and G. Kaplan-Meier plots of predictors built with the specific genes. Panel H. ROC curves of each parameters with AUC scores.
Interactions between the important prognostic factors and clinicopathologic parameters
We also analyzed the potential relationship between the three independent factors (ABL1, CD38 and ICOSLG) and the tumor characteristics, including comparison with normal esophageal tissues, N stage, TNM stage, tumor location, tumor grade and maximal tumor diameter. As shown in Fig. 6 Panel A-C, Expression of ABL1 and CD38 were significantly upregulated while expression of ICOSLG was significantly downregulated in ESCC tissues compared to normal tissues. Moreover, CD38 expression of ESCC tissues from different location was significantly different while ICOSLG expression was significantly associated with tumor differentiation grade. In addition, obvious association between ABL1 expression and tumor location or differentiation could also been seen, however, the difference was not statistical significant. Moreover, further correlation analysis also indicated weak links among these three genes (Fig. 6 Panel D). However, in the validation cohort which was detected by IHC staining method, significant correlation between ABL1 and ICOSLG was detected with Coef of 0.46 (P < 0.05) by correlation test (Fig. 6 Panel E). Different detection methods might account for the inconsistent results since that IHC was more suitable for grading and accurate quantification of IHC was still challenging which could affect the correlation tests. When comes to CD4 and CD8 expression in the validation cohort, correlation analysis revealed that CD4 and CD8 expression were both significantly correlated with CD38 and ICOSLG expression (Fig. 6 Panel E). In fact, CD4 expression was also significantly associated with CD8 expression (Fig. 6 Panel E). Thus, in the multivariate model with CD4 and CD8, only ABL1, ICOSLG and CD8 were independent prognostic factors for ESCC patients. Interactions between these genes detected by IHC might be the main reason that putting CD4 and CD8 into the prognostic model yield limited efficiency in the validation cohort. Taken together, interactions between these important prognostic factors and clinicopathologic parameters suggested dynamic involvement of these genes in ESCC and immune responses.
Figure 6.
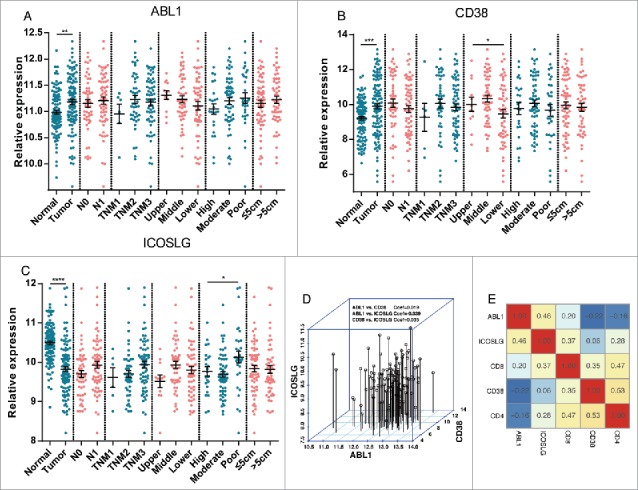
Interactions between important prognostic factors and clinicopathologic parameters. Panel A to C. Relationships between the three independent factors (ABL1, CD38 and ICOSLG) and the tumor characteristics, including comparison with normal esophageal tissues, N stage (N0 and N1), TNM stage (TNM1, TNM2 and TNM3), tumor location (Upper, Middle and Lower), tumor grade (High, Moderate, Low) and maximal tumor diameter (≤5 cm and >5 cm) were analyzed. Panel D. Correlations between ABL1, CD38 and ICOSLG expression in the microarray cohort. No significant correlation between these three genes was detected. Panel E. Correlations between ABL1, CD38, ICOSLG, CD4 and CD8 expression in the tissue array cohort. Correlation efficiencies were labeled in the matrix with scaled color indications. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Discussion
Growing evidences suggest that a comprehensive understanding of esophageal cancer requires attention to not only tumor cells but also the tumor microenvironment,29 which contains diverse cell populations that interact with cancer cells and participate in all stages of tumorigenesis. Tumor-infiltrating immune cells and immune responses within the tumor microenvironment draw a great attention to researchers and represent as promising therapeutic targets. Further study of how immune signatures relate to ESCC tumorigenesis and development will lead to development of novel and specific targeted therapeutic strategies, which offer considerable potential especially in the setting of combination therapy.29
The general role of local immune response status in ESCC progression and prognostication remains unknown. High-resolution microarray technology provides objective data for that purpose.30 In the present study, we curated immune signatures, including 708 immune related genes in five groups, from microarray data in 119 ESCC tissues and paired normal tissues. Dysregulated immune related genes were identified including significantly downregulated genes SPINK5 and IL1RN and significantly upregulated genes SPP1 and PLAU in ESCC tissues. These differentially expressed immune signatures highlighted important immune response processes including T cell activation and NF-kappa B signaling pathways, suggesting dynamic immune microenvironment and responses in ESCC which might account for tumorigenesis and development, shedding great potential influences on patients outcome. Meanwhile, survival analysis revealed three independent prognostic factors, which were ABL1, CD38 and ICOSLG, and a prediction model with high performance in distinguishing good or poor outcomes for ESCC patients. Accordingly, we performed IHC staining experiments to further validate these findings. For instance, IHC staining results from Human Protein Atlas showed obvious trends that SPINK5 and IL1RN were downregulated while SPP1 and PLAU were upregulated in ESCC tissues. Moreover, ABL1, CD38 and ICOSLG together with CD4+ and CD8+ T lymphocytes built up a more powerful survival predictor in an independent cohort by IHC staining. These results suggested that our findings might provide considerable and reliable novel insights in characterization of tumor microenvironment of ESCC.
Developing meaningful signatures to determine the immune status of a patient is appealing as they not only promise to be powerful predictive and prognostic biomarkers but, if correctly applied, also enable patient stratification for an increasing immunotherapeutic outcome. Notably, therapeutics targeting BCR-ABL fusion gene, such as Imatinib, have been used in chronic myeloid leukemia patients with dramatic benefits for years.31 Therefore, ABL1, high expression of which predicted poor outcome in ESCC patients, might serve as potential target despite it is different from BCR-ABL fusion genes. Meanwhile, daratumumab, a human IgGkappa monoclonal antibody that targets CD38, has been approved by the US. Food and Drug Administration (FDA) for the treatment of patients with multiple myeloma who have received at least three prior lines of therapy and significantly improves patient's survival.32,33 It is worth noting that high expression of CD38 predicts poor outcome in chronic lymphocytic leukemia34 while was a favorable prognostic factor in ESCC in our results. The underlying mechanisms of the controversy might be the great difference between hematologic cancer and solid tumor. For instance, low CD38 in prostate cancer is prognostic for biochemical recurrence and metastasis.35 Beyond these two target with approved therapeutics, ICOSLG and PLAU also showed great potential as targets in ESCC. ICOSLG (also known as ICOSL), is important for individual B cells to competitively participate in the germinal center reaction36 and together with ICOS, is critically involved in type 2 innate lymphoid cells function and homeostasis which can cause allergic asthma37; Correlation network identified PLAU as critical regulator for suppressor function of regulatory T cells.38 PLAU is overexpressed in many cancer cells, including breast cancer, and plays a crucial role in the metastatic process.39-41 These findings suggest important functions of ICOSLG and PLAU in immune responses, especially for PLAU which was not only significantly upregulated in ESCC tissues, but also significantly associated with overall survival in our results. Taken together, we uncovered considerable novel potential targets for ESCC immunotherapy in the future.
Tumor infiltrating lymphocytes (TILs) are increasingly involved in determining the progression and aggressiveness of tumors. TILs are composed of various lymphocytes with diverse activities. The most common lymphocytes are CD8+ and CD4+ T cells.42 Emerging evidence suggests that the amount of T lymphocyte infiltration of primary tumors consistently predicts favorable outcomes in a number of tumor types, including breast cancer,43 head and neck cancer,23 non-small cell lung cancer,44 colorectal cancer,45 and gastric cancer.46 A few initial studies examined prognostic implications of TILs in ESCC. The presence of TILs was associated with improved survival in ESCC patients.18,47 Our results also suggested that TILs were powerful prognostic factors in ESCC patient's outcome.
In summary, immune parameters, mainly tumor-infiltrating lymphocytes, have been previously investigated for patients with ESCC, while systemic analysis of immune signatures remain uncertain. For the first time, we established our predictive and prognostic factors by profiling of immune signatures in ESCC, which could be regarded as immune-related protective and risky patterns in ESCC microenvironment. These significantly dysregulated immune related genes and independent prognostic genes provides alternative targets alone or may be better in combination, attributing to their immune nature and prognostic significance. Therefore, our results may indicate a possible treatment strategy for ESCC patients by shaping the immune microenvironment, and thus improving the clinical outcome.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) under Grant 2016-I2M-1-001; National Key Basic Research Development Plan under Grant 2015CB553901; National Natural Science Foundation of China under Grant 81372219; Beijing Natural Science Foundation under Grant 7141011; and Chinese Academy of Medical Sciences Central Public-interest Scientific Institution Basal Research Fund under Grant 2016ZX310196.
References
- 1.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014; 371:2499-509. doi: 10.1056/NEJMra1314530. PMID:25539106 [DOI] [PubMed] [Google Scholar]
- 2.Gupta B, Kumar N. Worldwide incidence, mortality and time trends for cancer of the oesophagus. Eur J Cancer Prev. 2017;26:107-18. doi: 10.1097/CEJ.0000000000000249. PMID:27014938 [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32. doi: 10.3322/caac.21338. PMID:26808342 [DOI] [PubMed] [Google Scholar]
- 4.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-7. doi: 10.1136/gutjnl-2014-308124. PMID:25320104 [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. doi: 10.3322/caac.21262. PMID:25651787 [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-7. doi: 10.1038/nature01322. PMID:12490959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443-54. doi: 10.1056/NEJMoa1200690. PMID:22658127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al.. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938-45. doi: 10.1038/nm.3909. PMID:26193342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarhini AA, Lin Y, Lin HM, Vallabhaneni P, Sander C, LaFramboise W, Hamieh L. Expression profiles of immune-related genes are associated with neoadjuvant ipilimumab clinical benefit. Oncoimmunology. 2017;6:e1231291. doi: 10.1080/2162402X.2016.1231291. PMID:28344862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maccalli C, Giannarelli D, Capocefalo F, Pilla L, Fonsatti E, Di Giacomo AM, Parmiani G, Maio M. Immunological markers and clinical outcome of advanced melanoma patients receiving ipilimumab plus fotemustine in the NIBIT-M1 study. Oncoimmunology. 2016;5:e1071007. doi: 10.1126/scitranslmed.3000313. PMID:27057436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Sci Transl Med. 2010;2:14ps2. doi: 10.1126/scitranslmed.3000313. PMID:20371465 [DOI] [PubMed] [Google Scholar]
- 12.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147-57. doi: 10.1093/jnci/djk018. PMID:17227998 [DOI] [PubMed] [Google Scholar]
- 13.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: Ready for clinical use? J Natl Cancer Inst. 2010;102:464-74. doi: 10.1093/jnci/djq025. PMID:20233996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi L, Chen L, Li Y, Qin Y, Pan R, Zhao W, Gu Y, Wang H, Wang R, Chen X, et al.. Critical limitations of prognostic signatures based on risk scores summarized from gene expression levels: A case study for resected stage I non-small-cell lung cancer. Brief Bioinform. 2016;17:233-42. doi: 10.1093/bib/bbv064. PMID:26254430 [DOI] [PubMed] [Google Scholar]
- 15.Dalton WS, Friend SH. Cancer biomarkers–an invitation to the table. Science. 2006;312:1165-8. doi: 10.1126/science.1125948. PMID:16728629 [DOI] [PubMed] [Google Scholar]
- 16.Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017;27:96-108. doi: 10.1038/cr.2016.149. PMID:27981969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121:1-14. doi: 10.1111/j.1365-2567.2007.02587.x. PMID:17386080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Liu Y, Wang H, Wang H, Song Q, Sujie A, Huang J, Xu Y, Zeng H, Tan L, et al.. Tumour infiltrating lymphocytes correlate with improved survival in patients with esophageal squamous cell carcinoma. Sci Rep. 2017;7:44823. doi: 10.1038/srep44823. PMID:28322245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61. doi: 10.1016/j.cell.2014.12.033. PMID:25594174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, et al.. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. PMID:25853550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al.. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. PMID:27549193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang ZL, Lin ZR, Xiao YR, Cao X, Zhu LC, Zeng MS, Zhong Q, Wen ZS. High expression of TACC3 in esophageal squamous cell carcinoma correlates with poor prognosis. Oncotarget. 2015;6:6850-61. doi: 10.18632/oncotarget.3190. PMID:25760075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen N, Bellile E, Thomas D, McHugh J, Rozek L, Virani S, Peterson L, Carey TE, Walline H, Moyer J, et al.. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck. 2016;38:1074-84. doi: 10.1002/hed.24406. PMID:26879675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al.. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700-10. doi: 10.1136/gutjnl-2013-305806. PMID:24522499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091-3. doi: 10.1093/bioinformatics/btp101. PMID:19237447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337-44. doi: 10.1126/science.1260419. PMID:10877287 [DOI] [PubMed] [Google Scholar]
- 27.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. PMID:25613900 [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Lo AW, Wong A, Chen W, Wang Y, Lin L, Xu J. Prognostic significance of tumor-infiltrating immune cells and PD-L1 expression in esophageal squamous cell carcinoma. Oncotarget. 2017;8:30175-89. doi: 10.18632/oncotarget.15621. PMID:28404915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin EW, Karakasheva TA, Hicks PD, Bass AJ, Rustgi AK. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35:5337-49. doi: 10.1038/onc.2016.34. PMID:26923327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng W, Ren X, Zhang C, Cai J, Liu Y, Han S, Wu A. Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology. 2016;86:2226-34. doi: 10.1212/WNL.0000000000002770. PMID:27225222 [DOI] [PubMed] [Google Scholar]
- 31.Hochhaus A, Larson RA, Guilhot F, Radich JP, Branford S, Hughes TP, Baccarani M, Deininger MW, Cervantes F, Fujihara S, et al.. Long-term outcomes of Imatinib treatment for chronic myeloid leukemia. N Engl J Med. 2017;376:917-27. doi: 10.1056/NEJMoa1609324. PMID:28273028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker H. Daratumumab improves survival in multiple myeloma. Lancet Oncol. 2016;17:e480. doi: 10.1016/S1470-2045(16)30503-4. PMID:27746105 [DOI] [PubMed] [Google Scholar]
- 33.Palumbo A, Chanan-Khan A, Weisel K, Nooka AK, Masszi T, Beksac M, Spicka I, Hungria V, Munder M, Mateos MV, et al.. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375:754-66. doi: 10.1056/NEJMoa1606038. PMID:27557302 [DOI] [PubMed] [Google Scholar]
- 34.Durig J, Naschar M, Schmucker U, Renzing-Kohler K, Holter T, Huttmann A, Dührsen U. CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia. 2002;16:30-5. doi: 10.1038/sj.leu.2402339. PMID:11840260 [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Grogan TR, Hieronymus H, Hashimoto T, Mottahedeh J, Cheng D, Zhang L, Huang K, Stoyanova T, Park JW, et al.. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep. 2016;17:2596-606. doi: 10.1016/j.celrep.2016.11.010. PMID:27926864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, Xu H, Shih C, Wan Z, Ma X, Ma W, Luo D, Qi H. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517:214-8. doi: 10.1038/nature13803. PMID:25317561 [DOI] [PubMed] [Google Scholar]
- 37.Maazi H, Patel N, Sankaranarayanan I, Suzuki Y, Rigas D, Soroosh P, Freeman GJ, Sharpe AH, Akbari O. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42:538-51. doi: 10.1016/j.immuni.2015.02.007. PMID:25769613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He F, Chen H, Probst-Kepper M, Geffers R, Eifes S, Del Sol A, Schughart K, Zeng AP, Balling R. PLAU inferred from a correlation network is critical for suppressor function of regulatory T cells. Mol Syst Biol. 2012;8:624. doi: 10.1038/msb.2012.56. PMID:23169000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11:23-36. doi: 10.1038/nrm2821. PMID:20027185 [DOI] [PubMed] [Google Scholar]
- 40.Blasi F, Sidenius N. The urokinase receptor: Focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923-30. doi: 10.1016/j.febslet.2009.12.039. PMID:20036661 [DOI] [PubMed] [Google Scholar]
- 41.Moquet-Torcy G, Tolza C, Piechaczyk M, Jariel-Encontre I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014;42:11011-24. doi: 10.1093/nar/gku814. PMID:25200076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arens R, Schoenberger SP. Plasticity in programming of effector and memory CD8 T-cell formation. Immunol Rev. 2010;235:190-205. doi: 10.1111/j.0105-2896.2010.00899.x. PMID:20536564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian T, Ruan M, Yang W, Shui R. Evaluation of the prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers. Oncotarget. 2016;7:44395-405. doi: 10.18632/oncotarget.10054. PMID:27323808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brambilla E, Le Teuff G, Marguet S, Lantuejoul S, Dunant A, Graziano S, Pirker R, Douillard JY, Le Chevalier T, Filipits M, et al.. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol. 2016;34:1223-30. doi: 10.1200/JCO.2015.63.0970. PMID:26834066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al.. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960-4. doi: 10.1126/science.1129139. PMID:17008531 [DOI] [PubMed] [Google Scholar]
- 46.Liu K, Yang K, Wu B, Chen H, Chen X, Chen X, Jiang L, Ye F, He D, Lu Z, et al.. Tumor-infiltrating immune cells are associated with prognosis of gastric cancer. Medicine (Baltimore). 2015;94:e1631. doi: 10.1097/MD.0000000000001631. PMID:26426650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher K, Haensch W, Roefzaad C, Schlag PM. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer Res. 2001;61:3932-6. PMID:11358808 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


