ABSTRACT
To date, the exact impact of mast cells in tumor microenvironment is still controversial because of inconsistency in observations regarding the relationship between mast cell infiltrates and cancer development and prognosis. The discrepancies in previous studies have motivated us to examine the roles of mast cells in cancer pathology from different perspectives. Here, we investigated the impact of mast cells on transcriptomic profiles in the tissue microenvironment. Mice carrying the W-sh mutation in c-kit (KitW-sh) are deficient in mast cell production and were used to assess the influence of mast cells on gene expression. By examining the transcriptomic profile among wild-type mice, KitW-sh mice, and KitW-sh mice with mast cell engraftment, we identified a list of “mast cell–dependent genes,” which are enriched for cancer-related pathways. Utilizing whole-genome gene expression data from both mouse models and human cancer patients, we demonstrated that the expression profile of the mast cell–dependent genes differs between tumor and normal tissues from lung, breast, and colon, respectively. Mast cell infiltration is potentially increased in tumors compared with normal tissues, suggesting that mast cells might participate in tumor development. Accordingly, a prognostic molecular signature was developed based on the mast cell–dependent genes, which predicted recurrence-free survival for human patients with lung, breast, and colon cancers, respectively. Our study provides a novel transcriptomic insight into the impact of mast cells in the tumor microenvironment, though further experimental investigation is needed to validate the exact role of individual mast cell–dependent genes in different cancers.
KEYWORDS: mast cell, gene expression, lung cancer, breast cancer, colon cancer
Introduction
Mast cells are a type of white blood cell derived from bone-marrow haematopoietic progenitors. Immature mast cells circulate in blood until they migrate from vascular to peripheral tissues, where they reside close to blood vessels, nerves, and mucosal surfaces1 and mature with the help of stem-cell factor and other cytokines secreted by endothelial cells and fibroblasts.2 Mast cells are usually thought to be deeply involved in inflammatory processes. Once activated, mast cells can rapidly react to xenobiotics by either secreting or releasing mediators from their characteristic granules into the local microenvironment.3 Disorders of mast cell–activation lead to several immune diseases, such as asthma, eczema, itch, and allergic rhinitis.4
Mast cells may be important participants in regulating the tumor microenvironment. Firstly, mast cells are implicated in tumor angiogenesis.5 Angiogenesis is critical to tumor development. Enhanced vascular permeability and abnormal blood vessel development are often observed in tumors.6 Mast cells can facilitate tumor angiogenesis by secreting heparin-like molecules, angiogenesis factors (e.g., IL-8),7,8 and growth factors (e.g., VEGF).8-10 Decreased tumor angiogenesis has been observed in mast cell–deficient mice.11 Secondly, mast cells help tumor invasiveness. Several proteases released by mast cells, such as MMP-9,12,13 and the serine proteases chymase and tryptase,14 degrade components of the extracellular matrix and thus facilitate tumor invasiveness.6 Thirdly, mast cells may directly or indirectly interact with immunosuppressive and inflammatory cells in the tumor microenvironment, such as myeloid-derived suppressor cells, tumor-associated macrophages, and regulatory T-cells, to affect immunologic tolerance.1,15
Even though there is mounting evidence to indicate mast cell involvement in tumorigenesis, the exact impact of mast cells in the tumor microenvironment is still controversial.1,6 Particularly, there are several inconsistent observations regarding the relationship between mast cell infiltrates and human cancer development and prognosis. Here, we briefly review the discrepancies in previous studies regarding lung, breast, and colon cancers. For lung cancer, Imada et al. reported that the number of mast cells was positively correlated with angiogenesis and poor outcome in stage I lung adenocarcinoma,10 which was largely mirrored by the study conducted by Takanami et al.,16 However, Tomita et al. showed in a lung cancer study that the number of mast cells was significantly correlated with a favorable clinical outcome.17 The latter finding is consistent with the study by Welsh et al.,18 in which the authors claimed that mast cell–mediated invasion of tumor islets confers a survival advantage in lung cancer.18 For breast cancer, several studies have linked mast cells to a poor clinical outcome.19,20 However, a tissue microarray study containing 4,444 cases pointed out that stromal mast cell infiltration in invasive breast cancer is an independent marker of favorable prognosis,21 which is consistent with the observation of a significant increase in the number of mast cells in tumors from high hormone–receptive cancer cases compared with minimum hormone–receptive cancers.22 Similar contradictory findings also exist in colon cancer. Mast cell number was positively correlated with microvessel density and associated with a poor prognosis in colon cancer.23-25 These findings are apparently inconsistent with an earlier observation by Nielsen et al.,26 in which of 584 colon cancer patients a greater number of tryptase+ mast cells in a tumor specimen correlated significantly with better clinical outcomes.26
The discrepancies in these previous studies motivated us to look into the relationship between mast cells and cancer pathology from different perspectives. In this study, we investigated the impact of mast cells on transcriptomic profiles in the tissue microenvironment. Mast cell–deficient c-kit mutant rodents, C57BL/6-KitW-sh/W-sh (KitW-sh) mice,27 were used to assess the influence of mast cells on gene expression of tissue microenvironment. By examining the transcriptomic profile among wild-type (WT) mice, KitW-sh mice, and KitW-sh mice engrafted with mast cells derived from WT mice (KitW-sh+MC), we identified a list of “mast cell–dependent genes.” Gene ontology analysis indicates that the mast cell–dependent genes are enriched in cancer-related pathways. Utilizing whole-genome gene expression data from mouse models and human cancer patients, we demonstrated that the expression profile of the mast cell–dependent genes differentiates between tumor and normal tissues from lung, breast, and colon, respectively. Accordingly, a prognostic molecular signature was developed based on the mast cell–dependent genes. This signature successfully predicted recurrence-free survival for human patients with lung, breast, and colon cancers in a manner independent of standard clinical and pathological prognostic factors.
Results
Mast cell–dependent genes in mice
To assess the influence of mast cell on gene expression, we compared the gene expression pattern in 3 mouse groups: WT, KitW-sh, and KitW-sh+MC mice. We investigated a transcriptomic data set obtained from the Gene Expression Omnibus (GEO)28 database (GEO accession: GSE27066),29 which contains whole-genome gene expression data of WT, KitW-sh, and KitW-sh+MC mouse lung tissues. Gene expression fold changes were computed between KitW-sh and WT mice (expression in KitW-sh mice divided by that in WT mice) and between KitW-sh+MC and KitW-sh mice (expression in KitW-sh+MC mice divided by that in KitW-sh mice), respectively. A significant negative correlation (Spearman's rank correlation test: ρ = −0.413 and P < 10−10) was observed between the 2 sets of fold changes (Fig. 1A), which suggests that the deregulation caused by mast cell deficiency could be remarkably recovered by mast cell engraftment. At the specified significance level of false discovery rate <5% and fold change >1.5 (see Methods for details), the expression of 862 genes was downregulated in KitW-sh mice compared with that in WT mice but upregulated in KitW-sh+MC mice compared with that in KitW-sh mice, whereas 448 genes were upregulated in KitW-sh mice compared with that in WT mice but downregulated in KitW-sh+MC mice compared with that in KitW-sh mice (Fig. 1A). Because the expression pattern of all these deregulated genes showed a largely mast cell–dependent manner, we deemed these genes “mast cell–dependent genes.” The genes that were downregulated in mast cell–deficient mice but recovered by mast cell engraftment were deemed mast cell–positive (MC+) genes (Fig. 1B and Supplementary Table S1) whereas the genes that were upregulated in mast cell–deficient mice but restored after mast cell engraftment were considered as mast cell–negative (MC−) genes (Fig. 1B and Supplementary Table S2). We next searched the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG)30 physiologic pathways among the mast cell–dependent genes. Intriguingly, we found that the top 2 KEGG terms associated with the mast cell–dependent genes were “Pathways in cancer” and “Prostate cancer” (Fig. 1C), which support a significant role for mast cells in cancer pathology. To more precisely understand the biologic processes associated with the mast cell–dependent genes, we further performed pathway/ontology analysis for the MC+ and MC− genes separately from 3 tumor progression-related aspects: i) immunosuppression,31-33 ii) apoptosis,34 and iii) angiogenesis,35,36 in which mast cells were thought to be implicated. Firstly, we found that the KEGG terms, “T cell receptor signaling pathway” and “Natural killer cell mediated cytotoxicity,” were significantly enriched by the MC− genes but not the MC+ genes (Supplementary Fig. S1A), which suggests that increased mast cell infiltration potentially augments the suppression of T cells and natural killer cells in tumor microenvironment.31,32 Secondly, we found that the MC− genes, but not the MC+ genes, were significantly associated with the Gene Ontology (GO)37 term “Positive regulation of apoptotic process,” while the GO term “Negative regulation of apoptotic process” was significantly enriched by the MC+ genes instead of the MC− genes (Supplementary Fig. S1B), which suggests a potential anti-apoptotic role of mast cells in tumor microenvironment.34 Thirdly, we found that both the MC+ and MC− genes were significantly associated with the GO term “Angiogenesis” with a weaker significance level for the MC− genes, while the GO term “Blood vessel remodeling” was only significantly enriched by the MC+ genes but not the MC− genes (Supplementary Fig. S1C), which suggests a pro-angiogenic role of mast cells in tumor tissue.35 These observations further suggest the intrinsic feature of the mast cell–dependent genes regarding immunosuppression, apoptosis, and angiogenesis in tumor microenvironment.
Figure 1.
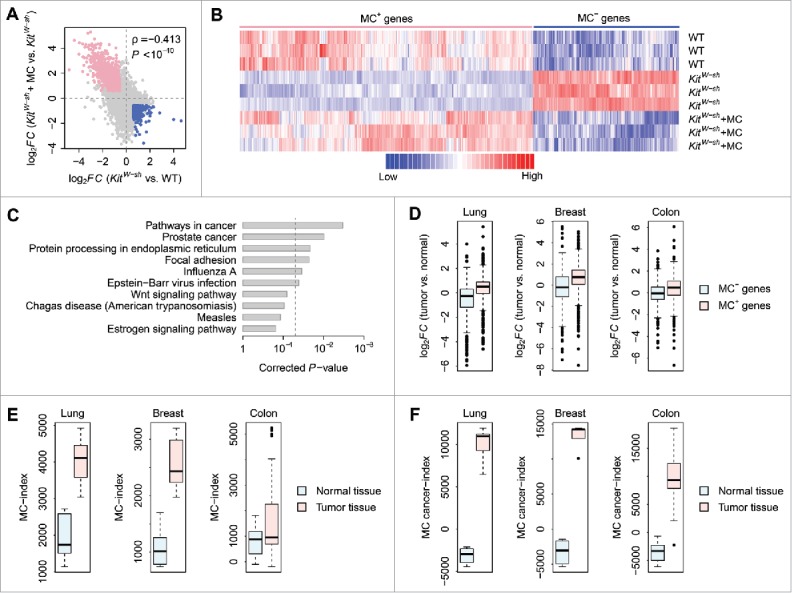
The mast cell–dependent mouse genes. (A) Correlation in log2-transformed gene expression fold change (log2FC) between KitW-sh and WT mice (X-axis) and between KitW-sh+MC and KitW-sh mice (Y-axis). Each dot stands for a gene. The log2FC between KitW-sh and WT mice is negatively correlated with the log2FC between KitW-sh+MC and KitW-sh mice. Only the genes differentially expressed between KitW-sh and WT mice and between KitW-sh+MC and KitW-sh mice in opposite direction were considered as mast cell–dependent genes. The pink dots denote the genes downregulated in mast cell–deficient mice but recovered after mast cell engraftment (MC+ genes). The blue dots represent the genes upregulated in mast cell–deficient mice but recovered after mast cell engraftment (MC− genes). (B) Gene expression heatmap of the MC+ and MC− genes. Each row in the heatmap denotes one mouse while each column denotes one gene. Red represents relatively increased gene expression whereas blue represents downregulation. (C) The top 10 KEGG pathways associated with the mast cell–dependent genes. The P-values were computed by Fisher's exact test and corrected by the Benjamini-Hochberg procedure. The vertical dash-line denotes the significance level of α = 0.05. (D) Gene expression fold change of the MC+ and MC− genes between mouse tumor and normal tissues. The expression pattern of both the MC+ and MC− genes in mouse lung, breast, and colon tumors was compared with normal lung, breast, and colon tissues, respectively. Y-axis denotes the log2FC between tumor and normal tissues. (E) Comparison of MC-index between mouse tumor and normal tissues. (F) Comparison of MC cancer-index between mouse tumor and normal tissues.
To determine to what extent the mast cell–dependent genes are involved in cancer pathology, we investigated the transcriptomic data in mouse lung (GEO accession: GSE31013),38 breast (GEO accession: GSE21444),39 and colon (GEO accession: GSE50794)40 tumors, respectively. Gene expression fold change in mouse lung, breast, and colon tumors were calculated over normal lung, breast, and colon tissues from control mice, respectively. Basically, we found that the log2-transformed gene expression fold change (log2FC) of the MC+ genes was significantly higher than that of the MC− genes (t-test: P < 10−10 for lung and breast; P = 1.2 × 10−7 for colon) (Fig. 1D). One-sample t-test indicates that the log2FC of the MC+ genes is statistically larger than zero (P < 10−10) in mouse lung, breast, and colon tumors, respectively (Fig. 1D). On the contrary, the log2FC of the MC− genes is statistically less than zero in lung and breast tumors, but not in colon tumor (one-sample t-test: P < 10−10 for lung and breast; P = 2.2 × 10−1 for colon) (Fig. 1D). Taken together, these results suggest that the MC+ genes, as compared with the MC− genes, are more likely to be overexpressed in mouse tumors, whereas the MC− genes, as compared with MC+ genes, show a higher chance to be downregulated in tumors.
We hypothesized that the mast cell–dependent tissue microenvironment could be delineated from expression deregulation profiles of the mast cell-dependent genes. Here, we developed a novel methodology to compute a mast cell index (MC-index) for individual tissue samples, based on the rank-weighted gene expression information of the MC+ and MC− genes (see Materials and Methods for details). We speculated that the MC-index could be used as a proxy of the impact of mast cells on shaping tissue microenvironment. Fig. 1E provides a comparison of MC-index between tumor and normal tissues from mouse lung, breast, and colon, respectively. The MC-index of tumor tissues was significantly higher than that of normal controls (t-test: P = 2.1 × 10−4 for lung; P = 1.2 × 10−3 for breast; P = 2.4 × 10−3 for colon), which suggests an active role for mast cells in tumor development. To more precisely assess the impact of mast cells on cancer pathology and tumor microenvironment, we made some modifications to the algorithm for computing MC-index: at the specified significance level of false discovery rate <5% and fold change >1.5, only the MC+ genes commonly upregulated and the MC− genes commonly downregulated in lung, breast, and colon tumors were considered. We deemed these genes “mast cell–dependent cancer genes.” A mast-cell cancer index (MC cancer-index) was calculated for individual tissue samples using the rank-weighted gene expression data of the mast cell-dependent cancer genes (see Materials and Methods for details). Fig. 1F indicates that the MC cancer-index of tumor tissues was significantly higher than that of normal controls (t-test: P = 1.6 × 10−6 for lung; P = 5.5 × 10−7 for breast; P < 10−10 for colon). In comparison with MC-index, the difference in MC cancer-index between tumor and normal tissues was even larger (Fig. 1F).
MC- and MC cancer-indices of human cancer patients
To assess the depth of involvement of mast cells in human cancers, we mapped the mast cell-dependent mouse genes to their distinct human orthologs. Next, we investigated the expression pattern of the mast cell-dependent human genes in 6 independent cancer cohorts: 2 lung cancer cohorts from Spain (Lung-ESP, GEO accession: GSE18842)41 and Taiwan (Lung-TWN, GEO accession: GSE19804),42 respectively; 2 breast cancer cohorts from the United States (Breast-USA1, GEO accession: GSE70947) and Malaysia (Breast-MYS, GEO accession: GSE15852),43 respectively; and 2 colon cancer cohorts from Japan (Colon-JPN, GEO accession: GSE22598)44 and Singapore (Colon-SGP, GEO accession: GSE10950),45 respectively. We chose these data sets based on the availability of paired transcriptomic data from both tumor and normal tissues. In total, paired tumor and normal tissues from 44 lung cancer patients from the Lung-ESP cohort, 60 lung cancer patients from the Lung-TWN cohort, 148 breast cancer patients from the Breast-USA1 cohort, 43 breast cancer patients from the Breast-MYS cohort, 17 colon cancer patients from the Colon-JPN cohort, and 24 colon cancer patients from the Colon-SGP cohort were investigated. Fig. 2A indicates that the MC-index of the tumor tissues was significantly higher than that of the matched normal tissues in all the 6 human cancer cohorts (paired t-test: P = 4.3 × 10−2 for Lung-ESP; P = 1.1 × 10−2 for Lung-TWN; P < 10−10 for Breast-USA1; P = 1.5 × 10−8 for Breast-MYS; P = 7.5 × 10−4 for Colon-JPN; P = 4.3 × 10−2 for Colon-SGP). An even more significant difference between tumor and normal tissues was observed for the MC cancer-index in all these cohorts (paired t-test: P < 10−10 for Lung-ESP; P < 10−10 for Lung-TWN; P < 10−10 for Breast-USA1; P = 5.6 × 10−8 for Breast-MYS; P = 1.3 × 10−7 for Colon-JPN; P < 10−10 for Colon-SGP) (Fig. 2B). All these results were highly consistent with our observations in mouse tumors, which suggests the similar significant impact of mast cells on human cancer development.
Figure 2.
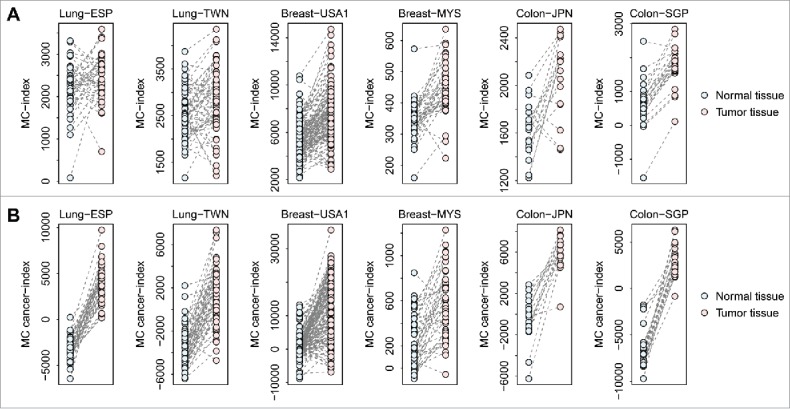
Comparison of MC-index and MC cancer-index between human tumor and normal tissues. Both the MC-index (Panel A) and MC cancer-index (Panel B) were compared between paired tumor and normal tissues from lung, breast, and colon cancer patients, respectively. Six independent human cancer cohorts were analyzed here.
Because the MC cancer-index was computed based on the mast cell dependent genes commonly deregulated in mouse lung, breast, and colon tumors, we further tested whether this computational model is applicable to other cancer types. Three human cancer cohorts were considered here: one liver cancer cohort from the United States (GEO accession: GSE14520),46 one prostate cancer cohort from the United States (GEO accession: GSE32448),47 and one thyroid cancer cohort from Belgium (GEO accession: GSE33630).48 In total, paired tumor and normal tissues from 214 liver cancer patients, 40 prostate cancer patients, and 44 thyroid cancer patients were investigated. Supplementary Fig. S2 indicates that the MC cancer-index of the tumor tissues was significantly higher than that of the matched normal tissues in liver, prostate, and thyroid cancers (paired t-test: P < 10−10 for liver; P = 3.5 × 10−4 for prostate; P < 10−10 for thyroid), which suggests the predictive power of MC cancer-index in these cancer types, resonating with our observations in lung, breast, and colon cancers.
Prognostic power of mast cell–dependent cancer genes
We hypothesized that the mast cell–dependent cancer genes would be predictive of cancer outcome and consequently designated these genes as the Mast Cell–Dependent Cancer (MCDC) signature (Table 1). To test the predictive power of the MCDC signature, we constructed a scoring system to assign each patient a risk score, representing a linear combination of the MCDC gene expression values weighted by the coefficients obtained from the training data sets (GEO accession: GSE8894, GSE21653, and GSE17536 for lung,49 breast,50 and colon51 cancers, respectively) (see Materials and Methods for details). We speculated that a higher MCDC-based risk score implies a poorer clinical outcome. MCDC-positive (MCDC+) patients were defined as those having risk scores larger than zero whereas the other patients were assigned as MCDC-negative (MCDC−).
Table 1.
The MCDC gene signature.
| Gene symbol | Gene title |
|---|---|
| ACP1 | acid phosphatase 1, soluble |
| AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 |
| ARGLU1 | arginine and glutamate rich 1 |
| BCAS2 | breast carcinoma amplified sequence 2 |
| BCR | breakpoint cluster region |
| BTRC | β-transducin repeat containing E3 ubiquitin protein ligase |
| CCDC59 | coiled-coil domain containing 59 |
| CEP57 | centrosomal protein 57kDa |
| CHD4 | chromodomain helicase DNA binding protein 4 |
| CNOT4 | CCR4-NOT transcription complex, subunit 4 |
| CPSF6 | cleavage and polyadenylation specific factor 6, 68kDa |
| CXCL12 | chemokine (C-X-C motif) ligand 12 |
| DDX39A | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| DDX6 | DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| DNAJC2 | DnaJ (Hsp40) homolog, subfamily C, member 2 |
| EIF3A | eukaryotic translation initiation factor 3, subunit A |
| EIF5 | eukaryotic translation initiation factor 5 |
| ELF2 | E74-like factor 2 (ets domain transcription factor) |
| ENY2 | enhancer of yellow 2 homolog (Drosophila) |
| FEZ2 | fasciculation and elongation protein zeta 2 (zygin II) |
| FYTTD1 | 42-three domain containing 1 |
| GAS2L3 | growth arrest-specific 2 like 3 |
| HDLBP | high density lipoprotein binding protein |
| HERPUD1 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| HNRNPU | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| HSPA8 | heat shock 70kDa protein 8 |
| IBTK | inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| IFNGR1 | interferon gamma receptor 1 |
| LIMS1 | LIM and senescent cell antigen-like domains 1 |
| LRRFIP1 | leucine rich repeat (in FLII) interacting protein 1 |
| LTN1 | listerin E3 ubiquitin protein ligase 1 |
| LUC7L3 | LUC7-like 3 (S. cerevisiae) |
| MCM4 | minichromosome maintenance complex component 4 |
| MRPL13 | mitochondrial ribosomal protein L13 |
| NAA15 | N(α)-acetyltransferase 15, NatA auxiliary subunit |
| NEMF | nuclear export mediator factor |
| NR1H3 | nuclear receptor subfamily 1, group H, member 3 |
| NUCKS1 | nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| ORC2 | origin recognition complex, subunit 2 |
| PDAP1 | PDGFA associated protein 1 |
| PDLIM5 | PDZ and LIM domain 5 |
| PFDN1 | prefoldin subunit 1 |
| PGGT1B | protein geranylgeranyltransferase type I, β subunit |
| PLLP | plasmolipin |
| POGZ | pogo transposable element with ZNF domain |
| PPAT | phosphoribosyl pyrophosphate amidotransferase |
| PPP1R12B | protein phosphatase 1, regulatory subunit 12B |
| PPTC7 | PTC7 protein phosphatase homolog (S. cerevisiae) |
| PRKG1 | protein kinase, cGMP-dependent, type I |
| PRPF40A | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| PSMC4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| RBM26 | RNA binding motif protein 26 |
| RBM4 | RNA binding motif protein 4 |
| RBM5 | RNA binding motif protein 5 |
| RNF169 | ring finger protein 169 |
| RNPC3 | RNA-binding region (RNP1, RRM) containing 3 |
| SDAD1 | SDA1 domain containing 1 |
| SERF1A | small EDRK-rich factor 1A (telomeric) |
| SKP2 | S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| SMARCA5 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| SMARCAD1 | SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| SNCG | synuclein, gamma (breast cancer-specific protein 1) |
| SOCS3 | suppressor of cytokine signaling 3 |
| SORBS1 | sorbin and SH3 domain containing 1 |
| SOX4 | SRY (sex determining region Y)-box 4 |
| SRSF3 | serine/arginine-rich splicing factor 3 |
| SSB | Sjogren syndrome antigen B (autoantigen La) |
| STAU2 | staufen, RNA binding protein, homolog 2 (Drosophila) |
| SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| SYNCRIP | synaptotagmin binding, cytoplasmic RNA interacting protein |
| TC2N | tandem C2 domains, nuclear |
| TGFBR3 | transforming growth factor, β receptor III |
| THOC1 | THO complex 1 |
| TMEM38B | transmembrane protein 38B |
| TOR1AIP1 | torsin A interacting protein 1 |
| TRA2B | transformer 2 β homolog (Drosophila) |
| TRIOBP | TRIO and F-actin binding protein |
| TRMT6 | tRNA methyltransferase 6 homolog (S. cerevisiae) |
| TTC3 | tetratricopeptide repeat domain 3 |
| TTC9C | tetratricopeptide repeat domain 9C |
| USP7 | ubiquitin specific peptidase 7 (herpes virus-associated) |
| ZC3H15 | zinc finger CCCH-type containing 15 |
We tested the prognostic power of the MCDC-based risk score in independent validation cohorts. For each cancer type, 2 validation data sets were collected: 2 lung cancer cohorts from Japan (Lung-JPN; GEO accession: GSE31210)52 and Sweden (Lung-SWE; GEO accession: GSE37745),53 respectively; 2 breast cancer cohorts from Singapore (Breast-SGP; GEO accession: GSE4922)54 and the United States (Breast-USA2; GEO accession: GSE2034),55 respectively; and 2 colon cancer cohorts from France (Colon-FRA; GEO accession: GSE39582)56 and Netherlands (Colon-NLD; GEO accession: GSE33113),57 respectively. These data sets were chosen based on the availability of recurrence-free survival information. Kaplan-Meier survival curves demonstrated a significantly reduced recurrence-free survival for the MCDC+ patients compared with the MCDC− ones in all the validation cohorts (log-rank test: P = 9.8 × 10−4 for Lung-JPN; P = 3.3 × 10−2 for Lung-SWE; P = 1.4 × 10−4 for Breast-SGP; P = 5.5 × 10−3 for Breast-USA2; P = 4.0 × 10−2 for Colon-FRA; P = 4.4 × 10−3 for Colon-NLD) (Fig. 3). Univariate Cox proportional hazards regression also confirmed the relationship between MCDC status and clinical outcome: the MCDC+ patients have a 2.35-, 1.83-, 2.23-, 1.70-, 1.36-, and 4.40-fold increased risk of recurrence in the Lung-JPN, Lung-SWE, Breast-SGP, Breast-USA2, Colon-FRA, and Colon-NLD cohorts, respectively (Table 2). These findings collectively indicate that the MCDC signature is predictive of recurrence-free survival in lung, breast, and colon cancers.
Figure 3.
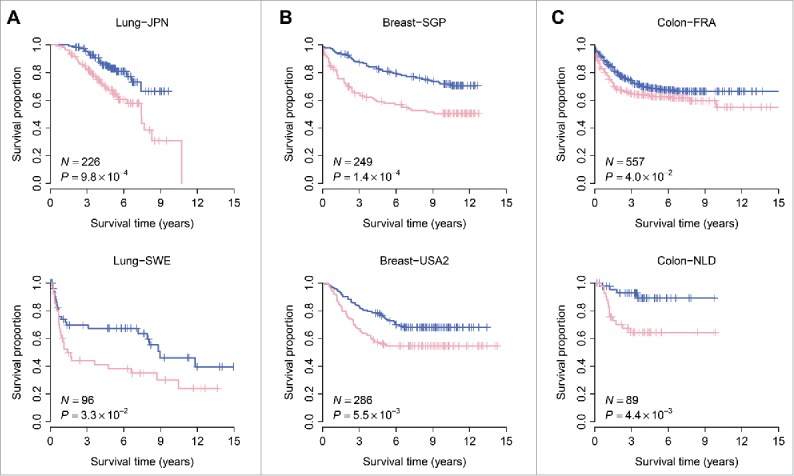
The MCDC signature predicts recurrence-free survival in lung, breast, and colon cancers. Kaplan-Meier curves were presented for lung (Panel A), breast (Panel B), and colon (Panel C) cancer, respectively. Six independent human cancer cohorts were analyzed here. The pink curves are for the MCDC+ patients whereas the blue curves are for the MCDC− patients. The P-values were calculated by log-rank test.
Table 2.
Cox proportional hazards regression of recurrence-free survival by MCDC status.
| Cohort | HR | 95% CI | P-value |
|---|---|---|---|
| Lung-JPN | 2.35 | (1.39, 3.98) | 1.4 × 10−3 |
| Lung-SWE | 1.83 | (1.04, 3.23) | 3.6 × 10−2 |
| Breast-SGP | 2.23 | (1.46, 3.40) | 2.0 × 10−4 |
| Breast-USA2 | 1.70 | (1.17, 2.49) | 5.9 × 10−3 |
| Colon-FRA | 1.36 | (1.02, 1.83) | 4.0 × 10−2 |
| Colon-NLD | 4.40 | (1.44, 13.38) | 9.1 × 10−3 |
Note – HR: hazard ratio; CI: confidence interval
Next, we investigated the performance of the MCDC signature in comparison with standard clinical and pathological factors associated with prognosis in human cancers. For the Lung-JPN cohort, patient age, gender, smoking history, stage, EGFR/KRAS/ALK gene mutation status, and MYC protein levels were considered. For the Lung-SWE cohort, we took age, gender, stage, and WHO performance status into account. For the Breast-SGP cohort, patient age, gender, grade, tumor size, lymph node status, estrogen receptor (ER) status, and TP53 mutation status were considered. For the Breast-USA2 cohort, ER status were included as covariate. For the Colon-FRA cohort, we considered factors including age, gender, stage, and BRAF, KRAS, and TP53 mutation status. For the Colon-NLD cohort, patient age and gender were considered as covariate. Multivariate Cox proportional hazards regression indicates that the MCDC status remained a significant covariate in relation to the clinical and pathological factors in each validation cohorts (P = 3.8 × 10−2 for Lung-JPN; P = 3.8 × 10−2 for Lung-SWE; P = 2.4 × 10−2 for Breast-SGP; P = 4.0 × 10−3 for Breast-USA2; P = 2.6 × 10−2 for Colon-FRA; P = 1.4 × 10−2 for Colon-NLD) (Table 3), which suggests that the MCDC signature is independent of standard clinical and pathological prognostic factors in lung, breast, and colon cancers.
Table 3.
Multivariate Cox proportional hazards regression of survival in the validation cohorts.
| Cohort | Covariate | HR | 95% CI | P-value |
|---|---|---|---|---|
| Lung-JPN | MCDC + vs. − | 1.79 | (1.01, 3.04) | 3.8 × 10−2 |
| Age (per year) | 1.04 | (1.00, 1.08) | 4.2 × 10−2 | |
| Gender male vs. female | 0.72 | (0.36, 1.42) | 3.4 × 10−1 | |
| Smoking + vs. − | 1.50 | (0.75, 2.85) | 2.7 × 10−1 | |
| Stage | 2.85 | (1.68, 4.84) | 1.0 × 10−4 | |
| EGFR/KRAS/ALK mutation + vs. − | 0.60 | (0.36, 1.01) | 5.3 × 10−2 | |
| MYC level high vs. low | 0.94 | (0.37, 2.40) | 9.0 × 10−1 | |
| Lung-SWE | MCDC + vs. − | 1.99 | (1.04, 3.81) | 3.8 × 10−2 |
| Age (per year) | 0.99 | (0.96, 1.03) | 7.3 × 10−1 | |
| Gender male vs. female | 1.01 | (0.54, 1.88) | 9.8 × 10−1 | |
| Stage | 1.81 | (0.86, 3.79) | 1.2 × 10−1 | |
| WHO performance | 1.33 | (0.97, 1.82) | 7.8 × 10−2 | |
| Breast-SGP | MCDC + vs. − | 1.84 | (1.08, 3.11) | 2.4 × 10−2 |
| Age (per year) | 1.01 | (0.99, 1.02) | 4.3 × 10−1 | |
| grade | 1.19 | (0.78, 1.81) | 4.1 × 10−1 | |
| Tumor size | 1.01 | (0.99, 1.02) | 3.1 × 10−1 | |
| Lymph node + vs. − | 1.50 | (0.94, 2.40) | 9.2 × 10−2 | |
| ER + vs - | 1.17 | (0.62, 2.23) | 6.2 × 10−1 | |
| TP53 mutation + vs. − | 1.14 | (0.66, 1.96) | 6.4 × 10−1 | |
| Breast-USA2 | MCDC + vs. − | 1.79 | (1.20, 2.66) | 4.0 × 10−3 |
| ER + vs - | 1.21 | (0.77, 1.91) | 4.1 × 10−1 | |
| Colon-FRA | MCDC + vs. − | 1.41 | (1.04, 1.91) | 2.6 × 10−2 |
| Age (per year) | 1.01 | (0.99, 1.02) | 3.7 × 10−1 | |
| Gender male vs. female | 1.49 | (1.10, 2.03) | 1.1 × 10−2 | |
| Stage | 2.71 | (2.16, 3.39) | < 10−10 | |
| BRAF mutation + vs. − | 0.86 | (0.47, 1.59) | 6.4 × 10−1 | |
| KRAS mutation + vs - | 1.28 | (0.93, 1.75) | 1.3 × 10−1 | |
| TP53 mutation + vs. − | 1.51 | (1.11, 2.04) | 7.7 × 10−3 | |
| Colon-NLD | MCDC + vs. − | 4.15 | (1.34, 12.87) | 1.4 × 10−2 |
| Age (per year) | 0.99 | (0.95, 1.02) | 4.8 × 10−1 | |
| Gender male vs. female | 0.78 | (0.29, 2.05) | 6.1 × 10−1 |
Note – HR: hazard ratio; CI: confidence interval
A bioinformatical study by Venet et al. points out that most published gene signatures are not significantly better than random gene sets of identical size that are randomly picked up from human genome.58 To address this issue, we further conducted resampling test for the MCDC signature. We obtained 1,000 random gene signatures by randomly selecting 82 genes from human genome. For each random set of genes, multivariate Cox proportional hazards regression was conducted. The association between each random gene signature and survival was measured by the mean of Cox regression Z-score. We found that the mean of Z-score of the MCDC signature is significantly larger than that of the random gene signatures (Right-tailed: P = 0.022) (Supplementary Fig. S3), which suggests the empirically non-random association between the MCDC signature and survival.
Discussion
For decades, there has been particular interest and speculation as to the physiologic function of mast cells in tumor biology. We know mast cells potentially influence many aspects of tumor biology, including tumor angiogenesis,5,11 tumor invasiveness,6 and immunosuppression1,15; however, the exact contributions of mast cells in tumorigenesis remain controversial. Particularly, there have been a considerable number of contradictory observations regarding the detrimental or protective roles of mast cells in tumor development.31 Although elucidating the detailed reasons for these discrepancies is beyond the scope of this study, we have presented a transcriptomic perspective to study the impact of mast cells on shaping tumor microenvironment. Based on the transcriptomic data from WT, KitW-sh, and KitW-sh+MC mice, we identified the mast cell–dependent genes, which were deregulated by mast cell deficiency but largely recovered upon mast cell engraftment. To quantify the transcriptomic impact caused by mast cells in tissue microenvironment, a computational algorithm was developed to assign each tissue sample a MC-index based on the rank-weighted expression profile of mast cell–dependent genes, which potentially serves as a proxy of mast cell infiltration level in the tissue microenvironment. We indicate that, in both mouse models and human patients, the MC-indices of tumors are statistically higher than those of normal tissues from lung, breast, and colon, respectively. To more precisely assess the contribution of mast cells in shaping the tumor microenvironment, the MC cancer-index was computed for each tissue sample, based on the expression profile of mast cell–dependent cancer (MCDC) genes commonly deregulated in mouse lung, breast, and colon tumors. The difference in MC cancer-index between tumor and normal tissues mirrors the pattern we observed for MC-index in both mouse and human. Based on the MCDC genes, the MCDC signature was developed, which predicts clinical outcomes as an independent covariate in lung, breast, and colon cancers, respectively.
Despite the debate over detrimental/protective roles for mast cells in tumorigenesis, we demonstrate a potential increased trend in MC- and MC cancer-indices in lung, breast, and colon tumors from a transcriptomic perspective, which suggests that, compared with normal tissues, the MC+ genes are more likely to be upregulated while the MC− ones tend to be downregulated in tumor microenvironment. In other words, the mast cell infiltration could be increased in lung, breast, and colon tumors compared with that in normal lung, breast, and colon tissues, respectively, suggesting that mast cells might be implicated in tumor development and progression.
The transcriptomic data from KitW-sh mice were applied in this study to infer the mast cell–dependent genes. Mice bearing c-kit mutations exhibit reduced c-kit tyrosine kinase–dependent signaling that results in not only mast cell deficiency but also other phenotypic abnormalities.27 Therefore, the differential gene expression between KitW-sh and WT mice might arise from other abnormalities, not solely attributed to mast cell deficiency. To address this issue, we used KitW-sh+MC mice to assess to what extent the abnormalities in gene expression of c-kit mutant mice can be recovered by mast cell engraftment.6 Hence, only the genes that were deregulated in mast cell–deficient mice but recovered upon mast cell engraftment were defined as mast cell–dependent genes.
The predictive power of the MCDC signature illustrates the link between the mast cell–dependent genes and prognosis of lung, breast, and colon cancers, which may be a common feature in various cancers. Indeed, ACP1,59 AKAP9,60 BCAS2,61 CEP57,62 CXCL12,63 EIF5,64 LRRFIP1,65 NUCKS1,66 PFDN1,67 RBM4,68 SKP2,69 SMARCA5,70 SMARCAD1,71 SNCG,72 SOCS3,73 SOX4,74 SVEP1,75 TGFBR3,76 THOC1,77 and USP778 from the MCDC signature are already under investigation regarding cancer pathology or treatment in some capacity. More work is needed to determine whether the other genes in our signature could be exploited for cancer therapy.
Recently, Dwyer et al. proposed a mast cell signature, which consists of 128 genes upregulated in mast cells compared with the other immunocytes.79 However, we didn't find any overlap between the 128 gene and MCDC signatures, which may be due to the difference in utility between the 2 signatures. The 128 gene signature was designed to differentiate mast cells from the other cells, which was developed upon cell-level gene expression data. In contrast, the MCDC signature was derived from tissue-level transcriptomic analysis and reflects the integrated signal of individual cell types, which implicitly correlates with mast cell infiltration and tumor development. Therefore, it's fairly reasonable that the 128 gene signature fails to differentiate tumors from normal tissues (Supplementary Fig. S4), while the MCDC signature works the other way around.
Although the MCDC signature potentially reflects some common mechanisms shared by different cancers, some mast cell–dependent genes differ substantially in expression pattern among different tumor types (Supplementary Fig. S5), which represent the intrinsic pathological difference among cancers. For example, the top GO terms associated with the mast cell–dependent genes differing between lung and breast tumors, between lung and colon tumors, and between breast and colon tumors are “tube development,” “positive regulation of nitrogen compound metabolic process” and “mammary gland development,” respectively (Supplementary Fig. S5).
Understanding the mast cell–dependent transcriptomic pattern may provide therapeutic benefit in cancer treatment. Our study provides a provocative insight into the role of mast cells in cancers. The expression profile of the mast cell–dependent genes potentially serves as a promising proxy of the impact of mast cells on tumor microenvironment although the molecular mechanisms remain unclear. When working cooperatively with known clinical and pathological prognostic factors, the MCDC signature might enhance the prediction accuracy for identifying patients at higher risk for recurrence. However, the real physiologic role of mast cells is more complicated than the transcriptomic data and appears to vary with cancer types. In future study, intensive experimental investigation is apparently needed to validate the exact role of individual mast cell–dependent genes in different cancers.
Materials and methods
Transcriptomic data
Four mouse transcriptomic data sets were included in this study. Firstly, the microarray data of lung RNA from WT, KitW-sh, and KitW-sh+MC mice were obtained from the GEO28 database (GEO accession: GSE27066; Affymetrix Mouse Genome 430 2.0 Array).29 We used this data set to filter out the mast cell–dependent mouse genes. Secondly, from the GEO database, we downloaded the gene expression data of both tumor and normal tissues in mouse lung (GEO accession: GSE31013; Affymetrix Mouse Genome 430 2.0 Array),38 breast (GEO accession: GSE21444; Affymetrix Mouse Genome 430 2.0 Array),39 and colon (GEO accession: GSE50794; Affymetrix Mouse Genome 430 2.0 Array).40 These data sets were used to examine the deregulation pattern of the mast cell–dependent genes in mouse tumors.
For human subjects, we applied 18 independent whole-genome gene expression data sets in this study. Firstly, we obtained the microarray data of paired normal and tumor tissues derived from lung, breast, colon, liver, prostate, and thyroid cancer patients from the GEO database. For lung cancer, we included the Lung-ESP (GEO accession: GSE18842; Affymetrix Human Genome U133 Plus 2.0 Array)41 and Lung-TWN (GEO accession: GSE19804; Affymetrix Human Genome U133 Plus 2.0 Array)42 cohorts; for breast cancer, we included the Breast-USA1 (GEO accession: GSE70947; Agilent-028004 SurePrint G3 Human GE 8 × 60K Microarray) and Breast-MYS (GEO accession: GSE15852; Affymetrix Human Genome U133A Array)43 cohorts; for colon cancer, we included the Colon-JPN (GEO accession: GSE22598; Affymetrix Human Genome U133 Plus 2.0 Array)44 and Colon-SGP (GEO accession: GSE10950; Illumina humanRef-8 v2.0 expression beadchip)45 cohorts; for liver, prostate, and thyroid cancers, the following 3 data sets were included respectively: GSE14520 (Affymetrix Human Genome U133A 2.0 Array),46 GSE32448 (Affymetrix Human Genome U133 Plus 2.0 Array),47 and GSE33630 (Affymetrix Human Genome U133 Plus 2.0 Array).48 These data sets were used to examine the deregulation profiles of the mast cell–dependent genes in human tumors. To investigate the prognostic power of the mast cell–dependent genes, we constructed training and validation cohorts for lung, breast, and colon cancers, respectively. From the GEO database, we first collected the training data sets with available information on recurrence-free survival for lung (GEO accession: GSE8894; Affymetrix Human Genome U133 Plus 2.0 Array),49 breast (GEO accession: GSE21653; Affymetrix Human Genome U133 Plus 2.0 Array),50 and colon (GEO accession: GSE17536; Affymetrix Human Genome U133 Plus 2.0 Array)51 cancers, respectively. Next, 2 validation cohorts with clinical outcome information were downloaded for each cancer type. For lung cancer, we collected the Lung-JPN (GEO accession: GSE31210; Affymetrix Human Genome U133 Plus 2.0 Array)52 and Lung-SWE (GEO accession: GSE37745; Affymetrix Human Genome U133 Plus 2.0 Array)53 cohorts; for breast cancer, we included the Breast-SGP (GEO accession: GSE4922; Affymetrix Human Genome U133A and U133B Arrays)54 and Breast-USA2 (GEO accession: GSE2034; Affymetrix Human Genome U133A Array)55 cohorts; for colon cancer, we considered the (Colon-FRA, GEO accession: GSE39582; Affymetrix Human Genome U133 Plus 2.0 Array)56 and Colon-NLD (GEO accession: GSE33113; Affymetrix Human Genome U133 Plus 2.0 Array)57 cohorts.
Detecting differential gene expression
Significance analysis of microarrays (SAM),80 implemented in the samr library of the R Statistical Package, was used to identify deregulated genes. False discovery rate was controlled using the q-value method.81 Transcripts with a fold-change >1.5 and false discovery rate <0.05 were deemed differentially expressed. We limited our analysis to the probes/probesets with unique annotations and removed genes on chromosomes X and Y to avoid the potential confounding sex factor.
Mast cell index and mast cell cancer index
Briefly, mast cell index (MC-index) is the difference in normalized centroid of rank-weighted gene expression between the MC+ and MC− genes, which is designed to utilize transcriptomic data to assess the impact of mast cells on shaping tissue microenvironment. For a transcriptomic data set with n genes, all genes in each sample are sorted in ascending order according to their expression values. If ri is the rank of gene i in a sample, the exponential weight (wi) of gene i can be calculated as:
| (1) |
For the MC+ genes, let n+ be the number of the genes and the normalized centroid (C+) can be calculated as the mean of gene weight across all the MC+ genes (Equation 2). For the complement gene set composed of all the other non-MC+ genes, let be the number of the genes and the normalized centroid () can be calculated as the mean of gene weight across all the non-MC+ genes (Equation 3). The index of the MC+ genes (I+) is simply the difference between the normalized centroid of MC+ and non-MC+ genes (Equation 4).
| (2) |
| (3) |
| (4) |
Similarly, for the MC− genes, let n− be the number of the genes and the normalized centroid (C−) can be calculated as the mean of gene weight across all the MC− genes (Equation 5). For the complement gene set composed of all the other non-MC− genes, let be the number of the genes and the normalized centroid () can be calculated as the mean of gene weight across all the non-MC− genes (Equation 6). The index of the MC− genes (I−) is the difference between the normalized centroid of MC− and non-MC− genes (Equation 7). Finally, the MC-index (I) of each sample is calculated as the difference between I+ and I− (Equation 8).
| (5) |
| (6) |
| (7) |
| (8) |
Mast cell cancer index (MC cancer-index) is designed to assess the impact of mast cells in tumor development. The method to compute MC cancer-index is the same as the procedure to compute MC-index, except for 2 modifications: i) replacing the MC+ genes with the MC+ genes commonly upregulated in mouse lung, breast, and colon tumors; and ii) replacing the MC− genes with the MC− genes commonly downregulated in mouse lung, breast, and colon tumors.
Risk score
Based on the gene expression and clinical outcome data from the training data sets (GEO accession: GSE8894, GSE21653, and GSE17536 for lung,49 breast,50 and colon51 cancers, respectively), we conducted univariate Cox proportional hazards regressions to evaluate the association between recurrence-free survival and gene expression for lung, breast, and colon cancers, respectively. A risk score was then calculated for each patient using a linear combination of gene expression weighted by the Wald statistic (ratio of regression coefficient to its standard error)82-84 as shown below:
| (9) |
Here, s is the risk score of the patient; n is the number of genes; wi denotes the Wald statistic of gene i; ei denotes the expression level of gene i; and μi and τi are the mean and standard deviation of the gene expression values for gene i across all samples, respectively. Patients were then divided into high-risk and low-risk groups with zero as the cutoff. We speculated that a higher risk score implies a poorer clinical outcome.
Supplementary Material
Disclosure of interest
The authors report no conflict of interest.
References
- 1.Liu J, Zhang Y, Zhao J, Yang Z, Li D, Katirai F, Huang B. Mast cell: Insight into remodeling a tumor microenvironment. Cancer Metastasis Rev. 2011;30(2):177-84. doi: 10.1007/s10555-011-9276-1. PMID:21267769. [DOI] [PubMed] [Google Scholar]
- 2.Urb M, Sheppard DC. The role of mast cells in the defence against pathogens. PLoS Pathog. 2012;8(4):e1002619. doi: 10.1371/journal.ppat.1002619. PMID:22577358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Silva EZ, Jamur MC, Oliver C. Mast cell function: A new vision of an old cell. J Histochem Cytochem. 2014;62(10):698-738. doi: 10.1369/0022155414545334. PMID:25062998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frieri M. Mast cell activation syndrome. Clin Rev Allergy Immunol. 2015;13:27. doi: 10.1007/s12016-015-8487-6. PMID:25944644. [DOI] [PubMed] [Google Scholar]
- 5.Crivellato E, Nico B, Ribatti D. Mast cells and tumour angiogenesis: New insight from experimental carcinogenesis. Cancer Lett. 2008;269(1):1-6. doi: 10.1016/j.canlet.2008.03.031. PMID:18450371. [DOI] [PubMed] [Google Scholar]
- 6.Marichal T, Tsai M, Galli SJ. Mast cells: Potential positive and negative roles in tumor biology. Cancer Immunol Res. 2013;1(5):269-79. doi: 10.1158/2326-6066.CIR-13-0119. PMID:24777963. [DOI] [PubMed] [Google Scholar]
- 7.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14(21):6735-41. doi: 10.1158/1078-0432.CCR-07-4843. PMID:18980965. [DOI] [PubMed] [Google Scholar]
- 8.Aoki M, Pawankar R, Niimi Y, Kawana S. Mast cells in basal cell carcinoma express VEGF, IL-8 and RANTES. Int Arch Allergy Immunol. 2003;130(3):216-23. doi: 10.1159/000069515. PMID:12660426. [DOI] [PubMed] [Google Scholar]
- 9.Sawatsubashi M, Yamada T, Fukushima N, Mizokami H, Tokunaga O, Shin T. Association of vascular endothelial growth factor and mast cells with angiogenesis in laryngeal squamous cell carcinoma. Virchows Arch. 2000;436(3):243-48. doi: 10.1007/s004280050037. PMID:10782883. [DOI] [PubMed] [Google Scholar]
- 10.Imada A, Shijubo N, Kojima H, Abe S. Mast cells correlate with angiogenesis and poor outcome in stage I lung adenocarcinoma. Eur Respir J. 2000;15(6):1087-93. doi: 10.1034/j.1399-3003.2000.01517.x. PMID:10885428. [DOI] [PubMed] [Google Scholar]
- 11.Starkey JR, Crowle PK, Taubenberger S. Mast-cell-deficient W/Wv mice exhibit a decreased rate of tumor angiogenesis. Int J Cancer. 1988;42(1):48-52. doi: 10.1002/ijc.2910420110. PMID:2455691. [DOI] [PubMed] [Google Scholar]
- 12.Almholt K, Johnsen M. Stromal cell involvement in cancer. Recent Results Cancer Res. 2003;162:31-42. doi: 10.1007/978-3-642-59349-9_3. PMID:12790319. [DOI] [PubMed] [Google Scholar]
- 13.Baram D, Vaday GG, Salamon P, Drucker I, Hershkoviz R, Mekori YA. Human mast cells release metalloproteinase-9 on contact with activated T cells: Juxtacrine regulation by TNF-alpha. J Immunol. 2001;167(7):4008-16. doi: 10.4049/jimmunol.167.7.4008. PMID:11564820. [DOI] [PubMed] [Google Scholar]
- 14.Huang C, Sali A, Stevens RL. Regulation and function of mast cell proteases in inflammation. J Clin Immunol. 1998;18(3):169-83. doi: 10.1023/A:1020574820797. PMID:9624576. [DOI] [PubMed] [Google Scholar]
- 15.Wasiuk A, de Vries VC, Hartmann K, Roers A, Noelle RJ. Mast cells as regulators of adaptive immunity to tumours. Clin Exp Immunol. 2009;155(2):140-46. doi: 10.1111/j.1365-2249.2008.03840.x. PMID:19077084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takanami I, Takeuchi K, Naruke M. Mast cell density is associated with angiogenesis and poor prognosis in pulmonary adenocarcinoma. Cancer. 2000;88(12):2686-92. doi: 10.1002/1097-0142(20000615)88:12%3c2686::AID-CNCR6%3e3.0.CO;2-6. PMID:10870050. [DOI] [PubMed] [Google Scholar]
- 17.Tomita M, Matsuzaki Y, Onitsuka T. Correlation between mast cells and survival rates in patients with pulmonary adenocarcinoma. Lung Cancer. 1999;26(2):103-08. doi: 10.1016/S0169-5002(99)00076-8. PMID:10568681. [DOI] [PubMed] [Google Scholar]
- 18.Welsh TJ, Green RH, Richardson D, Waller DA, O'Byrne KJ, Bradding P. Macrophage and mast-cell invasion of tumor cell islets confers a marked survival advantage in non-small-cell lung cancer. J Clin Oncol. 2005;23(35):8959-67. doi: 10.1200/JCO.2005.01.4910. PMID:16219934. [DOI] [PubMed] [Google Scholar]
- 19.Xiang M, Gu Y, Zhao F, Lu H, Chen S, Yin L. Mast cell tryptase promotes breast cancer migration and invasion. Oncol Rep. 2010;23(3):615-19. PMID:20126998. [DOI] [PubMed] [Google Scholar]
- 20.Ribatti D, Finato N, Crivellato E, Guidolin D, Longo V, Mangieri D, Nico B, Vacca A, Beltrami CA. Angiogenesis and mast cells in human breast cancer sentinel lymph nodes with and without micrometastases. Histopathology. 2007;51(6):837-42. doi: 10.1111/j.1365-2559.2007.02869.x. PMID:17944928. [DOI] [PubMed] [Google Scholar]
- 21.Rajput AB, Turbin DA, Cheang MC, Voduc DK, Leung S, Gelmon KA, Gilks CB, Huntsman DG. Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: A study of 4,444 cases. Breast Cancer Res Treat. 2008;107(2):249-57. doi: 10.1007/s10549-007-9546-3. PMID:17431762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.della Rovere F, Granata A, Familiari D, D'Arrigo G, Mondello B, Basile G. Mast cells in invasive ductal breast cancer: Different behavior in high and minimum hormone-receptive cancers. Anticancer Res. 2007;27(4B):2465-71. PMID:17695540. [PubMed] [Google Scholar]
- 23.Acikalin MF, Oner U, Topcu I, Yasar B, Kiper H, Colak E. Tumour angiogenesis and mast cell density in the prognostic assessment of colorectal carcinomas. Dig Liver Dis. 2005;37(3):162-69. doi: 10.1016/j.dld.2004.09.028. PMID:15888280. [DOI] [PubMed] [Google Scholar]
- 24.Yodavudh S, Tangjitgamol S, Puangsa-art S. Prognostic significance of microvessel density and mast cell density for the survival of Thai patients with primary colorectal cancer. J Med Assoc Thai. 2008;91(5):723-32. PMID:18672639. [PubMed] [Google Scholar]
- 25.Gulubova M, Vlaykova T. Prognostic significance of mast cell number and microvascular density for the survival of patients with primary colorectal cancer. J Gastroenterol Hepatol. 2009;24(7):1265-75. doi: 10.1111/j.1440-1746.2007.05009.x. PMID:17645466. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen HJ, Hansen U, Christensen IJ, Reimert CM, Brunner N, Moesgaard F. Independent prognostic value of eosinophil and mast cell infiltration in colorectal cancer tissue. J Pathol. 1999;189(4):487-95. doi: 10.1002/(SICI)1096-9896(199912)189:4%3c487::AID-PATH484%3e3.0.CO;2-I. PMID:10629548. [DOI] [PubMed] [Google Scholar]
- 27.Grimbaldeston MA, Chen CC, Piliponsky AM, Tsai M, Tam SY, Galli SJ. Mast cell-deficient W-sash c-kit mutant Kit W-sh/W-sh mice as a model for investigating mast cell biology in vivo. Am J Pathol. 2005;167(3):835-48. doi: 10.1016/S0002-9440(10)62055-X. PMID:16127161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207-10. doi: 10.1093/nar/30.1.207. PMID:11752295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Eckart MR, Morgan AA, Mukai K, Butte AJ, Tsai M, Galli SJ. Identification of an IFN-gamma/mast cell axis in a mouse model of chronic asthma. J Clin Invest. 2011;121(8):3133-43. doi: 10.1172/JCI43598. PMID:21737883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29-34. doi: 10.1093/nar/27.1.29. PMID:9847135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khazaie K, Blatner NR, Khan MW, Gounari F, Gounaris E, Dennis K, Bonertz A, Tsai FN, Strouch MJ, Cheon E, et al.. The significant role of mast cells in cancer. Cancer Metastasis Rev. 2011;30(1):45-60. doi: 10.1007/s10555-011-9286-z. PMID:21287360. [DOI] [PubMed] [Google Scholar]
- 32.Huang B, Lei Z, Zhang GM, Li D, Song C, Li B, Liu Y, Yuan Y, Unkeless J, Xiong H, et al.. SCF-mediated mast cell infiltration and activation exacerbate the inflammation and immunosuppression in tumor microenvironment. Blood. 2008;112(4):1269-79. doi: 10.1182/blood-2008-03-147033. PMID:18524989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blatner NR, Bonertz A, Beckhove P, Cheon EC, Krantz SB, Strouch M, Weitz J, Koch M, Halverson AL, Bentrem DJ, et al.. In colorectal cancer mast cells contribute to systemic regulatory T-cell dysfunction. Proc Natl Acad Sci U S A. 2010;107(14):6430-35. doi: 10.1073/pnas.0913683107. PMID:20308560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soucek L, Lawlor ER, Soto D, Shchors K, Swigart LB, Evan GI. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med. 2007;13(10):1211-18. doi: 10.1038/nm1649. PMID:17906636. [DOI] [PubMed] [Google Scholar]
- 35.Maltby S, Khazaie K, McNagny KM. Mast cells in tumor growth: Angiogenesis, tissue remodelling and immune-modulation. Biochim Biophys Acta. 2009;1796(1):19-26. doi: 10.1016/j.bbcan.2009.02.001. PMID:19233249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yano H, Kinuta M, Tateishi H, Nakano Y, Matsui S, Monden T, Okamura J, Sakai M, Okamoto S. Mast cell infiltration around gastric cancer cells correlates with tumor angiogenesis and metastasis. Gastric Cancer. 1999;2(1):26-32. doi: 10.1007/s101200050017. PMID:11957067. [DOI] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25-29. doi: 10.1038/75556. PMID:10802651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandiri AR, Sills RC, Ziglioli V, Ton TV, Hong HH, Lahousse SA, Gerrish KE, Auerbach SS, Shockley KR, Bushel PR, et al.. Differential transcriptomic analysis of spontaneous lung tumors in B6C3F1 mice: Comparison to human non-small cell lung cancer. Toxicol Pathol. 2012;40(8):1141-59. doi: 10.1177/0192623312447543. PMID:22688403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10(1):15. doi: 10.1186/1476-4598-10-15. PMID:21314937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belmont PJ, Budinska E, Jiang P, Sinnamon MJ, Coffee E, Roper J, Xie T, Rejto PA, Derkits S, Sansom OJ, et al.. Cross-species analysis of genetically engineered mouse models of MAPK-driven colorectal cancer identifies hallmarks of the human disease. Dis Model Mech. 2014;7(6):613-23. doi: 10.1242/dmm.013904. PMID:24742783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Farez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129(2):355-64. doi: 10.1002/ijc.25704. PMID:20878980. [DOI] [PubMed] [Google Scholar]
- 42.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, et al.. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590-97. doi: 10.1158/1055-9965.EPI-10-0332. PMID:20802022. [DOI] [PubMed] [Google Scholar]
- 43.Pau Ni IB, Zakaria Z, Muhammad R, Abdullah N, Ibrahim N, Aina Emran N, Hisham Abdullah N, Syed Hussain SN. Gene expression patterns distinguish breast carcinomas from normal breast tissues: The Malaysian context. Pathol Res Pract. 2010;206(4):223-28. doi: 10.1016/j.prp.2009.11.006. PMID:20097481. [DOI] [PubMed] [Google Scholar]
- 44.Okazaki S, Ishikawa T, Iida S, Ishiguro M, Kobayashi H, Higuchi T, Enomoto M, Mogushi K, Mizushima H, Tanaka H, et al.. Clinical significance of UNC5B expression in colorectal cancer. Int J Oncol. 2012;40(1):209-16. doi: 10.3892/ijo.2011.1201. PMID:21922135. [DOI] [PubMed] [Google Scholar]
- 45.Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, et al.. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13(6):529-41. doi: 10.1016/j.ccr.2008.04.019. PMID:18538736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, et al.. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202-12. doi: 10.1158/0008-5472.CAN-10-2607. PMID:21159642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derosa CA, Furusato B, Shaheduzzaman S, Srikantan V, Wang Z, Chen Y, Seifert M, Ravindranath L, Young D, Nau M, et al.. Elevated osteonectin/SPARC expression in primary prostate cancer predicts metastatic progression. Prostate Cancer Prostatic Dis. 2012;15(2):150-56. doi: 10.1038/pcan.2011.61. PMID:22343836. [DOI] [PubMed] [Google Scholar]
- 48.Dom G, Tarabichi M, Unger K, Thomas G, Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V, Maenhaut C. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107(6):994-1000. doi: 10.1038/bjc.2012.302. PMID:22828612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, Kim H, Lee HJ, Choi HY, Jung Y, et al.. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14(22):7397-404. doi: 10.1158/1078-0432.CCR-07-4937. PMID:19010856. [DOI] [PubMed] [Google Scholar]
- 50.Sabatier R, Finetti P, Cervera N, Lambaudie E, Esterni B, Mamessier E, Tallet A, Chabannon C, Extra JM, Jacquemier J, et al.. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126(2):407-20. doi: 10.1007/s10549-010-0897-9. PMID:20490655. [DOI] [PubMed] [Google Scholar]
- 51.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al.. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958-68. doi: 10.1053/j.gastro.2009.11.005. PMID:19914252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al.. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100-11. doi: 10.1158/0008-5472.CAN-11-1403. PMID:22080568. [DOI] [PubMed] [Google Scholar]
- 53.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Ponten F, et al.. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19(1):194-204. doi: 10.1158/1078-0432.CCR-12-1139. PMID:23032747. [DOI] [PubMed] [Google Scholar]
- 54.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al.. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292-301. doi: 10.1158/0008-5472.CAN-05-4414. PMID:17079448. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al.. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671-79. doi: 10.1016/S0140-6736(05)70933-8. PMID:15721472. [DOI] [PubMed] [Google Scholar]
- 56.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al.. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. PMID:23700391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Sousa EMF, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, et al.. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9(5):476-85. doi: 10.1016/j.stem.2011.10.008. PMID:22056143. [DOI] [PubMed] [Google Scholar]
- 58.Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7(10):e1002240. doi: 10.1371/journal.pcbi.1002240. PMID:22028643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spina C, Saccucci P, Bottini E, Gloria-Bottini F. ACP1 genetic polymorphism and colon cancer. Cancer Genet Cytogenet. 2008;186(1):61-62. doi: 10.1016/j.cancergencyto.2008.06.006. PMID:18786445. [DOI] [PubMed] [Google Scholar]
- 60.Frank B, Wiestler M, Kropp S, Hemminki K, Spurdle AB, Sutter C, Wappenschmidt B, Chen X, Beesley J, Hopper JL, et al.. Association of a common AKAP9 variant with breast cancer risk: A collaborative analysis. J Natl Cancer Inst. 2008;100(6):437-42. doi: 10.1093/jnci/djn037. PMID:18334708. [DOI] [PubMed] [Google Scholar]
- 61.Kuo PC, Huang CW, Lee CI, Chang HW, Hsieh SW, Chung YP, Lee MS, Huang CS, Tsao LP, Tsao YP, et al.. BCAS2 promotes prostate cancer cells proliferation by enhancing AR mRNA transcription and protein stability. Br J Cancer. 2015;112(2):391-402. doi: 10.1038/bjc.2014.603. PMID:25461807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mang J, Korzeniewski N, Dietrich D, Sailer V, Tolstov Y, Searcy S, von Hardenberg J, Perner S, Kristiansen G, Marx A, et al.. Prognostic significance and functional role of CEP57 in prostate cancer. Transl Oncol. 2015;8(6):487-96. doi: 10.1016/j.tranon.2015.11.004. PMID:26692530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen B, Zheng MQ, Lu JW, Jiang Q, Wang TH, Huang XE. CXCL12-CXCR4 promotes proliferation and invasion of pancreatic cancer cells. Asian Pac J Cancer Prev. 2013;14(9):5403-08. doi: 10.7314/APJCP.2013.14.9.5403. PMID:24175834. [DOI] [PubMed] [Google Scholar]
- 64.Lee NP, Tsang FH, Shek FH, Mao M, Dai H, Zhang C, Dong S, Guan XY, Poon RT, Luk JM. Prognostic significance and therapeutic potential of eukaryotic translation initiation factor 5A (eIF5A) in hepatocellular carcinoma. Int J Cancer. 2010;127(4):968-76. doi: 10.1002/ijc.25100. PMID:19998337. [DOI] [PubMed] [Google Scholar]
- 65.Ariake K, Ohtsuka H, Motoi F, Douchi D, Oikawa M, Rikiyama T, Fukase K, Katayose Y, Egawa S, Unno M. GCF2/LRRFIP1 promotes colorectal cancer metastasis and liver invasion through integrin-dependent RhoA activation. Cancer Lett. 2012;325(1):99-107. doi: 10.1016/j.canlet.2012.06.012. PMID:22750095. [DOI] [PubMed] [Google Scholar]
- 66.Kikuchi A, Ishikawa T, Mogushi K, Ishiguro M, Iida S, Mizushima H, Uetake H, Tanaka H, Sugihara K. Identification of NUCKS1 as a colorectal cancer prognostic marker through integrated expression and copy number analysis. Int J Cancer. 2013;132(10):2295-302. doi: 10.1002/ijc.27911. PMID:23065711. [DOI] [PubMed] [Google Scholar]
- 67.Wang P, Zhao J, Yang X, Guan S, Feng H, Han D, Lu J, Ou B, Jin R, Sun J, et al.. PFDN1, an indicator for colorectal cancer prognosis, enhances tumor cell proliferation and motility through cytoskeletal reorganization. Med Oncol. 2015;32(12):264. doi: 10.1007/s12032-015-0710-z. PMID:26553318. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Chen D, Qian H, Tsai YS, Shao S, Liu Q, Dominguez D, Wang Z. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374-89. doi: 10.1016/j.ccr.2014.07.010. PMID:25203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci U S A. 2001;98(9):5043-48. doi: 10.1073/pnas.081474898. PMID:11309491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gigek CO, Lisboa LC, Leal MF, Silva PN, Lima EM, Khayat AS, Assumpcao PP, Burbano RR, Smith Mde A. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29(2):162-66. doi: 10.3109/07357907.2010.543365. PMID:21261476. [DOI] [PubMed] [Google Scholar]
- 71.Al Kubaisy E, Arafat K, De Wever O, Hassan AH, Attoub S. SMARCAD1 knockdown uncovers its role in breast cancer cell migration, invasion, and metastasis. Expert Opin Ther Targets. 2016;20(9):1035-43. doi: 10.1080/14728222.2016.1195059. PMID:27232533. [DOI] [PubMed] [Google Scholar]
- 72.Wu K, Weng Z, Tao Q, Lin G, Wu X, Qian H, Zhang Y, Ding X, Jiang Y, Shi YE. Stage-specific expression of breast cancer-specific gene gamma-synuclein. Cancer Epidemiol Biomarkers Prev. 2003;12(9):920-25. PMID:14504205. [PubMed] [Google Scholar]
- 73.Kim G, Ouzounova M, Quraishi AA, Davis A, Tawakkol N, Clouthier SG, Malik F, Paulson AK, D'Angelo RC, Korkaya S, et al.. SOCS3-mediated regulation of inflammatory cytokines in PTEN and p53 inactivated triple negative breast cancer model. Oncogene. 2015;34(6):671-80. doi: 10.1038/onc.2014.4. PMID:24531711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, Ju HL, Yuan XY, Wang TJ, Lai BQ. SOX4 is a potential prognostic factor in human cancers: A systematic review and meta-analysis. Clin Transl Oncol. 2016;18(1):65-72. doi: 10.1007/s12094-015-1337-4. PMID:26250764. [DOI] [PubMed] [Google Scholar]
- 75.Glait-Santar C, Benayahu D. Regulation of SVEP1 gene expression by 17beta-estradiol and TNFalpha in pre-osteoblastic and mammary adenocarcinoma cells. J Steroid Biochem Mol Biol. 2012;130(1–2):36-44. doi: 10.1016/j.jsbmb.2011.12.015. PMID:22265959. [DOI] [PubMed] [Google Scholar]
- 76.Liu XL, Xiao K, Xue B, Yang D, Lei Z, Shan Y, Zhang HT. Dual role of TGFBR3 in bladder cancer. Oncol Rep. 2013;30(3):1301-08. doi: 10.3892/or.2013.2599. PMID:23835618. [DOI] [PubMed] [Google Scholar]
- 77.Chinnam M, Wang Y, Zhang X, Gold DL, Khoury T, Nikitin AY, Foster BA, Li Y, Bshara W, Morrison CD, et al.. The Thoc1 ribonucleoprotein and prostate cancer progression. J Natl Cancer Inst. 2014;106(11):dju306. doi: 10.1093/jnci/dju306. PMID:25296641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, et al.. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013;4:e867. doi: 10.1038/cddis.2013.400. PMID:24136231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dwyer DF, Barrett NA, Austen KF, Immunological Genome Project C . Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol. 2016;17(7):878-87. doi: 10.1038/ni.3445. PMID:27135604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116-21. doi: 10.1073/pnas.091062498. PMID:11309499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor J, Tibshirani R, Efron B. The ‘miss rate’ for the analysis of gene expression data. Biostatistics. 2005;6(1):111-17. doi: 10.1093/biostatistics/kxh021. PMID:15618531. [DOI] [PubMed] [Google Scholar]
- 82.Zhou T, Wang T, Garcia JG. Expression of nicotinamide phosphoribosyltransferase-influenced genes predicts recurrence-free survival in lung and breast cancers. Sci Rep. 2014;4:6107. doi: 10.1038/srep06107. PMID:25146220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang R, Gurguis CI, Gu W, Ko EA, Lim I, Bang H, Zhou T, Ko JH. Ion channel gene expression predicts survival in glioma patients. Sci Rep. 2015;5:11593. doi: 10.1038/srep11593. PMID:26235283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou T, Wang T, Garcia JG. Genes influenced by the non-muscle isoform of Myosin light chain kinase impact human cancer prognosis. PLoS One. 2014;9(4):e94325. doi: 10.1371/journal.pone.0094325. PMID:24714365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


