Abstract
The vertebrate-specific ESCC microRNA family arises from two genetic loci in mammals: miR-290/miR-371 and miR-302. The miR-302 locus is found broadly among vertebrates, whereas the miR-290/miR-371 locus is unique to eutheria, suggesting a role in placental development. Here, we evaluate that role. A knock-in reporter for the mouse miR-290 cluster is expressed throughout the embryo until gastrulation, when it becomes specifically expressed in extraembryonic tissues and the germline. In the placenta, expression is limited to the trophoblast lineage, where it remains highly expressed until birth. Deletion of the miR-290 cluster gene (Mirc5) results in reduced trophoblast progenitor cell proliferation and a reduced DNA content in endoreduplicating trophoblast giant cells. The resulting placenta is reduced in size. In addition, the vascular labyrinth is disorganized, with thickening of the maternal-fetal blood barrier and an associated reduction in diffusion. Multiple mRNA targets of the miR-290 cluster microRNAs are upregulated. These data uncover a crucial function for the miR-290 cluster in the regulation of a network of genes required for placental development, suggesting a central role for these microRNAs in the evolution of placental mammals.
KEY WORDS: miR-290, miR-371, microRNA, Placenta, Placentation, Trophoblast, Mouse
Highlighted Article: The miR-290 cluster plays multiple roles in placental development and function, and ultimately in embryonic survival, including regulating the mitotic division of trophoblast progenitors and endoreduplication of trophoblast giant cells.
INTRODUCTION
The placenta is a highly specialized organ that supports the normal growth and development of the embryo during pregnancy. Growth and function of the placenta are precisely regulated and coordinated, ensuring that the exchange of nutrients, gases and waste products between the maternal and fetal circulatory systems operates at maximal efficiency (Gude et al., 2004). Any inadequacy in placental function can lead to a wide range of pregnancy complications such as preeclampsia, intrauterine growth restriction (IUGR) and abruption (Chaddha et al., 2004). Both intrauterine growth restriction, which is defined as failure of an embryo to reach its growth potential (Dessi et al., 2012), and preeclampsia, which is the new onset of hypertension and significant proteinuria in a previously healthy woman after the twentieth week of gestation (Eiland et al., 2012), are among the most common conditions contributing to increased perinatal morbidity and mortality (Moran et al., 2015). These diseases are thought to predominantly arise from defects in placental development and function, but the mechanisms underlying these defects are largely unknown.
Given the diverse roles of microRNAs (miRNAs) in mammalian development, they are likely candidates to play a role in the growth and function of the placenta (Fu et al., 2013). A particularly interesting group of miRNAs is the mouse miR-290 cluster located on chromosome 7 and its human ortholog, the miR-371 cluster located on chromosome 19. Transcription of the miR-290 cluster is controlled by a super-enhancer that is bound by the core pluripotency transcription factors Oct4 (Pou5f1), Sox2 and Nanog in embryonic stem cells (Hnisz et al., 2013, 2015; Marson et al., 2008). Transcription of the cluster produces a long noncoding RNA that is then processed in the nucleus by the Microprocessor to produce seven hairpins. Following transport of these hairpins to the cytoplasm, they are further processed by Dicer to produce eleven mature miRNA species (Fig. S1A). These miRNA species can be grouped into four families based on shared seed sequences (Fig. S1A) (Houbaviy et al., 2005). Each miRNA binds and post-transcriptionally suppresses multiple mRNAs through partial base pairing with target sites in the 3′UTRs of the mRNAs. The seed sequence plays a central role in this pairing, and thus miRNAs within a family have overlapping targets (Bartel, 2009). One of the miRNA families within the miR-290 cluster comprises the embryonic stem cell-enriched cell cycle (ESCC) miRNAs, which have been shown to promote pluripotency in vitro (Greve et al., 2013). Much less is known about the other families in the cluster. Loss of the miR-290 cluster results in germ cell defects in mice (Medeiros et al., 2011). While the role of the miR-290 cluster in placental development has not been previously studied, its evolution is tightly correlated with the emergence of placental mammals (eutheria) (Houbaviy et al., 2005; Wu et al., 2014).
Given the evolutionary association between the emergence of the miR-290 cluster and placental mammals along with the known role of the ESCC miRNA family in pluripotency, we aimed to dissect the expression and requirement for this cluster in mouse placental development using a knock-in miR-290 cluster co-expressing reporter and a miR-290 knockout line (Fig. S1B,C).
RESULTS
The miR-290 cluster is uniquely expressed in extraembryonic tissues from E8.5 to birth
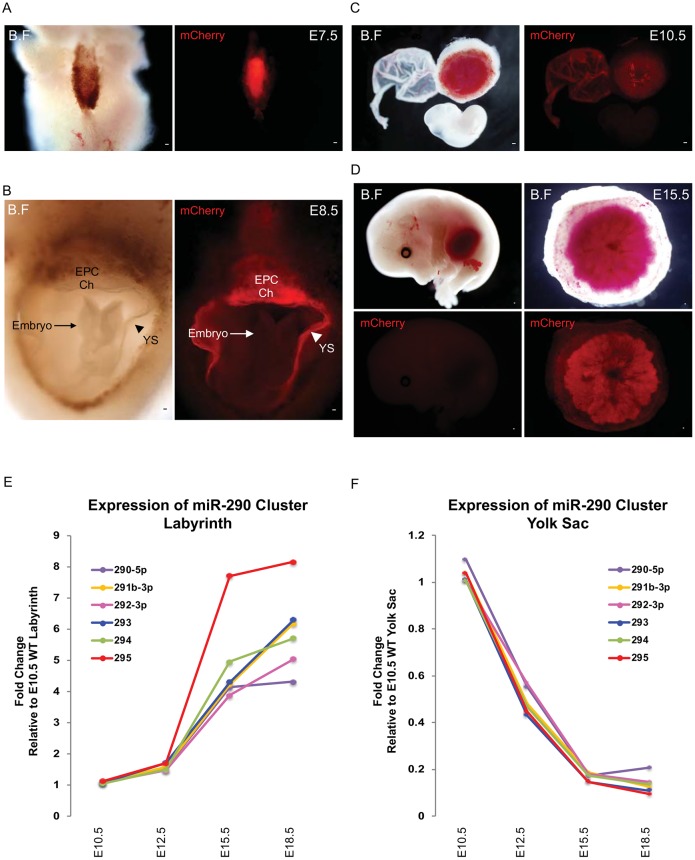
Given the evolutionary relationship between placentation and the emergence of the miR-290 cluster (Wu et al., 2014), we evaluated miR-290 cluster expression throughout mouse placental development using the miR-290-mCherry reporter (Parchem et al., 2014). As previously described, the miR-290 cluster was broadly expressed throughout the embryo from E2.5 to E6.5, but then began to diminish in the embryo proper at E7.5 (Parchem et al., 2014) (Fig. 1A). By E8.5, little to no expression was seen in the embryo proper and it remained absent throughout the rest of embryonic development (Fig. 1B-D, Fig. S1D-G). By contrast, expression remained high in extraembryonic tissues, including the yolk sac and placenta (Fig. 1B-D, Fig. S1D-G). Interestingly, qRT-PCR of mature miRNAs arising from the miR-290 cluster showed opposing expression patterns in the yolk sac and placenta, starting at E10.5 and extending throughout the remainder of development (Fig. 1E,F, Fig. S1H,I). Expression of the miRNAs increased within the placenta reaching maximal levels at birth, whereas they decreased in the yolk sac over time reaching minimal levels at birth. This switch coincides with the transition for the primary site of nutrient/waste transfer from the yolk sac to the placenta (Jollie, 1990; Zohn and Sarkar, 2010). This expression pattern of the miR-290 cluster is consistent with these miRNAs playing a central role in placental development.
Fig. 1.
miR-290 cluster expression becomes localized to extraembryonic tissues following gastrulation. (A) At E7.5, the miR-290-mCherry reporter (red) is expressed in both embryonic and extraembryonic tissue. (B) At E8.5, the miR-290 mCherry reporter (red) is strongly expressed in yolk sac, chorion and ectoplacental cone, but not in the embryo. (C,D) At E10.5 and E15.5, miR-290 mCherry reporter (red) continues to be expressed in placenta and yolk sac but not in the embryo. (E,F) qRT-PCR results showing that miR-290 cluster expression changes in yolk sac and placental labyrinth at different time points, normalized to Sno202. B.F, bright field; EPC, ectoplacental cone; Ch, chorion; YS, yolk sac. Scale bars: 100 μm.
The miR-290 cluster is expressed in syncytiotrophoblast cells and trophoblast giant cells throughout placental development
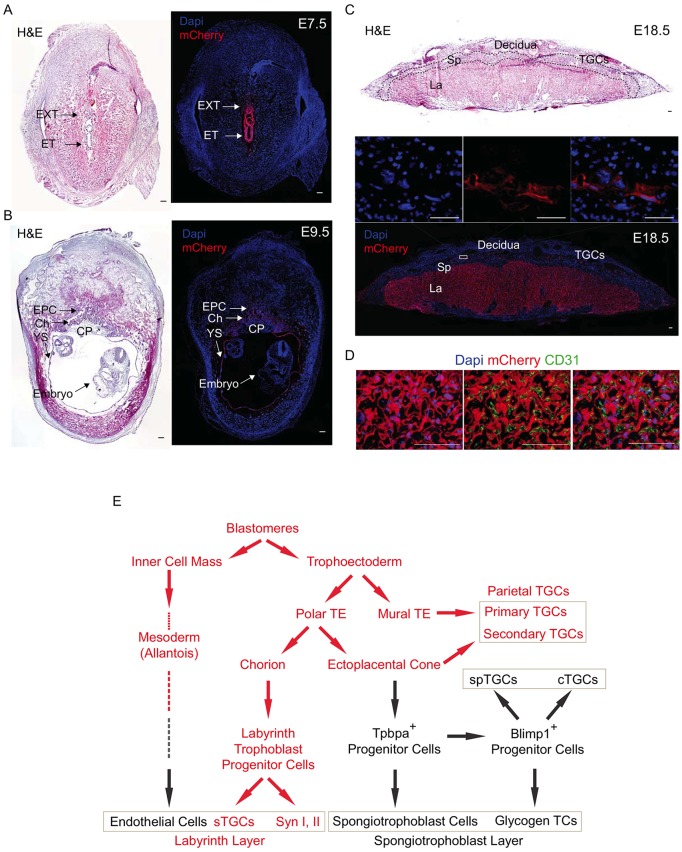
To gain an understanding of the ontogeny of cells expressing the miR-290 cluster during extraembryonic development, we performed detailed immunohistochemical analyses from E7.5 to E18.5 (Fig. 2, Fig. S2). Placental development starts with formation of the trophectoderm at E2.5, which becomes separated into the mural and polar trophectoderm with formation of the blastocoel at E3.5. The polar trophectoderm expands to form the inner chorion and outer ectoplacental cone (Gasperowicz and Natale, 2011). The miR-290 cluster was expressed in all of these cells through E9.5, although expression appeared to be slightly reduced in cells of the ectoplacental cone (Fig. 2A,B,E, Fig. S2A).
Fig. 2.
miR-290 cluster expression becomes localized to the trophoblast cells of the labyrinth and parietal TGC layers of the placenta. (A,B) H&E and immunofluorescent staining for miR-290-mCherry reporter at E7.5 and E9.5. At E7.5, the reporter is expressed strongly in extraembryonic tissue and also in embryonic tissue, whereas at E9.5 it is only expressed in extraembryonic tissues. (C) H&E and immunofluorescent staining for mCherry reporter of fully mature E18.5 placenta. It is expressed in labyrinth and parietal TGCs but not in the spongiotrophoblast cell layer. The boxed region is magnified above, showing single channels and merge. (D) The miR-290 cluster is expressed in trophoblast-derived cells of the labyrinth, whereas allantois-derived CD31+ endothelial cells are negative miR-290 cluster expression. DAPI (left), CD31 (middle) and merge (right) are shown with mCherry. (E) Ontogeny of cells expressing the miR-290 cluster during extraembryonic tissue formation. Red denotes expression of the miR-290 cluster in that cellular compartment. EXT, extraembryonic tissue; ET, embryonic tissue; EPC, ectoplacental cone; Ch, chorion; YS, yolk sac; CP, chorionic plate; La, labyrinth; Sp, spongiotrophoblast layer; TGCs, trophoblast giant cells; TE, trophectoderm; spTGCs, spiral artery TGCs; cTGCs, canal TGC; TCs, trophoblast cells; sTGCs, sinusoidal TGCs; Syn, syncytiotrophoblast cells. Scale bars: 100 μm.
With further development, the chorion develops into the labyrinth, the main site of maternal-fetal exchange, while the ectoplacental cone develops into the spongiotrophoblast layer, which is the site of nutrient storage (Malassine et al., 2003; Simmons and Cross, 2005). The miR-290 cluster was silenced within the spongiotrophoblast layer (Fig. 2C, Fig. S2B,C). In contrast, it remained highly expressed in the labyrinth throughout placental development (Fig. 2C, Fig. S2B,C). The labyrinth itself forms a thin four-cell layer barrier between maternal and fetal blood circulations consisting of sinusoidal TGCs, two layers of syncytiotrophoblasts, and fetal endothelium (Coan et al., 2005). Double staining for mCherry and the endothelial markers CD31 (Pecam1) and laminin showed that, within the labyrinth, all three trophoblast layers are miR-290 positive, whereas the mesoderm-derived fetal endothelial cells are miR-290 negative (Fig. 2D, Fig. S2D,E). Expression of the miR-290 cluster following formation of the labyrinth and spongiotrophoblast layers contrasted with expression of another marker, Tpbpa. Tpbpa became specifically localized in the spongiotrophoblast layer at the same time that the miR-290 cluster became localized in the labyrinth layer (Fig. S2C).
In addition to the labyrinth and spongiotrophoblast layers, there are the parietal TGCs that attach the embryo to the uterine lining or decidua. These cells are derived from both the mural trophectoderm (primary parietal TGCs) and the leading edge of the ectoplacental cone (secondary parietal TGCs) (Simmons and Cross, 2005; Simmons et al., 2007). Co-staining with placental lactogen-I (PL1; also known as Prl3d1), which is expressed in parietal TGCs (Faria et al., 1991), showed expression of the miR-290 cluster in these cells (Fig. S2F,G). The miR-290 cluster was also expressed in the yolk sac, specifically in the primitive endoderm layer, but not the mesoderm-derived layer (Fig. S2H). Together, these findings further detail the ontogeny of the cells that form the mammalian placenta (Hu, and Cross, 2011; Malassine et al., 2003; Mould et al., 2012; Simmons and Cross, 2005; Simmons et al., 2007; Ueno et al., 2013), as summarized in Fig. 2E.
miR-290 cluster knockout embryos show late embryonic lethality associated with defects in placenta growth
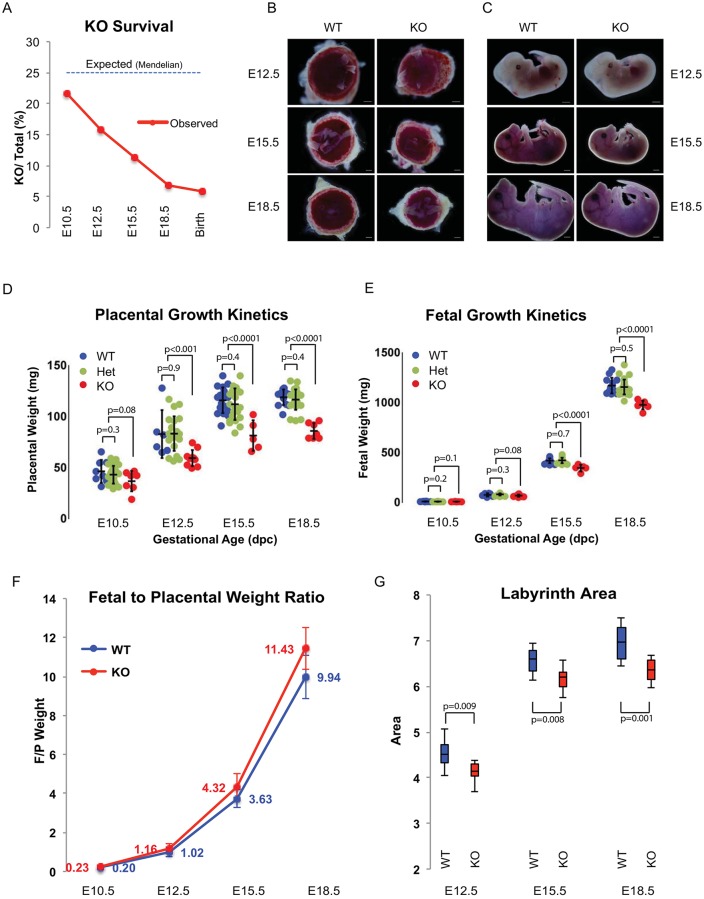
Given the striking expression pattern in, and evolutionary relationship to, the placenta, we next asked whether the miRNA cluster is essential for placental development. The miR-290 cluster knockout has a germline defect (Medeiros et al., 2011), the germline being another site of miR-290 cluster expression (Fig. S3A). In addition, the mice show a non-fully penetrant embryonic lethal phenotype, with most of the embryos dying late in development (Medeiros et al., 2011). Consistent with these previous findings, we found very few miR-290 cluster gene (Mirc5) homozygous knockout pups (7/120 births). Embryo loss was most evident starting at E10.5, with progressive loss throughout the remainder of embryonic development (Fig. 3A). Heterozygous animals showed no discernable lethality (Fig. S3B,C).
Fig. 3.
A decrease in placenta size precedes reduction in embryo size in miR-290 cluster knockouts. (A) Survival curve for miR-290 cluster knockout (KO) embryos during development determined as a ratio to all embryos (heterozygous mating). n>60 for each time point. (B,C) Bright-field images comparing the gross morphology of wild-type and miR-290 cluster knockout embryos and placentas at different developmental stages after mid-gestation. (D,E) Quantification of wild-type and miR-290 cluster knockout placenta and fetal weights. Error bars represent s.d. Line within the bars denotes mean. (F) Fetal-to-placental weight ratio during the second half of pregnancy. n>30 for each time point. Error bars represent mean±s.d. (G) Quantification of labyrinth area in wild-type and miR-290 cluster knockout placentas. n>3 placentas for each time point, and at least three sections per placenta. Upper and lower whiskers represent max and min, and middle line denotes the median. Scale bar: 1 mm.
Visual inspection of knockout versus heterozygous/wild-type embryos at E12.5, E15.5 and E18.5 suggested growth defects in both the placenta and embryo of the knockout animals (Fig. 3B,C). Careful measurements of the placenta and embryo showed a decrease in placental weight preceding that of the embryo. At E10.5 and E12.5, the mean weights of the knockout placentas were 83% and 72% of wild types, respectively. By contrast, no obvious difference was observed in the embryos. However, starting at E15.5, significant changes were also observed in the embryo (Fig. 3D,E, Fig. S3C). The difference in the fetal-to-placental weight ratio continued to increase throughout development, showing that the phenotype continued to be more severe in the placenta (Fig. 3F). These decreases in placental and embryonic size were not secondary to developmental delay as key morphological milestones including limb bud development, CNS development and closure of the abdominal midline all occurred at a similar time as in their wild-type and heterozygous counterparts (Fig. S3D,E). Area measurements of the labyrinth and spongiotrophoblast layers showed a more severe decline in the labyrinth layer, consistent with this being the site of miR-290 cluster expression (Fig. 3G, Fig. S3F,G). Together, these data strongly support a primary defect in the placenta, especially the labyrinth, associated with embryonic loss late in development.
miR-290 cluster knockout trophoblast progenitor cells exit the cell cycle prematurely and TGCs show reduced endoreduplication
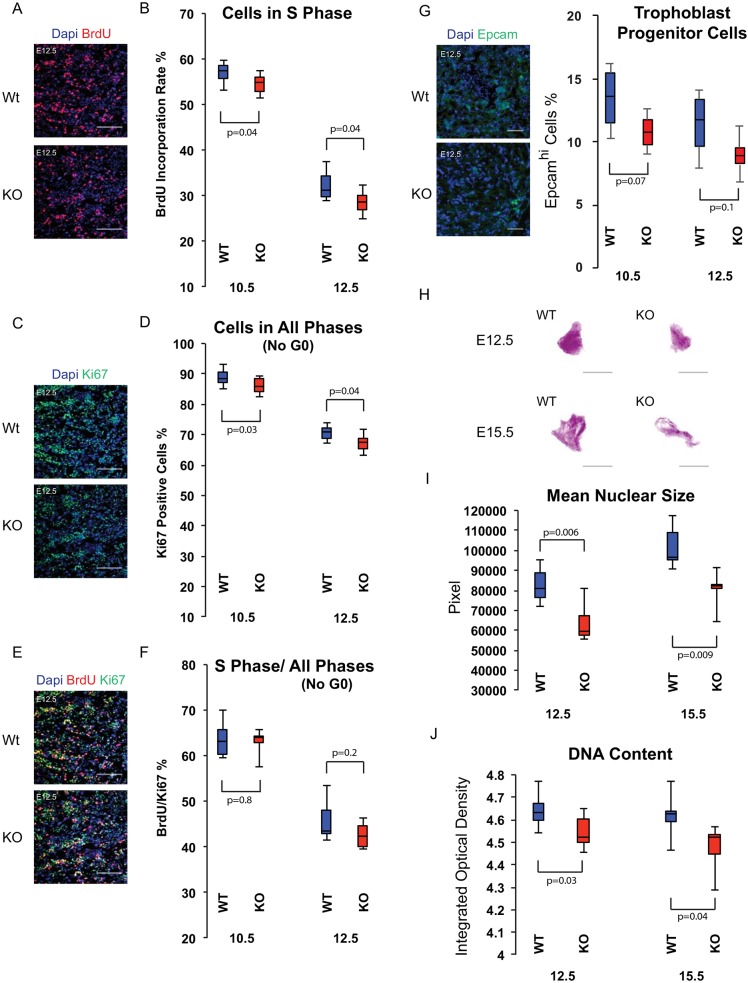
The increasing size of the wild-type placenta is regulated by a combination of trophoblast progenitor cell division and trophoblast cell growth (Ueno et al., 2013). Cell division primarily occurs in the ectoplacental cone and then later in the labyrinth, rapidly diminishing after E12.5, when placental growth largely depends on cell growth rather than on cell division (Iguchi et al., 1993). To determine the basis of the reduced size of the knockout placenta, we measured the number of cells in the cell cycle (Ki67) and passing through S phase (BrdU) at the height of placental cell division (E10.5 and E12.5). Changes in cell cycle rate are expected to show a change in the BrdU/Ki67 ratio, while exit from cell cycle is associated with the simultaneous loss of both markers (Farhy et al., 2013; Gonsalvez et al., 2013; Qu et al., 2013). Knockout placentas showed a small but reproducible reduction in the number of BrdU+ and Ki67+ cells, with the BrdU/Ki67 ratio remaining constant (Fig. 4A-F). These data suggest premature cell cycle exit and loss of progenitor cells. To directly measure the number of progenitor cells, we counted cells expressing high levels of Epcam, a marker for trophoblast progenitor cells in the labyrinth (Ueno et al., 2013). The fraction of Epcamhi cells was reduced in the knockout placenta at both E10.5 and E12.5, although variability in counts reduced the significance of these differences (Fig. 4G). Cleaved caspase 3 staining showed only extremely rare positive cells in the labyrinths of wild-type and knockout placentas (Fig. S4A). Therefore, the observed reduction in dividing progenitors was not secondary to apoptosis.
Fig. 4.
Labyrinth trophoblast progenitor cells exit the cell cycle prematurely and parietal TGCs show reduced endoreduplication in miR-290 cluster knockouts. (A) BrdU incorporation staining of E12.5 labyrinth. (B) Percentage of BrdU+ cells of the labyrinth in wild-type and miR-290 cluster knockout placentas. (C) Ki67 staining of E12.5 labyrinth. (D) Percentage of Ki67+ cells of the labyrinth in wild-type and miR-290 cluster knockout placentas. (E) BrdU and Ki67 co-staining of E12.5 labyrinth. (F) BrdU+/Ki67+ cell ratio in wild-type and miR-290 cluster knockout placentas. (B,D,F) n>3 placentas for each time point, at least three sections per placenta; cells are counted in at least 20 high-power fields (HPFs), randomly selected, in each section. (G) Epcam staining of E12.5 labyrinth. Epcamhi cells are arranged in clusters in labyrinths of wild-type and miR-290 cluster knockout placenta. n>3 placentas for each time point, at least three sections per placenta; cells are counted in at least 20 fields (20× objective lens), randomly selected, in each section. (H) Feulgen staining of parietal TGCs of wild-type and miR-290 cluster knockout placentas at E12.5 and E15.5. (I,J) Size and integrated optical density of parietal TGCs in wild-type and miR-290 cluster knockout placentas. n>3 placentas for each time point, at least three sections per placenta. All upper and lower whiskers represent max and min, and all middle lines denote median. Scale bars: 100 μm.
Another major source of growth of the placenta is the endoreduplication of TGCs. Parietal TGCs form particularly high DNA content nuclei associated with their overall large size (Hu and Cross, 2010; Sakaue-Sawano et al., 2013). We evaluated their DNA content in wild-type and miR-290 cluster knockout placentas using Feulgen DNA staining along with DNA image cytometry (Biesterfeld et al., 2011; Hardie et al., 2002). There was a significant reduction in both size and optical density in the knockout nuclei, consistent with a reduction in DNA content (Fig. 4H-J).
Together, these data show a crucial role for the miR-290 cluster in the cell cycle both in maintaining mitotic divisions of trophoblast progenitor cells and the endoreduplication of TGCs.
The labyrinth of miR-290 cluster knockout placenta is disorganized, with a reduced area of vasculature
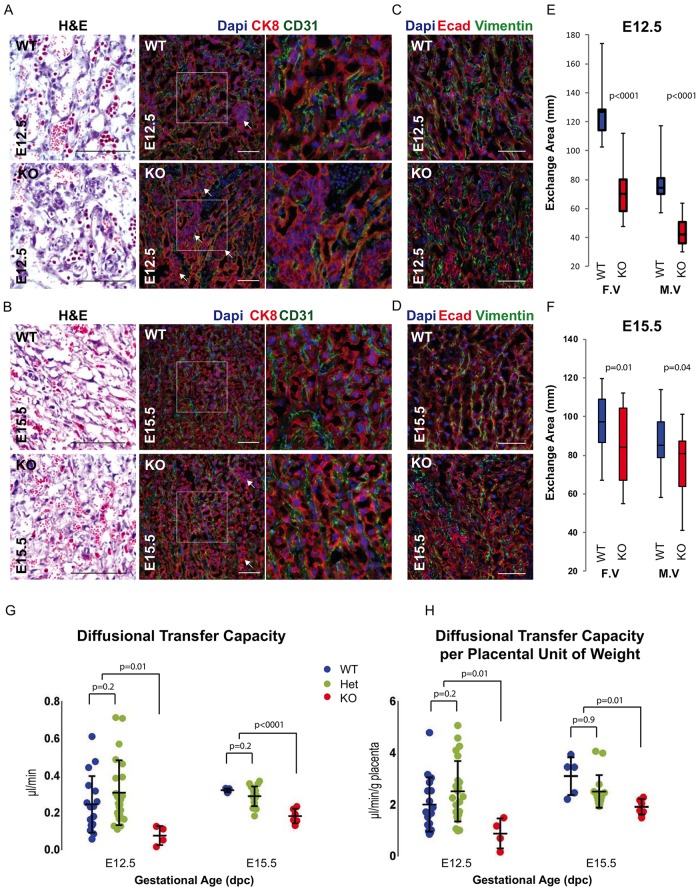
A key function of the placenta is nutrient, gas and waste exchange between fetal and maternal blood. This exchange occurs within the labyrinth across a relatively thin barrier consisting of the fetal endothelium, two layers of syncytiotrophoblast cells, and the sinusoidal TGCs, which are in direct contact with maternal blood. This thin barrier is essential for normal exchange (Rossant and Cross, 2001; Watson and Cross, 2005). Histological analysis of the labyrinth suggested disorganization, with thickening of the barrier in the knockout versus wild-type placenta. To better evaluate this phenotype, we stained with antibodies to cytokeratin 8 (CK8)/E-cadherin (trophoblast layers) and CD31/vimentin (endothelium). The knockout placentas had areas of trophoblast cell disorganization, with cells piled up leading to barrier thickening and resulting in increased intervascular space (Fig. 5A-D, Fig. S4B). Furthermore, measurements of fetal and maternal vasculature surface areas were significantly reduced in the labyrinths of the knockout placentas (Fig. 5E,F). Together, these data suggest a second phenotype in which the placenta is not only reduced in size but also, due to epithelial disorganization, the exchange surface between maternal and fetal blood is further reduced.
Fig. 5.
miR-290 cluster knockout labyrinth is disorganized and has reduced exchange area and diffusion capacity. (A,B) Staining of fetal and maternal vasculature in E12.5 and E15.5 labyrinths. CD31 stains endothelial cells surrounding fetal vasculature. CK8 stains the epithelial cells surrounding the maternal vasculature. Arrows indicate cell aggregates between vessels. The boxed region is magnified to the right. (C,D) Staining of fetal and maternal vasculature in E12.5 and E15.5 labyrinths. Vimentin stains endothelial cells surrounding fetal vasculature. E-cadherin stains the epithelial cells surrounding the maternal vasculature. (E,F) Quantification of exchange surface area in labyrinth of E12.5 and E15.5 placentas. n>3 placentas for each time point, at least five sections per placenta. Upper and lower whiskers represent max and min, and middle line denotes median. F.V, fetal vasculature; M.V, maternal vasculature. (G) Quantification of diffusion capacity of placenta as measured by transfer of 51Cr-EDTA from maternal blood to embryo. (H) Diffusion capacity normalized to placental weight. (G,H) Error bars represent s.d., and line within the bars denotes mean. dpc, days post coitus. Scale bars: 100 μm.
Diffusional exchange capacity is reduced in the miR-290 cluster knockout placenta
Reduced exchange surface area is expected to result in reduced diffusion between maternal and fetal blood. To study the directional maternal-fetal diffusional transfer capacity, we measured fetal accumulation of 51Cr-EDTA after its injection into the maternal circulation of pregnant mice at E12.5 and E15.5 (Constância et al., 2002; Sibley et al., 2004). Knockout embryos showed a significant reduction in the diffusional transfer capacity of the placenta compared with wild-type/heterozygous animals (27% and 61% of the mean of wild-type/heterozygous animals for E12.5 and E15.5, respectively; Fig. 5G). This decrease could be entirely due to a reduction in exchange area owing to the reduced size of the placenta. Therefore, to normalize the impact of the reduced placental size, we calculated the diffusional transfer capacity per unit of placental weight. This calculation still showed a reduction in the knockout placentas (38% and 72% of the mean of wild-type/heterozygous animals for E12.5 and E15.5, respectively; Fig. 5H). These data reveal a reduction in solute exchange between the mother and embryo due to both the reduced size of the maternal-fetal vasculature interface and the reduced efficiency of exchange across that interface.
Multiple targets are upregulated in miR-290 cluster knockout placenta
To explore the molecular basis underlying the miR-290 cluster placental defect, we analyzed and compared the transcript profile of wild-type and knockout placentas using RNA-sequencing (RNA-Seq) technology. As miRNAs negatively regulate both translation and mRNA stability, the abundance of mRNA targets should be increased in the miR-290 cluster knockout background. Although the gross phenotype of the knockout placentas is beginning to be evident at E12.5, the molecular defects underlying this outcome are likely to precede this event. Therefore, we performed RNA-seq on wild-type and knockout placentas at E10.5.
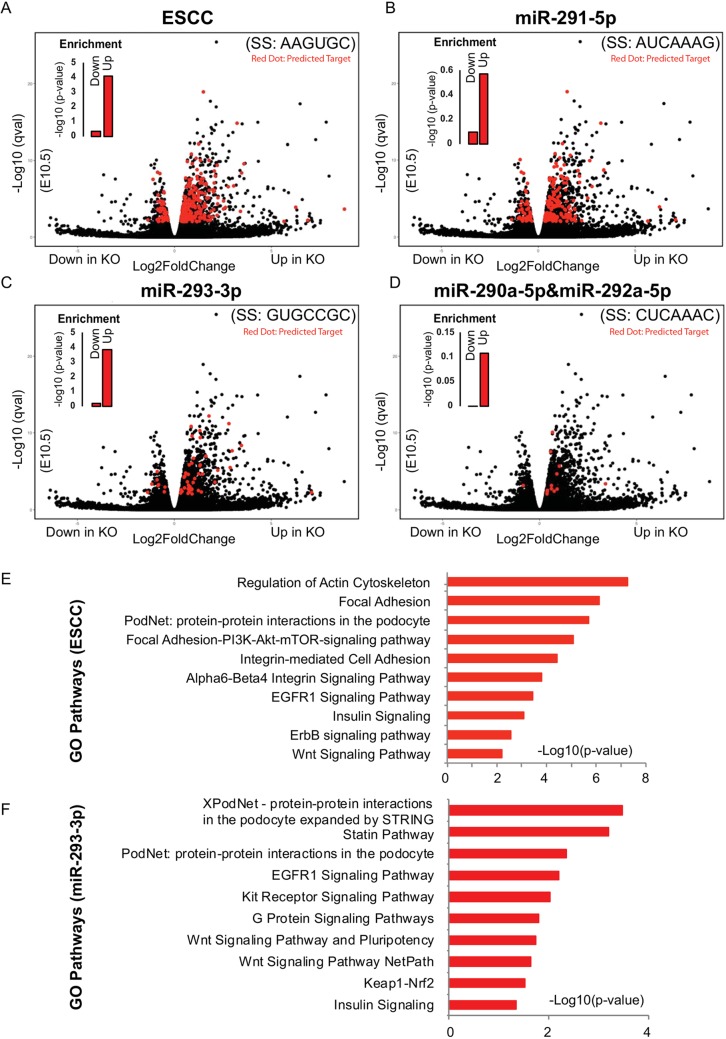
The results showed that hundreds of genes were significantly upregulated or downregulated. There was a highly significant enrichment among the upregulated genes for targets of the ESCC and miR-293-3p, but not the miR-291-5p and miR-290a-5p, seed families (Fig. 6A-D, Fig. S4C). The expression of eight representative genes that were upregulated and had seed sequence matches to the miR-290 cluster were validated by qRT-PCR (Fig. S4D). Gene ontology (GO) analysis on the upregulated ESCC and miR-293-3p targets showed enrichment for signaling and cell adhesion (Fig. 6E,F, Fig. S4E). Given the multiple targets per seed, it is unlikely that any single target is responsible for the observed placental phenotypes. Thus, these data suggest that the miRNAs of the miR-290 cluster, especially the ESCC and miR-293-3p miRNAs, regulate a network of genes that are responsible for the normal development of the mammalian placenta.
Fig. 6.
Many miR-290 cluster targets are upregulated in the knockout placental labyrinth at E10.5. (A-D) RNA-Seq results. Volcano plots showing fold change and significance values for the four distinct seed families produced by the miR-290 cluster. Targets for each seed family, as predicted by TargetScan, are highlighted in red. Inset bar charts show enrichment P-values for miRNA targets among the upregulated and downregulated genes. (E,F) Top ten GO terms identified by Enrichr for genes that are upregulated and have a seed match to the ESCC (E) or miR-293-3p (F) family. SS, seed sequence; Up, upregulated target; Down, downregulated targets.
DISCUSSION
The data presented here uncover a crucial role for a eutheria-specific miRNA cluster, miR-290/miR-371, in placental development. This process requires carefully coordinated growth of multiple cell types, which must parallel that of the embryo. The miR-290 cluster is expressed in all cells of the conceptus prior to gastrulation (Parchem et al., 2014), but then becomes localized to the germline and extraembryonic tissues including the yolk sac and developing placenta. Within the developing placenta, the miR-290 cluster becomes localized to the trophoblast cells of the labyrinth and parietal TGC layers. In general, there is a dearth of markers defining the different cells of the placenta and, to our knowledge, no other marker shows a similar expression pattern to the miR-290 cluster. Therefore, the miR-290 cluster provides new insights into the ontogeny of these cells. In particular, the expression pattern suggests an important bifurcation occurring within the ectoplacental cone, with one arm giving rise to TGCs of the parietal layer, while the other arm gives rise to the spongiotrophoblast cells, glycogen trophoblast cells, canal TGCs, and spiral artery TGCs. Future development of an inducible lineage-tracing model based on the miR-290 locus would enable further in-depth characterization of the timing and origin of these cell fate choices.
Our functional data complement the expression data. In particular, we uncover defects in trophoblast progenitor cell proliferation during early stages of placenta development followed by defects in endoreduplication in the later stages. These two defects correspond to the two major means of placental growth, with cell proliferation acting early versus cell growth being the major source late (post E12.5) (Iguchi et al., 1993; Ueno et al., 2013). In embryonic stem cells, the miR-290 cluster, specifically the ESCC miRNAs of this cluster, suppress the G1-S restriction point of the cell cycle resulting in a rapid G1 transition (Wang et al., 2008, 2013). By contrast, loss of the miR-290 cluster in the placenta does not appear to alter the structure of the cell cycle, but rather induces premature exit from the cell cycle as observed by the reduction in the number of Ki67+ cells. The combination of exit from cell cycle, decreased progenitor number, and no apparent increase in apoptosis is consistent with a premature differentiation of the progenitor cells. This phenotype is reminiscent of the miR-290 cluster paralog, miR-302, which is specifically expressed in the embryo proper. Loss of miR-302 results in premature differentiation of the neural epithelium first into progenitors and then into neurons, resulting in a failure to close the neural tube (Parchem et al., 2015). Therefore, a common role for these two miRNA clusters appears to be the regulation of the developmental timing of cell fate decisions.
The decrease in the number of cells in the cell cycle was specific to progenitor cells of the labyrinth. Notably, there was no significant change in the proliferation rate of cells in the spongiotrophoblast layer (Fig. S4F), consistent with the absence of miR-290 expression in this layer. However, there was a decrease in the DNA content of endoreduplicating TGCs at the periphery of the placenta, the parietal TGCs, which do express the miR-290 cluster. This reduction is once again likely to be secondary to premature exit from the cell cycle, albeit not a mitotic cell cycle. This phenotype could be another form of premature differentiation, although additional markers would be required to confirm such an interpretation. Of note, in primates there is another large cluster of miRNAs on chromosome 19 (C19MC), which is likely to have originated from the miR-290/miR-371 cluster (Morales-Prieto et al., 2012; Zhang et al., 2008). It is highly expressed in the placenta in the first trimester. Whether the cellular pattern of expression of this cluster and its function in the placenta are similar to those of the miR-290/miR-371 clusters is unknown.
Unlike the miR-290 cluster and its human ortholog miR-371, which are specific to placental mammals, miR-302 and its ortholog are found across vertebrate species. The miR-302 and miR-290 clusters are related in their production of the ESCC miRNAs, but they also express non-ESCC miRNAs that are not shared between the two loci (Gruber et al., 2014; Gu et al., 2016). Therefore, the miR-290 and miR-302 clusters not only differ in the timing and localization of expression, but also in the repertoire of miRNAs they produce. Our expression analysis of the miR-290 cluster knockout placenta at E10.5 shows a strong enrichment for predicted targets of the ESCC and miR-293-3p families (common seed) of the miR-290 cluster. However, for the other two families (miR-291-5p and miR-290a-5p/292a-5p), this enrichment is not significant, suggesting a less important role for these miRNAs. Separation of the roles for the different miRNAs will require intracluster deletions that remove one family at a time, similar to what has been done for the miR-17 cluster (Han et al., 2015).
Although it is unlikely that any single target is responsible for the observed placental phenotypes, considering the multiple seed sequences and multiple targets per seed, a number of the differentially expressed genes found by RNA-Seq are known to be involved in placental development and function. For example, CYR61 is essential for placental development and vascular integrity (Mo et al., 2002). Eomes is also required for mouse trophoblast development (Chen et al., 2013b; Russ et al., 2000). FOSL1 is a key downstream effector of the PI3K/AKT signaling pathway, which is responsible for development of trophoblast lineages integral to establishing the maternal-fetal interface (Kent et al., 2011). Insulin-like growth factor binding protein 1 and 2 regulate fetal growth (Jin et al., 2016; Madeleneau et al., 2015; Nawathe et al., 2016). In addition, gene set enrichment and pathway analysis revealed dysregulation of several pathways known to be involved in placenta development and function. For example, the Wnt signaling pathway is involved in implantation, trophoblast invasion and differentiation (Sonderegger et al., 2010). The epidermal growth factor receptor (EGFR) signaling and PI3K-Akt-mTOR pathways regulate trophoblast proliferation (Dackor et al., 2007; Ferretti et al., 2007; Pollheimer and Knöfler, 2005). By coordinating the activity of these genes and pathways, the miR-290 cluster might be striking the important balance between proliferation and differentiation required for the growth and function of the placenta.
The miR-290 knockout embryos were lost during a broad developmental window. Given that many conserved miRNAs are dispensable for animal development or viability (Vidigal and Ventura, 2015) and deletions of various miRNAs have been shown to confer partially penetrant phenotypes (Kuhnert et al., 2008; Li et al., 2006; Zhao et al., 2007), it has been suggested that miRNAs aid in conferring robustness to biological processes and suppressing random fluctuations in transcript copy number (Ebert and Sharp, 2012). Interestingly, expression analysis of later stage placentas showed many fewer transcriptional changes (Table S2). This might be due to increased noise and dilution of the primary molecular effects at later stages. Alternatively, knockout placentas with the most drastic changes in gene expression patterns might have arrested and been resorbed earlier, whereas mutants with gene expression patterns closer to those of their wild-type counterparts are likely to have survived to these later time points. Indeed, the later stage knockout embryos showed less severe differences in a number of measures than earlier embryos. In this regard, acute conditional knockouts at these later time points will be required to uncover specific molecular and phenotypic roles for the miR-290 cluster at different stages of placental development.
Although we see a significant impact of loss of the miR-290 cluster on placental size, with a likely secondary defect in fetal size, it is unclear how this defect relates to the demise of the majority of knockout animals prior to birth. Prior studies on other mutants suggest that the size reduction that we see here is not alone sufficient to result in embryonic death (Constância et al., 2002). Histological analysis also uncovered disorganization and thickening of the barrier between the maternal and fetal blood. Furthermore, physiological measurements showed a reduction of diffusion across this barrier even after normalization for the changes in placental size. This additional defect in maternal-fetal transport is likely to add to risk of fetal demise. However, it is likely that other processes are disrupted as well, which will require further physiological studies. One potential area of interest is defects in TGC secretion given the reduction in their endoreduplication. For example, the parietal TGCs are responsible for invasion/decidualization of the uterus as well as secretion of endocrine and paracrine factors. These factors, including steroid hormones and prolactin-related cytokines, play essential roles in pregnancy (Simmons et al., 2007).
In conclusion, our work establishes a crucial role for a eutheria-specific miRNA cluster in placental development and establishes a new model to dissect the ontogeny and distinct roles of the many cell types found in the placenta, an area currently understudied. In future experiments, it will be important to determine if the human orthologs of these miRNAs play a similar role in directing the timing of trophoblast cell fate decisions. If this is the case, their dysregulated expression could be involved in pregnancy complications that are associated with fetal growth restriction secondary to defects in placental structure and function.
MATERIALS AND METHODS
Husbandry and genotyping
Construction of miR-290-mCherry reporter was previously reported (Parchem et al., 2014). miR-290 heterozygous knockout mice were purchased from The Jackson Laboratories (B6;129S4-Mirc5tm1Jae/J). Reporter and heterozygous mice were maintained on a C57BL/6NCrSim background (N>10). Heterozygous mice were interbred to produce a combination of wild-type, heterozygous and knockout embryos. Genomic DNA was isolated from toes of postnatal mice or tails of embryos. Tissue was digested in lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 100 mM NaCl, 0.1% SDS, 5 mg/ml proteinase K) overnight at 55°C. DNA was isolated by the addition of an equal volume of isopropanol, gentle vortexing, and then a 15 min centrifugation (10,000 g). Isopropanol was removed, and samples were allowed to air dry. After the addition of water, samples were mixed and heated at 85°C for 5 min. PCR was performed using primer pairs (5′-3′, forward and reverse) to distinguish the miR-290 wild type (TCCAGGTTTCCTTCAGGTTG and GATGGCCGCTACATAGGTGT) and mutant (TCCAGGTTTCCTTCAGGTTG and CGTGCAATCCATCTTGTTCA). PCR conditions were 35 cycles at 94°C for 45 s, 52°C for 45 s, and 72°C for 60 s. Band sizes were: wild type, 391 bp; mutant, 697 bp. miR-290 mCherry primers were: wild type (GGTCTAGGGAGTCTATGCAG and CGGAGCCCTCCATGTGCA) and reporter (GGTCTAGGGAGTCTATGCAG and GGAAAGAACGTGGAGAAC). PCR conditions were as above except with an adjustment of the annealing temperature to step down 64°C, 62°C and 60°C. Band sizes were: wild type, 169 bp; mCherry, 219 bp.
Histopathology and immunohistochemistry
Tissue was collected from mice of various genotypes as described previously (Belair et al., 2015). Briefly, placentas and embryos were dissected in PBS, fixed in 4% paraformaldehyde (PFA) in PBS at 4°C overnight, dehydrated in 10%, 20% and 30% sucrose at 4°C, embedded in OCT (Thermo Scientific, Tissue-Plus O.C.T Compound) and stored at −80°C until use. Cryomicrotome sectioning was at 7-10 μm. Sections were stored at −80°C prior to Hematoxylin and Eosin (H&E) staining and immunohistochemistry. For H&E, briefly: 5 min in Hematoxylin (Sigma), 2 min wash (tap water), 1 min in Eosin Y (Richard Allan Scientific), 1 min wash (tap water) and dehydration through 80%, 90%, 95% and 100% ethanol (2 min each) before 30 min in Histo-Clear (National Diagnostics, HS-200) and mounting. For BrdU staining, 50 mg BrdU/kg body weight was injected 2 h prior to dissecting the mouse. BrdU-labeled cryosections were then steamed in 10 mM citrate buffer (pH 6) for 20 min at 99°C for antigen retrieval. Sections were blocked with 5% goat serum in PBS/0.1% Tween 20, and incubated in primary antibody in blocking solution at 4°C overnight: BrdU, 1:1000 (Abcam, ab6326); CD31, 1:50 (Abcam, ab28364); mCherry, 1:500 (Abcam, ab167453); CK8, 1:100 (Abcam, ab107115); vimentin, 1:100 (Cell Signaling, 5741s); Ki67, 1:500 (Thermo Scientific, RM-9106-S1); cleaved caspase 3, 1:400 (Cell Signaling, 9664s); laminin, 1:300 (Abcam, ab11575); Epcam, 1:250 (Abcam, ab71916); PL1, 1:100 (Santa Cruz, sc34713); E-cadherin, 1:200 (BD Biosciences, 610181). Secondary antibodies (AlexaFluor conjugated; Invitrogen, 1:500) in blocking buffer were applied for 2 h at room temperature.
Quantitative analysis of histological sections
To quantify BrdU and Ki67 expression, placentas of at least four biological replicates for each time point, and at least three sections for each placenta, were stained simultaneously for BrdU, Ki67 and with DAPI. The cells were evaluated and counted in at least 20 high-magnification fields (40× objective lens), randomly selected, in each section, blinded with respect to genotype.
To quantify Epcam expression, placentas of at least four biological replicates for each time point, and at least three sections for each placenta, were stained simultaneously for Epcam and with DAPI. The cells were evaluated and counted in at least 20 fields (20× objective lens), randomly selected, in each section, blinded with respect to genotype.
To measure the thickness of the spongiotrophoblast and labyrinth layers, we made serial sagittal sections through the entire placental tissue, mounting four sections per slide. We then stained every tenth slide with H&E. Slides were examined in order to find the midpoint of the placenta (site of umbilical cord attachment to placenta), which is used as the major reference point for comparisons between mutants and wild-type littermates (Natale et al., 2006). These layers have distinct morphological features and can be distinguished even in H&E-stained sections. Then, we used ImageJ software (NIH) to measure the labyrinth and spongiotrophoblast layer areas, blinded with respect to genotype (Jensen, 2013).
To measure the exchange surface and intervascular area in labyrinth, we performed immunofluorescence for CD31 and CK8, staining the epithelial and trophoblast cells, respectively. CD31 demarcates the fetal vessels, whereas CK8 demarcates the maternal vessels. CellProfiler (Jones et al., 2008) was used to measure the vascular perimeter (exchange surface) for both the fetal and maternal vessels as well as the intervascular area.
Feulgen staining and image analysis densitometry
Feulgen staining was performed as previously described (Hardie et al., 2002). Briefly, slides were thawed, fixed in 4% PFA for 10 min at room temperature and rinsed for 10 min in tepid running tap water. For hydrolysis, tissue sections were incubated in 55°C preheated 1 M HCL and, after staining by Schiff reagent (Sigma Aldrich, 1001579456), incubated in bisulfite solution (Sigma Aldrich, 1001521877) for 15 min at room temperature, rinsed in tepid running tap water, dried and mounted. Histological images were acquired on a Leica DM1000 (100× objective) with a DFC290 camera. CellProfiler software was used to measure nuclear area and integrated optical density.
Quantitative RT-PCR (qRT-PCR)
The labyrinth and yolk sac were isolated from E10.5, E12.5, E15.5 and E18.5 wild-type and mutant embryos. The labyrinth tissue was enriched by physically peeling off the decidua and junctional zone under a dissecting microscope. The isolated tissues were lysed in 1 ml Trizol (Invitrogen, 15596018) and RNA isolated and purified according to the manufacturer's instructions and stored at −80°C. cDNA was generated for miRNA and mRNA analyses by reverse transcription with oligo(dT) primers (SuperScript III Reverse Transcriptase kit, Invitrogen). Gene-specific primers (500 nM) and Power SYBR Green PCR Master Mix (Life Technologies) were used. PCR quality controls, experimental runs, and statistical methods were performed as described (Shi and Chiang, 2005). Primers are listed in Table S1.
Analysis of unidirectional maternal-fetal transfer
As previously described (Constância et al., 2002; Sibley et al., 2004), radiolabeled 51Cr-EDTA (50 mCi) in 100 ml PBS was injected at E15.5 into the jugular veins of miR-290 heterozygous female mice bred with heterozygous males. The females were sacrificed 4 min after injection of radioisotope. Embryos and placentas were weighed and a small section of tail was removed for genotyping. Embryos were lysed overnight at 55°C in Biosol (National Diagnostics, LS310). Then, liquid scintillation fluid (Bioscint, National Diagnostics, LS309) was added for γ-counting. Radioactive counts in each embryo were then used to calculate the amount of radioisotope transferred per whole placenta or per gram of placenta.
RNA-Seq library preparation
The labyrinth tissue was enriched as described above. Total RNA was isolated from four paired biological samples of dissected placentas using miRNeasy micro columns (Qiagen) according to the manufacturer's protocol. cDNA libraries were prepared using a KAPA Stranded mRNA-Seq Kit (07962142001, KK8400).
Statistical analysis
For small-scale experiments performed in three or more independent experiments, P-values were calculated using Student's t-test.
For RNA-Seq analysis the data were preprocessed using Kallisto (Bray et al., 2016). Differential expression analysis was performed using Sleuth (https://pachterlab.github.io/sleuth). Cutoffs for significance were set at an adjusted P<0.01, log2 fold change >0.3. Seed sequence enrichment was performed using Fisher's exact test (fisher.test function in R). GO terms were identified using Enrichr (Chen et al., 2013a). A list of all differentially expressed genes with fold change and significance value is provided in Tables S2 and S3. Raw, processed, fold change and significance values for all genes are provided at Gene Expression Omnibus (GSE95687).
Animal use
The Institutional Animal Care and Use Committee of the University of California, San Francisco, approved all animal experiments reported in this article.
Supplementary Material
Acknowledgements
We thank members of the University of California – San Francisco National Center of Translational Research in Reproduction and Infertility for their insightful comments throughout the design, execution and publication of this project. A special thanks to Drs Linda Giudice, Marco Conti and Susan Fisher.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.P., R.B.; Methodology: A.P., R.B.; Validation: A.P.; Formal analysis: A.P., C.B.; Investigation: A.P., D.S.; Resources: R.B.; Data curation: D.S.; Writing - original draft: A.P.; Writing - review & editing: A.P., R.B.; Visualization: A.P., C.B.; Supervision: R.B.; Project administration: R.B.; Funding acquisition: R.B.
Funding
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (P50HD055764). Deposited in PMC for release after 12 months.
Data availability
RNA-Seq data are available at Gene Expression Omnibus under accession number GSE95687.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.151654.supplemental
References
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belair C. D., Paikari A., Moltzahn F., Shenoy A., Yau C., Dall'Era M., Simko J., Benz C. and Blelloch R. (2015). DGCR8 is essential for tumor progression following PTEN loss in the prostate. EMBO Rep. 16, 1219-1232. 10.15252/embr.201439925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesterfeld S., Beckers S., Del Carmen Villa Cadenas M. and Schramm M. (2011). Feulgen staining remains the gold standard for precise DNA image cytometry. Anticancer Res. 31, 53-58. [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P. and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnol. 34, 525-527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Chaddha V., Viero S., Huppertz B. and Kingdom J. (2004). Developmental biology of the placenta and the origins of placental insufficiency. Semin. Fetal. Neonatal. Med. 9, 357-369. 10.1016/j.siny.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R. and Ma'ayan A. (2013a). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang K., Gong Y. G., Khoo S. K. and Leach R. (2013b). Roles of CDX2 and EOMES in human induced trophoblast progenitor cells. Biochem. Biophys. Res. Commun. 431, 197-202. 10.1016/j.bbrc.2012.12.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan P. M., Ferguson-Smith A. C. and Burton G. J. (2005). Ultrastructural changes in the interhaemal membrane and junctional zone of the murine chorioallantoic placenta across gestation. J. Anat. 207, 783-796. 10.1111/j.1469-7580.2005.00488.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constância M., Hemberger M., Hughes J., Dean W., Ferguson-Smith A., Fundele R., Stewart F., Kelsey G., Fowden A., Sibley C. et al. (2002). Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945-948. 10.1038/nature00819 [DOI] [PubMed] [Google Scholar]
- Dackor J., Strunk K. E., Wehmeyer M. M. and Threadgill D. W. (2007). Altered trophoblast proliferation is insufficient to account for placental dysfunction in Egfr null embryos. Placenta 28, 1211-1218. 10.1016/j.placenta.2007.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessi A., Ottonello G. and Fanos V. (2012). Physiopathology of intrauterine growth retardation: from classic data to metabolomics. J. Matern. Fetal Neonatal. Med. 25, 13-18. 10.3109/14767058.2012.714639 [DOI] [PubMed] [Google Scholar]
- Ebert M. S. and Sharp P. A. (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515-524. 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiland E., Nzerue C. and Faulkner M. (2012). Preeclampsia 2012. J. Pregnancy 2012, 586578 10.1155/2012/586578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy C., Elgart M., Shapira Z., Oron-Karni V., Yaron O., Menuchin Y., Rechavi G. and Ashery-Padan R. (2013). Pax6 is required for normal cell-cycle exit and the differentiation kinetics of retinal progenitor cells. PLoS ONE 8, e76489 10.1371/journal.pone.0076489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria T. N., Ogren L., Talamantes F., Linzer D. I. H. and Soares M. J. (1991). Localization of placental lactogen-I in trophoblast giant cells of the mouse placenta. Biol. Reprod. 44, 327-331. 10.1095/biolreprod44.2.327 [DOI] [PubMed] [Google Scholar]
- Ferretti C., Bruni L., Dangles-Marie V., Pecking A. P. and Bellet D. (2007). Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 13, 121-141. 10.1093/humupd/dml048 [DOI] [PubMed] [Google Scholar]
- Fu G., Brkić J., Hayder H. and Peng C. (2013). MicroRNAs in human placental development and pregnancy complications. Int. J. Mol. Sci. 14, 5519-5544. 10.3390/ijms14035519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperowicz M. and Natale D. R. C. (2011). Establishing three blastocyst lineages--then what? Biol. Reprod. 84, 621-630. 10.1095/biolreprod.110.085209 [DOI] [PubMed] [Google Scholar]
- Gonsalvez D. G., Cane K. N., Landman K. A., Enomoto H., Young H. M. and Anderson C. R. (2013). Proliferation and cell cycle dynamics in the developing stellate ganglion. J. Neurosci. 33, 5969-5979. 10.1523/JNEUROSCI.4350-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve T. S., Judson R. L. and Blelloch R. (2013). microRNA control of mouse and human pluripotent stem cell behavior. Annu. Rev. Cell Dev. Biol. 29, 213-239. 10.1146/annurev-cellbio-101512-122343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. J., Grandy W. A., Balwierz P. J., Dimitrova Y. A., Pachkov M., Ciaudo C., Nimwegen E. and Zavolan M. (2014). Embryonic stem cell-specific microRNAs contribute to pluripotency by inhibiting regulators of multiple differentiation pathways. Nucleic Acids Res. 42, 9313-9326. 10.1093/nar/gku544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K. L., Zhang Q., Yan Y., Li T.-T., Duan F.-F., Hao J., Wang X.-W., Shi M., Wu D.-R., Guo W. T. et al. (2016). Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 26, 350-366. 10.1038/cr.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gude N. M., Roberts C. T., Kalionis B. and King R. G. (2004). Growth and function of the normal human placenta. Thromb. Res. 114, 397-407. 10.1016/j.thromres.2004.06.038 [DOI] [PubMed] [Google Scholar]
- Han Y.-C., Vidigal J. A., Mu P., Yao E., Singh I., González A. J., Concepcion C. P., Bonetti C., Ogrodowski P., Carver B. et al. (2015). An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat. Genet. 47, 766-775. 10.1038/ng.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D. C., Gregory T. R. and Hebert P. D. N. (2002). From pixels to picograms: a beginners’ guide to genome quantification by Feulgen image analysis densitometry. J. Histochem. Cytochem. 50, 735-749. 10.1177/002215540205000601 [DOI] [PubMed] [Google Scholar]
- Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A. and Young R. A. (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934-947. 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Schuijers J., Lin C. Y., Weintraub A. S., Abraham B. J., Lee T. I., Bradner J. E. and Young R. A. (2015). Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362-370. 10.1016/j.molcel.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H. B., Dennis L., Jaenisch R. and Sharp P. A. (2005). Characterization of a highly variable eutherian microRNA gene. RNA 11, 1245-1257. 10.1261/rna.2890305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D. and Cross J. C. (2010). Development and function of trophoblast giant cells in the rodent placenta. Int. J. Dev. Biol. 54, 341-354. 10.1387/ijdb.082768dh [DOI] [PubMed] [Google Scholar]
- Hu D. and Cross J. C. (2011). Ablation of Tpbpa-positive trophoblast precursors leads to defects in maternal spiral artery remodeling in the mouse placenta. Dev. Biol. 358, 231-239. 10.1016/j.ydbio.2011.07.036 [DOI] [PubMed] [Google Scholar]
- Iguchi T., Tani N., Sato T., Fukatsu N. and Ohta Y. (1993). Developmental changes in mouse placental cells from several stages of pregnancy in vivo and in vitro. Biol. Reprod. 48, 188-196. 10.1095/biolreprod48.1.188 [DOI] [PubMed] [Google Scholar]
- Jensen E. C. (2013). Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec. 296, 378-381. 10.1002/ar.22641 [DOI] [PubMed] [Google Scholar]
- Jin M., Lv P.-P., Yu T.-T., Shen J.-M., Feng C. and Huang H.-F. (2016). IGFBP1 involved in the decreased birth weight due to fetal high estrogen exposure in mice. Biol. Reprod. 95, 96 10.1095/biolreprod.116.141242 [DOI] [PubMed] [Google Scholar]
- Jollie W. P. (1990). Development, morphology, and function of the yolk-sac placenta of laboratory rodents. Teratology 41, 361-381. 10.1002/tera.1420410403 [DOI] [PubMed] [Google Scholar]
- Jones T. R., Kang I. H., Wheeler D. B., Lindquist R. A., Papallo A., Sabatini D. M., Golland P. and Carpenter A. E. (2008). CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9, 482 10.1186/1471-2105-9-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L. N., Rumi M. A. K., Kubota K., Lee D.-S. and Soares M. J. (2011). FOSL1 is integral to establishing the maternal-fetal interface. Mol. Cell. Biol. 31, 4801-4813. 10.1128/MCB.05780-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F., Mancuso M. R., Hampton J., Stankunas K., Asano T., Chen C.-Z. and Kuo C. J. (2008). Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135, 3989-3993. 10.1242/dev.029736 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.-A. and Gao F.-B. (2006). MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20, 2793-2805. 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeleneau D., Buffat C., Mondon F., Grimault H., Rigourd V., Tsatsaris V., Letourneur F., Vaiman D., Barbaux S. and Gascoin G. (2015). Transcriptomic analysis of human placenta in intrauterine growth restriction. Pediatr. Res. 77, 799-807. 10.1038/pr.2015.40 [DOI] [PubMed] [Google Scholar]
- Malassine A., Frendo J.-L. and Evain-Brion D. (2003). A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 9, 531-539. 10.1093/humupd/dmg043 [DOI] [PubMed] [Google Scholar]
- Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J. et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521-533. 10.1016/j.cell.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros L. A., Dennis L. M., Gill M. E., Houbaviy H., Markoulaki S., Fu D., White A. C., Kirak O., Sharp P. A., Page D. C. et al. (2011). Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA 108, 14163-14168. 10.1073/pnas.1111241108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo F.-E., Muntean A. G., Chen C.-C., Stolz D. B., Watkins S. C. and Lau L. F. (2002). CYR61 (CCN1) is essential for placental development and vascular integrity. Mol. Cell. Biol. 22, 8709-8720. 10.1128/MCB.22.24.8709-8720.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Prieto D. M., Chaiwangyen W., Ospina-Prieto S., Schneider U., Herrmann J., Gruhn B. and Markert U. R. (2012). MicroRNA expression profiles of trophoblastic cells. Placenta 33, 725-734. 10.1016/j.placenta.2012.05.009 [DOI] [PubMed] [Google Scholar]
- Moran M. C., Mulcahy C., Zombori G., Ryan J., Downey P. and McAuliffe F. M. (2015). Placental volume, vasculature and calcification in pregnancies complicated by pre-eclampsia and intra-uterine growth restriction. Eur. J. Obstet. Gynecol. Reprod. Biol. 195, 12-17. 10.1016/j.ejogrb.2015.07.023 [DOI] [PubMed] [Google Scholar]
- Mould A., Morgan M. A., Li L., Bikoff E. K. and Robertson E. J. (2012). Blimp1/Prdm1 governs terminal differentiation of endovascular trophoblast giant cells and defines multipotent progenitors in the developing placenta. Genes Dev. 26, 2063-2074. 10.1101/gad.199828.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D. R., Starovic M. and Cross J. C. (2006). Phenotypic analysis of the mouse placenta. Methods Mol. Med. 121, 275-293. [DOI] [PubMed] [Google Scholar]
- Nawathe A. R., Christian M., Kim S. H., Johnson M., Savvidou M. D. and Terzidou V. (2016). Insulin-like growth factor axis in pregnancies affected by fetal growth disorders. Clin. Epigenetics 8, 11 10.1186/s13148-016-0178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R. J., Ye J., Judson R. L., LaRussa M. F., Krishnakumar R., Blelloch A., Oldham M. C. and Blelloch R. (2014). Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell 14, 617-631. 10.1016/j.stem.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R. J., Moore N., Fish J. L., Parchem J. G., Braga T. T., Shenoy A., Oldham M. C., Rubenstein J. L. R., Schneider R. A. and Blelloch R. (2015). miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 12, 760-773. 10.1016/j.celrep.2015.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollheimer J. and Knöfler M. (2005). Signalling pathways regulating the invasive differentiation of human trophoblasts: a review. Placenta 26 Suppl. A, S21-S30. 10.1016/j.placenta.2004.11.013 [DOI] [PubMed] [Google Scholar]
- Qu Q., Sun G., Murai K., Ye P., Li W., Asuelime G., Cheung Y.-T. and Shi Y. (2013). Wnt7a regulates multiple steps of neurogenesis. Mol. Cell. Biol. 33, 2551-2559. 10.1128/MCB.00325-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. and Cross J. C. (2001). Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2, 538-548. 10.1038/35080570 [DOI] [PubMed] [Google Scholar]
- Russ A. P., Wattler S., Colledge W. H., Aparicio S. A. J. R., Carlton M. B. L., Pearce J. J., Barton S. C., Surani M. A., Ryan K., Nehls M. C. et al. (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95-99. 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Hoshida T., Yo M., Takahashi R., Ohtawa K., Arai T., Takahashi E., Noda S., Miyoshi H. and Miyawaki A. (2013). Visualizing developmentally programmed endoreplication in mammals using ubiquitin oscillators. Development 140, 4624-4632. 10.1242/dev.099226 [DOI] [PubMed] [Google Scholar]
- Shi R. and Chiang V. L. (2005). Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 39, 519-525. 10.2144/000112010 [DOI] [PubMed] [Google Scholar]
- Sibley C. P., Coan P. M., Ferguson-Smith A. C., Dean W., Hughes J., Smith P., Reik W., Burton G. J., Fowden A. L. and Constancia M. (2004). Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc. Natl. Acad. Sci. USA 101, 8204-8208. 10.1073/pnas.0402508101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. G. and Cross J. C. (2005). Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284, 12-24. 10.1016/j.ydbio.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Simmons D. G., Fortier A. L. and Cross J. C. (2007). Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 304, 567-578. 10.1016/j.ydbio.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Sonderegger S., Pollheimer J. and Knöfler M. (2010). Wnt signalling in implantation, decidualisation and placental differentiation--review. Placenta 31, 839-847. 10.1016/j.placenta.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M., Lee L. K., Chhabra A., Kim Y. J., Sasidharan R., Van Handel B., Wang Y., Kamata M., Kamran P., Sereti K.-I. et al. (2013). c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev. Cell 27, 373-386. 10.1016/j.devcel.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidigal J. A. and Ventura A. (2015). The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 25, 137-147. 10.1016/j.tcb.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., Babiarz J. E., Baehner L. and Blelloch R. (2008). Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478-1483. 10.1038/ng.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Melton C., Li Y.-P., Shenoy A., Zhang X.-X., Subramanyam D. and Blelloch R. (2013). miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep. 4, 99-109. 10.1016/j.celrep.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E. D. and Cross J. C. (2005). Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20, 180-193. 10.1152/physiol.00001.2005 [DOI] [PubMed] [Google Scholar]
- Wu S., Aksoy M., Shi J. and Houbaviy H. B. (2014). Evolution of the miR-290-295/miR-371-373 cluster family seed repertoire. PLoS ONE 9, e108519 10.1371/journal.pone.0108519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang Y.-Q. and Su B. (2008). Molecular evolution of a primate-specific microRNA family. Mol. Biol. Evol. 25, 1493-1502. 10.1093/molbev/msn094 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J. F., Li A., Vedantham V., von Drehle M., Muth A. N., Tsuchihashi T., McManus M. T., Schwartz R. J. and Srivastava D. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129, 303-317. 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- Zohn I. E. and Sarkar A. A. (2010). The visceral yolk sac endoderm provides for absorption of nutrients to the embryo during neurulation. Birth Defects Res. A Clin. Mol. Teratol 88, 593-600. 10.1002/bdra.20705 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.