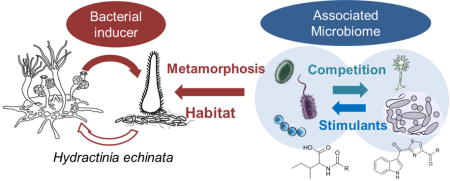
Abstract
Illumina 16S rRNA gene sequencing was used to profile the associated bacterial community of the marine hydroid Hydractinia echinata, a long-standing model system in developmental biology. 56 associated bacteria were isolated and evaluated for their antimicrobial activity. Three strains were selected for further in-depth chemical analysis leading to the identification of 17 natural products. Several γ-Proteobacteria were found to induce settlement of the motile larvae, but only six isolates induced the metamorphosis to the primary polyp stage within 24 h. Our study paves the way to better understand how bacterial partners contribute to protection, homeostasis and propagation of the hydroid polyp.
Keywords: Hydractinia echinate, Signaling molecules, Secondary metabolites, Indoles, Metamorphosis
Graphical abstract
1. Introduction
Marine microbes have an enormous biosynthetic potential to produce structurally diverse metabolites with a broad range of bioactivities, including antimicrobial, antiprotozoan, antiparasitic, and antitumour activities, as well as emulsifying, morphogenic or antifouling activities.1 In particular, marine Actinobacteria are known to be highly prolific sources of novel bioactive compounds,2 such as the potential anticancer drug leads saliniketal A (1)3 and salinosporamide A (2),4 both derived from the genus Salinispora. Marine γ-Proteobacteria, and in particular members of the Pseudoalteromonas genus,5 commonly associated with marine invertebrates, are also known to produce bioactive and potentially defensive secondary metabolites, such as the antifungal compound tambjamine (3),6 or the highly potent antibiotic thiomarinol A (4).7 Furthermore, steadily increasing numbers of genome sequencing projects have uncovered, that bacteria in general and marine host-associated bacteria in particular, habor an enormous biosynthetic capacity, which by far exceeds the numbers of currently reported natural products. However, most gene clusters remain “silent” (cryptic) under standard laboratory cultivation conditions,8 and require the application of optimized heterologous expression systems to enable the identification of the encoded metabolites. Only recently, Moore and co-workers isolated and identified the lipopeptides bromoalterochromide A and B (5–6)9 by transferring a ∼34 kb secondary metabolite pathway from Pseudoalteromonas piscicida JCM 20779 into E. coli as the expression host.
In addition to being prolific producers of natural products important to humankind, most bacterial-derived small molecules are likely to serve as chemical signals for the producing organism and the environment, and are therefore of ecological relevance.10 As an example, the intriguing small molecule tetrabromopyrrole (8) was identified from a host-associated Pseudoalteromonas strain as the first chemical mediator to induce larval metamorphosis of acroporid coral larvae (Acropora millepora).11 However, tetrabromopyrrole (8) did not induce larvae to settle on surfaces, indicating that additional, not yet identified, chemical mediators are likely important to replicate the complete morphogenic event. Later, tetrabromopyrrole (8) was found to induce both events in Caribbean corals, demonstrating that this compound might have widespread importance amongst coral species.12
In another study, Hadfield and co-workers reported that amongst other not yet identified chemical signals,13 phage tail-like bacteriocins derived from Pseudoalteromonas luteoviolacea induce larval settlement in the marine invertebrate Hydroides elegans.14,15 As a third example, Clardy and co-workers elucidated the absolute structure of a rosette-inducing factor (RIF-1, 7) produced by the prey bacterium Algoriphagus machipongonensis of the choanoflagellate Salpingoeca rosetta.16 RIFs induce the transition from the unicellular to the colonial phenotype of the predator S. rosetta, which allows the organism to improve predation.17
Despite the slowly increasing number of identified inter-kingdom signalling molecules, we are still far away from understanding the multidimensional (bio)chemical interactions between hosts und their associated microbes and the natural role of microbial derived natural products.10 To partially fill this knowledge gap, we set out to study the bacterial-induced larval morphogenesis of the marine model organism Hydractinia echinata (Cnidaria), a colony-forming hydrozoan.18 Although its life cycle and cell biology has been studied for decades, the structures of the bacterially produced morphogenic signals remain elusive.
Herein, we describe the first systematic characterization of the associated microbial community of H. echinata using deep 16S rRNA sequencing, the isolation of representative associated microbes and the assessment of their antimicrobial and morphogenic activity. Selected bacterial strains were genome sequenced,19 and analyzed for the production of bioactive secondary metabolites leading to the identification of 17 natural products, several of which have not yet been described from bacterial sources.
2. Results and discussion
2.1 Profiling of the bacterial communities associated with H. echinata using deep 16S rRNA sequencing
H. echinata colonies are mainly found growing on shells inhabited by hermit crabs (North Sea, Atlantic). To characterize the phylogenetic composition of its associated bacterial community, we purchased six freshly collected H. echinata colonies from Woods Hole Oceanographic Institution (Atlantic sea shore, Woods Hole, MA, US) and dissected 20 polyps per hydroid colony. All polyps derived from one colony were pooled and rinsed with sterile seawater to give samples 1 to 6. DNA of each sample was extracted using GenElute™ Bacterial Genomic DNA Kit and Illumina 16S rRNA gene sequencing was used to profile the bacterial community. We chose the V6 hypervariable region of the ribosomal small subunit 16S gene for amplification due to its high sensitivity towards diversity.20,21 The average number of 16S rRNA reads per sample was 86263 (± 44820 SD). Sequences used for analyses had a median length of 72 bp. Retrieved sequences were clustered to operational taxonomic units (OTUs) and classified to bacterial taxa. In total, 3405 unique OTUs were observed and 543 bacterial taxa classified (for details, see Table S1). The bacterial composition of all six H. echinata samples, resolved at the level of bacterial phyla und characteristic classes, is depicted in Figure 2A. The majority of detected 16S rRNA sequences belonged to Flavobacteria (mean relative abundance and SD: 25 ± 11%), α-Proteobacteria (24 ± 5%), γ-Proteobacteria (24 ± 4%) and Cyanobacteria (13 ± 4%). Eight taxa were present in all six samples with abundance above 1%, indicating a potential role of these taxa in symbiosis. These taxa include two members of Flavobacteriaceae (Bacteroidetes; 12.6 ± 6.7% and 7.6 ± 3.1%), a single Rhodobacteraceae (α-Proteobacteria; 6.8 ± 0.9%), Xenococcaceae (Cyanobacteria; 4.1 ± 2.1%) and Flammeovirgaceae (Bacteroidetes; 2.2 ± 0.3%), as well as two unknown α-Proteobacteria (3.9 ± 2.0% and 2.5 ± 0.8%) and one γ-Proteobacterium (1.7 ± 0.6%). Overall, the bacterial community composition of all six samples exhibited only small variations although individual colonies were sampled and analyzed. The results coincide with a global survey of oceans, where members of α- and γ-Proteobacteria, as well as Cyanobacteria were found to be the most abundant bacteria in seawater.22,23 In addition, members of the marine Bacteroidetes (including Flavobacteria, Cytophagia and Saprospiria) are known to colonize surfaces of marine organisms (e.g. algae), due to their ability to degrade a variety of high molecular weight polymers.24
Figure 2.
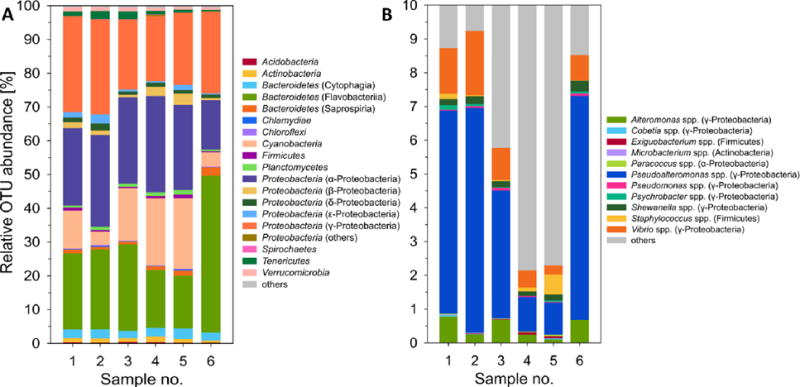
A) Bacterial community structure of the H. echinata associated microbiota showing the relative abundance of OTUs with taxonomic assignment to the level of phyla and characteristic classes. B) Relative OTU abundance of bacterial genera isolated from H. echinata. Data was generated by deep 16S rRNA amplicon sequencing and processed with QIIME.
However, our results are in contrast to other studies of related marine invertebrates, such as the hydroid Ectopleura crocea, which is related to H. echinata at the level of classes, and was found to be dominantly colonized by a single Flavobacteriaceae (Bacteroidetes and a Comamonadaceae (β-Proteobacteria).25
2.2 Profiling of bacterial communities using a culture-dependent approach
We then set out to chemically investigate representative members of the associated microbiome of H. echinata. Again, 20 polyps of each colony were dissected and pooled. The polyps were gently mixed in sterile sea water. After centrifugation, the resulting supernatant was collected and defined as SW-sample. The remaining polyps were then homogenized using a sterile pestle (P-sample). A dilution series of both samples, rinsed water (SW) and homogenized (P), were prepared and plated on MB and SWC agar plates. Plates were incubated at 20 °C for up to 14 days and distinct bacterial morphotypes were purified via subcultivation yielding in total 56 bacterial isolates. To analyze the phylogenetic diversity of our isolates, each strain was cultured in liquid media (SWC) and genomic DNA was extracted using GenElute™ Bacterial Genomic DNA Kit. The 16S rRNA gene was then amplified using the primer set 1492R/27F (accession no.: KY382774-KY382829, Figure 3 and Table S2).26 Subsequent phylogenetic analysis revealed that most bacterial isolates belonged to the Actinobacteria, Firmicutes, and Proteobacteria phylum. On the genus level, the majority of isolates belonged to the well-studied Pseudoalteromonas genus,27 which is known for its diverse secondary metabolite production and morphogenic activity.5,28 In addition, several members of the geographically widely distributed Cobetia,29 Exiguobacterium30 and Psychrobacter31 genera were obtained.
Figure 3.
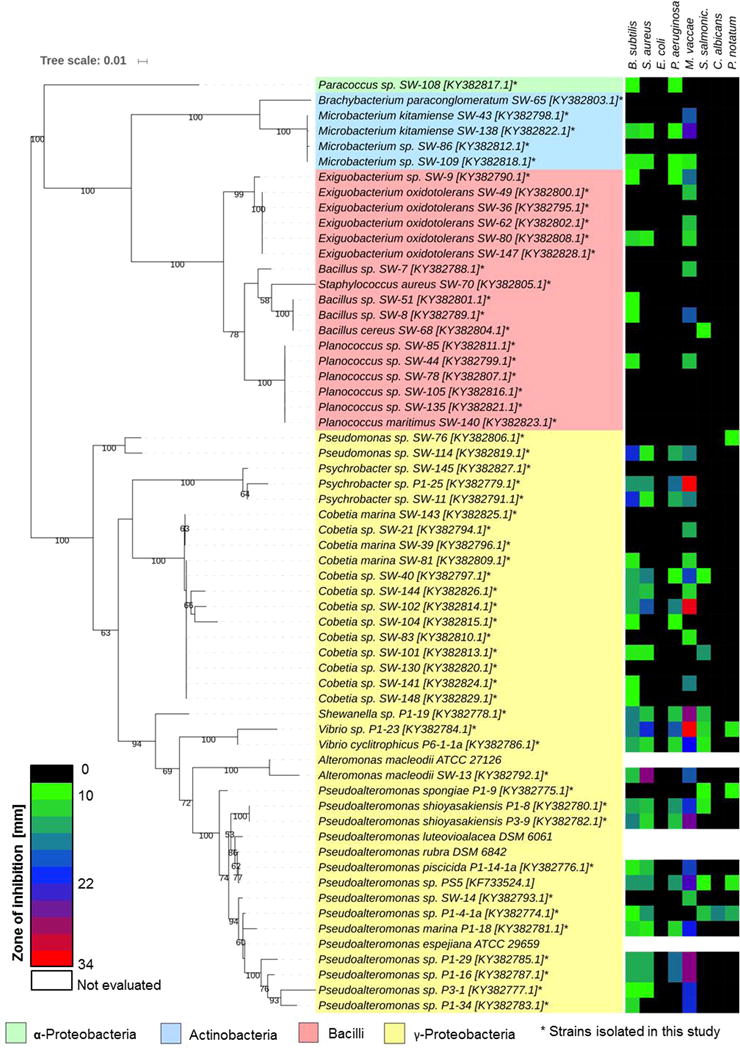
Right: phylogenetic tree based on 16S rRNA gene sequences of bacterial isolates, literature-reported bioactive strain Pseudoalteromonas sp. PS5,12 and phylogenetic-related and commercially available strains Pseudoalteromonas espejiana ATCC 29659, A. macleodii ATCC 27126, P. luteoviolacea DSM6061, P. rubra DSM6842 and P. tunicata DSM14096. Best DNA model was generated and the robustness of interfered tree topologies was evaluated after 1000 bootsraps (> 50% are shown). Left: correlated heatmap showing antimicrobial activities against test strains (zone of inhibition in mm in standardized assay).
We then compared the relative abundances of the isolated bacterial genera retrieved from the 16S deep sequencing data (Figure 2B). Interestingly, γ-proteobacterial Pseudoalteromonas spp. were found with a mean relative abundance of 4.2%; whereas, Vibrio spp. and Altermonas spp. were detected only in 1.0% and 0.5% respectively. Although several Cobetia strains (13 out of 56) were isolated, their isolation frequency was not reflected in the 16S sequencing data (abundance < 0.01%). In general, our results are in accordance with numerous observations that γ-proteobacterial Pseudoalteromonas spp. are frequently isolated from marine invertebrates. Pseudoalteromonas sp. A3, e.g., was isolated from the crustose coralline algae Hydrolithon onkodes and was shown to be responsible for larvae settlement and metamorphosis in the planula of coral Acropora species.32 Several Pseudoalteromonas strains, including P. luteoviolacea, were found to induce larval settlement of the sea urchin Heliocidaris erythrogramma,33 and the tubeworm Hydroides elegans.15 At this point, it’s noteworthy that several Pseudoalteromonas species produce pigments with antimicrobial, anti-fouling and algicidal properties and form phylogenetically distinct clades.34 Furthermore, Cobetia strains have also been investigated for their ability to produce bioactive metabolites, and were reported to secrete biosurfactant hydroxyl fatty acids and bioactive sulfated O-polysaccharides.29
To assess the antimicrobial activity and potential defensive role of our isolates and the reported bioactive strain Pseudoalteromonas sp. PS5,12 all bacterial strains were cultivated in either SWC or MBL broth (30 °C, 160 rpm). Cell-free culture supernatants were submitted to C-18 reverse phase solid phase extraction (C18-SPE) and stock solutions of the obtained SPE fractions were tested in a standardized disc diffusion assay against a panel of human pathogenic bacteria and fungi (Fig. 3 and Table S3).35 As depicted in Figure 3, most isolated γ-Proteobacteria, as well as isolates belonging to the Psychrobacter and Cobetia genus exhibited a broad range of antimicrobial activity. In contrast, non-pigmented Microbacterium isolates SW43 and SW86, as well as isolates belonging to the class Bacilli revealed only minor bioactivities. In a next step, we evaluated the overall encoded biosynthetic capacity of selected isolated genera and performed a web-based bioinformatic analysis (antiSMASH36 and SMIPS37: for details, see Table S4) of our sequenced marine isolates and related deposited genome sequences (data received from NCBI).19
In analogy to previous reports,9 we found that most members of the Pseudoalteromonas genus harbour biosynthetic gene clusters encoding for siderophores, as well as peptides of both ribosomal (NRPS) and non-ribosomal origin (RIPPs). Analyzed Cobetia spp. genomes revealed only a partially conserved T1-PKS cluster, and genes encoding for bacteriocins or other unspecified ribosomally synthesised and post-translationally modified peptide products (RiPPs). But we were unable to detect any PKS or NRPS-derived secondary metabolite gene cluster in the analyzed Psychrobacter genomes, which could be hold accountable for the observed antimicrobial activity.
3. Isolation of bioactive secondary metabolites
3.1 Growth factors from Cobetia sp. SW83
Although our genome survey of reported Cobetia genomes did not reveal secondary metabolite gene clusters, we decided to investigate isolate Cobetia sp. SW83 as it exhibited media-depended pigment production and weak antimicrobial activity. While standard cultivation conditions (SWCL and MBL media) resulted in the secretion of yellow pigments, nutrient-poor conditions (modified marine minimal medium (MMa)) induced the production of pinkish metabolites. Due to low production titers, we prepared a large-scale cultivation of SW83 in SWCL media (50 L, 30 °C, stirring, five days). Subsequent HP20 and SPE-C18 extraction, followed by UV-guided purification yielded two pinkish pigments. Based on the comparative analysis of their 1D NMR, HRMS and UV-Vis spectra (Antibase2014 and SciFinder), the structures of both compounds were assigned as the known coproporphyrin III (9) and zincphyrin (10) (Figure 4A).38 Furthermore, we were able to detect the coenzyme riboflavin and its photolytic product lumichrome (for details, see Figure S15–S18),39 as well as two proline-rich partially modified peptides with the following peptide sequences: Val-Tyr-Pro-Phe-Pro-Gly-Pro-Ile-Pro (11) and Glp-Val-Tyr-Pro-Phe-Pro-Gly-Pro-Ile-Pro (12) (Figure 4B).40 Coproporphyrin III (9) and zincphyrin (10) have been previously proposed to act as growth stimulants for other bacteria, while being toxic to the producing strain Sphingopyxis sp. GF9 at picomolar to nanomolar levels.38 As both compounds are secreted mainly under nutrient-depleted conditions, we speculate that they are likely to be secreted by Cobetia sp. SW83 in the natural environment and influence the bacterial community of H. echinata. In contrast, proline-rich peptides 11 and 12 are a result of the bacterial-induced hydrolysis and modification of the media component casein peptone (Figure 4B). Peptide (11) was just recently isolated from a protein hydrolysate and identified as a potential antioxidant peptide.40
Figure 4.
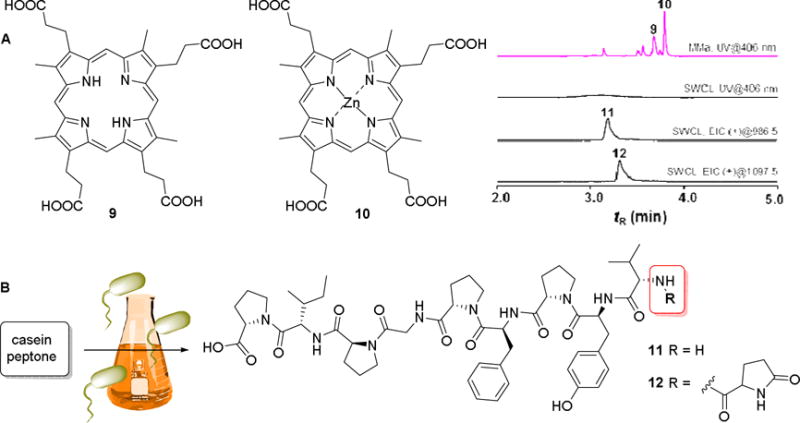
A) Structures of isolated coproporphyrin III (9) and zincphyrin (10) from Cobetia sp. SW83 and comparative UHPLC-MS chromatogram: UV trace of MeOH extract obtained from cultivation in liquid media (MMa and SWCL, 406 nm); EIC (+) mode of peptide 11 and 12. B) Structures of bacterially modified peptides (11) and (12).
3.2 N-acylamino acids from Cobetia sp. SW40
We then analyzed Cobetia sp., because of its promising antimicrobial activity against gram-positive bacteria. SW40 Large scale cultivation (50 L, 3d, SWCL), subsequent HP20 and SPE-C18 extraction and bioassay-guided fractionation resulted in the isolation of several antimicrobial N-acylamino acid derivatives (13–22, Figure 5), which were structurally characterized by comparative NMR and HRMS/MS analysis. Compounds 13 and 14 were obtained as an inseparable 1:3 mixture based on 1H NMR integration and found to have the identical molecular formula of C22H41O3N (ESI-HRMS: m/z 368.3151 [M+H]+, calcd. 368.3159 Δ = –2.17 ppm). LC-HRMS/MS fragmentation showed diagnostic molecular ions at m/z of 132.1018 (Ile and Leu) and 237.2213 (palmitenoyl moiety). Detailed 1D and 2D NMR analysis confirmed the presence of the amino acids leucine and isoleucine, which allowed the assignment of N-palmitenoyl-leucine (13) and N-palmitenoyl-isoleucine (14). According to the chemical shift of the allylic methylene 13C signals (δH 2.03 ppm, δC 28.6 ppm) a Z configuration for the double bond was proposed.41 The absolute configuration of the amino acids (Ile and Leu) was determined by acid hydrolysis and comparative Marfey’s analysis (for details, see Figure S19–S30, Table S5–S6). The hydrolyzed amino acids were assigned as both D/L-leucine and D/L-isoleucine based on retention time and mass spectra of authentic standards, indicating that both compounds (13, 14) are naturally present as racemic mixture.
Figure 5.
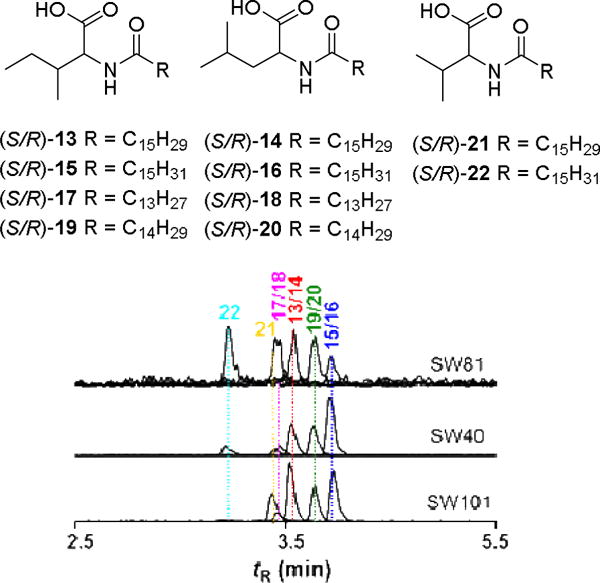
Isolated N-acylamino acids (13–22) from Cobetia sp. SW40; exemplary UHPLC-MS trace of SPE-C18 extracts of Cobetia sp. SW40, SW81 and SW101: EIC (+) mode of 13/14 at m/z 368.1; 15/16 at m/z 370.1; 17/18 at m/z 342.1; 19/20 at m/z 356.1; 21 at m/z 354.1; 22 at m/z 356.1
Structural N-acylamino acid analogs 15–22 were characterized using comparative NMR, HRMS/MS and Marfey analysis (see Supporting Information). We then compared N-acylamino acids production in four different Cobetia strains (Figure 5: SW-81, SW-40 and SW-101) using UHPLC-MS. All strains were found to produce the identified compounds 13–22, but in varying ratios (for details, see also Table S5 and S6). All isolated N-acylamino acids derivatives (13–22) were found to have antimicrobial activity and are therefore most likely accountable for the observed antimicrobial activity of the crude strain extracts (Figure 3).42 Because of the observed species-specific variation of N-acylamino acids production, we further speculate that N-acylamino acid production might be strongly regulated, thereby providing a unique tool for tuning antimicrobial activity in the highly competitive marine environment.
3.3 Indole-thiazole derivatives from Pseudoalteromonas sp. P1-9
In a third study, we analyzed the metabolome of Pseudoalteromonas sp. P1-9, because of its moderately anti-fungal activity and the presence of several genes assigned to NRPS within the genome.13 Strain P1-9 was cultivated on a 50 L scale using modified SWCL medium (stirring, 30 °C). Culture supernatant was extracted using XAD16 resin and the resulting crude extract was fractionated using an established C18-SPE purification protocol. HPLC/UV-Vis based analysis of SPE fractions revealed the presence of two distinct absorption bands (357 nm and 278 nm, Figure 6) indicative for indole-containing metabolites. Subsequent UV-guided semi-preparative HPLC purification led to the isolation of the two indole-thiazoles (23) and (24), as well as pseudoalteromone A (25).43 The structure of compound 23 was assigned based on 1D and 2D NMR analysis, as well as ESI-HRMS (m/z 330.0544 [M+H]+, calcd. 330.0543, C15H11O4N3S) and ESI-MS/MS fragmentation pattern.
Figure 6.
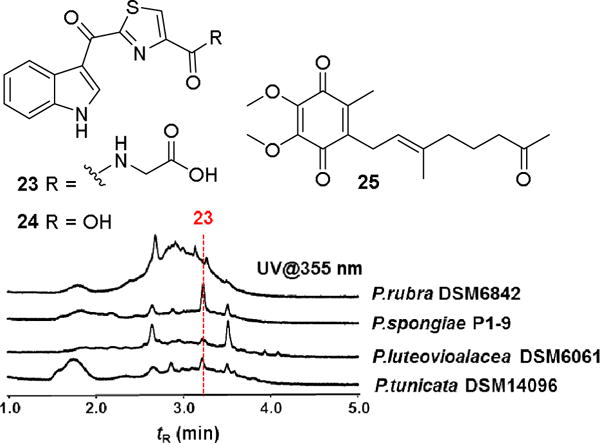
Isolated indole-thiazole derivatives (23, 24) and pseudoalteromone A (25) isolated from Pseudoalteromonas sp. P1-9; HPLC chromatogram (using indole-specific UV absorption (355 nm)) of SPE-C18 extracts from different Pseudoalteromonas strains cultivated in modified SWCL medium.
Diagnostic ion peaks at m/z 144.0446 (C9H6NO+), m/z 255.0226 (C13H7O2N2S+), and m/z 284.0491 (C14H10O2N3S+) were indicative for the indole and thiazole subfragments. Subsequent 1H-NMR analysis revealed four characteristic aromatic proton signals (δH 8.32, 7.27, 7.29 and 7.57) and an exchangeable NH-proton (δH 8.85). 1H-1H COSY, HSQC and HMBC analysis were consistent with a 3-carbonyl indole structure connected to a thiazole ring. The second derivative (24) and pseudoalteromone A (25) were identified by comparative ESI-HRMS and 2D NMR analysis (for details, see Figure S31–S41).44 We then pursued a comparative LC-MS and UV-based analysis of related Pseudoalteromonas strains and were able to observe stable production of indole-thiazol 23 in P. luteovioalacea DSM6061 and P. tunicata DSM14096 (Figure 6). Interestingly, we also detected the production of several so far unidentified indole-containing secondary metabolites in other Pseudoalteromonas strains, which are topic of current investigations.
Finally, we evaluated the antifungal actvities of compound 23 and 24. Both compounds showed no antimicrobial activity in our standardized assay, and were therefore not accountable for the observed antifungal activity of strain P1-9. However, it is noteworthy that bacterial-derived indole-containing metabolites are known to modulate important survival mechanisms in many bacteria, including biofilm formation, virulence, and drug resistance.1,10 Indole-acetic acid, e.g., is described to mediate interkingdom-communication and to serve as a growth factor for plants. Compound 23 was only recently characterized and reported in the literature as a metabolite of the plant model system Arabidopsis and proposed to be part of the plant defence mechanism.45
4. Bacterial-induced morphogenesis
Bacteria-induced metamorphosis is widespread among metazoans; in particular amongst marine invertebrates belonging to the phylum Cnidaria. Free-swimming larvae of many species settle upon sensing bacterial signals on suitable marine surfaces and transform into the benthic phenotype.1,10,14
Despite the fundamental importance of this transition, only few morphogenic cues have been identified and analyzed until today. As early as 1970s Leitz and Wagner reported that the bacterial isolate Alteromonas espejiana (reclassified as Pseudoalteromonas espejiana), amongst others, is responsible for the induction of metamorphosis in H. echinata larvae.46 We therefore assessed the morphogenic activity of our bacterial isolates belonging the γ-Proteobacteria class, as well as the literature-reported bioactive strain Pseudoalteromonas sp. PS5,12 and phylogenetic-related and commercially available strains P. espejiana ATCC 29659, A. macleodii ATCC 27126, P. luteoviolacea DSM6061, P. rubra DSM6842 and P. tunicata DSM14096 (for a phylogenetic tree including reference sequences, see Figure S.42).
First, selected strains were grown on agar plates for three days to enable biofilm formation. Single bacterial colonies were then suspended in sterile PBS and aliquots were transferred to 12-well plates. After adherence of bacterial cells to the well plate surface, 30–40 competent H. echinata larvae in sterile sea water were added to each well.46 The well volume was adjusted to 1 mL and bioassays kept at 20 °C. Settlement and metamorphosis rates were counted after 24 (Figure 7) and 48 h (not shown). All experiments were performed with three different larvae batches collected from different spawning events and each experiment was measured as triplicate. Morphologies of larvae and polyps were assessed according to Seipp et al.18 using the described normation of artificially induced metamorphosis (CsCl). Settlement was defined as the stage, when the anterior pole of the larvae was fixed to the well surface and no searching behavior was detectable. Morphogenesis was defined as the stage, when settled (surface attached) larvae had undergone apoptotic event and primary stolons and tentacles of a polyp were clearly visible. Toxic events were defined, when lysis of the larvae body was observed.
Figure 7.

Phylogenetic tree (left) based on 16S-rRNA gene sequences of bacteria evaluated for their morphogenic activities. Best DNA model was generated and the robustness of interfered tree topologies was evaluated after 1000 bootsraps (> 50% are shown on the trees). Heatmap (right) depicts settlement and metamorphosis (transformation of settled larvae) events counted after 24 h (in % as mean value of replicates).
Based on these definitions, six out of 21 strains induced settlement, directly followed by metamorphosis within 24 h; with Pseudoalteromonas strains P1-9 and P-29 exhibiting the highest morphogenic activity. Ten out of 21 tested strains, including the commercially obtained strain P. espejiana ATCC 29659,46 showed settlement activity of 60–80% within 24 h, but did not induce morphogenesis to the primary polyp even after 48 h. Only two strains showed neither induction of settling behavior nor morphogenesis. Four strains caused the lysis of larvae within 24 h, amongst them strain Pseudoalteromonas sp. PS5, which produces the metabolite bromopyrrol (8).11,12 Furthermore, preliminary data shows that none of the isolated and herein reported compounds (9-22) is responsible for the induction of morphogenesis of H. echinata larvae. Structure elucidation of potential morphogenic signals derived from bioactive strains is currently ongoing.
Overall, our results clearly support previous descriptions that morphogenesis of H. echinata larvae is induced by several species belonging to different bacterial genera of the γ-Proteobacteria class. The metamorphic transition of the motile larvae to the sessile polyp needs to be considered as an orchestrated multi-factorial or even synergistic event, which requires most likely different bacterial-produced signaling cues. Synergistic effects influencing the morphogenesis of marine organisms have already been described in several systems. Joint et al., e.g., showed that the presence of quorum sensing molecules (homoserine lactones) and the biofilm-forming bacterium (Vibrio anguillarum) induce optimal settlement of the zoospores of Ulva.47 Kitamura et al. reported in 2007 that the metamorphosis rates of the scleractinian coral Pseudosiderastrea tayamai is increased from ~30 % to ~90 % by application of a combination of bromo-substituted tyrosine derivatives and carotenoids.48 Similarly, metamorphosis rates in larvae of Leptastrae purpurea increased from 30% to almost 80%, when combinations of the macrodiolide luminaolide and methanolic extracts from the same producing crustose coralline algae were applied.49
5. Conclusion
This is the first systematic description of the associated microbiome of the marine hydroid polyp H. echinata using both, culture independent and dependent methods. Isolated bacterial species revealed a broad range of bioactivities, including antibacterial, antifungal, morphogenic and cytotoxic activities. Three bacterial isolates were chemically analyzed and 17 secondary metabolites characterized, amongst them antimicrobial molecules and potential growth stimulants. Assays evaluating the morphogenic activities of bacterial isolates showed that the transition from the motile larvae to primary polyp is divided in a two-step process, including a settlement and metamorphic event, which are most likely triggered by different bacterial-derived signals. Our studies pave the way to elucidate the natural mechanisms how morphogenesis in marine organisms is induced, and how microbes contribute to the protection of the organism.
6. Experimental section
NMR measurements were performed on a Bruker AVANCE III 500 MHz spectrometer, equipped with a Bruker Cryoplatform. The chemical shifts are reported in parts per million (ppm) relative to the solvent residual peak of DMSO-d6 (1H: 2.50 ppm, quintet; 13C: 39.52 ppm, heptet). LC-ESI-HRMS measurements were carried out on an Accela UPLC system (Thermo Scientific) coupled with a Accucore C18 column (100 × 2.1 mm, particle size 2.6 μm) combined with a Q-Exactive mass spectrometer (Thermo Scientific) equipped with an elecrospray ion (ESI) source. UHPLC-MS measurements were performed on a Shimadzu LCMS-2020 system equipped with single quadrupole mass spectrometer using a Phenomenex Kinetex C18 column (50 × 2.1 mm, particle size 1.7 μm, pore diameter 100 Å). Column oven was set to 40 °C; scan range of MS was set to m/z 150 to 2,000 with a scan speed of 10,000 u/s and event time of 0.25 s under positive and negative mode. DL temperature was set to 250 °C with an interface temperature of 350 °C and a heat block of 400 °C. The nebulizing gas flow was set to 1.5 L/min and dry gas flow to 15 L/min. Semi-preparative HPLC was performed on a Shimadzu HPLC system using a Phenomenex Luna C18(2) 250 × 10 mm column (particle size 5 μm, pore diameter 100 Å). IR spectra were recorded on an FT/IR-4100 ATR spectrometer (JASCO). Optical rotations were recorded in MeOH on a P-1020 polarimeter (JASCO). Solid phase extraction was carried out using Chromabond C18ec cartridges filled octadecyl-modified silica gel (Macherey-Nagel, Germany) using mixtures of MeOH or acetonitrile and ddH2O. Open column chromatography was performed on Sephadex LH20 (GE Healthcare, Sweden). Chemicals: Methanol and acetonitrile (VWR as HPLC grade, Germany); water for analytical and preparative HPLC (Millipore, Germany), formic acid (Carl Roth, Germany); acetonitrile (VWR as LC-MS grade), media ingredients (Carl Roth, Germany), sea salt (Instant ocean). Media cultures: Marine Broth Liquid (MBL): 40.1 g/L marine broth; Marine Broth Agar (MBA): 40.1 g/L marine broth, 20.0 g/L agar; Sea Water Complete Liquid (SWCL): 24 g/L sea salt, 5 g/L peptone, 3 g/L yeast extract, 3.0 mL/L glycerol; Sea Water Complete Agar (SWCA): 24 g/L sea salt, 5 g/L peptone, 3 g/L yeast extract, 3.0 mL/L glycerol, 20.0 g/L agar; Modified sea water complete liquid (mSWCL): 24 g/L sea salt, 10 g/L peptone from casein, and 2 g/L dextrin; Modified marine minimal media (MMa): 36.5 g/L sea salt, 12.8 g/L Na2HPO4•7H2O, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.0493 g/L MgSO4•7H2O, 0.4 g/L glucose, 0.00147 g/L CaCl2•2H2O, 0.0119 g/L KBr.
6.1 Laboratory maintenance of H. echinata
H. echinata colonies were obtained as single colonies on gastropod shells (common whelks (Buccinum spp.)) from the Marine Biological Laboratory in Woods Hole (MA 02543, USA) and Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research (Helgoland, Germany). Adult polyps were kept in artificial seawater (salinity of 33.2–33.7‰, pH 8.2–8.3 and 15 °C) in aerated tanks maintained in 16 h light/8 h dark cycle and were fed daily using 3–7 day old nauplii of Artemia salina. Fertilized eggs were collected in the two- to four-cell stage 3 hours after spawning event and transferred into freshly sterile seawater. Three to five-day old competent planula were used for morphogenic assay.
6.2 Deep 16S rRNA sequencing
Illumina 16S rRNA gene sequencing was used to profile the bacterial communities associated with freshly collected healthy H. echinata polyps. For each sample, twenty polyps of one colony were cut under a dissecting microscope and carefully rinsed with sterile seawater several times. The combined tissue was homogenized using a sterile pestile, and bacterial DNA was extracted using the GenElute™ Bacterial Genomic DNA Kit (Sigma Aldrich) according to manufacture protocol. DNA was eluted using 100 μL of deionized H2O and quantified using a NanoDrop Lite Spectrophotometer (Thermo Fisher). The average DNA yield was 80 ng per sample. All bacterial sequence data analysed in this study were obtained from the V6 hypervariable region of the 16S rRNA gene. Amplicons were generated using the same procedures as outlined in Gignoux-Wolfsohn et al.21 Briefly, 16S rRNA libraries were prepared in PCR reactions with DNA extracts obtained from six different H. echinata colonies. Primers binding to the V6 region of the 16S rRNA gene were used and contained a sample-specific five basepair as well as the barcode, Illumina sequencing adapter. PCR products were pooled and a second PCR reaction was conducted using the Illumina primers: OLJ139 and OLJ140.50 PCR products were cleaned with DNAmpure beads (Agencourt). Paired-end amplicon sequencing was done on an Illumina Hiseq 2000 system at the Tufts University. For sequence processing and analysis, forward and reverse reads were overlapped using FLASH.20 Sequences were processed with the QIIME pipeline v.1.9.1.51 Single samples were grouped according to their barcode. Primer sequences and barcodes were trimmed off and a Q30 quality filter was applied. Sequences were clustered to operational taxonomic units (OTUs) based on 97% sequence similarity against the Greengenes reference database52 using open-reference OTU picking and UCLUST.53 PyNast was used for alignment.54 Singleton OTUs and those that failed to align with PyNast were removed. Also removed were unassigned, non-bacterial 16S rRNA OTUs and those that belonged to cyanobacterial chloroplasts from algal contaminations. Sequence data were submitted to the European Nucleotide Archive (ENA) under the study accession number PRJEB19266 (accessible at http://www.ebi.ac.uk/ena/data/view/PRJEB19266).
6.3 Isolation of H. echinata surface-associated bacteria
Twenty polyps of each colony H. echinata were dissected and pooled. Samples were first washed with sterile sea water (300 μL) and centrifuged. The resulting supernatant was defined as SW-sample. The remaining poly tissue was suspended in sterile sea water (100 μL) and homogenized using a sterile pestle and the resulting suspension was defined as P-sample. SW and P-samples were serially diluted from 10−1 – 10−3 using filtered sterile seawater, and 100 μL of each dilution was used to inoculate SWC and MB agar plates supplemented with 0.05 mg/L cycloheximide. Plates were incubated at 20 °C for up to 14 days and monitored every day. Single colonies were transferred to new agar plates and subcultures until pure cultures were obtained. All bacterial isolates were cultured in SWC broth (30 °C, 160 rpm) for 1–3 days and maintained as 25% (v/v) glycerol suspensions at −80 °C.
6.4 DNA extraction, 16S rRNA sequencing, and phylogenetic analysis
Bacterial DNA was extracted from pure cultures using the Thermo Fischer GeneJET Genomic DNA Purification Kit according to the manufacturer’s instruction. DNA was eluted using 20 μL of deionized H2O and quantified using a NanoDrop Lite Spectrophotometer (Thermo Fisher). Amplification was performed in a 25 μL reaction volume with 5 μL of 10× HF buffer, 2.5 μM each primer, 0.25 μL Phusion polymerase, 10 mM dNTP mix and 1 μL of DNA with a concentration of 40 ng/μL. Cycling conditions were set as followed: initial denaturation 98°C for 38 s, 32 cycles of 98 °C for 30 s; 52 °C for 30 s; and 72 °C for 1.2 min and a final extension of 8 min at 72 °C was performed in a Peqstar 2× Gradient cycler. PCR products were separated by electrophoresis in 1 % agarose gel and detected under UV light, and target products (1500 bp) were purified using the GeneJET PCR Purification Kit (Thermo Fisher). The 16S rRNA PCR products were sequenced separately using the primer set (27F/1492R)26 at GATC Biotech AG (Konstanz). Sequences were first compared using the Basic Local Alignment Search Tool (BLAST),55 then manually compiled, assembled and trimmed using BioEdit Version 7.2.0 (1).56 Phylogenetic trees were generated using the Maximum likelihood algorithm of the MEGA 6.06 package.57
6.5 Antimicrobial activity testing
A total of 56 isolates were evaluated for their ability to inhibit growth of standard human pathogenic microorganisms (Jena Microbial Resource Collection, Jena, Germany). Isolated bacteria were grown in 100 mL of either SWCL medium for SW-strains or MBL medium for P-strains at 30 °C for 3 days (160 rpm). Supernatant was separated from cultures by centrifugation and the cell pellet was extracted using MeOH (10 mL). The mixture of supernatant and methanolic cell pellet extract was loaded on a activated SPE-C18 cartridge (500 mg) and eluted with 50% MeOH and 100% MeOH, respectively. The eluted fractions were dried under reduced pressure, and resuspended to concentrations of 1 mg/mL in 50% MeOH or 100% MeOH, respectively. Antimicrobial activity was evaluated against B. subtilis 6633; S. aureus SG511, E. coli SG458, P. aeruginosa K799/61, M. vaccae 10670, S. salmonicolor 549, C. albicans C.A., and P. notatum JP36 by measuring the inhibition zone in mm (disc diffusion assay) according to the NCCLS (National Committee for Clinical Laboratory Standards).35
6.6 Purification and identification of secondary metabolites
6.6.1 Cobetia sp. SW83
A three-day old culture of Cobetia sp. SW83 in MMa broth (150 mL, 30 °C, 160 rpm) was used for inoculation of 7 L MMa broth for five days (30 °C, 160 rpm) until the supernatant showed a typical pink color. The supernatant was separated from biomass by centrifugation and the cell pellet was extracted using MeOH. The methanolic cell extract was concentrated under reduced pressure. The culture supernatant was pooled and extracted using activated HP20 resin (20 g/L) overnight. The resin was first washed with ddH2O (200 mL each) and 10% MeOH (if not stated otherwise percentage refers to amount of MeOH (10%) in H20 (90%); 200 mL). Absorbed metabolites were eluted using first MeOH (1 L), followed by acetone (1 L). The combined organic extracts of cell pellet and HP20 eluent were concentrated under reduced pressure, resuspended in 10% MeOH (20 mL) and loaded onto an equilibrated SPE-C18 (10 g) cartridge. The crude extract was separated using a step gradient: 10% MeOH (400 mL), 30% MeOH (300 mL), 50% MeOH (200 mL), 80% MeOH (200 mL), 100% MeOH (100 mL) and acetone (100 mL). Fraction containing the pink pigment (80% MeOH) was concentrated and submitted to semipreparative HPLC to yield compound 9 (1.0 mg, tR = 13.9 min) and 10 (1.0 mg, tR = 16.9 min) using the following the gradient: 0–5 min, 45% MeCN; 5– 20 min, 45%–70% MeCN; 20–23 min, 70%–100% MeCN, 23– 28 min, 100% MeCN (A: dd H2O + 0.1% formic acid) with a flow rate of 2.0 mL/min. Due to the low production yield, a large-scale cultivation was performed. First, strain SW83 was was reached, cultured in 1.2 L SWCL until the stationary phase (OD > 1.2) and then transferred to a 75 L fermenter containing 50 L of SWCL broth. SW83 was cultivated for five days at 30 °C under stirring conditions (100 rpm). The supernatant and biomass was separated using a Westfalia CSA8 separator (9200 rpm), and then mixed with XAD16 resin (1 kg). After stirring for 1 h, the XAD16 resin was separated from the supernatant by filtration and washed with ddH2O (10 L) and 10% MeOH (5 L). Metabolites were then eluted using 100% MeOH (5 L), followed by and 100% acetone (5 L). The eluents were concentrated under reduced pressure, resuspended in 10% MeOH (100 mL), and loaded onto an equilibrated SPE-C18 (10 g) cartridge. Again, a step gradient was used to elute the respective compounds. Further UHPLC-MS analysis indicated that SPE fraction eluted with 50% MeOH contained peptidic compounds with an m/z of 986.5333 ([M+H]+) and 1097.5647 ([M+H]+). Both fractions were pooled, and further purified using Sephadex LH20 (50% MeOH elution), silica gel column chromatography (DCM/MeOH) and semipreparative HPLC to yield the pure compounds 11 and 12. Marfey’s reaction:57 Compounds 11 and 12 (0.1 mg each) were hydrolyzed using 6 N HCl (1.0 mL) at 110 °C for 15 h. HCl was removed under vacuum and 20 μL FDAA (1-fluoro-2,4-dinitrophenyl-5-L-alanine amide, 10 mg/mL in acetone) and 100 μL NaHCO3 (1 N aqueous solution) were added. The reaction was heated at 80 °C for 10 min, and then quenched by addition of 50 μL 2 N HCl. The reaction mixture was centrifuged (10 min, 13,000 rpm)) and the crude mixture (5 μL injection) was analyzed by UHPLC-MS (Figure S30) using the following conditions: gradient: 0–1 min, 10% B; 1–5 min, 10%–100% B; 5–7 min, 100% B; 7–7.1 min, 100%–10% B (A: dd H2O with 0.1% FA; B: MeCN with 0.1% FA) at a flow rate of 1.0 mL/min. Reference substrates using L- and D-proline, L- and D-tyrosine, L- and D-phenylalanine, glycine, L-isoleucine and D/L-isoleucine, and L- and D-valine references were converted accordingly.
6.6.2 Cobetia sp. SW40
A one-day old liquid culture of Cobetia sp. SW40 in SWCL media (1.2 L SWCL, 30 °C, 160 rpm) was used to inoculate a 50 L liquid SWCL culture in a 75 L fermenter and stirred for 3 days (100 rpm) at 30 °C. The supernatant was separated by centrifugation (9000 rpm), and subjected to XAD16 resin extraction and SPE-C18 purification as described above. Stock solutions of SPE fractions (1 mg/mL) were subjected to a standard antimicrobial disk-diffusion assay against S. epidermidis (STI10845). SPE-C18 fraction eluted with 80% MeOH showed antimicrobial activity and was submitted to preparative HPLC. Further bioassay-guided fractionation using reverse-phase semipreparative HPLC yielded compounds 13-22 in varying ratios. Marfey’s reaction: Compounds 13/14, 15/16, 17/18, 19/20, 21 and 22 (0.1 mg each) were hydrolyzed using 6 N HCl (1.0 mL) at 110 °C for 15 h. HCl was removed under vacuum and 20 μL FDAA (1-fluoro-2,4-dinitrophenyl-5-L-alanine amide, 10 mg/mL in acetone) and 100 μL NaHCO3 (1 N aqueous solution) were added. The reaction was heated at 80 °C for 10 min, and then quenched by addition of 50 μL 2 N HCl. The reaction mixture was centrifuged (10 min, 13,000 rpm) and the crude mixture (5 μL injection) was analyzed by UHPLC-MS (Figure S30) using the following conditions: gradient: 0–1 min, 10% B; 1–5 min, 10%–100% B; 5–7 min, 100% B; 7–7.1 min, 100%–10% B (A: dd H2O with 0.1% FA; B: MeCN with 0.1% FA) at a flow rate of 1.0 mL/min. Reference substrates using L-and D-leucine and L-isoleucine and D/L-isoleucine, and L- and D-valine references were converted accordingly.
6.6.3. Pseudoaltermonas sp. P1-9
A 2.5 L liquid culture of Pseudoalteromonas sp. P1-9 (SWCL, 30 °C, 200 rpm, 1 d) was used to inoculate 50 L SWCL broth kept in a 75 L fermenter. The culture was kept for five days at 30 °C while stirring (100 rpm). The culture supernatant was separated from the cell pellet, and subjected to XAD16 resin extraction. First, the resin was washed using 10% MeCN containing 0.1% FA and extracts gradually eluted using a step gradient from 20% and 100% MeCN containing 0.1% FA. UHPLC-MS analysis (characteristic UV adsorption of λmax 355 nm) was used to identify the indole-containing fraction (40–60% MeCN). Indole-containing fractions were submitted to Sephadex LH20 and silica gel column chromatography, followed by semi-preparative HPLC to yield compound 23 (3.0 mg) and 24 (2.0 mg) and compound 25. HPLC gradient: 0–5 min, 65% B; 5–20 min, 65%–100% B; 20–25 min, 100% B (A: dd H2O + 0.1% FA; B: MeOH) with a flow rate of 2.0 mL/min. Comparative UHPLC-MS analysis: Pseudoalteromonas strains were cultivated in 1 L modified SWCL (30 °C, 200 rpm, 1 d). The supernatant was separated and extracted using HP20 resin and SPE-C18 purification as described above. The concentrated indole containing fractions were submitted to LC-MS analysis using the following gradient: 0–1 min, 10% B; 1–5 min, 10%–100% B; 5– 7 min, 100% B; 7–7.1 min, 100%–10% B (A: dd H2O with 0.1% FA; B: MeCN with 0.1% FA).
6.7 Evaluation of morphogenic activity
Selected Pseudoalteromonas strains were grown on a MBA plate for three days. Single colonies were suspended in sterile PBS (10–50 μL, OD600 approx. 0.5–0.6), and transferred into a well of sterile 24-well plate. The suspension was incubated for 20 min at RT to allow bacterial surface attachment to the well plate. Approximately 30–40 competent larvae of H. echinata were added per well and each well filled up to 1 mL using sterile sea water (equal volume per well). The assays were kept at 20 °C for 48 h to evaluate the settlement and metamorphosis rates after 24 and 48 h, respectively.18 As positive control, 30–40 competent larvae were treated with 20 μl of a 120 mM CsCl solution. Two negative controls were applied: a) 30–40 competent larvae were treated with 100 μL PBS and kept in 1 mL sterile sea water; b) 30–40 competent larvae were kept in 1 mL sterile sea water without treatment. Experiments were performed using three different batches of larvae collected from different spawning events and each experiment measured as triplicate. The results were indicated as mean value of replicates.
Supplementary Material
Figure 1.
Examples of bioactive natural products derived from marine bacteria.
Acknowledgments
MR is generously supported by the Jena School for Microbial Communication (JSMC, DFG). CB is generously supported by the Leibniz Association and the Collaborative Research Centre 1127 “Chemical Mediators in complex Biosystems” (DFG). We thank Sarah Gignoux-Wolfsohn (Northeastern University, Boston, MA) for performing the sequencing, Mrs. Heike Heinecke (HKI) for recording NMR spectra, and Mrs. Andrea Perner (HKI) for HRMS measurements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary material (tables and figures giving oligonucleotide sequences and activity, NMR spectra, HPLC and ESI-MS traces) associated with this article can be found, in the online version.
References and Notes
- 1.a) Haefner B. Drug Discov Today. 2003;8:536–544. doi: 10.1016/s1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]; b) Zhang L, An R, Wang J, Sun N, Zhang S, Hu J, Kuai J. Curr Opin Microbiol. 2005;8:276–281. doi: 10.1016/j.mib.2005.04.008. [DOI] [PubMed] [Google Scholar]; c) Newman DJ, Hill RT. J Ind Microbiol Biotechnol. 2006;33:539–544. doi: 10.1007/s10295-006-0115-2. [DOI] [PubMed] [Google Scholar]; d) Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Nat Rev Drug Discov. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]; e) Penesyan A, Kjelleberg S, Egan S. Mar Drugs. 2010;8:438–459. doi: 10.3390/md8030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 3.Williams PG, Asolkar RN, Kondratyuk T, Pezzuto JM, Jensen PR, Fenical W. J Nat Prod. 2007;70:83–88. doi: 10.1021/np0604580. [DOI] [PubMed] [Google Scholar]
- 4.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew Chem Int Ed Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 5.a) Bowman JB. Mar Drugs. 2007;5:220–241. doi: 10.3390/md504220. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou C, Fleury Y. Mar Drugs. 2016;14:129. doi: 10.3390/md14070129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franks A, Haywood P, Holmström C, Egan S, Kjelleberg S, Kumar N. Molecules. 2005;10:1286–1291. doi: 10.3390/10101286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy AC, Gao S-S, Han L-C, Carobene S, Fukuda D, Song Z, Hothersall J, Cox RJ, Crosby J, Crump MP, Thomas CM, Willis CL, Simpson TJ. Chem Sci. 2014;5:397–402. [Google Scholar]
- 8.a) Machado H, Sonnenschein EC, Melchiorsen J, Gram L. BMC Genomics. 2015;16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA. Nat Rev Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross AC, Gulland LE, Dorrestein PC, Moore BS. ACS Synth Biol. 2015;4:414–420. doi: 10.1021/sb500280q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Romero D, Traxler MF, López D, Kolter R. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cantley AM, Clardy J. Nat Prod Rep. 2015;32:888–892. doi: 10.1039/c4np00141a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tebben J, Tapiolas DM, Motti CA, Abrego D, Negri AP, Blackall LL, Steinberg PD, Harder T. PLoS One. 2011;6:e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sneed JM, Sharp KH, Ritchie KB, Paul VJ. Proc Biol Sci. 2014:281. doi: 10.1098/rspb.2013.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freckelton ML, Nedved BT, Hadfield MG. Sci Rep. 2017;7:42557. doi: 10.1038/srep42557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadfield MG. Ann Rev Mar Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 15.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. Science. 2014;343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N. eLife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Beemelmanns C, Woznica A, Alegado RA, Cantley AM, King N, Clardy J. J Am Chem Soc. 2014;136:10210–10213. doi: 10.1021/ja5046692. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cantley AM, Woznica A, Beemelmanns C, King N, Clardy J. J Am Chem Soc. 2016;138:4326–4329. doi: 10.1021/jacs.6b01190. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Woznica A, Cantley AM, Beemelmanns C, Freinkman E, Clardy J, King N. Proc Natl Acad Sci U S A. 2016;113:7894–7899. doi: 10.1073/pnas.1605015113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dayel MJ, King N. PLoS One. 2014;9:e95577. doi: 10.1371/journal.pone.0095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Walther M, Ulrich R, Kroiher M, Berking S. Int J Dev Biol. 1996;40:313–322. [PubMed] [Google Scholar]; b) Seipp S, Schmich J, Kehrwald T, Leitz T. Dev Genes Evol. 2007;217:385–394. doi: 10.1007/s00427-007-0151-6. [DOI] [PubMed] [Google Scholar]
- 19.a) Klassen JL, Wolf T, Rischer M, Guo H, Shelest E, Clardy J, Beemelmanns C. Genome Announc. 2015;3:e01477–15. doi: 10.1128/genomeA.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Klassen JL, Rischer M, Wolf T, Guo H, Shelest E, Clardy J, Beemelmanns C. Genome Announc. 2015;3:e01380–15. doi: 10.1128/genomeA.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rischer M, Klassen J, Wolf T, Guo H, Shelest E, Clardy J, Beemelmanns C. Genome Announc. 2016;4:e00003–16. doi: 10.1128/genomeA.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a) Barriuso J, Valverde JR, Mellado RP. BMC Bioinformatics. 2011;12:473. doi: 10.1186/1471-2105-12-473. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Magoč T, Salzberg SL. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Caro-Quintero A, Ochman H. Genome Biol Evol. 2015;7:3416–3425. doi: 10.1093/gbe/evv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gignoux-Wolfsohn SA, Vollmer SV. PLoS One. 2015;10:e0134416. doi: 10.1371/journal.pone.0134416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Lynch MD, Neufeld JD. Nat Rev Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]; b) Pedrós-Alió C. Annu Rev Mar Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]; c) Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. ISME J. 2013;7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-Castillo FM, Costea PI, Cruaud C, d’Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C, Lima-Mendez G, Poulain J, Poulos BT, Royo-Llonch M, Sarmento H, Vieira-Silva S, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Tara Oceans coordinators. Bowler C, de Vargas C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Not F, Ogata H, Pesant S, Speich S, Stemmann L, Sullivan MB, Weissenbach J, Wincker P, Karsenti E, Raes J, Acinas SG, Bork P. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 24.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, Pedrós-Alió C. ISME J. 2013;7:1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Camillo CG, Luna GM, Bo M, Giordano G, Corinaldesi C, Bavestrello G. PLoS One. 2012;7:e39926. doi: 10.1371/journal.pone.0039926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumann PJ. Basic Microbiol. 1991;31:479–480. [Google Scholar]
- 27.Gauthier G, Gauthier M, Christen R. Int J Syst Evol Microbiol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 28.Holmström C, Egan S, Franks A, McCloy S, Kjelleberg S. FEMS Microbiol Ecol. 2002;41:47–58. doi: 10.1111/j.1574-6941.2002.tb00965.x. [DOI] [PubMed] [Google Scholar]
- 29.a) Arahal DR, Castillo AM, Ludwig W, Schleifer KH, Ventosa A. Syst Appl Microbiol. 2002;25:207–211. doi: 10.1078/0723-2020-00113. [DOI] [PubMed] [Google Scholar]; b) Kim MS, Roh SW, Bae JW. Int J Syst Evol Microbiol. 2010;60:620–626. doi: 10.1099/ijs.0.008847-0. [DOI] [PubMed] [Google Scholar]; c) Romanenko L, Tanaka N, Svetashev V, Falsen E. Int J Syst Evol Microbiol. 2013;63:288–297. doi: 10.1099/ijs.0.036863-0. [DOI] [PubMed] [Google Scholar]; c) Ibacache-Quiroga C, Ojeda J, Espinoza-Vergara G, Olivero P, Cuellar M, Dinamarca MA. Microb Biotechnol. 2013;6:394–405. doi: 10.1111/1751-7915.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kokoulin MS, Kuzmich AS, Kalinovsky AI, Tomshich SV, Romanenko LA, Mikhailov VV, Komandrova NA. Carbohydr Polym. 2016;154:55–61. doi: 10.1016/j.carbpol.2016.08.036. [DOI] [PubMed] [Google Scholar]
- 30.a) Vishnivetskaya TA, Kathariou S, Tiedje JM. Extremophiles. 2009;13:541–555. doi: 10.1007/s00792-009-0243-5. [DOI] [PubMed] [Google Scholar]; b) Carneiro AR, Ramos RT, Dall’Agnol H, Pinto AC, de Castro Soares S, Santos AR, Guimarães LC, Almeida SS, Baraúna RA, das Graças DA, Franco LC, Ali A, Hassan SS, Nunes CI, Barbosa MS, Fiaux KK, Aburjaile FF, Barbosa EG, Bakhtiar SM, Vilela D, Nóbrega F, dos Santos AL, Carepo MS, Azevedo V, Schneider MP, Pellizari VH, Silva A. J Bacteriol. 2012;194:6689–6690. doi: 10.1128/JB.01791-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dastager SG, Mawlankar R, Sonalkar VV, Thorat MN, Mual P, Verma A, Krishnamurthi S, Tang SK, Li WJ. Int J Syst Evol Microbiol. 2015;65:1611–1616. doi: 10.1099/ijs.0.000149. [DOI] [PubMed] [Google Scholar]
- 31.Azevedo JS, Correia A, Henriques I. FEMS Microbiol Ecol. 2013;84:451–460. doi: 10.1111/1574-6941.12075. [DOI] [PubMed] [Google Scholar]
- 32.Negri A, Webster NS, Hill RT, Heyward A. J Mar Ecol Prog Ser. 2001;223:121–131. [Google Scholar]
- 33.Huggett MJ, Williamson JE, de Nys R, Kjelleberg S, Steinberg P. Oecologia. 2006;149:604–619. doi: 10.1007/s00442-006-0470-8. [DOI] [PubMed] [Google Scholar]
- 34.Vynne NG, Månsson M, Nielsen KF, Gram L. Mar Biotechnol (NY) 2011;13:1062–1073. doi: 10.1007/s10126-011-9369-4. [DOI] [PubMed] [Google Scholar]
- 35.Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved Standard. 4th. Vol. 26 Clinical and Laboratory Standards Institute; Wayne, PA: 2006. [Google Scholar]
- 36.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. Nucleic Acids Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf T, Shelest V, Nath N, Shelest E. Bioinformatics. 2015;32:1138–1143. doi: 10.1093/bioinformatics/btv713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.a) Ilda K, Nakamura M, Hanamitsu H, Kajiwara M. Chem Pharm Bull. 2007;55:1067–1069. doi: 10.1248/cpb.55.1067. [DOI] [PubMed] [Google Scholar]; b) Bhuiyan MN, Takai R, Mitsuhashi S, Shigetomi K, Tanaka Y, Kamagata Y, Ubukata M. J Antibiot (Tokyo) 2016;69:97–103. doi: 10.1038/ja.2015.87. [DOI] [PubMed] [Google Scholar]
- 39.a) Müller F, Dudley KH. Helv Chim Acta. 1971;54:1487–1497. doi: 10.1002/hlca.19710540534. [DOI] [PubMed] [Google Scholar]; b) Yamamoto K, Asano Y. Appl Environ Microbiol. 2015;81:7360–7367. doi: 10.1128/AEM.02166-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bliumkin L, Dutta Majumdar R, Soong R, Adamo A, Abbatt JP, Zhao R, Reiner E, Simpson AJ. Environ Sci Technol. 2016;50:5506–16. doi: 10.1021/acs.est.6b00361. [DOI] [PubMed] [Google Scholar]
- 40.a) Vavrusova M, Pindstrup H, Johansen LB, Andersen ML, Andersen HJ, Skibsted LH. Int Dairy J. 2015;47:86–93. [Google Scholar]; b) Edens L, Harvey M, Hoeven RA. United States Patent Application Publication Pub. No.: US 2007/0031399 A1
- 41.Stothers JB. Carbon-13 NMR Spectroscopy. New York: Academic Press; 1972. [Google Scholar]
- 42.a) McKellar RC, Paquet A, Ma CY. Food Microbiol. 1992;9:67–76. [Google Scholar]; b) Bruns H, Thiel V, Voget S, Patzelt D, Daniel R, Wagner-Döbler I, Schulz S. Chem Biodivers. 2013;10:1559–1573. doi: 10.1002/cbdv.201300210. [DOI] [PubMed] [Google Scholar]
- 43.a) Jansen R, Mohr KI, Bernecker S, Stadler M, Müller R. J Nat Prod. 2014;77:1054–1060. doi: 10.1021/np500144t. [DOI] [PubMed] [Google Scholar]; b) Park JS, Yabe S, Shin-ya K, Nishiyama M, Kuzuyama T. J Antibiot (Tokyo) 2015;68:60–62. doi: 10.1038/ja.2014.93. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Lu M, Chang Y, Hwang T, Wang W, Weng C, Kuo J, Sung P. Tetrahedron Lett. 2012;53:1675–1677. [Google Scholar]
- 45.Rajniak J, Barco B, Clay NK, Sattely ES. Nature. 2015;525:376–379. doi: 10.1038/nature14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leitz T, Wagner T. Mar Biol. 1993;115:173–178. [Google Scholar]
- 47.Joint I, Tait K, Wheeler G. Philos Trans R Soc Lond B Biol Sci. 2007;362:1223–1233. doi: 10.1098/rstb.2007.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitamura M, Koyama T, Nakano Y, Uemura D. J Exp Mar Biol Ecol. 2007;340:96–102. [Google Scholar]
- 49.Kitamura M, Schupp PJ, Nakano Y, Uemura D. Tetrahedron Lett. 2009;50:6606. doi: 10.1016/j.tetlet.2009.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gloor GB, Hummelen R, Macklaim JM, Dickson RJ, Fernandes AD, MacPhee R, Reid G. PLoS One. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 54.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.http://blast.ncbi.nlm.nih.gov/Blast.cgi
- 56.Hall TA. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 57.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


