Abstract
Trisomy and triploidy, defined as the presence of a third copy of one or all chromosomes, respectively, are deleterious in many species including humans. Previous studies have demonstrated that Caenorhabditis elegans with a third copy of the X chromosome are viable and fertile. However, the extra X chromosome was shown to preferentially segregate into the first polar body during oocyte meiosis to produce a higher frequency of euploid offspring than would be generated by random segregation. Here, we demonstrate that extra autosomes are preferentially eliminated by triploid C. elegans and trisomy IV C. elegans. Live imaging of anaphase-lagging chromosomes and analysis of REC-8 staining of metaphase II spindles revealed that, in triploids, some univalent chromosomes do not lose cohesion and preferentially segregate intact into the first polar body during anaphase I, whereas other autosomes segregate chromatids equationally at anaphase I and eliminate some of the resulting single chromatids during anaphase II. We also demonstrate asymmetry in the anaphase spindle, which may contribute to the asymmetric segregation. This study reveals a pathway that allows aneuploid parents to produce euploid offspring at higher than random frequency.
The vast majority of eukaryotes have exactly two copies of each chromosome (homologous chromosomes) and reproduce sexually through the process of meiosis. Before meiosis, each homologous chromosome is replicated to produce a sister chromatid pair held together by REC-8 cohesin. During meiotic prophase, the sister chromatid pairs of two homologous chromosomes are connected by a crossover to generate a structure called a bivalent. During meiosis I, sister kinetochores must orient toward the same pole of a bipolar spindle while homologous kinetochore pairs must orient toward opposite spindle poles. Recognition of proper bipolar attachment of the bivalent requires the physical attachment generated by the crossover. Cleavage of REC-8 cohesin along a limited region of the bivalent allows homologous chromosomes to separate at anaphase I while maintaining cohesion between sister chromatids. During meiosis II, sister kinetochores orient toward opposite poles and the remaining sister cohesion is required to recognize this proper attachment (Nasmyth 2002; Miller et al. 2013). Because a physical attachment between homologous chromosomes is required for the normal mechanisms driving accurate chromosome segregation, a third copy of a chromosome presents a significant problem and might be expected to segregate randomly, resulting in 50% trisomic offspring.
In contrast with the expectation of random segregation, Caenorhabditis elegans with a third copy of the X chromosome have been found to produce twice as many haplo X ova as diplo X ova (Hodgkin et al. 1979). Female meiosis is asymmetric as only one gamete is produced and the remaining chromosomes are discarded in small polar bodies. The preferential elimination of an extra X in C. elegans occurs between metaphase I and metaphase II of female meiosis, and is likely due to preferential placement of the extra X in the first polar body (Cortes et al. 2015). Similar results were obtained with C. elegans him-8 mutants, which do not form a crossover between their two homologous X chromosomes (Phillips et al. 2005). These univalent X chromosomes are also preferentially placed in the first polar body during anaphase I of female meiosis (Hodgkin et al. 1979; Cortes et al. 2015). X univalents in him-8 mutants were found to biorient at metaphase I but failed to lose cohesion at anaphase I, most likely because they do not load the auroraB kinase, AIR-2, which must phosphorylate REC-8 to allow loss of sister cohesion (Rogers et al. 2002). These X univalents thus lag at anaphase I. Time-lapse imaging revealed that these lagging univalents resolve into the first polar body twice as often as they resolve into the embryo (Cortes et al. 2015).
Here, we set out to determine whether the mechanisms mediating preferential expulsion of univalent X chromosomes observed in triploX and him-8 worms might provide a general trisomy correction mechanism. Such a mechanism would allow triploid or trisomic worms to have a higher frequency of diploid progeny than would be generated by random segregation.
Materials and Methods
C. elegans strains
SP346: 4N tetraploid; AZ212: ruIs32 [pie-1p::GFP::H2B + unc-119(+)] III; AV494: XXX; and VS21: hjSi20 [myo-2p::mCherry::unc-54 3′UTR]. Single-copy transgene by mos1-mediated single copy insertion (MosSCI), inserted into cxTi10882; EG7911: oxTi705 [eft-3p::tdTomato::H2B::unc-54 3′UTR + Cbr-unc-119(+)]IV: miniMos insertion at 0.09. EG8299: snt-1(md290) II ; oxSi180[snt-1::gfp;unc-119(+)] IV: Single-copy transgene by MosSCI, inserted into cxTi10882; unc-41 (ox541[tagRFP::TEV::FLAG]) V; FM125: unc-119(ed3); ruIs57[pAZ147:pie-1/β-tubulin::GFP; unc-119(+)]; itIs37 [unc-119(+) pie-1::mCherry::H2B]); EU2933: itIs37[unc-119(+) pie-1::mCherry::H2B]; or1935[GFP::aspm-1]; N2: wild isolate; AB3: wild isolate; MY2: wild isolate; CB4856: wild isolate; ED3049: wild isolate; and JU258: wild isolate.
3N crosses
4N hermaphrodites at the L4 stage were mated to a male polymorphic strain (AB3, MY2, or CB4856) to obtain 3N worms. 3N hermaphrodites at the L4 stage were mated to a second male polymorphic strain (ED3049 or JU258). Strains and polymorphisms are listed in Supplemental Material, Table S1 in File S1. Only hermaphrodites with a mating plug and having 50% male progeny were considered to have mated. After 18–24 hr, the mated 3N hermaphrodites were picked onto single plates. They were moved onto new plates after 24 and 48 hr. 3N progeny (live hermaphrodites and males at the L3 or L4 stage and dead embryos) were singled into PCR tubes and digested in 6 μl of lysis buffer and proteinase K 48 hr after removing the mated 3N hermaphrodites from each plate. Lysis buffer was composed of the following: 1 M KCl, 1 M Tris pH 8.3, 1 M MgCl2, 0.45% IGEPAL, 0.45% Tween-20, and sterile water. The tubes were submerged in liquid N2 for 10 min and heated in a thermocycler at 60° for 60 min and 95° for 15 min. Each PCR reaction was 50 μl total. The following final concentrations of each reagent were used: 1× Standard Taq Reaction Buffer, 1 mM MgCl2, 300 μM dNTPs, 0.5 μM Forward Primer, 0.5 μM Reverse Primer, < 1000 ng template DNA (1/6th of a worm), 1 unit/50 μl PCR Hot Start Taq DNA Polymerase, and sterile water. Primers are listed in Figure S2 in File S1. All PCR reactions underwent an initial denaturation at 95° for 30 sec. For 32 cycles, the reactions went through a denaturation step at 95° for 30 sec, a primer-specific annealing step for 30 sec, and an extension step at 68° for 45 sec. There was a final extension at 68° for 5 min.
Statistics
Chi square and Fisher’s Exact tests of 2 × 2 contingency tables were calculated with GraphPad Prism. Fisher’s Exact Test of 2 × 3 contingency tables was calculated with http://www.danielsoper.com/statcalc/calculator.aspx?id=58.
Live imaging
4N hermaphrodites were mated with either AZ212 or EU2933 males to generate 3N progeny expressing either GFP::histone H2B, or mCherry::histone H2B and GFP::ASPM-1. 3N hermaphrodite progeny were anesthetized with tricaine/tetramisole as described (Kirby et al. 1990; McCarter et al. 1999) and gently mounted between a coverslip and a thin 3% agarose pad on a slide. Images in Figure 1, Figure 5, and Figure 7 were captured with a Solamere Spinning Disk Confocal equipped with a Yokogawa CSU10, Hammamatsu Orca FLASH 4.0 cMOS and an Olympus 100× 1.35 oil objective.
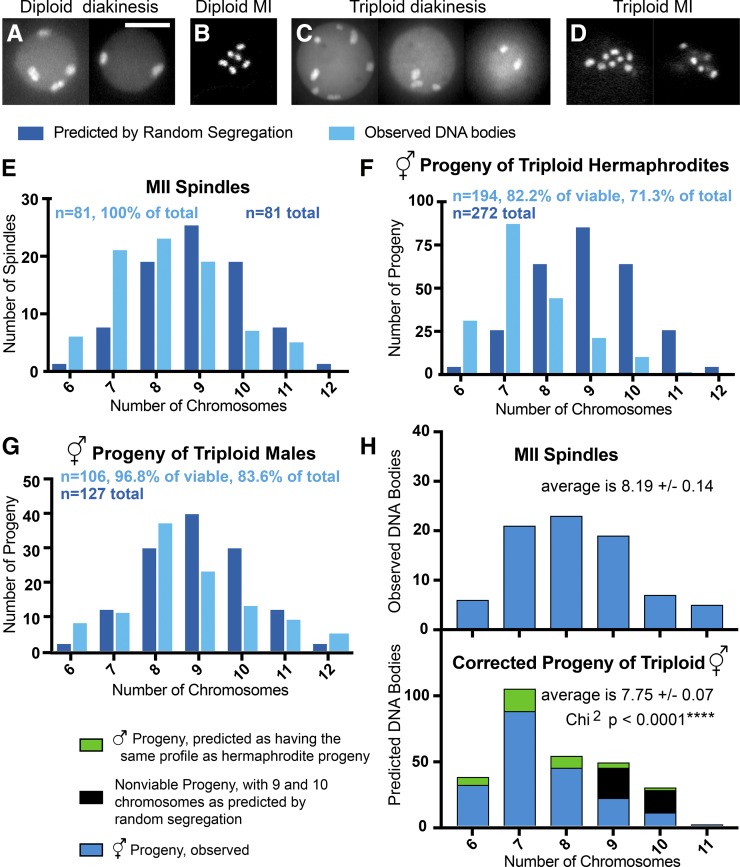
Figure 1.
Reduction of chromosome number occurs in triploid C. elegans. (A–D) Images of GFP::histone in a diakinesis oocyte of a diploid hermaphrodite (two focal planes (A), a metaphase I embryo from a diploid hermaphrodite (single focal plane) (B), a diakinesis oocyte of a triploid hermaphrodite (three focal planes) (C), and a metaphase I embryo from a triploid hermaphrodite (two focal planes) (D) Bar, 8 μm for (A–D). (E) DNA bodies were counted following three-dimensional (3D) fluorescence microscopy of metaphase II spindles in embryos of triploid hermaphrodites expressing GFP::H2B. (F and G) DNA bodies were counted following 3D fluorescence microscopy of the diakinesis oocytes in the adult hermaphrodite progeny of 3N hermaphrodites mated with 2N males (F), and of adult hermaphrodite progeny of 3N males mated with 2N hermaphrodites (G). (H) A corrected distribution of chromosomes in the total progeny of triploid hermaphrodites is compared with the distribution of chromosomes that is observed in MII spindles. Student’s t-test P = 0.0055; Chi square P < 0.0001.
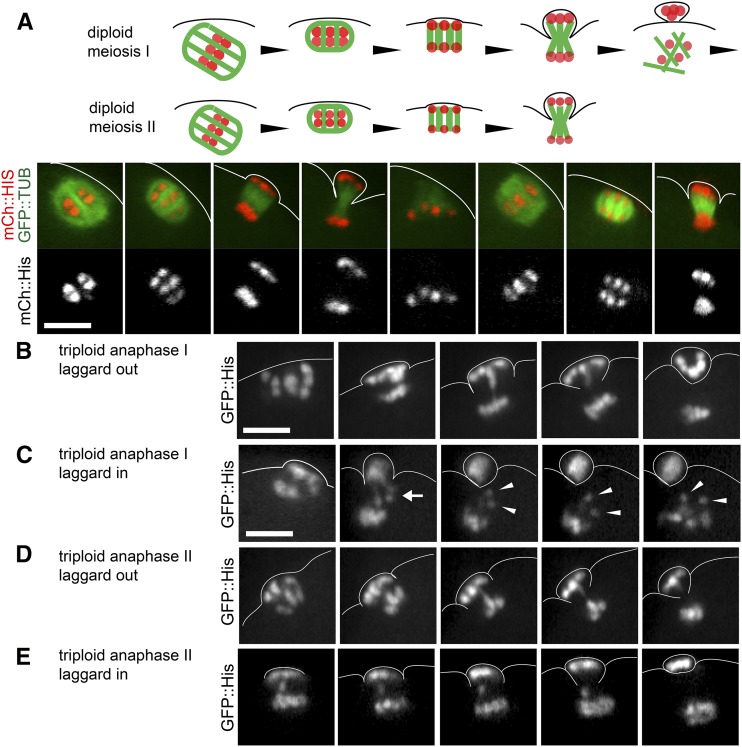
Figure 5.
Chromosomes lag during anaphase I and anaphase II in 3N embryos. (A) Chromosomes undergo two rounds of separation during anaphase I and anaphase II in a 2N control embryo expressing GFP::Tubulin (TUB) and mCh::His. (B) Anaphase I in a 3N embryo shows a chromosome that lags as it travels to the cortical pole. (C) Anaphase I in a 3N embryo shows two chromosomes which lag as they travel toward the interior pole. One of the lagging chromosomes appears to be a univalent, which separates into two chromatids as anaphase I ends and MII spindle formation begins. (D) Anaphase II in a 3N embryo shows a chromosome that lags as it travels to the cortical pole. (E) Anaphase II in a 3N embryo shows a chromosome that lags as it travels to the interior pole. Lagging chromosomes were observed in 14/15 anaphase I embryos and 17/20 anaphase II embryos from triploid hermaphrodites. Bar, 5 μM.
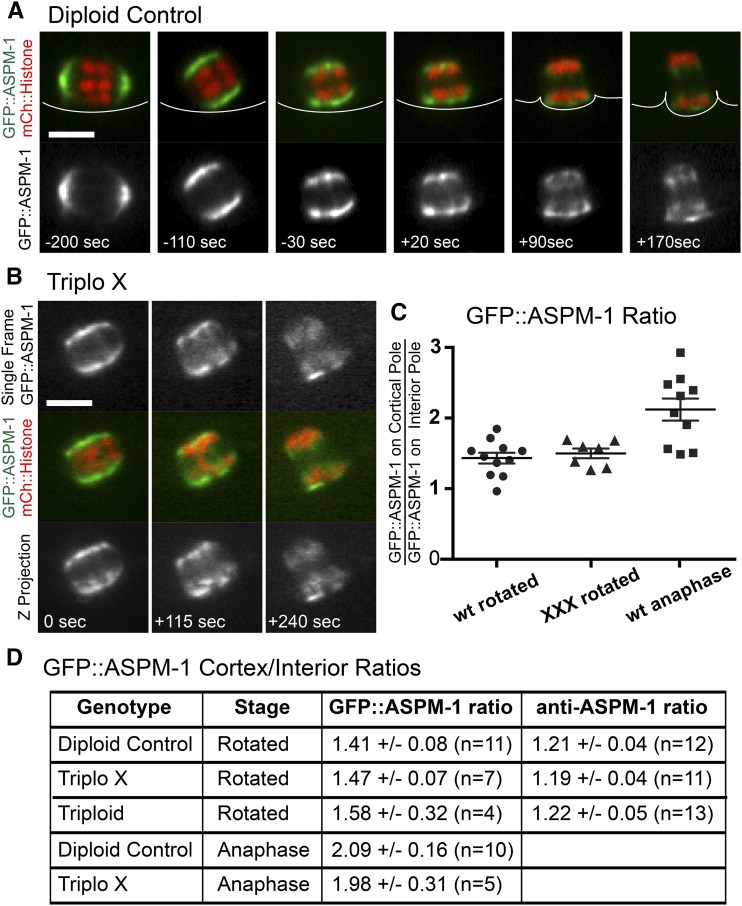
Figure 7.
ASPM-1 is enriched on the cortical spindle pole. (A) Single-plane time-lapse sequence of GFP::ASPM-1 and mCherry (mCh)::histone during meiosis I of a diploid. (B) Time-lapse sequence of a triploX anaphase I. (C and D) Ratios of three-dimensional pixel sums of GFP::ASPM-1 at the cortical pole/interior pole. “Rotated” spindles had a center-to-center homolog separation of 2.0 ± 0.3 μm (n = 16). “Anaphase” spindles had a center-to-center homolog separation of 3.4 ± 0.3 μm (n = 15). ± indicates SEM. Bar, 4 μm.
Fixed immunofluorescence and fluorescence in situ hybridization (FISH)
The chromosome V 5s rDNA repeat FISH probe was prepared as described in Phillips et al. (2009) and embryos were fixed and hybridized as described in Cortes et al. (2015). For immunofluorescence, C. elegans meiotic embryos were extruded from hermaphrodites in 0.8× egg buffer by gently squishing worms between coverslip and slide, flash frozen in liquid N2, permeabilized by removing the coverslip, and then fixed in cold methanol for 20 min, washed 1× in PBS and 2× in PBST for 10 min, and blocked in 4% BSA in PBST for 30 min before staining overnight with primary antibodies diluted in 4% BSA in PBST. Primary antibodies used in this work were mouse monoclonal anti-tubulin (DM1alpha; Sigma- Aldrich; 1:200), rabbit anti-ASPM-1 (van der Voet et al. 2009) and rabbit anti-REC-8 (Novus; 1:1000). Slides were washed 1× in PBST and 2× in PBS for 10 min and incubated for 1 hr in DAPI (1:200), Alexa Fluor 594 anti-rabbit (1:200), Alexa Fluor 488 anti-mouse (1:200), and 4% BSA in PBST. Slides were then washed 1× in PBST and 2× in PBS for 10 min and mounted with DABCO-Mowiol and a coverslip. Complete Z stacks were captured for each meiotic embryo. Images in Figure 3, Figure 4, and Figure 6 were captured on an Olympus IX81 microscope equipped with a 60× PlanApo 1.42 oil objective, an ORCA Flash 4.0 CMOS camera and an Olympus DSU (disk scanning unit).
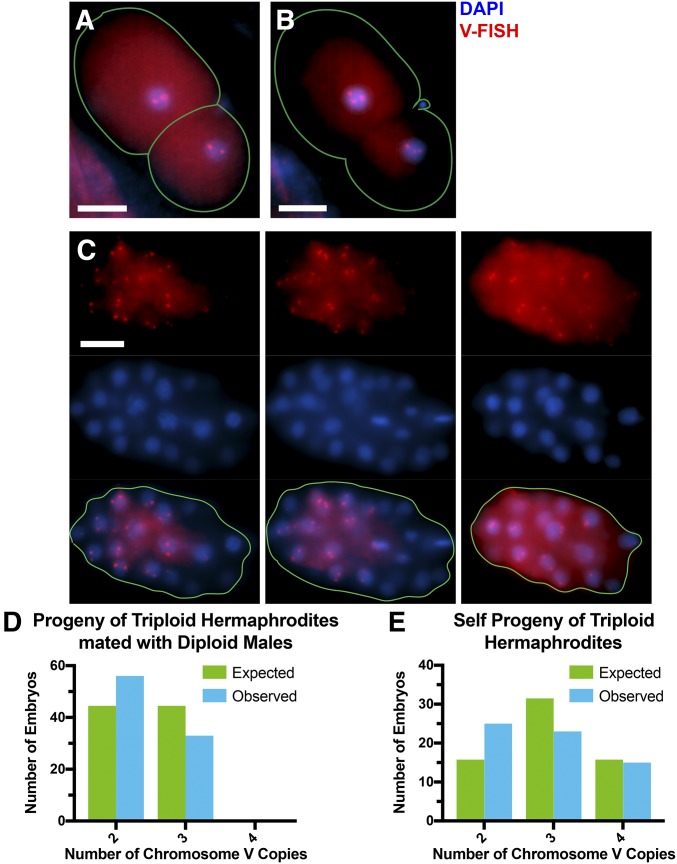
Figure 3.
Copy number of chromosome V is lower than expected from random segregation among the progeny of triploid hermaphrodites. (A) Example of chromosome V-FISH of a diploV two-celled embryo. (B) Example of chromosome V-FISH of a triploV two-celled embryo. (C) Three focal planes of a chromosome V-FISH-labeled diploV embryo. Bar, 10 μm in (A–C). (D–E) Number of embryos with two, three, or four copies of chromosome V among the cross-progeny (D) (n = 89) and self-progeny (E) (n = 63) of triploid hermaphrodites.
Figure 4.
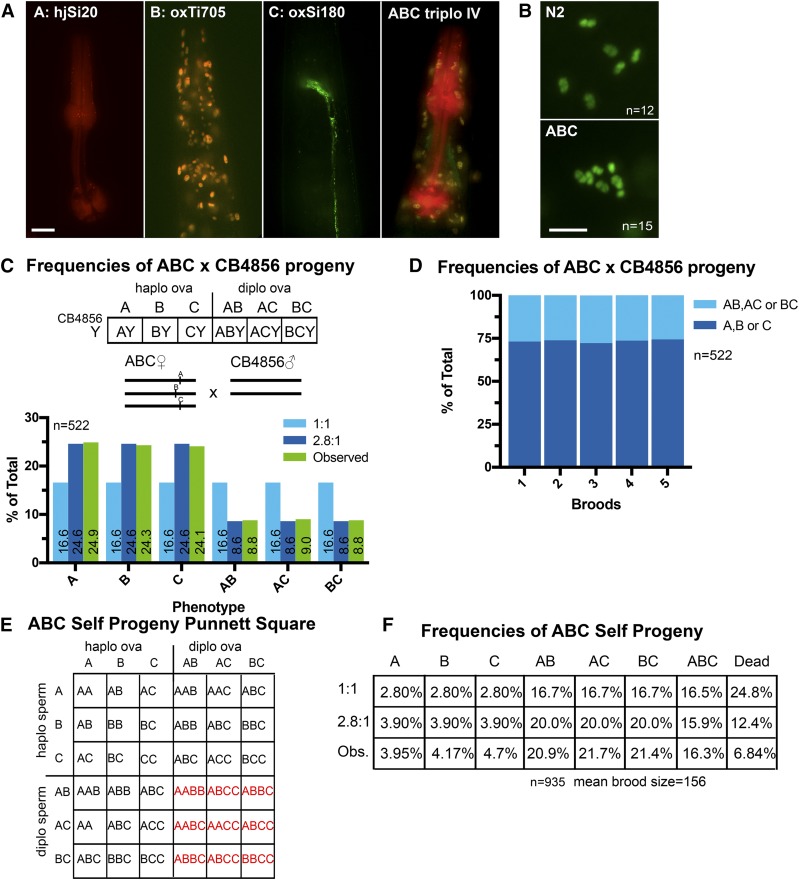
Trisomy IV is corrected. (A) Fluorescence images of the head/pharynx regions of adult worms bearing the transgenes used to generate the ABC trisomy IV strain. Bar, 10 μM. (B) Fluorescence images of DAPI-stained diakinesis nuclei showing six chromosome bodies in N2 wild-type and seven in ABC trisomy IV. Bar, 5 μM. (C) Punnett square showing expected genotypes resulting from a cross between an ABC hermaphrodite and a CB4856 diploid male without markers. Bar graph shows observed phenotypes of cross-progeny compared to expected frequencies from random (1:1) or 2.8:1 biased segregation. (D) F1’s from five broods of mated ABC hermaphrodites crossed with CB4856 males. Worms with single markers were observed 73.8% of the time, corresponding to a 2.8:1 bias of oocytes inheriting one copy:two copies of chromosome IV from the mother. (E) Punnett square showing expected genotypes resulting from ABC hermaphrodite self-progeny. (F) Observed frequencies of self-progeny, compared with expected frequencies with no correction in sperm, demonstrating that the phenotypically ABC worms are ABC trisomy IV rather than AABC tetrasomy IV. diplo, diploid; haplo, haploid; Obs., observed.
Figure 6.
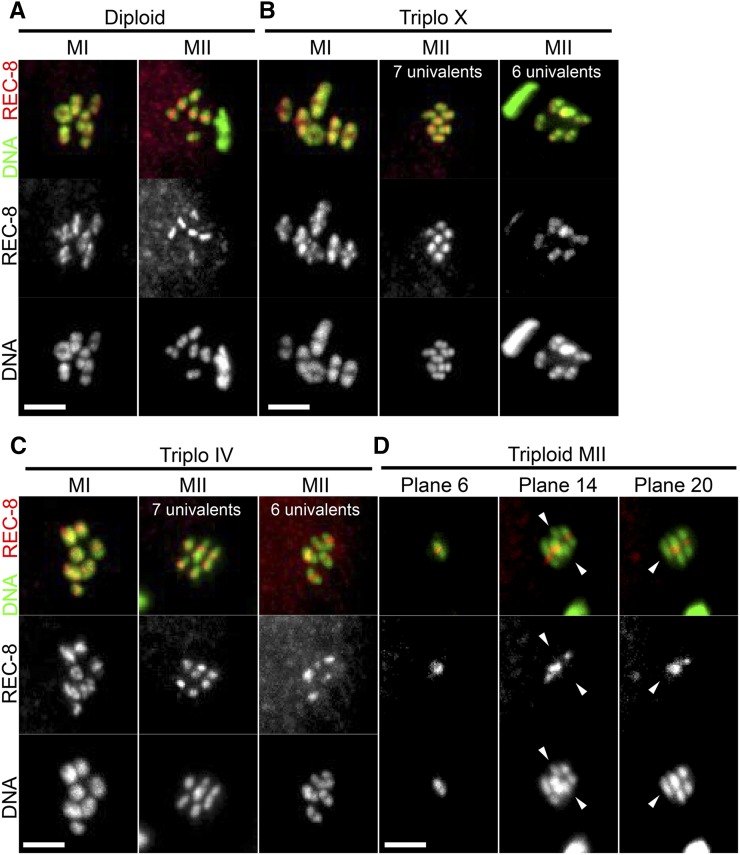
Single chromatids are present at metaphase II of triploids. z-projections of diploid embryos (A), triploX embryos (B), triploIV embryos (C), and triploid embryos (D) stained with REC-8 antibodies and DAPI. Metaphase I embryos: diploid control n = 27, triploX n = 38, triploIV n = 42, and triploid n = 12. Metaphase II embryos: diploid control n = 25, triploX n = 46, triploIV n = 39, and triploid n = 24. Arrowheads indicate chromatids with no REC-8 staining. Bar, 3 μm.
GFP::ASPM-1 asymmetry
Spinning disk confocal images of meiosis I spindles were captured in utero at focal planes separated by 0.5 μm. For each focal plane, spindle poles were identified by segmentation of pixels 1.4 times brighter than the adjacent cytoplasm and the three-dimensional (3D) pixel sum was determined for each pole. 3D pixel sums were obtained by combining the data from all the focal planes. Only spindles parallel to the plane of focus were analyzed because spherical aberration causes the further pole to be dimmer if there is any tilt in the z-axis.
Creating a trisomy IV strain
Chromosome IV was chosen because of a previous report that trisomy IV C. elegans are viable and fertile (Sigurdson et al. 1986). Males for crosses were generated by heat-shocking L4 worms at 30° for 8 hr. 4N hermaphrodites were crossed with VS21 males. F1’s were then selected and crossed with EG7911 males. Worms displaying both hjSi20(A) and oxTi705(B) were finally crossed with EG8299 males. At this point, worms displaying all three markers [hjSi20, oxTi705 and oxSi180(C)] were singled. Single worms with all three markers were picked for 18 successive generations. To maintain this strain, worms displaying all three markers were continually singled from the self-progeny of ABC worms using a fluorescence dissecting microscope. For test crosses with ABC hermaphrodites, we made CB4856 males by heat shocking L4 worms at 30° for 8 hr. Singled L4 hermaphrodites of our ABC strain were placed onto plates with CB4856 males in a 1:4 ratio for 24 hr and then singled. Only worms with mating plugs and 50% male progeny were used to score the segregation of different fluorescent markers, as this ensured that the worms had mated.
Male triploid analysis
4N hermaphrodites were mated with CB4856 males to generate triploid males. Sperm precursors were visualized by dissecting the male triploids on microscope slides in 10 μl of sperm media containing 100 mg/ml Hoechst 33342 (Miller and Shakes 1995). For the analysis of male triploid progeny, triploid males were mated with unc-5(e53) hermaphrodites expressing GFP::H2B. Wild-type progeny were counted, anesthetized, and visualized by live imaging (see above).
Data availability
Strains are available upon request. Strains and polymorphisms used in this study are listed in Table S1 in File S1. All potential deletion polymorphisms are listed in Table S2 in File S1. PCR primers used in this study are listed in Figure S2 in File S1. Representative images are presented within the article and complete imaging data sets are available upon request.
Results
To test whether a third copy of an autosome is lost during female meiosis at a frequency higher than the 50% expected from random segregation, tetraploid C. elegans hermaphrodites (SP346; Madl and Herman 1979) were mated with diploid males carrying a germline-expressed GFP::histone H2B transgene. Over 95% of the resulting triploid progeny had 12 discrete GFP::histone-labeled structures in their diakinesis oocytes (n = 36) and on the metaphase I plates of their meiotic embryos (n = 76) (Figure 1, C and D). This result contrasts with the six bivalents observed in diploid C. elegans (Figure 1, A and B) and is consistent with triploid hermaphrodites producing diakinesis oocytes and metaphase I embryos with six bivalents and six univalents as previously reported by Madl and Herman (1979). If each univalent maintained cohesion and segregated intact but randomly into the first polar body or into the embryo, chromosome number at metaphase II would fit a binomial distribution with the most abundant class of embryos having nine chromosomes (Figure 1E). Instead, the observed distribution was shifted with far more metaphase II embryos having six (fully diploid) or seven (single trisomy) GFP::histone-labeled chromosomal structures than expected from random segregation (Figure 1E). Using a Chi square test including all seven categories of chromosome number, the observed metaphase II distribution was significantly different from the binomial distribution expected from random segregation (P < 0.0001). To further test the significance of this apparent chromosome loss, we grouped chromosome number into three categories (6 or 7 chromosomes; 8 or 9 chromosomes; and 10, 11, or 12 chromosomes) to generate a 2 × 3 contingency table. By Fisher’s Exact Test, the observed metaphase II distribution of chromosome number was significantly different from that expected from random segregation (P = 0.00037). To test whether triploid chromosome number is reduced further after metaphase II, tetraploid hermaphrodites were first crossed with diploid males. The resulting triploid hermaphrodites were then crossed with diploid males carrying a germline-expressed GFP::histone H2B transgene, so that the female line was the sole contributor to aneuploidy in the progeny. Fifteen percent (229/1529) of the progeny arrested before hatching, 14% (211/1529) developed into adult males, and 71% (1089/1529) developed into adult hermaphrodites. The number of GFP::histone-labeled structures in the diakinesis oocytes of 194 adult hermaphrodites was then scored (Figure 1F). The distribution of chromosome number was shifted further toward six and seven chromosomes relative to the metaphase II chromosome counts in the triploid mother. This result might indicate further chromosome loss after anaphase I or it could be an artifact of aneuploids with greater chromosome numbers being overrepresented among the dead embryos and males whose chromosome number could not be determined.
To address the question of which chromosome numbers might be overrepresented among the nonhatching progeny of triploid hermaphrodites, we examined the progeny of triploid males crossed with diploid hermaphrodites. During C. elegans spermatogenesis, excess cytoplasm and organelles are eliminated in a large residual body. Thus meiotic correction might occur during male meiosis by leaving extra chromosomes in the residual body. Direct examination of spermatocytes from triploid males, however, did not reveal chromosomes in the residual body (Figure S1 in File S1), suggesting that segregation of univalents during triploid male meiosis is random. Nearly 100% of the viable progeny of triploid males were hermaphrodites. This allowed accurate scoring of chromosome number in the diakinesis oocytes of all but the inviable progeny of triploid males mated with diploid hermaphrodites (Figure 1G). Only worms with 9 or 10 chromosomes were significantly underrepresented relative to the binomial distribution expected from random segregation. This result suggested that only aneuploids with 9 or 10 chromosomes exhibit significant embryonic lethality. Therefore, we predicted that, among the nonviable progeny of triploid hermaphrodites, 57% would have 9 chromosomes and 43% would have 10 chromosomes, based on the binomial distribution expected from random segregation. We also predicted that the live male progeny of triploid hermaphrodites would have chromosome profiles similar to that of the live hermaphrodite progeny (16%: 6, 45%: 7, 22.5%: 8, 11%: 9, 5%: 10, and 0.5%: 11). Figure 1H shows a “corrected” chromosome profile in which the predictions for the nonviable embryos and live males have been added to the observed numbers for live hermaphrodites. This corrected distribution of chromosome numbers was significantly different from that observed for metaphase II spindles (Chi square P < 0.0001). The corrected average number of chromosomes was also significantly lower than the average chromosome number at metaphase II (Student’s t-test P = 0.0055). These results suggest that chromosome number may decrease further after metaphase II in triploids.
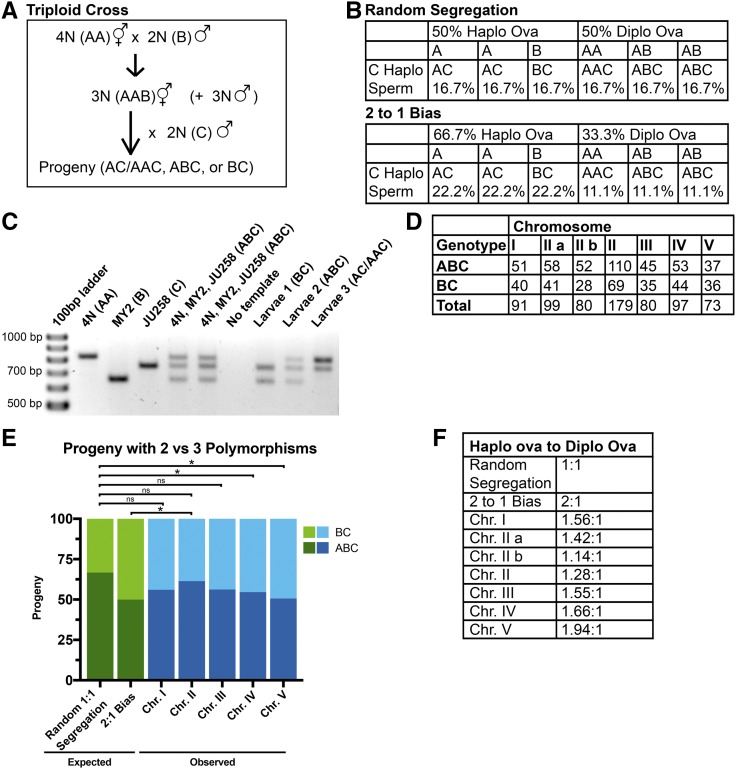
Because chromosome number cannot be easily scored in dead embryos or male progeny, and because GFP::histone labeling does not distinguish between different chromosomes, we developed a PCR-based strategy for quantifying trisomy correction of all five autosomes in 3N hermaphrodite × 2N male crosses (Figure 2A). We first wrote a code to identify small deletion/insertion polymorphisms for each autosome that would allow scoring of three alleles with a single PCR reaction from the whole-genome sequences of wild C. elegans isolates (Table S2 in File S1). The polymorphisms and strains used are listed in Table S1 in File S1. PCR primers and validation are shown in Figure S2 in File S1. Controls showing that these polymorphisms do not exhibit segregation bias in diploid × diploid crosses are shown in Figure S3 in File S1. 4N hermaphrodites, homozygous for the N2 Bristol allele designated AA, were mated to diploid polymorphic males carrying a second allele, designated B. The resulting AAB triploid hermaphrodites were mated to diploid males carrying a third allele, designated C (Figure 2A). Figure 2B shows the Punnett squares for random (1:1) segregation into the polar body vs. the embryo and for preferential 2:1 elimination of univalents in the polar body interpreted from triploX results (Cortes et al. 2015). An example of PCR results is shown in Figure 2C. For each chromosome, 18% dead embryo progeny, 14% L4 male progeny, and 68% L4 hermaphrodite progeny were subjected to PCR analysis. These percentages were within the SEM of the progeny counts described above. We concentrated our analysis on the BC and ABC genotypes for three reasons. First, these genotypes exclude any self-progeny from both the first and second cross. Second, the AC and AAC genotypes cannot be reliably distinguished by this assay. Inclusion of AC and AAC data therefore obscures differences between random (1:1) segregation and biased (2:1) segregation. Third, random (1:1) segregation should result in a 2:1 ABC:BC genotype ratio whereas 2:1 biased elimination of univalents would result in a 1:1 ABC:BC genotype ratio (Figure 2B). The frequencies of BC and ABC progeny are shown in Figure 2, D and E. Including the Holm–Bonferroni correction for multiple comparisons, the ABC:BC ratio was significantly different from that expected from random segregation only for chromosomes IV and V (P = 0.048 and 0.02, respectively). However, without the Holm–Bonferroni correction, the ABC:BC genotype ratio for chromosomes I, III, IV, and V was significantly different from that expected from random segregation (P = 0.031, 0.047, 0.012, and 0.004, respectively) and not significantly different from that expected from a 2:1 preferential univalent elimination (P = 0.25, 0.26, 0.36, and 0.9 respectively) by Chi square analysis. This result indicates that trisomy of some autosomes is preferentially corrected.
Figure 2.
Inheritance of three polymorphic alleles in a 3N × 2N cross. (A) Triploid progeny were generated by first crossing 4N hermaphrodites with a diploid polymorphic male strain to obtain 3N worms and then mating the 3N hermaphrodites to a second diploid polymorphic male strain. (B) Punnett squares of chromosome segregation in 3N worms. (C) An example of PCR results obtained for chromosome II. (D) Raw numbers of ABC and BC progeny produced from different crosses. (E) The frequencies of the ABC and BC genotypes were compared to the ABC:BC genotype ratios expected from random segregation and the 2:1 bias. * P < 0.05 by Chi square analysis after Holm-Bonferroni correction. (F) Haplo:diplo ova ratios that most closely fit the observed genotype ratios. Chr. chromosome; diplo, diploid; haplo, haploid; ns, not significant.
The ABC:BC genotype ratio for chromosome II was not significantly different from that expected from random segregation by Chi square analysis with or without Holm–Bonferroni correction (Figure 2E). Two different polymorphic loci on chromosome II were analyzed. The ABC:BC genotype ratios for each locus or the combined data for both loci were not significantly different from that expected from random segregation. This result suggested that third copies of different autosomes might be preferentially eliminated with different efficiencies. Punnett squares like those shown in Figure 2B were constructed for a continuous range of segregation biases and those with the ABC:BC genotypes most closely matching those observed for each chromosome are listed as “haplo to diplo ova” ratios in Figure 2F. Chromosome V had the highest apparent segregation bias (1.94 haplo ova: 1 diplo ova) and chromosome II had the lowest apparent segregation bias (1.28 haplo ova: 1 diplo ova). However, Fisher’s Exact test revealed no significant difference between the ABC:BC genotype data for chromosomes II and V (P = 0.12). Fisher’s Exact test also failed to reveal a significant difference between any pair of autosomes. Thus, we were unable to definitively demonstrate a significant difference in the efficiency of trisomy correction for different autosomes.
To test for preferential trisomy correction by a completely independent method, 2–50 celled embryos dissected from triploid hermaphrodites mated with diploid males or from selfed triploid hermaphrodites were fixed and labeled by FISH with a probe specific for the 5s rDNA repeat on chromosome V (Phillips and Dernburg 2006). Examples of two-celled embryos with two or three copies of chromosome V are shown in Figure 3, A and B and three focal planes of an older embryo with two copies of chromosome V in each nucleus are shown in Figure 3C. Random segregation of the third copy of chromosome V should result in equal frequencies of embryos with two or three copies from triploid hermaphrodites mated with diploid males. Instead, 63% of embryos had two copies of chromosome V and 37% had three copies (n = 89; Figure 3D). The observed frequencies were significantly different from random segregation by Chi square test (P = 0.0148). Among self-progeny, random segregation would generate offspring with two, three, or four copies of chromosome V at a ratio of 1:2:1. The observed frequencies were 25:23:15 (n = 63; Figure 3E), which, by Chi square test, is significantly different from random segregation (P = 0.02). The observed frequencies of self-progeny were not significantly different from what was expected from 63% haploV 37% diploV ova (from the cross-progeny results) and 50% haploV 50% diploV sperm by Fisher’s exact test (P = 0.39). These results confirm the preferential loss of a third copy of chromosome V during oocyte meiosis in a triploid hermaphrodite. Among the 152 total FISH-labeled embryos, none were obvious mosaics with > 10% of nuclei with a different number of copies of chromosome V. This result suggested that the reduction in total chromosome number between metaphase II and diakinesis of the grown progeny (Figure 1) occurs during anaphase II rather than during embryonic development.
To test whether trisomy correction occurs in a hermaphrodite with a true trisomy rather than triploidy, we generated a trisomy IV strain bearing three bright fluorescent transgenes inserted at the same locus. The “A” allele, a transgene expressing mCherry in the pharynx, and the “C” allele, consisting of a transgene expressing GFP in neurons, were both inserted at the exact same locus (cxTi10882) by MosSCI (Frøkjær-Jensen et al. 2012). The “B” allele, consisting of a transgene expressing nuclear tdTomato throughout the soma, was inserted by miniMos (Frøkjær-Jensen et al. 2014) into a site 0.14 map units from cxTi10882 (Figure 4A). Males homozygous for the A allele were first mated to nonfluorescent (XXXX) tetraploids. The resulting XXA triploids were mated to BB males and progeny bearing the A and B transgenes were mated to CC males. Progeny carrying all three fluorescent markers (Figure 4A) were picked for several generations. DAPI staining of diakinesis nuclei revealed seven rather than the normal six chromosomal bodies (Figure 4B).
To test for trisomy correction, ABC trisomy IV hermaphrodites were mated with diploid males with no fluorescent transgene. Of the progeny, 517/522 developed to adulthood. Random segregation should produce equal frequencies of progeny with one fluorescent transgene (A, B, or C) or two fluorescent transgenes (AB, AC, or BC) (Figure 4C). Instead, 73.8% of progeny had a single fluorescent marker and 26.2% of progeny had two fluorescent markers (Figure 4, C and D). This result is consistent with a 2.8:1 ratio of haplo IV ova to diplo IV ova and was significantly different from the 1:1 ratio expected from random segregation (P = 0.00001 by Chi square test, n = 522). Control crosses of A/C or A/B diploid hermaphrodites produced equal frequencies of each allele (Figure S4 in File S1) indicating that stochastic transgene silencing is not a significant contributing factor and that the transgenes do not cause non-Mendelian inheritance.
The DAPI staining could not reliably distinguish whether the extra chromosomal body in diakinesis oocytes was the univalent IV of a trisomy IV worm or the bivalent IV of a tetrasomy IV worm. However, an AABC tetrasomy IV worm, for example, would only have single-marker self-progeny with the A marker (Punnett square not shown), whereas a true ABC trisomy IV worm should have single-marker self-progeny for each marker (Figure 4E). Single-marker self-progeny were indeed observed with each of the three markers (Figure 4F) and dead embryos were observed among self-progeny. These results indicated that the phenotypically ABC worms were genotypically ABC trisomy IV worms and that tetrasomy IV is likely lethal. Self-progeny counts were also consistent with 2.8:1 biased elimination in ova and random 1:1 segregation in sperm (Figure 4F).
The apparent reduction in chromosome number between metaphase II and diakinesis of the adult progeny of triploid hermaphrodites (Figure 1H) suggested that extra chromosomes are lost both during anaphase I and at some time after metaphase II. Preferential loss of him-8 X univalents is associated with univalents that biorient but do not lose cohesion at anaphase I, resulting in lagging chromosomes during anaphase I in 90% of him-8 embryos (Cortes et al. 2015). Lagging chromosomes are extremely rare in control diploid embryos or in him-8 anaphase II embryos (Cortes et al. 2015; Figure 5A). We reasoned that if some univalents maintained cohesion while others lost cohesion during anaphase I of triploid embryos, then lagging univalents would be observed during anaphase I and lagging single chromatids would be observed during anaphase II. Time-lapse imaging of GFP::histone indeed revealed lagging chromosomes during anaphase I in 14/15 embryos from triploid mothers (Figure 5, B and C) and during anaphase II in 17/20 embryos from triploid mothers (Figure 5, D and E). These observations support the hypothesis that some univalents lose cohesion during anaphase I and the resulting single chromatids lag at anaphase II. Although 10/17 anaphase I laggards resolved into the polar body and 12/23 anaphase II laggards resolved into the polar body, we were not able to conclude whether this frequency is significantly different from random segregation. In the case of a 2:1 bias toward the polar body, as suggested by Figure 2F for chromosome V, detecting a significant difference from random segregation would require 75 time-lapse sequences. However, detecting a 1.6:1 bias, the average for all autosomes suggested by Figure 2F, would require > 150 time-lapse sequences. However, even random loss of single chromatids during anaphase II would result in a reduction in the number of chromosomal bodies after metaphase II.
To further test the hypothesis that single chromatids are present at metaphase II of triploids, meiotic embryos were fixed and stained with an antibody specific for the REC-8 cohesin subunit. REC-8 is present in a cruciform pattern on bivalents and in a single band on univalents (Pasierbek et al. 2001), and should be absent from single chromatids. One-hundred percent of metaphase I embryos from diploid control worms had six chromosomes with a cruciform pattern of REC-8 staining (n = 27) and 100% of metaphase II embryos from diploids had six chromosomes with a single band of REC-8 (n = 25; Figure 6A). One-hundred percent of metaphase I embryos from triploX worms had six chromosomes with cruciform labeling and one chromosome with a single band of REC-8 (n = 38). In addition, 67.4% of metaphase II embryos from triploX worms had six chromosomes, each with a single REC-8 band, and 32.6% of metaphase II embryos from triploX worms had seven chromosomes, each with a single REC-8 band (n = 46; Figure 6B). This result indicated that univalent X chromosomes in triploX always remain intact through anaphase I. Of the triploIV metaphase I embryos, 90.5% had six chromosomes with a cruciform and one chromosome with a single REC-8 band, while 9.5% of triploIV metaphase I embryos had seven chromosomes with REC-8 patterns that were difficult to score (n = 42). Of the triploIV metaphase II embryos, 66.7% had six chromosomes, each with a single REC-8 band, and 33.3% of triploIV metaphase II embryos had seven chromosomes, each with a single REC-8 band (n = 39; Figure 6C). This suggested that the X univalent and chromosome IV univalent are similar in that they both maintain sister chromatid cohesion during the first division. This result also suggested that X univalents and IV univalents are expelled into the first polar body with similar efficiency. Therefore, there is no conspicuous difference between an extra copy of a sex chromosome and an extra copy of an autosome throughout meiosis.
In triploid worms, 20/20 diakinesis oocytes and 12/12 metaphase I embryos had six chromosomes with a cruciform pattern and six chromosomes with a single band of REC-8 as expected. In 6/24 metaphase II embryos from triploids, all DAPI-staining bodies (1 × 6, 3 × 7, and 2 × 8) had a single REC-8 band similar to embryos from diploid or triploX worms. However, in 18/24 metaphase II embryos from triploid worms, one or more DAPI-staining bodies had no REC-8 staining while the remaining chromosomes had a single REC-8 band (Figure 6D). This result strongly supports the hypothesis that, in triploid worms, some univalents lose cohesion during anaphase I and the resulting single chromatids lag during anaphase II. Together with the observed reduction in chromosome number between metaphase II and adult progeny (Figure 1H), and the failure to detect mosaic embryos with clones of cells with different numbers of chromosome V (Figure 3), this result suggested that single chromatids are placed in the second polar body with at least random frequency.
In a previous study of preferential elimination of X univalents in a him-8 mutant, half of the preferential elimination was attributed to capture of late-lagging chromosomes by the ingressing polar body contractile ring. However, in half of him-8 time-lapse sequences, univalents were already associated with one anaphase chromosome mass before ingression of the polar body contractile ring (Cortes et al. 2015). During C. elegans meiosis, spindles are initially parallel to the cortex, spindle shortening then brings spindle poles into close proximity with metaphase chromosomes, one spindle pole then moves to the cortex, then homolog separation initiates on the extremely short, perpendicular spindle, which then elongates in an anaphase B-like process (Figure 7, A and B; McNally et al. 2016). Two closely related hypotheses might explain preferential positioning of univalent chromosomes proximal to the cortex before polar body ring ingression. First, univalent chromosomes offset toward one pole before rotation might cause accumulation of a molecule on that pole that caused the univalent-proximal pole to move to the cortex. Second, cortical contact might cause the cortex-proximal pole to accumulate a molecule that would cause more chromosome-motive force toward the cortex-proximal pole. Either model predicts asymmetric accumulation of some molecule on the cortical spindle pole relative to the interior pole. ASPM-1 is a microtubule minus end-binding protein (Jiang et al. 2017) that is concentrated at meiotic spindle poles and is required for spindle rotation (van der Voet et al. 2009), normal chromosome congression, and normal anaphase (Connolly et al. 2014). To test whether ASPM-1 is enriched on the cortical pole relative to the interior pole, we captured z-stacks of MI spindles that were parallel to the plane of focus in living embryos within a strain with the endogenous aspm-1 gene tagged with GFP (Connolly et al. 2015). Spindles were monitored by single-plane time-lapse imaging; one z-stack was captured just after spindle rotation and a second z-stack was captured during chromosome separation. The ratio of the GFP::ASPM-1 pixel sum for the cortical pole/interior pole (see Materials and Methods) was > 1.0 just after spindle rotation and increased during anaphase B for both diploid and triploX spindles (Figure 7, C and D). In principal, this apparent asymmetry could be due to spherical aberration making the more distant pole appear dimmer, or due to a number of other imaging artifacts. If the asymmetry were due to imaging artifacts, the cortical pole would appear brighter in 50% of embryos and the interior pole would appear brighter in 50% of embryos. In 36/37 GFP::ASPM-1 image stacks, the cortical pole was brighter than the interior pole with the single remaining spindle being almost perfectly symmetrical. This frequency was significantly different from the 50% expected from imaging artifacts (P < 0.0001 by Chi square test). Anti-ASPM-1 staining of fixed embryos after spindle rotation revealed that in 44/47 embryos, the cortical pole exhibited a greater pixel sum of ASPM-1 staining than the interior pole. This frequency was also significantly different from the 50% expected from imaging artifacts (P < 0.0001 by Chi square test). These results include diploid, XXX, and triploid spindles with no discernible difference between each type of spindle. This analysis demonstrates that ASPM-1 is present in a greater volume, a higher concentration, or both at the cortical pole relative to the interior pole. This is the first documentation of an asymmetrically-distributed molecule that might contribute either to preferential rotation of one pole to the cortex, greater pulling force toward the cortical spindle pole, or greater attachment of chromosomes to the cortical pole.
Discussion
Using multiple methods, we have shown that extra copies of autosomes are preferentially eliminated during anaphase I and are also eliminated during anaphase II of female meiosis in C. elegans. The efficiency of this elimination is far from perfect with the deviation from random segregation ranging from 1.6:1 to 2.8:1. We previously (Cortes et al. 2015) suggested that there is a selective advantage to a lower efficiency of correction if crossover failures are more common than trisomy. Perfect elimination of univalent autosomes would result in lethal monosomy in the case of autosomal crossover failure. The exact efficiency of univalent X expulsion increases the frequency of diploid offspring from triploX worms but does not reduce the frequency of diploid offspring below that expected from random segregation in him-8 worms.
Potential mechanisms of preferential elimination of autosomal univalents might be inferred from studies of him-8 univalent X chromosomes. A crossover is normally required to load the aurora B kinase, AIR-2, on MI bivalents (Rogers et al. 2002), thus him-8 MI X univalents lack AIR-2 (Cortes et al. 2015; Muscat et al. 2015), which is required for loss of cohesion (Rogers et al. 2002). him-8 univalents thus remain intact through anaphase I and frequently lag during anaphase I (Cortes et al. 2015; Muscat et al. 2015). The failure to detect chromatids without REC-8 in metaphase II spindles of triploX worms or trisomy IV worms (Figure 6) indicates that univalent chromosomes in triploX or trisomy IV worms also remain intact during anaphase I. Time-lapse imaging revealed that late-lagging X chromosomes are frequently captured by the polar body contractile ring (Cortes et al. 2015), which normally ingresses from the embryo surface to the midpoint of the late anaphase spindle (Fabritius et al. 2011). Inhibition of polar body ring ingression caused the frequency of late-lagging univalents moving toward the cortical pole to be randomized, and hyperactivation of polar body ring ingression with a myosin phosphatase mutant increased the frequency of elimination of univalent X chromosomes (Cortes et al. 2015). Thus, late-lagging autosomal univalents in triploid and trisomy IV embryos may also be captured by the asymmetrically-ingressing polar body. However, in half of him-8 anaphase I time-lapse sequences, lagging univalents moved into the cortical mass of chromosomes more frequently than into the interior mass of chromosomes before contractile ring ingression. The frequency of chromosome movement toward the cortex was unaffected by polar body inhibition for this subset of embryos (Cortes et al. 2015). Thus, additional mechanisms must generate biased movement of univalent chromosomes toward the cortex and future polar body.
Late-lagging chromosomes in 7/15 triploid anaphase I spindles (Figure 5B) and 4/20 triploid anaphase II spindles (Figure 5D) appeared elongated as if they were being stretched. This stretched appearance suggests that these chromosomes are under tension. Evidence for chromosome stretching has previously been reported for prometaphase monopolar spindles (Muscat et al. 2015) and bipolar spindles just before homolog separation (McNally et al. 2016). However, the mechanisms generating tension during late anaphase are unclear because outer kinetochore proteins dissociate from bivalents early in anaphase I (Dumont et al. 2010; Hattersley et al. 2016; McNally et al. 2016). Thus, several noncanonical anaphase mechanisms have been proposed for C. elegans female meiosis (Dumont et al. 2010; Muscat et al. 2015; McNally et al. 2016). The stretched appearance of lagging univalents supports mechanisms that generate a pulling force toward spindle poles. The higher concentration of ASPM-1 at the cortical pole during anaphase (Figure 7) might therefore support the idea of more pulling force toward the cortical pole. This hypothetical asymmetric force would have a minimal influence on bivalents that lose cohesion normally but would bias the inheritance of univalents or single chromatids that do not lose cohesion.
In many cases, trisomy or triploidy results in lethality or sterility due to checkpoint-mediated apoptosis of germ cells (e.g., Bhalla and Dernburg 2005). Consistent with this generality, triploid female oysters have reduced brood sizes but 57% of their progeny are diploid (Gong et al. 2004), suggesting trisomy correction during oogenesis. Women with trisomy 21 have been reported to have 60% diploid offspring (Bovicelli et al. 1982). Likewise, women with three copies of the X chromosome have also been reported to have normal fertility (Stochholm et al. 2013) and have far fewer than 50% XXX or XXY offspring (Neri 1984; Ratcliffe et al. 1990; Robinson et al. 1990; Stewart et al. 1990). However, diplotene or metaphase I oocytes of these trisomy 21 and triploX women were not examined and thus it is possible that these women were mosaics with diploid ovaries. Chromosomal mosaicism is extremely common in in vitro-fertilized human embryos (van Echten-Arends et al. 2011) and chromosomal mosaicism also occurs in adult humans (Conlin et al. 2010); however, its frequency is largely unknown because it is difficult to detect. We suggest that the meiotic trisomy correction reported here for C. elegans may allow trisomic invertebrates or mosaicly trisomic vertebrates to have a higher frequency of diploid offspring than would be generated by symmetric meiosis. This may be one of the selective advantages of asymmetric female meiosis.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300259/-/DC1.
Acknowledgments
We thank Michelle Panzica for critical reading of the manuscript. We thank Erik Jorgensen, Bruce Bowerman, Anne Villeneuve, and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440), for strains. This work was supported by National Institute of General Medical Sciences grant 1R01-GM-079421 and US Department of Agriculture National Institute of Food Agriculture Hatch project 1009162 (to F.J.M.).
Footnotes
Communicating editor: M. Colaiácovo
Literature Cited
- Bhalla N., Dernburg A. F., 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Bovicelli L., Orsini L. F., Rizzo N., Montacuti V., Bacchetta M., 1982. Reproduction in Down syndrome. Obstet. Gynecol. 59(6 Suppl.): 13S–17S. [PubMed] [Google Scholar]
- Conlin L. K., Thiel B. D., Bonnemann C. G., Medne L., Ernst L. M., et al. , 2010. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 19: 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly A. A., Osterberg V., Christensen S., Price M., Lu C., et al. , 2014. Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol. Biol. Cell 25: 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly A. A., Sugioka K., Chuang C. H., Lowry J. B., Bowerman B., 2015. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J. Cell Biol. 210: 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes D. B., McNally K. L., Mains P. E., McNally F. J., 2015. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. Elife 4: e06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., Desai A., 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabritius A. S., Flynn J. R., McNally F. J., 2011. Initial diameter of the polar body contractile ring is minimized by the centralspindlin complex. Dev. Biol. 359: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong N., Yang H., Zhang G., Landau B. J., Guo X., 2004. Chromosome inheritance in triploid Pacific oyster Crassostrea gigas Thunberg. Heredity 93: 408–415. [DOI] [PubMed] [Google Scholar]
- Hattersley N., Cheerambathur D., Moyle M., Stefanutti M., Richardson A., et al. , 2016. A nucleoporin docks protein phosphatase 1 to direct meiotic chromosome segregation and nuclear assembly. Dev. Cell 38: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Rezabkova L., Hua S., Liu Q., Capitani G., et al. , 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby C., Kusch M., Kemphues K., 1990. Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev. Biol. 142: 203–215. [DOI] [PubMed] [Google Scholar]
- Madl J. E., Herman R. K., 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93: 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- McNally K. P., Panzica M. T., Kim T., Cortes D. B., McNally F. J., 2016. A novel chromosome segregation mechanism during female meiosis. Mol. Biol. Cell 27: 2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., Shakes D. C., 1995. Immunofluorescence microscopy. Methods Cell Biol. 48: 365–394. [PubMed] [Google Scholar]
- Miller M. P., Amon A., Ünal E., 2013. Meiosis I: when chromosomes undergo extreme makeover. Curr. Opin. Cell Biol. 25: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat C. C., Torre-Santiago K. M., Tran M. V., Powers J. A., Wignall S. M., 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. Elife 4: e06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297: 559–565. [DOI] [PubMed] [Google Scholar]
- Neri G., 1984. A possible explanation for the low incidence of gonosomal aneuploidy among the offspring of triplo-X individuals. Am. J. Med. Genet. 18: 357–364. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., et al. , 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Dernburg A. F., 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11: 817–829. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., McDonald K. L., Dernburg A. F., 2009. Cytological analysis of meiosis in Caenorhabditis elegans. Methods Mol. Biol. 558: 171–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, S. G., G. E. Butler, and M. Jones, 1990 Edinburgh study of growth and development of children with sex chromosome abnormalities. IV. Birth Defects Orig. Artic. Ser. 26: 1–44. [PubMed] [Google Scholar]
- Robinson, A., B. G. Bender, M. G. Linden, and J. A. Salbenblatt, 1990 Sex chromosome aneuploidy: the Denver Prospective Study. Birth Defects Orig. Artic. Ser. 26: 59–115. [PubMed] [Google Scholar]
- Rogers E., Bishop J. D., Waddle J. A., Schumacher J. M., Lin R., 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson D. C., Herman R. K., Horton C. A., Kari C. K., Pratt S. E., 1986. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol. Gen. Genet. 202: 212–218. [DOI] [PubMed] [Google Scholar]
- Stewart, D. A., J. D. Bailey, C. T. Netley, and E. Park, 1990 Growth, development, and behavioral outcome from mid-adolescence to adulthood in subjects with chromosome aneuploidy: the Toronto Study. Birth Defects Orig. Artic. Ser. 26: 131–188. [PubMed] [Google Scholar]
- Stochholm K., Juul S., Gravholt C. H., 2013. Poor socio-economic status in 47,XXX–an unexpected effect of an extra X chromosome. Eur. J. Med. Genet. 56: 286–291. [DOI] [PubMed] [Google Scholar]
- van der Voet M., Berends C. W., Perreault A., Nguyen-Ngoc T., Gönczy P., et al. , 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat. Cell Biol. 11: 269–277. [DOI] [PubMed] [Google Scholar]
- van Echten-Arends J., Mastenbroek S., Sikkema-Raddatz B., Korevaar J. C., Heineman M. J., et al. , 2011. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum. Reprod. Update 17: 620–627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. Strains and polymorphisms used in this study are listed in Table S1 in File S1. All potential deletion polymorphisms are listed in Table S2 in File S1. PCR primers used in this study are listed in Figure S2 in File S1. Representative images are presented within the article and complete imaging data sets are available upon request.