Lesly, Bandura, and Calvi show that rapid DNA synthesis during early Drosophila embryogenesis is highly sensitive to mild mutations...
Keywords: Drosophila, DNA replication, Humpty dumpty, Donson, nuclear cleavage, microcephalic dwarfism
Abstract
Problems with DNA replication cause cancer and developmental malformations. It is not fully understood how DNA replication is coordinated with development and perturbed in disease. We had previously identified the Drosophila gene humpty dumpty (hd), and showed that null alleles cause incomplete DNA replication, tissue undergrowth, and lethality. Animals homozygous for the missense allele, hd272-9, were viable, but adult females had impaired amplification of eggshell protein genes in the ovary, resulting in the maternal effects of thin eggshells and embryonic lethality. Here, we show that expression of an hd transgene in somatic cells of the ovary rescues amplification and eggshell synthesis but not embryo viability. The germline of these mothers remain mutant for the hd272-9 allele, resulting in reduced maternal Hd protein and embryonic arrest during mitosis of the first few S/M nuclear cleavage cycles with chromosome instability and chromosome bridges. Epistasis analysis of hd with the rereplication mutation plutonium indicates that the chromosome bridges of hd embryos are the result of a failed attempt to segregate incompletely replicated sister chromatids. This study reveals that maternally encoded Humpty dumpty protein is essential for DNA replication and genome integrity during the little-understood embryonic S/M cycles. Moreover, the two hd272-9 maternal-effect phenotypes suggest that ovarian gene amplification and embryonic cleavage are two time periods in development that are particularly sensitive to mild deficits in DNA replication function. This last observation has broader relevance for interpreting why mild mutations in the human ortholog of humpty dumpty and other DNA replication genes cause tissue-specific malformations of microcephalic dwarfisms.
GENOMIC DNA must be fully and accurately replicated when a cell divides. To achieve this, multiple steps of DNA replication are coordinated with different phases of the eukaryotic cell cycle. Prereplicative complexes (pre-RC) assemble onto replication origins in G1 phase, and are then activated by cell cycle kinases to initiate replication during S phase (Diffley et al. 1994; Parker et al. 2017; Tocilj et al. 2017; Yuan et al. 2017; Zhai et al. 2017). Upon initiation, a large replisome complex of proteins assembles at replication forks, which then migrate bidirectionally outward from the origin to synthesize new DNA strands (Pellegrini and Costa 2016). Reassembly of new pre-RCs is then inhibited until after the next mitosis, ensuring that each genomic region is duplicated only once per cell division (Hills and Diffley 2014). Problems with any of these steps of DNA replication can cause “replication stress” and DNA damage, which is sensed and corrected by cell cycle checkpoints (Hills and Diffley 2014). However, high levels of replication stress and checkpoint failure are major contributors to genome instability and cancer (Hills and Diffley 2014; Zeman and Cimprich 2014). In addition, hypomorphic mutations in a number of human genes that encode pre-RC and other replication proteins are the genetic cause of a class of developmental malformations known as microcephalic primordial dwarfisms (MPDs) (Klingseisen and Jackson 2011; Khetarpal et al. 2016). While much has been learned, it is still not fully understood how the DNA replication program is coordinated with development and perturbed in disease.
An extreme example of developmental modification to the DNA replication program occurs during early Drosophila embryogenesis. Embryogenesis begins with division of nuclei in a common syncytium, and the first nine of these “nuclear cleavage” divisions consist of only S and M phases (Rabinowitz 1941; Foe and Odell 1993; O’Farrell et al. 2004). The duration of these first nine S/M cycles are rapid, ∼8 min, during which genomic DNA is replicated in as little as 3 min by initiating replication from numerous, closely spaced origins (∼10 kb apart) (Blumenthal et al. 1974; Sasaki et al. 1999). Because the majority of zygotic transcription does not begin until later during the mid-blastula transition (MBT), the early nuclear cleavage divisions are completely dependent on maternally encoded proteins (Edgar and O’Farrell 1989). Much remains unknown, however, about the regulation of the first nine S/M cycles, and how complete genome duplication exactly once per cycle is ensured. These early cycles lack a DNA replication or DNA damage checkpoint, although they do have a spindle assembly checkpoint (SAC) that monitors microtubule-kinetochore attachments (Raff and Glover 1988; Girdham and Glover 1991; Fogarty et al. 1997; Sibon et al. 1997, 1999, 2000; Takada et al. 2003, 2007, 2015; Perez-Mongiovi et al. 2005; Crest et al. 2007; Oliveira et al. 2010). While the early, rapid S/M cycles may be an advantage by shortening developmental time, they also increase the risk of propagating DNA mutation into multiple cells that are descended from these early founder nuclei (O’Farrell et al. 2004).
Another extreme example of developmental modification to the DNA replication program occurs during Drosophila oogenesis when genes required for eggshell (chorion) synthesis are selectively amplified in DNA copy number (Spradling 1981). This developmental gene amplification occurs through DNA rereplication from amplicon origins at six loci in somatic follicle cells of the ovary (Claycomb and Orr-Weaver 2005; Calvi 2006; Nordman and Orr-Weaver 2012). Similar to other origins, amplicon origins are bound by the pre-RC and activated by cell cycle kinases, and have been used as a model system to uncover conserved DNA replication mechanisms (Calvi et al. 1998; Asano and Wharton 1999; Royzman et al. 1999; Loebel et al. 2000; Aggarwal and Calvi 2004; Zhang and Tower 2004; Nordman et al. 2011; Alexander et al. 2016). Among other methodological advantages, gene amplification can be used for forward genetic identification of DNA replication genes. Mild, hypomorphic mutations in a number of pre-RC and other essential replication protein genes are homozygous viable, but defective for gene amplification in the ovary, resulting in a distinct thin eggshell phenotype (Landis et al. 1997; Landis and Tower 1999; Whittaker et al. 2000, Yamamoto et al. 2000; Claycomb et al. 2002; Schwed et al. 2002; Calvi 2006).
We used this forward genetic system to identify the gene humpty dumpty (hd) (Snyder et al. 1986; Bandura et al. 2005). Animals homozygous for a mild hypomorphic allele, hd272-9, are homozygous viable, but adult females are defective for amplification, resulting in eggs with thin shells that dehydrate and fail to develop. In contrast, animals homozygous for null alleles of hd die as pupae with severely underdeveloped brains and imaginal discs. Further analysis showed that hd expression is regulated by E2F1 and peaks at G1/S, and is essential in all proliferating cells to prevent incomplete DNA replication and DNA damage (Bandura et al. 2005). Hd protein sequence is conserved in multicellular eukaryotes, with one ortholog in each genome, suggesting a conserved essential role in genome integrity (Bandura et al. 2005). The mouse ortholog of Humpty dumpty was named Downstream of Son (Donson or Dons) because of its genomic position (Wynn et al. 2000). Consistent with evidence in flies, human Donson expression peaks at G1/S, and RNAi screens indicated that it is required for genome integrity (Whitfield et al. 2002; Fuchs et al. 2010).
It has been reported recently that human Dons associates with replication forks, and that Dons missense mutations are a genetic cause of microcephalic primordial dwarfisms (MPDs) (Evrony et al. 2017; Reynolds et al. 2017). MPDs are characterized by severe pre and postnatal undergrowth with microcephaly, and display variably expressive other phenotypes (Klingseisen and Jackson 2011; Khetarpal et al. 2016). Hypomorphic mutations in a number of replication proteins result in a subtype of MPD known as Meier-Gorlin Syndrome (MGS) (Khetarpal et al. 2016). It remains unclear, however, why different hypomorphic mutations in Dons and other essential replication proteins have different clinical presentations and affect some tissues more than others.
In this study we show that maternally encoded Hd protein is essential to support DNA synthesis during the rapid S phases of the enigmatic embryonic cleavage cycles, In addition, our results uncover that there are two time periods in Drosophila development that are sensitive to deficits of DNA replication proteins: ovarian developmental gene amplification and embryonic nuclear cleavage cycles. We discuss the broader implications of our findings for interpreting why mild deficits in the function of the hd ortholog, Donson, and other DNA replication proteins result in the tissue-specific phenotypes of microcephalic primordial dwarfisms.
Materials and Methods
Drosophila strains and genetics
All of the hd mutant strains have been described previously (Bandura et al. 2005), and other strains were obtained from the Bloomington Drosophila Stock Center (BDSC, Bloomington, IN). The plu3/CyO strain was a gift from T. Orr-Weaver (Shamanski and Orr-Weaver 1991). For the epistasis analysis of Figure 6, C323-Gal4; plu3/CyO-GFP; Df(3R)3-4 ru th st/TM6 Tb Hu females were created and crossed to P{w+mC UAS-hd}; plu3/SM1a; h th st hd272-9 e/TM3 Sb males. Different adult female progeny from this cross were either homozygous mutant for plu, or hd, both, or neither, and the nuclear cleavage phenotype of their embryos analyzed. Clonal analysis of hdFf in the ovary was performed by crossing males from the strain w1118; P{ry +t7.2 neor FRT}82B P{w +mC Ubi-GFP(S65T)nls}3R/TM6B Tb to virgin females that were either w; FRT82B hdFf/TM3, or w; FRT82B for control clones, as previously described (Bandura et al. 2005).
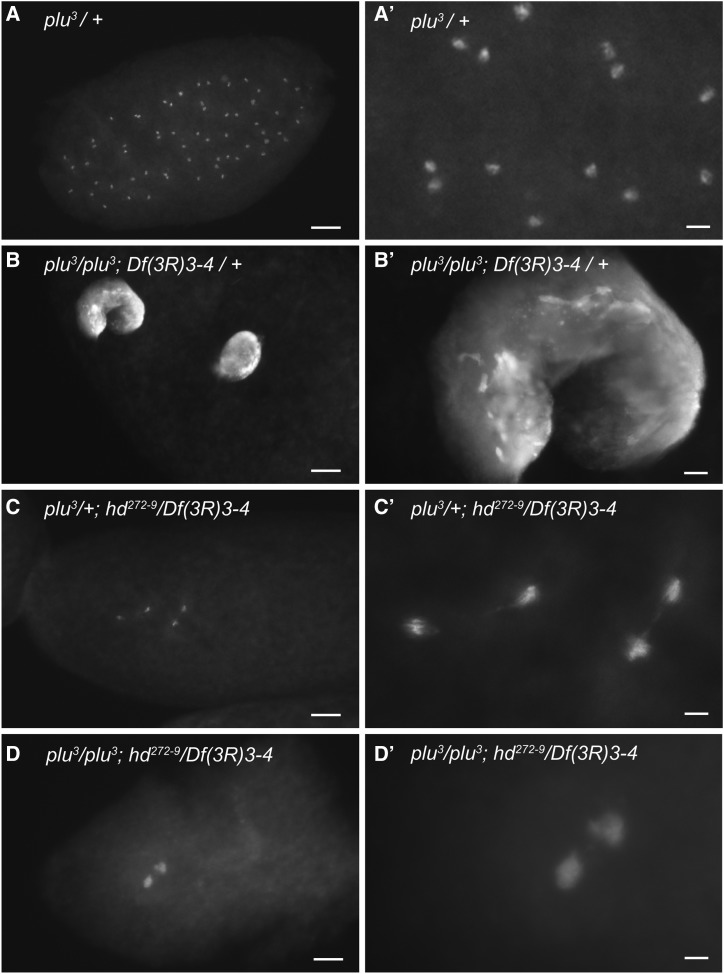
Figure 6.
hd is epistatic to plu and required for DNA replication. (A–D’) Embryos from mothers that were either single or double mutant for hd and plu were labeled with DAPI. Lower (A–D) and higher (A’–D’) magnifications of nuclear phenotypes are shown. (A and A’) An embryo with normal nuclear cleavage divisions from a mother heterozygous for the recessive plu3 mutation and wild type for hd. (B and B’) An embryo with two large polyploid nuclei from a mother that was mutant for plu but heterozygous for the recessive hd deletion Df(3R)3-4. (C and C’) An embryo with arrest at the second cleavage division and chromosome bridges from a hd SR mother who was also heterozygous for the recessive plu mutation. (D and D’) hd is epistatic to plu. An embryo from a hd SR mother that was also homozygous mutant for plu with a cleavage arrest/chromosome bridge phenotype similar to that of embryos from hd SR single mutant mothers. Bar, (A–D) 100 μm, (A’–D’) 20 μm.
Construction of hd rescue transgenes
The P-element transgene P{w+mC UAS-hd} expresses the hd cDNA under control of the UASt promoter, and was previously described (Bandura et al. 2005). The P-element transgene P{ w+mC hd-GFP} expresses an Hd-GFP fusion protein under control of the hd promoter. It contains a genomic subclone of the hd locus that is fused in frame on its 3′ end to eGFP, including 575 bp of genomic DNA upstream of the hd transcription start site, which contains a putative E2F binding site that responds to E2F1/Dp overexpression. The details of its construction using the Gateway Cloning System (ThermoFisher) are available upon request. An insertion of P{ w+mC hd-GFP} on the third chromosome was used for rescue experiments of Figure 4.
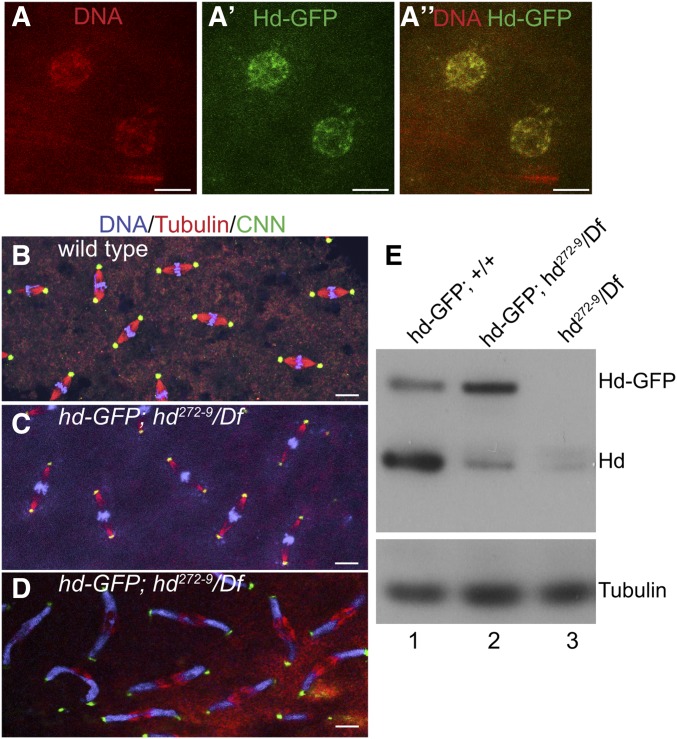
Figure 4.
Expression of hd-GFP in the maternal germline partially rescues embryonic nuclear cleavage cycles. (A–A”) Germline and somatic rescue (GSR) of hd mothers. Confocal images of nuclei of an early embryo from a hd-GFP; hd272-9/Df(3R)3-9 mother that expresses hd-GFP under control of the hd promoter in both germline and somatic cells of the ovary. Shown are DNA (A) Hd-GFP (A’) and merge (A”). Bar, 10 μm. (B–D) Maternally supplied Hd-GFP partially rescues the hd cleavage defect. Confocal images of embryonic mitoses from wild-type (B) and hd GRM mothers during early (C) and later (D) nuclear cleavage cycles. Nuclei in hd GSR embryos initially divide normally, partially migrate to cortex, but then eventually arrest with a highly expressive chromosome bridge phenotype during cycles 8–9. Bar for (B–D) are 10 μm. (E) Western Blot of 0–1 hr embryo extracts from mothers of genotypes indicated at the top of each lane. Blots were incubated with anti-Hd, which recognizes both endogenous Hd and Hd-GFP. Incubation with α-Tubulin was used as a loading control.
Embryo and ovary preparation and immunofluorescent analysis
Ovaries were dissected, fixed, and labeled as previously described (Bandura et al. 2005). Embryos were collected by allowing females to lay eggs on grape plates supplemented with fresh yeast paste for 1 hr. For nuclear counts in Figure 1, however, embryos were collected for 30 min, and fixed immediately or transferred to a humid chamber to develop at 25° for 30 or 60 min as indicated before fixing. Embryos were fixed by first dechorionating with a 1:1 mixture of household bleach: 0.2% NaCl/0.02% Triton-X 100, and then slow fixed using a modification of previous protocols (Rothwell and Sullivan 2000). Briefly, dechorionated embryos were brushed into a glass vial, and then 1 ml heptane was added followed by 1 ml of 3.7% formaldehyde in PEM buffer. After shaking the vial for 15 sec, the embryos were allowed to fix at room temperature for 15 min. The bottom layer was removed and 1 ml of methanol was added. After vortexing for 10 min, the embryos that were permeablized and sank were collected. Embryos were stored at −20°. Before antibody labeling, embryos were rehydrated through a PBTA/Methanol series as previously described (Rothwell and Sullivan 2000). Embryos that were labeled with anti-βTubulin were fixed in 500 μl heptane and 500 μl of room temperature 97% methanol with 15 mM EGTA for 10 min with constant shaking. The embryos were allowed to settle for up to 1 min and the fix removed. Embryos were washed 3× with methanol-EGTA solution, then fresh methanol-EGTA was added and the embryos stored at −20° for a minimum of 24 hr. Before fixation embryos were rehydrated using protocols appropriate for the fixation as described (Rothwell and Sullivan 2000).
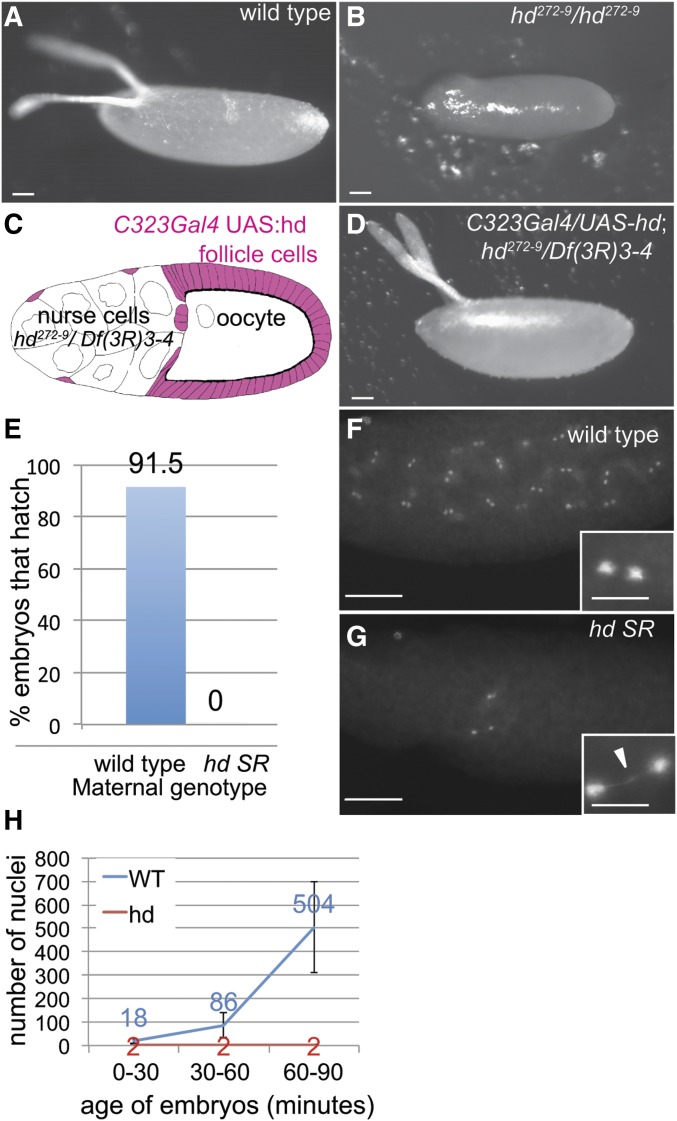
Figure 1.
Expression of hd in ovarian follicle cells rescues eggshell synthesis but not embryo survival. (A and B) Eggs laid by wild type (A) and hd272-9 mutant (B) mothers. Bar, 50 μm. (C) Somatic rescue (SR) of hd mutant mothers. Strategy for rescue of gene amplification defects in hd272-9/Df(3R)3-4 by driving expression of UAS-hd with c323GAL4 in somatic follicle cells (pink), but not germline nurse cells and oocytes. (D) Egg laid by hd SR female with fully rescued eggshell synthesis. Bar, 50 μm. (E) Eggs from hd SR mothers fail to hatch. Numbers indicate percent of wild type or hd SR embryos that hatched, n = 200 embryos. (F and G) Cleavage stage embryos from wild type (F) and hd SR (G) females labeled with DAPI, with higher magnifications shown in insets. Arrowhead in inset of (G) points to a chromosome bridge. Bar in (F and G) are 100 and 25 μm in the insets. (H) Quantification of the number of nuclei in wild type and hd SR embryos during three time windows after egg deposition (AED). Females were allowed to lay eggs on plates for 30 min, after which the population of embryos was either fixed immediately (0–30 min) or allowed to age for 30 min (30–60 min) or 60 min (60–90 min) before fixing and DAPI labeling. Nuclei were counted in >30 embryos, and the mean and SD of nuclear number per embryo are indicated. The error bars for hd SR embryos are not apparent because most embryos had two nuclei and the SD is very small.
The following primary antibodies were used for labeling fixed embryos or ovaries at the indicated concentrations: Rabbit anti-full length Hd antibody (1:1,000) (Bandura et al. 2005), mouse anti-βTubulin 1:500 [Developmental Studies Hybridoma Bank, (DSHB)], mouse anti-Cyclin B (1:100) (DHSB), rabbit polyclonal anti-GFP (1:50) (Invitrogen), rabbit anti-Cnn antibody was used at (1:200) (Eisman et al. 2009). Fluorescent secondary antibodies were anti-mouse or anti-rabbit Alexa 488 and Alexa 568 (Invitrogen), both used at 1:500–1:750. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) or TOTO-3 (Molecular Probes) as described (Calvi and Spradling 2001). Embryos and ovaries were mounted in Vectashield (VECTOR laboratories). Slides were analyzed using Leica SP5 laser scanning confocal microscope with LAS AF software, and a Leica DMRA2 epifluorescence microscope with Openlab software. Bright field images of the eggshell were taken with a Nikon SMZ1500 microscope and DMX1200 Nikon digital camera with Act-1 imaging software.
Protein preparation and Western blotting
Protein was prepared from 0 to 1 hr. dechorionated embryos by grinding in a microcentrifuge tube followed by boiling in Laemmli loading buffer (Adolphs et al. 1977; Thomer et al. 2004). Protein was transferred to membrane by electroblotting and Hd protein detected with rabbit anti- full-length Hd antibody at 1:10,000 as previously described (Bandura et al. 2005). Mouse anti-βActin (1:250) (Sigma A1978) or mouse anti-αTubulin (1:1000) (Eisman et al. 2009) were used as loading controls.
Data and reagent availability
More information about reagents used in this study can be found in the Supplemental Material, Reagent Table. All reagents from this study are freely available upon request.
Results
Expression of a humpty dumpty cDNA in ovarian somatic follicle cells rescues eggshell synthesis but not embryo viability
We previously showed that flies homozygous for the humpty dumpty (hd) hypomorphic allele, hd272-9 (G463D), are viable, but adult females have a severe defect in developmental gene amplification, and produce eggs with severely thin shells that dehydrate and collapse after they are laid (Figure 1, A and B and Table 1) (Bandura et al. 2005). As part of our initial identification of hd, we had used c323GAL4 to drive expression of a hd cDNA (UAS-hd) specifically in somatic follicle cells of the ovary, which completely rescued eggshell gene amplification and eggshell synthesis in these hd272-9 mutant females (Bandura et al. 2005). Despite rescue of eggshell synthesis, the embryos from these rescued females failed to hatch into larvae. Crossing these rescued females to wild-type males resulted in embryos heterozygous for the recessive hd272-9, but these embryos again failed to hatch, indicating that this failure in development is the result of a maternal effect. To ensure that this maternal-effect, embryonic lethality is not caused by homozygosity for an undefined mutation on the hd272-9 chromosome, we repeated this experiment by expressing UAS-hd in follicle cells of females who were trans-heterozygous for hd272-9 and a deletion allele of hd, Df(3R)3-4 (Figure 1C and Table 1) (Bandura et al. 2005). These rescued females laid eggs whose shells and overall morphology were indistinguishable from wild type, yet their embryos again failed to develop and hatch into larvae (Figure 1, D and E and Table 1). Given that the GAL4/UAS system that we used is active in somatic follicle cells but not the germline, we hypothesized that the embryonic lethality was the result of a deficit of hd that is supplied to the embryo through the maternal germline (Figure 1C). Because of this special genotype, we will hereafter refer to the c323GAL4/UAS-hd; hd272-9/Df(3R)3-4 females as hd Somatic Rescue (SR) mothers, and their offspring as hd SR embryos (Figure 1D).
Table 1. Summary of hd genotypes and phenotypes.
| Maternal Effects | |||
|---|---|---|---|
| Genotype | Viabilitya | Eggshell Synthesisb | Embryonic Cleavagec |
| hdFf/hdFf | — | N.A. | N.A. |
| hd272-9/hd272-9 | + | — | — |
| hd272-9/Df(3R)3-4 | + | — | — |
| c323Gal4/UAS-hd; hd272-9/Df(3R)3-4 | + | + | — |
| hd-GFP/hd-GFP; hd272-9/Df(3R)3-4 | + | + | +/−d |
| hd-GFP/hd-GFP; hdFf/Df(3R)3-4 | +e | n.d. | n.d. |
+, viable to adulthood; −, not viable to adulthood.
+, adult females have normal gene amplification and produce eggs with normal shells; −, defective amplification and thin eggshells.
+, embryos from mothers of indicated genotype have normal cleavage divisions; −, arrest during first few cleavage divisions.
+/−, hd-GFP expression in the maternal germline rescued the early embryonic cleavage arrest, but these embryos arrested during later cleavage division.
Rescue of viability was ∼100% relative to sibling controls.
Embryos from hd SR mothers arrest during the first few nuclear cleavage cycles
To explore why embryos from hd SR mothers failed to hatch, we examined embryonic development using fluorescence microscopy. A population of embryos from wild-type and hd SR females distributed from 0 to 1 hr of development were fixed and labeled with the fluorescent DNA dye DAPI. While wild-type embryos had up to several 100 nuclei, all hd embryos had, at most, a few nuclei, which often had long chromosome bridges between them (up to 30 μm) (Figure 1, F and G). These embryos were fertilized because arrested nuclei were in the interior of the embryo, indicating that maternal pronuclear migration had occurred on microtubules that are nucleated by sperm-derived centrioles (Figure 1G, see also centriole labeling in Figure 2). To quantify the arrest of hd SR embryos, we allowed wild-type and hd SR females to lay eggs on plates, and then aged the embryos for different amounts of time at 25° before fixing and DAPI labeling. Nuclear number increased exponentially during the rapid cleavage cycles of wild-type embryos, whereas hd SR embryos had an average of only two nuclei up to 90 min of development, with almost all nuclei connected by chromatin bridges (Figure 1H). Because expression of the zygotic genome does not begin until the MBT, these early nuclear cleavage cycles are completely dependent on the maternal supply of RNA and protein. These data suggested, therefore, that embryos from hd SR mothers arrest during the first few nuclear cleavage cycles with incomplete chromosome segregation because of a deficit in the maternal supply of Hd.
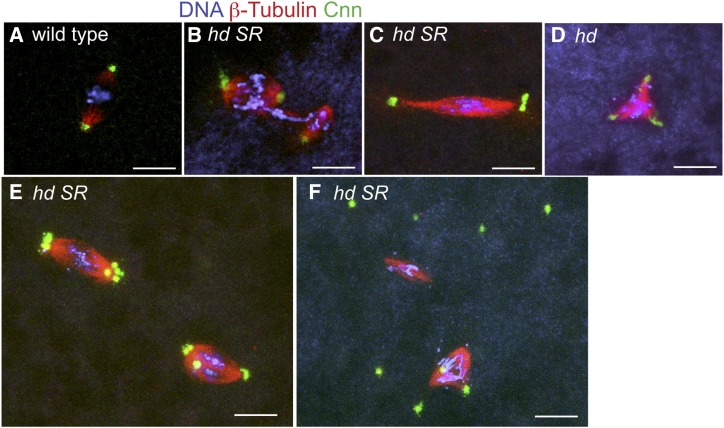
Figure 2.
Embryos from hd SR females have chromosome segregation errors and genome instability. (A–F) Confocal images of nuclear divisions in embryos from wild-type (A) and hd SR (B–F) mothers labeled with DAPI (DNA, blue), β-Tubulin (red), and centrosome protein Centrosomin (Cnn green). (A) Wild-type metaphase nucleus with a bipolar spindle and a single centrosome per pole. (B) hd SR embryo with a chromatin bridge that connects two nuclei arrested in metaphase, and of unequal DNA content, presumably a result of unequal and incomplete chromosome segregation in the previous cycle. (C) An example of an hd extended spindle with multiple centrosomes at the poles and stretched chromosomes. (D) An hd multipolar spindle. (E) Two spindles with incomplete clustering of excess centrosomes at the poles. (F) An example of free centrosomes that continue to duplicate uncoupled from the arrested nuclear division cycle. Bar, 10 μm.
hd SR embryos have severe chromosome segregation and spindle abnormalities
To define the nuclear division defect in hd SR embryos, we labeled chromosomes, spindles, and centrosomes with DAPI, anti-β-Tubulin, and anti-Centrosomin (Cnn) respectively (Chu and Klymkowsky 1989; Eisman et al. 2009). Compared to wild type, spindle morphology and chromosome segregation in hd SR embryos were severely aberrant (Figure 2, A–F). Spindles were variable in size and shape, with some appearing barrel-shaped while others were extremely narrow and long (Figure 2, B and C). In some cases, two spindles surrounded two closely adjacent chromatin masses of unequal size, and were connected by a chromatin bridge, indicative of incomplete and unequal chromosome segregation during the previous mitotic division (Figure 2B). Spindles often had more than two centrosomes, suggesting that centrosomes continued to duplicate despite nuclear division arrest, consistent with previous evidence that centrosome duplication can be uncoupled from mitotic divisions during embryonic cleavage stages (Raff and Glover 1988). Some of the spindles with extra centrosomes were bipolar, with evidence of complete or partial “centrosome clustering,” while other spindles with extra centrosomes were multipolar (Figure 2, C–E) (Kwon et al. 2008; Ganem et al. 2009). Many of the embryos had multiple centrosomes that were not associated with spindles or chromatin, further suggesting that centrosome duplication continued in the absence of nuclear division (Figure 2F). Together, these results suggest that a deficit of maternally supplied Hd results in severe chromosome instability and embryonic arrest during the first few nuclear cleavage divisions.
Maternal Hd protein is abundant during early embryonic cleavage cycles but severely reduced in embryos from hd SR mothers
The results suggested that maternally encoded Hd protein is required to support early cleavage cycles of the embryo. To examine maternal Hd protein abundance during these early cycles, we immunolabeled wild type embryos with an affinity-purified polyclonal Hd antibody during the first hour of embryogenesis before zygotic gene expression begins (Bandura et al. 2005). Beginning with the first division cycles, Hd protein was concentrated in the interphase (S phase) nucleus, with lower levels in the cytoplasm, and more evenly distributed during M phase after nuclear envelope breakdown (Figure 3, A–A”). In contrast, Hd protein was undetectable by immunofluoresence in embryos from hd272-9 SR mothers (Figure 3, B–B”). We further quantified the level of Hd protein in 0–1 hr wild-type and hd SR embryos by Western blotting, which confirmed that maternally encoded Hd protein is abundant during wild-type early embryonic cleavage stages, and reduced in embryos from hd272-9 SR mothers (Figure 3C). The ability to detect low levels of Hd protein by Western blotting but not immunofluoresence is because hd SR embryos arrest during M phase when Hd protein is dispersed throughout the embryoplasm, which is a limitation for detection by immunofluorescence but not Western blotting. The lower level of Hd protein in hd SR embryos is consistent with our previous evidence that the hd272-9 missense allele (G463D) results in lower levels of Hd protein in imaginal discs (Figure 3C) (Bandura et al. 2005). The maternal supply of Hd protein in early wild type embryos is also consistent with ModENCODE data that maternal hd mRNA is highly abundant in 0–2 hr embryos before the onset of zygotic transcription (Roy et al. 2010; Marygold et al. 2016; Gramates et al. 2017). Together, these data indicate that embryos from hd272-9 SR mothers have reduced amounts of maternally encoded Hd protein that are insufficient to support early nuclear cleavage cycles.
Figure 3.
Hd protein is abundant during embryonic nuclear cleavage cycles and reduced in embryos from hd SR females. Confocal images of embryos from wild type (A–A”) and hd SR (B and B’) mothers labeled with the DNA dye TOTO-3 (red) (A and B), anti-Hd (green) (A’ and B’), and merge (A” and B”). Bar, 10 μm. (C) Western blot of cleavage stage (0–1 hr) embryo extracts from hd SR and wild-type mothers incubated with anti-Hd antibody and anti-β-actin antibody as loading control.
Expression of humpty dumpty in the maternal germline and soma partially rescues the embryonic cleavage defects
To further address whether a deficiency of maternal hd is responsible for the early arrest phenotype in syncytial Drosophila embryos, we created a rescue transgene that expresses Hd protein in the soma and germline. This transgene contains a genomic subclone of the hd locus that is fused in frame on its 3′ end to GFP (hd-GFP), and includes 575 bp of genomic DNA upstream of the hd transcription start site, including a putative E2F binding site that responds to E2F1/Dp (Bandura et al. 2005). Unlike UAS-hd, hd-GFP is expressed in both germline and somatic cells of the ovary. We used this hd-GFP transgene to rescue hd272-9/Df(3R)3-4, hereafter hd272-9 Germline and Somatic Rescue (GSR) mothers. Embryos from these hd272-9 GSR mothers had high levels of maternally encoded Hd-GFP in the nucleus during early cleavage cycles (Figure 4, A–A”). Early cleavage divisions in these hd272-9 GSR embryos appeared normal, with no evidence of the chromosome bridges that were seen in hd SR embryos, indicating that maternal hd-GFP rescues the early nuclear cleavage arrest (Figure 4, B and C). During later nuclear division cycles, however, aberrant spindles and chromatin bridges began to accumulate as nuclei migrated to the cortex, and all hd272-9 GSR embryos arrested before cellular blastoderm (Figure 4, B–D). Thus, the expression of Hd-GFP in the maternal germline partially rescues embryonic nuclear cleavage divisions. The progressive accumulation of chromosome bridges and aberrant spindles during later cleavage cycles may result from depletion of insufficient Hd-GFP. Consistent with this, Western blotting indicated that Hd-GFP protein was expressed to lower levels than endogenous Hd protein, perhaps explaining why rescue was not complete, although fusion to GFP may also compromise Hd function (Figure 4E). In contrast, hd-GFP completely rescued imaginal disc cell proliferation and adult viability of hd null larvae (hdFf/Df(3R)3-4) (Table 1). The ability of the hd-GFP transgene to fully rescue zygotic hd null animals but not embryos from hd hypomorph mothers is consistent with the interpretation that cleavage cycles are sensitive to small deficits of hd function. The penetrant bridging in the partially rescued embryos indicates that a deficit of maternal Hd results in a failure of chromosome segregation and a nuclear cleavage cycle arrest.
hd mutant cells arrest during G2 and early M phase of canonical mitotic cell cycles
The results indicated that a deficit in the maternal supply of Hd results in chromosome instability and mitotic division arrest during the first few embryonic nuclear S/M cleavage divisions. The penetrant chromosome bridging in these embryos may reflect a requirement for hd for proper sister chromatid segregation during M phase. Alternatively, since early nuclear cleavage cycles lack DNA replication or damage checkpoints, the chromosome bridges could result from an attempt to segregate sister chromatids that were incompletely replicated during the previous S phase.
To begin to evaluate whether the chromosome bridge phenotype reflects a function of Hd during S or M phase, we examined the impact of hd mutations during a canonical mitotic cell cycle that has DNA replication and damage checkpoints. We chose proliferating somatic follicle cells of the ovary because they have a well-characterized cell cycle and form an epithelial sheet that is amenable to imaging. These follicle cells mitotically proliferate up to stage 6 of oogenesis, after which they switch to G/S endocycles (Mahowald et al. 1979; Klusza and Deng 2011). We used a heat inducible FLP/FRT system to make homozygous mutant clones of the null allele hdFf, and then examined follicle cell cycle behavior by antibody labeling for Cyclin B (Knoblich and Lehner 1993; Xu and Rubin 1993). During canonical mitotic cell cycles, Cyclin B protein begins to appear in the cytoplasm during late S phase, increases in the cytoplasm during G2 phase, becomes both nuclear and cytoplasmic during the G2 to M phase transition, and is then rapidly degraded at metaphase/anaphase (Knoblich and Lehner 1993; Gavet and Pines 2010). Labeling of wild-type follicle cells with anti-Cyclin B antibody reflected this known cell cycle behavior of Cyclin B protein during the mitotic proliferation of follicle cells (Figure 5A). As expected, anti-Cyclin B did not label follicle cells after stage 6 when they are in the endocycle, and the expression of Cyclin B is repressed (Figure 5A) (Sun and Deng 2005). In contrast, clones of homozygous hdFf cells had high levels of Cyclin B in the cytoplasm only, and this labeling persisted past stage 6 when neighboring wild type follicle cells had switched to endocycles (Figure 5B). This observation is consistent with our previous clonal analysis in wing discs, which showed that hd mutant cells can divide a limited number of times as stable Hd protein is slowly depleted, but then eventually arrest with incomplete DNA replication and DNA damage (Bandura et al. 2005). The accumulation of hd mutant follicle cells with high levels of Cyclin B in the cytoplasm suggest that these cells are arresting at the G2/M DNA damage checkpoint, perhaps because of incomplete DNA replication in the previous S phase. Unlike hd mutant cleavage stage embryos, we saw no evidence of chromosome bridging in hd mutant follicle cells. This difference between hd follicle cells and embryos is not because of different alleles or levels of Hd protein because we saw no evidence of bridging or arrest in the follicle cells of hd272-9 SR mothers, despite a highly penetrant bridging phenotype in their embryos. These data are consistent with the idea that the hd chromosome bridge phenotype during cleavage cycles is caused by a sensitivity to replication defects combined with an absence of a G2/M checkpoint, which results in a failed attempt to segregate incompletely replicated sister chromatids.
Figure 5.
Depletion of hd during canonical mitotic cell cycles results in a G2/M arrest. (A) An ovariole from a control hsp70-FLPase; FRT hdFf/FRT hd+ GFP+ female that was not heat induced. Labeling of this ovariole with anti-GFP (green) and anti-cyclin B (red) reveals oscillating levels of Cyclin B in follicle cells during mitotic division cycles up until stage 6 (S6), but not in endocycling follicle cells of stage 7 (S7) and later egg chambers. The large polyploid nurse cells in the interior of the egg chamber that switch into endocycles before stage 1 do not express mitotic cyclins during these stages. Inset: A higher magnification of a follicle cell with Cyclin B in cytoplasm and nucleus. (B) An ovariole from an hsp70-FLPase; FRT hdFf/FRT hd+ GFP+ female, in which an hdFf/hdFf null mutant clone (arrow, GFP−) was heat induced 2 days earlier. After initial cell divisions, slow depletion of Hd protein in the hdFf null cells results in their arrest with high levels of Cyclin B in the cytoplasm but not nucleus, suggestive of a G2/M arrest, which persists after stage 6. Inset: A higher magnification of a hd mutant follicle cell arrested with Cyclin B in the cytoplasm but not nucleus. Bar, 25 μm for main panels, 6 μm for insets.
Maternal Hd is required for DNA rereplication in embryos from plutonium mutant mothers
We wanted to test directly whether maternal Hd protein is required for DNA replication during early cleavage cycles. To do this would require separating the potential function of Hd during S phase vs. M phase, which is challenging during the rapid S/M cycles. We therefore took a genetic approach by conducting an epistasis experiment with the gene plutonium (plu) (Shamanski and Orr-Weaver 1991). The Plu protein is a member of the trimeric giant nucleus complex, which is composed of Plu, Giant nucleus (Gnu), and Pan Gu (Png) (Lee et al. 2003). This maternally encoded complex is required for normal Cyclin B levels and mitosis during embryonic nuclear cleavage cycles (Lee et al. 2001). Embryos from mothers who are homozygous mutant for the null allele plu3 undergo multiple rounds of S phase without mitosis, and eventually arrest with one or a few giant polyploid nuclei (Axton et al. 1994; Lee et al. 2003). We reasoned that if Hd is required for mitosis and not S phase, embryos from hd; plu double mutant mothers should have the plu mutant phenotype: one to several giant nuclei. Conversely, if hd is required for DNA replication, embryos form hd; plu double mutant mothers should have the hd phenotype: several small nuclei.
We created female siblings that had different combinations of genotypes at the plu and hd loci, and that were somatically rescued for hd eggshell defects using c323GAL4; UAS-hd. As expected, females heterozygous for the recessive plu3 allele gave rise to embryos in which S/M cleavage cycles occurred normally, whereas females that were homozygous mutant for plu3 gave rise to embryos with the giant nucleus phenotype, which was not genetically modified when these mothers were heterozygous for the hd deletion Df(3R)3-4 (Figure 6, A–B’). Females that were heterozygous for the recessive plu3 allele (plu3/+) and trans-heterozygous mutant for hd (hd272-9/Df(3R)3-4) gave rise to embryos with the hd early cleavage arrest and chromosome bridge phenotypes (Figure 6, C and C’). Importantly, the plu3; hd272-9/Df(3R)3-4 double mutant females had embryos that arrested with the hd single mutant phenotype: several small nuclei (Figure 6, D and D’). This last result indicates that hd is epistatic to plu, and that hd is required for the multiple rounds of DNA rereplication that occur in plu mutant embryos in the absence of mitosis. Thus, in embryos from hd single mutant mothers, the long chromosome bridges likely result from insufficient Hd function during S phase and subsequent stretching of incompletely replicated sister chromatids by the mitotic spindle. Altogether, the data suggest that maternal Hd is essential for DNA replication and genome integrity during early Drosophila development.
Discussion
We previously showed that animals homozygous for the hypomorphic genotype hd272-9/Df(3R)3-4 are viable, but adult females have severe defects in developmental gene amplification, produce eggs with thin shells, and are sterile. In this study, we found that rescue of this amplification defect is not sufficient for female fertility, and that mothers with a hd272-9/Df(3R)3-4 mutant germline provide insufficient Hd function to their embryos, resulting in arrest during the first few nuclear cleavage cycles with long chromosome bridges and genome instability. Although our previous findings were consistent with a role of Hd in DNA replication, it remained formally possible that Hd had a different essential function in genome maintenance (Bandura et al. 2005). Our current genetic and cell biological evidence show that maternally encoded Hd protein is essential for DNA replication during rapid nuclear cleavage cycles of early embryogenesis. Together with our previous analysis of the hd272-9 allele, these findings suggest that embryonic nuclear cleavage cycles and ovarian developmental gene amplification represent two time periods in Drosophila development that are sensitive to deficits in DNA replication protein function. These observations have important broader relevance for understanding how hypomorphic mutations in the human ortholog of hd and other essential replication proteins cause tissue-specific phenotypes in different types of microcephalic primordial dwarfisms (MPDs).
How DNA replication is coordinated with mitosis during early, rapid S/M cycles is not completely understood. Our data indicate that maternal Hd is essential for DNA synthesis during these early cycles. Embryos from hd hypomorphic mothers arrested in the first or second mitosis of nuclear cleavage division, often with pairs of condensed chromatin masses of unequal size and connected by chromatin bridges. In contrast, depletion of humpty dumpty in cells with canonical mitotic cell cycles and checkpoints resulted in a cell cycle arrest during G2 and early M phase, consistent with a replication checkpoint arrest. Together with our finding that hd is epistatic to the rereplication phenotype of plu mutants, these data suggest that the long chromosome bridges in hd mutant embryos are likely the result of spindle forces pulling on incompletely replicated and attached sister chromatids. This interpretation is consistent with the known absence of a DNA replication checkpoint during these early S/M cycles (Raff and Glover 1988). Although early cycles lack a DNA replication checkpoint, they do have a spindle assembly checkpoint (SAC), which likely contributed to the preponderance of M phase arrest that we observed (Oliveira et al. 2010). The hd bridging phenotype is similar to that after injection of the DNA polymerase alpha inhibitor aphidicolin into early embryos and that of maternal effect mutations in several DNA replication and repair genes (Raff and Glover 1988; Girdham and Glover 1991; Apger et al. 2010; Gosnell and Christensen 2011; Sakurai et al. 2011). Among this group of mutants, the hd arrest is the earliest, during the first or second cleavage cycles, suggesting that these early cycles are extremely sensitive to levels of Hd function. This early arrest is not a reflection of a unique requirement for Hd protein during the first few cleavage cycles because partial rescue with hd-GFP resulted in a similar chromosome bridging and arrest during later S/M cleavage cycles. Thus, sufficient maternal Hd function is essential to ensure DNA replication and genome integrity during early embryogenesis.
The hd embryos had supernumerary centrosomes that nucleated multipolar spindles, while other extra centrosomes clustered together to form a bipolar spindle. These supernumerary centrosomes of multipolar and bipolar spindles likely contribute to chromosome instability (CIN), and the unequal DNA content of arrested nuclei, similar to that documented in cancer cells with amplified centrosomes (Kwon et al. 2008; Ganem et al. 2009; Godinho and Pellman 2014). It was previously reported that overexpression of the human ortholog of hd, Donson (Dons), results in its association with centrosomes in human cells, but we did not observe Hd localization to centrosomes using either Hd antibodies or Hd-GFP fusions (Fuchs et al. 2010). Therefore, our interpretation is that the extra centrosome phenotype is an indirect consequence of nuclear cycle arrest with continued centrosome duplication, consistent with the known independence of centrosome and nuclear division cycles in early embryos (Raff and Glover 1988; Archambault and Pinson 2010).
Consistent with our findings, while this manuscript was in preparation, Reynolds and colleagues reported that human Dons protein associates with replication forks and is required for replication fork progression and the replication stress checkpoint in human cells (Reynolds et al. 2017). It remains unclear, however, whether Hd associates with replication forks in Drosophila (Bandura et al. 2005). Further work is needed in flies and human cells to define the precise molecular function(s) of Hd/Dons proteins in DNA replication and checkpoint signaling.
Our analysis of different hd genotypes has revealed insights into how DNA replication programs are modified during development. We previously showed that animals homozygous for null alleles of hd (e.g., hdFf) die as pupae with DNA damage and severe undergrowth of brains and imaginal discs (Table 1) (Bandura et al. 2005). Our evidence for abundant Hd protein in the early embryo suggests that hdFf homozygous mutant embryos and larvae likely survive on maternally supplied Hd, with severe defects in the mitotic proliferation of imaginal discs manifesting as pupal lethality when it is time for these tissues to metamorphose into adult structures. In contrast, animals homozygous for the hypomorphic allele hd272-9 are viable, but adult females have severely reduced gene amplification resulting in thin eggshells (Figure 7) (Snyder et al. 1986; Bandura et al. 2005). The hd272-9 amplification defect is similar to that caused by hypomorphic mutations in other essential DNA replication genes (i.e., Orc1, Orc2, Cdt1, Dbf4, Mcm6, and mus101), indicating that thin eggshells are a sensitized phenotype for small reductions in DNA replication protein function (Calvi and Spradling 1999; Calvi 2006). Through rescuing the amplification defect of hd272-9 in somatic follicle cells of the ovary, we have uncovered that early S/M cleavage cycles of the embryo are also sensitive to reductions in maternal hd function. Thus, early embryonic cleavage cycles and developmental gene amplification are two time periods in Drosophila development that are highly sensitive to small deficits in replication protein function (Figure 7). Both of these times in development require high levels of DNA replication. During S/M cycles, the genome is duplicated in ∼3 min by initiating replication from numerous, closely spaced origins (Figure 7) (Blumenthal et al. 1974; Sasaki et al. 1999). During ovarian gene amplification, repeated reinitiation from specific origins is required to rapidly increase eggshell protein gene copy number during a several hour time window (Figure 7) (Calvi 2006). The genetic evidence suggests that these two rapid DNA replication programs require high levels of DNA replication protein function. Moreover, early S/M cycles and developmental gene amplification have compromised checkpoints and lack backup dormant origins that normally respond to replication stress during canonical cell cycles. Thus, differences in DNA replication demand and replication stress response among cell types likely explain why hypomorphic alleles of essential DNA replication genes cause specific developmental phenotypes.
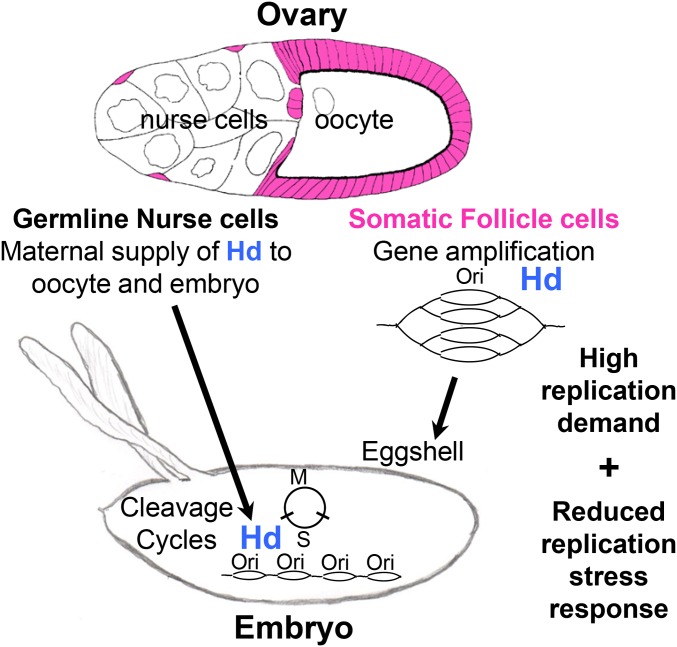
Figure 7.
Developmental gene amplification and embryonic cleavage cycles are sensitive to reductions in Hd function. A model of Hd function in germline and somatic cells of the Drosophila ovary. Hd is required in somatic follicle cells (pink) for DNA rereplication to amplify the copy number of genes that are required for eggshell synthesis. Ovarian germline nurse cells provide maternal Hd to the oocyte and future embryo to support rapid DNA synthesis from closely spaced origins during nuclear cleavage cycles. Females homozygous or hemizygous for the hypomorphic allele hd272-9 are viable but manifest severe maternal effects on both ovarian gene amplification and DNA synthesis during embryonic nuclear cleavage, suggesting that these two time periods in Drosophila development are especially sensitive to reductions in the function of maternally encoded DNA replication proteins. We propose that these two time periods are sensitive because of a high replication demand and a reduced replication stress response. Please see the Discussion for a full description of this model.
Two recent reports indicate that hypomorphic mutations in human Dons are a genetic cause of MPD (Evrony et al. 2017; Reynolds et al. 2017). The clinical presentation of children with some Dons mutations overlaps that of hypomorphic mutations in a number of other replication proteins (Orc1, Orc4, Orc6, Cdt1, Cdc6, MCM5, Cdc45, and Geminin) that cause a subtype of microcephalic dwarfism known as MGS, characterized by pre and postnatal undergrowth, microcephaly, small ears (microtia), and underdeveloped or absent knee caps (knee patella syndrome), as well as other variably expressive phenotypes (Bicknell et al. 2011a,b; Balasov et al. 2015; Burrage et al. 2015; Fenwick et al. 2016; Khetarpal et al. 2016; Vetro et al. 2017). Thus, it appears that mild mutations in hd gene family members result in phenotypes that are similar to hypomorphic alleles of these other essential replication protein genes in both flies (thin eggshell) and humans (microcephalic dwarfism).
While some alleles of Dons resemble MGS with relatively mild effects on growth, other alleles resemble other classes of MPDs with more severe pleiotropic phenotypes of impaired skeletal, brain, and lung development, which, in some cases, result in perinatal lethality (Khetarpal et al. 2016; Evrony et al. 2017; Reynolds et al. 2017). It remains unclear, however, why different mutations in Dons, and other replication genes, have different pleiotropic clinical presentations, and affect some tissues more than others. Our finding that specific time periods of Drosophila development are more sensitive to hypomorphic alleles of hd raises the possibility that the range of tissue-specific clinical presentations of different Dons alleles may reflect a difference among developing tissues in the speed of DNA synthesis, availability of dormant origins, or effectiveness of DNA replication checkpoints. A further examination of Hd–Dons function in specific tissues of fly and human should shed light on the developmental and molecular mechanisms of microcephalic primordial dwarfisms.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300318/-/DC1.
Acknowledgments
We would like to thank R. Eisman and T. Kaufman for advice and Cnn antibody, and T. Orr-Weaver for plu flies. We also thank the Bloomington Drosophila Stock Center (BDSC) and Flybase for flies and critical information, J. Powers of the Indiana University (IU) Light Microscopy Imaging Center (LMIC) for help and advice, and M.-Y. Cho for Hd-GFP construction. This work was supported by funding from National Institutes of Health (NIH) 1R01GM113107 to B.R.C.
Footnotes
Communicating editor: G. Bosco
Literature Cited
- Adolphs K. W., Cheng S. M., Paulson J. R., Laemmli U. K., 1977. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc. Natl. Acad. Sci. USA 74: 4937–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal B. D., Calvi B. R., 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376. [DOI] [PubMed] [Google Scholar]
- Alexander J. L., Beagan K., Orr-Weaver T. L., McVey M., 2016. Multiple mechanisms contribute to double-strand break repair at rereplication forks in Drosophila follicle cells. Proc. Natl. Acad. Sci. USA 113: 13809–13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apger J., Reubens M., Henderson L., Gouge C. A., Ilic N., et al. , 2010. Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics 185: 1151–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V., Pinson X., 2010. Free centrosomes: where do they all come from? Fly (Austin) 4: 172–177. [DOI] [PubMed] [Google Scholar]
- Asano M., Wharton R. P., 1999. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 18: 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axton J., Shamanski F., Young L., Henderson D., Boyd J., et al. , 1994. The inhibitor of DNA replication encoded by the Drosophila gene plutonium is a small, ankyrin repeat protein. EMBO J. 13: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Akhmetova K., Chesnokov I., 2015. Drosophila model of Meier-Gorlin syndrome based on the mutation in a conserved C-terminal domain of Orc6. Am. J. Med. Genet. A. 167A: 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura J. L., Beall E. L., Bell M., Silver H. R., Botchan M. R., et al. , 2005. Humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila. Curr. Biol. 15: 755–759. [DOI] [PubMed] [Google Scholar]
- Bicknell L. S., Bongers E. M., Leitch A., Brown S., Schoots J., et al. , 2011a Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 43: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell L. S., Walker S., Klingseisen A., Stiff T., Leitch A., et al. , 2011b Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 43: 350–355. [DOI] [PubMed] [Google Scholar]
- Blumenthal A. B., Kriegstein H. J., Hogness D. S., 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38: 205–223. [DOI] [PubMed] [Google Scholar]
- Burrage L. C., Charng W. L., Eldomery M. K., Willer J. R., Davis E. E., et al. , 2015. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am. J. Hum. Genet. 97: 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B. R., 2006. Developmental DNA amplification, pp. 233–255 in DNA Replication and Human Disease, edited by DePamphilis M. L. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Calvi B. R., Spradling A. C., 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18: 407–417. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Spradling A. C., 2001. The nuclear location and chromatin organization of active chorion amplification origins. Chromosoma 110: 159–172. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., Spradling A. C., 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. T., Klymkowsky M. W., 1989. The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Dev. Biol. 136: 104–117. [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Orr-Weaver T. L., 2005. Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet. 21: 149–162. [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., MacAlpine D. M., Evans J. G., Bell S. P., Orr-Weaver T. L., 2002. Visualization of replication initiation and elongation in Drosophila. J. Cell Biol. 159: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crest J., Oxnard N., Ji J. Y., Schubiger G., 2007. Onset of the DNA replication checkpoint in the early Drosophila embryo. Genetics 175: 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J., Cocker J., Dowell S., Rowley A., 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316. [DOI] [PubMed] [Google Scholar]
- Edgar B. A., O’Farrell P. H., 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell 57: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman R. C., Phelps M. A., Kaufman T. C., 2009. Centrosomin: a complex mix of long and short isoforms is required for centrosome function during early development in Drosophila melanogaster. Genetics 182: 979–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony G. D., Cordero D. R., Shen J., Partlow J. N., Yu T. W., et al. , 2017. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly-micromelia syndrome. Genome Res. 27: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick A. L., Kliszczak M., Cooper F., Murray J., Sanchez-Pulido L., et al. , 2016. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and Craniosynostosis. Am. J. Hum. Genet. 99: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., Odell G., Edgar B. A., 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint., pp. 149–300 in The Development of Drosophila melanogaster, edited by Bate M., Martinez Arias A. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Fogarty P., Campbell S. D., Abu-Shumays R., Phalle B. S., Yu K. R., et al. , 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7: 418–426. [DOI] [PubMed] [Google Scholar]
- Fuchs F., Pau G., Kranz D., Sklyar O., Budjan C., et al. , 2010. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 6: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Godinho S. A., Pellman D., 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O., Pines J., 2010. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189: 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdham C. H., Glover D. M., 1991. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 5: 1786–1799. [DOI] [PubMed] [Google Scholar]
- Godinho S. A., Pellman D., 2014. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosnell J. A., Christensen T. W., 2011. Drosophila Ctf4 is essential for efficient DNA replication and normal cell cycle progression. BMC Mol. Biol. 12: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J. M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills S. A., Diffley J. F., 2014. DNA replication and oncogene-induced replicative stress. Curr. Biol. 24: R435–R444. [DOI] [PubMed] [Google Scholar]
- Khetarpal P., Das S., Panigrahi I., Munshi A., 2016. Primordial dwarfism: overview of clinical and genetic aspects. Mol. Genet. Genomics 291: 1–15. [DOI] [PubMed] [Google Scholar]
- Klingseisen A., Jackson A. P., 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusza S., Deng W. M., 2011. At the crossroads of differentiation and proliferation: precise control of cell-cycle changes by multiple signaling pathways in Drosophila follicle cells. Bioessays 33: 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A., Lehner C. F., 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO 12: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Godinho S. A., Chandhok N. S., Ganem N. J., Azioune A., et al. , 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22: 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G., Tower J., 1999. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126: 4281–4293. [DOI] [PubMed] [Google Scholar]
- Landis G., Kelley R., Spradling A., Tower J., 1997. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosphila homolog of yeast origin recognition complex subunit 2. Proc. Natl. Acad. Sci. USA 94: 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. A., Elfring L. K., Bosco G., Orr-Weaver T. L., 2001. A genetic screen for suppressors and enhancers of the Drosophila PAN GU cell cycle kinase identifies cyclin B as a target. Genetics 158: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. A., Van Hoewyk D., Orr-Weaver T. L., 2003. The Drosophila cell cycle kinase PAN GU forms an active complex with PLUTONIUM and GNU to regulate embryonic divisions. Genes Dev. 17: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel D., Huikeshoven H., Cotterill S., 2000. Localisation of the DmCdc45 DNA replication factor in the mitotic cycle and during chorion gene amplification. Nucleic Acids Res. 28: 3897–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald A., Caulton J., Edwards M., Floyd A., 1979. Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster. Exp. Cell Res. 118: 404–410. [DOI] [PubMed] [Google Scholar]
- Marygold S. J., Crosby M. A., Goodman J. L., FlyBase Consortium , 2016. Using FlyBase, a database of Drosophila genes and genomes. Methods Mol. Biol. 1478: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J., Orr-Weaver T. L., 2012. Regulation of DNA replication during development. Development 139: 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J., Li S., Eng T., Macalpine D., Orr-Weaver T. L., 2011. Developmental control of the DNA replication and transcription programs. Genome Res. 21: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P. H., Stumpff J., Su T. T., 2004. Embryonic cleavage cycles: how is a mouse like a fly? Curr. Biol. 14: R35–R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R. A., Hamilton R. S., Pauli A., Davis I., Nasmyth K., 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. W., Botchan M. R., Berger J. M., 2017. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 52: 107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Costa A., 2016. New insights into the mechanism of DNA duplication by the eukaryotic replisome. Trends Biochem. Sci. 41: 859–871. [DOI] [PubMed] [Google Scholar]
- Perez-Mongiovi D., Malmanche N., Bousbaa H., Sunkel C., 2005. Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 132: 4509–4520. [DOI] [PubMed] [Google Scholar]
- Rabinowitz M., 1941. Studies on the cytology and early embryology of the egg of Drosophila melanogaster. J. Morphol. 69: 1–49. [Google Scholar]
- Raff J. W., Glover D. M., 1988. Nuclear and cytoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited with aphidicolin. J. Cell Biol. 107: 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J. J., Bicknell L. S., Carroll P., Higgs M. R., Shaheen R., et al. , 2017. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat. Genet. 49: 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell W. F., Sullivan W., 2000. Fluorescent Analysis of Drosophila Embryos, Drosophila Protocols, edited by Sullivan W., Ashburner M., Hawley R. S. Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Austin R. J., Bosco G., Bell S. P., Orr-Weaver T. L., 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Okado M., Ito F., Kawasaki K., 2011. Anaphase DNA bridges induced by lack of RecQ5 in Drosophila syncytial embryos. FEBS Lett. 585: 1923–1928. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Sawado T., Yamaguchi M., Shinomiya T., 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwed G., May N., Pechersky Y., Calvi B. R., 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanski F. L., Orr-Weaver T. L., 1991. The Drosophila plutonium and pan gu genes regulate entry into S phase at fertilization. Cell 66: 1289–1300. [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Stevenson V. A., Theurkauf W. E., 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388: 93–97. [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Laurencon A., Hawley R., Theurkauf W. E., 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9: 302–312. [DOI] [PubMed] [Google Scholar]
- Sibon O. C., Kelkar A., Lemstra W., Theurkauf W. E., 2000. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2: 90–95. [DOI] [PubMed] [Google Scholar]
- Snyder P., Galanopoulos V., Kafatos F., 1986. Trans-acting amplification mutants and other eggshell mutants of the third chromosome in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 83: 3341–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., 1981. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell 27: 193–201. [DOI] [PubMed] [Google Scholar]
- Sun J., Deng W. M., 2005. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Kelkar A., Theurkauf W. E., 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113: 87–99. [DOI] [PubMed] [Google Scholar]
- Takada S., Kwak S., Koppetsch B. S., Theurkauf W. E., 2007. grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development 134: 1737–1744. [DOI] [PubMed] [Google Scholar]
- Takada S., Collins E. R., Kurahashi K., 2015. The FHA domain determines Drosophila Chk2/Mnk localization to key mitotic structures and is essential for early embryonic DNA damage responses. Mol. Biol. Cell 26: 1811–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomer M., May N. R., Aggarwal B. D., Kwok G., Calvi B. R., 2004. Drosophila double-parked is sufficient to induce re-replication during development and is regulated by Cyclin E / CDK2. Development 131: 4807–4818. [DOI] [PubMed] [Google Scholar]
- Tocilj A., On K. F., Yuan Z., Sun J., Elkayam E., et al. , 2017. Structure of the active form of human origin recognition complex and its ATPase motor module. Elife 6: e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetro A., Savasta S., Russo Raucci A., Cerqua C., Sartori G., et al. , 2017. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur. J. Hum. Genet. 25: 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., et al. , 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13: 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker A. J., Royzman I., Orr-Weaver T. L., 2000. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 14: 1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wynn S. L., Fisher R. A., Pagel C., Price M., Liu Q. Y., et al. , 2000. Organization and conservation of the GART/SON/DONSON locus in mouse and human genomes. Genomics 68: 57–62. [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M., 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yamamoto R. R., Axton J. M., Yamamoto Y., Saunders R. D., Glover D. M., et al. , 2000. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics 156: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Riera A., Bai L., Sun J., Nandi S., et al. , 2017. Structural basis of Mcm2–7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat. Struct. Mol. Biol. 24: 316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M. K., Cimprich K. A., 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Cheng E., Wu H., Li N., Yung P. Y., et al. , 2017. Open-ringed structure of the Cdt1-Mcm2–7 complex as a precursor of the MCM double hexamer. Nat. Struct. Mol. Biol. 24: 300–308. [DOI] [PubMed] [Google Scholar]
- Zhang H., Tower J., 2004. Sequence requirements for function of the Drosophila chorion gene locus ACE3 replicator and ori-beta origin elements. Development 131: 2089–2099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.