Abstract
Diversity of the founding population of Human Immunodeficiency Virus Type 1 (HIV-1) transmissions raises many important biological, clinical, and epidemiological issues. In up to 40% of sexual infections, there is clear evidence for multiple founding variants, which can influence the efficacy of putative prevention methods, and the reconstruction of epidemiologic histories. To infer who-infected-whom, and to compute the probability of alternative transmission scenarios while explicitly taking phylogenetic uncertainty into account, we created an approximate Bayesian computation (ABC) method based on a set of statistics measuring phylogenetic topology, branch lengths, and genetic diversity. We applied our method to a suspected heterosexual transmission case involving three individuals, showing a complex monophyletic-paraphyletic-polyphyletic phylogenetic topology. We detected that seven phylogenetic lineages had been transmitted between two of the individuals based on the available samples, implying that many more unsampled lineages had also been transmitted. Testing whether the lineages had been transmitted at one time or over some length of time suggested that an ongoing superinfection process over several years was most likely. While one individual was found unlinked to the other two, surprisingly, when evaluating two competing epidemiological priors, the donor of the two that did infect each other was not identified by the host root-label, and was also not the primary suspect in that transmission. This highlights that it is important to take epidemiological information into account when analyzing support for one transmission hypothesis over another, as results may be nonintuitive and sensitive to details about sampling dates relative to possible infection dates. Our study provides a formal inference framework to include information on infection and sampling times, and to investigate ancestral node-label states, transmission direction, transmitted genetic diversity, and frequency of transmission.
Keywords: coalescent, phylogeny, approximate Bayesian computation, co-infection, superinfection, ancestral node state
MOST HIV-1 infections are the result of sexual transmission (Shattock and Moore 2003), where 20–40% involve transmission of multiple genetic variants (Keele et al. 2008; Salazar-Gonzalez et al. 2009; Li et al. 2010; Rieder et al. 2011). Transmitting more than one variant raises many important biological, clinical, and epidemiological issues. Biologically, successful transmission of more than one variant means that many viruses in a donor have the capacity to establish infection, and, further, that they had similar fitness as they did not outcompete each other in the new host. Following establishment of infection, the existence of multiple lineages may also generate virus with higher relative fitness than when single lineages establish infection (Carrillo et al. 2007), due either to recombination or competition after transmission (Sanborn et al. 2015). Clinically, transmission of several virus variants may make it harder for the immune system to combat the virus (Grobler et al. 2004; Yang et al. 2005; Smith et al. 2006), easier for the virus to evade antiviral treatment (Smith et al. 2004), and may accelerate disease progression (Gottlieb et al. 2004). Epidemiologically, the establishment of more than one genetic variant can occur simultaneously at one time, or sequentially over a long period of time, which is defined as co-infection or superinfection, respectively (van der Kuyl and Cornelissen 2007). This has further impact on whether one infection protects against another (Altfeld et al. 2002; Ronen et al. 2013), or if later superinfections may induce drug resistance (Smith et al. 2005), and if a potential vaccine to one form would protect against another.
Phylogenetics reconstructs evolutionary history, and, for an organism like HIV-1 that evolves very rapidly, the joint pathogen phylogeny from hosts that have infected each other reveals details about the host-to-host transmission. Recently, coalescent-based simulations showed that the resulting phylogeny may reveal both direction and directness in epidemiologically linked hosts, i.e., who infected whom, and whether missing host-links were likely (Romero-Severson et al. 2016). Furthermore, it has previously been shown that there exists a pretransmission interval that describes the bias toward the past when using phylogenetic trees to estimate transmission times (Leitner and Albert 1999; Leitner and Fitch 1999; Romero-Severson et al. 2014). Importantly, when multiple phylogenetic lineages have been transmitted from one host to another, the resulting tree opens up alternative interpretations of whether all lineages were transmitted at one or several occasions. Thus, while simulations have shown that phylogenies carry detailed information about who infected whom, and within-host models predict the pretransmission interval, a single framework to determine the evidence for the various possible transmission scenarios between two infected hosts is lacking.
The objective of this study was to create a unified framework to investigate the nature of an epidemiological link, and to apply that to a real HIV-1 transmission case. Based on previous theoretical work, the tree topology should probabilistically indicate direction and directness, whether more than one lineage were transmitted, as well as when transmission occurred. Here, we also intended to determine the evidence for whether the infection was established by a single transmission event or an ongoing process of reinfections. In addition, we show how conflicting statements about when transmission(s) could have occurred can be evaluated as alternative priors. We also wanted to avoid basing our inferences on a single (best) phylogenetic tree as many trees with different topology and distance properties may be nearly as likely as the best tree. Basing our method on the entire posterior distribution of trees allows us to consider the full range of solutions that the data may support, and to propagate uncertainty in phylogenetic reconstruction onto the parameter estimates. Thus, we extended our previous within-host coalescent methods to simulate trees corresponding to different transmission scenarios and parameterizations, and analyzed a previously unpublished HIV-1 transmission chain. To test and compare alternative scenarios of the epidemiological link, i.e., when and how transmission(s) occurred, we developed and applied an approximate Bayesian computation (ABC) method based on tree topology, root host-assignment, and patristic tree distance measures. The ABC method also allowed us to estimate the diversity at the time of transmission rather than at time of sampling.
Materials and Methods
Motivating case
The analysis developed in this paper was motivated by a complex transmission case involving three persons referred to as MP1, MP2, and MP3. MP1 (woman) and MP2 (man) had been married sometime in the past; after their divorce, MP2 was found to be infected with HIV-1 prompting an accusation that he was infected by his ex-wife. Clones were sampled from MP2 and MP3 as part of an ongoing investigation into those accusations. Approximately 1.5 years after MP2 was diagnosed, MP3, the current girlfriend of MP2, was also diagnosed with HIV-1, and clones from MP3 were sequenced at that time as well. MP1 and MP2 subjects had a history of intravenous drug use, but MP3 did not. Thus, based on the epidemiological record, MP1 and MP2 could potentially have infected each other via either sexual contact or needle injection, but transmission between MP2 and MP3 could only have been through sexual interaction.
Based on maximum likelihood (ML) phylogenetic reconstruction of HIV-1 env DNA sequences, MP1 taxa were separated from MP2 taxa by multiple local control and database sequences (Supplemental Material, Figure S1 and File S1). Hence, MP1 was highly unlikely to have infected MP2 or MP3. However, the phylogenetic reconstruction was consistent with HIV-1 transmission between MP2 and MP3. The criminal investigation concluded that MP1 had not infected MP2, in part based on the phylogenetic evidence (Figure S1). That investigation used the case sequences in this paper plus 119 env sequences selected from Portuguese and publicly available databases. The motivation for the analysis presented in this paper was to quantify the evidence that MP2 and MP3 infected one another given that their combined virus sequences displayed a complex poly-/para-phyletic phylogenetic topology.
Joint linear within-hosts population model
We considered three alternative sexual transmission scenarios: (1) a “singular” transmission model where some number of virus is transmitted at a single occasion, (2) a “co-infection” model with ongoing unidirectional transmissions over a fixed 90-day window, and (3) a “superinfection” model with ongoing bidirectional transmissions for the duration of the infectious period (Figure 1). In the singular transmission scenario the within-host effective population size, is a linear function of time, where is the population size at the time of infection, and is the linear increase in population size per day. The linear population size growth is motivated by the empirical observation that HIV-1 diversity typically grows linearly over the first 7–8 years of an infection in absence of antiviral treatment and AIDS (Shankarappa et al. 1999; Zanini et al. 2015). Expanding this model to a transmission pair, we assume that all times and parameters are defined along a single forward time axis such that the population size in the donor is simply given by while the population size in the recipient is given by where subscript d indicates the donor and subscript r indicates the recipient. The time of transmission is indicated as when the population size is in the donor and in the recipient.
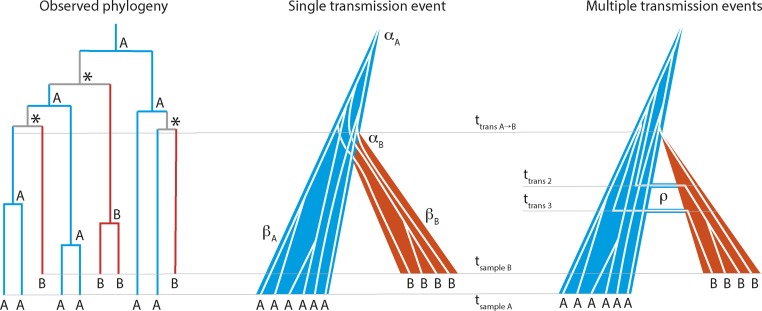
Figure 1.
Phylogenetic assessment of transmission scenario. Given a joint donor-recipient HIV-1 phylogenetic tree that suggests transmission of multiple lineages, two transmission scenarios are possible: (1) transmission of multiple lineages at a single transmission event, or (2) transmission of single lineages at multiple events (unidirectional during 90 days, co-infection; or bidirectional from initial transmission until sampling, superinfection). In this example, host A (blue) is donor and B is recipient (red). In the observed phylogeny the root host-label [A, B, or equivocal (*)] is derived by standard maximum parsimony. At time of transmission (ttrans A->B) either multiple lineages are transmitted (single transmission event with αB lineages) or the initial transmission takes place (in multiple transmission events). Additional transmissions (migration) occur at later time points (ttrans 2 and ttrans 3) at rate ρ. The effective populations grow at βA and βB in donor and recipient, respectively. Samples with individual HIV-1 clonal sequences are taken at tsample A and tsample B, respectively.
In the co- and superinfection models, we assume that infection occurs over a specific window. In the co-infection model, lineages are assumed to migrate from the donor to the recipient at rate when the donor is male, and rate when the donor is female (Boily et al. 2009). In the superinfection model, we assume the same migration rates, but bidirectional migration (i.e., lineages can freely move between hosts). The population sizes are given by the same equations as in the singular transmission scenario, but where 0. We assume that is small enough that is not significantly affected by the migration of lineages between the donor and recipient. We also assume that all extant lineages are equally probable to migrate.
Simulating trees from the joint coalescent model
All of the coalescent models that we used can be thought of as versions of the same model. This model is stochastic and has two possible actions: (1) coalescence of two sampled lineages in either the donor or recipient populations into one lineage, and (2) migration of a sampled lineage between the two hosts. Because we assume that the migration of lineages does not affect the population dynamics in either host, these processes are independent of one another conditional on the sampled number of lineages being constant. First, we deal with the time to coalesce in a population model where the population size varies over time. These equations are a modified version of a model that we presented in previous work (Romero-Severson et al. 2014).
We can obtain a density for the time to the next coalescent event in a time variable model by mapping the changing population size to the changing rate of coalescence (Nordborg 2001) and then performing a transformation of variables from the standard n-coalescent. Assuming extant lineages existing at time such that the population size is our approach is to get an expression for the changing rate of coalescence as a function of the current time and number of extant lineages. Over an infinitesimal time period along the reverse time axis, the change in the coalescent rate is therefore, for our linear growth model
is the changing rate of coalescence for lineages sampled at time We can use this equation to obtain a density of the time to the next coalescent event under the linear growth model by a simple transformation of variables. Starting with the density of the time to the next coalescent event in Kingman’s n-coalescent for extant lineages, (Wakeley 2009), we have the transformation:
for .
Migration is assumed to be a homogenous process where lineages migrate in the male-to-female direction at rate and the female-to-male direction at rate The time to the next migration event of one of the lineages in the sample is Because we assume that migration does not affect the population dynamics in either population, we only need to model migration events that occur in sampled lineages. Therefore, we have to account for the fact that, as the population size decreases along the reverse time axis, the probability of a migration event being in the sample increases (assuming constant ). As before, the mapping from the change in time to the change in migration rate of a single lineage in the sample is given by We can perform the same transformation of variables as before, substituting for yielding
To generate random variates from Z and M, we use the inverse cumulative functions
and, to simulate the time to the next migration event using the inverse cumulative function,
where is a unit uniform random variate.
In the singular transmission model, a coalescent process was simulated in each of the “derived populations” of the donor and recipient up to the time of transmission. We define a “derived population” as a population that exists in each host after transmission has occurred (in forward time), as illustrated in Figure 2. This involved drawing a random time to the next coalescent event given the current number of extant lineages and the current index time (i.e., the time of the previous coalescent event or the sampling time). If the time of the next event occurred in the derived population, then two lineages in the appropriate derived population were selected with uniform random probability to coalesce. Once the next coalescent event crossed over into the “source population” (Figure 2), we extended all extant lineages up to the transmission time and merge both sets of lineages into a single population. From there, the simulation proceeds as before but with all lineages now being in the donor’s source population.
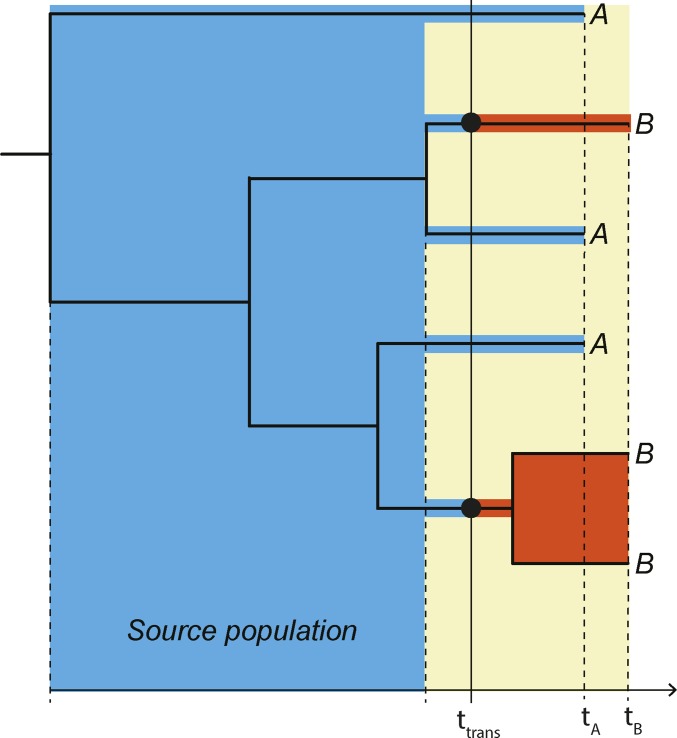
Figure 2.
Principle joint donor-recipient time-scaled phylogeny. When a donor (A, blue) infects a recipient (B, red), the possible time-interval when transmission could have occurred (yellow field) is restricted in a time-scaled topology of when the most recent donor-recipient (A–B) coalescence occurred among the sampled lineages and when the recipient was sampled at tB. The actual transmission (ttrans) must have occurred in this interval. The “source population” in direct transmission exists in the donor (blue field), from which at least two lineages were transmitted in this example to the donor (red fields). We refer to the populations that exist in each host after transmission as the “derived populations.” Note that if ttrans occurred later at least three lineages could have been transmitted.
To simulate migration in the super and co-infection models, we first simulated a coalescent process in MP3 up to the point of sampling for MP2 such that the two populations are at the same calendar time index. Then, we drew random times for all four possible events (migration from MP2 to MP3, migration from MP3 to MP2, coalescence in MP2, and coalescence in MP3). The next event was taken to be the minimum of the set of the random times. If the next event was a migration event, a random lineage from the appropriate population was moved to the other population; if the next event was a coalescence, then two random lineages from the appropriate population were merged into a single lineage. Random times were drawn again and the process was repeated until the infection time of the donor was reached.
To test the validity of our method, we simulated 100,000 genealogies from the superinfection model at the maximum posterior parameter values conditional on the observed data. We then treated the set of simulated genealogies in the same manner as the MrBayes posterior phylogenies. The true parameter values were all covered by the 50% credible intervals. The simulations with the correct donor were seven times more likely to be sampled. This is lower support than we observed for the real data. This is due to the fact the simulated data included stochasticity in the realization of the genealogy, while, for the observed data, the actual genealogy is fixed; that is, the simulated data are more variable than the observed data due to additional stochasticity in the simulations.
Model priors and constraints
The singular-infection model is specified by five parameters: the duration of MP2’s infection, the duration of MP3’s infection, the bottleneck size at transmission, the growth rates in MP2 and MP3, and respectively. The co- and superinfection models introduce two additional parameters: the migration rate, and the duration of the infection window, which is fixed at 90 days in the co-infection model, or from the initiation of infection to the sampling time of MP2 for the superinfection model.
There are several hard constraints based on the known epidemiological parameters: (1) the time difference between the sampling of MP2 and MP3 is 588 days, (2) MP2 was diagnosed 508 days before being sampled, (3) the sexual relationship between MP2 and MP3 began either after MP2’s divorce (according to MP3) or some time before then (according to MP2’s ex-wife). We operationalize constraint three as two priors either assuming that the sexual relationship between MP2 and MP3 began at the finalization of MP2’s divorce (prior 1) or some time before then (prior 2).
To our knowledge, both MP2 and MP3 were treatment naïve and did not have an AIDS diagnosis at the time of sampling. Based on that, and a lack of other relevant information that could constrain the infection times, we assumed a uniform distribution of infection durations of ≤12 years in the donor. We assume that the population growth rate in both subjects is drawn from βd ∼ Exponential(20−1) units per day. This distribution includes growth rates that correspond to most of the published estimates of the HIV within-host effective population numbers (Leigh Brown 1997; Nijhuis et al. 1998; Pennings et al. 2014). In the case of a singular transmission event, we assume that the donor transmits percent of their extant population at the time of transmission. We assume and in the co- and superinfection models, respectively. The values of were selected by trial and error to give the approximately correct average number of unique ancestors in the donor in each model.
Phylogenetic measures for ABC
For a tree with taxa from two hosts, “A” and “B,” we used the following statistics to define the probability that a simulation should be accepted: (1) the root label, (2) the topological class, (3) the number of unique ancestors of one of the host labels, (4) the total number of nucleotide substitutions in the tree, the average pairwise distance between (5) tips with mismatched labels, (6) tips with “A” labels, and (7) tips with “B” labels. We also considered both the (8) mean and (9) SD of the tree height [normalized to be in (0,1)] at which each unique ancestor occurred.
We chose these statistics because they are either known or believed to be related to aspects of the models that we want to infer. The root label has been shown to be related to the identity of the donor in previous work (Romero-Severson et al. 2016); however, we show below that this relationship is more complex than previously discussed; the topological class is known to be strongly correlated to the directness of transmission (Romero-Severson et al. 2016); the number of unique ancestors is related to the number of transmitted variants in the singular-transmission model, and the migration rate in the co- and superinfection models in addition to the population growth rates in each population; the total number of substitutions in the tree acts as a scaling factor for the infection and transmission times; the diversity measures are related to the within-host population dynamics in each host; the mean ancestor height is related to the transmission time/window; and the SD of the ancestor heights is related to the mode of transmission.
The root label is defined as the maximum parsimony host assignment of the root (“A”; “B”; or ambiguous, “?”) using the rules: A,A->A; B,B->B; A,B->?; A,?->A; B,?->B; ?,?->?. The topological relationship can be one out of three classes: MM (both host sets of taxa are monophyletic), PM (taxa from one host forms a monophyletic clade that inserts into the sample of the other host forming a paraphyletic clade), and PP (taxa from one host are paraphyletic to the other host’s taxa that are polyphyletic, or both host’s taxa are polyphyletic). Root label and topological class have been demonstrated to be associated with the epidemiologic relationship between two sampled hosts (Romero-Severson et al. 2016). The number of unique ancestors is counted by applying Dollo’s law (Dollo 1893), which logically follows from the irreversible fact that the donor was infected before the recipient. In principle, this translates on the tree to first assigning the “A” label to each node on a root to “A”-tip path, and then counting the minimum “A” to “B” transformations needed to observe the tip labels. We call each resulting “B” clade a unique ancestor, including clades with only one “B” taxon. Assuming we can interpret the phylogeny as a genealogy, the number of unique ancestors places a strict lower bound on the number of transmitted lineages.
Statistics 4, 5, 6, and 7 are based on the observed number of mutations and require rescaling the coalescent simulations, which are measured in units of time, to expected numbers of substitutions. To do this, we assume that a molecular clock with evolutionary rate operates on the whole tree, and multiply the simulated genealogy by the evolutionary rate to obtain the expected number of mutations on each branch. We assume that is Gamma distributed with mean 0.0080 and SD 0.0014 substitutions/site per year (Zanini et al. 2015).
Distance function and posterior sampling
We define the distance function between the simulated and observed data in a nonstandard way to integrate the joint distribution of the statistics over the set of phylogenies. We first used MrBayes (details below) to obtain a large sample of trees from the posterior over which we calculated the joint density of the nine phylogenetic statistics defined above. Our distance function considered four statistical probes as multiplicative factors based on the density of the measured statistics and assuming partial independence. The first three probes are defined by the density of the first three statistics (the ancestral root label, the topological class, and the number of unique ancestors). The final probe consisted of the joint distribution of the remaining statistics modeled as a multivariate normal using the mvtnorm R library (Genz et al. 2017). Thus, for a simulation with parameter set giving a vector of statistics the distance function would be where is the empirical density of from the posterior sample of trees. For example, because 94% of the posterior trees had MP2 as the root label, the simulation with MP2 as the root label followed
To obtain parameter estimates, we calculated for 20 million parameters drawn from both priors for each model (assuming equal prior probability of MP2 or MP3 being the donor). We then sampled 20 million parameters from the prior with probability proportional to Point estimates and credible intervals were obtained by measuring the mean and appropriate quantiles in the resampled data. We considered the effective sample size to be the number of unique parameters comprising the resampled posterior, and the marginal approximate evidence for each as the sum over Approximate Bayes factors, aBF, were calculated as the ratio of the marginal evidence.
DNA sequencing
Chromosomal DNA was extracted from infected peripheral blood mononuclear cells of each subject using Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer recommendations. Nested PCR was done to obtain a 534 bp fragment from the C2V3 env region (HXB2 positions 6858–7392). Thermal cycling conditions were as previously described (Bartolo et al. 2009). PCR products were cloned into the pCR4-TOPO vector (Invitrogen), using the TOPO TA Cloning Kit (Invitrogen) according to the manufacturer’s instructions. DNA sequencing was performed using the BigDye Terminator V3.1 Cycle sequencing Kit (Applied Biosystems, Foster City, CA) and an automated sequencer (3100-Avant Genetic Analyzer; Applied Biosystems). We derived 31, 20, and 19 sequences from MP1, MP2, and MP3, respectively.
Phylogenetic reconstruction
HIV-1 sequences were aligned using MAFFT with the L-INS-i algorithm (Katoh and Toh 2008). Maximum likelihood phylogenetic trees were inferred using PhyML (Guindon et al. 2005) under a GTR+I+G substitution model, four categories Gamma optimization, with a Bio-NJ starting tree and best of NNI and SPR search, and aLRT SH-like branch support (Anisimova and Gascuel 2006). The posterior distribution of trees was sampled using MrBayes (Ronquist and Huelsenbeck 2003) under the same model parameterization as the PhyML trees. Two Markov chains were run for 20 million steps each. Removing 25% of the chain as burn-in, combining the chains, and sampling every 1000th tree, we obtained 30,000 independent trees from the posterior distribution of trees.
Data availability
Sequences have been deposited in GenBank under accession numbers KT123041–KT123171.
Results
Tree statistics in the ML and posterior trees
Using the MP1 population as outgroup, the inferred rooted ML tree was paraphyletic in MP2 and polyphyletic in MP3, with the root label being MP2 (Figure 3). The number of apparent unique ancestors is seven regardless of who the assumed donor is. That is, in either case, the donor transmitted a minimum of seven lineages to the recipient, implying either a highly diverse founding population that was transmitted once, or that there was an ongoing transmission process over some time.
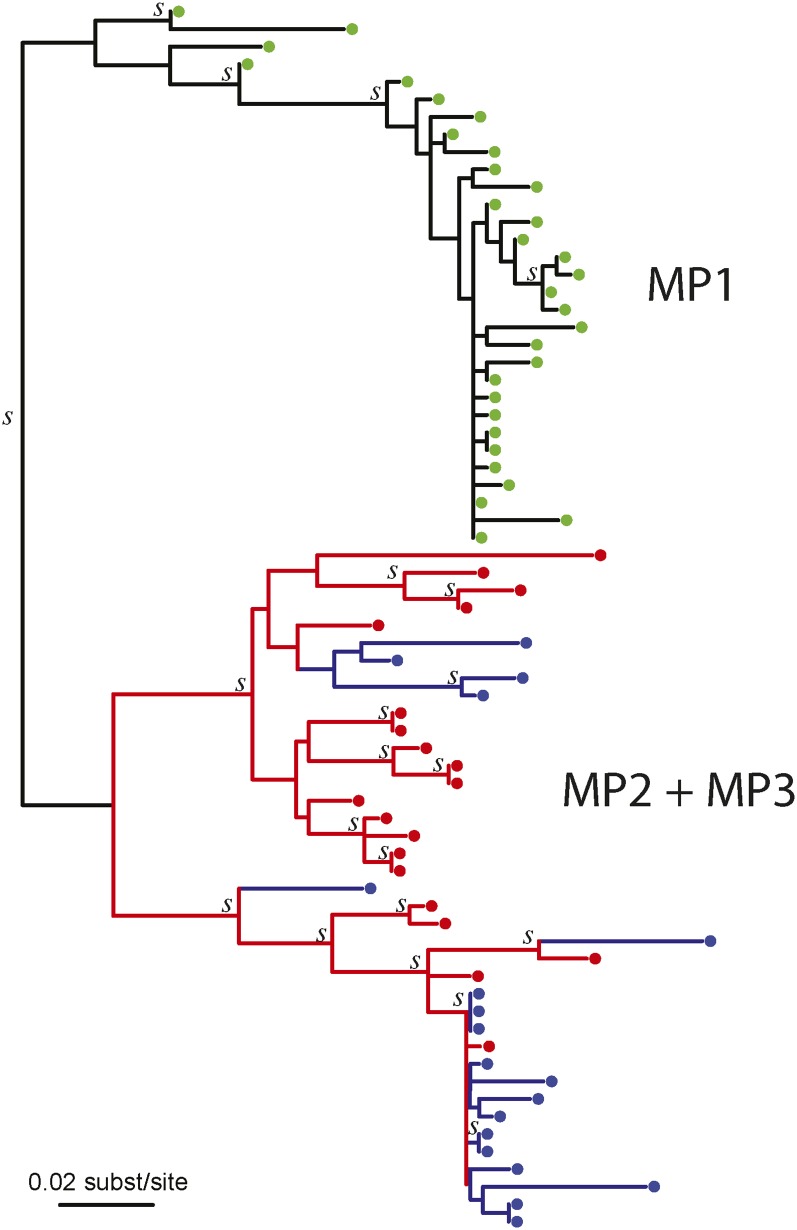
Figure 3.
Maximum likelihood reconstruction of the MP2-MP3 joint HIV-1 env phylogeny. MP1 (yellow) did not infect either MP2 or MP3 (Figure S1), and is used to root the MP2 (red) and MP3 (blue) HIV-1 tree. Clades with aLTR support (>0.90) are indicated with a “s.” The topology of this tree suggested that at least seven lineages were transmitted between MP2 and MP3. Because the branch lengths were zero or near zero in the bottom clade, we added a small distance for readability purpose to show the four possible transmitted lineages that the topology suggested in this clade. Partially to avoid depending on this single (best) tree, we evaluated a large collection of posterior trees in the main analyses of this case.
The topological statistics from the ML tree are very close to the posterior mean values calculated on the posterior distribution of trees. In the empirical posterior distribution of phylogenies, 94% had MP2 as the root label while <1% had MP3 as the root label (Figure 4A; ABC probe 1), 100% had a Para-phyletic/Poly-phyletic (PP) topology (ABC probe 2), and almost all trees had either seven (75%) or six (23%) unique ancestors assuming MP3 was the donor (Figure 4B; ABC probe 3). Interestingly, comparing the distribution of unique ancestors in MP2 and MP3 as recipients, respectively, thus assuming that the other was the donor, shows a broad Poisson-like distribution in MP3, and a sharp single peak at seven unique ancestors in MP2 (Figure 4B). It is important to note that the statistic underlying the number of unique ancestors is only interpretable in the recipient of a donor-recipient pair; in the donor, the statistic becomes a meaningless measure of tree shape. The fact that the distribution of this statistic is narrow when assuming that MP3 is the donor but broad if MP2 is the donor possibly suggests that the narrow distribution represents biological signal while the broad distribution is simply noise in the phylogenetic reconstruction.
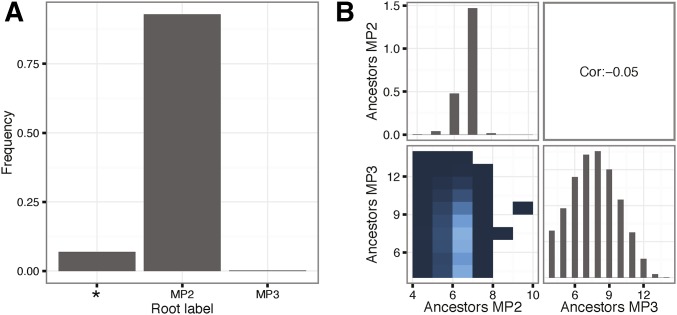
Figure 4.
Root label and number of unique ancestors. (A) shows the density of the three possible root labels [MP2, MP3, or equivocal (*); statistical probe 1], and (B) shows the joint and marginal distributions of the number of unique ancestors assuming MP2 or MP3 as the donor. Lighter color in the joint distribution indicates higher density, and white indicates no data. The overall Pearson correlation between MP2 and MP3 number of ancestors was very low at −0.05. Statistics were calculated on a set of 232,000 phylogenies sampled from the posterior distribution after burn-in based on the real sequence data.
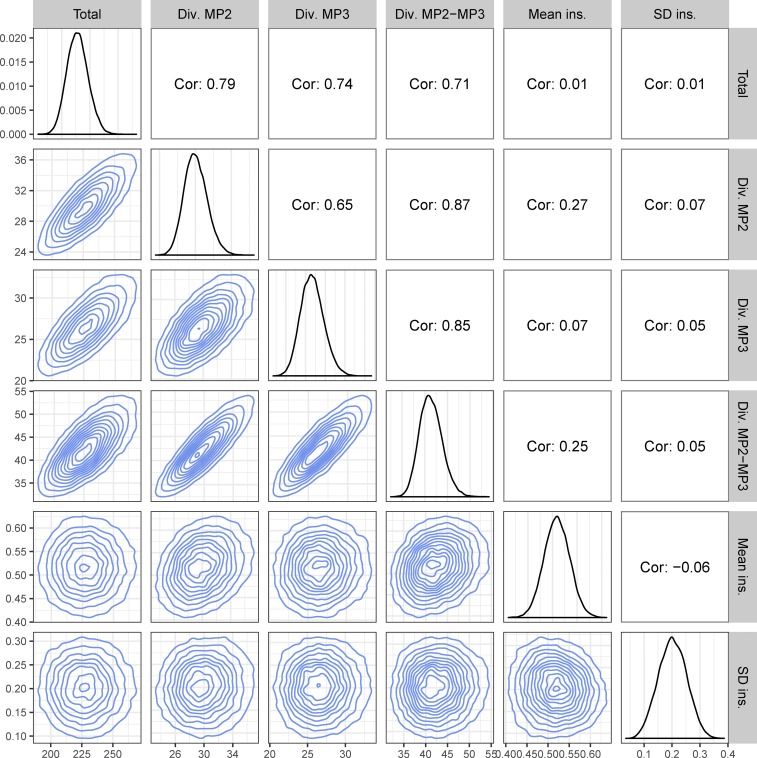
Figure 5 shows the pair-wise joint distributions of the other tree statistics (combined in ABC probe 4), clearly showing Normal-like distributions in the marginal and pairwise joint distributions. As expected, some statistics were closely correlated to each other. Because at least one of the patients must have been infected for a long time, and transmitted much diversity to the other, the total number of substitutions in the tree was strongly correlated (R > 0.71) to both MP2 and MP3 within-host diversity as well as between-host diversity. Similarly, MP2 and MP3 within-host diversities were also strongly correlated (R > 0.65). More interestingly, the mean ancestor heights showed some correlation (R = 0.27) with MP2 within-host diversity, but not with that of MP3 (R = 0.07). We hypothesize that this too might be an indication of transmission direction as a recipient’s population diversity at sampling will be influenced by the donor’s diversity at the time of transmission when multiple lineages are transmitted.
Figure 5.
Diversity measures and ancestor heights. This figure shows the marginal (diagonal) and pairwise joint (lower triangle) distributions for the diversity and ancestors height statistics (statistical probe 4). The upper triangle shows the pairwise Pearson correlations. Diversity (Div.) and sum of all tree branches (Total) are in units of number of nucleotide substitutions, and the mean ancestor insertion height (Mean ins.) is on a relative root-to-tip 0–1 scale. Statistics were calculated on a set of 232,000 phylogenies sampled from the posterior distribution after burn-in based on the real the sequence data.
Evidence for direction and frequency of transmission
To evaluate how so much diversity could be transferred among MP2 and MP3, we considered three models (singular-, co-, and superinfection) and two formulations of the prior [describing when transmission(s) could have occurred].
Overall, the model with the highest approximate marginal evidence was the superinfection model under prior 2, i.e., the model that assumes a long period of ongoing transmissions between MP2 and MP3, and that the relationship between MP2 and MP3 started before the divorce of MP1 and MP2. Jointly considering all models, we calculated an aBF of 22 favoring MP3 as the donor of MP2’s infection. That is, regardless of the model and prior formulation, the evidence clearly favors MP3 as the donor.
In detail, comparing the best fitting model to the next best (superinfection model under prior 2 vs. singular-infection under prior 1) we obtained an aBF of 10, suggesting clear, but not overwhelming, evidence for ongoing transmission. However, the aBF for superinfection compared to co-infection is overwhelming (aBF > 100) in favor of superinfection, suggesting that ongoing transmission only fits the data well if the transmission window is >90 days. The supremacy of the superinfection model comes from the fact that in the singular-infection model the number of unique ancestors is correlated with an ambiguous root label that is rarely observed in the data. That is, to get seven unique ancestors in the singular-infection model, many more lineages have to survive into the source population; however, when there are many “MP2” and “MP3” lineages in the source population, the root label will be ambiguous ∼50% of the time. Ongoing transmission resolves this issue by limiting the number of lineages from the donor that exist in the source population at any given time, both allowing for coalescences between the donor and recipient lineages that define unique ancestors while maintaining a high probability of a nonambiguous root label. Finally, prior 2, which assumed that the relationship between MP2 and MP3 started before the divorce of MP2 and MP1, is only slightly favored over the less permissive prior 1 (aBF = 3.5).
Model choice decomposition
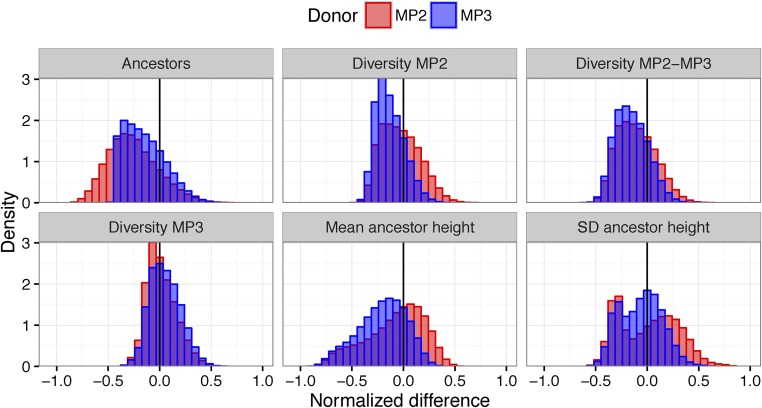
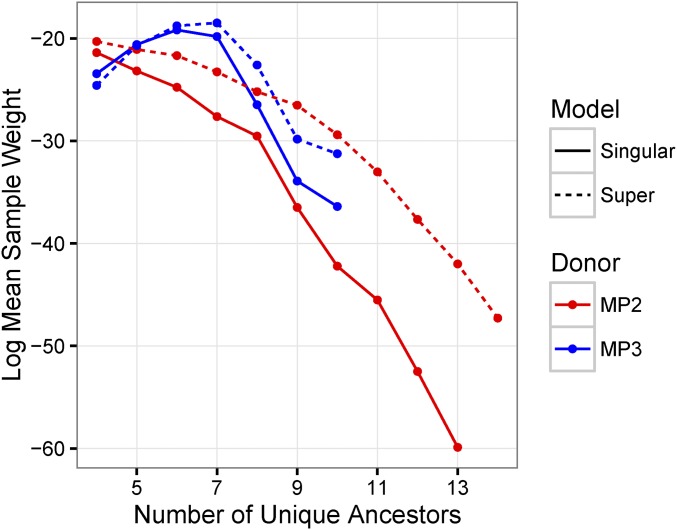
The difference between the empirical and simulated distributions of the statistics for the superinfection model stratified by the identity of the donor is shown in Figure 6. Considering only marginal distributions gives the impression that the preference for MP3 as the donor is driven by the number of ancestors and the SD of the insertion heights, which are both closer to the empirical distribution when MP3 is the donor. In fact, the marginal empirical density of the statistics is generally higher when MP3 is the donor (Figure S2) for many of the statistics; however, when MP3 is the donor, a random draw from the posterior only has 13% probability of having MP2 as the root label. To understand the preference for MP3 as the donor, we need to consider the joint distribution of statistics in both the simulations and the data. Figure 7 shows the log mean sample weight as a function of the number of unique ancestors, donor identity, and model. Assuming MP3 is the donor, >95% of the empirical trees have six or seven ancestors. In the simulation, when MP3 is the donor and the simulation gives six or seven ancestors, the values of the remaining statistics are approximately correct, leading to a high sample weight. Hence, the narrow distribution of number of ancestors assuming MP3 is the donor (Figure 4B) filters out simulations that also have low densities of the remaining statistics. However, when MP2 is the donor, the broad distribution of ancestors does not produce a similar effect, leading to an overall preference for MP3 as the identified donor.
Figure 6.
Normalized difference in empirical and simulated statistics stratified by donor in the superinfection model. Each panel shows the estimated difference in a statistic (Number of unique Ancestors, Diversity in MP2, Diversity between MP2 and MP3 taxa, Diversity in MP3, Mean ancestor height, and SD of the ancestor height). The distributions show the results from randomly selecting a phylogeny from the posterior conditional on the observed sequences and a random simulation from the prior for the superinfection model. Values are normalized to be in [0,1] so they can all be plotted on the same axis. Blue distributions are from simulations with MP3 as donor, and red with MP2 as donor. Densities closer to zero mean that the simulation tends to give values of the statistics that are probabilistically close to the empirically observed values. Typically, simulations with MP3 as donor better reflected the empirically observed trees.
Figure 7.
Log mean sample weight stratified by number of unique ancestors. This figure shows the natural log of the mean sample weight stratified by the number of unique ancestors in samples from the prior distribution for the singular-infection model (solid lines), the superinfection model (dashed lines), with donor being either MP2 (red lines), or MP3 (blue lines). The mean sample weight is highest in the superinfection model when MP3 is the donor and there are seven unique ancestors. This is due to the fact that, in this stratum, the simulation tends to get higher density values of the remaining statistics.
Probability of the root label matching the donor in poly/paraphyletic trees
In previous work (Romero-Severson et al. 2016), we suggested that the root label would be “inconsistent” (i.e., root label is not the donor’s label) only rarely. Here, we performed a set of simulations to determine how improbable it is to obtain a label other than the donor’s at the root when there is multiple transmission and a poly/paraphyletic tree topology under a variety of counterfactual situations. The situation under analysis in this paper is quite different from what we had previously considered in that the samples are taken at different times and the within-host population parameters are allowed to vary between the donor and recipient. To study the effects of the differential sampling times and population dynamic parameters, we simulated 36 parameter sets with instances each assuming (1) MP2 or MP3 as the donor, (2) the singular-infection or superinfection models, (3) different sampling times, and (4) different population dynamic parameters.
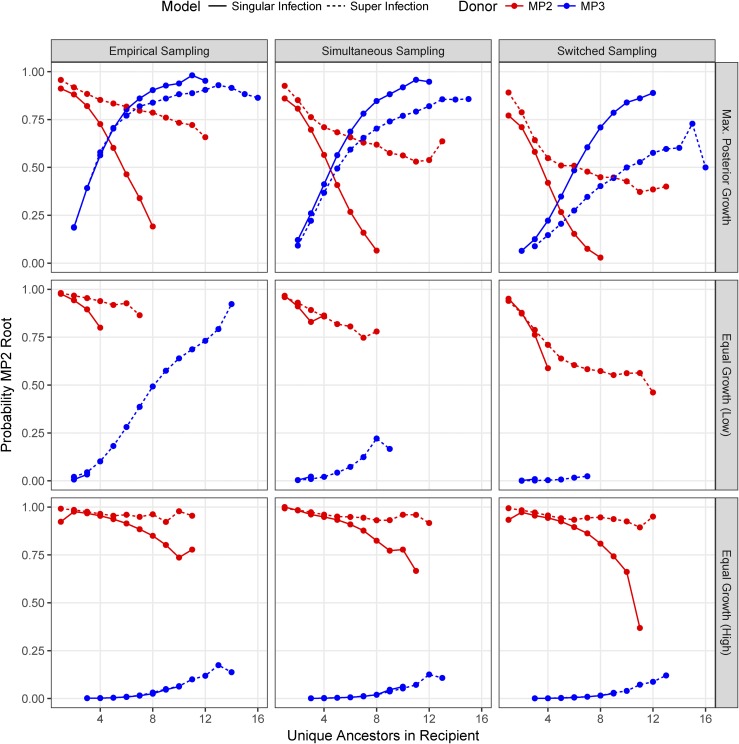
Figure 8 shows the probability of getting an MP2 root label stratified by the number of unique ancestors for each simulated parameter set. The upper left panel assumes the maximum posterior parameter values and the empirical sampling times. When MP3 is the donor, the probability of an MP2 root label grows with increasing number of unique ancestors in the recipient. This is due to the fact that the number of unique ancestors is the minimum number of lineages in the recipient that must have survived into the donor’s source population on the reverse time scale; as more lineages from the recipient survive into the donor’s population, the higher the probability of obtaining the recipient’s label at the root. At seven unique ancestors in the superinfection model, the probability of getting an MP2 label at the root is about equal regardless of who the donor was. That is, in this particular case, the relationship between the root label and the donor is complicated.
Figure 8.
Probability of obtaining MP2 root label stratified by number of unique ancestors in the recipient given alterative sampling and population growth assumptions. Each panel represents the results of simulations. The red lines indicate that the donor was MP2 while blue indicates that the donor was MP3. Solid lines show the single-infection model while dashed lines show the superinfection model. Panels in the “Empirical Sampling” column assumed the same sampling times as was actually observed (MP3 sampled 588 days before MP2), the “Simultaneous Sampling” column assumed that sampling of MP2 and MP3 occurred at the same time at the midpoint of the actual sampling times, and the “Switched Sampling” column assumed that the sampling times are switched. Panels in the “Max. Posterior Growth” row have and equal to the mean values from the posterior distribution, the “Equal Growth (Low)” row have the “Equal Growth (High)” row have
In poly/paraphyletic trees, the relationship between the root label and the donor is determined by the distribution of lineages from the donor and recipient that survive into the donor’s source population. This is influenced by the mode of transmission, the population dynamics in each host, and the sampling times. If we assume that the sampling times are switched (i.e., that MP2 is assumed to be sampled 588 days after MP3), we observed a large decrease in the probability of observing an MP2 root label when MP3 is the donor (upper row, right column, Figure 8). This is due to the fact that fewer MP2 lineages now survive into the source population, as they are lost to coalescence in the period from sampling to the transmission event. Likewise, setting the population growth rates equal in MP2 and MP3 shows a strong effect on the probability of obtaining an MP2 root label; we observed a large difference in the probability of obtaining an MP2 root label given the identity of the donor regardless of the sampling time. That is, both the differential population growth rates inferred for MP2 and MP3 and the difference in sampling times contribute to the “inconsistency” of the root label in this analysis.
Model parameter estimates
The point estimates and 95% CIs for the superinfection model are days, (2072, 3590) days, (0.3, 3,9) day−1, (2.2, 30) day−1, (0.2, 1.7) day−1. These values imply an ongoing infection window of 1464 days. In the singular transmission model, we have (518, 688) days, (2174, 3592) days, (8, 58), (16, 95) day−1, (0.5, 2.7) day−1. Thus, the infection duration of MP3 was robust to model formulation and prior assumptions (∼7–8 years), while the infection duration of MP2 was model dependent (about double in the superinfection vs. singular-infection model).
Discussion
In this study, we show how to apply previously described theoretical evaluations of epidemiological linkage to a real HIV-1 transmission case that involved a highly diverse founding HIV-1 population. We show that one can simultaneously estimate direction and diversity, and evaluate frequency of the transmission event(s). We used a previously developed within-host coalescent framework (Romero-Severson et al. 2014), and expanded it by allowing additional transmission events (migration) between the hosts. Inference was achieved using an ABC method informed by topological and distance-based tree statistics, which allowed approximate Bayes factor comparisons between alternative epidemiological hypotheses.
The transmission between MP3 and MP2 involved many lineages, certainly more than we could observe in the limited sample of HIV-1 sequences derived from the patients. It is impossible to know exactly how many lineages were transmitted with these data. However, comparing the singular-, co-, and superinfection transmission scenarios, we found that most likely there had been ongoing transmissions between MP3 and MP2 for a long time, where MP3 initially infected MP2. The evidence that MP3 infected MP2 is surprising in more than one way: first, because MP2 accused MP1 of transmission, MP2 must have assumed that MP3 was uninfected. Second, because the root label was MP2 in 94% of the posterior trees, this result is also surprising as our previous simulation analyses suggested that the root label is strongly associated with the donor (Romero-Severson et al. 2016). This previous analysis assumed, however, that the donor and recipient were sampled at the same time (like in the simulations in Figure 8, simultaneous sampling and equal growth), whereas in the MP2-MP3 case we have the somewhat unusual scenario where the donor was sampled 588 days after the recipient. This result highlights that a simplistic interpretation of a multi-sample phylogeny could be misleading, and that the exact details of the epidemiological scenario must be taken into account when assessing who-infected-whom and when. Similarly, this argues against simplistic use of ancestral state reconstruction in other research fields such as phylogeographic reconstruction of infection origins. Clearly, phylogenetic patterns can be unintuitive and must be statistically interpreted using additional data on when sampling and possible migration events occurred in time.
Our study provides the first results of modeling single vs. ongoing transmission events to explain how multiple lineages could end up in a recipient. A possible extension to our framework could be to allow for transmission of more than lineage at multiple times, but without additional data, e.g., frequent longitudinal and deep sampling; there would not be enough power to identify how many variants that were transmitted at each possible occasion. Our ABC framework can, however, estimate the diversity that was transmitted, and arguably this measure is more important from a clinical perspective as it may relate to how difficult it is to combat the incoming virus for the immune system, antiviral drugs, and future vaccines.
The initial transmission date from MP3 to MP2 was model dependent ( days, singular-infection model; and 1464 days, superinfection model). This difference in transmission duration estimation suggests that measuring clinical markers, such as BED (Parekh et al. 2002; Skar et al. 2013), could be used to calculate prior distributions of infection times, which potentially could help to discriminate between alternative transmission hypotheses. In our case, the number of transmitted lineages in the singular transmission model needs to be more than three times as large as the number of unique ancestors. Under a neutral coalescent model, this implies a large diversity in the founding population. In general, for any tree where the number of unique ancestors is more than one, the founding population must be highly diverse in the singular transmission model. Likewise, the migration rate under the ongoing transmission model is quite high, averaging thousands of migration events over a 4-year period. The migration rate should be interpreted with caution, however, as it measures a hypothetical rate of separate lineage migrations rather than a real number of transmitted unique variants that would end up as detected ancestors to the population, and cannot inform about the number of actual transmission events as more than one lineage could potentially be transmitted per contact. Likewise, the superinfection model could be picking up the signal of multiple discrete transmission events rather than a constant migration process.
HIV-1 co-infection has been defined as infection of several HIV-1 genetically diverse virions before seroconversion [typically 21 days after infection (Cohen et al. 2011)] or within a somewhat longer time (3–6 months) when an immune response has developed to the initial inoculum, and superinfection as additional infections after a strong immune response has been established (van der Kuyl and Cornelissen 2007; Ronen et al. 2013). In addition, superinfection is often thought of as an additional infection from another donor than the initial one. In the transmission case we studied here, both co- and superinfection was evaluated involving only the original donor and recipient—a stable heterosexual couple. Thus, with repeated contacts over time, transmissions may span and blur the defined periods of co- and superinfection. Furthermore, because HIV-1 evolves significantly during any period of >1 month (Skar et al. 2011), variants transmitted later from the same donor also blur the transmitted genetic diversity possible in co- and superinfections. Thus, while superinfection involving multiple donors appears rare (van der Kuyl and Cornelissen 2007), given the fact that 20–40% of sexual infections involve more than one genetic variant (Keele et al. 2008; Salazar-Gonzalez et al. 2009; Li et al. 2010; Rieder et al. 2011), ongoing transmission between stable couples as investigated here may be more common than previously realized.
In conclusion, taking phylogenetic uncertainty into account, we have created a framework that can evaluate how much diversity is transmitted, and whether transmission occurs once or over a period of time. We show that it is important to take epidemiological information into account when analyzing support for one transmission scenario over another, as results may be nonintuitive, and sensitive to details about sampling dates relative to possible infection dates.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300284/-/DC1.
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases/National Institutes of Health (NIAID/NIH) under award number R01AI087520, and by grants PTDC/SAU-EPI/122400/2010, VIH/SAU/0029/2011 and UID/Multi/04413/2013 from Fundação para a Ciência e Tecnologia (FCT), Portugal. I.B. was supported by a post-doctoral fellowship (SFRH/BPD/76225/2011) from FCT, Portugal. I.B. was supported by a post-doctoral fellowship (BU 2685/4-1) from the Deutsche Forschungsgemeinschaft.
Footnotes
Communicating editor: R. Nielsen
Literature Cited
- Altfeld M., Allen T. M., Yu X. G., Johnston M. N., Agrawal D., et al. , 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420: 434–439. [DOI] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O., 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. [DOI] [PubMed] [Google Scholar]
- Bartolo I., Rocha C., Bartolomeu J., Gama A., Marcelino R., et al. , 2009. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: new insights into the origins of the AIDS pandemic. Infect. Genet. Evol. 9: 672–682. [DOI] [PubMed] [Google Scholar]
- Boily M. C., Baggaley R. F., Wang L., Masse B., White R. G., et al. , 2009. Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect. Dis. 9: 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo F. Y., Sanjuan R., Moya A., Cuevas J. M., 2007. The effect of co- and superinfection on the adaptive dynamics of vesicular stomatitis virus. Infect. Genet. Evol. 7: 69–73. [DOI] [PubMed] [Google Scholar]
- Cohen M. S., Shaw G. M., McMichael A. J., Haynes B. F., 2011. Acute HIV-1 infection. N. Engl. J. Med. 364: 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollo L., 1893. Les lois de l’évolution. Bull. Soc. Belge Géol. Pal. Hydr. 7: 164–166. [Google Scholar]
- Genz A., Bretz F., Miwa T., Mi X., Leisch F., et al. , 2017. mvtnorm: multivariate normal and t distributions, R package version 1.0–6. [Google Scholar]
- Gottlieb G. S., Nickle D. C., Jensen M. A., Wong K. G., Grobler J., et al. , 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363: 619–622. [DOI] [PubMed] [Google Scholar]
- Grobler J., Gray C. M., Rademeyer C., Seoighe C., Ramjee G., et al. , 2004. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J. Infect. Dis. 190: 1355–1359. [DOI] [PubMed] [Google Scholar]
- Guindon S., Lethiec F., Duroux P., Gascuel O., 2005. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33: W557–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H., 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Keele B. F., Giorgi E. E., Salazar-Gonzalez J. F., Decker J. M., Pham K. T., et al. , 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 105: 7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh Brown A. J., 1997. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc. Natl. Acad. Sci. USA 94: 1862–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T., Albert J., 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96: 10752–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T., Fitch W. M., 1999. The phylogenetics of known transmission histories, in The Evolution of HIV, edited by Crandall K. A. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Li H., Bar K. J., Wang S., Decker J. M., Chen Y., et al. , 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhuis M., Boucher C. A., Schipper P., Leitner T., Schuurman R., et al. , 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95: 14441–14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., 2001. Coalescent Theory. Wiley Online Library, Hoboken, NJ. [Google Scholar]
- Parekh B. S., Kennedy M. S., Dobbs T., Pau C. P., Byers R., et al. , 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res. Hum. Retroviruses 18: 295–307. [DOI] [PubMed] [Google Scholar]
- Pennings P. S., Kryazhimskiy S., Wakeley J., 2014. Loss and recovery of genetic diversity in adapting populations of HIV. PLoS Genet. 10: e1004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder P., Joos B., Scherrer A. U., Kuster H., Braun D., et al. , 2011. Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin. Infect. Dis. 53: 1271–1279. [DOI] [PubMed] [Google Scholar]
- Romero-Severson E., Skar H., Bulla I., Albert J., Leitner T., 2014. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol. Biol. Evol. 31: 2472–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Severson E. O., Bulla I., Leitner T., 2016. Phylogenetically resolving epidemiologic linkage. Proc. Natl. Acad. Sci. USA 113: 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen K., McCoy C. O., Matsen F. A., Boyd D. F., Emery S., et al. , 2013. HIV-1 superinfection occurs less frequently than initial infection in a cohort of high-risk Kenyan women. PLoS Pathog. 9: e1003593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez J. F., Salazar M. G., Keele B. F., Learn G. H., Giorgi E. E., et al. , 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206: 1273–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn K. B., Somasundaran M., Luzuriaga K., Leitner T., 2015. Recombination elevates the effective evolutionary rate and facilitates the establishment of HIV-1 infection in infants after mother-to-child transmission. Retrovirology 12: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R., Margolick J. B., Gange S. J., Rodrigo A. G., Upchurch D., et al. , 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73: 10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattock R. J., Moore J. P., 2003. Inhibiting sexual transmission of HIV-1 infection. Nat. Rev. Microbiol. 1: 25–34. [DOI] [PubMed] [Google Scholar]
- Skar H., Gutenkunst R. N., Wilbe Ramsay K., Alaeus A., Albert J., et al. , 2011. Daily sampling of an HIV-1 patient with slowly progressing disease displays persistence of multiple env subpopulations consistent with neutrality. PLoS One 6: e21747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skar H., Albert J., Leitner T., 2013. Towards estimation of HIV-1 date of infection: a time-continuous IgG-model shows that seroconversion does not occur at the midpoint between negative and positive tests. PLoS One 8: e60906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. M., Wong J. K., Hightower G. K., Ignacio C. C., Koelsch K. K., et al. , 2004. Incidence of HIV superinfection following primary infection. JAMA 292: 1177–1178. [DOI] [PubMed] [Google Scholar]
- Smith D. M., Wong J. K., Hightower G. K., Ignacio C. C., Koelsch K. K., et al. , 2005. HIV drug resistance acquired through superinfection. AIDS 19: 1251–1256. [DOI] [PubMed] [Google Scholar]
- Smith D. M., Strain M. C., Frost S. D., Pillai S. K., Wong J. K., et al. , 2006. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 355: 1–5. [DOI] [PubMed] [Google Scholar]
- van der Kuyl A. C., Cornelissen M., 2007. Identifying HIV-1 dual infections. Retrovirology 4: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction. Roberts and Company Publishers, Greenwood Village, CO. [Google Scholar]
- Yang O. O., Daar E. S., Jamieson B. D., Balamurugan A., Smith D. M., et al. , 2005. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J. Virol. 79: 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F., Brodin J., Thebo L., Lanz C., Bratt G., et al. , 2015. Population genomics of intrapatient HIV-1 evolution. Elife 4: e11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences have been deposited in GenBank under accession numbers KT123041–KT123171.