Abstract
N6-methyladenosine (m6A) is an abundant nucleotide modification in mRNA that is required for the differentiation of mouse embryonic stem cells. However, it remains unknown whether m6A controls differentiation of normal and/or malignant myeloid hematopoietic cells. Here we show that shRNA-mediated depletion of the m6A-forming enzyme METTL3 in human hematopoietic stem/progenitor cells promotes differentiation coupled with reduced proliferation. Conversely, overexpression of wild-type METTL3, but not the catalytic-dead form of METTL3, inhibits differentiation and increases cell growth. METTL3 mRNA and protein is expressed more abundantly in acute myeloid leukemia (AML) cells compared to healthy hematopoietic stem/progenitor cells and other types of tumors. Furthermore, METTL3 depletion in humanmyeloid leukemia cell lines induces differentiation and apoptosis and delays leukemia in recipient mice in vivo. Single-nucleotide resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of c-MYC, BCL2 and PTEN mRNAs in human myeloid leukemia MOLM13 cells. Moreover, loss of METTL3 leads to increased levels of pAKT, which contributes to the differentiation effects of METTL3 depletion. Overall these results provide a rationale for therapeutic targeting of METTL3 in myeloid leukemia.
A recently identified regulator of differentiation is N6-methyladenosine (m6A), a reversible nucleotide modification detected in at least 20% of mouse and human cellular mRNAs in diverse cell types 1,2. Embryonic stem (ES) cells deficient in Mettl3 – the m6A RNA methyltransferase – remain pluripotent despite treatment with diverse stimuli that promote differentiation3,4. Mettl3−/− ES cells retain pluripotency markers and fail to acquire gene expression patterns seen in differentiated cells. In contrast, more differentiated stem cells, such as primed stem cells, a cell type that is unable to contribute to embryonic development, show enhanced and abnormal expression of differentiation markers upon depletion of m6A3. Overall these data suggest that alterations in m6A levels can markedly influence cell fate and differentiation status but the effects are dependent on the cellular context.
Hematopoiesis provides an ideal model system to explore differentiation states as hematopoietic stem cells (HSCs) have well-defined developmental trajectories, and differentiation of hematopoietic stem/progenitor cells (HSPCs) can be monitored and quantified. Additionally, abnormal or blocked differentiation is a common feature of myeloid hematological malignancies. Whether m6A influences normal hematopoiesis or the abnormal differentiation seen in AML is not known. Elevated expression of FTO, a demethylase originally thought to target m6A, was reported to enhance leukemic oncogene-mediated cell transformation and inhibit all-trans-retinoic acid-induced differentiation in AML5. However, FTO is now known to demethylate N6-2′-O-dimethyladenosine (m6Am) in mRNA 5′ caps and has negligible effects on m6A6. Thus, the role for m6A in the balance of normal and malignant differentiation of myeloid cells remains unclear.
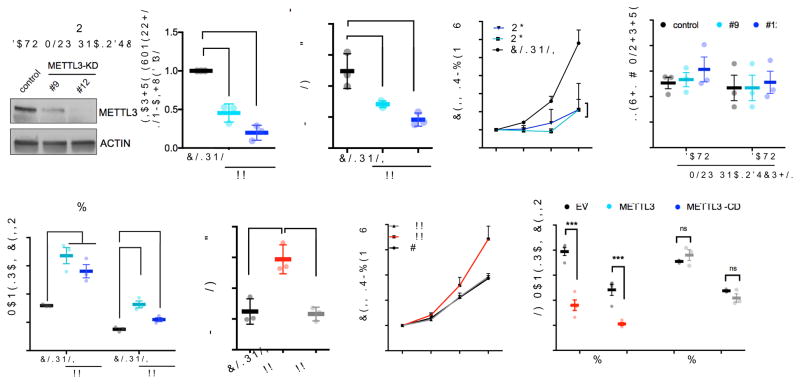
To examine the function of m6A in hematopoietic stem cells, we depleted the m6A “writer,” METTL3, using shRNA in human cord blood (CB)-derived CD34+ HSPCs. Two independent shRNAs showed efficient knockdown of METTL3 (shRNA #9 and #12) (Fig. 1a) and resulted in a reduction of global m6A levels (Fig. 1b and Supplementary Fig. 1a). METTL3-depleted cells gave rise to significantly less colony-forming units (CFUs) in all lineages when plated in methylcellulose (Supplementary Fig. 1b). Loss of METTL3 also resulted in inhibition of cell growth (Fig. 1c) without evidence for apoptosis (Fig. 1d). Additionally, we observed an increase in myeloid differentiation in METTL3-depleted cells after 7 days compared to control cells expressing a scrambled shRNA based on both flow cytometric markers (CD11b and CD14) and morphology (Fig. 1e and Supplementary Fig. 1c–e). Increased differentiation was also confirmed by the rapid loss of the stem/progenitor cell surface marker CD34 in METTL3-knockdown cells (Supplementary Fig. 1f,i). Taken together, these data suggest that METTL3 depletion enhances myeloid differentiation.
Figure 1. m6A inhibits myeloid differentiation of human stem/progenitor cells (HSPCs).
(a–e) Human cord blood CD34+ (HSPCs) cells were transduced with lentiviruses expressing either a scramble (control) shRNA or two independent shRNAs targeting METTL3 (#9 and #12; METTL3-KD). (a) Cells were selected for puromycin resistance and immunoblotted at four days post-transduction. Left, representative immunoblot. Right, the quantitative summary n=3 independent experiments; error bars, s.e.m. **p<0.01,***p<0.001 two-tailed t test.
(b) m6A levels in poly(A) purified mRNA were quantified by two-dimensional thin layer chromatography (2D-TLC, see methods). n=3 independent experiments; error bars, s.e.m. * p<0.05, two-tailed t test.
(c) The number of viable cells was measured over the course of seven days beginning four days post-transduction of shRNAs. n=3 independent experiments; error bars, s.e.m. * p<0.05, two-tailed t test.
(d) The percentage of apoptotic cells was determined at day four and five post-transduction. Cells were stained for Annexin V and DAPI and quantified by flow cytometry.
(e) Myeloid differentiation was measured using CD11b and CD14 as markers of myeloid differentiation. Cells were stained and expression of each surface marker was quantified by flow cytometry seven days after plating. error bars, s.e.m. * p<0.05, **p<0.001, two-tailed t test.
(f–h) Human cord blood CD34+ (HSPCs) cells were transduced with retroviruses expressing GFP together with empty vector (EV) or wild type METTL3 or catalytically dead METTL3 (METTL3-CD). Cells were sorted based on GFP positivity two days post transduction.
(f) At XX time point cells were analyzed by XXX method. Immunoblots at two days post transductions n=3 independent experiments; error bars, s.e.m. ** p<0.01, two-tailed t test.
(g) Sorted cells were plated in basic media (See Supplementary methods). Cells were counted for seven days after plating. EV: Empty vector (black line), METTL3 (red line), catalytically dead METTL3-CD (gray line). n=4 independent experiments; error bars, s.e.m. * p<0.05, two-tailed t test.
(h) Myeloid differentiation was evaluated as in (e) seven days after plating in myeloid differentiation conditions. n=4 independent experiments; error bars, s.e.m. * p<0.05, *** p <0.0001 two-tailed t test.
Conversely, we examined whether METTL3 overexpression can inhibit differentiation. To test this, we transduced CB-CD34+ cells with retroviruses expressing GFP alone or with wild-type METTL3. To directly address the requirement of the catalytic activity of METTL3, we also overexpressed a catalytically dead mutant of METTL3 (METTL3-CD; residues 395–399: DPPW→APPA)7,8) (Supplementary Fig. 1j). METTL3 but not METTL3-CD overexpression increased m6A levels compared to control cells (Fig. 1f and Supplementary Fig. 1g). Overexpression of METTL3 wild type, but not METTL3-CD, promoted proliferation and colony formation (Fig. 1g and Supplementary Fig. 1h) and significantly inhibited myeloid differentiation of HSPCs (Fig. 1h and Supplementary Fig. 1i and k). Additionally, Mettl3 mRNA, which was abundant in hematopoietic stem cells and progenitor cells, was expressed in lower amounts in mature differentiated myeloid cells (Supplementary Fig. 1l). These data indicate that the level of METTL3 and its enzymatic activity is negatively correlated with the differentiation of normal myeloid cells.
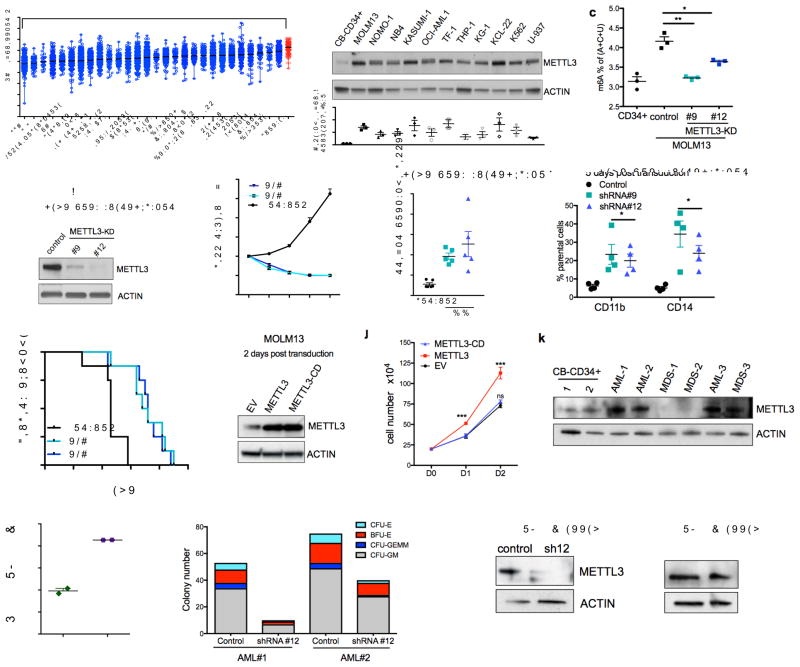
Since myeloid differentiation is frequently dysregulated in leukemia, we next determined if METTL3 expression is altered in leukemia. METTL3 mRNA expression in human acute myeloid leukemia (AML) samples is significantly higher than in other cancer types (Fig. 2a). To further assess the relative abundance of METTL3 in myeloid leukemia, we examined METTL3 mRNA and protein levels in multiple leukemia cell lines in comparison to primary HSPCs cord blood derived CD34+ cells. METTL3 mRNA was more abundant in AML cell lines (8/11) (Supplementary Fig. 2a) as was METTL3 protein (11/11) (Fig. 2b). We found no significant difference in METTL3 expression across multiple subtypes of AML in the BloodPool database9 (Supplementary Fig. 2b).
Figure 2. m6A promotes leukemogenesis.
(a) METTL3 mRNA expression in acute myeloid leukemia (AML) compared to other cancers (The Cancer Genome Atlas database). Data are presented as mean log2 expression with range. AML: orange dots, **** p<0.00001, ** p<0.01 ANOVA with multiple comparisons,
(b) METTL3 protein expression in AML cell lines compared to normal HSPCs. Top: An immunoblot for METTL3 and loading control (ACTIN) in the indicated myeloid leukemia cell lines and cord blood (CB) CD34+ cells. Bottom: quantitative summary of the immunoblots. n=3 independent experiments; error bars, s.e.m. * p<0.05, **p<0.01,***p<0.001 two-tailed t test.
(c) Global m6A levels in AML cells versus normal HSPCs. m6A levels from poly(A) purified mRNA were quantified in CB-CD34+ and MOLM-13 AML cells by two-dimensional thin layer chromatography (2D-TLC, see methods). n=3 independent experiments, two-tailed t test.
(d–h) MOLM13 cells were transduced with lentiviruses expressing either a scramble (control) shRNA or two independent shRNAs targeting METTL3 (#9 and #12; METTL3-KD). Cells were selected for puromycin resistance and assayed four days post transduction.
(d) Representative immunoblot for METTL3 depletion four days post-transduction.
(e) Proliferation assay of MOLM13 control cells versus METTL3 knockdown. The number of viable cells was measured daily beginning four days post-transduction of shRNAs: shRNA#9 and #12 (light blue and dark blue lines) and control shRNA (black line).
(f) The percent of apoptotic cells was determined five days post-transduction by flow cytometry analysis for Annexin V positivity.n=5, independent experiments; error bars, s.e.m. * p<0.05, **p<0.01 two-tailed t test.
(g) Cells were stained for myeloid differentiation markers CD11b and CD14 five days post-transduction. Quantification of positive cells was performed by flow cytometry. METTL3 knockdown cells (light and dark blue bars) versus control cells (black bars). n = 4 independent experiments; error bars, s.e.m. * p<0.05, **p<0.01 two-tailed t test.
(h) Leukemia-free survival of mice transplanted with MOLM13 cells transduced with either a control shRNA or METTL3-targeting shRNAs. MOLM13 cells at four days post transduction were injected into sub-lethally irradiated mice (n=8 for each group). Mantel-Cox test ****p<0.0001.
(i–j) MOLM13 cells were retrovirally transduced with vectors expressing METTL3 and METTL3-CD. Cells were sorted two days post-transduction based on GFP positivity.
(i) Representative immunoblot analysis of METTL3 expression. ACTIN serves as loading control.
(j) Proliferation assay of MOLM13 cells transduced with empty vector (EV), METTL3, or METTL3-CD overexpressing vectors. The number of viable cells was measured daily beginning two days post-transduction. n = 3 independent experiments; error bars, s.e.m. ***p<0.001 two-tailed t test.
(k) Immunoblot analysis of METTL3 protein expression in primary MDS and AML patient cells. ACTIN serves as loading control. Quantitative summary is shown.
(l) Global m6A levels in primary AML patient cells versus normal HSPCs.
(m) Colony forming ability of primary AML patient cells depleted of METTL3. Primary AML patient cells were transduced with control and shRNA targeting METTL3. Cells were sorted based on GFP positivity after 3 days. Cells were plated in methycellulose and colonies were scored after 14 days.
(n) Immunoblot showing METTL3 expression before (D0) and 14 days after (D14) plating of primary AML patient cells in (m). ACTIN serves as loading control. Quantitative summary is shown.
METTL3, the enzymatic unit, forms an obligate heterodimer with METTL14 to activate its methyltransferase activity and bind RNA10–12. METTL14 mRNA expression in AML was also significantly higher compared to most other cancers, but not significantly different from thyroid, clear cell renal carcinoma, and lower than breast (Supplemental Fig. 2c). METTL14 protein showed no increase in expression in most, but not all, AML cell lines relative to primary CD34+ cells (Supplementary Fig. 2d).
With increased METTL3 in leukemic cells versus normal cells and the abundant levels of METTL14, we measured m6A abundance in poly(A) mRNA and found a significant increase in the MOLM13 AML cell line compared to CD34+ control cells (Fig. 2c). These data suggest that elevated m6A might be critical to maintaining an undifferentiated state in myeloid leukemia.
To directly address the role of m6A in human myeloid leukemia cells, we examined the effect of METTL3 knockdown in MOLM13 cells. METTL3 knockdown significantly decreased m6A levels, blocked cell growth, induced differentiation, and resulted in an increased in apoptosis (Fig. 2d–g and Supplementary Fig. 2e–g). Similar effects of shRNA-mediated METTL3 depletion were seen in NOMO-1 and NB4 AML cell lines (Supplementary Fig. 2h–o).
We next asked if METTL3 was required for in vivo leukemogenesis. MOLM13 cells were transduced with METTL3-shRNA and transplanted into immunodeficient recipient mice. These cells exhibited delayed leukemia development compared to MOLM13 cells transduced with the scrambled-shRNA (Fig. 2h). Additionally, when we examined leukemia cells that were able to grow in mice transplanted with METTL3-shRNA-transduced cells, we found that these cells showed equivalent METTL3 abundance as seen in the scrambled-shRNA transduced cells. This indicates that METTL3-depleted cells were negatively selected, and cells that escaped knockdown account for the development of leukemia (Supplementary Fig. 2p).
We next sought to address whether the effects of METTL3 shRNA could be due to off-target effects. To test this, we used a shRNA-resistant form of METTL3. MOLM13 cells transduced with METTL3 shRNA exhibited significant rescue of both the apoptotic and differentiation phenotypes when the shRNA-resistant form of METTL3 was expressed (Supplementary Fig. 3a–f). Additionally, we utilized an independent approach to deplete METTL3 using CRISPR/Cas9-mediated mutation of METTL3 in MOLM13 cells. As we observed in the shRNA experiments, METTL3 depletion lead to increased differentiation and apoptosis with two independent guide RNAs (Supplementary Fig. 3g–j). These data support a requirement for METTL3 for maintaining the undifferentiated state and survival of myeloid leukemia cell lines.
We next asked if the enzymatic activity of METTL3 was required for its effects in leukemia cell lines. To test this, we overexpressed wild-type METTL3 and METTL3-CD in MOLM13 cells (Fig. 2i). Here, we observed that the enzymatic activity is required for growth advantage of METTL3-overexpressing cells (Fig. 2j). To further evaluate the role of METTL3 enzymatic activity in the context of murine leukemia, we utilized a CRISPR/Cas9 strategy that specifically targets functional protein domains within a gene and infers functional importance based on negative selection13. We tested gRNAs targeting the transcriptional start site (TSS) and the enzymatic domain of METTL3 in mouse leukemia RN2 (MLL-AF9, NRASG12D) cells. Only cells transduced with gRNAs targeting the methyltransferase domain were negatively selected over time, indicating that METTL3 enzymatic activity is critical for survival of leukemia cells (Supplementary Fig. 3k).
To provide additional evidence for requirement of m6A and the RNA methylation complex for survival of leukemia cells, we examined the rankings of m6A “writers” and “erasers” in a genome-wide CRISPR-based screen for genes essential for survival in 14 acute myeloid leukemia cell lines14. Whereas all members of the “writer” complex including METTL3, METTL14, WTAP and KIAA1429 were scored highly (at the top 10%) in the essential gene list, the sole m6A eraser ALKBH5 showed no essentiality for leukemia cell survival (Supplementary Fig. 3i–m). Additionally, depletion of FTO, which was previously shown to be required for AML survival5, did not show a substantial effect on AML cell viability across most cell lines. Overall, these data strongly implicate the entire m6A writer complex in leukemia cell survival, but not the m6A eraser ALKBH5 or the m6Am eraser FTO.
Next we examined METTL3 expression in primary cells from patients with AML. We observed higher METTL3 expression in 3/3 AML patient samples and 1/3 MDS patient samples, compared to CB-CD34+ HSPCs (Fig. 2k). In addition, we detected a significant elevation in global m6A levels in primary AML cells versus healthy HSPCs (Fig. 2i). METTL3 depletion by shRNA inhibited the colony formation by primary AML cells (Fig. 2m–n and Supplementary Table 1). Depletion of METTL3 has been previously shown to correlate with m6A levels in a diverse set of contexts1,2. In these experiments, the only colonies that survived at day 14 were comprised of cells in which METTL3 was not depleted. Together, these data indicate a requirement for m6A and METTL3 activity in AML patient cells.
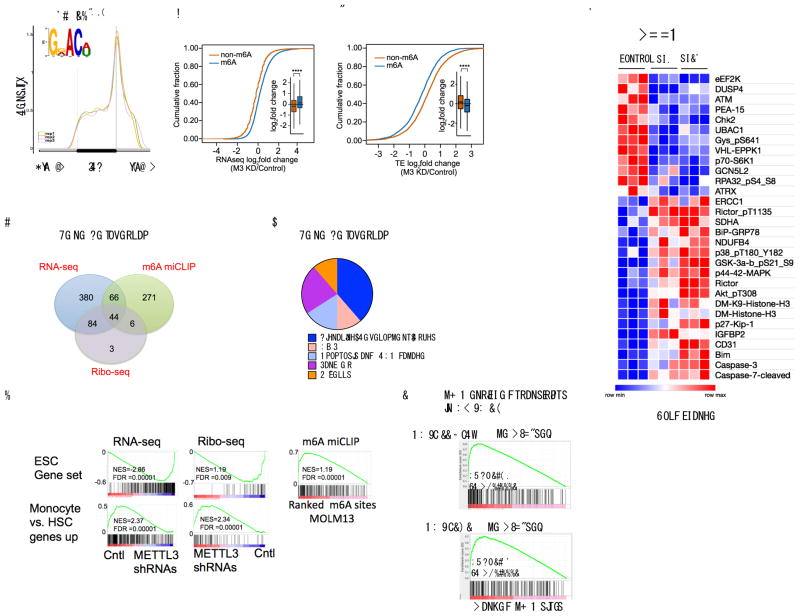
To identify the mRNAs that are methylated in MOLM13 cells, we mapped m6A at single-nucleotide resolution using m6A individual-nucleotide resolution cross-linking and immunoprecipitation (miCLIP)15. In line with our previous analysis, m6A sites were primarily found in both the exons and 3′UTRs (52.4% and 32.4% respectively) with the highest enrichment of m6A residues located near the stop codon (Fig. 3a and Supplementary Fig. 4a). As in other cell types15, m6A was most commonly found in the canonical m6A motif DRACH (D = A, G, U; R = G, A; H = A, C, U), with GGACU being the most frequent sequence context. The frequency of m6A per gene ranged from a single m6A in 2034 genes to 10 or more called m6A residues in 117 genes (Supplementary Fig. 4b and Supplementary Table 2).
Figure 3. m6A is required for maintaining the translation of target mRNAs that control cell fate.
(a) m6A distribution pattern in human leukemia MOLM13 cells. The distribution of m6A in MOLM13 cells was determined at single-nucleotide resolution using miCLIP. The identified m6A sites were plotted in triplicate for their relative distribution across the 5′UTR, coding sequence, and 3′UTR. The m6A DRACH (D= A/G/U, R=A or G, and H= A/C/U) motif was enriched as expected (inset) (E p-value: 2.9*10^973).
(b) The abundance of methylated transcripts upon METTL3 knockdown. RNA-Seq of MOLM13 cells was performed four days post transduction with shRNAs. mRNAs were classified as either m6A-containing (m6A, blue) or non-m6A-containing (non-m6A, orange) based on miCLIP analysis. n=3 per condition, **** p<2.2e−16, two-sided Kolmogorov–Smirnov test.
(c) The translational efficiency of methylated transcripts upon METTL3 knockdown. Ribo-Seq was performed to determine ribosome-protected fragments in MOLM13 METTL3 knockdown compared to control shRNA cells as in (c). Translational efficiency was calculated as the number of ribosome-protected fragments divided by mRNA expression and mRNAs stratified as either m6A-containing (blue) or non-m6A-containing (orange) as described above. n=3 per condition, **** p<2.2e−16, two-sided Kolmogorov–Smirnov test.
(d) Gene set overlap analysis depicted in a Venn diagram (1) enriched for m6A targets, (2) negatively enriched for transcripts upregulated after METTL3 depletion and (3) gene sets negatively enriched for transcripts with reduced translation efficiency in METTL3-depleted cells (For specific gene sets see Supplementary Tables 5, 6, 9 and 10).
(e) Pie chart of the 44 overlapping gene sets from (e) manually curated into general pathway categories (see also Supplementary Table 11).
(f) Gene set enrichment analysis for the RNA-Seq, translational efficiency and m6A ranked lists. The core ESC gene set is enriched as described in (e and f), (top panels). The gene set promoting monocyte differentiation is enriched as indicated after METTL3 knockdown in MOLM13 cells (bottom panels and see also Supplementary Table 10).
(g) Gene set enrichment analysis for the m6A-enriched transcripts in AML patients compared to MOLM13 cells.
(h) Heatmap depicting the fold changes in proteins with significantly altered expression (p<0.01) in a reverse phase protein array (RPPA). Cell lysates from MOLM13 cells four days post-transduction with shRNAs were used for RPPA. The arrays consisted of 304 different antibodies for specific proteins and protein modifications. Fold change was determined by averaging biological replicates (n=3) averaged then divided by the control shRNA expression.
To determine how METTL3 depletion affects m6A -containing transcripts, we performed RNA-Seq on MOLM13 cells following METTL3 knockdown (Supplementary Fig. 4c). Consistent with the idea that m6A destabilizes mRNA16, transcripts with at least one called m6A site showed increased abundance following METTL3 depletion compared to transcripts with no called m6A sites (Fig. 3b). Moreover, the change in abundance was directly correlated with the number of m6A sites per transcript (Supplementary Fig. 4c and Supplementary Table 3).
To determine if m6A depletion alters translation of mRNA in MOLM13 cells, we performed Ribo-Seq17 to determine the ribosome occupancy on mRNAs (Supplementary Fig. 4d). We measured the translational efficiency for each mRNA, which is defined as the number of ribosome protected fragments divided by the mRNA expression17. Although m6A-containing transcripts became more abundant upon METTL3 depletion, m6A depletion reduced their translational efficiency (Fig. 3c, Supplementary Fig. 4d and Supplementary Table 4). A similar increase in mRNA abundance coupled with reduced translation was found upon depletion of METTL3 in HeLa cells (Supplementary Fig. 4e–f)18.
To determine the functional pathways associated with m6A-marked mRNAs, we performed Gene Set Enrichment Analysis (GSEA)19 on all 4733 curated gene sets in the Molecular Signatures Database (MSigDB, http://www.broadinstitute.org/msigdb) combined with 92 additional relevant gene sets from our experimentally derived or published hematopoietic self-renewal and differentiation signatures19,20. We performed GSEA using the ranked lists of m6A sites, differentially expressed transcripts, and translational efficiency upon METTL3 depletion. This resulted in 44 overlapping gene sets (Fig. 3d and Supplementary Tables 5–11). These common gene sets were (1) enriched for m6A targets, (2) negatively enriched for transcripts upregulated upon METTL3 depletion, and (3) negatively enriched for transcripts with reduced translation efficiency in METTL3 depleted cells. We then grouped these pathways into general categories that included apoptosis and DNA damage, cancer, and MYC-associated gene sets (Fig. 3e–f and Supplementary Tables 11–12). Interestingly, the monocytic differentiation gene set was enriched with transcripts that were reduced in their mRNA abundance but increased in their translation efficiency upon METTL3 depletion (Fig 3f and Supplementary Table 11). The effect is likely to be an indirect effect, as this gene set was not statistically significantly (p=0.259) enriched for transcripts containing m6A (Supplementary Fig. 4g). Nevertheless, this analysis suggests the genes associated with differentiation, cell survival and MYC are particularly affected by METTL3 in AML cells through either direct or indirect mechanisms.
To assess clinical relevance of the MOLM13 m6A profile, we mapped m6A in two AML patient samples and compared the m6A-enriched transcripts found in MOLM13 cells to the m6A-enriched transcripts detected in primary AML patient samples. We found that the mRNAs with the highest levels of m6A (top 300 targets) from two independent AML patient samples (AML_118_Dx21 and AML_141 22) were enriched with the m6A targets from the MOLM13 cells (Fig. 3g).
We next sought to understand how METTL3 depletion influences signaling pathways in AML cells. We utilized a reverse phase protein array (RPPA) to look for changes in the expression of proteins that are involved in signaling pathways. In this experiment, we compared MOLM-13 cells expressing control shRNA and either of two METTL3-specific shRNAs. The most significant increases in protein abundance included apoptosis mediators: CASPASE-7, CASPASE-3 and BIM (Fig. 3h and Supplementary Table 13). The other significant effect was an increase in phosphorylated AKT (AKT-pT308 and AKT-pS473) and RICTOR (Rictor-pT1135). These data indicate that m6A depletion may activate the PI3K/AKT pathway (Fig. 3h and Supplementary Table 13).
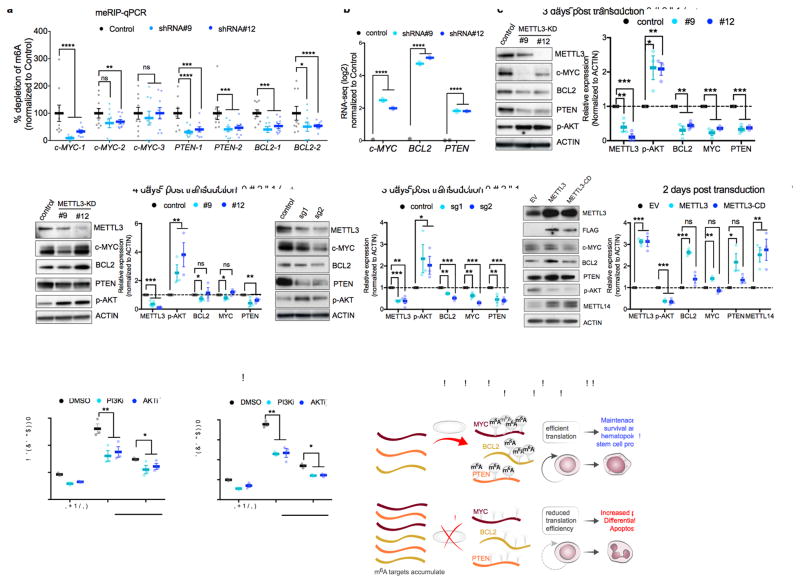
Based on the enrichment of gene sets associated with c-MYC and apoptosis, and our finding that c-MYC and BCL2 contained among the highest called m6A sites in the leukemia transcriptome (20 and 16 sites, respectively, Supplementary Fig. 4a–b and Supplementary Table 2). PTEN, a negative regulator of pAKT– which has six called m6A sites and could explain the increase in pAKT observed in the RPPA was also examined with MYC and BCL2. We first validated that m6A levels in c-MYC, BCL2, and PTEN are reduced following METTL3 knockdown. In these experiments, we used MeRIP-qPCR, which is a measure of transcript levels in m6A immunoprecipitates. We found that knockdown of METTL3 reduced enrichment of m6A at mapped sites in c-MYC, BCL2 and PTEN (Fig. 4a and Supplementary Fig. 5a–b).
Figure 4. m6A directly controls expression of c-MYC, BCL-2 and PTEN.
(a) m6A enrichment at transcripts of c-MYC, BCL-2 and PTEN. Poly(A)+ RNA was isolated from control and METTL3 knockdown MOLM13 cells. In vitro transcribed A and m6A-containing mRNA was then immunoprecipitated with an anti-m6A antibody. Enrichment of m6A at multiple sites (boxes highlighted in Supplementary figure 5a) at transcripts of c-MYC, BCL-2 and PTEN was determined by qPCR.
(b) MYC, BCL2, and PTEN mRNA expression in MOLM13 METTL3 knockdown cells. Plotted is the average change in expression from RNA-Seq of METTL3 knockdown (light and dark blue) compared to control knockdown cells (black) from Figure 3b.
(c–d) Immunoblot analysis for proteins that were associated with the m6A program, e.g., c-MYC, BCL2 and PTEN (based on global genomic approaches or RPPA in Figure 3d–h) The panels are representative blots from three days (c) or four days (d) post-transduction of MOLM13 cells with shRNAs targeting METTL3. ACTIN serves as loading control. Top: representative immunoblot images. Bottom: quantitative summary of the immunoblots. n=3 for 3 days, n=6 for METTL3 expression and n=3 for target genes expression for 4 days independent experiments; error bars, s.e.m. * p<0.05, **p<0.01,***p<0.001 two-tailed t test.
(e) Immunoblot analysis for proteins that were associated with the m6A program three days post transduction of MOLM13 cells with sgRNAs targeting METTL3. ACTIN serves as loading control. Top: representative immunoblot images. Bottom: quantitative summary of the immunoblots. n=4 independent experiments; error bars, s.e.m. * p<0.05, **p<0.01,***p<0.001 two-tailed t test
(f) Immunoblot analysis for proteins that were associated with the m6A program two days post transduction of MOLM13 cells overexpressing wild type METTL3 or METTL3 catalytically dead (METTL3-CD). ACTIN serves as loading control. Top: representative immunoblot images. Bottom: quantitative summary of the immunoblots. n=3 independent experiments; error bars, s.e.m. * p<0.05, **p<0.01,***p<0.001 two-tailed t test
(g–h) Inhibition of the AKT pathway inhibits myeloid differentiation of MOLM13 METTL3 knockdown cells. MOLM13 cells were transduced with shRNAs for three days followed by treatment with DMSO (gray bars), GDC-0068 AKT inhibitor (1 μM, purple bars), or GDC-0032 PI3K inhibitor (0.1 μM, pink bars) for 48 h before myeloid differentiation. Differentiation was assessed by flow cytometry as previously described (Fig. 1m) (n=3 independent experiments; error bars, s.e.m. * p<0.05, **p<0.001, *** p <0.0001 two-tailed t test.
(i) A proposed model of METTL3-dependent myeloid differentiation. METTL3 upregulation in AML cells results in methylation of specific mRNAs critical for regulating apoptosis and differentiation, including MYC, BCL2, and PTEN. Methylated targets are efficiently translated resulting in survival, proliferation, and maintenance of the hematopoietic stem cell program (top panel). Depletion of METTL3 in AML cells reduces translation associated with m6A transcripts resulting in AKT activation, differentiation, and apoptosis.
Consistent with m6A sites controlling translational efficiency, we observed a reduction of c-MYC, BCL2 and PTEN protein expression despite a 2–5 log2 fold increase in RNA expression after METTL3 depletion using METTL3-specific shRNA (Fig 4b–d). Similarly, MYC, BCL2 and PTEN were reduced at day 3 using sgRNAs (Fig 4e). The effect on MYC and BCL2 was either diminished or reversed on day 4 as observed with hairpin #12, indicating that these proteins are translated in a m6A-independent manner at this time point. Translation at day 4 may be mediated by an alternative IRES translational mechanism that induces MYC and BCL2 protein synthesis during apoptosis and stress23–25. Indeed, the cells exhibit commitment into an apoptotic program upon depletion of METTL3 based on the increase in BIM expression at 3 days and 4 days post-transduction (Fig. 4c–d and Supplementary 5c).
Conversely, overexpressing wild-type METTL3, but not the METTL3-CD, leads to an increase in c-MYC, BCL2 and PTEN protein expression abundance in cells (Fig. 4f), indicating that the translational activation of target genes by METTL3 is dependent on its ability to catalyze the formation of m6A. We also found that overexpression of METTL3 and METTL3-CD resulted in increased levels of METTL14. This likely reflects stabilization of METTL14 by binding METTL3 as described previously26.
We also examined the effect of METTL3 depletion on phosphorylated AKT (pAKT), which was detected in the RPPA (Fig. 3h). METTL3 depletion using either METTL3-specific shRNA or sgRNA lead to a marked increase in pAKT (Fig. 4c–e). Conversely, overexpression of METTL3 reduced pAKT (Fig. 4f). Interestingly, pAKT levels were reduced in both the METTL3 and METTL3-CD overexpressing cells. Therefore, it is possible that m6A-independent mechanisms may contribute to reduction in pAKT levels after METTL3 overexpression.
Previous studies found that forced AKT activation in mouse hematopoietic cells and MLL-AF9 transformed cells results in differentiation and a loss of self-renewal27,28. We therefore tested if AKT activation induced by METTL3 knockdown contributes to the rapid differentiation phenotype in m6A-depleted cells. To test this, we treated cells with small molecule inhibitors of PI3K (GDC0032) and AKT (GDC0068). In both cases, inhibition of this pathway partially blocked the differentiation effects elicited by METTL3 depletion (Fig. 4g–h and Supplementary Fig. 4d)).
Overall our study is the first to demonstrate that m6A is critical for maintaining the differentiation program in the hematopoietic system and that this process is dysregulated in myeloid leukemia. Depletion of the m6A writer METTL3 in normal CD34-enriched cord blood cells resulted in reduced proliferation and increased differentiation. In myeloid leukemia cell lines, METTL3 depletion similarly caused differentiation and was also associated with an increase in apoptosis (Fig. 4i).
Unlike in glioblastoma where m6A depletion promotes tumorigenesis29, 30, we find that METTL3 and m6A maintains the undifferentiated leukemic phenotype. Our finding of a requirement of METTL3 is consistent with previous findings that METTL3 is critical for the proliferation of lung cancer cells8. Additionally, genetic alterations of genes encoding m6A writer complex proteins correlate with poorer prognosis in AML patients based on an analysis of the Cancer Genome Atlas Research Network (TCGA) datasets31. Future studies will determine whether elevated METTL3 expression is sufficient to drive initiation of leukemia. Taken together, our data suggest that AML cells regulate their translational state through methylation of specific transcripts, and disruption of this process by depletion of METTL3 yields a translational program that induces differentiation and apoptosis.
Our data suggest that inhibition of METTL3 could be exploited as a therapeutic strategy for myeloid malignancies. It is notable that leukemia cells show elevated abundance of METTL3 compared to normal hematopoietic cells. Furthermore, we find that METTL3 depletion shows markedly increased levels of apoptosis in leukemia cells compared to normal cells. These findings suggest a possible therapeutic index. Future studies that target METTL3, potentially in combination with current and emerging therapeutic agents such as chemotherapy, FLT3 and IDH inhibitors should be explored.
Online Methods
Purification, viral transduction and culture of cord blood (CB) derived HSPC-CD34+ cells
All primary patient samples unless otherwise noted were collected under Biospecimen collection/banking study 09-141. The use of the samples for research purposes is covered under biospecimen research protocol 16-354. CD34+ HSPCs were purified from at least 5 or 10 mixed CB units (each unit from one healthy donor) in each purification. Mononuclear cells were first isolated from CB using Hespan and Ficoll-Hypaque Plus density centrifugation, followed by positive selection using the Auto MACS Pro Seperator and isolation Kit (Miltenyi). CD34+ cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM, Cellgro) 20% BIT 9500 medium (Stem Cell Technologies) supplemented with SCF (100 ng/ml), FLT-3 ligand (10 ng/ml), IL-6 (20 ng/ml) and TPO (100 ng/ml) as the basic culture. CD34+ cells were transduced with high-titer retroviral and lentiviral supernatant and 8μg/ml polybrene. To differentiate HSPCs, cells were cultured under the myeloid-promoting conditions: SCF (100 ng/ml), FLT-3 ligands (10 ng/ml), IL-3 (20 ng/ml), IL-6 (20 ng/ml), GM-CSF (20 ng/ml) and G-CSF (20 ng/ml) and the erythroid-promoting conditions: Epo (6 IU/ml) and SCF (100 ng/ml). Cytokines were purchased from Peprotech, NJ.
Colony forming unit (CFU) assay
104 CD34+ transduced cells were plated (in duplicate) in methylcellulose (MethoCult TMH4434 Classic – Stem Cell Technologies). CFU colonies: erythroid progenitor cells (BFU-E and CFU-E), granulocyte-macrophage progenitor cells (CFU-GM, CFU-G and CFU-M), and multipotent granulocyte, erythroid, macrophage and megakaryocyte progenitor cells (CFU-GEMM) were scored 14 days after seeding.
Proliferation assay
Cells were transduced with viruses by spinfection of leukemia cells in RPMI with 10% FBS or CD34+ cells in basic media. After 48 hours of infection, cells transduced with shRNAs against METTL3 or control scramble shRNA were treated with 3μg/ml puromycin for leukemia cells and 2μg/ml puromycin for CD34+ cells. Two days after puromycin selection, cells were plated at 250,000 cells/ml for proliferation assay. After 48 hours of infection, cells expressing METTL3 or METTL3-CD were sorted based on GFP positivity and plated at 250,000 cells/ml. Cells were counted at indicated time points using hemocytometer. All cell lines were purchased from ATCC, authenticated by Genetica and tested negative for mycoplasma contamination.
Flow cytometry
To monitor the differentiation status, cells were stained with the following antibodies: APC-CD11b (ThermoFisher, CD11b 05), APC-CD13 (ThermoFisher, MHCD1305), PE-CD14 (BD Pharmingen, 555398), PE-CD33 (BD Pharmingen, 555450). To measure apoptosis, cells were washed with PBS and incubated with anti– PE-Annexin V (BD Pharmingen, 556421) in the ANNEXIN-V binding buffer in a reaction volume of 100 μl for 15 minutes based on manufacturer’s instruction. DAPI was added prior to analysis. Cells were analyzed on a BD FACS LSR Fortessa instrument.
To monitor human leukemia cell engraftment, bone marrow cells from recipient mice were stained with APC-mouse CD45.1 (Biolegend 110720) and PerCP-Cyanine 5.5-human CD45 (eBioscience, 45-9459-42). Immunoblot analysis of engrafted human cells was performed using sorted human CD45 positive cells.
PI3K/AKT inhibitor experiment
Prior to drug treatment, MOLM13 cells were transduced with high-titer lentiviral expressing control or METTL3 targeting short hairpins (shRNA #9 and #12). 48hrs after viral transduction, cells were selected by 3μg/ml puromycin. After 24h selection, cells were treated with DMSO (vehicle), GDC0068 (1μM) and GDC0032 (0.1μM) for 48h. Flow cytometry was then performed to evaluate cell differentiation by using CD11b and CD14 as markers as described above.
Morphological analysis
2x105 cells were centrifuged onto slides for 5 minutes at 500 RMP and air-dried prior to GIEMSA staining. The cell morphology was evaluated by light microscopy by three people. Evaluators were not blinded to sample identity
Immunoblot analysis
Cells were counted and washed twice with cold PBS prior to collection. ~ 250,000 were resuspended and lysed in 40μl 1X Lamine protein running buffer and boiled for 5 minutes. Whole cell lysates were run on 4%–15% gradient SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blotted with indicated antibodies: METTL3 (Proteintech, 15073-1-AP), p-AKT(s473) (193H12 Cell Signaling, 4058s), c-MYC (D84C12 Cell Signaling, 5605s), PTEN (138G6 Cell Signaling, 9559s), BIM (C34C5 Cell Signaling, 2933), BCL2 (C-2 Santa Cruz, SC-7382) and ACTIN (AC-15 Sigma, A3854). For miCLIP and MeRIP-qPCR anti-N6-methyladenosine (Abcam, ab151230).
CRISPR deletion
For CRISPR deletion in human cells, MOLM13-Cas9 cells (MOLM13 cells were engineered to constitutively express Cas9) were transduced with lentivirus pLKO5.sgRNA.EFS.GFP expressing sgRNAs targeting METTL3 (sgRNA#1 and sgRNA#2) or control empty vector GFP. sgRNAs were chosen for the highest score and least predicted off targets as designed by MIT CRISPR Design (cripr.mit.edu). The sequences of sgRNAs are provide below: #1: TGTTAAGGCCAGATCAGAG; #2: CTGGGCTTAGGGCCACCAG. sgRNAs were chosen. Five days after transduction, the pool of GFP positive cells was gated by flow cytometry to assess apoptosis and differentiation as described for shRNAs transduced cells.
For CRISPR domain targeting assay, mouse leukemia cells constitutively expressing Cas9 (RN2c cells) were transduced with lentivirus pLKO5.sgRNA.EFS.GFP expressing sgRNAs targeting transcription start site (TSS) of Mettl3 or Mettl3 enzymatic domain or control GFP empty vector (EV). sgRNAs were chosen for the highest score and least predicted off targets as designed by MIT CRISPR Design (cripr.mit.edu). Sequences of sgRNAs provided below. Percentages of GFP positive cells were measured by flow cytometry two days post transduction (D0) and 17 days after plating (D17).
| mettl3- TSS-g#1 | CACCGACTCGGCTCGAATCAGGCG |
| mettl3-TSS-g#2 | CACCGATTCGAGCCGAGTCCGCGC |
| mettl3- TSS-g#3 | CACCGGACACGTGGAGCTCTATCC |
| mettl3-enzymatic domain-g#1 | CACCGTGTTGTGATATCCGCTACC |
| mettl3-enzymatic domain-g#2 | CACCGTCACATGGAGCTACCGTAT |
| mettl3-enzymatic domain-g#3 | CACCGCCACCTTGGGATATTCACA |
Sample preparation for miCLIP
m6A miCLIP was performed exactly as previously described15, with the following modifications: total RNA from MOLM13 cells (N= 3 biological replicates) was extracted using TRIzol (ThermoFisher) and treated with RQ1 RNase-free DNase I (Promega). Poly(A) RNA was then isolated using two rounds of selection with oligo(dT)25 magnetic beads (New England Biolabs). This resulted in 30 μg of poly(A) RNA for each replicate used in this study. Poly(A) RNA was subjected to fragmentation using RNA Fragmentation Reagents (ThermoFisher) for exactly 12 minutes at 75°C. Fragmented RNA was then incubated with 15 μg of m6A antibody (Abcam) per replicate and the antibody-RNA complexes were processed by crosslinking, immunoprecipitation, RNA 3′ linker ligation, purification, and reverse transcription exactly as previously described15.
Mapping of m6A sites
After sequencing, we used the miCLIP pipeline as previously described. In summary, raw miCLIP reads were trimmed of their 3′ adaptor and demultiplexed. Then, PCR-duplicated reads of identical sequences were removed using the pyDuplicateRemover.py script of the pyCRAC tool suite (version 1.2.2.3). Unique reads were mapped with Novoalign (v3.02.12) using hg19 as reference genome. Further analysis including the identification of CITS was performed as previously described15. A custom script was then used to filter the identified putative m6A residues from the CITS file. This script filters C to T transitions based on the number of transitions (m ≥ 2) and the transition frequency (1% ≥ m/k ≤ 50%) at any given coordinate. Additionally, we filtered for those called sites that have a DRACH sequence. These sites will be referred as m6A sites throughout this study. To allow comparing across replicates, we used unique tag counts obtained from CIMS analysis. Each iCLIP tag represents a unique antibody–m6 A binding event. The number of unique events from a million such events is proportional within replicates. For this, the number of miCLIP tags per million uniquely mapped tags (unique tags per million, uTPM) was calculated as reported previously33. The total uTPM were summed across the entire gene length and used to generate a rank list of miCLIP reads per gene.
Metagene analysis and motif enrichment analysis
Metagenes were constructed for the called m6 A sites using MetaPlotR34. In brief, DRACH sites were mapped to different RNA features (5′ UTR, CDS and 3′ UTR) of the human genome (hg19). The position of the sites was normalized to the median feature length of the transcripts to which the sites mapped. A frequency distribution plot was generated by counting the number of sites in contiguous bins on a virtual mRNA transcript, sites whose feature lengths represent the median feature lengths of transcripts under analysis. A Gaussian estimate of kernel density was then plotted as a metagene. Analysis of motif enrichment was performed on the sense DNA sequence ±2 nucleotides up- and down-stream of the called truncation sites using the MEME suite.
Determination of relative m6A levels by two-dimensional thin layer chromatography
Relative levels of internal m6A was determined by thin layer chromatography (TLC) as described previously35. m6A measured using TLC does not have the problem of potential contamination by ubiquitous ribosomal RNA m6A and snRNA m6A since these m6A sites are found in a consensus site that prevents its detection by TLC35. 100 ng of poly(A) purified RNA was digested with 2U ribonuclease T1 for 2h at 37°C in the presence of RNAseOUT (Invitrogen). Digested RNA was 5′ labeled with 0.4 mBq [γ-32P] ATP for 30 min at 37°C followed by removal of the γ– phosphate of ATP by incubation with 10U apyrase (NEB) for 30 min at 30°C. RNA was purified by phenol-chloroform extraction and ethanol precipitation and resuspended in 10 μl of DEPC-H2O and digested to single nucleotides with 2U of P1 nuclease for 3h at 37°C. 1 μl of released 5′ monophosphates were analyzed by on glass-backed PEI-cellulose plates (MerckMillipore) as described previously36. Plates were exposed to a storage phospho screen (GE Healthcare Life Sciences) below saturation and processed on a Typhoon Trio imager (GE Healthcare Life Sciences) at 200 μm resolution. Quantification of individual nucleotides was done with ImageJ (1.49v). The relative amount of m6A was calculated as a percent of the total of A, C, and U spots, as described previously35.
Ribo-seq and RNA-seq in MOLM13 cells
Translation efficiency was determined essentially as described previously37. Briefly, MOLM13 cells were transduced with control shRNA or two independent shRNAs targeting METTL3. Four days post-transduction 3x106 MOLM13 cells were washed twice with ice-cold PBS pelleted by centrifugation. After pelleting, cells were resuspended in 300 μl lysis buffer (25 mM HEPES-KOH pH 7.2, 200 mM KOAc, 1% NP-40, 10 mM MgCl2, 4 mM CaCl2) for 5 minutes on ice then centrifuged at 8000 x g for 5 minutes. 100 μl of lysate was mixed with TRIzol and saved as input. To generate ribosome protected fragments the lysates were first mixed with 200 μl DEPC-H2O then incubated with 10 μg/ml micrococcal nuclease for 30 min at 37°C. RNA was extracted with TRIzol LS. T4 PNK was used to generate RNA fragments with a 5′ phosphate and 3′ hydroxyl. The RNA was then run on a 15% 8M urea TBE gel, stained with SYBR Gold, and a gel fragment between 27–35 nucleotides corresponding to ribosome protected RNA was excised. RNA was eluted overnight in 400 μl TE after crushing the gel fragment. RNA was ethanol precipitated and resuspended in 7 μl water. To generate libraries from RNA protected fragments 3 μl of the RNA was used with the NEBNext small RNA library prep kit (NEB) according to the manufacturer’s instructions. Due to the similarity in size between ligated and unligated adapters, prepared libraries (~155 nt) were gel purified. Ribosomal RNAs were removed from inputs using NEBNext rRNA Depletion Kit (NEB). Input RNA libraries were generated using the NEBNext Ultra Directional RNA library prep kit for Illumina (NEB). Libraries were sequenced using an Illumina HiSeq 2500 platform with 50 nucleotide reads. Adapters were trimmed using FLEXBAR tool (--min-read-length 12, --pre-trim-phred 32, 3′ adaptor: AGATCGGAAGAGCACACGTCT). Ribosomal RNA reads were removed using STAR aligner38. Gene count tables were generated using STAR aligned to hg19 transcriptome and normalized using DESeq239. Translation efficiency values were calculated by dividing normalized Riboseq read counts by normalized RNAseq read counts for each replicate.
MeRIP-qPCR
Total RNA was isolated from MOLM-13 cells by Trizol and poly(A)+ RNA isolated using Dynabeads Oligo-(dT)25 magnetic beads (ThermoFisher Scientific). Five micrograms of anti-m6A antibody (Abcam, ab151230) was pre-bound to Protein A/G magnetic beads (Pierce) in IP buffer (20 mM Tris pH 7.5, 140 mM NaCl, 1%% NP-40, 2 mM EDTA) for one hour. A total of 1–3 μg of poly(A)+ RNA and 100 ng of in vitro transcribed A and m6A-containing RNA was mixed with 400 μl of IP buffer and added to the Protein A/G beads for 2 hours at 4°C. Samples were washed twice in low salt wash buffer (10 mM Tris pH 7.5, 5 mM EDTA), twice with high salt wash buffer (20 mM Tris pH 7.5, 1M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA), and twice with RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA). RNA was eluted from the beads by incubating with 200 μl of 0.5 mg/ml N6-methyladenosine 5′-monophosphate sodium salt (Sigma) for one hour at 4°C. Following ethanol precipitation, input and eluted poly(A)+ RNA were reverse transcribed using Superscript III with random hexamers and enrichment determined by quantitative PCR.
Patient Sample Processing and MeRIP
AML patients (AML_118_Dx33 and AML_141) provided informed consent according to the Declaration of Helsinki for collection and use of sample materials in research protocols at the University of Pennsylvania (IRB protocol number 703185), also described here40 Samples were subjected to Ficoll separation on the day of collection, depleted of CD3+ and CD19+ cells using magnetic beads per the manufacturer’s recommendations (Miltenyi Biotec). Samples were then subjected to nucleic acid extractions using standard techniques. Control (non-MeRIP) RNA-seq libraries from patient RNA were prepared using the TruSeq RNA-Seq by poly(A) enrichment (Illumina) kit and sequenced on HiSeq2500 (Illumina) using a 50-bp paired-end method per the manufacturer’s recommendations, with Q-values >30 for all bases called by CASAVA. The remainder of the RNA was processed with the MeRIP protocol41 with two rounds of immunoprecipitation with the m6A antibody (Synaptic Systems) and sequenced to 1x50 reads.
We identified peaks using the open-source tool MeRIPPeR (https://sourceforge.net/projects/meripper/), which defines peaks in m6A-IP over input control read counts using Fisher’s Exact test, with a minimum peak size of 100 bases. The false discovery rate (FDR) was set to <0.05 using a Benjamini-Hochberg correction. Intersections between the peaks called by two replicates provided the final set of peak calls.
In vivo transplantation of human leukemia cells
MOLM13 human leukemia cells were transduced with lentiviruses expressing puromycin resistance gene and shRNAs against METTL3 or a control scramble shRNA. Transduced cells were selected by 3μg/ml puromycin for 2 days. 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6–8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. All animal studies were performed on animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Memorial Sloan Kettering Cancer Center.
Statistical analysis
Student’s t test was used for significance testing in the graphs, except where stated otherwise. A two-sample equal variance with normal distribution was used. The investigators were not blinded to the sample groups for all experiments. P values less than 0.05 were considered significant. Graphs and error bars reflect mean + s.e.m., except where stated otherwise.
For animal studies, survival probabilities were estimated using the Kaplan-Meier method and compared with the log-rank test. Eight mice per group were chosen to have an estimated 80% power in detecting a greater than 1.3 s.d. difference in means at a significance level of α= 0.05 using a two-sided test. All animals were randomly assigned to the experimental groups.
All statistical analyses were carried out using GraphPad Prism 4.0 and the R statistical environment.
Data Availability
RNA-Seq, Ribo-Seq, and miCLIP next-generation sequencing datasets and processed data were deposited to the Gene Expression Omnibus under accession ID: GSE98623. For reviewers see link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ytmhoiaazxmxpqf&acc=GSE98623
Data from these patient samples are available under the Database of Genotypes and Phenotypes (dbGaP) accession number phs001027.v1.p1.
Accession code
GSE98623. For reviewers see link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ytmhoiaazxmxpqf&acc=GSE98623
Supplementary Material
Acknowledgments
We thank Daniel Bachovchin (MSKCC) for the constitutively expressing CRISPR/Cas9 MOLM13 cell line and Christopher Vakoc (Cold Spring Harbor Laboratory) for the constitutively expressing Cas9 - RN2c cell line. We would like to thank the members of the Scaltriti laboratory for providing us PI3K/AKT inhibitors. MGK was supported by the US National Institutes of Health National Institute of Diabetes Digestive and Kidney Diseases Career Development Award, NIDDK NIH R01-DK101989-01A1, NCI 1R01CA193842-01, Kimmel Scholar Award, V-Scholar Award, Geoffrey Beene Award and Alex’s Lemonade Stand A Award, Tri-Institutional Stem Cell Award (MGK and SJ), R01CA186702 (S.R.J), T32CA062948 (B.F.P.), Ruth L. Kirschstein National Research Service Award 1F32CA22104-01 (B.F.P.), a Damon Runyon-Sohn Pediatric Cancer Fellowship Award DRSG10-14 (L.P.V.), the American-Italian Cancer Foundation (S.Z.) The research was funded in part through the NIH/NCI Cancer Support Core Grant P30 CA08748 MGK. RPPA core facility is funded by NCI #CA16672.
Footnotes
Competing Financial Interest Statement
There is no competing interest.
Author Contributions
L.P.V. led the project, performed experiments, analyzed data and wrote the manuscript. B.F.P., YC, DN, SZ performed experiments, analyzed data and edited the manuscript. GM, TC and AC provided experimental supports. C.E.M., YS, MM performed and analyzed MeRIP-seq experiments on patient samples. JS, CF, MP, F.E.G.-B., AM, V.M.K., and MC provided patient samples. SRJ supervised the project and wrote the manuscript. M.G.K. directed the project, analyzed data and wrote the manuscript.
References
- 1.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 4.Batista PJ, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mauer J, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, et al. Corrigendum: Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2017;542:260. doi: 10.1038/nature21073. [DOI] [PubMed] [Google Scholar]
- 8.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagger FO, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic acids research. 2016;44:D917–924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5 doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 13.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nature biotechnology. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903e815. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer S, Lavi U, Darnell JE., Jr The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol. 1978;124:487–499. doi: 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- 17.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SM, et al. Musashi-2 controls cell fate, lineage bias, and TGF-beta signaling in HSCs. The Journal of experimental medicine. 2014;211:71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nature medicine. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarry JE, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rgammac-deficient mice. The Journal of clinical investigation. 2011;121:384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoneley M, et al. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Molecular and cellular biology. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey S, Wang E. Cells en route to apoptosis are characterized by the upregulation of c-fos, c-myc, c-jun, cdc2, and RB phosphorylation, resembling events of early cell-cycle traverse. J Cell Biochem. 1995;58:135–150. doi: 10.1002/jcb.240580203. [DOI] [PubMed] [Google Scholar]
- 25.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. The Journal of biological chemistry. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 26.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharas MG, et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–1415. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Q, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell reports. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606e596. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patil DP, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olarerin-George AO, Jaffrey SR. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Vasseur J-J, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2016 doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruse S, et al. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Scientific reports. 2011;1:126. doi: 10.1038/srep00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid DW, Shenolikar S, Nicchitta CV. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods. 2015;91:69–74. doi: 10.1016/j.ymeth.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li S, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nature medicine. 2016;22:792–799. doi: 10.1038/nm.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq, Ribo-Seq, and miCLIP next-generation sequencing datasets and processed data were deposited to the Gene Expression Omnibus under accession ID: GSE98623. For reviewers see link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ytmhoiaazxmxpqf&acc=GSE98623
Data from these patient samples are available under the Database of Genotypes and Phenotypes (dbGaP) accession number phs001027.v1.p1.