Summary
Spatiotemporal variations of neurogenesis are thought to account for the evolution of brain shape. In the dorsal telencephalon (pallium) of vertebrates, it remains unresolved which ancestral neurogenesis mode prefigures the highly divergent cytoarchitectures that are seen in extant species. To gain insight into this question, we developed genetic tools to generate here the first 4-dimensional (3D + birthdating time) map of pallium construction in the adult teleost zebrafish. Using a Tet-On-based genetic birthdating strategy, we identify a “sequential stacking” construction mode where neurons derived from the zebrafish pallial germinal zone arrange in outside-in, age-related layers from a central core generated during embryogenesis. We obtained no evidence for overt radial or tangential neuronal migrations. Cre-lox-mediated tracing, which included following Brainbow clones, further demonstrates that this process is sustained by the persistent neurogenic activity of individual pallial neural stem cells (NSCs) from embryo to adult. Together, these data demonstrate that the spatiotemporal control of NSC activity is an important driver of the macroarchitecture of the zebrafish adult pallium. This simple mode of pallium construction shares distinct traits with pallial genesis in mammals and non-mammalian amniotes such as birds or reptiles, suggesting that it may exemplify the basal layout from which vertebrate pallial architectures were elaborated.
Keywords: pallium, neural stem cell, teleost, neuronal birthdating, clonal analysis
Graphical Abstract
Highlights
-
•
Neurons of the teleost pallium are arranged in concentric age-dependent layers
-
•
Neurons of the central pallial domain, Dc, are born during embryogenesis
-
•
Most pallial neurons are generated from ventricular her4-positive radial glia
-
•
The majority of individual pallial radial glia are neurogenic throughout life
Furlan et al. investigate the spatiotemporal events building the zebrafish pallium. Their “3D + birthdating time” map reveals an outside-in organization where neurons order in age-dependent sheets and where most individual neural stem cells are neurogenic life long. This strategy suggests a possible basal layout for pallial diversification.
Introduction
The dorsal telencephalon (pallium) of vertebrates is involved in complex sensory processing and cognitive operations. It derives from homologous developmental territories between species, which also partly share pallial organization and connectivity but display huge variations in pallial morphology. How this diversity in morphology has arisen such that species-specific adult cytoarchitectures can be constructed from the same set of developmental genes and fields to achieve similar functions is not fully understood.
There are several potential drivers of species-specific variations in brain size and architecture, such as the spatiotemporal control of progenitors’ neurogenic activity, neurogenesis lineages, neuronal migration, or neuronal death. The relative contribution of these events to pallium construction has been best studied in the neocortex of mammals and its homologous pallial territory in birds and reptiles [1, 2]. In mammals, neural progenitor cells (NPCs) along the neocortical ventricular zone (VZ) are multipotent and sequentially produce distinct excitatory neuronal subtypes, either directly or through transient amplifying progenitors (TAPs) [3], following a chronological switch of fate determinants inside single NPCs [4, 5, 6, 7]. When complemented by radial migration, this process generates a layered functional organization of excitatory neurons reflecting neuronal birthdate, ordered from deep (old) to upper (young) layer identities in an “inside-out” pattern [8] (Figure S1Aa). Classically, layer 4 (L4) neurons receive thalamic afferents and, together with upper-layer neurons, are involved in intra-cortical circuitries, whereas deep layers (L5) host projection neurons.
The temporal regulation of encoding neuronal subtypes by NPCs is shared between amniote species [9, 10]. However, the macro-organization of the pallium differs strikingly between mammalian and non-mammalian amniotes. In birds and reptiles, the pallial domain develops from a progenitor field homologous to the neocortex [11, 12], contains identical neuronal subtypes, is involved in similar circuitries [13, 14, 15], and is largely organized in functional fields or nuclei (Figures S1Ab and S1Ac) [2]. In birds, distinct regulation of neurogenesis timing (on- and offset times) was observed along the medio-lateral axis of the developing pallial VZ [9]. Because neuronal identities are birthdate dependent, this could generate differently sized and functionally specialized nuclei as opposed to layers [9] (Figure S1Ab)—although whether this occurs within individual functional networks is unclear [15]. The reptilian neocortex, in its dorsal sector, exhibits a three-layered laminar structure, generated in a radial outside-in pattern during a brief temporal window [10, 16]. The relative positions of L4 and L5 neuronal fields are also suggestive of spatial restrictions in NPC activities along the VZ (Figure S1Ab) [10, 15]. Overall, it has proven difficult to infer from these comparisons what could be the ancestral strategy involved in building neocortical (or, more generally, pallial) architecture.
We identify here the spatiotemporal events driving pallium construction in the teleost zebrafish as a representative of a more distant anamniote non-tetrapod vertebrate. This first “3D + birthdating time” pallial neuronal map reveals an outside-in organization where neurons order in age-dependent sheets and where most individual NPCs are neurogenic throughout their lifetime. This globally simple strategy of pallium construction bears reminiscence with mechanisms in amniotes, and its simplicity suggests a possible basal layout for the elaboration of pallial diversification.
Results
Proliferation and Neurogenic Activities in the Zebrafish Pallium Are Largely Restricted to Ventricular Radial Glia
Ray-finned fish (actinopterygians) all have an everted pallium leading to positioning the VZ, which contains NPC bodies, at the surface of the hemispheres [17]. The ventricle is enclosed by a stretching sheet of cells derived from the roof plate, the tela choroida, attached to the base of the everted folds (Figure S1B). To analyze the ontogeny of the zebrafish adult pallium, we first characterized the proliferation and neurogenic activities of NPCs along the pallial VZ from embryonic to adult stages. The neurogenic population in this territory is essentially composed of radial glial cells (RGs), identified by their expression of Glial Fibrillary Acidic Protein (gfap) [18], glutamine synthase (GS) [19, 20], and the transcription factor-encoding gene her4/Hes5 [21]. As previously documented, NPCs cover the entire VZ from embryonic to adult stages (Figures S1D–S1G), and exhibit life-long proliferative activity, measured with the Proliferating Cell Nuclear Antigen (PCNA) marker (Figure S2) [18]. This differs from pallial neurogenesis in amniotes [22, 23, 24]. RG proliferation along the zebrafish pallial VZ nevertheless decreases from juvenile stages to adulthood (Figures S2B–S2E) [21, 25]. During neocortex developmental neurogenesis in mammals, but not in most non-mammalian amniotes, there is an amplification step via intermediate progenitors, forming a recognizable sub-VZ [10, 26, 27]. These NPCs are non-glial but proliferative. To examine this issue in zebrafish, we compared PCNA expression with RG markers from 2 days post-fertilization (dpf) to 3 months post-fertilization (mpf). At all times, PCNA expression in the pallium was largely limited to RGs themselves, failing to indicate a non-RG, proliferating sub-VZ (Figure S2, insets). We previously reported that lineage amplification from RGs is limited in adults [28]; thus, this property extends to the stages of pallium construction.
In the following section, “NPCs” (versus RGs) will refer to the population of neural progenitors as a whole (RG + intermediate progenitors), or to a neural progenitor whose nature was not precisely defined.
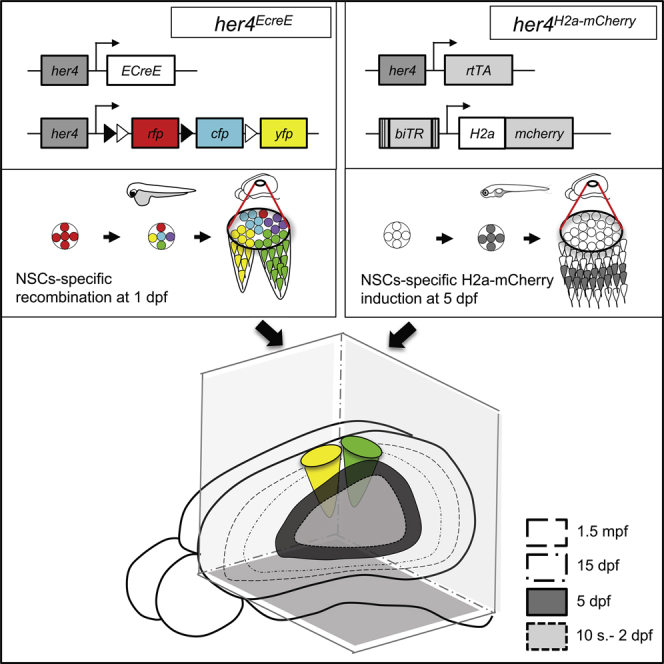
H2a-mCherry Retention Is a Suitable Birthdating Strategy in the Zebrafish Pallium
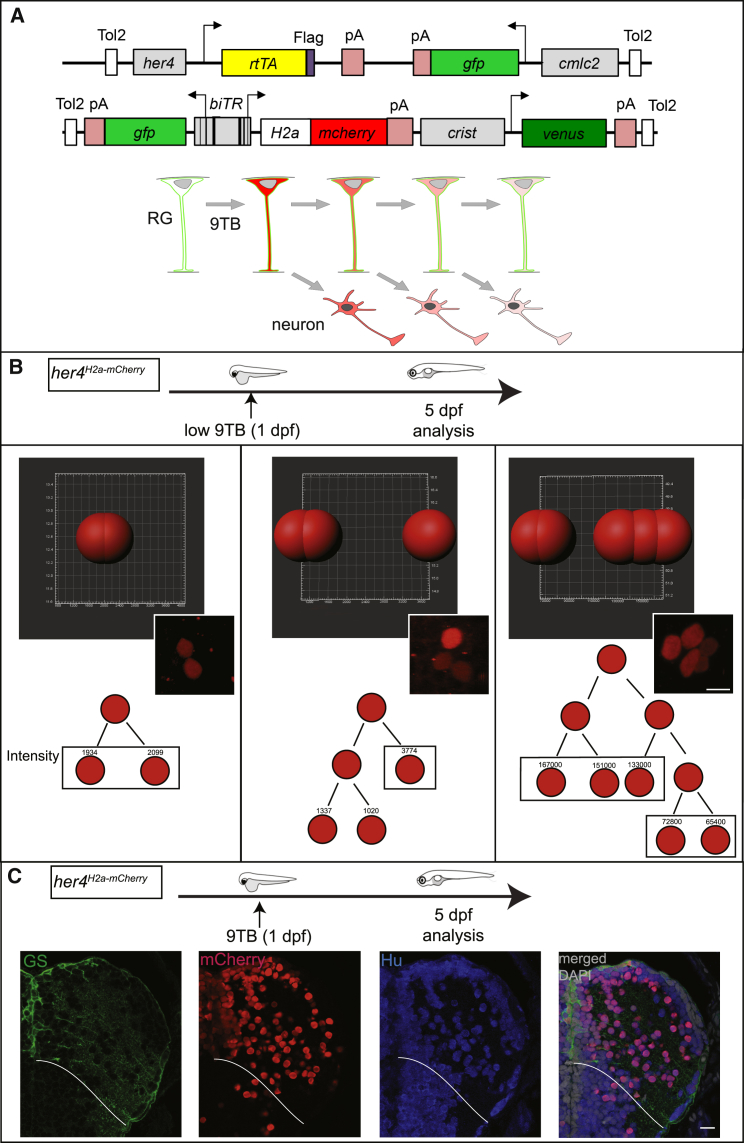
We reasoned that, in light of the minimal amplification exhibited by neurogenic lineages in the zebrafish pallium, we should be able to use a retention assay as a neuronal birthdating strategy. The Histone-fluo retention assay (a fluorescent reporter fused to stable Histone2a or H2b proteins) was efficiently used to study stem cell divisions in mammals [29, 30]. We thus developed an in vivo birthdating method based on Tet-On elements [31] that would lead to the expression of H2a-mCherry upon induction. In the driver line Tg(her4:rtTA, GFP:cmlc2), the her4/Hes5 NPC-specific promoter drives expression of reverse tetracycline-controlled transcriptional transactivator rtTA with a Flag epitope (Figure 1A). This line faithfully drives rtTA expression in all pallial RGs (Figure S3A). In the reporter line Tg(GFP:biTRE:H2amCHERRY, crist:Venus), the bidirectional tetracycline responsive element biTRE drives expression of both GFP and an H2a-mCherry fusion protein (Figure 1A).
Figure 1.
Neuronal Birthdating from her4-Expressing RGs In Vivo
(A) Genetic strategy. Top: 9-tert-butyldoxycycline (9TB) triggers stable H2a-mCherry and transient GFP activation in her4-positive RGs in double-transgenic animals for the depicted constructs. Bottom: neurons generated by the first RG (top, green) divisions following 9TB induction inherit detectable levels of H2a-mCherry (red). The label is lost upon successive divisions.
(B) Imaris quantification of mCherry immunostaining in 2- (left; n = 6), 3- (middle; n = 2), or 5- (right; n = 1) cell clones from different her4H2a-mCherry animals pulsed with 9BT at 1 dpf and analyzed at 5 dpf. Top: Imaris segmentation of mCherry-positive cells in individual clones. Middle: corresponding photomicrograph. Bottom: reconstructed lineage trees and fluorescence intensities. Scale bar, 7 μm.
(C) Neuronal fate of the mCherry-labeled daughter cells of her4-positive RGs in her4H2a-mCherry,9BT(1dpf) fish analyzed at 5 dpf. Triple immunocytochemistry for GS, mCherry, and HuC/D on a pallium cross-section (one hemisphere). White lines indicate pallial-subpallial boundary. Scale bar, 10 μm.
See also Figures S1–S3. Abbreviation definitions can be found in Figure S1B.
We induced rtTA activity in double-transgenic animals by a 9-tert-butyldoxycycline (9TB) treatment [32]. No leaky H2A-mCherry expression was detected without 9TB (not shown). Upon 9TB application, H2A-mCherry was selectively induced in RGs at all stages (Figures S3C–S3G). One day after treatment, most RGs (ventricular GFP-positive, Flag-positive cells) expressed high levels of H2a-mCherry, indicating efficient induction (Figures S3C–S3G). Induction frequency was homogeneous along the antero-posterior axis, 80% of VZ cells being H2a-mCherry positive at anterior, middle, and posterior pallial levels after a treatment at 5 dpf (Figure S3B). At 1.5 mpf, however, induction appeared less efficient in the medial domain of Dl (Figure S3G, red asterisk). Together, this strategy allowed us to pulse-label RGs with H2a-mCherry at all stages, and perform fate tracking at the population level.
A birthdating method implies that the tracer highlights cells generated within a restricted number of cell divisions following induction. Therefore, a non-dividing RG should retain its initial mCherry staining, whereas daughter cells of a dividing RG should inherit half its mCherry content, progressively diluting out the label. Low amounts of 9TB and short chase times were used to validate these dilution properties on polyclones induced at 1 dpf and analyzed at 5 dpf (Figure 1B). The amount of mCherry per nucleus was inferred from summing mCherry fluorescence staining intensity over the whole nucleus volume, measured using Imaris software. We observed a 2-fold dilution of the mCherry content after each division (Figure 1B). Finally, we verified that label-retaining cells in our system were indeed mainly neurons: after 9TB treatment at 1 dpf and a 4-day chase, all mCherry-positive cells were positive for the neuronal marker HuC/D (Figure 1C). Thus, the neurons generated after few divisions from transiently induced her4-positive pallial RGs retain H2a-mCherry, and the time of 9TB application is a proxy for their birthdate.
Pallial Neurogenesis Follows a Sequential Stacking Process
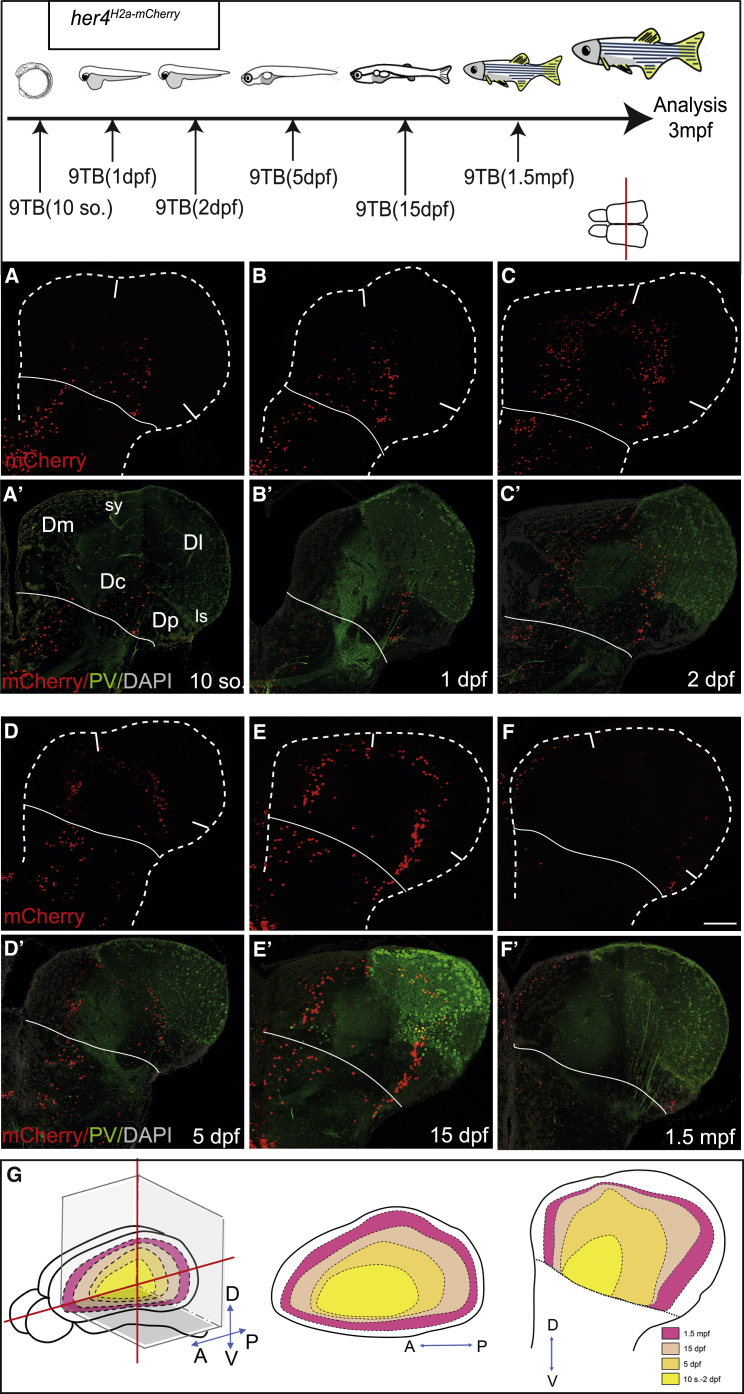
We timed neuron generation from her4-positive pallial RGs at embryonic, larval, and juvenile stages (10 somites, 1–2–5–15 dpf and 1.5 mpf) in Tg(her4:rtTA);Tg(GFP:biTRE:H2a-mCherry) double-transgenic animals (hereafter referred to as her4H2a-mCherry,9TB(t) for a 9TB induction at time t). To position these neurons within the adult pallium, we first analyzed a mid-antero-posterior level, identified by the joint presence of the dorsal sulcus ypsiloniformis (sy) and the lateral sulcus (ls) (Figure S1B). Co-immunostaining of mCherry with parvalbumin (PV) together with the sulci landmark helped identify neuroanatomical territories: the lateral pallium (Dl) (containing PV-positive neuronal cell bodies), the medial pallium (Dm) (located medial to sy), and a deep domain recognized as Dc (enriched in PV projections) [33] (Figure S1B; Figure 2A′).
Figure 2.
Zebrafish Pallial Neurogenesis Follows a Sequential Stacking Process: Medio-lateral Analysis
Top: experimental design.
(A–F′) Distribution of mCherry-positive neurons (A–F) born from her4-positive RGs in her4H2a-mCherry induced with 9TB at the stages indicated. Cross-sections at 3 mpf at mid-antero-posterior levels, the level indicated by a red line on telencephalon dorsal view, are co-labeled for parvalbumin (PV; A′–F′). Solid white lines indicate pallium-subpallium boundary. Scale bar, 50 μm.
(G) Color-coded map of the position in the adult brain (left: “open” whole-mount view; middle: horizontal section; right: cross-section) of the neurons born from her4-positive RGs at the stage indicated.
See also Figure S4.
Early 9TB treatments (10 somites, 1–2 dpf) generated mCherry-positive neurons positioned deep into the pallial parenchyma, close to the pallial-subpallial boundary (Figures 2A–2B′), in a domain overlapping with Dc. In her4H2a-mCherry,9TB(5dpf) animals, the mCherry-positive domain partly overlapped with this earlier-generated territory in medial locations but was otherwise positioned more dorsally and laterally and precisely surrounded Dc, including neurons in the deepest regions of Dm, Dl, and Dp (Figures 2C–2D′; Figures S4M and S4N). With increasing induction stages (15 dpf and beyond), concentric horseshoe domains were again progressively displaced to more medial, dorsal, and lateral positions (Figures 2C–2E′). The neurons generated at late stages (1.5 mpf) formed the most superficial area of the pallium (Figures 2F and 2F′). The paucity of neurons generated at 1.5 mpf in the most dorsal aspect of the pallium is consistent with a lower efficacy of 9TB induction in this area at this late stage (Figure S3G). The narrow territory built by pallial neurons between 1.5 and 3 mpf also shows that neurogenesis slows down after 1.5 mpf.
These data demonstrate the existence of an overall centrifugal gradient of neurogenesis in the zebrafish pallium, with the following characteristics: (1) it originates from the deepest territory of the adult pallium, in the position of Dc and the deepest neurons of Dm, Dl, and Dp: Dc and ventrally located pallial neurons form a “core” generated at embryonic stages; (2) it progresses in the dorsal and lateral directions (and medially at least starting at 15 dpf), neurons assembling around the core domain in progressively more superficial layers with time; and (3) it deposits neurons that arrange in an outside-in manner according to their birthdate with little or no radial mixing dorsally and laterally from the core. We postulate that pallial neurons simply sequentially stack over time as they delaminate from the VZ, contributing to pallial growth, in parallel to the enlargement of the germinal zone through symmetric RG divisions [28].
To understand pallium construction in 3D, systematic analyses were conducted at different antero-posterior levels, as well as on horizontal sections and whole-mount cleared preparations, with similar conclusions (Figure 3;Figures S4A–S4L and S5A). For example, at most anterior levels, mCherry-positive neurons only appeared in her4H2a-mCherry,9TB(15dpf) animals and later (Figures 3A–3D′). Taken together, our data indicate that the general logic of pallium construction is a 3-dimensional sequential stacking process where neurogenesis progresses dorsally and laterally, but also toward the anterior and posterior, from the central “core” domain over time (Figure 2G).
Figure 3.
Zebrafish Pallial Neurogenesis Follows a Sequential Stacking Process: Antero-posterior Analysis
Top: experimental design.
(A–D′) Horizontal sections are shown and the level is indicated by a red line on telencephalon lateral view; same stainings as in Figures 2A–2F′. Red and orange arrows indicate anterior and posterior limits of the mCherry-positive neuronal layers, respectively. Red asterisks in (D) and (D′) indicate RGs maintaining the mCherry label.
(E–F′) Transparent whole-mount preparation of a her4H2a-mCherry,9TB(15dpf) pallium at 3 mpf. The pallium is observed from different angles (E: dorsal anterior left; F: lateral oblique).
Scale bars, 100 μm.
The her4-Positive RG Population Is Neurogenic throughout Life
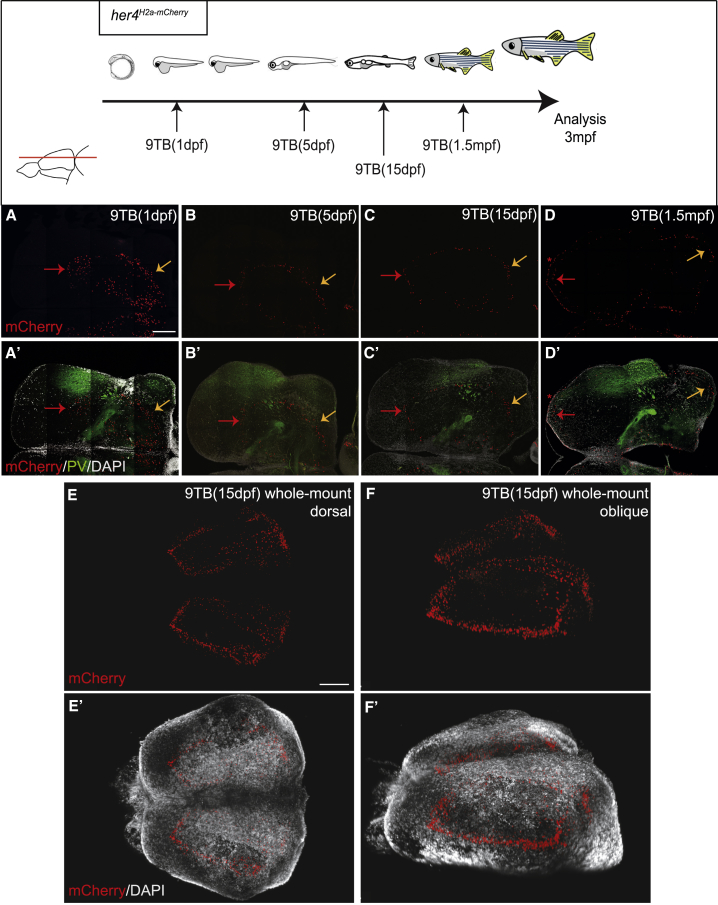
To determine whether the same her4-positive RG population is neurogenic at all stages—or, in contrast, whether distinct RG subsets expressing her4 at the time of induction are transiently neurogenic—we used conditional Cre-mediated lineage tracing [21]. Double-transgenic fish for her4:ERT2CreERT2 [34] and the ubiquitous reporter ubi:switch (Tg(−3.5ubi:loxP-GFP-loxP-mCherry)) [35] were pulsed with 4-hydroxy-tamoxifen (4OHT) (hereafter her4switch,T(t) fish for tamoxifen treatment at time t) at increasing developmental stages from 10 somites to 1.5 mpf. This approach labels with mCherry all the neurons produced by RGs expressing her4 at the time of 4OHT application, and tracks their neurogenic potential from the onset of treatment.
mCherry distribution was analyzed in the 3-mpf pallial parenchyma on median sections (equivalent to Figure 2). mCherry-labeled neurons occupied large domains extending from the VZ to a precise parenchymal boundary below which no neurons were stained (red line, Figure 4). The latter position recedes from deep to superficial as recombination is induced at later times, and is comparable to the layer hosting neurons born at the 4OHT treatment time (compare Figures 2A–2F′ with Figures 4A–4F). In the labeled domains, most neurons express mCherry (see also [21]). These observations indicate that, at the population level, pallial her4-positive RGs are constitutively neurogenic.
Figure 4.
Continuous Neurogenesis from the Pallial her4-Positive RG Population
Top: experimental design. Distribution of mCherry-positive neurons (A–F) born from RGs that were her4 positive at the time of 4OHT induction (stages at the bottom right of each panel). Cross-sections at 3 mpf are at identical mid-antero-posterior levels and the level is indicated by a red line on telencephalon dorsal view (A′–F′: mCherry and DAPI staining). Red lines indicate boundaries between territories with mCherry-positive and -negative neuronal cell bodies (mCherry is cytoplasmic in the ubi:switch line [35], and thus also stains processes). White lines indicate pallial-subpallial boundaries. Green lines delimit the recombined (Dm, medial part of Dl) and un-recombined (lateral part of Dl, Dp) pallial territories in her4switch animals treated with 4OHT (see [21]; at any given time point, all pallial RGs are her4 positive [Figure S2]; however, the pallial VZ expands laterally over time through the addition of RGs originating from a her4-negative source; therefore, the green lines shift toward the lateral with later recombination times). The her4switch fish in (F) and (F′) was subjected to two consecutive inductions, at 1 dpf (medial neurons only, left to green bar limit) and then at 1.5 mpf (superficial neurons, right to green bar limit). Scale bar, 50 μm.
Individual her4-Positive Pallial RGs Are Neurogenic throughout Life
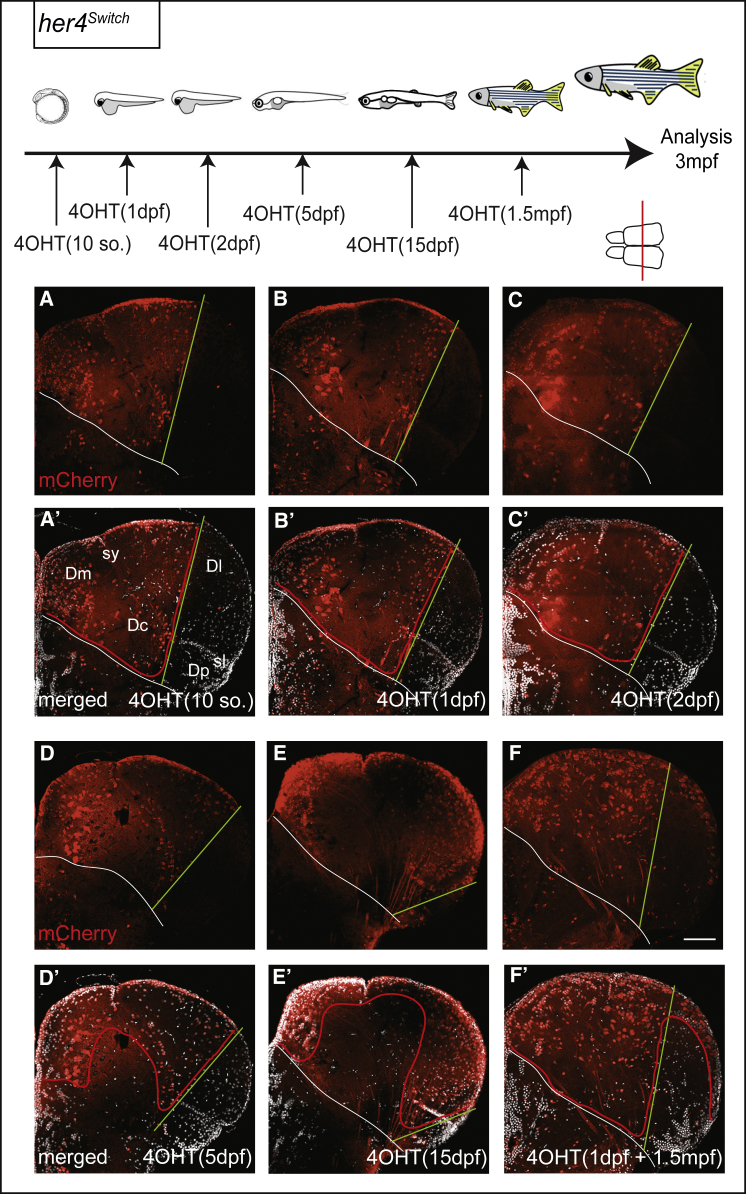
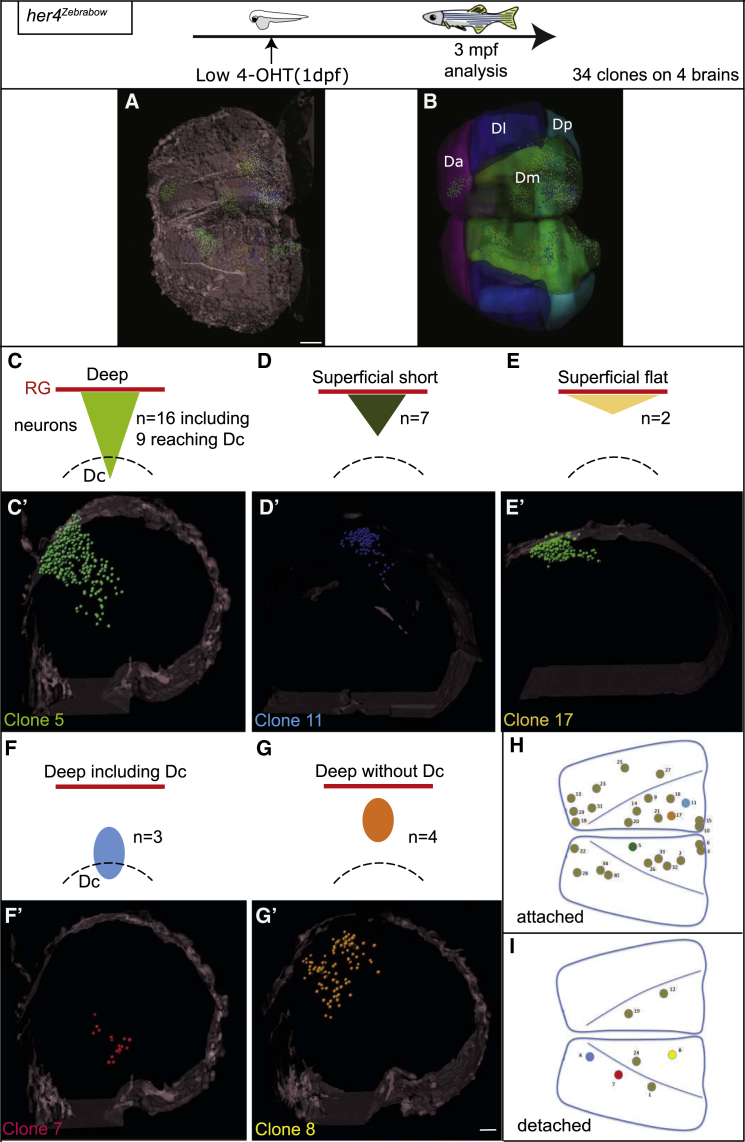
Neurogenesis in the mammalian neocortex is in part achieved by the constitutive neurogenic activity of individual RG progenitor cells throughout embryogenesis [4]. To further challenge the parallels between the mammalian and zebrafish neocortex, we asked whether individual zebrafish pallial RGs could sustain the entire neurogenesis process, here from 1 dpf to 3 mpf. We adapted Brainbow technology [36] [37] to visualize, in 3D, the contribution of unique embryonic RGs to the adult pallium. her4:ERT2CreERT2;ubi:Zebrabow double-transgenic embryos were mosaically recombined at 1 dpf. The corresponding adult telencephali were cleared and imaged in whole-mount view using a long-distance multiphoton confocal microscope, and clones were semi-automatically analyzed with single-cell resolution based on color ratios using Imaris (Figure 5A; Movie S1). 34 Brainbow clones (7,509 cells, from 8 pallial hemispheres) could be unambiguously mapped, including the coordinates of every constituent cell. For each individual clone, VZ cells versus neurons were counted—providing information on division mode and neurogenic activity—(Table S1), and the position of each clone was reported relative to neuroanatomical domains (schematically reconstructed in 3D from the manual segmentation of optical sections; Figure 5B) and on pallium dorsal views (Figures 5H and 5I).
Figure 5.
Individual Pallial RGs Are Neurogenic Lifelong
Top: experimental design.
(A) Whole-mount dorsal view (anterior left) of a 3-mpf pallium from a her4Zebrabow,4OHT(1dpf) fish, cleared and scanned. The position of individual cells of each clone is plotted (color coded). Brain surface (gray) was digitally added to help visualization. Scale bar, 150 μm.
(B) Corresponding location of pallial subdivisions.
(C–G′) Schematics (C–G) (red lines: RG layer; dashed lines: Dc limit) and thick optical cross-sections (C′–G′) of brains as in (A) highlighting the different categories of attached clones (C–E′) or detached clones (F–G′) (Table S1). “Deep” versus “superficial” clones contain neurons born before or at 5 dpf (Table S2).
(F–G′) Schematics (F and G) and thick optical cross-sections (F′ and G′) of brains as in (A) with examples of detached clones.
(H and I) Position of the 34 clones attached (H) or detached (I) on flattened dorsal views of the 3-mpf pallium.
Clones illustrated in (C′)–(G′) are colored. Scale bar, 80 μm. See also Figure S5, Tables S1 and S2, and Movies S1 and S2.
Overall, the pallium appeared composed of clones of different sizes (29–654 cells), shapes, and compositions (Figures 5C–5G; Table S1). We identified 5 clone categories based on qualitative and quantitative criteria related to the neurogenic activity of their constituent NPCs. First, we distinguished clones “attached to” versus “detached from” the pallial VZ (Figures 5C–5E versus Figures 5F and 5G). Based on the sequential stacking model, the former clones identify active RGs whose neurogenic activity was continuous since its onset, whereas the latter identify RGs whose neurogenic activity was terminated before adulthood. Second, we quantitatively correlated pallial neuronal positions and neuronal birthdates (Table S2) and distinguished clones contributing to deep territories (containing neurons generated until 5 dpf) or, in contrast, confined to more superficial layers (Figures 5C, 5F, and 5G and Figures 5D and 5E, respectively). We found that “attached” clones all contained a significant number of VZ cells (Table S1), consistent with the frequent occurrence of RG-amplifying divisions already described in the adult [28]. Third, “attached” clones were largely predominant (n = 27, versus n = 7 “detached” clones). Finally, the majority of “attached” clones reached into deep pallial layers (Figure 5C; n = 16 clones of 25), 9 of them (56%) including Dc neurons (Table S1; Figure S5; Movie S2). Thus, there is a strong tendency for the neurogenic activity of individual pallial RGs to be continuous once initiated and, most remarkably, the majority of these RGs are neurogenic over an extended period of time, from embryonic stages until adulthood.
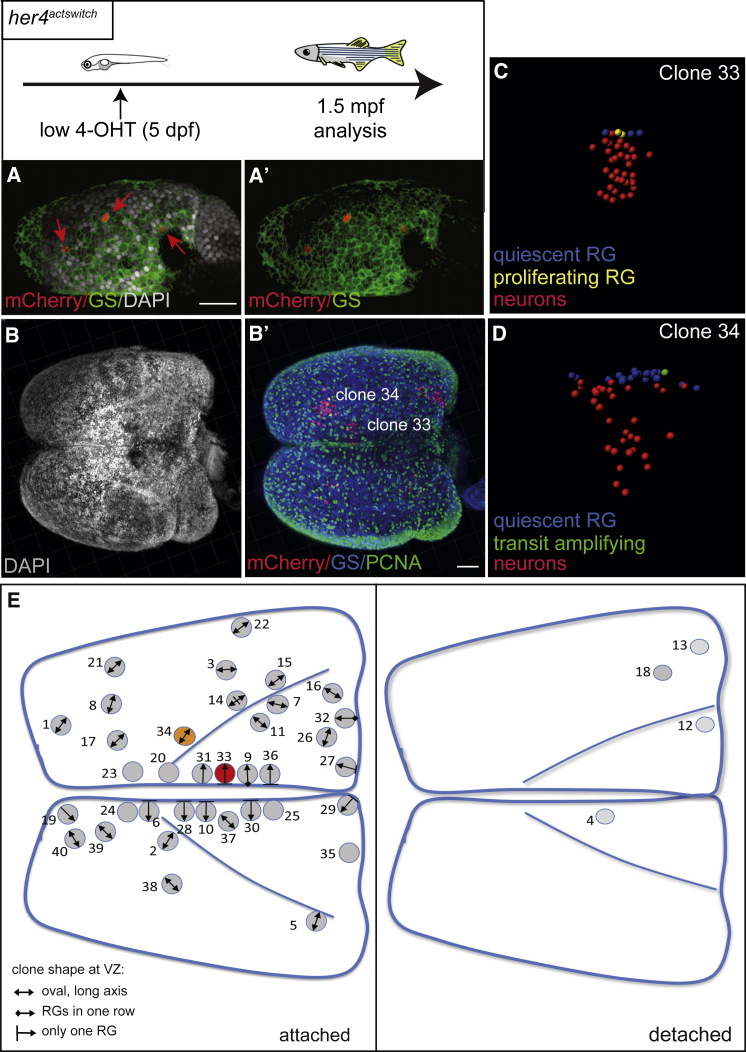
For confirmation, we backed up these results using classical Cre-lox tracing with low doses of 4OHT to obtain sparse labeling with 1–5 clones per telencephalic hemisphere, ensuring unequivocal clonality (Figures 6A and 6A′; Figure S5E–S5G). Double-transgenic fish for her4:ERT2CreERT2 and the ubiquitous reporter Tg(βactin:lox-stop-lox-hmgb1-mcherry) [38] were pulsed at 5 dpf (her4actswitch,T(5dpf) fish) and analyzed at 1.5 mpf (Figures 6B–6D), allowing scoring 40 additional clones (Table S3) from 26 hemispheres. mCherry-positive clones were segmented in 3D using Imaris following triple immunofluorescence to detect quiescent RGs (GS-positive, PCNA-negative), proliferating RGs (GS-positive, PCNA-positive), and the few intermediate progenitors (GS-negative, PCNA-positive) [18, 39] (Figures 6C and 6D). The recombination at 5dpf in this case precludes the recovery of “deep” clones (as defined in Figure 5); however, the same general clone types as defined in her4Zebrabow fish were found using this method, with a large majority of “attached” clones (90%).
Figure 6.
Clonal Analysis with Cell-Type Identification Confirms Long-Lasting Neurogenesis from Individual Pallial RGs
Top: experimental scheme.
(A and A′) Whole-mount dorsal view (anterior left) of a typical 6-dpf pallial hemisphere from a her4actswitch,4OHT(5dpf) fish stained at 1 day post-treatment for mCherry and GS (counterstained with DAPI). Red arrows point to individual recombined RGs.
(B and B′) Whole-mount dorsal view (anterior left) of a typical 1.5-mpf pallium from a her4actswitch,4OHT(5dpf) fish stained for mCherry, GS, and PCNA (B′).
(C and D) Segmented image of clone 33 (C) and clone 34 (D) highlighted in (B′) (color coded for cell types).
(E) Position of the 40 clones in flattened dorsal views of the 1.5-mpf pallium (clones 33 and 34 in B and B′ are colored). The shape of the VZ component of each clone is indicated (code bottom left).
We next asked whether pallial neurogenesis modes from her4-positive RGs were subject to spatial variations along the antero-posterior and medio-lateral axes over time. We analyzed the spatial distribution across the VZ of the clone subtypes defined above (Figures 5H, 5I, 6E, and 6F). We also considered the proportion of symmetric versus neuron-generating divisions, inferred from the RG/neuron ratio within clones, and the presence or absence of intermediate progenitors (Tables S1 and S3). Although important clone-to-clone differences were observed, we found no bias in spatial distribution for either parameter, indicating that the net product of neurogenesis from individual her4-positive RGs is spatially homogeneous during pallium construction.
Birthdate-Independent Expression of Neuronal Subtype Markers in the Adult Zebrafish Pallium
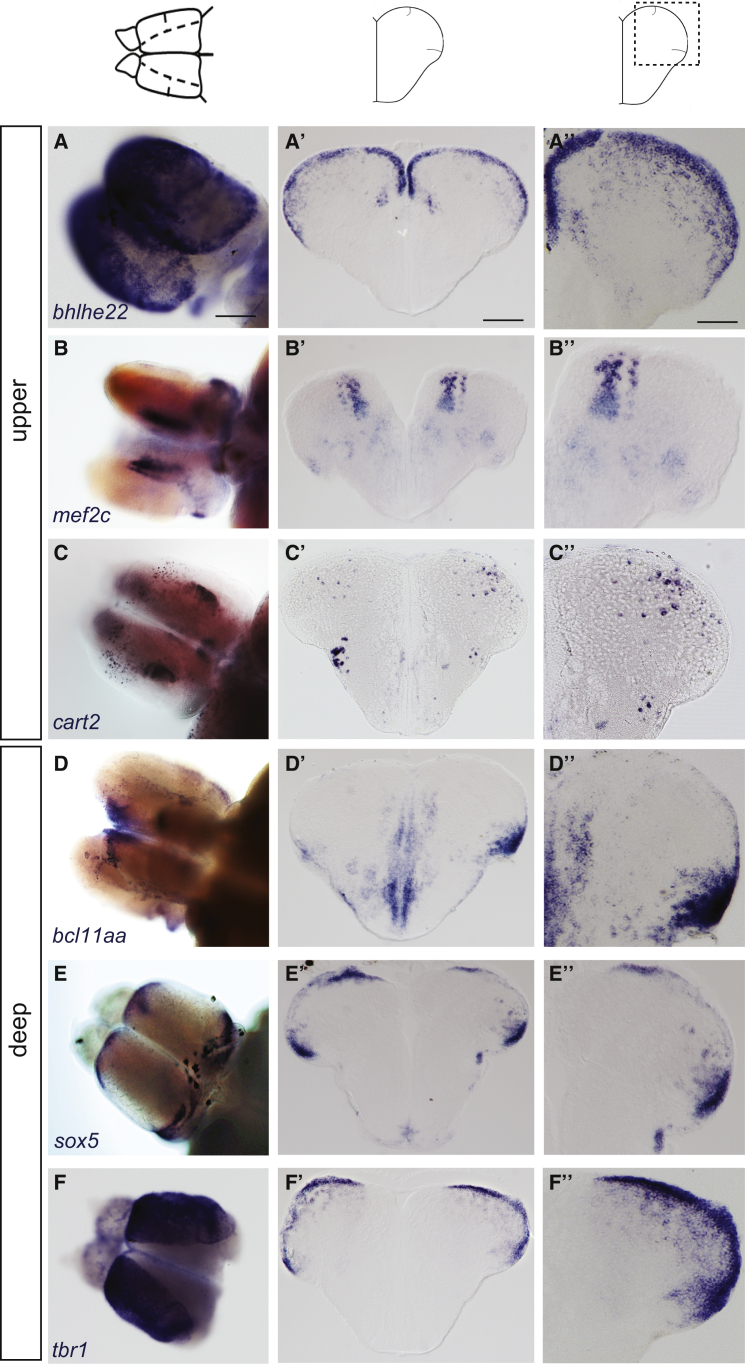
Finally, we addressed the relationship between the topological organization of lineages and birthdates and neuronal identity. Except for the expression of regional markers in large areas [20, 33], the molecular identities of zebrafish adult pallial neurons are unknown. In rodent and human neocortex, several molecular markers are expressed in a lamina-specific manner [40]. For example, Ctip2, Fezf2, and ER81 are predominantly expressed by early-born L5 projection neurons [9, 41, 42], RORβ by L4 neurons [15], and Satb2, Mef2c, and Cux2 by late-born L2–L4 neurons [9, 15, 42, 43]. We established a list of “most informative candidates” based on shared expression in rodents and humans [44], notably at adult stage, and/or expression in functionally homologous pallial territories in birds [15], and conducted an in situ hybridization screen of adult brain slices (Figure 7; Figure S6; Table S4). Most marker genes were expressed within the zebrafish adult pallium, but their expression patterns were neither obviously layered nor did they form nuclei or cone-like clones. We could classify expression patterns into two broad categories. In the first, genes were expressed in neurons underlying the VZ (Figures 7D–7F″; Figures S6D–S6F) (some of them only regionally; e.g., Figures 7E–E″). These genes could reveal a maturation stage (ongoing or recent neurogenesis) or, alternatively, a specific neuronal identity(ies) born around the stage of analysis (2–3 mpf). Expression at 15 dpf and 1 mpf remained confined to a stripe underlying the VZ (not shown), arguing for the first interpretation. The second category included genes expressed in a scattered pattern across the depth of the pallial parenchyma, in some cases in specific pallial subdivisions (e.g., Dm or Dl) (Figures 7A–C″; Figures S6A–S6C). Expression of these genes therefore overlaps with multiple neuronal ages, suggesting that the corresponding identities are shared by neurons born across the fish lifespan. Finally, we carefully analyzed the Dc area to determine, based on the sequential stacking model, whether some of the neocortical identities of amniotes were exclusively attributed/maintained in adult zebrafish pallial neurons born during embryogenesis. For all the markers considered, however, Dc was devoid of expression (Figure 7; Figure S6G). Within the limit of the selected marker genes chosen, these data suggest that the attribution of neuronal identities in the zebrafish pallium does not follow a simple scheme based on neuronal age or clonal lineage. To add support to this conclusion, we finally considered neurons expressing the neurotransmitters glutamate and GABA. Adult pallia from her4H2a-mCherry,9TB(5dpf) larvae were jointly processed for mCherry immunocytochemistry and in situ hybridization of vglut or gad transcripts. In both cases, positive neurons distributed within the neuronal layer born at 5 dpf, but also deeper as well as more superficially (Figure S7), confirming a broad age range for the generation of neurons of these phenotypes from the pallial VZ.
Figure 7.
Expression in the Adult Zebrafish Pallium of Genes Identifying Neocortical Identities in Mammals and/or Birds
In situ hybridization of 2-mpf pallia (A–F: whole-mount views from the top; A′–F′: low and high magnifications of cross-sections at mid-antero-posterior levels, respectively) for 6 genes (Table S4) displaying scattered expression spanning age layers (A–C) or expression restricted to the neurogenic domain (D–F). Scale bars, 200 μm (A–F); 100 μm (A′–F′); and 50 μm (A″–F″). See also Figures S6 and S7.
Discussion
Pallium Construction in Zebrafish Shares Distinct Traits with Mammalian and Non-mammalian Amniotes, Suggesting a Basal Pallial Layout
Developmental, hodological, functional, and lineage studies have unambiguously identified a pallial entity in jawed vertebrates, including teleosts. The dorsal telencephalons in mammals, birds, reptiles, amphibians, and teleosts share developmental markers [11, 12, 45, 46, 47], network, and functions at adult stage [48]. In teleosts, the preglomerular complex of the posterior tuberculum, a diencephalic nucleus relaying sensory modalities [49], is the primary source of ascending projections to a large Dm/Dl region, like the dorsal thalamus of tetrapods. The teleost pallium hosts functional fields processing spatial learning/memory (Dl; similar to the mammalian hippocampus), reward (Dm; similar to amygdalar nuclei), and cognitive tasks (Dm/Dl; resembling neocortical processing) [50, 51, 52]. Finally, Cre-mediated tracing demonstrated that these functional domains originate from the embryonic dorsal telencephalon, and their relative position fits their developmental origin when taking into account pallial eversion. These observations leave no doubt for the teleost pallium sharing developmental origin, neuroanatomical fields, and functions with that of tetrapods. From this starting point, we addressed here the key question of the pallial macroarchitecture and its generation from NSCs in zebrafish.
At the cell-population level, our results highlight the widespread and persistent neurogenic activity of zebrafish pallial RGs from embryonic stages until early adulthood (3 mpf), without major spatiotemporal gradients. A distinction should be made for the hippocampal area, where the generation of neurogenic RGs itself is delayed until early juvenile stages [21]. In all pallial territories, we failed to find evidence for major transit amplification, both from proliferation (this study) and clonal analyses [28], bringing the zebrafish pallial neurogenesis mode globally closer to that operating in amphibians, reptiles, and birds than mammals [10, 16, 47, 53, 54, 55]. Collectively, these species exhibit a net decrease in neurogenic output per NSC. The proportion of neurons per clone generated from pallial NSCs in gecko is lower than in mouse after 1 day [10]. Likewise, we find clones of 10–160 (on average 49) neurons per induced RG after 37 days (Table S3), compared to an average of 160 neurons per NSC in 11–20 days during neocortical neurogenesis in mouse [4].
Our work identifies for the first time the relationship between NSC activity and the spatiotemporal generation of the adult zebrafish pallial structure. By combining genetic birthdating and clonal analyses, we reached several important conclusions: (1) pallial neurons originating from pallial VZ RGs are organized in an outside-in age-related disposition (taking as a landmark neuronal distribution relative to the position of the VZ, a more stable reference than parenchymal orientation when comparing species with an everted pallium such as teleosts); (2) neuronal deposition expands in all directions over time from a deep central core generated during embryogenesis; (3) the spatial refinement of age-related layers and centrifugal lineage clones argue, respectively, against massive neuronal migrations along the radial and horizontal (parallel to the VZ surface) dimensions; and (4) neuronal deposition is largely driven by continuous neurogenic activity from individual pallial RGs. These results reveal a sequential stacking mode of pallium construction, where the spatiotemporal control of RG activity is an important mechanism of structure generation—although we cannot formally exclude the contribution of local neuronal migrations to the final architecture.
This simple strategy shares traits with neocortical organization in mammals, birds, and reptiles. Common denominators with mammals include the disposition of pallial VZ-derived neurons in age-related layers (although lamination is cryptic in zebrafish—only artificially revealed by our Tet-on induction time points—and mostly visible in neurons generated after early larval stages), the generation of clonally related neuronal “columns,” and the neurogenic contribution of individual NSCs to all cortical layers. The latter point is particularly remarkable in zebrafish, where neurogenic RG activity extends over months. Columnar organization in mammals is proposed to reflect local functional circuitries [56]. We observed minimal inter-clone dispersion in the zebrafish pallium, also highlighting the existence of ontogenetic columns, the relevance of which needs to be studied. Finally, the inside-out disposition of excitatory neurons in the mammalian neocortex results from the radial outward migration of newborn neurons, notably following Reelin-dependent cues. The broad distribution of Reelin pathway components in the developing and adult zebrafish pallium may be non-permissive for oriented migration [57, 58].
The neocortex equivalent of other amniotes consists of a thin dorsal cortex (“Wulst” in birds) and an enlarged, laterally positioned “dorsal ventricular ridge” (DVR). The cortex shows layered arrangements in all phyla but very different cytoarchitectures: in birds, L4 and L5 neurons [15, 59, 60] support a vertical functional organization, which does not fully correspond to neuronal birthdates, as outside-in development is partially blurred by radial migrations [61]. In reptiles and turtles, the 3-layer cortex develops in an outside-in manner [16] but excitatory L4 and L5 neurons only position within layer 2, occupying different fields along the antero-posterior axis [15, 62, 63]. Finally, in all sauropsids studied, the DVR is organized in large nuclei of distinct L4 or L5 identities, at least in part corresponding to distinct birthdates [9, 15, 61]. Several mechanisms were postulated to account for generating these structures: the segregation of VZ territories concomitantly neurogenic but generating different neuronal fates in the reptilian cortex [15] and, in the bird DVR, the segregation of VZ territories with different neurogenic periods [9] or continuous VZ neurogenesis accompanied by neuronal migration and the coalescence of age-related neurons [61]. A precise comparison with the zebrafish pallium will obviously require knowledge of neuronal identities and/or analyses of connectivity. A common trait with non-mammalian amniotes is, however, the generation of outside-in age-related organizations and the most likely minimal contribution of neuronal migration events. Recently, cortical neuron generation was studied in the amphibian Xenopus laevis through larval stages and was also shown to display an outside-in organization [47], although the neurogenic activity of individual NPCs was not addressed.
In the absence of a fossil record of the brain, and because we are not working with ancestral species, evolutionary considerations should be made with caution. Our results first further highlight the amazing divergence in pallium construction modes of extant vertebrates, but also stress the apparent simplicity of pallial neurogenesis in the teleost zebrafish and indicate that it shares several traits with different amniote phyla. We propose that the spatiotemporal mode of zebrafish pallium construction may mimic a basal layout, from which the generation of amniote pallial structures could have diverged, with mammals evolving radial migration and layering and birds and reptiles applying regional constraints on NSC neurogenic activity. The fact that anamniote tetrapods such as Xenopus may use a structure generation process comparable to zebrafish [47] further supports this hypothesis.
Ontogeny of Presumed Neuroanatomical Domains in the Zebrafish Pallium
The cytoarchitectonic subdivisions of the zebrafish adult pallium [64] are based on the spatial organization of neuronal cell bodies within the adult parenchyma. In this regard, the present work is also important as it solves, with genetic arguments, long-standing and controversial issues on the significance of these subdivisions. We found that Dc is the sole territory matching a temporal ontogenetic window, its constituent neurons being all generated prior to 5 dpf—in a manner reminiscent of the deep domains of Dm, Dl, and Dp. We further show that Dc is generated from the VZ of Dm, Dl, and Da (Table S1). These results contradict the recent conclusion that Dc possesses its own germinative domain [33], and formally provide evidence for the hypothesis of Braford that Dc neurons do not reflect a primary pallial subdivision but correspond to the deep neurons of overlying subdivisions [65, 66]. In contrast to Dc, we find that neither Dm, Dl, nor Dp corresponds to defined temporal or clonal units.
Encoding Pallial Neuronal Identities
The chronological generation of different neuronal subtypes by pallial RGs is observed in all amniotes [9, 10, 15]. It is unknown, however, whether it extends to other vertebrates, and whether all neuronal subtypes are shared between species. Classical anatomical studies in teleosts suggest that superficial and deep (Dc) neurons are of different identities, with periventricular pallial zones composed of small stellate neurons with widely branching dendrites receiving ascending sensory input, whereas deep pallial territories host large efferent neurons receiving most input from the more superficial neurons and, in the case of Dc, projections to the optic tectum [67, 68, 69]. The present work demonstrates that these different neuronal subtypes are generated at different stages, periventricular neurons being “young,” whereas deep pallial neurons are “old.” Thus, at the population level, the bulk of neuronal identities most likely vary over time during development to adulthood. Because we also show that individual RGs are capable of generating, sequentially, these old then young neuronal subtypes (Figure 5), our findings suggest that the sequential encoding of neuronal identities by pallial NSCs is a shared feature extending to teleosts.
A correlated question is how neuronal identities in the zebrafish adult pallium compare with those of amniotes. Neurogenesis being lifelong in zebrafish, it is difficult to hypothesize on the expected expression of mammalian identifiers of cortical layers in the zebrafish adult pallium. Moreover, only a few of these “identifiers” in fact share expression patterns among amniotes, and are maintained in the adult brain. For this reason, we largely relied on markers expressed in similar neocortical layers in adult mouse and human [44] (Table S4) or mouse and ferret [15]. Although a majority of markers were expressed in the zebrafish adult pallium, superimposing birthdates and clones failed to extract a simple rule based on age or lineage that could account for their expression profile: there was no link between a subventricular or deep expression in the zebrafish adult pallium and mammalian markers of upper versus deep layers, and none of the markers tested identified adult neurons generated at embryonic or early larval stages. This could either indicate transient expression or neuronal subtypes that are not found in zebrafish. In support of the first hypothesis, cux1b, cux2a, satb2, sox5, bcl1aa (ctip1), and fezf2 were expressed in rather broad parenchymal domains of the 5-dpf pallium but either absent (cux1b, cux2a, satb2) or confined to the neurogenic zone (sox5, bcl1aa, fezf2) in the adult pallium (Table S4 and not shown). In addition, markers such as mef2c and cart, expressed across a broad neuronal age range in the zebrafish pallium, identify functions rather than identities in rodents [70, 71]. In support of the second hypothesis, we never found expression of bcl1bb (ctip2) or ER81, diagnostic markers of L5 neurons in amniotes, or of the conserved L4 marker Rorβ in the zebrafish pallium at either stage. Projections from the preglomerular complex argue for the existence of neurons functionally equivalent to amniote L4 neurons. Their molecular identity will need to be precisely assessed.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-mCherry 1/300 | Clontech | Cat#632496; RRID: AB_10013483 |
| Chicken polyclonal anti-GFP 1/1000 | Aves lab | Cat#GFP-1020; RRID: AB_10000240 |
| Mouse monoclonal (IgG2a) anti-PCNA | Santa Cruz | Cat# sc-56 AB-16.0387; RRID: AB_628110 |
| Rabbit polyclonal (IgG2a) anti-PCNA 1/500 | GeneTex | Cat#GTX124496 AB-16.0386; RRID: AB_11161916 |
| Mouse monoclonal (IgG1) anti-Flag 1/500 | Sigma | Cat#F1804 AB-16.0331; RRID: AB_262044 |
| Mouse monoclonal (IgG2a) anti-GS 1/1000Clone GS-6 | Millipore | Cat#MAB302 AB-16.0342 |
| Human polyclonal anti-Hu 1/10000 | Gift from B. Zalk | AB-16.0356 |
| Anti-Parvalbumine 1/2000 | Millipore | Cat#MAB1572 AB-16.0382 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Hydroxytamoxifen | Sigma-Aldrich | T176; CAS: 68392-35-8 |
| 9-tert-ButylDoxycycline, hydrocloride salt | Echelon Biosciences | B-0801; CAS: 233585-94-9 |
| DAPI | Sigma-Aldrich | D9542; CAS: 28718-90-3 |
| Critical Commercial Assays | ||
| Taq DNA polymerase | EmeraldAmp Takara | Cat# 330A |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: Tg(T2Kβactin:lox-stop-lox-hmgb1-mcherry)jh15 | [38] | ZFIN ID: ZDB-TGCONSTRCT-110221-4 |
| Zebrafish: Tg(her4.1:EGFP)y83 | [25] | ZFIN ID: ZDB-FISH-150901-14830 |
| Zebrafish: Tg(−3her4.3:ERT2-Cre-ERT2)vu298a | [33] | N/A |
| Zebrafish: Tg(-3.5ubi:loxP-GFP-loxP-mCherry) | [72] | ZFIN ID: ZDB-ALT-070612-3 |
| Zebrafish: Tg(ubb:LOX2272-LOXP-RFP-LOX2272-CFP-LOXP-YFP)a131, ubi:Zebrabow-M | [73] | ZFIN ID: ZDB-ALT-110825-11 |
| Zebrafish: Tg(her4:rtTA, cmlc2:GFP) | [71] | ZFIN ID: ZDB-PUB-101209-26 |
| Zebrafish: Tg(GFP:biTRE:H2amCherry,crist:Venus) | [36] | ZFIN ID: ZDB-ALT-130816-2 |
| Oligonucleotides | ||
| rtTA probe fw: ATGTCTAGACTGGACAAGAGCAAAGTC, rv:GGATCCATTAACCCTCACTAAAGGGACTAACTGTCGACCTTGTC. | N/A | N/A |
| her4AttB4-fw: GGGGACAACTTTGTATAGAAAAGTTGTTTTGCATTATTTCCCTAATTTTAAATGTC (−999-1028bp from AY691485.1) her4AttB1-Rv: GGGGACTGCTTTTTTGTACAAACTTGGTCAGGATCAGATCTGAGCTG (−3389-3409bp from AY691485.1) | N/A | N/A |
| Recombinant DNA | ||
| her4:rtTA,GFP:cmlc2 | This paper | N/A |
| GFP:biTRE:H2amCherry,crist:Venus | This paper | N/A |
| tetOn Toolkit | [31] | https://www.bio.umass.edu/biology/jensen/node/5 |
| Tol2kit | [72] | http://tol2kit.genetics.utah.edu/index.php/Main_Page |
| Software and Algorithms | ||
| Imaris (Spot Segmentation) | Bitplane | http://www.bitplane.com/imaris/imaris |
| Photoshop CS6 | N/A | N/A |
| ZEN Black software | Carl Zeiss, Germany | https://www.zeiss.fr/microscopie/produits/microscope-software/zen.html |
| MATLAB 2014b, 2015b (Data analysis) | MathWorks | http://www.mathworks.com |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Laure Bally-Cuif (laure.bally-cuif@pasteur.fr). Sharing of zebrafish lines and constructs are subject to MTA request from Institut Pasteur.
Experimental Model and Subject Details
Zebrafish Lines
The her4:rtTA, GFP:cmlc2 and the GFP:biTRE:H2amCherry, crist:Venus constructs were obtained with the gateway strategy and the TetOn Toolkit [31]. The her4.1 regulatory region was amplified as a 2411bp fragment by PCR using AB wild-type fish genomic DNA as template, followed by a BP reaction with pDONR P4-P1R (Tol2 kit) [72]. The Tg(her4:rtTA, GFP:cmlc2) and the Tg(GFP:biTRE:H2amCherry, crist:Venus) transgenic lines were made by injecting 1-cell embryos with a mix containing 15ng/μL of plasmid and 15ng/μL of transposase capped RNA.
Fish maintenance and staging
Embryos/larvae up to 5dpf were maintained and staged as described [74]. Juvenile fish were staged by size as described in [21]. Adult zebrafish (3-6-month old) were maintained using standard fish husbandry protocols. Animals of both sexes were used indiscriminately.
Ethical approval
All experiments were carried out in accordance with the official regulatory standards of the Department of Essonne (agreement number A 91-577 to L.B.-C.) and Department of Paris (agreement number B75-15-22 to L.B.-C.).
Method Details
4OHT and 9TB treatments
4OHT treatments were performed as described [35] (10so.: 5μM, 4 hr; 1 or 2 dpf: 5μM, 6 hr; 5 dpf: 5μM, 30 hr; 1.5mpf: 5μM, 100 hr). Fish were then washed, transferred into fresh water, and grown as usual. For clonal analyses, 1dpf Tg(her4:ERT2CreERT2;ubi:zebrabow) embryos were recombined for 1 hr with 5μM 4OHT and 5dpf Tg(her4:ERT2CreERT2;-3.5ubi:loxP-GFP-loxP-mCherry) larvae for 10min with 0.5μM 4OHT. 9TB was dissolved in water at a final stock concentration of 5mg/mL, then diluted in embryo medium prior to use (≤2 dpf: 2.5μg/mL, 6 hr; 5 dpf – 15 dpf: 10μg/mL, 6 hr; 1.5 mpf: 10μg/mL, 96 hr). 9TB treatments were performed in the dark at 28°C. In the Tg(GFP:biTRE:H2amCherry, crist:Venus) line, a small GFP cluster was visible in the parenchyma at posterior levels even in the absence of treatment (not shown); it did not interfere with our analysis. For polyclones analysis, 9TB was applied at 0.5μg/mL for 10min onto 1dpf embryos. The embryos were then rinsed, grown and analyzed at 5dpf. Control experiments to assess clonality were analyzed on embryos treated with 0.5μM 4OHT for 10min at 5dpf, followed by a one- to two-day chase (for mCherry to be visible). Nearest neighbor distances were calculated per hemisphere (Imaris spot to spot closest distance on 20 larval hemispheres) and their cumulative probability distribution was plotted. We chose induction conductions such that in more than 85% of cases, induced cells were distant from each other of more than 15μm (equivalent to 4 cells diameters). Our brainbow analysis indicates very few cases, if any, of clone fragmentations. We therefore estimated that an initial distance of 4-cell diameters in the majority of cases should be sufficient to avoid this issue. To assess the occurrence of clone fusion, we also compared the estimated numbers of clones at time t+2days and at the analysis time point, 1.5mpf, and found that both were equivalent (Figure S5). Three hemispheres with ambiguous cases were discarded, and isolated mCherry cells, which were often observed, were not analyzed.
Immunohistochemistry and In Situ Hybridization
Immunohistochemistry and in situ hybridization were performed as described previously [18, 75]. The gad probes were a mix of gad65/67a/67b; the vglut probes were a mix of vglut1/2.1/2.2, as published in [76].
Whole-mount brain clearing
Whole zebrabow dissected adult brains were cleared using the SCALE approach [77] and mounted in glycerol 35%. Whole her4H2a-mCherry,9TB(15dpf) mCherry immunostained brains were cleared using the CUBIC approach [78] with the following incubations: CUBIC-1, 37°C, overnight; primary antibody, 37°C, 48 hr; secondary antibody, 37°C, 24 hr, CUBIC-2, overnight. Mounting was done in CUBIC-2.
Confocal microscopy, image acquisition and processing
All images except Figures 3E–3F′, 5, S5, S6, and 7 were taken using an inverted confocal microscope (Zeiss LSM700) and processed with the ZEN 2011 software (Carl Zeiss MicroImaging). Figures 2, 3A–3D, and S4A–S4L images are tile scans followed by maximum intensity projections of 8 squares and around 20 confocal optical sections. Images were then processed with Imaris or Photoshop CS6. Figures 3E–3F′, S7M, and S7N were processed with a median filter 3x3x3 followed or not by removal of object bigger than 250μm to remove background in the vessels or surface background. In situ hybridization pictures (Figure 7; Figure S6) were photographed with a Zeiss Axiozoom V6 Macroscope. CUBIC cleared brain was imaged using an upright confocal microscope (Zeiss LSM710) with a Plan-Apochromat 20X/0,8 M27 objective.
Multiphoton microscopy and related image analysis
Large-volume multicolor two-photon microscopy was performed using the wavelength mixing method described in [79]. Imaging was performed on a lab-built multiphoton point-scanning microscope constructed around an inverted frame (IX-70, Olympus, Japan) and integrating galvo scanners (VM500+, GSI, USA), a high-index immersion objective with 4mm working distance (XLPN25XSVMP, Olympus) and a motorized sample stage for mosaic acquisition (PS3H122 and ProScan H117, Prior Scientific). Excitation was provided by a Titanium-sapphire oscillator (Chameleon Ultra2, Coherent, USA) and an optical parametric oscillator (compact OPO, Coherent, USA). For simultaneous excitation of CFP, YFP and RFP signals, TiS, OPO and two-color equivalent excitation λ3 = 2/(1/ λTiS + 1/ λOPO) wavelengths were set to 850 nm, 1100 nm and 959 nm, respectively. Nonlinear signals were selected with appropriate dichroics (Semrock FF520-Di02 FF560-Di01) and filters (Semrock FF01-475/64 FF01-538/40 FF01-607/70), and epidetected on three separate channels by photomultiplier modules (P25PC SensTech UK and H7422P-40 Hamamatsu Japan) and lab-designed photon counting electronics. The pixel dwell time was 12 μs, and the voxel size was 0.8 × 0.8 × 2 μm3. For whole pallium imaging, cleared brains were mounted on Scale media between two 150μm-thick glass coverslips separated by a spacer, and a mosaic of 9 × 6 volumes each encompassing 260 × 285 × 1000 μm3 with a 20% lateral overlap was recorded.
Multicolor multiphoton stacks were preprocessed for flat-field correction and stitched with the open-source FIJI Image Stitching plugin using the Max Intensity fusion method [80]. Unlike cells, blood vessels exhibited intense fluorescence in all three channels and appeared “white.” These signals were removed using MATLAB by zeroing pixels having this characteristic. This processing step removed the vessels images without affecting cell signals. Semi-automatic cell detection was then performed using the Imaris Spot detection tool separately in the three channels. The automated detection exhibited an error rate (missed cells and false positives) of 50%–60%. Manual correction was then performed, which led to an uncertainty rate of 3%–5% for cell detection. Finally, clones were manually identified, based on cell colors, spatial clustering and cell sizes. The sparsity of labeling led to only a few ambiguous cases, which were discarded.
Quantification and Statistical Analysis
All details on the number of brains, hemispheres or cells processed can be found in the text and legends for figures (Figures S5E and S5F). n = 20 pallial hemispheres at 1dpt and 24 pallial hemispheres at 1.5mpt. The numbers in bracket indicate the number of hemispheres concerned for each number of induced cells/clones (Figure 5F). Data are presented as mean ± SEM, and statistical differences were determined using t test, p < 0,25. sem: 0.32 at 1dpt, 0.28 at 1.5mpt. For Figure S3B, n = 3 brains, t-test, non significant.
Author Contributions
G.F., L.D., V.C., and I.F. conducted all experiments; Table S4 and corresponding expression patterns were analyzed in collaboration with A.J.M. and C.H.; N.V. and E.B. conducted the segmentation and image analysis of Brainbow clones (Figure 5; Figure S5; Movies S1 and S2); N.D. provided expertise with whole-mount image generation and analyses with Imaris; M.C. generated the Tet-on fish DNA constructs; and S.B. maintained the fish necessary to this study. L.B.-C. and I.F. directed the work, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank members of the L.B.-C. laboratory for their critical input, W. Supatto for advice on image analysis, and M. O’Connell for critical proofreading. Work in the L.B.-C. laboratory was funded by the Agence Nationale de la Recherche (grant ANR-2012-BSV4-0004-01), Ecole des Neurosciences de Paris (ENP), European Research Council (AdG 322936), and Labex Revive. E.B.’s contribution was supported by Agence Nationale de la Recherche (contracts ANR-10-INBS-04 France BioImaging and ANR-11-EQPX-0029 Morphoscope2). G.F. was recipient of a fellowship from the Erasmus Programme. C.H. was supported by the Medical Research Council (G0901525) and European Union ZF-Health Programme.
Published: October 26, 2017
Footnotes
Supplemental Information includes seven figures, four tables, and two movies and can be found with this article online at https://doi.org/10.1016/j.cub.2017.09.052.
Contributor Information
Isabelle Foucher, Email: isabelle.foucher@pasteur.fr.
Laure Bally-Cuif, Email: laure.bally-cuif@pasteur.fr.
Supplemental Information
References
- 1.Suzuki I.K., Hirata T. Neocortical neurogenesis is not really “neo”: a new evolutionary model derived from a comparative study of chick pallial development. Dev. Growth Differ. 2013;55:173–187. doi: 10.1111/dgd.12020. [DOI] [PubMed] [Google Scholar]
- 2.Dugas-Ford J., Ragsdale C.W. Levels of homology and the problem of neocortex. Annu. Rev. Neurosci. 2015;38:351–368. doi: 10.1146/annurev-neuro-071714-033911. [DOI] [PubMed] [Google Scholar]
- 3.Molnár Z. Evolution of cerebral cortical development. Brain Behav. Evol. 2011;78:94–107. doi: 10.1159/000327325. [DOI] [PubMed] [Google Scholar]
- 4.Gao P., Postiglione M.P., Krieger T.G., Hernandez L., Wang C., Han Z., Streicher C., Papusheva E., Insolera R., Chugh K. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco S.J., Gil-Sanz C., Martinez-Garay I., Espinosa A., Harkins-Perry S.R., Ramos C., Müller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcamo E.A., Chirivella L., Dautzenberg M., Dobreva G., Fariñas I., Grosschedl R., McConnell S.K. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Toma K., Hanashima C. Switching modes in corticogenesis: mechanisms of neuronal subtype transitions and integration in the cerebral cortex. Front. Neurosci. 2015;9:274. doi: 10.3389/fnins.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noctor S.C., Martinez-Cerdeño V., Kriegstein A.R. Neural stem and progenitor cells in cortical development. Novartis Found. Symp. 2007;288:59–73. [PubMed] [Google Scholar]
- 9.Suzuki I.K., Kawasaki T., Gojobori T., Hirata T. The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev. Cell. 2012;22:863–870. doi: 10.1016/j.devcel.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Nomura T., Gotoh H., Ono K. Changes in the regulation of cortical neurogenesis contribute to encephalization during amniote brain evolution. Nat. Commun. 2013;4:2206. doi: 10.1038/ncomms3206. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez A.S., Pieau C., Repérant J., Boncinelli E., Wassef M. Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: implications for the evolution of telencephalic subdivisions in amniotes. Development. 1998;125:2099–2111. doi: 10.1242/dev.125.11.2099. [DOI] [PubMed] [Google Scholar]
- 12.Puelles L., Kuwana E., Puelles E., Bulfone A., Shimamura K., Keleher J., Smiga S., Rubenstein J.L.R. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Csillag A. Striato-telencephalic and striato-tegmental circuits: relevance to learning in domestic chicks. Behav. Brain Res. 1999;98:227–236. doi: 10.1016/s0166-4328(98)00088-6. [DOI] [PubMed] [Google Scholar]
- 14.Zeier H., Karten H.J. The archistriatum of the pigeon: organization of afferent and efferent connections. Brain Res. 1971;31:313–326. doi: 10.1016/0006-8993(71)90185-5. [DOI] [PubMed] [Google Scholar]
- 15.Dugas-Ford J., Rowell J.J., Ragsdale C.W. Cell-type homologies and the origins of the neocortex. Proc. Natl. Acad. Sci. USA. 2012;109:16974–16979. doi: 10.1073/pnas.1204773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goffinet A.M., Daumerie C., Langerwerf B., Pieau C. Neurogenesis in reptilian cortical structures: 3H-thymidine autoradiographic analysis. J. Comp. Neurol. 1986;243:106–116. doi: 10.1002/cne.902430109. [DOI] [PubMed] [Google Scholar]
- 17.Folgueira M., Bayley P., Navratilova P., Becker T.S., Wilson S.W., Clarke J.D.W. Morphogenesis underlying the development of the everted teleost telencephalon. Neural Dev. 2012;7:32. doi: 10.1186/1749-8104-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapouton P., Skupien P., Hesl B., Coolen M., Moore J.C., Madelaine R., Kremmer E., Faus-Kessler T., Blader P., Lawson N.D., Bally-Cuif L. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grupp L., Wolburg H., Mack A.F. Astroglial structures in the zebrafish brain. J. Comp. Neurol. 2010;518:4277–4287. doi: 10.1002/cne.22481. [DOI] [PubMed] [Google Scholar]
- 20.Ganz J., Kroehne V., Freudenreich D., Machate A., Geffarth M., Braasch I., Kaslin J., Brand M. Subdivisions of the adult zebrafish pallium based on molecular marker analysis. F1000Res. 2014;3:308. doi: 10.12688/f1000research.5595.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dirian L., Galant S., Coolen M., Chen W., Bedu S., Houart C., Bally-Cuif L., Foucher I. Spatial regionalization and heterochrony in the formation of adult pallial neural stem cells. Dev. Cell. 2014;30:123–136. doi: 10.1016/j.devcel.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Lim D.A., Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci. 2014;37:563–571. doi: 10.1016/j.tins.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urbán N., Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 2014;8:396. doi: 10.3389/fncel.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Götz M., Nakafuku M., Petrik D. Neurogenesis in the developing and adult brain—similarities and key differences. Cold Spring Harb. Perspect. Biol. 2016;8:a018853. doi: 10.1101/cshperspect.a018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo S.Y., Kim M., Kim H.S., Huh T.L., Chitnis A.B. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Cheung A.F.P., Pollen A.A., Tavare A., DeProto J., Molnár Z. Comparative aspects of cortical neurogenesis in vertebrates. J. Anat. 2007;211:164–176. doi: 10.1111/j.1469-7580.2007.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charvet C.J., Owerkowicz T., Striedter G.F. Phylogeny of the telencephalic subventricular zone in sauropsids: evidence for the sequential evolution of pallial and subpallial subventricular zones. Brain Behav. Evol. 2009;73:285–294. doi: 10.1159/000230673. [DOI] [PubMed] [Google Scholar]
- 28.Rothenaigner I., Krecsmarik M., Hayes J.A., Bahn B., Lepier A., Fortin G., Götz M., Jagasia R., Bally-Cuif L. Clonal analysis by distinct viral vectors identifies bona fide neural stem cells in the adult zebrafish telencephalon and characterizes their division properties and fate. Development. 2011;138:1459–1469. doi: 10.1242/dev.058156. [DOI] [PubMed] [Google Scholar]
- 29.Qiu J., Papatsenko D., Niu X., Schaniel C., Moore K. Divisional history and hematopoietic stem cell function during homeostasis. Stem Cell Reports. 2014;2:473–490. doi: 10.1016/j.stemcr.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y., Gotoh Y. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 2015;18:657–665. doi: 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- 31.Campbell L.J., Willoughby J.J., Jensen A.M. Two types of Tet-On transgenic lines for doxycycline-inducible gene expression in zebrafish rod photoreceptors and a Gateway-based Tet-On toolkit. PLoS ONE. 2012;7:e51270. doi: 10.1371/journal.pone.0051270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halterman M.W. An improved method for the study of apoptosis-related genes using the Tet-On system. J. Biomol. Screen. 2011;16:332–337. doi: 10.1177/1087057110397355. [DOI] [PubMed] [Google Scholar]
- 33.Mueller T., Dong Z., Berberoglu M.A., Guo S. The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei) Brain Res. 2011;1381:95–105. doi: 10.1016/j.brainres.2010.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boniface E.J., Lu J., Victoroff T., Zhu M., Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosimann C., Kaufman C.K., Li P., Pugach E.K., Tamplin O.J., Zon L.I. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Y.A., Livet J., Sanes J.R., Lichtman J.W., Schier A.F. Multicolor Brainbow imaging in zebrafish. Cold Spring Harb. Protoc. 2011;2011 doi: 10.1101/pdb.prot5546. pdb.prot5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y.A., Freundlich T., Weissman T.A., Schoppik D., Wang X.C., Zimmerman S., Ciruna B., Sanes J.R., Lichtman J.W., Schier A.F. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Rovira M., Yusuff S., Parsons M.J. Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing β-cells. Development. 2011;138:609–617. doi: 10.1242/dev.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alunni A., Krecsmarik M., Bosco A., Galant S., Pan L., Moens C.B., Bally-Cuif L. Notch3 signaling gates cell cycle entry and limits neural stem cell amplification in the adult pallium. Development. 2013;140:3335–3347. doi: 10.1242/dev.095018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillemot F., Molnár Z., Tarabykin V., Stoykova A. Molecular mechanisms of cortical differentiation. Eur. J. Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- 41.Arlotta P., Molyneaux B.J., Chen J., Inoue J., Kominami R., Macklis J.D. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Cobos I., Seeley W.W. Human von Economo neurons express transcription factors associated with layer V subcerebral projection neurons. Cereb. Cortex. 2015;25:213–220. doi: 10.1093/cercor/bht219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leifer D., Li Y.L., Wehr K. Myocyte-specific enhancer binding factor 2C expression in fetal mouse brain development. J. Mol. Neurosci. 1997;8:131–143. doi: 10.1007/BF02736778. [DOI] [PubMed] [Google Scholar]
- 44.Molyneaux B.J., Arlotta P., Menezes J.R.L., Macklis J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 45.Mueller T., Wullimann M.F., Guo S. Early teleostean basal ganglia development visualized by zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (eomesa), and GAD67 gene expression. J. Comp. Neurol. 2008;507:1245–1257. doi: 10.1002/cne.21604. [DOI] [PubMed] [Google Scholar]
- 46.Wullimann M.F., Mueller T. Teleostean and mammalian forebrains contrasted: evidence from genes to behavior. J. Comp. Neurol. 2004;475:143–162. doi: 10.1002/cne.20183. [DOI] [PubMed] [Google Scholar]
- 47.Moreno N., González A. Pattern of neurogenesis and identification of neuronal progenitor subtypes during pallial development in Xenopus laevis. Front. Neuroanat. 2017;11:24. doi: 10.3389/fnana.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karten H.J. Neocortical evolution: neuronal circuits arise independently of lamination. Curr. Biol. 2013;23:R12–R15. doi: 10.1016/j.cub.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 49.Northcutt R.G. Forebrain evolution in bony fishes. Brain Res. Bull. 2008;75:191–205. doi: 10.1016/j.brainresbull.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 50.von Trotha J.W., Vernier P., Bally-Cuif L. Emotions and motivated behavior converge on an amygdala-like structure in the zebrafish. Eur. J. Neurosci. 2014;40:3302–3315. doi: 10.1111/ejn.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Portavella M., Torres B., Salas C. Avoidance response in goldfish: emotional and temporal involvement of medial and lateral telencephalic pallium. J. Neurosci. 2004;24:2335–2342. doi: 10.1523/JNEUROSCI.4930-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Portavella M., Vargas J.P., Torres B., Salas C. The effects of telencephalic pallial lesions on spatial, temporal, and emotional learning in goldfish. Brain Res. Bull. 2002;57:397–399. doi: 10.1016/s0361-9230(01)00699-2. [DOI] [PubMed] [Google Scholar]
- 53.Nomura T., Takahashi M., Hara Y., Osumi N. Patterns of neurogenesis and amplitude of Reelin expression are essential for making a mammalian-type cortex. PLoS ONE. 2008;3:e1454. doi: 10.1371/journal.pone.0001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura T., Ohtaka-Maruyama C., Yamashita W., Wakamatsu Y., Murakami Y., Calegari F., Suzuki K., Gotoh H., Ono K. The evolution of basal progenitors in the developing non-mammalian brain. Development. 2016;143:66–74. doi: 10.1242/dev.127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taverna E., Götz M., Huttner W.B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 56.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 57.Costagli A., Kapsimali M., Wilson S.W., Mione M. Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J. Comp. Neurol. 2002;450:73–93. doi: 10.1002/cne.10292. [DOI] [PubMed] [Google Scholar]
- 58.Imai H., Oomiya Y., Kikkawa S., Shoji W., Hibi M., Terashima T., Katsuyama Y. Dynamic changes in the gene expression of zebrafish Reelin receptors during embryogenesis and hatching period. Dev. Growth Differ. 2012;54:253–263. doi: 10.1111/j.1440-169X.2012.01327.x. [DOI] [PubMed] [Google Scholar]
- 59.Shimizu T., Cox K., Karten H.J. Intratelencephalic projections of the visual wulst in pigeons (Columba livia) J. Comp. Neurol. 1995;359:551–572. doi: 10.1002/cne.903590404. [DOI] [PubMed] [Google Scholar]
- 60.Wild J.M., Williams M.N. Rostral wulst in passerine birds. I. Origin, course, and terminations of an avian pyramidal tract. J. Comp. Neurol. 2000;416:429–450. [PubMed] [Google Scholar]
- 61.Striedter G.F., Keefer B.P. Cell migration and aggregation in the developing telencephalon: pulse-labeling chick embryos with bromodeoxyuridine. J. Neurosci. 2000;20:8021–8030. doi: 10.1523/JNEUROSCI.20-21-08021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ulinski P. The cerebral cortex of reptiles. In: Jones E., Peters A., editors. Cerebral Cortex. Plenum; 1990. pp. 139–216. [Google Scholar]
- 63.Connors B.W., Kriegstein A.R. Cellular physiology of the turtle visual cortex: distinctive properties of pyramidal and stellate neurons. J. Neurosci. 1986;6:164–177. doi: 10.1523/JNEUROSCI.06-01-00164.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wullimann M.F., Rupp B., Reichert H. Birkhäuser; 1996. Neuroanatomy of the Zebrafish Brain: A Topological Atlas. [Google Scholar]
- 65.Nieuwenhuys R. The comparative anatomy of the actinopterygian forebrain. J. Hirnforsch. 1963;7:171–192. [PubMed] [Google Scholar]
- 66.Braford M.R., Jr. Comparative aspects of forebrain organization in the ray-finned fishes: touchstones or not? Brain Behav. Evol. 1995;46:259–274. doi: 10.1159/000113278. [DOI] [PubMed] [Google Scholar]
- 67.Demski L., Beaver J. Brain and cognitive function in teleost fishes. In: Roth G., Wullimann M., editors. Brain Evolution and Cognition. Wiley; 2001. pp. 297–332. [Google Scholar]
- 68.Northcutt R.G. Connections of the lateral and medial divisions of the goldfish telencephalic pallium. J. Comp. Neurol. 2006;494:903–943. doi: 10.1002/cne.20853. [DOI] [PubMed] [Google Scholar]
- 69.Northcutt R.G. Do teleost fishes possess a homolog of mammalian isocortex? Brain Behav. Evol. 2011;78:136–138. doi: 10.1159/000330830. [DOI] [PubMed] [Google Scholar]
- 70.Harrington A.J., Raissi A., Rajkovich K., Berto S., Kumar J., Molinaro G., Raduazzo J., Guo Y., Loerwald K., Konopka G. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. eLife. 2016;5:e20059. doi: 10.7554/eLife.20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subhedar N.K., Nakhate K.T., Upadhya M.A., Kokare D.M. CART in the brain of vertebrates: circuits, functions and evolution. Peptides. 2014;54:108–130. doi: 10.1016/j.peptides.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. The Tol2kit: a multisite Gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 73.Bernardos R.L., Raymond P.A. GFAP transgenic zebrafish. Gene Expr. Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 74.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 75.Bosco A., Bureau C., Affaticati P., Gaspar P., Bally-Cuif L., Lillesaar C. Development of hypothalamic serotoninergic neurons requires Fgf signalling via the ETS-domain transcription factor Etv5b. Development. 2013;140:372–384. doi: 10.1242/dev.089094. [DOI] [PubMed] [Google Scholar]
- 76.Higashijima S., Mandel G., Fetcho J.R. Distribution of prospective glutamatergic, glycinergic, and GABAergic neurons in embryonic and larval zebrafish. J. Comp. Neurol. 2004;480:1–18. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- 77.Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 78.Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- 79.Mahou P., Zimmerley M., Loulier K., Matho K.S., Labroille G., Morin X., Supatto W., Livet J., Débarre D., Beaurepaire E. Multicolor two-photon tissue imaging by wavelength mixing. Nat. Methods. 2012;9:815–818. doi: 10.1038/nmeth.2098. [DOI] [PubMed] [Google Scholar]
- 80.Preibisch S., Saalfeld S., Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.