Abstract
Transforming growth factor β1 (TGF-β1) has been associated with poor outcomes in patients with breast cancer. However, the functions and underlying molecular mechanisms of TGF-β1 in breast cancer remain unknown. Therefore, the present study aimed to identify the effects of components of the TGF-β/microRNA (miR-)21/phosphatase and tensin homolog (PTEN) signaling axis in breast cancer. TGF-β1 was identified to upregulate the expression of miR-21, and miR-21 was demonstrated to be significantly upregulated in breast cancer tissues compared with benign proliferative breast disease. In addition, the expression of miR-21 was significantly associated with increased TGF-β1 and clinical characteristics in patients, including tumor grade and lymph node metastasis (all P<0.05). Furthermore, in the breast cancer MCF-7 cell line, TGF-β1 was revealed to induce the expression of miR-21 in a dose- and time-dependent manner. The results of the present study additionally demonstrated that increased miR-21, in response to TGF-β1 signaling, was associated with tumor invasion and chemoresistance in vitro. In addition, suppression of PTEN was mediated by TGF-β1-induced expression of miR-21 in breast cancer cells and using a miR-21 inhibitor revitalized the expression of PTEN. The results of the present study explored the functions of TGF-β1-stimulated expression of miR-21 to suppress the PTEN axis, which promotes breast cancer progression and chemoresistance.
Keywords: transforming growth factor-β1, microRNA 21, phosphatase and tensin homolog, chemoresistance, invasion, breast cancer
Introduction
Breast cancer is the most common cancer among women in the world (1). In recent years, advanced developments in treatment options have been proposed for patients with breast cancer, including neoadjuvant chemotherapy, hormone therapy and targeted therapy (2). However, a number of patients exhibited poor survival due to chemoresistance and metastasis (3). Transforming growth factor β1 (TGF-β1) is a well-known factor in regulating breast epithelial cell development, differentiation and carcinogenesis, and tumor progression (4). TGF-β1 was initially identified as a regulator of breast cancer over two decades ago (5). Previous studies have demonstrated that TGF-β1 facilitates breast carcinoma metastasis by promoting epithelial-mesenchymal transition (EMT) in tumor cells (6,7). Additionally, TGF-β1 was previously associated with chemoresistance, and increased TGF-β1 levels led to poor clinical outcomes in a number of types of cancer, including breast cancer (8,9). Multiple pathways are involved in TGF-β1 signaling (10); however, the downstream molecular pathways for TGF-β1-mediated breast cancer metastasis and chemoresistance remain unknown.
MicroRNA (miR) are short (between 21 and 25 nucleotides) non-coding RNAs involved in regulating various cell activities. miR-21 is overexpressed in a number of types of cancer including breast cancer (11), glioblastoma (12) and lung cancer (13). Furthermore, numerous studies (11–13) suggest that miR-21 promotes tumor growth, migration and invasion by interacting with tumor suppressor genes, including PTEN (14); therefore, miR-21 is considered an important oncogenic miRNA. A previous study revealed that expression of miR-21 was induced by an increased level of TGF-β during smooth muscle cell development (15). Furthermore, Yu et al (8) demonstrated that TGF-β1 induced miR-21, which targeted the DNA damage sensors ATM serine/threonine kinase and mutS homolog 2 to regulate cancer responses to genotoxic chemotherapy (8). Therefore, the function of TGF-β1 in simulating miR-21 to promote metastasis and chemoresistance requires additional research.
In the present study, the associations between miR-21 and breast cancer characteristics, and between TGF-β1 and miR-21 were investigated in breast cancer cells and tissue. Furthermore, the results of the present study demonstrated that miR-21 regulates cell invasion and chemoresistance induced by TGF-β1 in breast cancer cells by targeting the tumor suppressor gene PTEN. The present study indicates that miR-21 may be a regulator of tumor metastasis and chemoresistance, induced by TGF-β1 in breast cancer, and a promising target for cancer therapy.
Materials and methods
Breast cancer samples
A total of 37 breast cancer tissue and 11 benign proliferative breast disease (BPBD) specimens were selected from the First Affiliated Hospital of Jinan University (Guangzhou, China) between October 2009 and March 2011. All patients were female. The patient age ranged between 37 to 68 years and the median age was 47 years. Samples obtained from surgery were fixed with 4% formalin for 24 h at room temperature and then were embedded in paraffin. Sections (4-µm) were prepared from each formalin-fixed, paraffin-embedded specimen. According to the World Health Organization histological classification (16), 29 patients were diagnosed with invasive ductal carcinoma, 2 were diagnosed with invasive lobular carcinoma (ILC) and 4 exhibited mixed type carcinoma. In addition, 1 patient was diagnosed as medullary carcinoma (MC) and 1 patient was considered a neuroendocrine tumor. The 11 BPBD specimens included 6 breast adenosis and 5 breast fibroadenomas. All protocols in the present study were approved by the Ethics Committee of the Jinan School of Medicine (Guangzhou, China), and written informed consent was obtained from all patients.
Cell culture
In the present study, the human breast cancer MCF-7 cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum at 37°C in a humidified incubator containing 5% CO2. Human recombinant TGF-β1 (PeproTech, Inc., Rocky Hill, NJ, USA) was dissolved in DMEM for the in vitro experiments.
Cell transfection
A total of 50 nM miR-21 inhibitors (anti-miR-21 sequence: 5′-TCAACATCAGTCTGATAAGCTA-3′) and the scramble miRNA control (anti-miR-NC sequence: 5′-CAGUACUUUUGUGUAGUACAA-3′) (Guangzhou RiboBio Co., Ltd., Guangzhou, China) were transiently transfected into MCF-7 cells using the Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol.
MTT assay
Cell proliferation was determined using an MTT assay, as previously described (16). A total of 1×104 cells were plated onto 96-well plates and transfected for 24 h, as aforementioned. At 6, 12, 24, 48 and 72 h after this, DMEM was removed and replaced with DMEM containing 10 µl of sterile MTT dye (5 mg/ml). Negative control was untreated cells plus solubilizing buffer. After 4 h incubation at 37°C, the MTT solution was removed and 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well, followed by measuring the absorbance at 570 nm on an enzyme immunoassay analyzer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
In vitro assay of chemosensitivity
MCF7 cells (1×104) were seeded into 96-well plates and then transfected for 24 h, followed by exposure to 5 ng/ml TGF-β1 with different concentrations of doxorubicin (DOX) (0, 1, 10, 50, 100, 200, 1,000 nM (Sigma-Aldrich; Merck KGaA) for 24 h. A Negative control was untreated cells plus solubilizing buffer. After 48 h incubation, cell viability was determined using an MTT assay, as aforementioned. Subsequently, the half maximal inhibitory concentration (IC50) values were calculated as previously described (17).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Small RNA was extracted from the specimen using RecoverAll™ Total Nucleic Acid Isolation kit (cat no. AM1975, Ambion; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA synthesis was performed using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The first chain of cDNA was reverse transcribed from 2 µl of total RNA using a specific stem-loop structure RT primer for miR-21 (5′-GAAAGAAGGCGAGGAGCAGATCGAGGAAGAAGACGGAAGAATGTGCGTCTCGCCTTCTTTC-3′). The reverse transcription reaction was carried out at 37°C for 60 min and at 95°C for 5 min. Subsequently, PCR was performed using the first chain of cDNA as the template. The reaction system contained 2 µl of cDNA, 12.5 µl of 1× SYBR Green I Master mix, 2.5 µl of miRNA specific primers, and 2.5 µl of upstream universal primer. The reaction conditions were as follows: Denatured at 95°C for 15 min, 40 cycles of 95°C for 15 sec, 57°C for 30 sec and 70°C for 30 sec. The small nuclear RNA U6 housekeeping gene was used for normalization. U6 RT primer: 5′-CGCTTCACGAATTTGCGTGTCA-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, CGCTTCACGAATTTGCGTGTCAT; miR-21 forward, primer was 5′-UAGCUUAUCAGACUGAUGUUGA-3′; reverse, 5′-CGAGGAAGAAGACGGAAGAAT-3′. To estimate miR-21 expression, an expression index (EI) was calculated as previously described (18) and the following equation was used: EI=1000×2(−ΔΔCq); where ΔΔCq is the difference between Cq values of the target and reference miRs (ΔΔCq=CqmiR-21-CqU6). Cq is the predetermined threshold of amplification cycle number.
Immunohistochemical (IHC) staining
Sections (4 µm) of breast cancer specimens were stained with IHC using a streptavidin-peroxidase technique (Gene Company, Ltd., Hong Kong, China) as previously described (19). Briefly, the sections were incubated in methanol/H2O2 for 30 min to inhibit endogenous peroxidase activity, washed with PBS for 5 min, and blocked with 10% normal goat serum (Thermo Fisher Scientific, Inc.) for 20 min at room temperature. The sections were subsequently incubated with antibodies against TGF-β1 (dilution 1:1,000, cat no. sc-130348; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or PTEN (dilution 1:1,000, cat no. 9188; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature for 1 h. Subsequently, the sections were treated with biotinylated secondary antibody (dilution, 1:500; cat no. E0466; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) for 1 h at room temperature and avidin-conjugated peroxidase for 45 min. Subsequent to washing the sections with PBS three times, peroxidase was stained with diaminobenzidine (1 mg/ml). The sections were counterstained with hematoxylin for 1 min at room temperature. PBS was used as a negative control instead of as a primary antibody. After staining, the slides were photographed by light microscope under ×200 magnification.
Western blot analysis
Proteins were determined using western blot analysis. Cells were washed with PBS and lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 0.05% deoxycholate, 1% SDS and 50 mM TrisCl; pH, 7.5). Cell lysates were quantified for protein content using the BCA method. Subsequently, 20-µg protein/lane was resolved on SDS-PAGE (10% gel) and transferred onto a nitrocellulose membrane. Membranes were blocked in 5% bovine serum album in Tris-buffered saline with Tween-20 for 1 h at room temperature. Subsequently, membranes were probed with primary antibodies against PTEN (dilution 1:1,000, cat no. 9188; Cell Signaling Technology, Inc.), and GAPDH (dilution 1:5,000, cat no. 5174; Cell Signaling Technology, Inc.) at 4°C overnight. Subsequent to washing, the membranes were incubated with secondary antibodies (dilution 1:5,000, cat no. 14708; Cell Signaling Technology, Inc.) for 2 h at room temperature and visualized using the enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.).
Transwell assays (cell invasion assays)
Transwell assays were performed in 6.5-mm diameter Boyden chambers with pore sizes of 8.0 µm (Corning Incorporated, Corning, NY, USA). Following transfection with miR-21 inhibitor or scramble anti-miRNA for 24 h, cells were cultured in serum-free DMEM alone or with 5 ng/ml TGF-β1 for 24 h at 37°C. Subsequently, 2.5×104 cells were seeded on an 8-µm pore size upper chamber Transwell filter coated with 20 µl culture medium containing 6.7% Matrigel™, with DMEM supplemented with 10% FBS present in the lower chamber; cells were incubated for 24 h. The invaded cells were attached to the lower surface of the membrane and stained with crystal violet for 30 min at room temperature. The number of cells was counted under a light microscope (magnification, ×100). Data were collected from at least three experiments performed in triplicate.
Statistical analysis
Data were expressed as the mean ± standard deviation of mean from at least three independent experiments and analyzed using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). Statistical analysis between two samples was performed using a Student's unpaired two-tailed t-test. For more than two groups, one-way analysis of variance, followed by Tukey's post-hoc test, were performed. P<0.05 was considered to indicate a statistically significant difference.
Results
TGF-β1 induces the expression of miR-21 in vitro
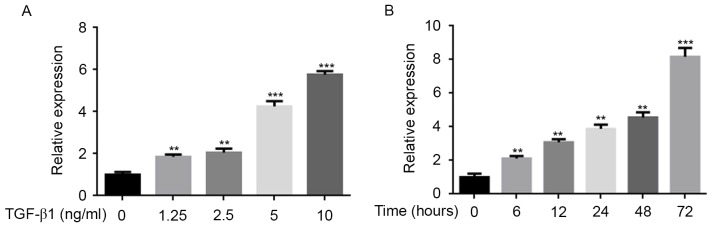
To validate the association between TGF-β1 and miR-21, the miR-21 expression levels in MCF-7 cells were analyzed using the qPCR method. As the concentration of TGF-β1 increased, the expression of miR-21 increased (Fig. 1A). Furthermore, it was demonstrated that the effects of TGF-β1 on miR-21 expression was time-dependent (Fig. 1B). This result indicated that the miR-21 expression is significantly associated with the dose and induction time of TGF-β1 in vitro.
Figure 1.
MCF-7 cells treated with TGF-β1 at distinct concentrations or time-points to determine the effect of TGF-β1 on miR-21. (A) Dose response. MCF-7 cells were treated with 0, 1.25, 2.5, 5 or 10 ng/ml TGF-β1 for 24 h. Expression levels of miR-21 were examined using qPCR, showing a dose-dependent association between TGF-β1 treatment and relative miR-21 expression. Data are presented as the mean ± standard error of the mean of three independent assays. (B) Time course. MCF-7 cells were treated with 10 ng/ml TGF-β1 for 0, 6, 12, 24, 48 and 72 h. Expression levels of miR-21 were examined using qPCR, showing a time-dependent association between TGF-β1 treatment and relative miR-21 expression. Data are expressed as the mean ± standard error of the mean of three independent assays. **P<0.01 vs. control; ***P<0.001 vs. control. TGF-β1, transforming growth factor β1; miR, microRNA; qPCR, quantitative polymerase chain reaction.
miR-21 expression is increased in breast cancer specimens and associated with TGF-β1
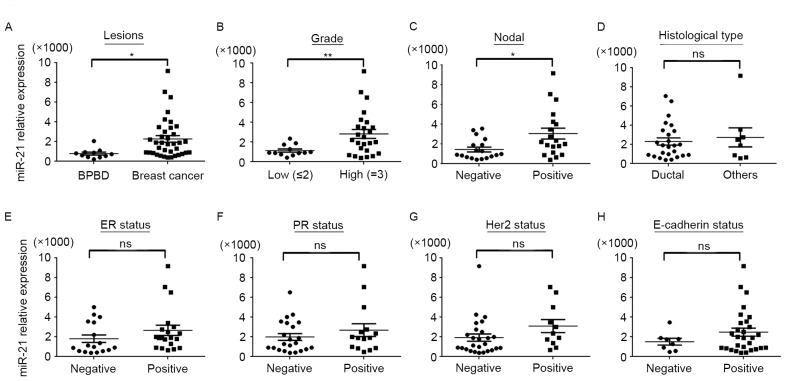
To analyze the function of miR-21 in human breast cancer, 37 specimens of primary breast cancer and 11 BPBD specimens were investigated using RT-qPCR). As presented in Fig. 2A, the expression of miR-21 in breast cancer was significantly increased, compared with BPBD. Subsequently, the association between miR-21 expression and clinical characteristics of patients was analyzed. As presented in Fig. 2B and C and Table I, miR-21 was significantly associated with tumor grade and lymph node metastasis (P<0.01); however, there was no association determined between miR-21 expression and histological type, estrogen receptor, progesterone receptor, human epidermal growth factor 2 and epithelial-cadherin status (Fig. 2D-H and Table I).
Figure 2.
Association between miR-21 expression and clinical and pathological features of breast cancer. Expression of miR-21 in breast cancer tissue was assessed using quantitative polymerase chain reaction to determine the association between miR-21 expression in terms of (A) benign and cancerous lesions; (B) grade of breast cancer lesion; (C) lymph node status; (D) histological type of lesion; (E) ER status; (F) PR status; (G) Her2 status; and (H) E-cadherin status. The expression of miR-21 in breast cancer was significantly increased, compared with BPBD, and miR-21 expression was significantly associated with tumor grade and lymph node metastasis. No association was determined between miR-21 expression and histological type, ER, PR, Her-2 and E-cadherin status. Data are presented as the mean ± standard error of the mean of three independent assays. U6 was used as an internal control. *P<0.05, **P<0.01. miR, microRNA; ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor 2; E-cadherin, epithelial cadherin; BPBD, benign proliferative breast disease; ns, non-significant.
Table I.
Association between miR-21 expression and the clinical and pathological features of breast cancer.
| Variable | n | Median expression of miR-21, arbitrary units | P-value |
|---|---|---|---|
| Grade | <0.01 | ||
| ≤2 | 12 | 1,130 | |
| 3 | 25 | 2,375 | |
| Histological type | 0.651 | ||
| Ductal | 29 | 2,470 | |
| Othera | 8 | 1,960 | |
| Lymph node status | 0.013 | ||
| Negative | 18 | 1,390 | |
| Positive | 19 | 3,090 | |
| ER status | 0.209 | ||
| Negative | 20 | 2,085 | |
| Positive | 17 | 2,470 | |
| PR status | 0.319 | ||
| Negative | 15 | 2,080 | |
| Positive | 22 | 2,375 | |
| Her2 status | 0.108 | ||
| Negative | 11 | 1,780 | |
| Positive | 16 | 2,375 | |
| E-cadherin status | 0.236 | ||
| Negative | 29 | 2,370 | |
| Positive | 8 | 2,170 |
Of these, two cases were invasive lobular carcinoma, four cases were mixed type carcinoma, one case was medullary carcinoma and one case was neuroendocrine tumor. miR, microRNA; ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor 2; E-cadherin, epithelial cadherin.
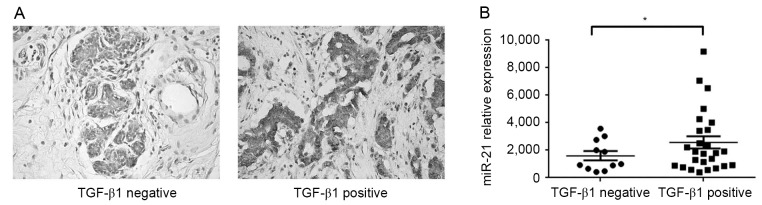
To investigate the potential association between TGF-β1 and miR-21 in human breast cancer, the expression of TGF-β1 in 37 breast cancer specimens was determined using IHC (Fig. 3A), and subsequently the expression of miR-21 in TGF-β1-positive and negative specimens were compared. The expression of TGF-β1 was positive in 26 breast cancer specimens and negative in 11 cases. Notably, miR-21 expression was significantly increased in TGF-β1 positive tumors, compared with negative tumors (Fig. 3B; P<0.05). These results suggested that miR-21 may act as a downstream mediator of TGF-β1 in breast cancer.
Figure 3.
miR-21 expression is increased in breast cancer tissue and associated with TGF-β1. (A) Representative images from immunohistochemical labeling for TGF-β1 in breast cancer. The cytoplasm of tumor cells was stained (magnification, ×200). TGF-β1 protein was identified to be negative (11/37 cases) or positive (26/37) in breast cancer specimens. (B) Expression of miR-21 in TGF-β1 negative and positive breast cancer tissues was determined using quantitative polymerase chain reaction. Data are presented as the mean ± standard error of the mean of three independent assays. miR-21 expression was significantly increased in TGF-β1 positive tumors, compared with negative tumors. U6 was used as an internal control. *P<0.05. miR, microRNA; TGF-β1, transforming growth factor β1.
Downregulation of miR-21 inhibits TGF-β-induced invasion in MCF-7 cells
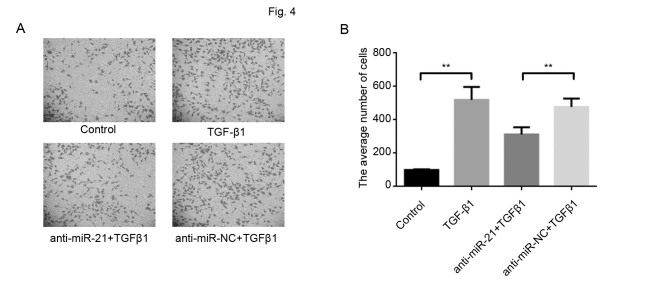
The results of the present study demonstrated that miR-21 expression was positively associated with TGF-β1 levels and a previous study revealed that TGF-β1 promoted cancer metastasis (20). Therefore, in the present study, the function of miR-21 in TGF-β1-mediated breast cancer cell invasion was subsequently investigated. The downregulation of miR-21, using anti-miR-21, in MCF-7 cells significantly inhibited invasion induced by TGF-β1 (Fig. 4A and B), which suggested that miR-21 has an active role in TGF-β1 induced metastasis.
Figure 4.
miR-21 effect on cell invasion of breast cancer cells induced by TGF-β1. Cell invasion was determined in MCF-7 cells by the Transwell chamber assay following transfection with anti-miR-21 and scramble anti-miR-NC alone or in combination with TGF-β1. (A) Representative images from invasion assays (magnification, ×100). (B) The average number of cells that invaded through the filter was counted. The downregulation of miR-21, using anti-miR-21, in MCF-7 cells significantly inhibited invasion induced by TGF-β1. Data are presented as the mean ± standard error of the mean of three independent assays. **P<0.01. miR, microRNA; TGF-β1, transforming growth factor β1; NC, negative control.
Downregulation of miR-21 decreases resistance of breast cancer cells to DOX
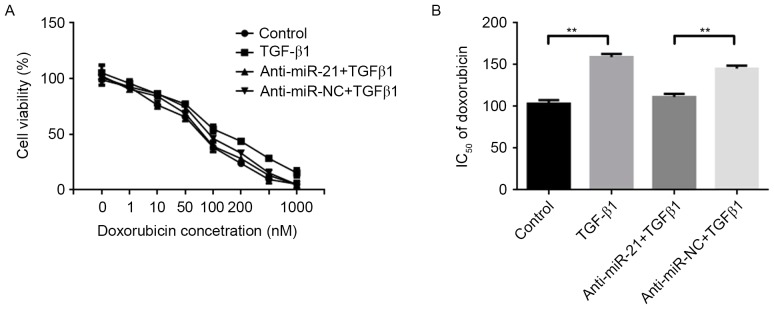
Previous studies have demonstrated that TGF-β1 decreased the sensitivity of cancer cells to chemotherapeutic drugs (8,9). In addition, miR-21 may modulate the chemosensitivity of breast cancer cells to DOX (21). Therefore, miR-21 may mediate DOX resistance in breast cancer cells, induced by TGF-β1. MCF-7 cells were transfected with anti-miR-21 or scramble anti-miR-NC, and subsequently, cells were exposed to distinct concentrations of DOX alone or a combination with 5 ng/ml TGF-β1 for 48 h. Cell viability was determined using an MTT assay. The results revealed that the downregulation of miR-21 decreased resistance in MCF-7 cells to DOX induced by TGF-β1 (Fig. 5A). Compared with the IC50 values of DOX in the control group (103.21±4.34 nM), TGF-β1 increased the IC50 value of DOX in MCF-7 cells (159.75±3.84 nM), whereas the downregulation of miR-21 decreased the IC50 value of DOX in MCF-7 cells (111.48±3.65 nM), compared with the anti-miR-NC and TGF-β1 group (145.38±3.78 nM) (Fig. 5B). These results suggested that TGF-β1 increased miR-21 to increase breast cancer cell resistant to DOX.
Figure 5.
Effect of miR-21 on DOX chemoresistance in breast cancer cells induced by TGF-β1. (A) Cell viability was determined using an MTT assay in MCF-7 cells transfected with anti-miR-21 or scramble anti-miR-NC at 48 h following exposure to 5 ng/ml TGF-β1 in combination with different concentrations of DOX (0, 1, 10, 50, 100, 200 and 1,000 nM). Downregulation of miR-21 decreased resistance in MCF-7 cells to DOX induced by TGF-β1. (B) MTT analyzed alterations in IC50 values of DOX in MCF-7 cells, following exposure to 5 ng/ml TGF-β1 alone or transfection with anti-miR-21 or scramble anti-miR-NC. Compared with the IC50 values of DOX in the control group, TGF-β1 increased the IC50 value of DOX in MCF-7 cells. Downregulation of miR-21 decreased the IC50 value of DOX in MCF-7 cells, compared with the anti-miR-NC and TGF-β1 group. Data are presented as the mean ± standard error of the mean of three independent assays. **P<0.01. miR, microRNA; DOX, doxorubicin; TGF-β1, transforming growth factor β1; NC, negative control; IC50, half maximal inhibitory concentration.
miR-21 mediates the suppression of PTEN induced by TGF-β in MCF-7 cells
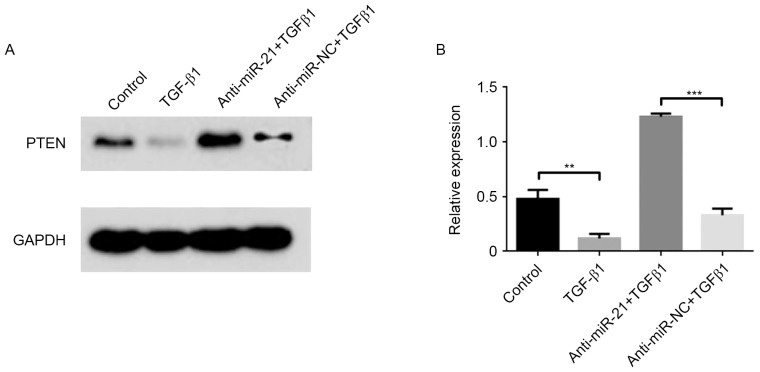
Previous studies have demonstrated that TGF-β1 suppressed the expression of PTEN proteins to enhance tumor metastasis and chemotherapy resistance (22,23). Furthermore, PTEN was a direct target of miR-21 (24). Therefore, the effects of miR-21 on the downregulation of PTEN induced by TGF-β1 were investigated in the present study. As presented in Fig. 6, the PTEN protein was significantly decreased (P<0.01) in the presence of TGF-β1. In addition, following the downregulation of miR-21, using anti-miR-21, in MCF-7 cells, the repression of PTEN by TGF-β1 was brought back to original levels. These results indicated that TGF-β1 stimulated the expression of miR-21 and suppressed the expression of PTEN in breast cancer MCF-7 cells.
Figure 6.
Effect of miR-21 on the downregulation of PTEN induced by TGF-β1. (A) PTEN protein from MCF-7 cells was exposed with 5 ng/ml TGF-β1 alone or transfected with anti-miR-21 or scramble anti-miR-NC, and subsequently determined using western blot analysis and normalized to GAPDH. PTEN protein was significantly decreased in the presence of TGF-β1. (B) The relative fold changes were calculated. Following the downregulation of miR-21, using anti-miR-21, in MCF-7 cells, the repression of PTEN by TGF-β1 was rescued. Data are presented as the mean ± standard error of the mean of three independent assays. **P<0.01; ***P<0.001. miR, microRNA; PTEN, phosphatase and tensin homolog; TGF-β1, transforming growth factor β1; NC, negative control.
Discussion
In the present study, miR-21 was identified to be upregulated in breast cancer cells, compared with BPBD, and its expression was significantly associated with elevated levels of TGF-β1 and clinicopathological features, including lymph node metastasis and tumor grade. In addition, increased concentrations of TGF-β1 stimulated the expression of miR-21, which mediated breast cancer invasion and chemoresistance. Furthermore, the results of the present study demonstrated that TGF-β1 promoted the expression of miR-21 to suppress PTEN, which facilitated breast cancer metastasis and chemoresistance. These results may provide novel insights into the underlying molecular mechanisms of TGF-β1 and suggest a novel therapy target for breast cancer.
A previous study revealed that the expression of miR-21 was regulated by increased TGF-β1 during the development of smooth muscle cells (15). A number of studies have demonstrated that TGF-β1 regulated miR-21 to promote renal (25,26), liver (14) and cardiac fibrosis (27). The results of the present study identified that TGF-β1 simulated miR-21 in a dose- and time-dependent manner in breast cancer cells. Furthermore, the expression of miR-21 was positively associated with TGF-β1 in breast cancer specimens. This result supports previous studies in breast cancer (11,18). Davis et al (15) demonstrated that TGF-β promoted the processing of primary transcripts of miR-21 into precursor miR-21 by the DROSHA complex in smooth muscle cells. However, the underlying molecular mechanism of TGF-β1 that results in the promotion of miR-21 in breast cancer remains unknown.
It is well-known that TGF-β1 is important in breast cancer (28). TGF-β1 is a tumor suppressor in the early stages of carcinogenesis and acts as a tumor promoter in advanced stages of cancer progression (29). Previous studies have demonstrated that increased TGF-β1 in breast cancer led to chemoresistance and was associated with poor outcomes in patients (8,9,30). The present study revealed that miR-21 mediated breast cancer invasion and chemoresistance stimulated by TGF-β1. Furthermore, a previous study demonstrated that miR-21 mediated TGF-β-dependent EMT in cancer cells, which promotes metastasis and chemoresistance (31). Therefore, miR-21 may be a key regulatory molecule of TGF-β signaling in breast cancer.
As a non-coding RNA, miRNA controls its target genes to regulate a number of important physiological and pathological processes. The tumor suppressor PTEN is a key target gene of miR-21 in breast cancer (32). Furthermore, the results of the present study, which were consistent with a previous study (33), indicated that the expression of PTEN is regulated by TGF-β. Therefore, PTEN suppresses tumor progression by its interaction with p53 and dephosphorylation of phosphatidylinositol phosphates (34,35). Wu et al (36) demonstrated that miR-21 induced gemcitabine resistance via the PTEN/Akt signaling pathway in breast cancer. Therefore, downregulation of PTEN by TGF-β1/miR-21 may cause metastasis and chemoresistance in breast cancer. The TGF-β/miR-21/PTEN signaling axis was identified to be responsible for non-cancer diseases (37). Dey et al (37) demonstrated that TGF-β-stimulated expression of miR-21 inhibits PTEN protein levels, which led to mesangial cell hypertrophy. The results of the present study suggested that TGF-β1/miR-21/PTEN may be an important pathway for breast cancer and requires additional study.
The results of the present study demonstrated that miR-21 was upregulated in breast cancer tissue and its expression level was significantly associated with increased TGF-β1, tumor grade and lymph node metastasis. Upregulation of miR-21 in breast cancer cells mediated the effects of TGF-β1 for cell invasion and chemoresistance by directly downregulating PTEN. These results suggested that TGF-β1/miR-21/PTEN signaling is involved in breast cancer and represents a potential target for future breast cancer therapies.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 31201028), the Natural Science Foundation of Guangdong Province (grant no. 2014A030313382), the Fundamental Research Fund for the Central Universities (grant no. 21617462), the Guangzhou Science Technology and Innovation Commission (grant nos. 2013J4100035 and 201707010099) and the Guangzhou City-belonged Universities Scientific Research Program (grant no. 2012C202).
References
- 1.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. 2016;17:43–46. doi: 10.7314/APJCP.2016.17.S3.43. [DOI] [PubMed] [Google Scholar]
- 2.Santa-Maria CA, Gradishar WJ. Changing treatment paradigms in metastatic breast cancer: Lessons learned. JAMA Oncol. 2015;1(528–534):549. doi: 10.1001/jamaoncol.2015.1198. [DOI] [PubMed] [Google Scholar]
- 3.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moses H, Barcellos-Hoff MH. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silberstein GB, Daniel CW. Reversible inhibition of mammary gland growth by transforming growth factor-beta. Science. 1987;237:291–293. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 6.Muraoka RS, Dumont N, Ritter CA, Dugger TC, Brantley DM, Chen J, Easterly E, Roebuck LR, Ryan S, Gotwals PJ, et al. Blockade of TGF-beta inhibits mammary tumor cell viability, migration, and metastases. J Clin Invest. 2002;109:1551–1559. doi: 10.1172/JCI0215234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie L, Law BK, Aakre ME, Edgerton M, Shyr Y, Bhowmick NA, Moses HL. Transforming growth factor beta-regulated gene expression in a mouse mammary gland epithelial cell line. Breast Cancer Res. 2003;5:R187–R198. doi: 10.1186/bcr640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Wang Y, Ren X, Tsuyada A, Li A, Liu LJ, Wang SE. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor β contributes to chemoresistance in breast cancer cells. Mol Cancer Res. 2010;8:1633–1642. doi: 10.1158/1541-7786.MCR-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Jia L, Liu C, Gong Y, Ren D, Wang N, Zhang X, Zhao Y. Axl as a downstream effector of TGF-β1 via PI3K/Akt-PAK1 signaling pathway promotes tumor invasion and chemoresistance in breast carcinoma. Tumour Biol. 2015;36:1115–1127. doi: 10.1007/s13277-014-2677-3. [DOI] [PubMed] [Google Scholar]
- 10.Wieser R. The transforming growth factor-beta signaling pathway in tumorigenesis. Curr Opin Oncol. 2001;13:70–77. doi: 10.1097/00001622-200101000-00014. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 12.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 13.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakhani SR, Ellis IO, Schnitt SJ, Tan PJ, van de Vijver MJ. WHO Classification of Tumours of the Breast. Fourth. IARC; Lyon, France: 2012. [Google Scholar]
- 17.Zhang Y, Wang SX, Ma JW, Li HY, Ye JC, Xie SM, Du B, Zhong XY. EGCG inhibits properties of glioma stem-like cells and synergizes with temozolomide through downregulation of P-glycoprotein inhibition. J Neurooncol. 2015;121:41–52. doi: 10.1007/s11060-014-1604-1. [DOI] [PubMed] [Google Scholar]
- 18.Xiao X, Liao X, Qiu S, Liu Z, Du B, Xu S. Synthesis, cytotoxicity and apoptosis induction in human tumor cells by galaxamide and its analogues [corrected] Mar Drugs. 2014;12:4521–4538. doi: 10.3390/md12084521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 20.Du B, Yang ZY, Zhong XY, Fang M, Yan YR, Qi GL, Pan YL, Zhou XL. Metastasis-associated protein 1 induces VEGF-C and facilitates lymphangiogenesis in colorectal cancer. World J Gastroenterol. 2011;17:1219–1226. doi: 10.3748/wjg.v17.i9.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padua D, Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 22.Giunti L, da Ros M, Vinci S, Gelmini S, Iorio AL, Buccoliero AM, Cardellicchio S, Castiglione F, Genitori L, de Martino M, et al. Anti-miR21 oligonucleotide enhances chemosensitivity of T98G cell line to doxorubicin by inducing apoptosis. Am J Cancer Res. 2014;5:231–242. [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 cluster regulates TGF-β-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. doi: 10.1158/1535-7163.MCT-13-0448. [DOI] [PubMed] [Google Scholar]
- 24.Singha PK, Pandeswara S, Geng H, Lan R, Venkatachalam MA, Saikumar P. TGF-β induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes Cancer. 2014;5:320–336. doi: 10.18632/genesandcancer.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung AC, Dong Y, Yang W, Zhong X, Li R, Lan HY. Smad7 suppresses renal fibrosis via altering expression of TGF-β/Smad3-regulated microRNAs. Mol Ther. 2013;21:388–398. doi: 10.1038/mt.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front Physiol. 2012;3:49. doi: 10.3389/fphys.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang H, Zhang C, Ban T, Liu Y, Mei L, Piao X, Zhao D, Lu Y, Chu W, Yang B. A novel reciprocal loop between microRNA-21 and TGFβRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. 2012;44:2152–2160. doi: 10.1016/j.biocel.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Katsuno Y, Hanyu A, Kanda H, Ishikawa Y, Akiyama F, Iwase T, Ogata E, Ehata S, Miyazono K, Imamura T. Bone morphogenetic protein signaling enhances invasion and bone metastasis of breast cancer cells through Smad pathway. Oncogene. 2008;27:6322–6333. doi: 10.1038/onc.2008.232. [DOI] [PubMed] [Google Scholar]
- 29.Imamura T, Hikita A, Inoue Y. The roles of TGF-β signaling in carcinogenesis and breast cancer metastasis. Breast Cancer. 2012;19:118–124. doi: 10.1007/s12282-011-0321-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Wu H, Zheng J, Yu P, Xu L, Jiang P, Gao J, Wang H, Zhang Y. Transforming growth factor β1 signal is crucial for dedifferentiation of cancer cells to cancer stem cells in osteosarcoma. Stem Cells. 2013;31:433–446. doi: 10.1002/stem.1298. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Pan T, Zhong X, Cheng C. Nicotine upregulates microRNA-21 and promotes TGF-β-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014;35:7063–7072. doi: 10.1007/s13277-014-1968-z. [DOI] [PubMed] [Google Scholar]
- 32.Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, Su F, Yao H, Song E. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 34.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/S1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 36.Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J, Zhang JF, Hu XC. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumour Biol. 2016;37:7245–7254. doi: 10.1007/s13277-015-4604-7. [DOI] [PubMed] [Google Scholar]
- 37.Dey N, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. TGFβ-stimulated microRNA-21 utilizes PTEN to orchestrate AKT/mTORC1 signaling for mesangial cell hypertrophy and matrix expansion. PLoS One. 2012;7:e42316. doi: 10.1371/journal.pone.0042316. [DOI] [PMC free article] [PubMed] [Google Scholar]