Abstract
Peptide cyclization is a strategy used to improve stability and/or activity of peptides. The most commonly used cyclization method is disulfide bridge formation of cysteine-containing peptides as typically found in nature. Over the years, an increasing number of alternative chemistries for peptide cyclization with improved efficiency, kinetics, orthogonality, and stability have been reported. However, there has been less appreciation for the opportunity to fine-tune peptide activity via the diverse chemical entities introduced at the site of linkage by different cyclization strategies. Here, we demonstrate how cyclization optimization of an M2 “anti-inflammatory” macrophage-binding peptide (M2pep) resulted in a significant increase in binding affinity of the optimized analog to M2 macrophages while maintaining binding selectivity compared to M1 “pro-inflammatory” macrophages. In this study, we report synthesis and evaluation of four cyclic M2pep(RY) analogs with diverse cyclization strategies; 1) Asp-[Amide]-Lys, 2) Azido-Lys-[Triazole (Copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC))]-Propargyl-Gly, 3) Cys-[Decafluorobiphenyl (DFBP)]-Cys, and 4) Cys-[Decafluorobiphenyl sulfone (DFS)]-Cys, whereby the chemical entity/linker at the linkage site is shown in the square bracket and is between the residues involved in cyclization. These peptides are compared to a disulfide-cyclized M2pep(RY) that we previously reported as a serum-stable, affinity enhanced analog to the original linear M2pep. DFBP-cyclized M2pep(RY) exhibits the highest binding activity to M2 macrophages with apparent dissociation constant (KD) about 2.03 μM compared to 36.3 μM for the original disulfide-cyclized M2pep(RY) and 220 μM for the original linear peptide. DFS-cyclized M2pep(RY) also binds more strongly than the original cyclized analog whereas Amide- and Triazole-cyclized M2pep(RY) analogs bind less strongly. We verified that DFBP alone has negligible binding to M2 macrophages and the incorporation of diphenylalanine to the original sequence improves binding activity at the expense of solubility and increased toxicity. In conclusion, we report development of cyclic M2pep(RY) analogs with diverse cyclization strategies leading to the discovery of DFBP-cyclized M2pep(RY) with enhanced M2 macrophage-binding activity.
Graphical abstract
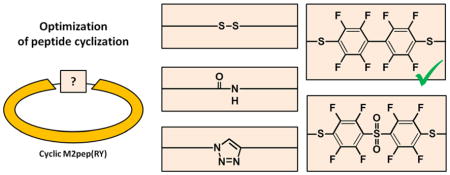
INTRODUCTION
Peptides are an important class of biomolecules for biomedical applications. The diversity of available amino acids and their different side chains allow for unique peptide sequences with specific bio-recognition to target receptors. The discovery of targeting peptides has been greatly accelerated by high-throughput combinatorial screening technologies such as peptide phage display library and one-bead-one-compound (OBOC) peptide library as well as by advanced computational design.1,2 Hence, peptides have been increasingly used as targeting ligands for diagnostics and drug delivery systems and also as therapeutic entities. Peptides have advantages of being smaller in size than proteins, allowing for application-dependent advantages such as higher diffusivity and lower manufacturing cost.3,4 However, peptides are also more susceptible to proteolytic degradation in serum and fast renal clearance.3 In addition, peptide sequences are sometimes too short to engage with the target receptors with sufficient affinity.5 As a result, most first generation peptides discovered from library screening need to be engineered or further optimized to improve serum stability and binding affinity before successful clinical translation.6,7 Common approaches in peptide optimization include alanine scanning and/or truncation analysis to identify key residues for subsequent biased library selection,8–11 incorporating unnatural or D-amino acids,12 altering the amide backbone,13–15 and stapling or cyclizing peptides to enhance stability, activity, and in some cases, cellular permeability.16,17
Conventional as well as newly developed bioconjugation chemistries have broadened the repertoire of cyclization strategies beyond disulfide bridge formation between cysteine pair(s) or lactamization between lysine and aspartic acid/glutamic acid.18,19 Copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) between non-natural amino acids with azide and alkyne side chains allows for orthogonal cyclization of cysteine-containing peptides without affecting the free cysteine residue which is sometimes important for peptide biological activity.19,20 Ruthenium-catalyzed ring-closing olefin metathesis (RCM) and molybdenum-catalyzed ring-closing alkyne metathesis (RCAM) enable orthogonal formation of bicyclic peptides containing non-natural amino acids with alkene and alkyne side chains, respectively, in a one-pot reaction.21 A family of perfluoroaryl compounds such as decafluorobiphenyl (DFBP) and decafluorobiphenyl sulfone (DFS) has been developed for selective cyclization of cysteine-containing peptides with fast reaction kinetics whereas DFS has also been successfully used to cyclize lysine-containing peptides that are absent in cysteine residues.22–24
Optimization of peptide activity via cyclization has focused on selecting the right amino acid positions for modification as well as the appropriate length of the linker.19,21,25 However, there have been few studies investigating the effect of cyclization chemistry on peptide bioactivity or stability, which may be an important parameter since the linker chemistry may affect peptide/receptor binding affinity or provide structural reinforcement to the peptide. In this study, we demonstrate that cyclization strategies significantly affect peptide activity using an M2 macrophage-binding peptide (M2pep) as an example.26 We previously reported a disulfide-cyclized M2pep(RY) as a serum stable, affinity-enhanced M2pep analog.27 This peptide binds to murine M2 (IL4-polarized, “anti-inflammatory”) macrophages with an apparent dissociation constant (KD) of about 36.3 μM and with high selectivity over M1 (IFN-γ and LPS-polarized, “pro-inflammatory”) macrophages. The disulfide-cyclized M2pep(RY) peptide was also successfully used to target tumor-promoting M2-like tumor associated macrophages (M2-TAMs) in CT-26 colon carcinoma and 4T1 breast cancer mouse models making it a useful targeting platform for development of M2-TAM-targeting diagnostics or therapeutics for cancer treatments.27 Here, we report synthesis of four additional cyclic M2pep(RY) analogs (Figure 1) with focus on the development of on-resin synthesis strategy for eventual manufacturing scalability. The effect of cyclization strategies on biological activity of these analogs were also compared through binding studies with M1 and M2 murine macrophages.
Figure 1.

Schematics of M2pep and cyclic M2pep(RY) analogs. RY substitution to the original M2pep sequence for affinity improvement is shown in green.27 Modified amino acids for cyclization and serum stability improvement are shown in red.
RESULTS AND DISCUSSION
Peptide synthesis
In this work, five cyclic M2pep(RY) peptides were synthesized using various cyclization chemistries (Figure 1). Successful synthesis of all peptides was verified by MALDI-ToF MS (Table S1). On-resin cyclization of Amide-, Triazole-, and DFBP-cyclized M2pep(RY)Biotin proceeded efficiently based on HPLC traces during purfication. Full schematics for on-resin synthesis of these analogs as well as their associated HPLC traces and methods are shown in Figure S1. Successful CuAAC-mediated cyclization of Triazole-cyclized M2pep(RY)Biotin was confirmed by insensitivity to TCEP-mediated azide reduction and inactivity to subsequent CuAAC reaction with propargyl alcohol (Figure S2). Attempts in on-resin cyclization of DFS-cyclized M2pep(RY)Biotin following the developed strategy for on-resin synthesis of DFBP-cyclized M2pep(RY)Biotin were not successful and instead, a major side product of -346 Da was observed (Figure S3A). Nonetheless, in-solution cyclization of DFS-cyclized M2pep(RY)Biotin was optimized to proceed in high yield with minimal formation of an unidentified “-20 Da” side product (Figure S3B). In addition, we found that addition of TCEP to the DFBP and DFS cyclization reactions in solution significantly increases yield of the desired product by suppressing formation of the disulfide-cyclized peptide. Three of the cyclized variants (Disulfide, DFBP, and DFS analogs) can be readily synthesized from the same linear precursor, thus saving time for the optimization process. Although not investigated in this study, it should also be possible to synthesize Amide- and Triazole-cyclized analogs from a common linear precursor (e.g. GYEQDPWGVRYWYGX2kkk(K-Biotin-resin) where X2 is azidolysine), followed by coupling with Boc-Propargylglycine-OH for CuAAC-mediated cyclization and coupling with Boc-Asp(OAll)-OH followed by reduction of azide to amine30 and OAll deprotection before on-resin lactamization, respectively.
In vitro binding study
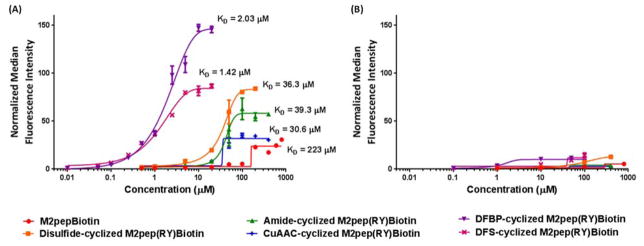
M1 and M2 polarized murine macrophages were incubated with various concentrations of biotinylated M2pep analogs and probed for peptide binding using streptavidin-FITC with flow cytometry analysis. Binding curves were acquired by determining the median fluorescence intensity (MFI) of streptavidin-FITC associated with cells at various peptide concentrations, and apparent KD values were calculated based on half-maximal MFI of each individual peptide analog. All M2pep analogs bind preferentially to M2 macrophages over M1 macrophages (Figure 2). For comparison, previously-reported binding curves for M2pepBiotin and Disulfide-cyclized M2pep(RY)biotin are also included.27 Disulfide-, Amide-, and Triazole-cyclized M2pep(RY)Biotin have similar KD values, between 30–40 μM, although varying maximum MFI values were observed for each construct. Strikingly, DFBP- and DFS-cyclized M2pep(RY)Biotin have significantly improved affinity with about 15–30 fold lower KD values (2.03 μM and 1.42 μM, respectively) than the other cyclic analogs. Comparing these two analogs, DFS-cyclized M2pep(RY)Biotin has lower maximal MFI than the DFBP analog. The maximum MFI correlates to the maximal extent at which peptides can engage with surface receptors on plasma membrane of target cells. It is surprising to us that different cyclic analogs exhibit different maximal MFI values even though they are expected to bind to the same target receptor on M2 macrophages. However, since the identity of the M2pep receptor remains unknown, we are not able to make further conclusions regarding the mechanism of binding of these peptide constructs. Nonetheless, our binding data demonstrate that optimization of cyclization chemistry is important in maximizing the cyclic peptide activity.
Figure 2.
Binding curves of M2pep analogs on (A) M2 macrophages and (B) M1 macrophages.
Due to the low KD and high maximal MFI obtained for DFBP-cyclized M2pep(RY)Biotin, we next verified whether the RY modification, originally reported to improve binding affinity of M2pep27, remains a relevant modification for this cyclic analog. DFBP-cyclized M2pepBiotin without RY modification was synthesized and evaluated for binding activity. Binding studies confirmed that the RY modification enhances binding activity of the cyclized-M2pep peptide to M2 murine macrophages by about 4-fold compared to the original sequence (Figure 3). Hence, we confirmed the discovery of DFBP-cyclized M2pep(RY)Biotin as an affinity-enhanced M2 macrophage-targeting peptide which could be useful for future development of diagnostics or drug delivery systems targeting M2 macrophages or M2-like tumor-associated macrophages.
Figure 3.
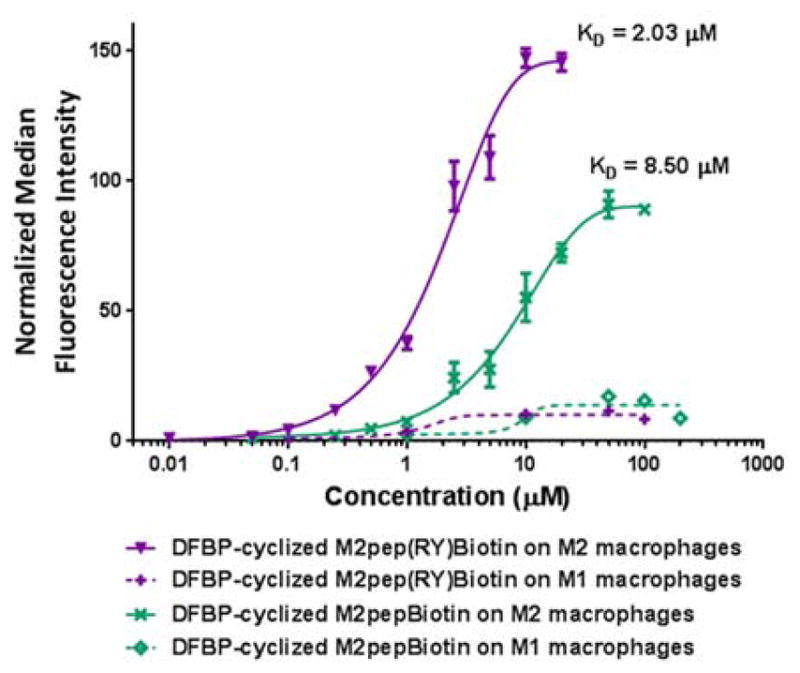
Binding curves of DFBP-cyclized M2pep(RY)Biotin and DFBP-cyclized M2pepBiotin with M1 and M2 macrophages.
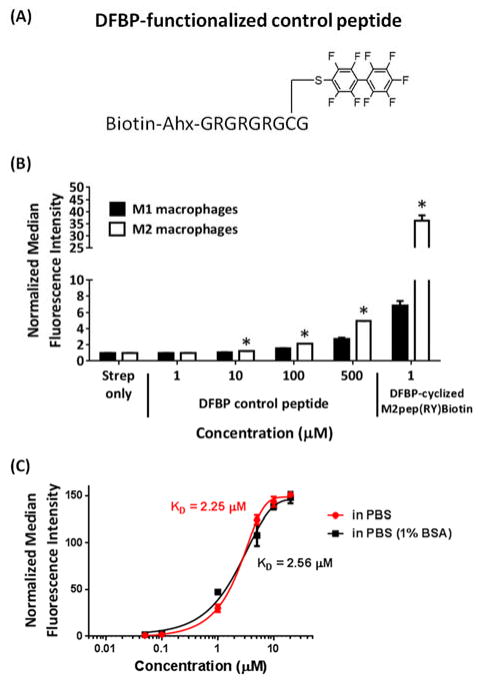
In order to understand the affinity enhancement of DFBP-cyclized M2pep(RY)Biotin, we first assessed the effect of DFBP on macrophage binding by synthesizing a DFBP-functionalized linear control peptide (Biotin-Ahx-GRGRGRG(C-DFBP)G) where DFBP is conjugated to a single cysteine and the GR region serves as a spacer and solubility enhancer for DFBP (Figure 4A). This peptide binds very weakly to macrophages even at 100 μM whereas DFBP-cyclized M2pep(RY)Biotin exhibits strong binding activity even at 1 μM (Figure 4B). This data suggests that DFBP itself does not increase binding to the unknown M2pep receptor and that DFBP may instead increase binding by an alternative mechanism such as via altering or stabilizing peptide conformation. Since DFBP is a small hydrophobic molecule, we speculated that DFBP might bind to some hydrophobic pockets on BSA present in our binding solutions and promote enhanced binding via a multivalent effect. This possibility was eliminated based on binding studies of DFBP-cyclized M2pep(RY)Biotin performed in both PBS and PBSA (PBS with 1% albumin) that showed no difference in binding (Figure 4C).
Figure 4.
(A) DFBP-functionalized linear control peptide. (B) Binding study of DFBP-functionalized linear control peptide on M1 and M2 macrophages. Stars denote statistical significance between M2 and M1 macrophages. *P < 0.05. (C) Binding study of DFBP-cyclized M2pep(RY)Biotin in PBS and PBSA on M2 macrophages.
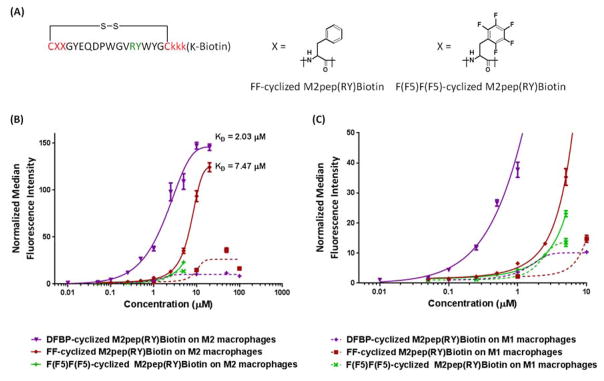
Next, we investigated whether the higher binding activity of DFBP-cyclized M2pep(RY)Biotin could be recapitulated by an addition of a dipeptide with aromatic side chain into the original disulfide-cyclized M2pep(RY)Biotin peptide. Two additional disulfide-cyclized M2pep(RY)Biotion analogs were synthesized incorporating either diphenylalanine (FF-cyclized M2pep(RY)Biotin) or dipentafluorophenylalanine (F(F5)F(F5)-cyclized M2pep(RY)Biotin) before the N-terminal cysteine to assess the effect of aromatic ring and fluorine substitution on binding activity to macrophages (Figure 5A). Introduction of these dipeptides significantly reduces solubility of the peptide analogs with the F(F5)F(F5)-cyclized analog being less soluble than the FF-cyclized analog. Notably, despite being highly fluorinated, DFBP-cyclized M2pep(RY)Biotin has higher solubility than FF-cyclized M2pep(RY)Biotin. Addition of diphenylalanine indeed improves binding activity of the peptide on M2 macrophages implying that the presence of aromatic structure near the site of cyclization may contribute to the enhanced binding (Figure 5B). F(F5)F(F5)-cyclized M2pep(RY)Biotin was very toxic to macrophages even at low micromolar concentrations (Figure S4), so we were not able to accurately construct a binding curve for this analog. Nonetheless, based on the binding data at the lower concentrations (Figure 5C), this analog seems to exhibit similar binding activity as FF-cyclized M2pep(RY)Biotin with M2 macrophages implying that fluorine substitution may not have a significant effect towards binding activity. However, since FF-cyclized M2pep(RY)Biotin still has lower binding activity than DFBP-cyclized M2pep(RY)Biotin, there are likely additional factors involved in the enhanced binding phenomenon observed with the DFBP-cyclized peptide.
Figure 5.
(A) FF- and F(F5)F(F5)-cyclized M2pep(RY)Biotin. (B) Binding curves of DFBP-, FF-, and F(F5)F(F5)-cyclized M2pep(RY)Biotin with M1 and M2 macrophages. (C) Enlarged binding curves at the lower concentration range.
Circular Dichroism (CD) measurement
Improvement in peptide activity via cyclization can sometimes be attributed to reinforcement in the peptide’s secondary structure.31 Hence, we performed a preliminary assessment on peptide conformation of the M2pep peptides via CD measurement. None of the CD spectra of these M2pep analogs matches characteristic spectra of 100% α-helix, β-sheet, or random coil structures32 implying that the peptides may only be partially structured or unstructured (Figure 6A–D). The effect of RY modification results in an inversion from a predominantly negative CD signal with two minima at about 200 and 225 nm to a predominantly positive signal with two maxima at about 200 and 230 nm (Figure 6A). A decline in the CD signal at 200 and 230 nm maxima for DFBP- and DFS-cyclized M2pep(RY)Biotin analogs was observed (Figure 6C) compared to the other cyclic M2pep(RY)Biotin analogs which may account for the affinity improvement of the former. However, this trend is not observed in FF-cyclized M2pep(RY)Biotin which also exhibits enhanced affinity. Unlike stapled peptides of helical nature whose secondary structure reinforcement may be estimated in term of helicity content, it is more challenging to elucidate the conformational reinforcement of non-helical peptides like cyclic M2pep(RY) analogs. Future characterization studies on identification of M2pep receptor as well as its interaction with M2pep peptides will be needed in order to better understand the observed enhanced affinity to M2 macrophage.
Figure 6.
CD spectra of (A) M2pepBiotin and Disulfide-cyclized M2pep(RY)Biotin, (B) Amide- and Triazole-cyclized M2pep(RY)Biotin, (C) DFBP- and DFS-cyclized M2pep(RY)Biotin, and (D) DFBP-cyclized M2pepBiotin and FF-cyclized M2pep(RY)Biotin.
In vitro serum stability study
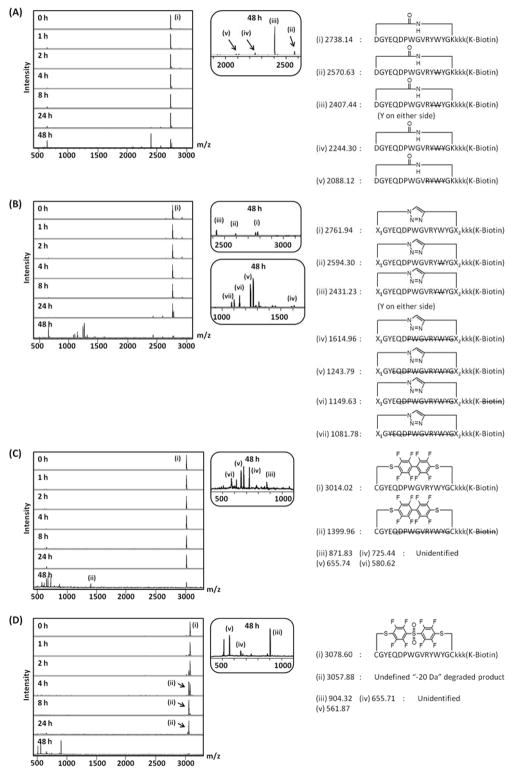
The original M2pepBiotin is degraded in mouse serum within 4 h via N-terminal degradation and endolytic cleavages at the W/W site in the binding region and at the S/K site in the spacer region.27 By replacing L-lysines in the spacer region with D-lysines and cyclizing the binding region of the peptide, we demonstrate significant improvement in serum stability of M2pep.27 Nonetheless, the cyclized peptide was eventually degraded by prolonged incubation in mouse serum for more than 1 day. Since cyclization of the first generation cyclized M2pep(RY)Biotin was based on a disulfide bridge which may be prone to linearization over time due to changes in the redox environment, we investigated whether the alternative cyclization chemistries can further improve serum stability over disulfide cyclization. Serum stability of the newly-synthesized cyclic M2pep(RY)Biotin peptides were evaluated by incubation in mouse serum and analysis for the presence of peptides at different time points by MALDI-ToF MS. As observed for Disulfide-cyclized M2pep(RY)Biotin27, Amide-, Triazole-, and DFBP-cyclized M2pep(RY)Biotin were stable in serum for at least 24 h after which degradation products were observed (Figure 7A–C). Multiple attempts to quantify the extent of degradation were unsuccessful due to variability in the amount of extracted peptides following the protein precipitation with ACN, thus preventing quantitative comparisons regarding serum stability. Nonetheless, by identifying the degradation products, it is clear that degradation was initiated at either Y/W or W/Y which corresponds to the W/W site of the original M2pepBiotin. Even though endolytic cleavage at the Y/W/Y site seems to be the rate-determining step towards peptide degradation of cyclic M2pep(RY)Biotin, the new generation analogs are expected to be more favorable than the disulfide-cyclized analog due to insensitivity to reduction. Surprisingly for DFS-cyclized M2pep(RY)Biotin, formation of the “-20 Da” side product was observed over time (Figure 7D). This side product was also observed when cyclization reaction with DFS was left for too long (Figure S3) but was not observed in the purified peptide stock solution in water, PBS, or PBSA for at least one day (Figure S5). Similarly, the Pentelute lab has recently reported an observation on oxidative degradation when DFS is used to cyclize cysteine-containing peptides.24 The group has also demonstrated superior stability when DFS is instead used to cyclize lysine-containing peptides. Hence, studies by others as well as ours emphasize the need to evaluate any newly developed cyclization chemistries for stability towards physicochemical and physiological stresses such as redox environment and enzymatic degradation. Since we have shown here a higher serum stability of DFBP-cyclized M2pep(RY)Biotin as well as a similar improvement in binding affinity of DFBP- and DFS-cyclized M2pep(RY), it is possible to utilize the DFS-stapled peptide phage library reported by the Derda lab23 for initial screening of high affinity peptide candidates before synthesizing the peptide with DFBP cyclization for the follow-up evaluation. All in all, different cyclization chemistries as well as robust and flexible synthetic strategies greatly expand our toolbox for peptide optimization that is needed for the development of new effective targeting agents or therapeutics.
Figure 7.
Serum stability of (A) Amide-, (B) Triazole-, (C) DFBP-, and (D) DFS-cyclized M2pep(RY)Biotin. Left: MALDI-ToF MS spectra. Middle: Selected enlarged regions of the spectra. Right: Identified degraded peptide fragments.
CONCLUSIONS
We have reported synthesis and evaluation of cyclic M2pep(RY)Biotin analogs with different cyclization chemistries. Our binding studies of these cyclic M2pep(RY)Biotin on macrophages identify DFBP-cyclized M2pep(RY)Biotin as an affinity-enhanced M2 macrophage-binding analog and highlight the possibility of optimizing peptide activity via modulation of cyclization chemistry. The on-resin synthesis strategy of DFBP-cyclized peptide reported here allows for manufacturing scalability, an important consideration in the development of peptide therapeutics.
MATERIALS AND METHODS
Materials
Protected amino acids, 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU), (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBOP), and 1-hydroxybenzotriazole hydrate (HOBt) were purchased from AAPPTec (Louisville, KY), AnaSpec (Fremont, CA), and Chem-Implex International (Wood Dale, IL). Triisopropylsilane (TIPS), 1,2-ethanedithiol (EDT), 1,3-dimethoxybenzene (DMB), tetrakis(triphenylphosphine)palladium(0) (Pd(PPh3)4), sodium diethyldithiocarbamate trihydrate, copper(II) sulfate pentahydrate (CuSO4•5H2O), and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO). Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) was purchased from TCI America (Portland, OR). Normal mouse serum (Catalog number 10410), phenylsilane, decafluorobiphenyl, and other reagents were purchased from Thermo Fisher Scientific (Waltham, MA). Quant*Tag Biotin quantification kit was purchased from Vector Laboratories (Burlingame, CA). Imidazole-1-sulfonyl azide hydrochloride and decafluorobiphenyl sulfone (DFS) were synthesized following the previously reported protocols.28,29 For the synthesis of DFS, an additional purification step was performed by dissolving the partially purified light yellow product in DMF and precipitating in H2O twice before lyophilizing to obtain the product as white powder.
Synthesis of biotinylated M2pep analogs
General peptide synthesis strategy
Peptides were synthesized using a PS3 automated peptide synthesizer (Protein Technologies, Phoenix, AZ) via standard Fmoc/tbu solid-phase peptide synthesis unless noted in Table 1. Manual coupling of amino acids was performed by incubating the peptide resin in a solution of amino acid (4 eq.) and HCTU (3.9 eq.) in 0.4 M N-methylmorpholine in DMF for 3 h. Fmoc deprotection was performed by two 30-min incubations in 20% (v/v) piperidine in DMF. Biotinylated resin was prepared from NovaPEG rink amide resin as reported previously27 and was used for the synthesis of all biotinylated peptides. Peptides were cleaved off the resin by incubating in a cleavage cocktail containing TFA/DMB/TIPS/EDT (90:5:2.5:2.5 v/v/v/v) for 2.5 h followed by double precipitation in cold ether. EDT was included in the cleavage cocktail only for peptides containing a free cysteine after cleavage. The crude peptides were purified by RP-HPLC (Agilent 1200, Santa Clara, CA) to 95% purity using a Phenomenex Fusion-RP C18 semi-preparative column (Torrance, CA) at the flow rate of 5 mL/min with H2O (0.1% TFA) and ACN (0.1% TFA) as a mobile phase. Molecular weights of the purified peptides were confirmed by MALDI-ToF Mass Spectrometry (MS).
Table 1.
Protected amino acids used for cyclization.
| Linear precursor: X1GYEQDPWGVRYWYGX2kkk(K-Biotin) | ||
|---|---|---|
|
| ||
| M2pep analogs | Protected X1 | Protected X2 |
| Disulfide-cyclized M2pep(RY)Biotin | Fmoc-Cys(tbu)-OH | Fmoc-Cys(tbu)-OH |
| Amide-cyclized M2pep(RY)Biotin | Fmoc-Asp(OAll)-OH | Fmoc-Lys(Alloc)-OH |
| Triazole-cyclized M2pep(RY)Biotin | Boc-Lys(Fmoc)-OH | Fmoc-Propargyl-Gly-OH |
| DFBP-cyclized M2pep(RY)Biotin | Fmoc-Cys(Stbu)-OH | Fmoc-Cys(Mmt)-OH |
| DFS-cyclized M2pep(RY)Biotin | Fmoc-Cys(tbu)-OH | Fmoc-Cys(tbu)-OH |
Disulfide-cyclized M2pep(RY)Biotin
The purified linear precursor (10–20 mg) was dissolved in H2O (1–3 mL) and added to a deaerated 0.1 M ammonium bicarbonate buffer (pH 8) at a final peptide concentration of 0.14 mg/mL. The solution was left to stir for 2 days for oxidation, then acidified with TFA and desalted with a HyperSep™ C18 cartridge. The desalted cyclized peptide was concentrated with rotary evaporator and lyophilized.
Amide-cyclized M2pep(RY)Biotin
The N-terminus-Fmoc-protected precursor peptide on resin was incubated in a solution of Pd(PPh3)4 (0.2 eq.) and phenylsilane (20 eq.) in DCM for 20 min twice to selectively deprotect OAll and Alloc protecting groups. The resin in the deprotection solution was purged with nitrogen for 5 min prior to each incubation. Following deprotection, the resin was washed with 5 mM sodium diethyldithiocarbamate solution in DMF to remove the palladium catalyst. On-resin lactamization follows the optimized protocol by Thakkar et al.33 by incubating the resin in a solution of HOBt (5 eq.), PyBOP (5 eq.), and DIPEA (10 eq.) in DCM/DMF/MNP (3:2:2 v/v/v) with 1% (v/v) triton X100 for 24 h. A small amount of peptide on resin was cleaved to monitor for completion of the cyclization via MALDI-ToF MS. Upon completion, the cyclized peptide was Fmoc-deprotected, cleaved, and purified by RP-HPLC.
Triazole-cyclized M2pep(RY)Biotin
The epsilon amine on the N-terminal lysine of the precursor peptide on resin was Fmoc-deprotected and converted to azide by an overnight incubation in a solution of imidazole-1-sulfonyl azide hydrochloride (3 eq.), CuSO4•5H2O (0.01 eq. pre-dissolved in a small amount of H2O), and DIPEA (9 eq.) in DMSO as reported previously.34 The completion of the diazotransfer reaction was monitored by the Kaiser test for the absence of amine. Alternatively, Fmoc-Lys(N3)-OH may be used instead of Boc-Lys(Fmoc)-OH. On-resin CuAAC cyclization was performed by incubating the peptide resin in a solution of tetrakis(acetonitrile)copper(I) hexafluorophosphate (1 eq.), TBTA (1 eq.), and DIPEA (2 eq.) in DMF for 24 h. The resin was then washed with 5 mM sodium diethyldithiocarbamate solution in DMF, and the cyclized peptide was cleaved and purified by RP-HPLC.
DFBP-cyclized M2pep(RY)Biotin
In-solution cyclization of the linear peptide precursor with DFBP follows the previously described protocol22 modified with an addition of TCEP to suppress disulfide formation during DFBP cyclization. In brief, the crude peptide precursor (20 mg) was dissolved in DMF (2.5 mL) in a 5-mL microcentrifuge tube. A fresh TCEP stock solution in H2O (5 eq.) was added to the peptide solution, and an additional volume of H2O was added to make up a total H2O volume of 0.5 mL. The reaction was left for 10 min followed by an addition of 50 mM tris base in DMF (1.5 mL). DFBP predissolved in DMF (2 eq.) was then added, and the reaction was vortexed vigorously. On the next day, the reaction was diluted with 3-fold volume of H2O (0.1% TFA) and desalted with a Sep-Pak C18 cartridge. The desalted cyclized peptide solution was concentrated and purified by RP-HPLC.
On-resin cyclization of DFBP utilizes two cysteines with orthogonal protecting groups (Table 1). First, the Stbu protecting group was deprotected by an overnight incubation in a solution of DTT (10 eq.) and TEA (20 eq.) in DMF. The free cysteine was functionalized by incubating in a solution of DFBP (4 eq.) and TEA (8 eq.) in DMF for 3 h. Mmt protecting group was then removed by 20-min incubations in TFA/TIPS/DCM (2:5:93 v/v/v) until yellow-colored solution disappeared (3–4 incubations). The DFBP cyclization was then proceeded by incubating the peptide resin in a TEA solution (8 eq.) in DMF for 24 h. The cyclized peptide was cleaved and purified by RP-HPLC. All deprotection and DFBP functionalization steps were monitored by the modified on-resin Ellman’s assay following the previously reported protocol.35
DFS-cyclized M2pep(RY)Biotin
In-solution cyclization of the linear peptide precursor with DFS was performed as described for DFBP cyclization with an exception that the reaction was quenched by an addition of 3-fold volume of H2O (0.1% TFA) after a 10-min incubation to prevent formation of a side product.
Bone marrow harvest and macrophage culture
All animal handling protocols were approved by the University of Washington Institutional Animal Care and Use Committee. Bone marrow-derived cells were harvested from female c57bl/6 mice as previously described.36 In brief, femurs and tibias of the mice were excised and flushed with RPMI 1640 medium via an 18G needle. The cells were cultured on petri dishes containing RPMI 1640 medium supplemented with 20% donor horse serum, 1% antibiotic-antimycotic (AbAm), and 20 ng/mL M-CSF. After 7 d in culture, the medium was replaced with an activation medium whereby M-CSF was replaced with 25 ng/mL IFN-γ and 100 ng/mL LPS for M1 activation or with 25 ng/mL IL-4 for M2 activation. For M1 activation medium, AbAm was replaced with 1% penicillin-streptomycin (Pen-Strep). After 2 d of activation, the activated macrophages were scraped off the petri dishes for binding studies.
In vitro binding study
Biotinylated peptide stock solutions were prepared in H2O at 10 mg/mL concentration and quantified for the exact concentration using Quant*Tag Biotin quantification kit. The stock solutions were diluted in PBS with 1% BSA (PBSA) to different concentrations for binding studies. Activated macrophages were seeded on a black 96-well plate (50,000 cells/well) and incubated with the peptide solutions for 20 min on ice. The cells were washed twice with PBSA and subsequently incubated with streptavidin-FITC for 15 min on ice. After two additional washes, the cells were resuspended in PBS for analysis via MACSQuant Flow Cytometer (Miltenyi Biotec, San Diego, CA). Propidium iodide (PI) was added before running each sample to assess for cell viability. Data analysis was processed on FlowJo Analysis Software (Tree Star, Ashland, OR). Construction of binding curves and determination of apparent KD were performed using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA) based on an average normalized median fluorescence intensity value of triplicate samples in the same experiment.
CD measurement
M2pep analogs were prepared in H2O at 50 μM concentration. CD measurement of the peptides was performed on Jasco 720 Circular Dichroism Spectrophotometer (Jasco Inc., Easton, MD) at 25 °C with an average from 8 scans. The CD spectra were corrected for baseline and smoothened on Spectra Manager Software (Jasco Inc., Easton, MD).
In vitro serum stability study
Different M2pep analogs (30 μL from 10 mg/mL stock solution in H2O) were incubated in normal mouse serum (300 μL) at 37 °C in an incubator. An aliquot of the serum (40 μL) was drawn at different time intervals and precipitated with an equal volume of cold ACN. The mixture was centrifuged at 15,000 rpm for 5 min, and the supernatant was collected. A solution of H2O/ACN (1:1 v/v, 80 μL) was added to the pellet with a 10-min sonication to further extract residual peptides. The mixture was then centrifuged. The supernatant was pooled with the former and dried under vacuum on a Speedvac machine. The peptide pellet was resuspended in H2O (50 μL) with a 10-min sonication before analysis by MALDI-ToF MS.
Supplementary Material
Acknowledgments
This work was supported by NIH 1R01CA177272. Chayanon Ngambenjawong was supported by an Anandamahidol Foundation Fellowship. Julio Marco B. Pineda was supported by a Mary Gates Research Scholarship. We thank Professor Niels H. Anderson and Kalkena Sivanesam for consultation on interpretation of the CD spectra.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS publication website at DOI:
Calculated/Observed Mw of the synthesized peptides, synthesis schematics and characterization, toxicity study, and additional peptide stability data
References
- 1.Falciani C, Lozzi L, Pini A, Bracci L. Bioactive Peptides from Libraries. Chem Biol. 2005;12:417–426. doi: 10.1016/j.chembiol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Kliger Y. Computational approaches to therapeutic peptide discovery. Biopolymers. 2010;94:701–710. doi: 10.1002/bip.21458. [DOI] [PubMed] [Google Scholar]
- 3.Zhang XX, Eden HS, Chen X. Peptides in cancer nanomedicine: drug carriers, targeting ligands and protease substrates. J Control Release. 2012;159:2–13. doi: 10.1016/j.jconrel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson F, Tarli L, Viti F, Neri D. The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev. 2000;43:165–196. doi: 10.1016/s0169-409x(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 6.Brown KC. Peptidic tumor targeting agents: the road from phage display peptide selections to clinical applications. Curr Pharm Des. 2010;16:1040–1054. doi: 10.2174/138161210790963788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodziej AF, Zhang Z, Overoye-Chan K, Jacques V, Caravan P. Peptide optimization and conjugation strategies in the development of molecularly targeted magnetic resonance imaging contrast agents. Methods Mol Biol. 2014;1088:185–211. doi: 10.1007/978-1-62703-673-3_13. [DOI] [PubMed] [Google Scholar]
- 8.Boersma MD, Sadowsky JD, Tomita YA, Gellman SH. Hydrophile scanning as a complement to alanine scanning for exploring and manipulating protein-protein recognition: application to the Bim BH3 domain. Protein Sci. 2008;17:1232–1240. doi: 10.1110/ps.032896.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatanaka T, Ohzono S, Park M, Sakamoto K, Tsukamoto S, Sugita R, Ishitobi H, Mori T, Ito O, Sorajo K, Sugimura K, Ham S, Ito Y. Human IgA-binding peptides selected from random peptide libraries: affinity maturation and application in IgA purification. J Biol Chem. 2012;287:43126–43136. doi: 10.1074/jbc.M112.389742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larimer BM, Thomas WD, Smith GP, Deutscher SL. Affinity maturation of an ERBB2-targeted SPECT imaging peptide by in vivo phage display. Mol imaging Biol MIB Off Publ Acad Mol Imaging. 2014;16:449–458. doi: 10.1007/s11307-014-0724-5. [DOI] [PubMed] [Google Scholar]
- 11.Santoso B, Murray BW. Maleimide-based method for elaboration of cysteine-containing peptide phage libraries. Methods Mol Biol. 2015;1248:267–276. doi: 10.1007/978-1-4939-2020-4_18. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Gfeller D, Buth SA, Michielin O, Leiman PG, Heinis C. Improving binding affinity and stability of peptide ligands by substituting glycines with D-amino acids. Chembiochem. 2013;14:1316–1322. doi: 10.1002/cbic.201300228. [DOI] [PubMed] [Google Scholar]
- 13.Valverde IE, Bauman A, Kluba CA, Vomstein S, Walter MA, Mindt TL. 1,2,3-Triazoles as amide bond mimics: triazole scan yields protease-resistant peptidomimetics for tumor targeting. Angew Chem Int Ed Engl. 2013;52:8957–8960. doi: 10.1002/anie.201303108. [DOI] [PubMed] [Google Scholar]
- 14.Harris KS, Casey JL, Coley AM, Karas JA, Sabo JK, Tan YY, Dolezal O, Norton RS, Hughes AB, Scanlon D, Foley M. Rapid optimization of a peptide inhibitor of malaria parasite invasion by comprehensive N-methyl scanning. J Biol Chem. 2009;284:9361–9371. doi: 10.1074/jbc.M808762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilimire TA, Bennett RP, Stewart RA, Garcia-Miranda P, Blume A, Becker J, Sherer N, Helms ED, Butcher SE, Smith HC, Miller BL. N-Methylation as a Strategy for Enhancing the Affinity and Selectivity of RNA-binding Peptides: Application to the HIV-1 Frameshift-Stimulating RNA. ACS Chem Biol. 2016;11:88–94. doi: 10.1021/acschembio.5b00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldflam M, Ullman CG. Recent Advances Toward the Discovery of Drug-Like Peptides De novo. Front Chem. 2015;3:69. doi: 10.3389/fchem.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walensky LD, Bird GH. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 19.Lau YH, de Andrade P, Wu Y, Spring DR. Peptide stapling techniques based on different macrocyclisation chemistries. Chem Soc Rev. 2015;44:91–102. doi: 10.1039/c4cs00246f. [DOI] [PubMed] [Google Scholar]
- 20.Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides in protein chemical and physical stability. Curr Protein Pept Sci. 2009;10:614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cromm PM, Schaubach S, Spiegel J, Furstner A, Grossmann TN, Waldmann H. Orthogonal ring-closing alkyne and olefin metathesis for the synthesis of small GTPase-targeting bicyclic peptides. Nat Commun. 2016:7. doi: 10.1038/ncomms11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spokoyny AM, Zou Y, Ling JJ, Yu H, Lin YS, Pentelute BL. A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling. J Am Chem Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalhor-Monfared S, Jafari MR, Patterson JT, Kitov PI, Dwyer JJ, Nuss JM, Derda R. Rapid biocompatible macrocyclization of peptides with decafluoro-diphenylsulfone. Chem Sci. 2016;7:3785–3790. doi: 10.1039/c5sc03856a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lautrette G, Touti F, Lee HG, Dai P, Pentelute BL. Nitrogen Arylation for Macrocyclization of Unprotected Peptides. J Am Chem Soc. 2016;138:8340–8343. doi: 10.1021/jacs.6b03757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oddo A, Thomsen TT, Britt HM, Løbner-Olesen A, Thulstrup PW, Sanderson JM, Hansen PR. Modulation of Backbone Flexibility for Effective Dissociation of Antibacterial and Hemolytic Activity in Cyclic Peptides. ACS Med Chem Lett. 2016 doi: 10.1021/acsmedchemlett.5b00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cieslewicz M, Tang J, Yu JL, Cao H, Zavaljevski M, Motoyama K, Lieber A, Raines EW, Pun SH. Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc Natl Acad Sci U S A. 2013;110:15919–15924. doi: 10.1073/pnas.1312197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngambenjawong C, Gustafson HH, Pineda JM, Kacherovsky NA, Cieslewicz M, Pun SH. Serum Stability and Affinity Optimization of an M2 Macrophage-Targeting Peptide (M2pep) Theranostics. 2016;6:1403–1414. doi: 10.7150/thno.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goddard-Borger ED, Stick RV. An efficient, inexpensive, and shelf-stable diazotransfer reagent: imidazole-1-sulfonyl azide hydrochloride. Org Lett. 2007;9:3797–3800. doi: 10.1021/ol701581g. [DOI] [PubMed] [Google Scholar]
- 29.Xu L, Cheng J, Trudell ML. Chromium(VI) Oxide Catalyzed Oxidation of Sulfides to Sulfones with Periodic Acid. J Org Chem. 2003;68:5388–5391. doi: 10.1021/jo030031n. [DOI] [PubMed] [Google Scholar]
- 30.Lohani CR, Rasera B, Scott B, Palmer M, Taylor SD. α-Azido Acids in Solid-Phase Peptide Synthesis: Compatibility with Fmoc Chemistry and an Alternative Approach to the Solid Phase Synthesis of Daptomycin Analogs. J Org Chem. 2016;81:2624–2628. doi: 10.1021/acs.joc.5b02882. [DOI] [PubMed] [Google Scholar]
- 31.Pelay-Gimeno M, Glas A, Koch O, Grossmann TN. Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes. Angew Chem Int Ed Engl. 2015;54:8896–8927. doi: 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 33.Thakkar A, Trinh TB, Pei D. Global Analysis of Peptide Cyclization Efficiency. ACS Comb Sci. 2013;15:120–129. doi: 10.1021/co300136j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro V, Blanco-Canosa JB, Rodriguez H, Albericio F. Imidazole-1-sulfonyl Azide-Based Diazo-Transfer Reaction for the Preparation of Azido Solid Supports for Solid-Phase Synthesis. ACS Comb Sci. 2013;15:331–334. doi: 10.1021/co4000437. [DOI] [PubMed] [Google Scholar]
- 35.Badyal JP, Cameron AM, Cameron NR, Coe DM, Cox R, Davis BG, Oates LJ, Oye G, Steel PG. A simple method for the quantitative analysis of resin bound thiol groups. Tetrahedron Lett. 2001;42:8531–8533. [Google Scholar]
- 36.Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5080. pdb.prot5080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.