Summary
Amyotrophic lateral sclerosis (ALS) is a progressive, fatal neurodegenerative disease characterized by degeneration of upper and lower motor neurons in the brain and spinal cord. The hallmark pathological feature in most cases of ALS is nuclear depletion and cytoplasmic accumulation of the protein TDP-43 in degenerating neurons. Consistent with this pattern of intracellular protein redistribution, impaired nucleocytoplasmic trafficking has emerged as a mechanism contributing to ALS pathology. Dysfunction in nucleocytoplasmic transport is also an emerging theme in physiological aging and other related neurodegenerative diseases, such as Huntington’s and Alzheimer’s diseases. Here we review transport through the nuclear pore complex, pointing out vulnerabilities that may underlie ALS and potentially contribute to this and other age-related neurodegenerative diseases.
Keywords: ALS, aging, nucleocytoplasmic transport, nuclear pore, C9ORF72, dipeptide repeats
Introduction
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease in the United States and as motor neurone disease in the United Kingdom, is a progressive and uniformly fatal neurodegenerative disease characterized by progressive degeneration of motor neurons in the brain and spinal cord (Taylor et al., 2016). The dysfunction and ultimate demise of motor neurons lead to secondary denervation, muscle wasting, and weakness, typically culminating in death due to respiratory failure or bulbar dysfunction. Recent advances in defining the genetics of ALS, combined with clinical reports and pathological observations, have led to our current understanding of ALS as one disease that falls within a spectrum of a broader degenerative disorder that also includes frontotemporal dementia (FTD), inclusion body myopathy, and the overlap syndrome called multisystem proteinopathy (Taylor et al., 2016). Nearly 50% of patients with ALS demonstrate mild to moderate cognitive or behavioral symptoms, and as many as 20% of patients meet the clinical criteria for FTD (Ringholz et al., 2005; Wheaton et al., 2007). Less frequently, ALS occurs together with inclusion body myopathy or Paget’s disease of bone (Taylor et al., 2016).
A significant proportion (~10%) of patients with ALS are defined as having “familial ALS,” which is typically inherited as a highly penetrant, dominant trait. The remaining 90% of cases are classified as “sporadic” because they lack a clear family history, although this term does not preclude a genetic contribution in these cases. More than 100 potential ALS genes have been reported (http://alsod.iop.kcl.ac.uk), although the evidence supporting a causative role for these individual genes and variants varies. Currently, 16 genes are accepted with certainty as being implicated in ALS pathogenesis, largely falling into three functional categories (Taylor et al., 2016). The first group, consisting of VCP, OPTN, UBQLN2, SQSTM1, TBK1 and SOD1, influence cellular proteostasis and protein quality control. The second group, consisting of TARDBP, FUS, HNRNPA1, MATR3, and ANG, play various roles in the life cycle of RNAs, affecting RNA processing, assembly into membrane-less organelles (e.g., granules), and intracellular transport. The third group consists of DCTN1, TUBA4A, and PFN1, all of which play roles in cytoskeletal dynamics within the motor neuron axon and distal terminal. Two ALS genes (CHCHD10 and C9ORF72) defy this simplified classification or have less clear function. ALS-causing mutations are typically missense substitutions, with the notable exception of C9ORF72, in which ALS is caused by a massive expansion of an intronic hexanucleotide repeat (DeJesus-Hernandez et al., 2011; Renton et al., 2011).
A pathological hallmark of motor neuron degeneration in ALS is the accumulation of rounded or thread-like deposits of aggregated proteins termed neuronal cytoplasmic inclusions. The main component of these cytoplasmic inclusions in most familial and sporadic cases of ALS is TAR DNA-binding protein 43 (TDP-43), encoded by TARDBP (Neumann et al., 2006). Although TDP-43 undergoes nucleocytoplasmic shuttling, it is normally a predominantly nuclear protein (Ayala et al., 2008); thus, the accumulation of cytoplasmic pathology in ALS is quite striking. The significance of TDP-43 as a driver of ALS was cemented by the identification of rare ALS-causing mutations in TARDBP in familial cases. Cases of ALS caused by mutations in other genes such as FUS, HNRNPA1, and MATR3 can also feature abnormal cytoplasmic localization of predominantly nuclear proteins (Dormann et al., 2010; Gallego-Iradi et al., 2015; Kim et al., 2013).
Multiple Routes to Nucleocytoplasmic Transport Defects in ALS
The wide variety of genetic insults that culminate in motor neuron degeneration would appear to suggest that multiple, distinct mechanisms may contribute to ALS. Nonetheless, clinical and pathological evidence collected from both familial and sporadic ALS has highlighted surprising consistencies, including similarities in the pattern of cell type vulnerability, onset, and progression, as well as pathological hallmarks such as cytoplasmic inclusions of TDP-43, as described above. This congruity suggests that despite significant heterogeneity in initiating genetic events, a substantial portion of ALS may arise through common mechanisms. A dominant theme that has emerged over several years is disturbance of cellular structures that arise through biological phase transitions of proteins with low complexity sequence domains (LCDs) (Taylor et al., 2016). LCDs with a variety of compositions are found in up to one third of the proteome and contribute to the assembly of dynamic, higher order structures such as nucleoli, RNA granules, and the central channel of the nuclear pore through the process of liquid-liquid phase transition (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Molliex et al., 2015; Nott et al., 2015; Patel et al., 2015; Schmidt and Gorlich, 2015). Until recently, however, within the broad spectrum of biological phase transitions, a point of intersection for the various genetic etiologies of ALS has remained elusive, with a central unresolved question regarding which cellular structures that arise through phase separations are impacted in ALS. Insight into a potential common pathway has recently been highlighted by a series of studies that have implicated defects in nucleocytoplasmic transport as a shared consequence downstream of a variety of ALS-initiating mutations. These include the identification of ALS-causing mutations in the nuclear localization signals (NLSs) of FUS and hnRNPA1 (Dormann et al., 2010; Gal et al., 2011; Liu et al., 2016), evidence of impaired nucleocytoplasmic transport downstream of C9ORF72-related ALS-FTD (Boeynaems et al., 2016; Freibaum et al., 2015; Jovičić et al., 2015; Shi et al., 2017; Zhang et al., 2015), and recognition that cytoplasmic protein aggregates of TDP-43, and cytoplasmic deposition of amyloids more generally, interfere with nucleocytoplasmic transport of protein and RNA (Woerner et al., 2016). In this review, we provide an overview of the current understanding of nucleocytoplasmic transport through the nuclear pore complex (NPC) in mammalian cells. We then describe the evidence that defects in nucleocytoplasmic transport contribute to ALS. Finally, we explore studies suggesting that defects in nucleocytoplasmic transport contribute to other neurodegenerative diseases and physiologic aging, thus connecting the most important risk factor for neurodegenerative disease—aging—to the most common pathologic feature of neurodegenerative disease—pathological deposition of protein aggregates in the cytoplasm.
Architecture of the Nuclear Pore Complex
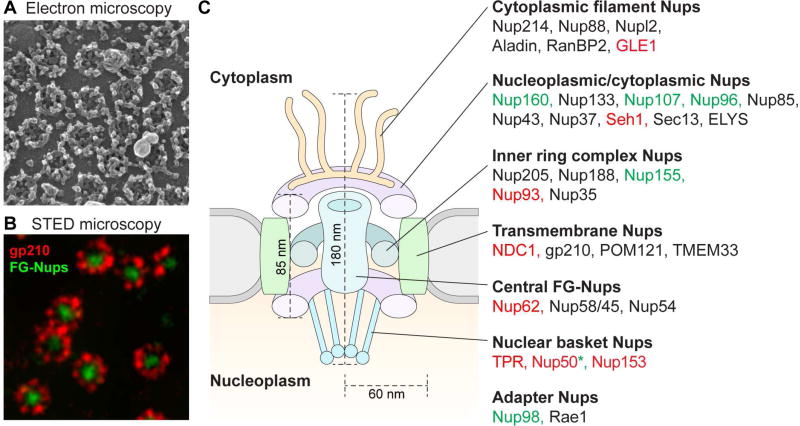
Central to the process of nucleocytoplasmic transport is the NPC, which provides communication between the nucleoplasm and cytoplasm through regulated nucleocytoplasmic transport. NPCs are morphologically defined structures composed of evolutionarily conserved proteins termed nucleoporins (Nups) (Hoelz et al., 2011; Kabachinski and Schwartz, 2015). The NPC is among the largest protein complexes in the cell, with an estimated molecular mass of 110 MDa (Hoelz et al., 2016). A single NPC is assembled from approximately 30 Nups, each present in multiple copies (Cronshaw et al., 2002; Davis and Blobel, 1986; Rout et al., 2000; Schmidt and Gorlich, 2016). Based on their approximate location within the NPC and their structural role, Nups may be organized into seven general categories (Figure 1): (1) cytoplasmic filament Nups, (2) nucleoplasmic and cytoplasmic ring Nups, (3) inner ring complex Nups, (4) transmembrane Nups, (5) central Nups containing phenylalanine-glycine (FG) repeats, (6) nuclear basket Nups, and (7) adaptor Nups (Hoelz et al., 2016; Schwartz et al., 2015; Schwartz, 2016). The role of the NPC in ALS has entered the spotlight due to several recent lines of evidence, including the finding that genes encoding NPC constituents are genetic modifiers of disease in ALS model systems, the demonstration that disease-related proteins may directly poison the NPC, and the identification of ALS-causing mutations in at least one component of the NPC, as described in detail below.
Figure 1. Spatial organization of nucleoporins in the nuclear pore complex.
(A) Field emission scanning electron micrograph of an isolated Xenopus laevis oocyte nuclear envelope showing the outer surface the outer nuclear membrane and the NPCs. (B) Stimulated emission depletion (STED) image of nuclear pores in X. laevis cells. The protein gp210 (red) is arranged symmetrically around the central pore channel. In green, various FG repeat nucleoporins were labeled with a pan-specific antibody. (C) A schematic cut-away view showing half of an NPC embedded in the nuclear envelope. Nups that modify C9ORF72-mediated phenotypes in G4C2-repeat expanded flies and yeast are represented by color-coded text: components in which loss of function (LOF) suppresses or in which gain of function (GOF) enhances degeneration are labeled green, components in which LOF enhances or GOF suppresses degeneration are labeled red. *Different studies have observed opposite modifying effects of Nup50. Images reproduced with permission from (A) Martin W. Goldberg and (B) Stefan W. Hell.
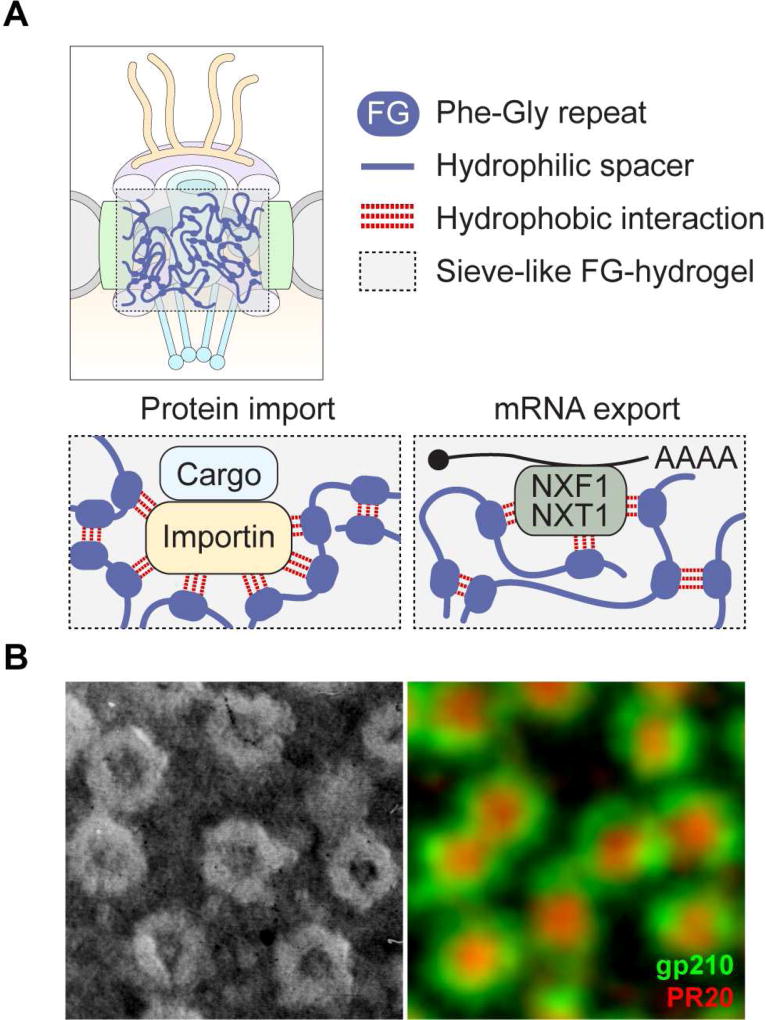
The symmetrical nucleoplasmic and cytoplasmic rings, together with the inner ring complex, form an essential scaffold for the NPC. Contained within this scaffold, the central channel of the nuclear pore is assembled by Nups containing conserved LCDs that have the notable feature of enrichment in interspersed phenylalanine-glycine repeats (FG-Nups). The flexible LCDs in the FG-Nups project into the central channel to create a selectively permeable sieve controlling transport between the cytoplasm and the nucleoplasm (Bestembayeva et al., 2015; Schmidt and Gorlich, 2016) (Figure 2). Various models have been proposed to account for how the FG-Nups create this selective barrier (Knockenhauer and Schwartz, 2016; Schmidt and Gorlich, 2016). One compelling model based on recent evidence, the selective phase model, proposes that cohesive weak hydrophobic interactions between FG repeats mediate phase separation to create a hydrogel meshwork within the NPC (Frey and Gorlich, 2007; Mohr et al., 2009; Ribbeck and Gorlich, 2001; Schmidt and Gorlich, 2016) (Figure 2). Small molecules can passively diffuse through numerous randomly distributed fenestrations (≤ 2.6 nm in diameter) in this mesh, but as the size of the diffusing molecules approaches or exceeds ~30 kDa in mass (typically ~5 nm in diameter), the mesh becomes increasingly restrictive. Nuclear transport receptors, which enhance the transport efficiency of cargos through the NPC, directly interact with the FG repeats of FG-Nups and dissolve into and through the FG-hydrogel network (Figure 2). Importantly, FG-Nups emerged as genetic modifiers in a Drosophila model of C9ORF72-related neurodegeneration and were identified as direct binding targets of toxic poly-dipeptides produced from mutant C9ORF72 (Freibaum et al., 2015; Lee et al., 2016; Lin et al., 2016; Shi et al., 2017).
Figure 2. Schematic model for the structural assembly of FG-Nups in the NPC channel.
(A) According to the selective phase model, highly cohesive FG domains in FG-Nups interact multivalently with each other to form a sieve-like hydrogel. The mesh size sets an upper size limit for free passage of cargos. Nuclear transport receptors (importins for proteins and NXF1-NXT1 for mRNPs) not only bind FG motifs but also disengage repeat-repeat interactions. This transiently opens meshes in the immediate vicinity of nuclear transport receptors and allows a nuclear transport receptor with its cargo to pass through the FG hydrogel. (B) Left panel: Metal-shadowed electron microscopy image of the nuclear envelope of the newt Notophthalmus viridescens. Image courtesy of Joseph G. Gall. Right panel: Image of the nuclear envelope of X. laevis as obtained by super-resolution stimulated emission depletion microscopy. Nuclear pore outer ring complexes are visualized by staining for gp210 (green); central channels of the nuclear pores are shown by staining for poly(PR)20 (red). Image courtesy of Zehra F. Nizami.
The Basis of Selective Nucleocytoplasmic Transport of Proteins
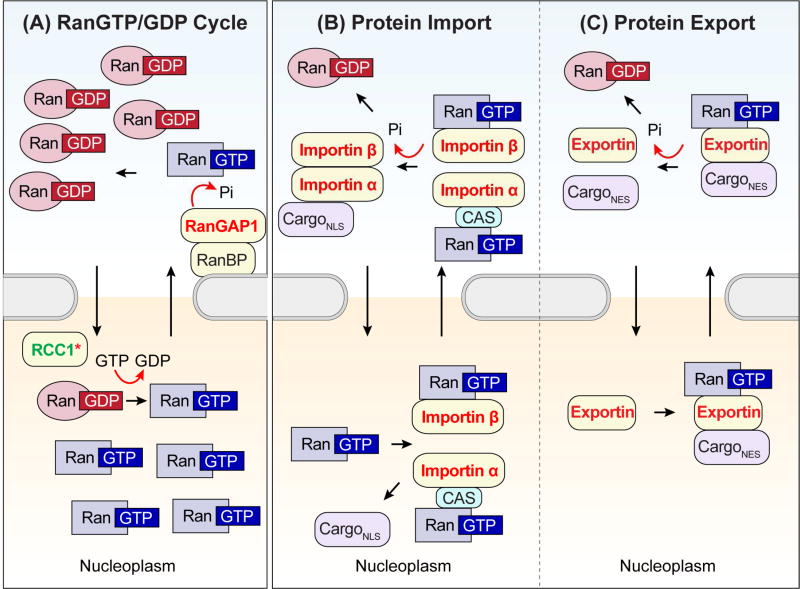
Proteins are imported or exported through the NPC via nuclear transport receptors, most of which are members of the karyopherin-β family, also known as importins and exportins (Cautain et al., 2015; Soniat and Chook, 2015). The small GTPase Ras-related nuclear protein (Ran) regulates the ability of both importins and exportins to transport their cargo across the nuclear membrane depending on the bound nucleotide state. RCC1, the guanine nucleotide exchange factor (GEF) for Ran, is located preferentially in the nucleus, whereas the Ran GTPase-activating proteins (GAPs) and Ran-binding proteins (RanBPs) that catalyze the hydrolysis of GTP to GDP are localized exclusively to the cytoplasm (Stewart, 2007) (Figure 3A). This intracellular arrangement leads to concentration of RanGTP in the nucleus and RanGDP in the cytosol, forming a gradient harnessed by importins and exportins to provide direction to nucleocytoplasmic transport (Figure 3B–C). Importantly, as with the NPC components described above, nuclear transport receptors, Ran, and RanBPs have emerged as genetic modifiers in yeast and Drosophila models of C9ORF72-related neurodegeneration (Figure 3) (Freibaum et al., 2015; Jovičić et al., 2015; Zhang et al., 2015).
Figure 3. Schematics of protein movement through the nuclear pore complex.
(A) The Ran gradient arises due to asymmetric distribution of Ran regulators, which dictate whether GTP or GDP is bound to Ran. Nuclear RanGEF (RCC1) promotes the dissociation of GDP from Ran and allows the binding of GTP. Ran in the nucleus is predominantly in the GTP-bound form. When RanGTP leaves the nucleus, the cytosolic RanGAP induces GTP hydrolysis by Ran in cooperation with RanBP. In the cytoplasm, the RanGTP concentration is low and the RanGDP concentration is high. Pi, inorganic phosphate. (B) In the cytoplasm, importin-β binds cargo via the adaptor protein importin-α, forming a trimeric complex. Importin-β then carries cargo through the NPC. Some importin-βs directly bind cargo, forming a dimeric complex that transports cargo into the nucleoplasm. In the nucleus, binding of RanGTP induces a conformational change in the importin, causing it to dissociate from its cargo. This process results in two complexes: a trimeric complex composed of importin-α, its nuclear export factor CAS, and RanGTP, and a dimeric complex composed of importin-β and RanGTP. These complexes translocate into the cytoplasm, where RanGTP is hydrolyzed. Importin-α and importin-β are released from CAS-RanGDP and RanGDP, respectively, and are available to transport the next cargo. (C) Exportin recruits its cargo when bound to RanGTP in the nucleus. This ternary export complex crosses the NPC to the cytoplasm, where GTP undergoes hydrolysis, triggering the release of the cargo. Proteins known to modify C9ORF72-mediated phenotypes in G4C2-repeat expanded flies and yeast are represented by color-coded text as follows: components in which LOF suppresses or in which GOF enhances degeneration are colored green, components in which LOF enhances or GOF suppresses degeneration are colored red. *Different studies have observed opposite modifying effects of RCC1.
The Basis of RNA Export through the Nuclear Pore Complex
The mechanisms underlying export of transfer RNA, microRNA, small nuclear RNA, and ribosomal RNA (rRNA) are similar to those of proteins, involving exportins and the RanGTP–RanGDP cycle. Small transfer RNAs, microRNAs, and small nuclear RNAs follow relatively simple export routes by binding directly to their export receptors exportin-t, exportin-5, and exportin-1, respectively. rRNAs first assemble into large (60S) and small (40S) ribosomal subunits, which then translocate into the cytoplasm following separate export routes. These routes are known to involve the RanGTP–RanGDP cycle and exportin-1, although the precise mechanisms remain poorly understood. For an in-depth review of rRNA export, see (Kohler and Hurt, 2007).
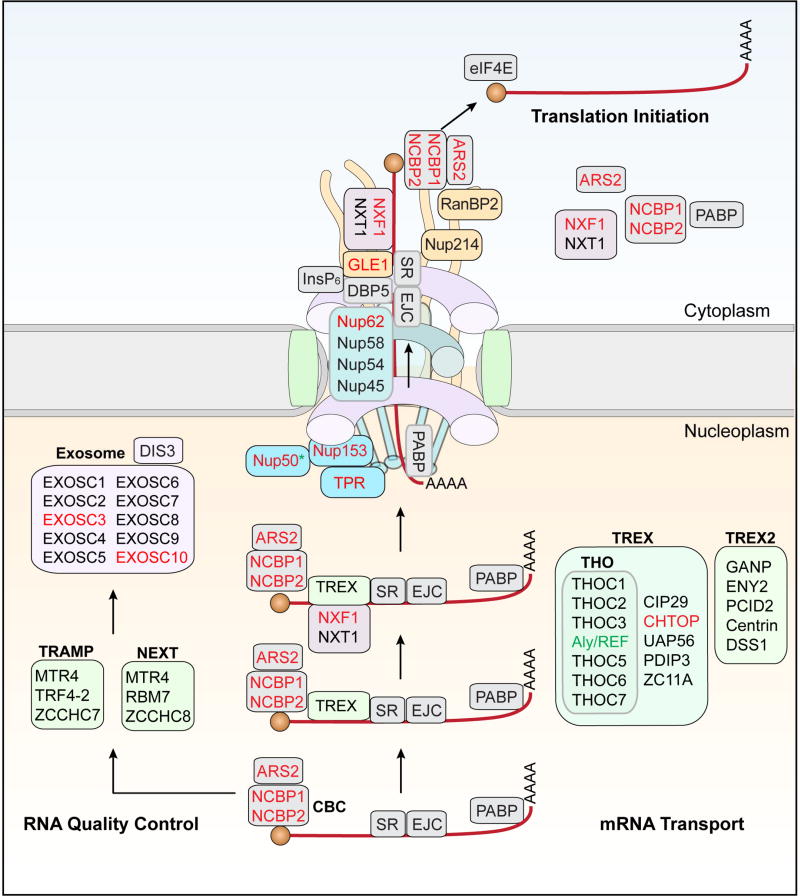
The export of mRNAs into the cytoplasm is mechanistically distinct from other RNAs, as this process uses a non-karyopherin transport receptor, the NXF1-NXT1 heterodimer that works in concert with the TRanscription-Export (TREX) complex, and does not depend on the RanGTP–RanGDP cycle (Kohler and Hurt, 2007). The export of mRNAs is coordinated with transcript processing and assembly into messenger ribonucleoprotein (mRNP) complexes (Figure 4). In this process, multiple RNA-modifying proteins bind to a single mRNA synthesized by Pol II as a precursor mRNA (pre-mRNA). These assembled proteins form a large mRNP complex, which then translocates through the NPC. Once inside the central channel, an mRNP can move forward and backward, a phenomenon observed for many cargos that traverse the nuclear pore (Ma et al., 2013; Tu et al., 2013). Only 15–36% of all nuclear mRNPs are successfully exported to the cytoplasm, with most mRNPs returned to the nucleoplasm (Grunwald and Singer, 2010; Ma et al., 2013; Siebrasse et al., 2012). For those mRNPs that reach the cytoplasm, extensive remodeling of mRNPs occurs at the cytoplasmic face to ensure that these mRNPs do not reenter the nucleus. This remodeling of mRNPs at the cytoplasmic side of the NPC is crucial for maintaining the direction of mRNA export and preparing mRNAs for translation by the ribosome. RNA export factors have also emerged as genetic modifiers in yeast and Drosophila models of C9ORF72-related neurodegeneration (Figure 4) (Freibaum et al., 2015; Jovičić et al., 2015; Zhang et al., 2015).
Figure 4. Export of mRNPs through the nuclear pore complex.
A single pre-mRNA is bound to multiple RNA-modifying proteins, including the cap-binding complex (CBC), serine–arginine-rich (SR) proteins, and the exon-junction complex (EJC). An mRNA containing an error in 3′ end processing, splicing, or packaging into mRNPs is directed to the exosome complex by the exosome cofactor complexes TRAMP and NEXT. The TREX and TREX2 complexes are recruited to a mature mRNP that passes quality control. NXF1 and NXT1 are then recruited to the mRNP, and the cargo mRNP is transferred to the nuclear basket, which initiates the translocation of the cargo mRNP through the NPC. In the cytoplasm, GLE1 and InsP6 activate DBP5, which binds to the mRNA and alters the structure of the mRNP, thereby removing the NXF1–NXT1 complex in an ATP-dependent manner.
Initial Clues Linking Nucleocytoplasmic Transport and Motor Neuron Disease
Perhaps the earliest clue suggesting a nucleocytoplasmic transport defect in ALS arose from the study of a congenital form of motor neuron disease first characterized in 1995, when 15 infants from 11 Finnish families were described with a phenotype of lethal arthrogryposis and anterior horn motor neuron loss (OMIM: 611890, (Vuopala et al., 1995)). Thirteen years later, the cause of this disease was definitively shown to be biallelic, loss-of-function mutations in GLE1, which encodes a component of the cytoplasmic face of the NPC that facilitates the export of mRNAs from the nucleus (Nousiainen et al., 2008) (Figures 1 and 4). More recently, a large screening effort to identify ALS-causing mutations in French and French-Canadian patients identified rare, heterozygous loss-of-function mutations in GLE1 that were not present in controls, suggesting that mild defects in mRNA export could contribute to adult-onset motor neuron disease (Kaneb et al., 2015). Subsequently, GLE1 was identified as a dominant modifier in Drosophila models of C9ORF72-related ALS, further linking this gene, and defects in mRNA export more generally, to ALS (Freibaum et al., 2015). Moreover, recent reports indicate that GLE1 is sequestered within polyglutamine inclusions in Huntington’s disease, broadening the spectrum of diseases that may involve nucleocytoplasmic transport defects (Gasset-Rosa et al., 2017).
Circumstantial Evidence for a Nucleocytoplasmic Transport Defect in ALS
The most prominent pathological feature of ALS, redistribution of nuclear proteins to cytoplasmic inclusions, also intimated dysfunction of nucleocytoplasmic transport in ALS and related neurodegenerative diseases. As mentioned above, the major component of cytoplasmic inclusions in approximately 97% of ALS patients and 45% of FTD patients is TDP-43 (Ling et al., 2013), a ubiquitously expressed RNA-binding protein that shuttles between the nucleus and cytoplasm, but is predominantly located in the nucleus in healthy neurons (Ayala et al., 2008). Despite the fact that genetic mutations in TDP-43 account for only ~2% of familial ALS (Kabashi et al., 2008; Sreedharan et al., 2008; Van Deerlin et al., 2008) and rare cases of FTD (Borroni et al., 2009; Kovacs et al., 2009), recognition that the vast majority of ALS cases exhibit cytoplasmic TDP-43 pathology raised the question of whether impaired nuclear import of TDP-43 was a pathological feature common to many forms of ALS (Winton et al., 2008). Mislocalization of TDP-43 to the cytoplasm is a frequent secondary feature in other neurodegenerative diseases as well, including Huntington’s disease, Alzheimer’s disease, corticobasal degeneration, and Lewy body dementia, suggesting that impaired nuclear import may be a feature common to a broad spectrum of diseases (Nakashima-Yasuda et al., 2007; Schwab et al., 2008; Uryu et al., 2008; Wilson et al., 2011).
Mutations in FUS, an RNA-binding protein that shares structural and functional properties with TDP-43, cause rare cases of ALS and FTD. In these cases, FUS is the major pathological protein in neuronal cytoplasmic inclusions. Remarkably, approximately half of ALS- and FTD-associated FUS mutations are located in its NLS and disrupt its normal nuclear localization (Ling et al., 2013). Notably, the degree to which these FUS NLS mutations affect the nucleocytoplasmic transport of FUS robustly correlates with the severity of disease in these patients (Bosco et al., 2010; Chio et al., 2009; DeJesus-Hernandez et al., 2010; Dormann et al., 2010). For example, one patient with a C-terminal truncation mutation that entirely removed the NLS in FUS showed dramatic redistribution of FUS into the cytoplasm and very early age of onset (DeJesus-Hernandez et al., 2010). More recently, a novel missense mutation in the NLS of hnRNPA1, which causes redistribution of hnRNPA1 from the nucleus to the cytoplasm, was identified as a cause of familial ALS (Liu et al., 2016), expanding the repertoire of NLS mutations in ALS-associated proteins. Interestingly, mutations that disrupt the NLS in a related protein, hnRNPH2, cause neurodevelopmental delay and autism (Bain et al., 2016).
The nuclear depletion and cytoplasmic mislocalization of TDP-43, FUS, and hnRNPA1, together with the finding of frequent NLS mutations in ALS-linked proteins, adds to the evidence that dysfunction in nucleocytoplasmic transport through the NPC may contribute to disease initiation or progression, either through loss of vital nuclear functions or toxic gain of cytoplasmic function through increased concentration or residence time of these proteins in cytoplasmic assemblies. Consistent with this concept, early studies in post-mortem tissue from ALS and FTD patients revealed a considerable reduction in expression and/or mislocalization of nuclear import factors such as importin β1 (also known as karyopherin β1) and importin β2 (also known as karyopherin β2 and transportin 1) in brain and spinal cord (Neumann et al., 2012; Nishimura et al., 2010; Takeuchi et al., 2013), further implicating defective nuclear transport as a contributor to the pathogenesis of these diseases.
The Emergence of Nucleocytoplasmic Transport Defects in C9 ALS-FTD
The first mechanistic links between nucleocytoplasmic transport and ALS emerged from a series of studies focusing on chromosome 9-linked ALS/FTD (C9 ALS-FTD). The disease-causing mutation in these cases is expansion of an intronic hexanucleotide repeat (G4C2) in the gene C9ORF72 (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Healthy individuals generally carry between two and 23 G4C2 repeats, whereas individuals with C9 ALS-FTD have hundreds or thousands of such repeats. Expansion of the repeat in C9ORF72 is highly prevalent among the ALS-FTD population, causing up to 40% of familial cases and ~5–10% of sporadic cases (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Although patients with C9 ALS-FTD show a modest reduction in C9ORF72 expression that may contribute to disease progression, the disease is most likely driven by a toxic gain of function. At the source of this gain of function are repeat-containing transcripts of C9ORF72 that accumulate in affected neurons and perhaps other cells in the brain and spinal cord (Taylor, 2017), although the specific pathogenic mechanism has not been precisely defined. Limited evidence suggests that these repeat-containing transcripts form RNA foci that sequester and deplete RNA-binding proteins. More compelling evidence argues for toxicity arising from poly-dipeptides produced by unconventional translation of repeat-containing transcripts of mutant C9ORF72 (Kwon et al., 2014; Mizielinska et al., 2014; Wen et al., 2014; reviewed in (Freibaum and Taylor, 2017). The expanded repeat in the mutant C9ORF72 gene is bidirectionally transcribed, and both the sense and antisense transcripts undergo repeat-associated non-AUG (RAN) translation in all reading frames (Zu et al., 2011). RAN translation of expanded C9ORF72 transcripts produces five different poly-dipeptide repeats (DPRs): poly-Gly-Ala [poly(GA)] and poly-Gly-Arg [poly(GR)] from sense transcripts, poly-Pro-Ala [poly(PA)] and poly-Pro-Arg [poly(PR)] from antisense transcripts, and poly-Gly-Pro [poly(GP)] from both sense and antisense transcripts, all of which are found to accumulate in the brain and spinal cords of C9 ALS-FTD patients (Ash et al., 2013; Gendron et al., 2013; Mackenzie et al., 2013; Mori et al., 2013; Zu et al., 2013).
To develop an animal model that captures G4C2-mediated toxicity, Freibaum and colleagues generated a Drosophila model of C9ORF72-associated ALS/FTD by expressing 8 or 58 G4C2 repeats [(G4C2)8 or (G4C2)58], demonstrating length- and dosage-dependent degeneration phenotypes (Freibaum et al., 2015). Importantly, both G4C2 repeat-expanded RNA and DPRs [poly(GP) and poly(GR)] from the sense-strand were expressed in flies expressing (G4C2)58 (Freibaum et al., 2015). Using (G4C2)58-expressing flies, the investigators performed a large-scale unbiased genetic screen to identify dominant modifiers of G4C2 repeat–mediated toxicity. This approach identified two genes, Nup50 and Ref1 (Drosophila orthologs of human NUP50 and ALYREF), whose loss strongly enhanced or suppressed, respectively, neurodegeneration in (G4C2)58-expressing flies (Freibaum et al., 2015) (Table S1).
NUP50 is a component of the nuclear basket (Figure 1C) and plays a critical role in promoting the nuclear import of proteins by interacting with importin α (also known an karyopherin α) and Ran (Makise et al., 2012). Aly/REF export factor (Aly/REF) is a component of the TREX complex (Figure 4), which associates with the 5′ cap of mRNAs and facilitates delivery of mRNPs to NXF1, which then mediates mRNP export through the NPC (Reed, 2003; Viphakone et al., 2012; Zhou et al., 2000). Further dissection of these two pathways resulted in the identification of 18 strong genetic modifiers of neurodegeneration in (G4C2)58-expressing flies, all of which encode components of the NPC and nucleocytoplasmic transport machineries (Freibaum et al., 2015) (Figures 1, 3, and 4, Table S1). Consistent with these findings, G4C2-expressing flies exhibited nuclear envelope abnormalities and nuclear RNA retention in a G4C2 length- and dose-dependent manner. Mirroring the results from the Drosophila model, nuclear retention of RNA was also demonstrated in human cells and iPSC-derived cortical neurons from patients with C9 ALS-FTD (Freibaum et al., 2015).
Working independently, Zhang and colleagues also found a role for defective nucleocytoplasmic transport and C9 ALS-FTD (Zhang et al., 2015). These investigators reasoned that stable, G4C2 repeat-containing RNA species that accumulate in patient tissues might sequester RNA-binding proteins and deplete their function. Therefore, they conducted a targeted modifier screen in a Drosophila model that focused on genes encoding proteins reported to physically interact with G4C2 repeat-containing RNAs in vitro. In this manner, Zhang and colleagues identified a dominant gain-of-function allele of RanGAP that suppressed neurodegeneration in flies expressing expanded G4C2 repeats. Although this model system does not distinguish between RNA-mediated and DPR-mediated toxicity, biochemical and cellular experiments revealed that RanGAP1 binds to the RNA G-quadruplex structures formed by G4C2-repeat expansions in vitro, raising the possibility that repeat-expanded RNA species are a direct mediator of toxicity. Of note, this report also showed that G4C2 repeat expansion in C9ORF72 significantly shifted the nuclear-to-cytoplasmic ratio of TDP-43 in patient-derived cells that had been differentiated to neurons (Zhang et al., 2015), mimicking the nuclear depletion and cytoplasmic aggregation of TDP-43 found in the affected tissues of patients with ALS or FTD (Neumann et al., 2006). Finally, RanGAP1 and two NPC components, Nup205 and Nup107, abnormally localized to nuclear aggregates not only in iPSC-derived neurons derived from patients with C9ORF72-mediated ALS but also in cortical tissue from C9ORF72 ALS patients (Zhang et al., 2015). Similar results were subsequently found in a mouse model in which the DPR poly(GA) was exogenously expressed in the mouse brain (Zhang et al., 2016).
In contrast to the approach by Zhang and colleagues, Jovičić and colleagues focused specifically on DPR-mediated toxicity (Jovičić et al., 2015). In this study, the investigators engineered expression in yeast cells of each of the five DPR species produced by RAN translation of expanded G4C2 repeats in C9 ALS-FTD patients. Consistent with prior studies (Kwon et al., 2014; Mizielinska et al., 2014; Wen et al., 2014), they found that the arginine-containing DPRs [poly(GR) and poly(PR)] were highly cytotoxic. Subsequently, they performed a large-scale, unbiased genetic screen to identify modifiers of poly(PR)-associated toxicity in yeast, resulting in the identification of 11 genes that regulate nucleocytoplasmic transport (Table S1). Consistent with the studies mentioned above, these investigators documented a disturbance in the nuclear-to-cytoplasmic ratio of RanGEF (RCC1) in fibroblasts from C9 ALS-FTD patients that were transdifferentiated into neurons (Jovičić et al., 2015). A follow-up study that re-evaluated genes identified in the yeast screen corroborated these results in a Drosophila model (Boeynaems et al., 2016).
Taken together, these studies provided strong evidence that impairment of nucleocytoplasmic transport is an important driver of toxicity in simple genetic models of C9 ALS-FTD. The identification of similar defects in patient-derived neurons, and associated histopathological defects in patient-derived CNS tissue, supports the contention that impairment of nucleocytoplasmic transport is also a contributor to the initiation or progression of ALS-FTD in human patients.
Toward a Mechanism of DPR-Mediated Neurodegeneration
A major unanswered question regarding C9 ALS-FTD is the extent to which the toxic gain of function is mediated directly by the toxicity of repeat-expanded RNA versus the toxicity of DPRs derived from these RNAs. Of the three studies that first identified a defect in nucleocytoplasmic transport, the study by Jovičić et al. demonstrated defects in nucleocytoplasmic transport in the context of expressing poly(PR) exclusively. Although it is possible that repeat-expanded RNA contributes to or exacerbates nucleocytoplasmic transport defects in C9 ALS-FTD, the Jovičić et al. study implies that poly(PR) is sufficient to initiate this defect.
But how might DPRs produce a defect in nucleocytoplasmic transport? Two recent studies addressed this issue. First, Lee et al. used affinity purification to identify protein interactors of each of the five DPRs produced in C9 ALS-FTD patients (Lee et al., 2016). Whereas the relatively non-toxic species [poly(PA), poly(GP), and poly(GA)] bound very few proteins, the toxic arginine-containing species [poly(GR) and poly(PR)] each bound about 140 proteins and these interactors showed considerable overlap. Importantly, the list of interacting proteins was highly enriched (~68%) in proteins containing LCDs, including many RNA-binding proteins (e.g., hnRNPA1, hnRNPA2B1, FUS, and TDP-43) and other components whose phase separation contributes to the assembly and liquid properties of membrane-less organelles (e.g., NPM1, G3BP1, and G3BP2). Also enriched among these interactors were multiple FG-Nups that undergo phase separation in the assembly of the central channel of the nuclear pore. Importantly, Lee et al. showed that poly(GR) and poly(PR) insinuated into phase-separated assemblies in purified systems and in living cells, and altered their material properties toward more rigid and immobile entities. Moreover, normal functions of these membrane-less organelles (e.g., nucleolus and stress granules) were impaired by poly(GR) and poly(PR). Independently, Lin et al. used a chemical cross-linking approach to capture direct interactors of poly(PR) and obtained highly consistent results (Lin et al., 2016). This study presented evidence that poly(PR) interacts directly with LCDs of RNA-binding proteins that are the constituents of membrane-less organelles and intermediate filaments, and favors interaction with polymeric forms of these protein segments. An intermediate filament is critical for proper deposition of RNA granules. In particular, spatial partitioning and trafficking of RNA granules (e.g., neuronal granules) to the synapses are important for nerve cells to survive. Thus, the author proposed that interactions between the LCDs of RNA granule proteins and intermediate filaments may be centrally involved in the control of localized translation. Disturbance of such interactions by poly(PR) may interfere with dynamic behavior of intermediate filaments and proper deposition of RNA granules that will lead to blockage of information flow from gene to message to protein. Together, these studies revealed that DPR toxicity involves broad impairments to dynamic cellular assemblies that arise from phase separation of LCDs. However, an important unanswered question remains: which dynamic cellular assemblies that arise from phase separation are most vulnerable to insults that drive ALS and related diseases?
DPRs Produced by Mutant C9ORF72 Directly Plug the Nuclear Pore
Arginine-containing DPRs accumulate in several subcellular structures, including nucleoli, stress granules, and other membrane-less organelles that assemble through phase separation (Kwon et al., 2014; Mizielinska et al., 2014; Wen et al., 2014; Lee et al., 2016). By examining the distribution of poly(PR) in nuclei of mammalian cells and oocytes of the frog Xenopus laevis in greater detail with super-resolution microscopy, Shi et al. recently found that poly(PR) peptide also accumulates in the central channel of the nuclear pore complex, consistent with prior reports that the arginine-containing DPRs bind to nuclear pore proteins, including FG-Nups that form the central channel (Figure 2) (Lee et al., 2016; Lin et al., 2016). Shi and colleagues showed that the low-complexity FG domains of two Nups (Nup54 and Nup98) readily assemble into labile polymers, a property that may relate to selective transport through the dynamic barrier within the central channel of the pore. Further, they provided evidence that poly(PR) binds and stabilizes the fibrillar forms of Nup54 and Nup98, which would be expected to reduce dynamics of the central channel and impair nucleocytoplasmic transport. Moreover, when poly(PR) peptide was added to cells, it accumulated in the central channel of the nuclear pore, resulting in a defect in nuclear transport of RNA and protein, consistent with prior reports (Lee et al., 2016). The study not only provides a satisfying explanation for how nucleocytoplasmic transport may be impaired in C9 ALS-FTD, but may also illuminate a biophysical feature (disturbance of a biological phase transition) that is relevant to the molecular basis of ALS-FTD pathogenesis more broadly.
An Unanticipated Link Between the ESCRT Complex and Nucleocytoplasmic Transport in ALS-FTD
Recent studies of the endosomal-sorting complexes required for transport (ESCRT) complex identified an unexpected connection to nucleocytoplasmic transport that may contribute to ALS-FTD. The ESCRT complex machinery is composed of five modules of protein subunits: ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and the AAA-ATPase VPS4. Mutations in the ESCRTIII-complex subunit CHMP2B are causative of ALS-FTD (Cox et al., 2010; Parkinson et al., 2006; Skibinski et al., 2005; van der Zee et al., 2008). The best characterized function of the ESCRT complex is sorting ubiquitinated membrane protein receptors into multivesicular bodies (McCullough et al., 2013). Importantly, recent discoveries indicate that the sorting activity of the ESCRT complex may extend beyond the endosomal system. Indeed, a recent study in yeast revealed a broad interaction network between the NPC and the ESCRT complex machinery (Webster et al., 2014). Genetic ablation of ESCRT-III and VPS4 in yeast led to clustering of defective NPCs due to the mis-assembly of NPC proteins, suggesting that the ESCRT complex may be responsible for eliminating defective NPC assembly intermediates before they mature into a nonfunctional NPC (Webster et al., 2014). In addition to a surveillance role in NPC assembly, ESCRT-III may also play a role in the control of post-mitotic nuclear envelope reformation in mammalian cells and in repair of nuclear envelope-rupturing events that are associated with cellular migration or cellular aging (Denais et al., 2016; Olmos et al., 2015; Raab et al., 2016). The link between the ESCRT complex and nuclear pore function was recent and unanticipated, and it remains to be determined whether mutations in ESCRT-III that are causative of ALS-FTD impact nucleocytoplasmic transport.
Nucleocytoplasmic Transport Defects in Other Neurodegenerative Diseases
Although extensively studied in the context of C9ORF72-mediated ALS and FTD, the role of the NPC and nuclear transport has also been investigated in a number of other types of neurological disease, particularly Huntington’s disease and Alzheimer’s disease. A hallmark of Huntington’s disease is increased levels of polyglutamine-expanded huntingtin protein (Htt) in the nucleus. Some evidence suggests that reduced nuclear export of Htt may be one mechanism resulting in accumulation of the protein in the nucleus (Truant et al., 2007). The N-terminal 17 amino acids of Htt compose a consensus exportin-1–dependent nuclear export signal, which promotes shuttling between the cytoplasm and the nucleus (Maiuri et al., 2013; Zheng et al., 2013). Interestingly, the N-terminal Htt fragment directly interacts with nucleoprotein TPR (Figure 1), and this interaction is inhibited by the presence of a polyglutamine-expanded repeat and Htt aggregation, thus decreasing Htt export to the cytoplasm and increasing nuclear accumulation of mutant Htt (Cornett et al., 2005). Similar results were reported in spinocerebellar ataxia type 7, another polyglutamine expansion disease (Taylor et al., 2006).
Although these earlier reports largely focused on nucleocytoplasmic transport of the mutant Htt protein itself, two recent studies have advanced the concept that mutant Htt is sufficient to induce general defects in nucleocytoplasmic transport and that these deficits are indeed central components of Huntington’s disease in humans (Gasset-Rosa et al., 2017; Grima et al., 2017). Using a variety of models, including mouse, fly, iPSC-derived neurons, and postmortem human tissues, these studies revealed highly abnormal NPC pathology in cells expressing mutant Htt, including mislocalization and aggregation of Nups; GLE1 and RanGAP1 were also mislocalized in mouse models expressing mutant Htt, apparently the result of sequestration by intranuclear mutant Htt, leading to defects in mRNA export. Consistent with these findings, mislocalization of RanGAP1 and nuclear accumulation of mRNA were also observed in postmortem tissue of patients with Huntington’s disease. Given the parallels between these Htt-related findings and observations from wild-type aging neurons, which include a general decline in nucleocytoplasmic transport (see below), the authors hypothesized that mutant Htt may accelerate cellular aging through its effects on nucleocytoplasmic transport (Gasset-Rosa et al., 2017).
Notably, similar to hexanucleotide repeat expansion in C9 ALS-FTD, it was recently demonstrated that the CAG repeat expansion in Huntington’s disease is associated with RAN translation to produce four homopolymeric expansion proteins in addition to mutant Htt itself (polyAla, polySer, polyLeu, and polyCys) (Banez-Coronel et al., 2015). These sense and antisense HD-RAN proteins are also toxic to neural cells. Grima and colleagues demonstrated that these RAN translation peptides impair nuclear import of protein in primary neurons, suggesting that they may contribute to a nucleocytoplasmic transport defect in Huntington’s disease. Remarkably, the toxic effects of mutant Htt and/or RAN translation products could be mitigated by overexpression of Nups or pharmacological treatment of neurons using small molecules that target the nucleocytoplasmic transport pathway, supporting a potential role for these processes in pathogenesis (Grima et al., 2017).
More indirect evidence has been reported for Alzheimer’s disease. For example, neurons from patients with Alzheimer’s disease have exhibited irregular nuclear morphology of FG-Nup62 and mislocalization of importin-α1 to inclusions (Lee et al., 2006; Sheffield et al., 2006). Moreover, the prosurvival nuclear transcription factor NRF2, which is imported into the nucleus by importin-α5 and importin-β1 (Theodore et al., 2008), was predominantly cytoplasmic in hippocampal neurons from patients with Alzheimer’s disease, suggesting that nuclear import of NRF2 is impaired in the brains of these patients (Ramsey et al., 2007).
Cerebral Aging and Nucleocytoplasmic Transport
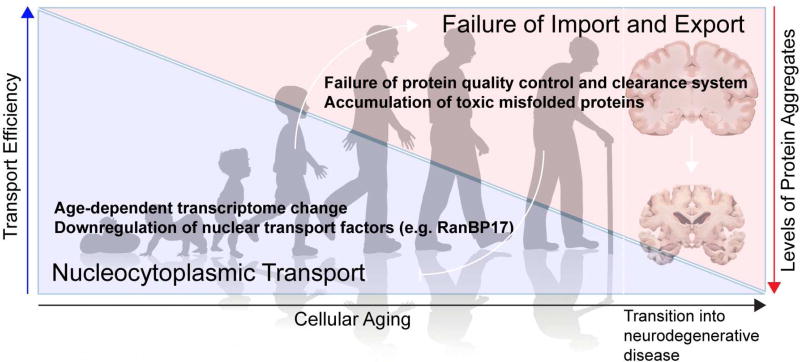
Connections have been drawn between age-related neurodegenerative diseases and physiologic aging. Prominent among the many cellular processes that decline with advanced age is nucleocytoplasmic transport. The precise assembly, maintenance, and repair of the NPC is critical to maintaining the health of the cell. Among the aspects of nucleocytoplasmic transport that diminish or are altered with age, the strongest evidence highlights alterations in the integrity of the NPC, although changes in expression of transport factors and indirect effects from the accumulation of aggregated proteins have also been implicated (Figure 5).
Figure 5. Cellular aging, disease, and nucleocytoplasmic transport.
The dysfunction of nucleocytoplasmic transport is observed during aging and neurodegenerative diseases. Age-related transcriptome changes, in particular the downregulation of nuclear transport factors (e.g., karyopherins and RanBP1), lead to dysfunction of nucleocytoplasmic transport that results in the accumulation and aggregation of proteins in the cytoplasm. Cytosolic aggregates, in turn, cause structural abnormalities in the NPCs and further compromise nuclear import and export of proteins and mRNAs. Disease-associated proteins take the same route as aging to target and perturb the function of the NPC.
In dividing cells, NPCs disassemble during mitosis and reassemble with newly synthesized proteins in newly forming nuclei (Rabut et al., 2004). However, in postmitotic cells such as neurons, complete disassembly of all NPCs in a synchronous manner is impractical. Instead, individual subcomplexes are exchanged slowly over time as new copies are synthesized. Strikingly, scaffold Nups (nucleoplasmic/cytoplasmic rings and inner ring complex Nups, Figure 1C) are extremely long-lived and are reported to remain incorporated in the NPC during the entire cellular life span in Caenorhabditis elegans and in rat brains (D'Angelo et al., 2009; Savas et al., 2012; Toyama et al., 2013). These long-lived proteins may represent cumulative vulnerability of the NPC over time, as they are exposed to harmful metabolites and/or chemicals throughout their lifespan. In fact, a subset of scaffolding Nups is oxidatively damaged in aged cells, and this age-related deterioration of NPCs leads to increased nuclear permeability and the leaking of cytoplasmic proteins into the nucleus (D'Angelo et al., 2009). This observation raises the intriguing possibility that defective NPCs and dysregulated nucleocytoplasmic transport may contribute to aging and age-associated diseases.
Changes in the expression levels of nucleocytoplasmic transport factors also occur with age. For instance, nucleocytoplasmic transport factors such as importin α1, CAS, and RanBP1 were found to be downregulated in human fibroblasts obtained from old donors compared to young donors, accompanied by a reduction in the protein import rate in fibroblasts from older donors, suggesting an age-dependent nucleocytoplasmic transport defect (Pujol et al., 2002).
Modeling studies suggest that age-dependent alterations eventually emerge as decreased nucleocytoplasmic transport function. In one study involving human donors from a broad range of ages, fibroblasts and induced neurons derived from aged donors exhibited impaired nucleocytoplasmic transport compared with neurons derived from young or middle-aged donors (Mertens et al., 2015). Comparison of the transcriptomes obtained from fibroblasts, induced neurons, and cortex samples led to the identification of Ran-binding protein 17 (RanBP17), an importin-β family member, as a substantially downregulated gene in aged cells. Remarkably, the knockdown of RanBP17 in young fibroblasts resulted in transcriptional changes similar to those found in aged cells (Mertens et al., 2015).
Recently, Woerner and colleagues provided evidence of a different, indirect impact of aging on nucleocytoplasmic transport caused by accumulation of misfolded proteins (Woerner et al., 2016). As proteostasis machineries become compromised during aging, aberrant protein aggregates accumulate as toxic species (Taylor and Dillin, 2011), a process associated with numerous neurodegenerative diseases including ALS and FTD. Woerner and colleagues employed artificial β-sheet proteins to form cross-β structures with no evolved biological function and demonstrated that their aggregation in the cytoplasm, but not in the nucleus, led to defects in nuclear protein import and mRNA export. Similar nucleocytoplasmic transport defects occurred in response to overexpression of mutant Htt protein and a C-terminal fragment of TDP-43, both of which are pathogenic β-sheet proteins that form cytoplasmic aggregates. Quantitative interactome analysis of these β-proteins identified the THO complex, a member of the TREX complex (Figure 4), as a highly enriched β-protein interactor. Indeed, THO complex subunit 2, importin-α1, and importin-α3 mislocalized in cells containing cytoplasmic β-protein aggregates (Woerner et al., 2016). Taken together, these reports suggest that the NPC and nucleocytoplasmic transport machinery may be targets of cellular aging in models of normal and pathologic neuronal senescence, and provide a mechanistic link between cerebral aging and neurodegenerative disease (Figure 5).
Concluding Remarks
Perturbation of nucleocytoplasmic transport has been identified as a central mechanism underlying ALS-FTD, particularly those cases caused by mutations in C9ORF72, but also other neurodegenerative diseases as well as normal aging. Although the observation that most cases of ALS are associated mislocalization of TDP-43 was, in hindsight, the first clue that the pathomechanism of ALS might involve an abnormality in nuclear transport, the temporal relationship between mislocalization of TDP-43, nuclear transport defects, and neurotoxicity in ALS remains unclear. It is possible that a primary defect in nuclear transport causes mislocalization and subsequent aggregation of TDP-43. It is also possible that accumulation of cytoplasmic aggregates of TDP-43 drive a secondary defect in nuclear transport.
Many additional questions remain. Despite the accumulation of experimental data indicating impairment of nucleocytoplasmic transport through the NPC in disease models, evidence of such defects in patients with ALS and other related diseases is thus far limited to redistribution of NPC components in end-stage disease. In addition, the relative pathogenic contribution of nucleocytoplasmic transport defects compared to impairments in other cellular processes has not been fully evaluated. Moreover, the relative roles of the toxic species causing damage to nucleocytoplasmic transport machinery – whether toxic RNA or toxic DPRs – remains unresolved. While challenging, we anticipate that these issues will be addressed soon in this rapidly moving field.
Supplementary Material
Acknowledgments
We thank Nisha Badders and Natalia Nedelsky for editorial assistance. This work was supported by grants from NIH grant R35NS097974, The Packard Center for ALS Research at the Johns Hopkins University, the ALS Association, the Clinical Research in ALS and related disorders for Therapeutic Develoment (CReATe) Consortium, the American-Lebanese-Syrian Associated Charities, and the Howard Hughes Medical Institute to J.P.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- Bain JM, Cho MT, Telegrafi A, Wilson A, Brooks S, Botti C, Gowans G, Autullo LA, Krishnamurthy V, Willing MC, et al. Variants in HNRNPH2 on the X Chromosome Are Associated with a Neurodevelopmental Disorder in Females. Am J Hum Genet. 2016;99:728–734. doi: 10.1016/j.ajhg.2016.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. RAN Translation in Huntington Disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestembayeva A, Kramer A, Labokha AA, Osmanovic D, Liashkovich I, Orlova EV, Ford IJ, Charras G, Fassati A, Hoogenboom BW. Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes. Nat Nanotechnol. 2015;10:60–64. doi: 10.1038/nnano.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovicic A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, Padovani A. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat. 2009;30:E974–983. doi: 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr, Sapp P, McKenna-Yasek D, Brown RH, Jr, Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cautain B, Hill R, de Pedro N, Link W. Components and regulation of nuclear transport processes. FEBS J. 2015;282:445–462. doi: 10.1111/febs.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Restagno G, Brunetti M, Ossola I, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Mandrioli J, et al. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol Aging. 2009;30:1272–1275. doi: 10.1016/j.neurobiolaging.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, Li XJ. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- Cox LE, Ferraiuolo L, Goodall EF, Heath PR, Higginbottom A, Mortiboys H, Hollinger HC, Hartley JA, Brockington A, Burness CE, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5:e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat. 2010;31:E1377–1389. doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015 doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Taylor JP. The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD. Front Mol Neurosci. 2017;10:35. doi: 10.3389/fnmol.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol Aging. 2011;32:2323 e2327–2340. doi: 10.1016/j.neurobiolaging.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. Subcellular Localization of Matrin 3 Containing Mutations Associated with ALS and Distal Myopathy. PLoS One. 2015;10:e0142144. doi: 10.1371/journal.pone.0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, Lagier-Tourenne C. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron. 2017;94:48–57 e44. doi: 10.1016/j.neuron.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron. 2017;94:93–107 e106. doi: 10.1016/j.neuron.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- Hoelz A, Glavy JS, Beck M. Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol. 2016;23:624–630. doi: 10.1038/nsmb.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabachinski G, Schwartz TU. The nuclear pore complex--structure and function at a glance. J Cell Sci. 2015;128:423–429. doi: 10.1242/jcs.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Murrell JR, Horvath S, Haraszti L, Majtenyi K, Molnar MJ, Budka H, Ghetti B, Spina S. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Ueda M, Miyamoto Y, Yoneda Y, Perry G, Smith MA, Zhu X. Aberrant localization of importin alpha1 in hippocampal neurons in Alzheimer disease. Brain Res. 2006;1124:1–4. doi: 10.1016/j.brainres.2006.09.084. [DOI] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788 e717. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL. Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers. Cell. 2016;167:789–802 e712. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Shu S, Wang RR, Liu F, Cui B, Guo XN, Lu CX, Li XG, Liu MS, Peng B, et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology. 2016;87:1763–1769. doi: 10.1212/WNL.0000000000003256. [DOI] [PubMed] [Google Scholar]
- Ma J, Liu Z, Michelotti N, Pitchiaya S, Veerapaneni R, Androsavich JR, Walter NG, Yang W. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat Commun. 2013;4:2414. doi: 10.1038/ncomms3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, Neumann M. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- Maiuri T, Woloshansky T, Xia J, Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum Mol Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem. 2012;287:38515–38522. doi: 10.1074/jbc.M112.378893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola AC, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Nakashima-Yasuda H, Uryu K, Robinson J, Xie SX, Hurtig H, Duda JE, Arnold SE, Siderowf A, Grossman M, Leverenz JB, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Neumann M, Valori CF, Ansorge O, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ishihara K, Ang LC, Bilbao JM, Mackenzie IR. Transportin 1 accumulates specifically with FET proteins but no other transportin cargos in FTLD-FUS and is absent in FUS inclusions in ALS with FUS mutations. Acta Neuropathol. 2012;124:705–716. doi: 10.1007/s00401-012-1020-6. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Zupunski V, Troakes C, Kathe C, Fratta P, Howell M, Gallo JM, Hortobagyi T, Shaw CE, Rogelj B. Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain. 2010;133:1763–1771. doi: 10.1093/brain/awq111. [DOI] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Pujol G, Soderqvist H, Radu A. Age-associated reduction of nuclear protein import in human fibroblasts. Biochem Biophys Res Commun. 2002;294:354–358. doi: 10.1016/S0006-291X(02)00492-8. [DOI] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, Piel M. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- Rabut G, Lenart P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr Opin Cell Biol. 2004;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, Hamilton RL, Chu CT, Jordan-Sciutto KL. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66:75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringholz GM, Appel SH, Bradshaw M, Cooke NA, Mosnik DM, Schulz PE. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–590. doi: 10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. Elife. 2015;4 doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends in biochemical sciences. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Travesa A, Martell SW, Forbes DJ. Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin. Nucleus. 2015;6:40–54. doi: 10.1080/19491034.2015.1004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz TU. The Structure Inventory of the Nuclear Pore Complex. J Mol Biol. 2016;428:1986–2000. doi: 10.1016/j.jmb.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield LG, Miskiewicz HB, Tannenbaum LB, Mirra SS. Nuclear pore complex proteins in Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:45–54. doi: 10.1097/01.jnen.0000195939.40410.08. [DOI] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, McKnight SL. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrasse JP, Kaminski T, Kubitscheck U. Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc Natl Acad Sci U S A. 2012;109:9426–9431. doi: 10.1073/pnas.1201781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- Soniat M, Chook YM. Nuclear localization signals for four distinct karyopherin-beta nuclear import systems. Biochem J. 2015;468:353–362. doi: 10.1042/BJ20150368. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Takeuchi R, Toyoshima Y, Tada M, Shiga A, Tanaka H, Shimohata M, Kimura K, Morita T, Kakita A, Nishizawa M, Takahashi H. Transportin 1 accumulates in FUS inclusions in adult-onset ALS without FUS mutation. Neuropathol Appl Neurobiol. 2013;39:580–584. doi: 10.1111/nan.12022. [DOI] [PubMed] [Google Scholar]
- Taylor J, Grote SK, Xia J, Vandelft M, Graczyk J, Ellerby LM, La Spada AR, Truant R. Ataxin-7 can export from the nucleus via a conserved exportin-dependent signal. J Biol Chem. 2006;281:2730–2739. doi: 10.1074/jbc.M506751200. [DOI] [PubMed] [Google Scholar]
- Taylor JP. A PR plug for the nuclear pore in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2017;114:1445–1447. doi: 10.1073/pnas.1621085114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore M, Kawai Y, Yang J, Kleshchenko Y, Reddy SP, Villalta F, Arinze IJ. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J Biol Chem. 2008;283:8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Burtnik A. Nucleocytoplasmic trafficking and transcription effects of huntingtin in Huntington's disease. Prog Neurobiol. 2007;83:211–227. doi: 10.1016/j.pneurobio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Tu LC, Fu G, Zilman A, Musser SM. Large cargo transport by nuclear pores: implications for the spatial organization of FG-nucleoporins. EMBO J. 2013;32:3220–3230. doi: 10.1038/emboj.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uryu K, Nakashima-Yasuda H, Forman MS, Kwong LK, Clark CM, Grossman M, Miller BL, Kretzschmar HA, Lee VM, Trojanowski JQ, Neumann M. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J Neuropathol Exp Neurol. 2008;67:555–564. doi: 10.1097/NEN.0b013e31817713b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, Urwin H, Engelborghs S, Bruyland M, Vandenberghe R, Dermaut B, De Pooter T, Peeters K, Santens P, De Deyn PP, et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum Mol Genet. 2008;17:313–322. doi: 10.1093/hmg/ddm309. [DOI] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuopala K, Ignatius J, Herva R. Lethal arthrogryposis with anterior horn cell disease. Hum Pathol. 1995;26:12–19. doi: 10.1016/0046-8177(95)90109-4. [DOI] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton MW, Salamone AR, Mosnik DM, McDonald RO, Appel SH, Schmolck HI, Ringholz GM, Schulz PE. Cognitive impairment in familial ALS. Neurology. 2007;69:1411–1417. doi: 10.1212/01.wnl.0000277422.11236.2c. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Dugger BN, Dickson DW, Wang DS. TDP-43 in aging and Alzheimer's disease - a review. Int J Clin Exp Pathol. 2011;4:147–155. [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, Hipp MS. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Li A, Holmes BB, Marasa JC, Diamond MI. An N-terminal nuclear export signal regulates trafficking and aggregation of Huntingtin (Htt) protein exon 1. J Biol Chem. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.